Abstract
Tim23p is a mitochondrial inner membrane protein essential for the import of proteins from the cytosol. Tim23p contains an amino-terminal hydrophilic segment and a carboxyl-terminal hydrophobic domain (Tim23Cp). To study the functions and interactions of the two parts of Tim23p separately, we constructed tim23N, encoding only the hydrophilic region of Tim23p, and tim23C, encoding only the hydrophobic domain of Tim23p. Only the Tim23C protein is imported into mitochondria, indicating that the mitochondrial targeting information in Tim23p resides in its membrane spans or intervening loops. Tim23Cp, however, cannot substitute for full-length Tim23p, suggesting that the hydrophilic portion of Tim23p also performs an essential function in mitochondrial protein import. We found that overexpression of Tim23Cp is toxic to yeast cells that carry the tim23-1 mutation. Excess Tim23Cp causes Tim23-1p to disappear, leaving tim23-1 cells without a full-length version of the Tim23 protein. If Tim17p, another inner membrane import component, is overexpressed along with Tim23Cp, the toxicity of Tim23Cp is largely reversed and the Tim23-1 protein no longer disappears. In coimmunoprecipitations from solubilized mitochondria, Tim17p associates with the Tim23C protein. In addition, we show that Tim23p and Tim17p can be chemically cross-linked to each other in intact mitochondria. We conclude that the hydrophobic domain encoded by tim23C targets Tim23p to the mitochondria and mediates the direct interaction between Tim23p and Tim17p. In contrast, Tim23Cp cannot be coimmunoprecipitated with Tim23p, raising the possibility that the hydrophobic domain of Tim23p does not interact with other Tim23 molecules.
Most mitochondrial proteins are encoded in the nucleus, translated in the cytosol, targeted to the mitochondria, and then translocated across one or both mitochondrial membranes to their final destinations (reviewed in references 50, 56, 59, and 60). Mitochondrial proteins are usually synthesized as precursors, with positively charged amino-terminal presequences that contain targeting information (23–25, 74). On the mitochondrial surface, precursors encounter several proteins proposed to act as receptors, including Tom70p, Tom37p, Tom22p, and Tom20p (16, 21, 31, 44, 62, 68, 69). Cytosolic chaperones bind precursors to prevent premature folding or aggregation, and one chaperone also plays a role in targeting the precursor to the mitochondrial surface (17, 18, 34). The outer membrane receptors, along with several other proteins, including Tom40p, Tom6p, Tom7p, and Tom8p, make up the TOM complex, which translocates precursors across the mitochondrial outer membrane (32, 33, 46, 70). Translocation of the precursor across the outer membrane is proposed to occur via a transfer of the presequence from a binding site on the outside of the mitochondria to a site located on the inside of the membrane (45). The interaction between the positively charged presequence and the TOM components appears to be at least partly electrostatic (6, 45).
At least three essential yeast proteins located in the mitochondrial inner membrane, Tim23p, Tim17p, and Tim44p, are required for protein import. Tim23p was first identified in a screen of temperature-sensitive yeast mutants for those that accumulated mitochondrial precursors (13). TIM17 was initially isolated as a multicopy suppressor of the temperature-sensitive tim23-1 mutant (57). Tim44p was identified by genetics (40) and as a protein that could be chemically cross-linked to a precursor arrested in transit across the mitochondrial inner membrane (61). The bulk of the Tim44 protein is located on the inside face of the inner membrane, where it associates with a matrix-localized member of the Hsp70 family (mt-Hsp70 [35, 53, 63]). mt-Hsp70 and Tim44p are proposed to drive the translocation of the precursor across the inner membrane (15, 50, 72, 75). In the matrix, the presequence is removed by a two-subunit processing protease (20, 28, 77, 79). Imported proteins then fold into their final conformations with the help of several matrix-localized chaperones (42).
It is not yet known how precursors cross the mitochondrial inner membrane. Tim23p and Tim17p are integral membrane proteins and have been proposed to form part of a protein-translocating channel in the inner membrane (11, 13, 41, 57). The Tim23 protein, formerly called Mas6p or MIM23 (51), has a 9-kDa amino-terminal hydrophilic segment which faces the intermembrane space, followed by a 14-kDa hydrophobic domain containing four predicted membrane spans. The Tim17 protein, formerly called Sms1p or MIM17 (51), contains four potential membrane-spanning segments flanked by short amino- and carboxyl-terminal segments. The C terminus of Tim17p faces the intermembrane space (35). Tim17p is 46% homologous (25% identical) in amino acid sequence to the carboxyl-terminal hydrophobic portion of Tim23p, and Tim17p is also similar to the Tim23p hydrophobic domain in the number of predicted membrane spans and the size of intervening loops.
