Abstract
In heat-stressed (HS) tomato (Lycopersicon peruvianum) cell cultures, the constitutively expressed HS transcription factor HsfA1 is complemented by two HS-inducible forms, HsfA2 and HsfB1. Because of its stability, HsfA2 accumulates to fairly high levels in the course of a prolonged HS and recovery regimen. Using immunofluorescence and cell fractionation experiments, we identified three states of HsfA2: (i) a soluble, cytoplasmic form in preinduced cultures maintained at 25°C, (ii) a salt-resistant, nuclear form found in HS cells, and (iii) a stored form of HsfA2 in cytoplasmic HS granules. The efficient nuclear transport of HsfA2 evidently requires interaction with HsfA1. When expressed in tobacco protoplasts by use of a transient-expression system, HsfA2 is mainly retained in the cytoplasm unless it is coexpressed with HsfA1. The essential parts for the interaction and nuclear cotransport of the two Hsfs are the homologous oligomerization domain (HR-A/B region of the A-type Hsfs) and functional nuclear localization signal motifs of both partners. Direct physical interaction of the two Hsfs with formation of relatively stabile hetero-oligomers was shown by a two-hybrid test in Saccharomyces cerevisiae as well as by coimmunoprecipitation using tomato and tobacco whole-cell lysates.
The vast majority of eukaryotic heat stress (HS)-inducible genes share conserved palindromic promoter elements with the consensus motif (AGAAn)(nTTCT) (2, 23, 32). They represent the recognition sites for the corresponding HS transcription factors (Hsfs), which are characterized by an N-terminal DNA-binding domain with a central helix-turn-helix motif (11, 46, 52), an adjacent domain with heptad hydrophobic repeats (HR-A/B) involved in oligomerization, and a C-terminal activator domain (for reviews, see references 27, 30, and 54).
Hsfs are encoded by small multigene families (see the summary by Nover et al. [30]). So far, four types of Hsfs have been characterized for plants (5, 8, 12, 43, 44) and vertebrates (19, 21, 34, 41, 45). In tomato, a constitutively expressed HsfA1 is accompanied by two HS-inducible forms, HsfA2 and HsfB1. By use of tobacco protoplasts as a transient-expression system, all three were shown to function as transcriptional activators (15, 51).
In contrast to those in plants, none of the four Hsfs in vertebrates is expressed in a stress-dependent manner. Hsf1 is the major form expressed in all cells. Its activity and intracellular localization are under stress control. Hsf2 is evidently involved in developmental control of chaperone gene expression, whereas Hsf3 may be considered a cell-specific variant of Hsf1 (13, 16, 19, 20, 40, 49). A new member of the vertebrate Hsf family is the recently described Hsf4 (21). It lacks an activator region and probably functions as a repressor of the basal-level expression of HS genes. This multiplicity may be increased by additional variants of Hsf1 and Hsf2 created by alternative splicing adding or eliminating a small, 66-bp exon close to the C-terminal HR-C region (6, 9).
Two reports from R. Morimoto’s group present evidence for a functional cooperation of different Hsfs in vertebrate cells. (i) Both Hsf1 and Hsf2 are expressed in human erythroleukemia cells; Hsf1 is activated by HS, whereas Hsf2 is probably involved in chaperone gene expression during hemin-induced differentiation of these cells. Both proteins trimerize and translocate into the nucleus. Interestingly, a synergistic effect on hsp70 gene transcription was observed if hemin treatment was followed by HS (48). (ii) In the avian erythroblast HD6 cell line, Hsf1 and Hsf3 can be activated by HS to undergo trimerization and transport to the nucleus. However, Hsf1 activation precedes that of Hsf3 (20). In both studies, the active Hsf forms were found to be homotrimers. There is no evidence for a physical interaction of different Hsfs.
In the course of our investigations of the intracellular localization of Hsfs in tobacco protoplasts and the characterization of putative nuclear localization signal (NLS) motifs in tomato HsfA1 and HsfA2, we observed an unexpected property of HsfA2. Despite having a functional NLS, it is defective in nuclear import, forming large cytoplasmic aggregates under HS conditions. This defect can be relieved by deletion of short C-terminal peptide motifs, and efficient nuclear import of this truncated versions is connected with a considerable increase of their activator potential as determined by an Hsf-dependent reporter assay (15).
In order to mimic the physiological basis for the HsfA2 function and to reconstruct the situation of the native tomato cells, we coexpressed HsfA2 with HsfA1 and/or HsfB1 in tobacco protoplasts. The results presented in this paper demonstrate efficient nuclear import of HsfA2 in the presence of HsfA1 but not of HsfB1. Using immunofluorescence and immunoelectron microscopy, we document the dynamic changes of the intracellular localization of HsfA2 in tomato cells and present evidence for the direct physical interaction of HsfA1 and HsfA2 by means of coimmunoprecipitation and a Saccharomyces cerevisiae two-hybrid test.
MATERIALS AND METHODS
General materials and methods.
On the basis of an international agreement (30), the nomenclature of Hsfs and of their functional parts was revised. Following this, tomato Hsf8, Hsf30, and Hsf24 (44) are now designated HsfA1, HsfA2, and HsfB1, respectively.
For the culture conditions and properties of the tomato cell suspension culture (Lycopersicon peruvianum), see reference 24. The use of tobacco mesophyll protoplasts (Nicotiana plumbaginifolia) for transient expression and functional characterization of Hsfs as well as the corresponding plant expression vectors was described previously (15, 51). After polyethylene glycol-mediated transformation with the plasmids indicated below, protoplasts were incubated for 10 h at 25°C under dim light (control) or, after 6-h incubation at 25°C, were shifted for 2 h to 35°C and then allowed 2 h of recovery (HS). For the Gus assay, cells were harvested after 10 h. Relative Gus activity of 100 represents the cleavage of 154 pmol of methylumbelliferyl glucuronide in 1 min by an aliquot of cell extract corresponding to 4,000 protoplasts. For immunofluorescence, cells were fixed immediately after the HS treatment. Cell fractionation and preparation of HS granules (HSG) were done as described by Nover and Scharf (26) and Nover et al. (29).
Antisera.
Rabbit and chicken antisera against tomato HsfA1, HsfA2, and HsfB1 and pea Hsp18 class I were generated by Eurogentec SA (Seraing, Belgium) by using purified recombinant proteins prepared in our laboratory. For detection of Hsp90 and the Hsp-Hsc70 complex and detection of tubulin, commercial antisera from StressGen and Amersham, respectively, were used.
Coimmunoprecipitation.
Tomato cells (200 mg [fresh weight]) were collected on filter discs and homogenized by sonication in 400 μl of lysis buffer containing 25 mM HEPES (pH 8.0), 500 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 10 mM NaF, 0.2% Nonidet P-40, 10 mM β-mercaptoethanol, and protease inhibitors (10 μg of Pefabloc per ml, 1 μg of pepstatin per ml, 2 μg of aprotinin per ml, 1 μg of leupeptin per ml, 50 μg of TLCK [Nα-p-tosyl-l-lysyl chloromethyl ketone] per ml, and 100 μg of TPCK [Nα-p-tosyl-l-phenylalanyl chloromethyl ketone] per ml). Prior to addition of antiserum, protein levels were adjusted to about 10 μg/μl, and 50-μl samples were diluted 10-fold with modified lysis buffer containing 100 mM NaCl but no β-mercaptoethanol. Tobacco protoplast lysates from 4 × 105 cells were prepared by sonication in 400 μl of modified lysis buffer containing 150 mM NaCl and 1 mM β-mercaptoethanol. The resulting lysates were precleared with protein A-Sepharose and incubated for 1 h at 4°C with 3 μl of HsfA2 antiserum in bovine serum albumin (BSA)-coated reaction tubes. Protein A-Sepharose beads (Pharmacia) were added as a 1:1 slurry in lysis buffer, and samples were further incubated at 4°C for 1 h with gentle agitation. The beads were pelleted and washed three times with 500 μl of 10 mM Tris buffer (pH 8.0) supplied with 140 mM NaCl and 500 mM LiCl and once with 10 mM Tris buffer (pH 8.0) containing 140 mM NaCl. Bound proteins were eluted with 30 μl of sodium dodecyl sulfate (SDS) sample buffer by being heated for 5 min to 95°C. Ten-microliter samples were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and processed for Western analysis using a chicken HsfA1 antiserum.
Yeast plasmids.
Plasmids were constructed and transformed into Escherichia coli DH5α by standard techniques (38). The 2μm vectors pADGal4 (Gal4p activator domain [Gal4p-AD] amino acids [aa] 768 to 881; LEU2) and pBDGal4 (Gal4p DNA-binding domain [Gal4p-DBD] aa 1 to 147; TRP1) were obtained from Stratagene, La Jolla, Calif. Yeast transformation procedures were performed according to the Stratagene two-hybrid manual.
For two-hybrid screening and protein interaction studies, the bait plasmid pRL 123 was constructed by ligating an EcoRI/SalI fragment coding for aa 23 to 448 of HsfA1 to the Gal4p-DBD fragment of pBDGal4. The insert for bait plasmid pRL125 coding for HsfB1 (aa 1 to 301) fused to Gal4p-DBD was generated by PCR amplification introducing the appropriate restriction sites. The PCR product was inserted into the SalI/PstI sites of pBDGal4. The CEN plasmid pAS1 encodes a hybrid protein made of Gal4p-DBD and the HR-A/B part of HsfA2 (aa 122 to 209). The insert of pAS1 coding for PADC1–Gal4p-DBD–HR-A/B(LpHsfA2) was inserted into the ApaI/SpeI sites of pPC86 (4).
Plasmids coding for fusion proteins between Gal4p-AD and HsfA1 (aa 131 to 527) and HsfA2 (aa 231 to 351) were generated by subcloning the corresponding SalI-XbaI fragments derived from plasmid pHSFA1K/R1 (M2) or pHSFA2K/R2 (M3) (15) into pADGal4.