Both genetic and biochemical data indicate that the inner membrane import components Tim23p, Tim17p, and Tim44p function in a complex. The high-copy suppression of tim23-1 by TIM17 suggests that Tim17p and Tim23p physically interact (57). In addition, mutant alleles of TIM23, TIM17, and TIM44 are synthetically lethal in all pairwise combinations (4). Preliminary biochemical evidence for a translocation complex came from a study showing that Tim23p could be coimmunoprecipitated with two other proteins, each of which was cross-linked to an arrested precursor (55). Subsequently, it was shown that Tim23p, Tim17p, and Tim44p could all be cross-linked to a mitochondrial precursor arrested at the same point in the import pathway (36). Recent studies have now shown that Tim23p and Tim17p can be coimmunoprecipitated from detergent-solubilized mitochondria (2, 3, 7). All these results indicate that there is an inner membrane translocation complex (TIM complex) analogous to the TOM complex in the outer membrane. However, many questions about the TIM complex remain. For example, its composition is unclear, since one study found that Tim23p and Tim17p were associated with proteins of 33 and 14 kDa (3) while another found that Tim23p and Tim17p were associated with proteins of 55 and 20 kDa (4). The structure of the TIM complex is also unknown, since the interactions between individual proteins within the complex have not been defined. Finally, the functions of individual inner membrane components in the translocation pathway have yet to be determined.
In this study, we have focused on the function of Tim23p and its interaction with Tim17p. We show here that the hydrophobic region of Tim23p, called Tim23Cp, is imported into mitochondria and associates with Tim17p. Therefore, the C-terminal hydrophobic half of Tim23p is a distinct domain which targets Tim23p to the mitochondria and mediates the direct interaction with Tim17p. However, we found that Tim23Cp cannot substitute for the full-length Tim23p, suggesting that the hydrophilic amino terminus of Tim23p performs an essential function in mitochondrial protein import. We also found that the truncated Tim23C protein does not associate with full-length Tim23p in the mitochondrial inner membrane.
MATERIALS AND METHODS
Yeast strains and relevant genotypes.
The haploid tim23-1 ura3 trp1 leu2 his3 strain KRR131 was obtained by crossing the MATa mas6-1 strain JE14-5b (13) to the MATα ura3-52 trp1Δ63 his3Δ200 leu2Δ1 strain RJ500 (76). The MATα tim23::URA3 ura3 trp1 leu2 cyh2 strain KRR123 was obtained by crossing the tim23::URA3 strain RJ341 (12a) to YPH858 (71). KRR123 also carries the TIM23-LEU2-CYH2 plasmid pKR1 (see below). The wild-type strain D273-10b (64) and the tim17::TRP1 strain KRR111, which carries the 2μm-TIM17-URA3 plasmid, pKR7 (57), have been described previously. Yeast transformations were performed as described previously (30). Standard yeast media (65) and genetic techniques (54) were used.
Plasmid constructions.
pKR14, a plasmid that expresses Tim23Np, was constructed as follows. A fragment encoding the 9-kDa amino-terminal domain of Tim23p followed by a NotI site was amplified from plasmid pJE2 (13) with oligonucleotide 83 (5′-GGGGCGGCCGCCGTCATCGGTCCACCCACG-3′), oligonucleotide 21 (5′-ATTAACCCTCACTAAAG-3′), and PCR (58). This fragment carrying tim23N was digested with SacI and NotI and ligated into SacI-NotI-digested pJE9 (13) to create pKR14. Tim23Np consists of the first 96 amino acids of Tim23p, followed by Gly-Gly-Arg encoded by the NotI site.
pKR15, which expresses the 14-kDa Tim23C protein, was constructed as follows. We used oligonucleotide 79 (5′-GGGGCGGCCGCGATGACCTATGTTACGG-3′), oligonucleotide 20 (5′-AATACGACTCACTATAG-3′), and pJE2 to obtain by PCR a fragment encoding the C-terminal domain of Tim23p, preceded by a NotI site. The tim23C fragment was digested with NotI and BamHI and ligated into NotI-BamHI-digested pJE9 to create pKR15. Tim23Cp consists of amino acids 95 to 222 of Tim23p and begins with Met-Gly-Gly-Arg as a result of our manipulations. pKR16, which encodes the Tim23C-HA protein, was constructed in several steps as follows. First, pJE7 (13) was digested with NotI, and a 114-bp NotI fragment containing three tandem copies of the hemagglutinin (HA) epitope gene (73) was inserted to create pM37 (12a). We then digested pM37 with NcoI and HindIII and inserted the fragment encoding the Tim23 C terminus and HA epitope into NcoI-HindIII-digested pKR15.
pKR17, which carries tim23C under the control of the inducible GAL1 promoter, was constructed as follows. A PCR fragment containing tim23C with a XhoI site upstream of the start codon was isolated with oligonucleotide 125 (5′-GGGCTCGAGTTAACAGATCACACACAATC-3′), oligonucleotide 98 (5′-AACAGCTATGACCATG-3′), and pKR15. The PCR fragment was digested with XhoI and BamHI and inserted behind the GAL1 promoter region of pRS314GU (47). pKR18, which carries GAL1-tim23C-HA, was constructed by first amplifying tim23C-HA from pKR16 with oligonucleotide 125 and oligonucleotide 21 and then inserting the XhoI-EcoRI-digested PCR fragment into pRS314GU.
pKR19, a multicopy plasmid which carries tim23C (2μm-tim23C), was constructed by digesting pKR15 with SacI and BamHI and inserting the fragment containing the tim23C open reading frame into SacI-BamHI-digested pRS425 (67). pKR20, a centromere-containing plasmid which carries TIM23 (CEN-TIM23), was constructed by digesting pJE11 (13) with XhoI and BamHI and inserting the DNA fragment carrying TIM23 into the LEU2 plasmid pRS315 (67). pKR21, a multicopy plasmid which carries TIM23 (2μm-TIM23), was produced by inserting the TIM23-containing SacI fragment from pJE11 into the LEU2 plasmid pRS425. The 2μm-LEU2-TIM17 plasmid pKR3 and the GAL1-TIM17 plasmid have been described previously (57).
pKR1, a TRP1-CYH2-TIM23 plasmid, was constructed in several steps as follows. First, a 1.1-kbp BamHI fragment containing the CYH2 gene was isolated from pK64 (a gift from J. Boeke). The ends of the fragment were filled in with Klenow, and the fragment was ligated into the NaeI site of pRS314 (67) to produce pKS1 (58a). Next, pM39 (12a) was digested with SacI and BamHI, and the fragment containing TIM23 was inserted into SacI-BamHI-digested pKS1 to produce pKR1.
For in vitro synthesis, genes were inserted into the SP6-containing plasmid pSP64 or pSP65 (Promega Corp., Madison, Wis.). pSP64-tim23C was constructed by first amplifying the tim23C fragment by PCR with pJE2 and oligonucleotides 79 and 20 and then digesting the tim23C fragment with NotI and AvaI and inserting it into NotI-AvaI-digested pT41 (30a). pSP64-tim23N was constructed by first amplifying the tim23N fragment by PCR with pJE2 and oligonucleotides 83 and 21 and then digesting the tim23N fragment with XbaI and NotI and inserting it into XbaI-NotI-digested pT42 (30a). pJE29, which carries TIM23, was constructed by inserting the 1.7-kbp HpaI-BamHI TIM23 fragment from pJE2 into SmaI-BamHI-digested pSP65. pKR13, which carries TIM17 in pSP64, has been described previously (57).
Imports into isolated mitochondria.
Mitochondria were isolated from wild-type strain D273-10b as described previously (10), except that SEH buffer (250 mM sucrose, 1 mM EDTA, 20 mM HEPES-KOH [pH 7.4]) was used in place of breaking buffer. Radiolabeled proteins were produced from the SP6-containing plasmids with 1.5 mCi of [35S]methionine per ml (1,000 Ci/mmol; Amersham) in a coupled transcription-translation system (SP6 TNT system; Promega Biotech) as specified by the manufacturers. For import reactions, mitochondria were suspended in import buffer (61) to a final concentration of 1 mg of protein per ml. A 200-μg portion of mitochondria was used per reaction. A 10-μl volume of lysate containing the radiolabeled protein was added to each assay mixture, and the samples were incubated at 30°C for 30 min. Imports were stopped by transferring the tubes to ice and adding carbonyl cyanide m-chlorophenylhydrazone (Sigma Chemical Co.) to a final concentration of 30 μM. The reaction mixtures were treated with 100 μg of proteinase K (Sigma) per ml on ice for 30 min, and then 1 mM phenylmethylsulfonyl fluoride (PMSF; Sigma) was added. After all the manipulations, the mitochondria were pelleted by centrifugation at 12,500 × g for 10 min, resuspended in 1× sample buffer (125 mM Tris-HCl [pH 6.8], 2% sodium dodecyl sulfate [SDS], 20% glycerol), and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) (37). Radiolabeled proteins were detected by fluorography (8).
Localization of the Tim23C-HA protein.
The tim23-1 leu2 strain KRR131 expressing Tim23C-HA from pKR16 was grown in supplemented S medium containing 2% galactose to an optical density at 600 nm (OD600) of 2.0. The cells were homogenized and separated into a mitochondrial pellet and a postmitochondrial supernatant as described previously (10), except that SEH buffer was used. Proteins from the cell fractions were separated by SDS-PAGE and transferred to Immobilon filters (Millipore, Bedford, Mass.) (19). To detect Tim23C-HA, the filters were decorated with a 1:25 dilution of culture supernatant from 12CA5 cells, which produce a monoclonal antibody against the influenza virus HA epitope (48). The filters were also probed with antisera against the β subunit of the F1-ATPase and against hexokinase (1:10,000 dilutions; gifts from M. Yaffe, University of California, San Diego, Calif.). Immunocomplexes were visualized with a 1:10,000 dilution of horseradish peroxidase-conjugated antibody (Amersham, Arlington Heights, Ill.) followed by chemiluminescence (Amersham).
Production of antibodies against Tim17p.
A peptide based on the carboxyl-terminal region of Tim17p (CEAPSSQPLQA; a gift from K. Kinnally, New York State Department of Health, Albany, N.Y.) was coupled to keyhole limpet hemocyanin (Sigma) with m-maleimidobenzoic acid-N-hydroxysuccinimide (Pierce) as the cross-linking agent (12). Injection of the antigen plus adjuvant into rabbits and collection of antiserum were performed by Covance, Inc. (Denver, Pa.).