Library construction, screening, and interaction studies.
A library of hybrid proteins between the yeast Gal4p-AD and oligo(dT)-primed cDNA fragments, derived from an HS tomato cell culture, was constructed in pADGal4 with a HybriZAP Two-Hybrid cDNA Gigapack cloning kit (Stratagene). The average insert size was 1.5 kb. The S. cerevisiae YRG-2 reporter strain carrying the HIS3 and lacZ genes, both under control of a Gal4p-inducible promoter, was sequentially transformed with the bait plasmid (pRL123) and this plasmid library. Of the estimated 4 × 106 transformants, 174 were histidine prototrophs. They were recovered and tested by retransformation. Of 69 hybrid constructs proved to be positive, 25 were representative of L. peruvianum HsfA2 (LpHsfA2).
Two-hybrid interaction studies were performed by sequential transformation of both two-hybrid expression plasmids and selection of cotransformants on medium lacking leucine and tryptophan. The cotransformants were tested for histidine prototrophy.
Quantification of β-galactosidase activity.
Yeast cultures were grown overnight in 20 ml of yeast extract-peptone-dextrose medium. The cells were washed with ice-cold 100 mM potassium phosphate (pH 6.5) (KPP) and centrifuged for 5 min at 4°C at 4,500 × g. The pellet was resuspended in 6 volumes of KPP buffer and vortexed for 10 min at 4°C with 0.3 to 0.5 g of glass beads (Sigma). Aliquots (20 μl) of the cell extracts in the wells of a microtiter plate were incubated with 100 μl of Galacton-Star substrate (Galacto-Star kit; Tropix, Bedford, Mass.). Immediately after addition of the substrate, the light emission was registered with a luminometer (Microlumat 2400; EG&G Berthold). The activity is expressed as relative light units (RLU) per second and milligram of protein.
Indirect immunofluorescence.
Cells from rapidly growing tomato cultures, partially protoplasted by a 2-h incubation with 1% mazerozyme–1% cellulase (22), or tobacco protoplasts were fixed for 30 min in 3% paraformaldehyde–50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 6.0). After sedimentation, the cells were washed twice in phosphate-buffered saline (PBS) containing 1% Nonidet NP-40 and transferred to poly-l-lysine-coated coverslips. After being washed twice with PBS and once with PBS–1% BSA (Sigma), the coverslips were incubated for 1.5 h at 37°C with the indicated antiserum diluted 1:1,000 in PBS–1% BSA. After being washed with PBS and PBS–1% BSA, the cells were incubated with fluorescein isothiocyanate-labeled polyclonal goat anti-rabbit immunoglobulin G (Sigma) diluted 1:200 in PBS–1% BSA and then subjected to extensive washing with PBS and mounting in PBS-glycerol (1:3) containing 0.1% phenylenediamine. The immunostaining was analyzed with a Zeiss Axiophot microscope, and micrographs were taken with Ilford HP5 film. For a control, all samples were also stained with 4′,6-diamidino-2-phenylindole (DAPI) to localize the nuclei (pictures not shown) (see Fig. 1).
FIG. 1.
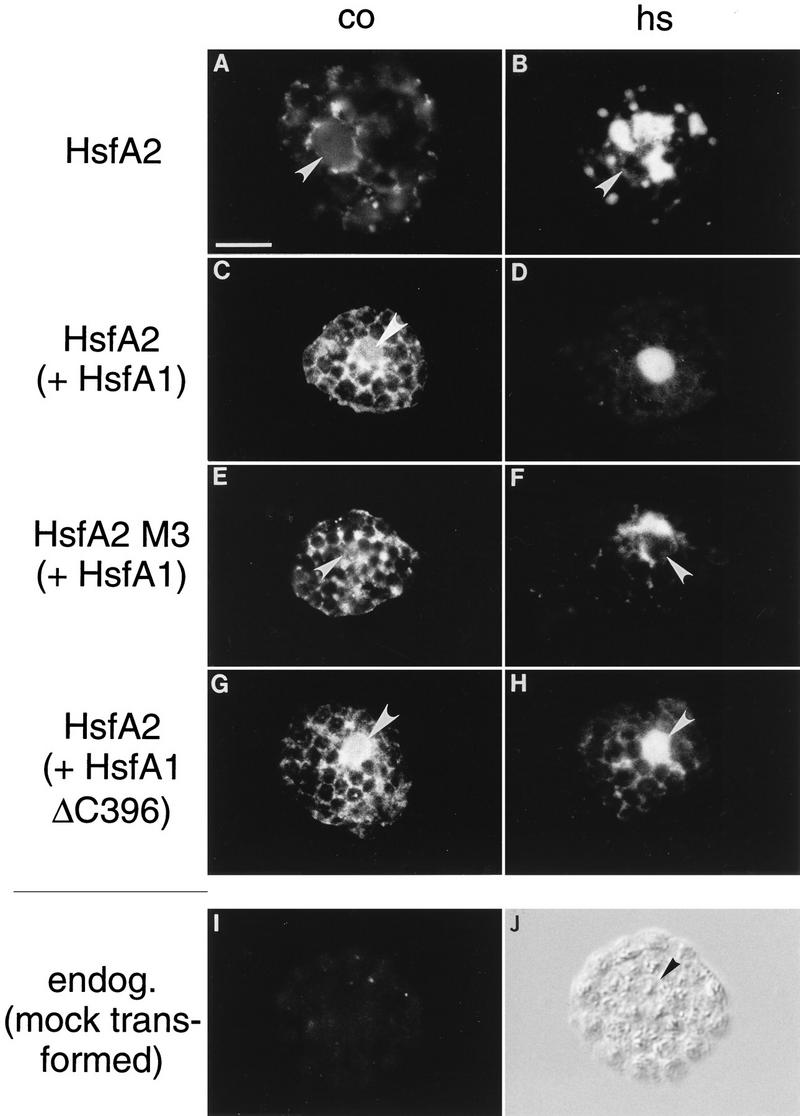
Immunofluorescence of tobacco protoplasts demonstrating nuclear transport of HsfA2 by coexpression with HsfA1. A total of 105 tobacco protoplasts were transformed with the indicated Hsf expression plasmids and incubated for 10 h under control (CO) (A, C, E, and G) and HS (B, D, F, and H) conditions (see Materials and Methods). Samples were processed for immunofluorescence with an antiserum against HsfA2. HsfA2 was expressed alone (A and B) or with wild-type HsfA1 (C and D), the NLS-defective mutant M3 of HsfA2 was expressed with HsfA1 (E and F), and wild-type HsfA2 was coexpressed with a C-terminally truncated version of HsfA1 (ΔC396) (G and H). The position of the nucleus as deduced from the corresponding staining with DAPI is indicated (arrowheads). A control sample showing the background fluorescence of protoplasts transformed with the empty vector only is also shown (I and K). Bar, 10 μm. endog., endogenous.
Electron microscopy.
L. peruvianum cells were initially fixed in 50 mM cacodylate buffer (pH 7.4) containing 1.5% glutaraldehyde (4 h at 4°C) and then fixed with 1% OsO4 in 50 mM cacodylate buffer, pH 7.4 (2 h at 4°C). After a washing, the cell material was en bloque contrasted overnight with 2% uranyl acetate, washed again, dehydrated in ethanol and propylene oxide, and embedded in Spurr (TAAB Laboratories, Aldermaston, United Kingdom) according to standard procedures. Blocks were sectioned with a Reichert Ultracut S ultramicrotome. Micrographs were taken with a Zeiss 902 electron microscope using Agfa Scientia EM film.
For postembedding immunogold labeling, colloidal gold particles were prepared as described by Mühlpfordt (17) for 5-nm-diameter gold particles and by Frens (7) for 15-nm-diameter gold particles. Protein A (Sigma) was coupled to the gold particles according to the protocol of Roth et al. (37).
L. peruvianum cells were prefixed in 20 mM cacodylate buffer (pH 7.4) containing 0.25% glutaraldehyde (30 min at room temperature) and then fixed for 3 h at 25°C in 50 mM cacodylate buffer (pH 7.4) containing 3% formaldehyde and 0.25% glutaraldehyde. The cells were dehydrated in a graded ethanol series with the temperature progressively lowered to −20°C and were finally transferred to LR Gold. After UV light polymerization at −20°C, ultrathin sections were prepared, mounted on Cu grids, and immunolabeled with the indicated antisera (1:200 dilutions in PBS–1%BSA). Before and after treatments with the antisera, sections were washed repeatedly with PBS–1%BSA. Protein A-gold was applied as a solution in PBS–1% BSA diluted to a final optical density at 520 nm of 0.2. Finally, the preparations were extensively washed with PBS and water and stained for 10 min with 2% uranyl acetate.
RESULTS
Nuclear cotransport of HsfA1 and HsfA2 in tobacco protoplasts.
During earlier experiments on the functional characterization of tomato Hsfs in a transient-expression system using tobacco protoplasts, an interesting peculiarity of the HS-inducible HsfA2 was noticed (15). Despite the presence of a functional NLS, HsfA2 is defective in nuclear transport irrespective of HS or control conditions. Instead, it forms large cytoplasmic complexes during HS (Fig. 1A and B). As demonstrated previously (15), this cytoplasmic retention can be overcome by deletion of 8 or 28 aa residues from the C-terminal activator domain (HsfA2ΔC343 or HsfA2ΔC323).