Immunoprecipitations from detergent-solubilized mitochondria.
The tim23-1 strain KRR131 and the wild-type strain RJ500 carrying either the GAL1-tim23C-HA plasmid (pKR18) or an empty vector (pRS314GU) were grown in S medium containing 2% raffinose to an OD600 of 1.0. Then 2% galactose was added to each culture to induce expression from the GAL1 promoter, and the cells were grown for 6 h to an OD600 of approximately 2. Mitochondria were isolated from each culture as described previously (10), except that SEH buffer was used. Immunoprecipitations were carried out as follows. Mitochondria were solubilized as described previously (3). Solubilization buffer (0.5% digitonin, 50 mM NaCl, 30 mM HEPES-KOH [pH 7.4], 1 mM PMSF, 1 μg of aprotinin [Calbiochem] per ml, 1 μg of leupeptin [Calbiochem] per ml) was added to the mitochondria to a final protein concentration of 1 mg/ml, and the suspension was incubated at 4°C with gentle agitation for 30 min. After centrifugation at 12,500 × g for 10 min, 20 μl of antiserum against Tim23p (13) or 20 μl of ascites fluid containing antibodies to the HA epitope (BABCO, Berkeley, Calif.) was added to a 1-ml aliquot of the supernatant. The samples were incubated at 4°C with gentle agitation for 5 h. Then 150 μl of a 1:1 slurry of protein A-Sepharose (Sigma Chemical) in solubilization buffer was added to each sample, the tubes were incubated at 4°C for 1.5 h, and the beads were collected by centrifugation. Then 340 μl of 4× sample buffer was added to each supernatant, and the supernatants were heated at 95°C for 5 min. Pellets containing the protein A-Sepharose were washed four times with 1.0 ml of solubilization buffer, and the bound proteins were eluted by two sequential extractions with 75 μl of 2× sample buffer at 65°C for 5 min. The proteins were immunoblotted with antibodies to Tim23p, Tim17p, the HA epitope, and Tim44p (a gift from G. Schatz, Biocenter, Basel, Switzerland).
Chemical cross-linking of mitochondrial proteins.
Mitochondria were isolated from a tim17::TRP1 strain carrying the TIM17-HA plasmid pKR11 (57). Then 500 μg of mitochondria was resuspended in 500 μl of breaking buffer (0.6 M sorbitol, 20 mM HEPES-KOH [pH 7.4], 1 μg of PMSF per ml, 1 mM aprotinin, 1 mM leupeptin), and the cross-linker succinimidyl 4-(p-maleimidophenyl)butyrate (SMPB; Pierce) was added to a final concentration of 0.5 mM. After incubation on ice for 30 min, the reactions were quenched by adding 550 μl of 0.6 M sorbitol–100 mM Tris-HCl (pH 7.2)–100 mM cysteine, and the mixtures were incubated on ice for a further 10 min. For immunoprecipitations, the mitochondria were solubilized in 200 μl of boiling buffer (1% SDS, 50 mM Tris-HCl [pH 7.5], 1 mM EDTA) and heated at 95°C for 5 min. Then 2.5 ml of TNET* (1% Triton X-100, 150 mM NaCl, 5 mM EDTA, 50 mM Tris-HCl [pH 7.5], 1 μg of PMSF per ml, 1 mM aprotinin, 1 mM leupeptin) was added, and the sample was centrifuged at 12,500 × g for 10 min. A 25-μl volume of antiserum to Tim23p was added to the supernatant, and the mixture was incubated overnight at 4°C. Immunocomplexes were collected with protein A-Sepharose as described above.
RESULTS
The carboxyl-terminal hydrophobic domain of Tim23p contains mitochondrial targeting information.
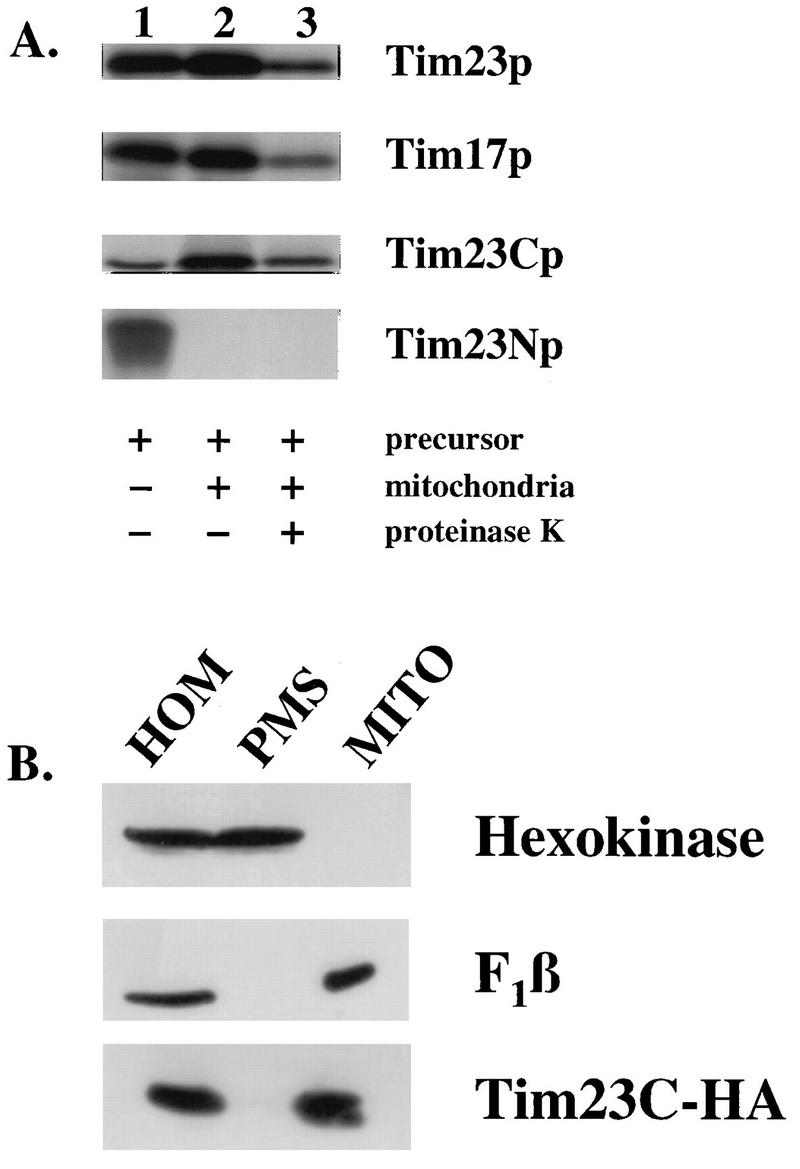
To determine the roles of the hydrophobic and hydrophilic segments of Tim23p, we constructed two truncated versions of the TIM23 gene. tim23N encodes the hydrophilic, amino-terminal 9 kDa of Tim23p (Tim23Np), while tim23C encodes the 14-kDa hydrophobic carboxyl terminus (Tim23Cp). Tim23p is a member of a small family of mitochondrial proteins that lack an amino-terminal presequence and thus contain internal mitochondrial targeting information. We therefore asked if either Tim23Np or Tim23Cp was correctly targeted to the mitochondria. Tim23Np, Tim23Cp, and the full-length Tim23 and Tim17 proteins were synthesized in vitro (Fig. 1A, lane 1). After incubation with mitochondria, Tim23Cp, Tim23p, and Tim17p were each found in the mitochondrial pellet (lane 2) and were protected from proteinase K added to the mitochondria after the import reaction (lane 3). Tim23Cp appears to be imported to the same extent as the full-length Tim23 and Tim17 proteins. In contrast, Tim23Np did not bind to mitochondria and was not imported (lanes 2 and 3).
FIG. 1.
The carboxyl-terminal domain of Tim23p is imported into mitochondria. (A) [35S]methionine-labeled proteins were synthesized by in vitro transcription and translation and incubated with mitochondria isolated from wild-type strain D273-10b as described in Materials and Methods. After the import reaction, mitochondria were reisolated by centrifugation. Mitochondrial proteins were separated by SDS-PAGE, and the radiolabeled proteins were detected by fluorography. Lanes: 1, 30% of the translation lysate added to each assay; 2, the mitochondrial pellet after the import reaction; 3, the mitochondrial pellet after the import reaction, following treatment with 100 μg of proteinase K per ml. (B) tim23-1 cells carrying pKR16, which expresses the Tim23C-HA protein, were grown at 24°C to an OD600 of 2. The homogenate (HOM) was separated into a postmitochondrial supernatant (PMS) and a mitochondrial pellet (MITO) by centrifugation at 9,600 × g for 10 min. Aliquots of homogenate, mitochondria, and postmitochondrial supernatant representing equal numbers of cells were analyzed by SDS-PAGE and immunoblotting with antibodies to hexokinase, the F1-ATPase β subunit (F1β), or the HA epitope tag on Tim23C-HA.
To confirm the import of Tim23Cp in vivo, we localized Tim23Cp in both tim23-1 mutant and wild-type yeast cells. Since our antibodies to Tim23p recognize only the amino-terminal hydrophilic domain, we tagged Tim23Cp with the influenza virus HA epitope (73) and transformed the Tim23C-HA construct into yeast cells. Cells expressing Tim23C-HA were homogenized and separated into a mitochondrial pellet and a postmitochondrial supernatant. In both the tim23-1 mutant (Fig. 1B) and its wild-type parent (data not shown), we found that Tim23C-HA was located in the mitochondrial fraction, along with the mitochondrial marker, the β subunit of the F1-ATPase (F1β). Since Tim23Cp, but not Tim23Np, is targeted to the mitochondria, the following experiments focused on the function of the C-terminal hydrophobic domain of Tim23p.
Tim23Cp cannot substitute for Tim17p or Tim23p.