The experimental procedure to create C-terminal deletions for study of the functional significance of putative NLS motifs (15) indicates that the molecular basis of the cytoplasmic retention of HsfA2 may be an intramolecular shielding of the NLS by the C terminus. For the wild-type protein in its native surrounding (tomato cells), an evident possibility would be a conformational change of the protein induced by interaction with the constitutively expressed HsfA1. Under normal conditions, HsfA2 always coexists with HsfA1, which is presumably responsible for the HS-induced expression of the former. To mimic the natural situation, we performed coexpression experiments with both Hsfs in tobacco protoplasts. Indeed, under these conditions, a substantial part of HsfA2 is detected in the nucleus (Fig. 1C and D). The cotransport does not require the full-length HsfA1; it is also achieved by a C-terminally truncated version lacking the entire activator region (HsfA1ΔC396 [Fig. 1G and H]). However, no nuclear import was detected (Fig. 1E and F) if the NLS of HsfA2 was made defective by mutation (15). In the combination of HsfA2 M3 with HsfA1, even part of the carrier HsfA1 is retained in the cytoplasm (data not shown). This effect indicates that the two Hsfs form relatively stable hetero-oligomers. The endogenous Hsf of tobacco protoplasts does not cross-react with our antisera. This is shown by a control protoplast sample mock transformed with the empty vector (Fig. 1I and J) as well as by Western analysis (e.g., see Fig. 8).
FIG. 8.
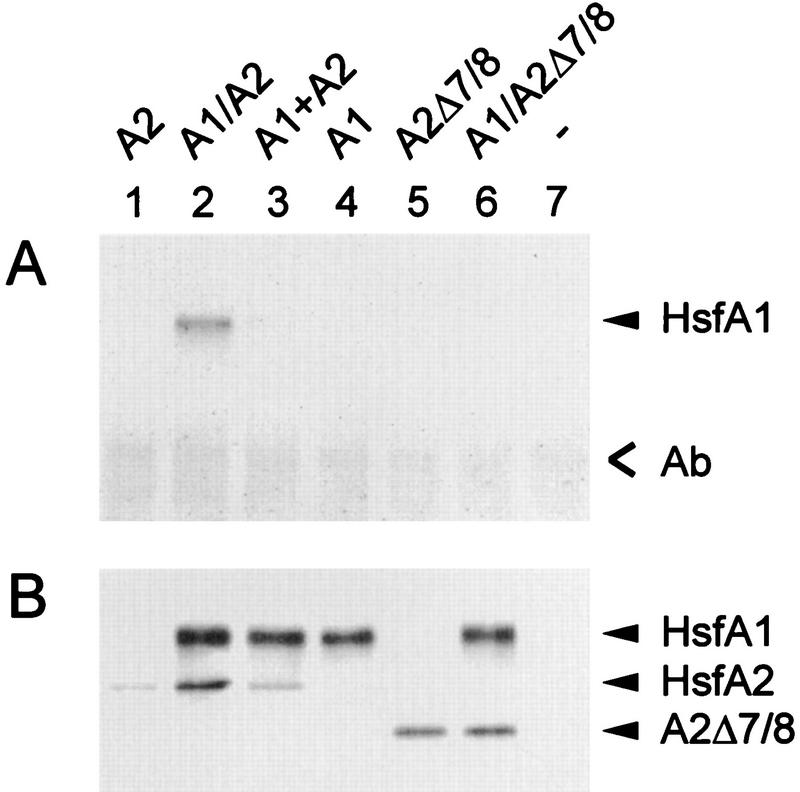
Coimmunoprecipitation experiment with anti-HsfA2 using tobacco protoplast lysates. Samples of 4 × 105 protoplasts transformed with 5 μg of the indicated expression plasmids for HsfA2 (lane 1), HsfA1 (lane 4), and HsfA2Δ7/8 (lane 6) and for neomycin phosphotransferase as a control (lane 7) per 105 protoplasts were harvested after 20 h of expression at 25°C and processed for immunoprecipitation as described in Materials and Methods. Results for protoplasts coexpressing HsfA1 with HsfA2 (lane 2) and HsfA2Δ7/8 (lane 6) are also shown. For lane 3, lysates from samples 1 and 4 were mixed 5 min prior to the addition of the anti-HsfA2. Ab, position of immunoglobulins.
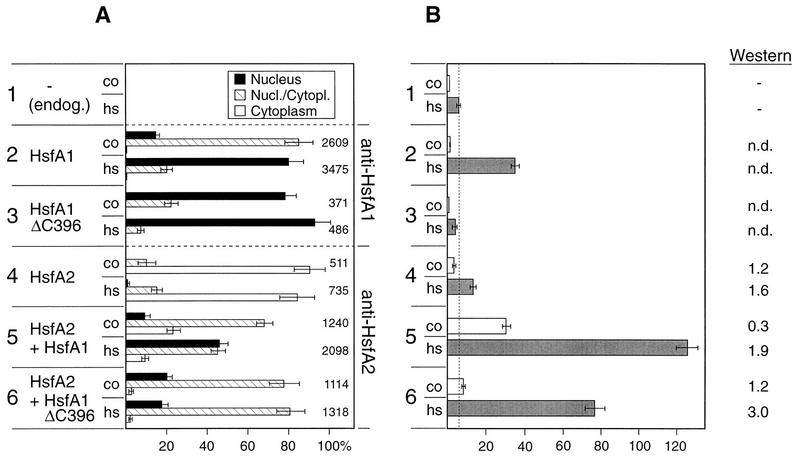
The selective immunofluorescence pictures are complemented by quantitative data compiled in Fig. 2A. Several hundred to thousands of HS and control protoplasts were evaluated and divided into three classes with (i) dominant or exclusive staining of the nucleus (see examples in Fig. 1D and H), (ii) nuclear and cytoplasmic staining (examples in Fig. 1C and G), and (iii) dominant cytoplasmic localization of Hsf (examples in Fig. 1A, B, E, and F). Samples 2 and 3 in Fig. 2A show results obtained with the full-length HsfA1 and its C-terminally truncated version (HsfA1ΔC396), respectively. During HS, both are detected almost exclusively in nuclei. Samples 4 to 6 represent corresponding analyses for HsfA2. If expressed alone (sample 4), most of the HsfA2 is found in the cytoplasm. In contrast to this, coexpression with either form of HsfA1 (samples 5 and 6) results in substantial nuclear import of HsfA2.
FIG. 2.
Interaction of HsfA1 and HsfA2 in tobacco protoplasts: intracellular localization (A) and transactivation assay (B). Protoplasts were transformed with 5 μg of the phsp17gus reporter (51) and the indicated Hsf expression plasmids. After 10 h of incubation under control (co) and HS (hs) conditions (see Materials and Methods), cells were harvested and processed for immunofluorescence (A) and the Gus assay (B). For evaluation of the intracellular localization of Hsfs, protoplasts were assigned to three classes (see examples in Fig. 1) with dominant nuclear (Nucleus), nuclear-cytoplasmic (Nucl./Cytopl.), and dominant cytoplasmic (Cytoplasm) localization of the Hsf. Samples 2 and 3 were probed with antiserum against HsfA1, and samples 4 to 6 were probed with antiserum against HsfA2. The total number of protoplasts evaluated is given to the right of the bars in panel A. The effective nuclear import of HsfA2 in the presence of HsfA1 (samples 5 and 6) is also evident from the increased relative Gus expression levels shown in panel B. To optimize the detection of the synergistic effect, we reduced the amount of the HsfA1 expression plasmids in this assay to 0.25 μg per 100,000 protoplasts. Under these conditions, the intracellular level of the carrier Hsf is barely detectable by Western analysis (n.d., not detected). In contrast to this, HsfA2 expression in samples 4 to 6 was monitored by Western blotting. endog., endogenous Hsf of tobacco protoplasts.
As is to be expected, cotransport has important consequences for the activator potential of HsfA2, as deduced from the expression of the corresponding gus reporter construct (Fig. 2B). Samples 1 to 3 represent essential control and reference values. The low Gus expression in sample 1 transformed with the gus reporter plasmid alone reflects the evidently very low level of endogenous Hsfs. It was considerably increased in the presence of HsfA1 (Fig. 2B, sample 2). The inactive HsfA1ΔC396 probably competes with the endogenous Hsf for binding sites on the DNA. Hence, Gus expression is reduced below the level obtained with the reporter alone (Fig. 2B, sample 3). Important information derives from the results for samples 4 to 6. The modest stimulation of Gus expression by HsfA2 alone (Fig. 2B, sample 4) is markedly increased by coexpression with either HsfA1 or HsfA1ΔC396 (samples 5 and 6). The synergistic effects of HsfA1 and HsfA2 in this transactivation assay are most prominent when a natural reporter construct (phsp17gus) containing a promoter fragment from the soybean hsp17 gene is used instead of the more active pHSE9gus with a synthetic oligonucleotide with nine adjacent heat shock element (HSE) modules used previously (15, 51). The overall expression levels achieved with the former construct are only about 10% of those with the latter, but the expression system with the phsp17gus reporter reacts more sensitively to changes of the Hsf levels in the nucleus. Interestingly, the data from Western blots (Fig. 2B) indicate that the presence of HsfA1 results in increased levels of HsfA2, especially under HS conditions. The reason for this is unclear, but it is tempting to speculate that the structure-bound forms of HsfA2 in the nucleus and the HSG are more stable than the cytoplasmic soluble form prevailing in cells at control temperatures. Clearly, this matter requires further investigation. At any rate, an increase of the HsfA2 level alone, e.g., by using larger amounts of the corresponding expression plasmid for transformation, does not result in markedly higher levels of Gus expression (data not shown).
The evident functional module for a physical interaction between the two Hsfs is the oligomerization domain HR-A/B. Both Hsfs belong to the class A type of plant Hsfs (see the review by Nover et al. [30]). In contrast to the class B type, e.g., tomato HsfB1, and all non-plant Hsfs, HsfA1 and HsfA2 contain a 21-aa residue insertion in their HR-A/B regions. Because of the exclusive nuclear localization of HsfB1, we tested coexpression of HsfA2 with HsfB1 as well. As is to be expected from the specificity of the HR-A/B regions, HsfB1 does not function as a mediator of nuclear import of HsfA2 (data not shown).