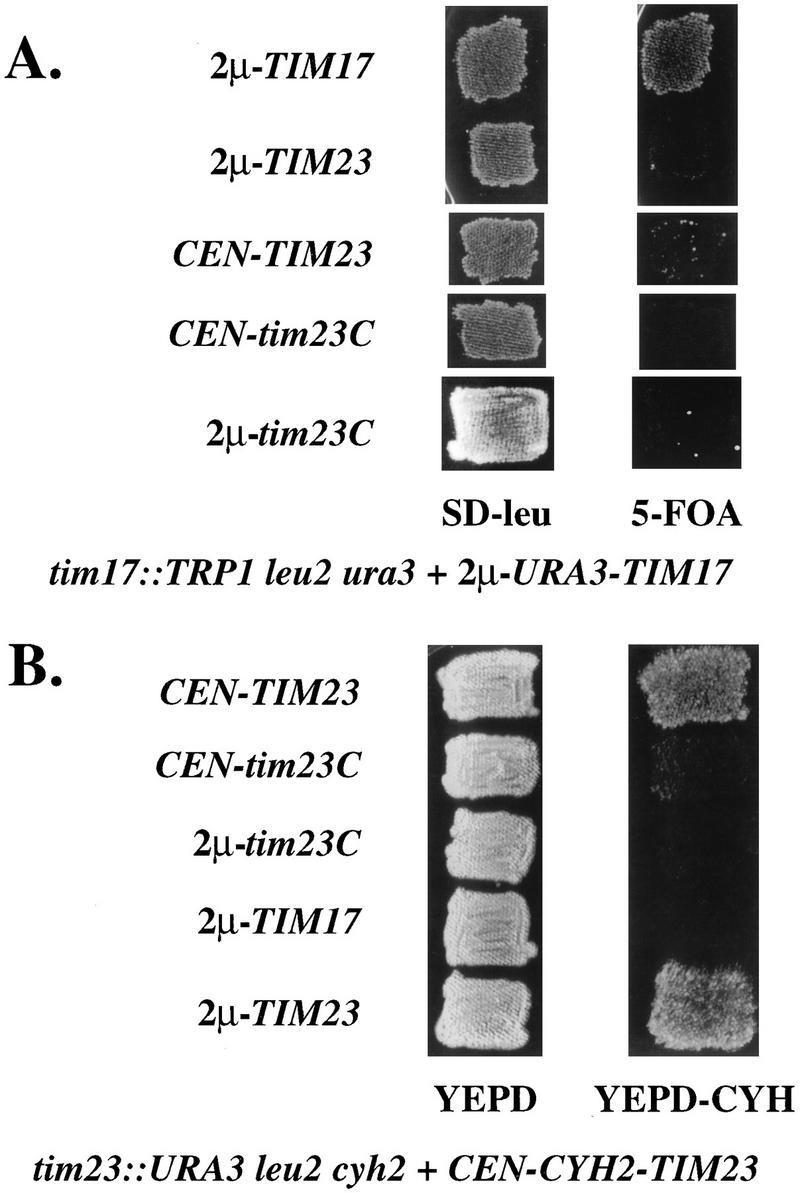
Since Tim17p is homologous to the carboxyl-terminal region of Tim23p, we asked if Tim23Cp could substitute for Tim17p in yeast cells. Since both TIM23 and TIM17 are essential genes, we used a plasmid shuffle scheme to examine the activity of Tim23Cp. The ura3 leu2 strain KRR111 carries the tim17::TRP1 chromosomal disruption and a wild-type TIM17 gene on the URA3-containing plasmid pKR7 (57). The URA3 gene on pKR7 confers sensitivity to 5-fluoroorotic acid (5-FOA) (5). If this strain is transformed with a second plasmid that provides Tim17p activity, the cells can lose pKR7 and become resistant to 5-FOA. We transformed KRR111 with LEU2-containing plasmids expressing Tim23Cp, wild-type Tim23p, or Tim17p. We used two types of plasmids to express our constructs: plasmids containing a yeast centromere (CEN), which are present in 1 or 2 copies per cell, or plasmids that carry the 2μm origin of replication, which are present in multiple copies (usually 30 to 50 per cell) (53a). To select for cells that have lost the TIM17-URA3 plasmid, we replica plated the transformants to medium containing 5-FOA. tim17::TRP1 cells containing either CEN-tim23C or 2μm-tim23C did not grow on 5-FOA and were therefore unable to lose the TIM17-URA3 plasmid (Fig. 2A). Likewise, cells expressing full-length TIM23 could not grow on medium containing the inhibitor. Only cells transformed with an additional copy of TIM17 were able to grow on 5-FOA. Therefore, neither Tim23Cp nor Tim23p can substitute for Tim17p.
FIG. 2.
Tim23Cp cannot substitute for Tim23p or Tim17p. (A) tim17::TRP1 leu2 ura3 cells carrying the URA3-TIM17 plasmid pKR7 were transformed with the indicated CEN-LEU2 or 2μm-LEU2 plasmids as described in Materials and Methods. Transformants were patched onto medium lacking leucine (SD-leu) and then replica plated onto medium containing 5-FOA to detect the loss of pKR7. (B) tim23::URA3 leu2 cyh2 cells carrying a CYH2-TIM23 plasmid (pKR1) were transformed with the indicated CEN-LEU2 or 2μm-LEU2 plasmids as described in Materials and Methods. Transformants were patched onto YEPD medium and then replica plated onto YEPD-CYH medium to detect the loss of pKR1.
In similar experiments, we examined whether Tim23p requires its hydrophilic amino terminus for function. Plasmids expressing Tim23Cp, Tim23p, or Tim17p were transformed into the tim23::URA3 leu2 cyh2 strain KRR123. KRR123 carries TIM23 on the CYH2 plasmid pKR1. Since CYH2-containing cells are unable to grow in the presence of cycloheximide (66), we tested our transformants for their ability to lose the TIM23-CYH2 plasmid by replica plating them onto YEPD medium containing 10 mg of cycloheximide per liter (YEPD-CYH). Only transformants carrying a second copy of wild-type TIM23 were able to grow on YEPD-CYH. Tim23Cp was unable to substitute for the full-length Tim23 protein, indicating that the amino-terminal hydrophilic domain of Tim23p is essential (Fig. 2B). Furthermore, Tim17p overexpressed from a 2μm plasmid cannot rescue the lethality of tim23::URA3 strains (Fig. 2B), confirming our previous results (57) showing that TIM17 and TIM23 do not encode overlapping activities.
Overexpression of the carboxyl-terminal domain of Tim23p is toxic to tim23-1 cells.
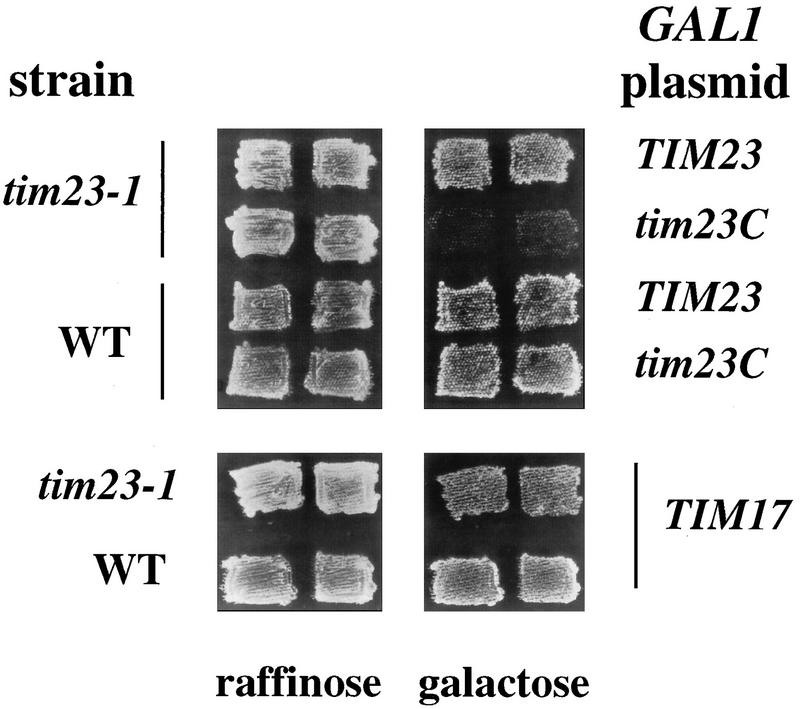
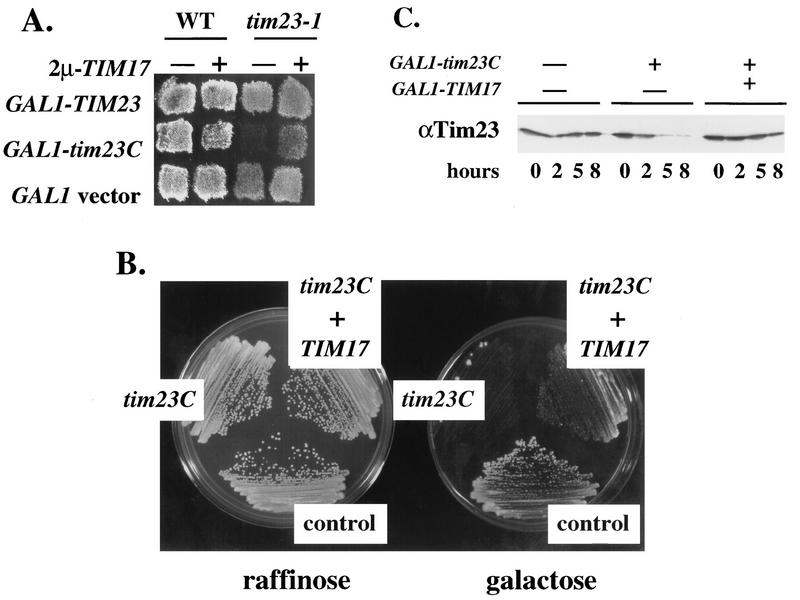
A surprising observation during our studies with Tim23Cp suggested that overproduction of this domain may be deleterious to yeast cells. While tim23-1 cells carrying the CEN-tim23C plasmid grew normally at the permissive temperature (24°C), tim23-1 cells transformed with the 2μm-tim23C plasmid grew extremely slowly. Tim23Cp therefore appeared to be toxic to the tim23-1 strain when present at a high level. To confirm the toxicity of Tim23Cp, we constructed a plasmid encoding Tim23Cp under the control of the inducible GAL1 promoter (GAL1-tim23C). We previously showed that full-length Tim23p and Tim17p are overproduced 10- to 20-fold when expressed from GAL1 (13, 57). Wild-type and tim23-1 transformants containing GAL1-tim23C were patched onto raffinose medium, which does not induce expression from the GAL1 promoter, and then replica plated onto galactose medium to induce the expression of Tim23Cp. As controls, we used plasmids expressing Tim23p or Tim17p under the control of GAL1. We found that tim23-1 cells expressing high levels of Tim23p or Tim17p were able to grow on galactose-containing medium, whereas tim23-1 cells expressing Tim23Cp failed to grow (Fig. 3). Thus, overexpression of the carboxyl-terminal domain of the Tim23 protein is toxic to the tim23-1 mutant. We noted, however, that wild-type TIM23 strains are able to grow in the presence of high levels of Tim23Cp (Fig. 3).