Expression and localization of Hsfs in tomato cell cultures.
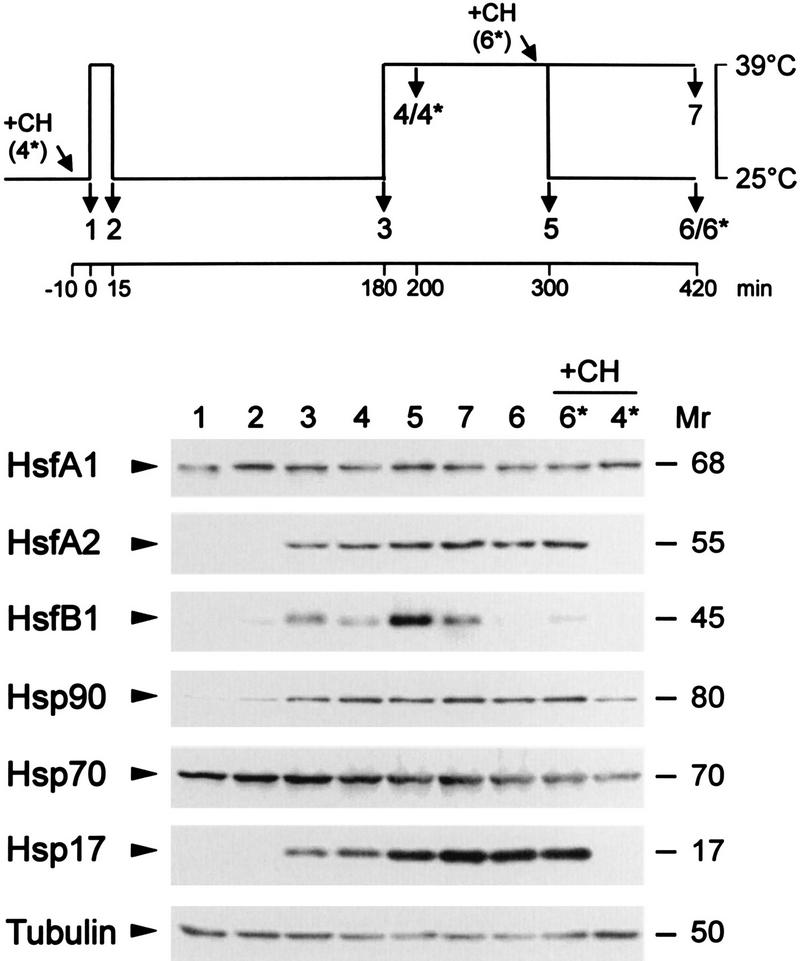
The intriguing observation of the interaction of HsfA1 and HsfA2 during nuclear transport in tobacco protoplasts led us to investigate the natural Hsf system in tomato cell cultures. In contrast to the former, the latter is characterized by a continuously changing composition and intracellular distribution of Hsfs in the course of an HS and recovery period. As described earlier (26, 29), our standard procedure for study of such dynamic changes comprises temperature shift experiments, as indicated in the pictograph of Fig. 3. Samples of tomato cell suspension cultures were harvested at the indicated time points and analyzed for expression levels of Hsfs and Hsps (Fig. 3).
FIG. 3.
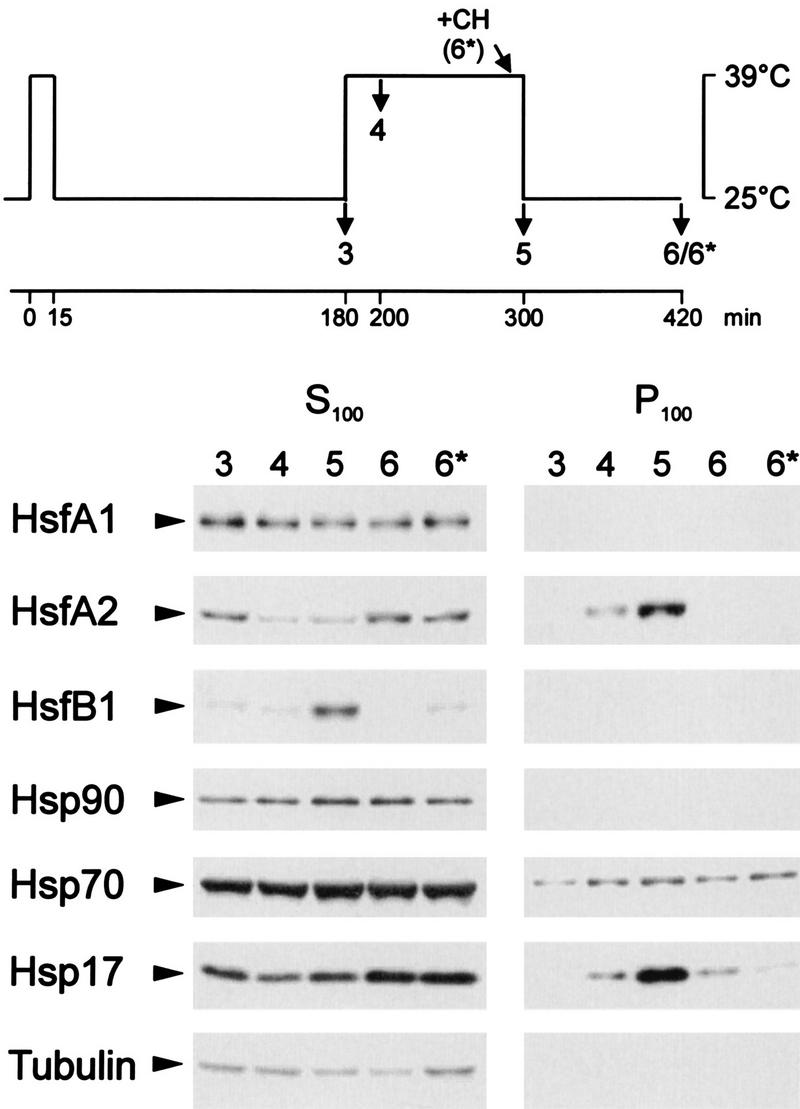
Expression levels of Hsfs and Hsps in tomato cell cultures. Cell suspension cultures were subjected to the HS treatment indicated in the pictograph at the top. Total protein extracts were analyzed by Western blotting using the indicated antisera. Two samples were supplemented with 10 μg of cycloheximide (CH) at the beginning of the HS regimen (4*) or before the recovery (6*). Mrs are indicated in thousands.
In agreement with the Northern analyses reported earlier (42, 44), the HsfA1 level does not change markedly in the course of the HS regimen, similar to the results for the housekeeping protein tubulin and proteins of the Hsp-Hsc70 complex used as a control. After the initial 15-min pulse HS, increasing levels of HsfA2 and HsfB1 complement the constitutively expressed HsfA1 (Fig. 3, samples 2 and 3). The ongoing accumulation of HsfA2 during the HS parallels that of Hsp17 (Fig. 3, samples 4 and 5), which represents a typical cytosolic Hsp of tomato absent in nonstressed cells (26, 28). In contrast to the relative stability of the HsfA2 level in cells under long-term HS (Fig. 3, samples 5 and 7) or in the recovery period (samples 6 and 6*), the HsfB1 level rapidly declines. Interestingly, this decay is retarded if cycloheximide is added prior to the recovery (Fig. 3, sample 6*). None of the three HS-induced proteins (HsfA2, HsfB1, and Hsp17) are formed if cycloheximide is added prior to the first HS treatment (Fig. 3, sample 4*).
The intracellular localization of the three Hsfs in tomato was studied by immunofluorescence (Fig. 4). To this aim, we compared cell culture samples from time points 1, 3, 4, 5, and 6 of the HS regimen used in Fig. 3. (i) Similar to the results with tobacco protoplasts (Fig. 1), the constitutively expressed HsfA1 is distributed between the nuclei and the cytoplasm at 25°C (Fig. 4A and B), migrates to the nucleus during HS (Fig. 4C and D), and gradually returns to the cytoplasm during the recovery period (Fig. 4E). (ii) Irrespective of the temperature conditions, HsfB1 is always found in the nucleus (Fig. 4K to O). (iii) The most complex pattern is observed for HsfA2 (Fig. 4F to J). It is not detected in control cells (Fig. 4F), but after the 15-min pulse induction, it is rapidly synthesized and localized mainly in the cytoplasm (Fig. 4G). Shortly after the onset of the second HS, practically all HsfA2 is found in the nucleus (Fig. 4H). We assume that this effective nuclear transport reflects the interaction with HsfA1. With the ongoing accumulation of HsfA2 during the second HS and the following recovery, most of it is again detected in the cytoplasm, giving rise to a speckled fluorescence staining (Fig. 4I and J).
FIG. 4.
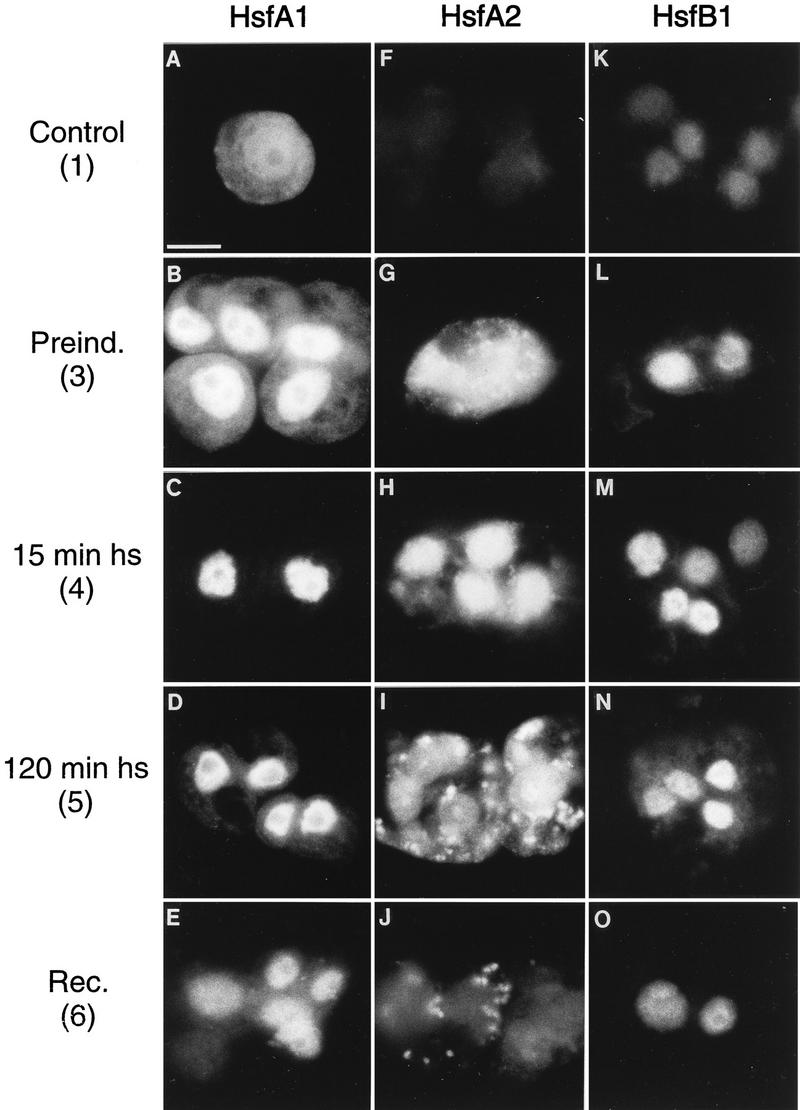
Intracellular localization of HsfA1 and HsfA2 in tomato cell cultures. Cells were harvested in the course of an HS treatment as indicated by the numbers, which refer to the pictograph in Fig. 3. Samples were processed for immunofluorescence with antiserum against HsfA1 (A to E), antiserum against HsfA2 (F to J), and antiserum against HsfB1 (K to O). The speckled cytoplasmic fluorescence in panels I and J results from the accumulation of HsfA2 in HSG (see Fig. 5). Bar, 10 μm.
HsfA2 becomes part of the HSG.
The coarse, speckled fluorescence generated by antibody staining of HsfA2 (Fig. 4I and J) indicates formation of large Hsf-containing protein aggregates in the cytoplasm. As described previously, a general peculiarity of HS plants is the formation of 40-nm-diameter ribonucleoprotein (RNP) aggregates in the cytoplasm (HSG) which represent major sites of Hsp accumulation (25, 28, 29). Using immunogold labeling and cell fractionation techniques, we investigated the possible colocalization of Hsp17 as a marker of the HSG fraction with HsfA2. Figure 5 shows ultrathin sections of tomato cells after a preinduction period followed by a 2-h HS. The typical, highly contrasted 40-nm-diameter particles in the cytoplasm (Fig. 5A and B) can be immunolabeled with antibodies against HsfA2 (Fig. 5A) and Hsp17 (Fig. 5C). In Fig. 5D, we show the result of a double immunolabeling using anti-Hsp17–protein A–15-nm-diameter gold in the first step and anti-HsfA2–protein A–5-nm-diameter gold in the second step. Both proteins are evidently part of the same type of HSG complex. In fact, as a control for specificity, the ultrathin section shown in Fig. 5C was also treated with the double labeling procedure, but with preimmune serum used for the second step. In this case, no protein A–5-nm-diameter gold was bound.
FIG. 5.
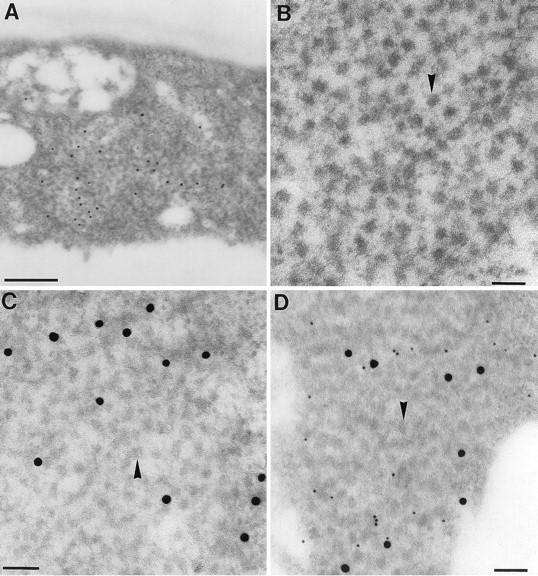
Colocalization of HsfA2 and Hsp17 in cytoplasmic HSG. HS tomato cell cultures corresponding to sample 5 in Fig. 3 were processed for electron microscopy as described in Materials and Methods. (A, C, and D) Cells embedded in LR Gold. Ultrathin sections were used for immunolabeling with anti-HsfA2 (A) and with anti-Hsp17 (C); both were detected with protein A–15-nm-diameter gold. The ultrathin section in panel D was double labeled, first with anti-Hsp17–protein A–15-nm-diameter gold and then with anti-HsfA2–protein A–5-nm-diameter gold. (B) Cells embedded in Epon and contrasted with uranyl acetate show the 40-nm-diameter particles (arrowheads) characteristic for HSG. Bars, 500 (A) and 100 (B to D) nm.
As reported earlier (28, 29), an alternative method to demonstrate the HS-dependent structural binding of proteins to the HSG is a simple cell fractionation procedure using sonication of tomato cells in a HEPES buffer supplemented with 500 mM NaCl, 30 mM EDTA, 1% Nonidet P-40, and 0.2% sarcosyl (for details, see Materials and Methods). Under these stringent buffer conditions, most cell components are disrupted effectively, but HSG remain intact and can be sedimented by 1 h of centrifugation at 100,000 × g. The protein compositions of corresponding HSG fractions (Fig. 6, P100) and of the supernatant (S100) were analyzed by Western blotting. Irrespective of the temperature conditions and the pretreatment, no HsfA1, HsfB1, tubulin, or Hsp90 is detectable in the P100 fraction. However, as expected, the HSG fraction from HS cells contains HsfA2 and Hsp17 (Fig. 6, samples 4 and 5), and these proteins are released in the recovery period (sample 6). The intracellular redistribution of HsfA2 and Hsp17 between the structure-bound and soluble forms is independent of new synthesis of these proteins (Fig. 6, sample 6* with cycloheximide added before the recovery). This is reflected by the inversely changing levels of the two proteins in the S100 fraction.
FIG. 6.
Cofractionation of HsfA2 and Hsp17 in tomato cell cultures. Cells were harvested at the indicated time points of an HS regimen. After disruption by sonication, the crude homogenate was fractionated by centrifugation to give fractions with soluble proteins (S100) and structure-bound proteins (P100). The proteins were dissolved in SDS sample buffer and processed for Western analysis using the indicated antisera for detection. Under HS conditions (samples 4 and 5), substantial proportions of the HsfA2 and Hsp17, but not of the other proteins tested, are found in a high-molecular-weight sedimentable form (HSG), which dissociates in the recovery (samples 6 and 6*).
Physical interaction of HsfA1 and HsfA2.
We confirmed the direct physical interaction of the two Hsfs by two independent methods and expression systems: (i) coimmunoprecipitation of whole-cell protein extracts from tomato cells and tobacco protoplasts and (ii) two-hybrid interaction tests in yeast.
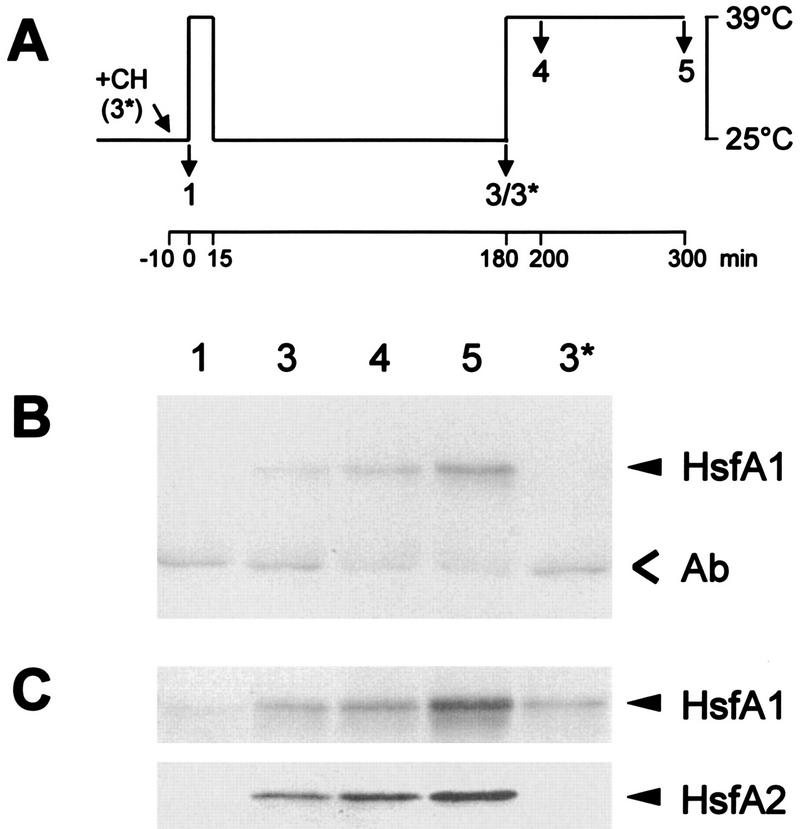
For coimmunoprecipitation (Fig. 7), tomato cell cultures were HS induced to trigger expression of HsfA2 (Fig. 7A). Total cell lysates were prepared, separated by SDS-PAGE, and blotted for detection of HsfA1 and HsfA2 (Fig. 7C). Similar to the results presented in Fig. 3, HsfA1 is present in all samples irrespective of control (Fig. 7C, sample 1) or HS (samples 3 to 5) conditions, whereas HsfA2 is detected only in the samples from HS cells (samples 3 to 5). It was not formed when 10 μg of cycloheximide per ml was added at the onset of the first HS treatment (Fig. 7C, sample 3*). Figure 7B shows the results obtained by coimmunoprecipitation with a rabbit antiserum against HsfA2. The coprecipitated HsfA1 was detected by immunoblotting with the corresponding chicken antiserum. Clearly, a substantial amount of HsfA1 can be precipitated with the HsfA2 antiserum whenever HsfA2 is present, but not in the two samples lacking HsfA2 (samples 1 and 3*).
FIG. 7.
Coimmunoprecipitation of HsfA1 and HsfA2 from lysates of tomato cell suspension cultures. Total cell lysates were prepared for coimmunoprecipitation with HsfA2 antiserum (see Materials and Methods). Protein samples were from control cells expressing HsfA1 only (sample 1) and from cultures subjected to the HS treatment indicated at the top (A) (samples 3 to 5). Western blots of the selected samples with antisera detecting HsfA1 and HsfA2 (C) and the results from the corresponding coimmunoprecipitation with anti-HsfA2 using chicken anti-HsfA1 for detection (B) are shown. HsfA1 is precipitated only in samples 3 to 5 containing HsfA2. Ab, position of immunoglobulins.
Similar results were obtained with lysates from tobacco protoplasts after expression of HsfA2 or HsfA1 alone or together (Fig. 8, lanes 1 and 4 versus lane 2). The experimental flexibility of this transient-expression system allows two important additional controls to be included. (i) Coprecipitation of HsfA1 by anti-HsfA2 is observed only after coexpression of both Hsfs (Fig. 8, lane 2) but not when two lysates containing HsfA2 or HsfA1 were mixed (lane 3). (ii) Interaction between the two Hsfs requires the oligomerization domain (HR-A/B region). There is no coprecipitation of HsfA1 if coexpressed with an HR-A/B deletion form of HsfA2 (HsfA2Δ7/8) (Fig. 8, lane 5 versus lane 6).
Two-hybrid test.
Initially, we used a C-terminally truncated HsfA1 fused to Gal4p-DBD (Fig. 9, bait B) as bait for detection of interacting proteins expressed from an oligo(dT)-primed HS tomato library. Among the positive clones, we identified a considerable number of clones coding for C-terminal parts of HsfA2. The longest clone started at codon 95 (Fig. 9, clone 3); the shortest started at codon 168 (clone 7). Evidently, clone 7 defines the minimum part of the HR-A/B region required for interaction with HsfA1. To verify this conclusion and extend the results, we created an additional clone (clone 8) lacking the entire HR-A/B region. As expected, it was unable to interact with baits B and C. The latter contains the HR-A/B region of HsfA2. It interacts with all constructs of HsfA2 identified by the initial screening (clones 3 to 7). In this context, it is surprising and unexplained that all heterologous interactions (A1-A2) as well as the homologous combination A2-A2 are positive in the two-hybrid test on the basis of His prototrophy (Fig. 9). In contrast to this, yeast strains with the homologous combination A1-A1 are nonviable on His-free medium. In support of this, we did not find any HsfA1-specific clone in the initial screening. Essential controls for the specificity of the protein interactions are the nonviable strains containing Gal4p-DBD alone (bait A) and the combinations of baits B and C with Gal4p-AD alone (clone 1) or with the fusion construct of HsfA2 lacking the entire HR-A/B region (clone 8).
FIG. 9.
Yeast two-hybrid test for interaction between HsfA1 and HsfA2. (Top) Basic structure of the two tomato Hsfs. AHA1 and AHA2, activator modules (for details, see reviews by Nover et al. [30] and Nover and Scharf [27]). Sequence details for the HR-A/B regions with the conserved heptad repeat pattern of hydrophobic amino acid residues are given. For the two-hybrid constructs, we used the usual N-terminal (aa 1 to 147) and C-terminal (aa 768 to 881) parts of the yeast Gal4 activator protein (see Materials and Methods). (Bottom) For the two-hybrid test, the baits were composed of the DBD or HR region of Gal4p (aa 1 to 147) alone (bait A) or fused to the indicated parts of the tomato Hsfs, i.e., HsfA1 (aa 23 to 448 [bait B]) and HsfA2 (aa 122 to 209 [bait C]). The activators combined with them were Hsf fragments fused to the C-terminal Gal4p activator domain (Gal4p-AD) (samples 2 to 8). Interaction is indicated by growth (+) or nongrowth (−) of the cultures on His-free synthetic dextrose minimal (SD) medium and by the activity measured in the lacZ assay (RLU per second and milligram of protein [see Materials and Methods]). β-Gal, β-galactosidase; prot., protein.
The yeast strain used for the two-hybrid test contains an additional, less sensitive chromosomal marker with a Gal4p-dependent promoter and the lacZ gene as a reporter. This allows estimation of the efficiency of interaction between two protein fragments. For this purpose, we used cell extracts of the given yeast strains and the Galacto-Star detection kit (Tropix), which is based on the cleavage of a luminescent substrate. The results are given as RLU per milligram of protein (see Materials and Methods). The expression levels of baits B and C and of hybrid activator proteins 2 to 8 were similar in all combinations as detected by Western blotting (data not shown).
Basically, the results support the conclusions from the assay for His prototrophy. All heterologous combinations (A1-A2 or A2-A1) result in significant lacZ expression levels, whereas the homologous combinations A1-A1 and A2-A2 are inactive or weakly active, even if they allow survival on His-free medium. However, at present, it is not reasonable to extend the discussion beyond this point because only additional constructs with defined point mutations rather than deletions can help to elaborate the basis for the quantitative differences as indicated by the lacZ assay.
DISCUSSION
On the basis of structural peculiarities of their oligomerization domains (HR-A/B region), the plant Hsfs are assigned to classes A and B. In contrast to class B and all non-plant Hsfs so far investigated, the HR-A/B region of the class A Hsfs is extended by an insertion of 21 aa residues creating a structure of the putative coiled-coil region with two overlapping heptad hydrophobic repeats (see sequences given in Fig. 9 and the summary by Nover et al. [30]). The consequences for the structural organization of this domain and the functional significance of the differences between class A and class B Hsfs are unclear. This reflects also our limited knowledge about the detailed structure of this domain, which is important for oligomerization and control of activity of Hsfs (33, 35, 50, 55).
Here, we present experimental evidence that the tomato Hsf system is characterized by an intriguing interaction between two Hsfs (HsfA1 and HsfA2) which is essential for the nuclear import and probably physical stabilization of HsfA2. The latter is synthesized only in HS cells and accumulates to fairly high levels (Fig. 3). Following the preinduction protocol, we can distinguish three major states of HsfA2 in tomato cell cultures. They are evident from the immunofluorescence (Fig. 1 and 4) and cell fractionation (Fig. 6) data. (i) Shortly after the temperature upshift from 25 to 40°C, most of the HsfA1 and HsfA2 is found in the nucleus (Fig. 4C and H). This corresponds to the behavior of HsfA2 if coexpressed with HsfA1 in tobacco protoplasts (Fig. 1D). (ii) After 2 h of HS with ongoing synthesis, a considerable proportion of the HsfA2, together with Hsp17, can be detected in large cytoplasmic aggregates representing the well-known HSG (Fig. 5). The structural binding of HsfA2 is specific; HsfA1 and other cytosolic proteins (tubulin, Hsp90) are not bound to the HSG fraction (Fig. 6). (iii) The HS-specific, high-salt-resistant structural binding of HsfA2 in the nuclear and HSG fractions is reversible. At the end of a 2-h recovery period, most of the HsfA2 is found in soluble form in the cytoplasm (Fig. 4G and sample 6 in Fig. 6).
Formation of HSG is a general phenomenon in all plant tissues with sufficiently high levels of Hsp synthesis (25). They can readily be detected as large aggregates of electron-dense material composed mainly of 40-nm-diameter particles (Fig. 5A and B). There is experimental evidence that they may function as sites for transient storage and protection of housekeeping mRNP (29). Incorporation of HsfA2 into HSG and its rapid release in the recovery period are new aspects which may indicate an additional function as storage sites for excess Hsf and probably other HS-induced proteins. Our knowledge about the fine structure of the HSG complexes is still very limited. We previously reported on the electron microscopic and biochemical characterization of 10-nm-diameter hollow-core particles (precursor HSG) with a sedimentation coefficient of about 15S (29). We are currently investigating whether HsfA2 is already incorporated into the pre-HSG particles or, more likely, associates with the HSG complexes only during the HS-induced aggregation of the pre-HSG. Transient-expression assays with individual components (Hsp17, HsfA2) in tobacco protoplasts, which are capable of forming HSG from the endogenous precursor particles, are valuable tools in this context.
The HsfA1-assisted nuclear import of HsfA2 (Fig. 1 and 4) depends on a direct physical interaction between the two proteins. This is documented by the coimmunoprecipitation experiments (Fig. 7 and 8) and by the yeast two-hybrid tests (Fig. 9). As expected, the interaction is mediated by the A-type-specific HR-A/B region only (Fig. 8 and 9). HsfB1 does not function as a mediator of nuclear import of HsfA2 or as bait in the two-hybrid test (data not shown). A series of partial clones resulting from the two-hybrid screening allowed definition of the part of the oligomerization domain which is indispensable for the interaction (sequence data in Fig. 9). The strongest interaction is actually found for a HsfA2 fragment lacking the whole N-terminal half of the HR-A/B region. Thus, structural prerequisites for the HsfA1-mediated nuclear import of HsfA2 are the C-terminal half of the HR-A/B region and a functional NLS (Fig. 1E and F). On the other hand, HsfA1 as a carrier does not need its activator domain. Truncated versions, inactive as transcription factors (e.g., HsfA1ΔC396), are equally effective and represent valuable tools with which to demonstrate the increased activator potential of HsfA2 if the cytoplasmic retention is released in coexpression experiments (Fig. 2). It is worth mentioning that the two-hybrid test indicates an unexpected peculiarity. The heterologous combinations of HsfA1 with HsfA2 are more stable (efficient) than the homologous combinations. In fact, the A1-A1 combination cannot be detected at all. However, this matter requires further investigation, since (i) only Hsf fragments with unknown conformational abnormalities were tested in the two-hybrid system and (ii) a positive two-hybrid effect probably requires dimer formation, whereas the natural interaction of Hsfs by their oligomerization domains leads preferentially to trimers.
The pronounced negative effect of the C-terminal HR-C region on the activator function of HsfA2 can be released by deletion of the last 8 aa (15). HsfA2ΔC343 is transported to the nucleus without need for interaction with HsfA1. Though not directly comparable with the particular situation of HsfA2, the role of the C-terminal HR-C region for maintenance of the inactive state and the cytoplasmic localization was also demonstrated for the Drosophila melanogaster, chicken, and human Hsfs (1, 19, 20, 35, 47). The model of an intramolecular interaction between the HR-A/B and HR-C regions in the monomeric Hsf proposed by Rabindran et al. (35) is very suggestive but, unfortunately, not supported by direct experimental evidence, e.g., a two-hybrid test. Probably, the generation and/or maintenance of the inactive state requires not only the HR-C region but additional factors, e.g., corepressors of the chaperone type interacting with it (18, 31, 36, 39, 55, 56).
The time course of formation and the stability of the putative hetero-oligomers of HsfA1 and HsfA2 as well as their composition are unknown. They are evidently not formed readily upon mixing protoplast lysates containing the two proteins separately (Fig. 8, lane 3). However, the procedure of coimmunoprecipitation can be used to search for factors or conditions allowing postsynthetic formation of the HsfA1-HsfA2 hetero-oligomers. Unfortunately, we were never able to demonstrate the existence of monomeric Hsf forms in tomato cells or in tobacco protoplasts during transient expression, nor is there any real evidence for HS-dependent changes of the oligomeric state of plant Hsfs (12, 15, 41a). The HS-induced transition between monomeric, inactive Hsf and the active trimeric form as an inherent part of the activation-deactivation cycle in Drosophila and vertebrates (10, 35, 53, 55, 56) appears to be lacking in plants. It is tempting to speculate that the HsfA2 form in the cytoplasm with a shielded NLS motif represents a homo-oligomeric form, whereas the transport-competent form may represent hetero-oligomers with one or two subunits of HsfA1. Oligomerization itself is evidently not sufficient to open the shielded conformation of HsfA2.
Our findings about a physical interaction of HsfA1 and HsfA2 as prerequisite for efficient nuclear import of the latter help to understand the dynamic changes of HsfA2 distribution in tomato cells. The accumulation and stability of this transcription factor during a prolonged stress period (Fig. 4) are connected with its aggregation in the cytoplasmic HSG together with other HS-induced proteins. Monitoring the Hsf levels by Western blotting, though valuable, may have limited value for the evaluation of data about Hsf-mediated reporter gene expression. In particular, the more-than-additive increase of Gus levels in protoplast samples coexpressing HsfA2 with HsfA1 (Fig. 2) may reflect the increased availability of HsfA2 homo-oligomers in the nucleus. Alternatively, the HsfA1-HsfA2 hetero-oligomers represent not only the transport-competent form but also the more stable and transcriptionally active form. In vivo footprinting data with a defined set of different HS reporter constructs can help to clarify this problem.
One point of concern in the evaluation of our results obtained with tobacco protoplasts is the ill-defined role and expression of the endogenous Hsfs. Because of the evident conservation of Hsf types between distantly related plant species such as maize, Arabidopsis thaliana, soybean, and tomato (30), we can assume that N. plumbaginifolia also contains Hsfs of the A1, A2, and B1 types. Fortunately for the application of our antisera used for immunofluorescence (Fig. 1), Western blotting (Fig. 8 and reference 15), and coimmunoprecipitation (Fig. 8), there is essentially no detectable cross-reactivity of Hsfs or cross-reacting Hsf material in the tobacco protoplasts. This situation probably reflects the low level of endogenous Hsfs and the well-known high degree of sequence variability in the C-terminal parts of plant Hsfs. However, even a low level of the putative tobacco HsfA1 might be sufficient to cause some nuclear import and thus transcriptional activity of the tomato HsfA2 if expressed in the absence of the homologous HsfA1 (Fig. 2, sample 4). Formally, we cannot exclude this possibility at present. It may affect the sensitivity of the detection system by reducing the synergistic effects by coexpression (Fig. 2), but it would not influence the general conclusions.
ACKNOWLEDGMENTS
We thank Gisela Englich and Daniela Bublak for excellent technical assistance and Karsten Melcher, Christoph Forreiter, Raffaella Caligaris, and Roger Stuger for helpful discussions and comments during the preparation of the manuscript. We thank Eckhardt Treuter for guidance and technical advice during the early steps of the two-hybrid screening.
This work was supported by grants from the Deutsche Forschungsgemeinschaft to L.N. and K.D.S., by the Fonds der Chemischen Industrie, and by a grant from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie to K.D.S.
REFERENCES
- 1.Baler R, Dahl G, Voellmy R. Activation of human heat shock genes is accompanied by oligomerization, modification, and rapid translocation of heat shock transcription factor. Mol Cell Biol. 1993;13:2486–2496. doi: 10.1128/mcb.13.4.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bienz M, Pelham H R B. Expression of Drosophila heat-shock protein in Xenopus oocytes: conserved and divergent regulatory signals. EMBO J. 1982;1:1583–1588. doi: 10.1002/j.1460-2075.1982.tb01359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boscheinen O, Lyck R, Queitsch C, Treuter E, Zimarino V, Scharf K-D. Heat stress transcription factors from tomato can functionally replace Hsf1 in the yeast Saccharomyces cerevisiae. Mol Gen Genet. 1997;255:322–331. doi: 10.1007/s004380050503. [DOI] [PubMed] [Google Scholar]
- 4.Chevray P M, Nathans D. Protein interaction cloning in yeast: identification of mammalian proteins that react with the leucine zipper of Jun. Proc Natl Acad Sci USA. 1992;89:5789–5793. doi: 10.1073/pnas.89.13.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czarnecka-Verner E, Yuan C X, Foxand P C, Gurley W B. Isolation and characterization of six heat shock transcription factor cDNA clones from soybean. Plant Mol Biol. 1995;29:37–51. doi: 10.1007/BF00019117. [DOI] [PubMed] [Google Scholar]
- 6.Fiorenza M T, Farkas T, Dissing M, Kolding D, Zimarino V. Complex expression of murine heat shock transcription factors. Nucleic Acids Res. 1995;23:467–474. doi: 10.1093/nar/23.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frens G. Controlled nucleation for the regulation of the particle size in monodisperse gold solution. Nat Phys Sci. 1973;241:20–22. [Google Scholar]
- 8.Gagliardi D, Breton C, Chaboud A, Vergne P, Dumas C. Expression of heat shock factor and heat shock protein 70 genes during maize pollen development. Plant Mol Biol. 1995;29:841–856. doi: 10.1007/BF00041173. [DOI] [PubMed] [Google Scholar]
- 9.Goodson M L, Park-Sarge O K, Sarge K D. Tissue-dependent expression of heat shock factor 2 isoforms with distinct transcriptional activities. Mol Cell Biol. 1995;15:5288–5293. doi: 10.1128/mcb.15.10.5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodson M L, Sarge K D. Heat-inducible DNA binding of purified heat shock transcription factor 1. J Biol Chem. 1995;270:2447–2450. doi: 10.1074/jbc.270.6.2447. [DOI] [PubMed] [Google Scholar]
- 11.Harrison C J, Bohm A A, Nelson H C M. Crystal structure of the DNA binding domain of the heat shock transcription factor. Science. 1994;263:224–227. doi: 10.1126/science.8284672. [DOI] [PubMed] [Google Scholar]
- 12.Hübel A, Schöffl F. Arabidopsis heat shock factor: isolation and characterization of the gene and the recombinant protein. Plant Mol Biol. 1994;26:353–362. doi: 10.1007/BF00039545. [DOI] [PubMed] [Google Scholar]
- 13.Kroeger P E, Sarge K D, Morimoto R I. Mouse heat shock transcription factor 1 and factor 2 prefer a trimeric binding site but interact differently with the hsp70 heat shock element. Mol Cell Biol. 1993;13:3370–3383. doi: 10.1128/mcb.13.6.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J H, Hübel A, Schöffl F. Derepression of the activity of genetically engineered heat shock factor causes constitutive synthesis of heat shock proteins and increased thermotolerance in transgenic Arabidopsis. Plant J. 1995;8:603–612. doi: 10.1046/j.1365-313x.1995.8040603.x. [DOI] [PubMed] [Google Scholar]
- 15.Lyck R, Harmening U, Höhfeld I, Scharf K-D, Nover L. Intracellular distribution and identification of the nuclear localization signals of two tomato heat stress transcription factors. Planta. 1997;202:117–125. doi: 10.1007/s004250050110. [DOI] [PubMed] [Google Scholar]
- 16.Mezger V, Rallu M, Morimoto R I, Morange M, Renard J P. Heat shock factor 2-like activity in mouse blastocysts. Dev Biol. 1994;166:819–822. doi: 10.1006/dbio.1994.1361. [DOI] [PubMed] [Google Scholar]
- 17.Mühlpfordt H. The preparation of colloidal gold particles using tannic acid as an additional reducing agent. Experientia. 1982;38:1127–1128. [Google Scholar]
- 18.Nair S C, Toran E J, Rimerman R A, Hyermstad S, Smithgall T E, Smith D F. A pathway of multi-chaperone interactions common to diverse regulatory proteins: estrogen receptor, Fes tyrosine kinase, heat shock transcription factor Hsf 1, and the aryl hydrocarbon receptor. Cell Stress Chap. 1996;1:237–250. doi: 10.1379/1466-1268(1996)001<0237:apomci>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakai A, Morimoto R I. Characterization of a novel chicken heat shock transcription factor, heat shock factor 3, suggests a new regulatory pathway. Mol Cell Biol. 1993;13:1983–1997. doi: 10.1128/mcb.13.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakai A, Kawazoe Y, Tanabe M, Nagata K, Morimoto R I. The DNA-binding properties of two heat shock factors, Hsf1 and Hsf3, are induced in the avian erythroblast cell line HD6. Mol Cell Biol. 1995;15:5268–5278. doi: 10.1128/mcb.15.10.5268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakai A, Tanabe M, Kawazoe Y, Inazawa J, Morimoto R I, Nagata K. HSF4, a new member of the human heat shock factor family which lacks properties of a transcriptional activator. Mol Cell Biol. 1997;17:469–481. doi: 10.1128/mcb.17.1.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neumann D, zur Nieden U, Manteuffel R, Walter G, Scharf K-D, Nover L. Intracellular localization of heat-shock proteins in tomato cell cultures. Eur J Cell Biol. 1987;43:71–81. [Google Scholar]
- 23.Nover L. Expression of heat shock genes in homologous and heterologous systems. Enzyme Microb Technol. 1987;9:130–144. [Google Scholar]
- 24.Nover L, Kranz E, Scharf K-D. Growth cycle of suspension cultures of Lycopersicon esculentum and L. peruvianum. Biochem Physiol Pflanzen. 1982;177:483–499. [Google Scholar]
- 25.Nover L, Neumann D, Scharf K-D, editors. Heat shock and other stress response systems of plants. Results and problems in cell differentiation. Vol. 16. Berlin, Germany: Springer-Verlag; 1990. [PubMed] [Google Scholar]
- 26.Nover L, Scharf K-D. Synthesis, modification and structural binding of heat shock proteins in tomato cell cultures. Eur J Biochem. 1984;139:303–313. doi: 10.1111/j.1432-1033.1984.tb08008.x. [DOI] [PubMed] [Google Scholar]
- 27.Nover L, Scharf K-D. Heat stress proteins and transcription factors. Cell Mol Life Sci. 1997;53:80–103. doi: 10.1007/PL00000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nover L, Scharf K-D, Neumann D. Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol Cell Biol. 1983;3:1648–1655. doi: 10.1128/mcb.3.9.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nover L, Scharf K-D, Neumann D. Cytoplasmic heat shock granules are formed from precursor particles and are associated with a specific set of mRNAs. Mol Cell Biol. 1989;9:1298–1308. doi: 10.1128/mcb.9.3.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nover L, Scharf K D, Gagliardi D, Vergne P, Czarnecka-Verner E, Gurley W B. The Hsf world: classification and properties of plant heat stress transcription factors. Cell Stress Chap. 1996;1:215–223. doi: 10.1379/1466-1268(1996)001<0215:thwcap>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nunes S L, Calderwood S K. Heat shock factor-1 and the heat shock cognate 70 protein associate in high molecular weight complexes in the cytoplasm of NIH-3T3 cells. Biochem Biophys Res Commun. 1995;213:1–6. doi: 10.1006/bbrc.1995.2090. [DOI] [PubMed] [Google Scholar]
- 32.Pelham H R B. A regulatory upstream promoter element in the Drosophila hsp 70 heat-shock gene. Cell. 1982;30:517–528. doi: 10.1016/0092-8674(82)90249-5. [DOI] [PubMed] [Google Scholar]
- 33.Peteranderl R, Nelson H C M. Trimerization of the heat shock transcription factor by a triple-stranded alpha-helical coiled-coil. Biochemistry. 1992;31:12272–12276. doi: 10.1021/bi00163a042. [DOI] [PubMed] [Google Scholar]
- 34.Rabindran S K, Giorgi G, Clos J, Wu C. Molecular cloning and expression of a human heat shock factor. Proc Natl Acad Sci USA. 1991;88:6906–6910. doi: 10.1073/pnas.88.16.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabindran S K, Haroun R I, Clos J, Wisniewski J, Wu C. Regulation of heat shock factor trimerization: role of a conserved leucine zipper. Science. 1993;259:230–234. doi: 10.1126/science.8421783. [DOI] [PubMed] [Google Scholar]
- 36.Rabindran S K, Wisniewski J, Li L, Li G C, Wu C. Interaction between heat shock factor and Hsp70 is insufficient to suppress induction of DNA-binding activity in vivo. Mol Cell Biol. 1994;14:6552–6560. doi: 10.1128/mcb.14.10.6552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roth J, Bendayan M, Orci L. Ultrastructural localization of intracellular antigens by the use of protein A-gold complex. J Histochem Cytochem. 1978;26:1074–1081. doi: 10.1177/26.12.366014. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 39.Sarge K D, Murphy S P, Morimoto R I. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sarge K D, Park-Sarge O K, Kirby J D, Mayo K E, Morimoto R I. Expression of heat shock factor 2 in mouse testis: potential role as a regulator of heat-shock protein gene expression during spermatogenesis. Biol Reprod. 1994;50:1334–1343. doi: 10.1095/biolreprod50.6.1334. [DOI] [PubMed] [Google Scholar]
- 41.Sarge K D, Zimarino V, Holm K, Wu C, Morimoto R I. Cloning and characterization of two mouse heat shock factors with distinct inducible and constitutive DNA binding ability. Genes Dev. 1991;5:1902–1911. doi: 10.1101/gad.5.10.1902. [DOI] [PubMed] [Google Scholar]
- 41a.Scharf, K.-D. Unpublished results.
- 42.Scharf K D, Materna T, Treuter E, Nover L. Heat stress promoters and transcription factors. In: Nover L, editor. Plant promoters and transcription factors. Berlin, Germany: Springer-Verlag; 1994. pp. 121–158. [Google Scholar]
- 43.Scharf K D, Rose S, Thierfelder J, Nover L. Two cDNAs for tomato heat stress transcription factors. Plant Physiol. 1993;102:1355–1356. doi: 10.1104/pp.102.4.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scharf K D, Rose S, Zott W, Schöffl F, Nover L. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA-binding domain of the yeast HSF. EMBO J. 1990;9:4495–4501. doi: 10.1002/j.1460-2075.1990.tb07900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuetz T J, Gallo G J, Sheldon L, Tempst P, Kingston R E. Isolation of a cDNA for HSF2: evidence for two heat shock factor genes in humans. Proc Natl Acad Sci USA. 1991;88:6911–6915. doi: 10.1073/pnas.88.16.6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schultheiss J, Kunert O, Gase U, Scharf K-D, Nover L, Rüterjans H. Solution structure of the DNA-binding domain of the tomato heat stress transcription factor HSF24. Eur J Biochem. 1996;236:911–921. doi: 10.1111/j.1432-1033.1996.00911.x. [DOI] [PubMed] [Google Scholar]
- 47.Sheldon L A, Kingston R E. Hydrophobic coiled-coil domains regulate the subcellular localization of human heat shock factor-2. Genes Dev. 1993;7:1549–1558. doi: 10.1101/gad.7.8.1549. [DOI] [PubMed] [Google Scholar]
- 48.Sistonen L, Sarge K D, Morimoto R I. Human heat shock factors 1 and 2 are differentially activated and can synergistically induce Hsp70 gene transcription. Mol Cell Biol. 1994;14:2087–2099. doi: 10.1128/mcb.14.3.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sistonen L, Sarge K D, Phillips B, Abravaya K, Morimoto R I. Activation of heat shock factor 2 during hemin-induced differentiation of human erythroleukemia cells. Mol Cell Biol. 1992;12:4104–4111. doi: 10.1128/mcb.12.9.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sorger P K, Nelson H C M. Trimerization of a yeast transcriptional activator via a coiled-coil motif. Cell. 1989;59:807–813. doi: 10.1016/0092-8674(89)90604-1. [DOI] [PubMed] [Google Scholar]
- 51.Treuter E, Nover L, Ohme K, Scharf K-D. Promoter specificity and deletion analysis of three tomato heat stress transcription factors. Mol Gen Genet. 1993;240:113–125. doi: 10.1007/BF00276890. [DOI] [PubMed] [Google Scholar]
- 52.Vuister G W, Kim S-J, Orosz A, Marquardt J, Wu C, Bax A. Solution structure of the DNA-binding domain of Drosophila heat shock transcription factor. Nat Struct Biol. 1994;1:605–614. [PubMed] [Google Scholar]
- 53.Westwood J T, Clos J, Wu C. Stress-induced oligomerization and chromosomal relocalization of heat shock factor. Nature. 1991;353:822–827. doi: 10.1038/353822a0. [DOI] [PubMed] [Google Scholar]
- 54.Wu C. Heat stress transcription factors. Annu Rev Cell Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 55.Zuo J, Baler R, Dahl G, Voellmy R. Activation of the DNA-binding ability of human heat shock transcription factor 1 may involve the transition from an intramolecular to an intermolecular triple-stranded coiled-coil structure. Mol Cell Biol. 1994;14:7557–7568. doi: 10.1128/mcb.14.11.7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zuo J, Rungger D, Voellmy R. Multiple layers of regulation of human heat shock transcription factor 1. Mol Cell Biol. 1995;15:4319–4330. doi: 10.1128/mcb.15.8.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]