FIG. 3.
High levels of Tim23Cp are toxic to tim23-1 cells. The tim23-1 strain KRR131 and the wild-type (WT) strain RJ500 were transformed with each of the following plasmids: GAL1-TIM23 (13), GAL1-TIM17 (57), and GAL1-tim23C (pKR17). Transformants were patched onto selective medium containing 2% raffinose as the sole carbon source. The patches were then replica plated onto selective medium containing 2% galactose to induce the expression of the plasmid-borne genes. All the cells were grown at 24°C, the permissive temperature for tim23-1.
High-level expression of Tim23Cp causes the loss of the Tim23-1 protein and a defect in protein import.
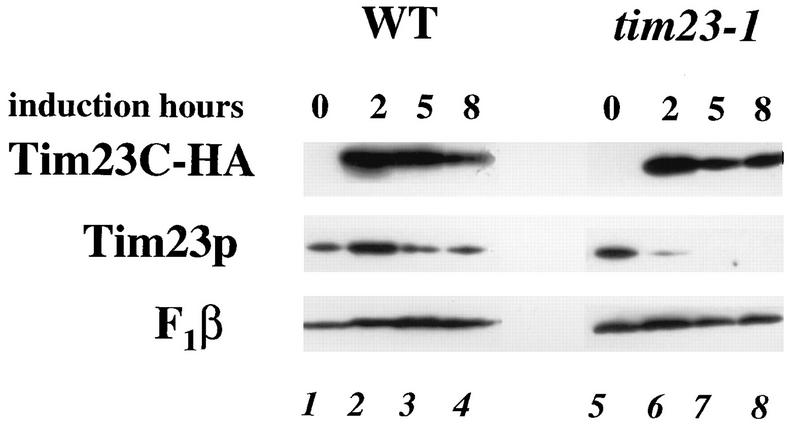
To determine why Tim23Cp is toxic to tim23-1 strains, we examined the level of the Tim23-1 protein in cells overproducing the carboxyl-terminal domain of Tim23p. We placed the HA epitope-tagged version of Tim23Cp (Tim23C-HA) under the control of GAL1 and transformed this plasmid into tim23-1 and TIM23 (wild-type) strains. The cells were pregrown on raffinose-containing medium, and the expression of Tim23C-HA was induced by adding galactose. Total proteins were then isolated and analyzed by immunoblotting. As shown in Fig. 4, Tim23C-HA was not expressed on raffinose medium in either wild-type or tim23-1 cells, but similar levels of Tim23C-HA were produced in wild-type and tim23-1 cells within 2 h of the addition of galactose. The Tim23 protein in the wild-type strain remained relatively constant during the 8 h of galactose induction (Fig. 4, lanes 1 to 4). In contrast, the amount of the Tim23-1 protein in the tim23-1 mutant strain was significantly reduced after 2 h of Tim23C-HA induction (lane 6). By 5 h, Tim23-1p was completely gone (lane 7). The amount of F1β, a mitochondrial inner membrane protein, was not affected by the induction of Tim23C-HA in either wild-type or tim23-1 cells. Our results suggest that overexpression of the carboxyl-terminal domain of Tim23p is toxic to tim23-1 cells because it causes the loss of the full-length Tim23-1 protein.
FIG. 4.
Tim23-1p disappears when Tim23Cp is overexpressed. The tim23-1 strain KRR131 and the wild-type (WT) strain RJ500, each carrying the GAL1-tim23C-HA plasmid pKR18, were grown to an OD600 of 0.5 in selective medium containing 2% raffinose. Galactose was added to a final concentration of 4%, and aliquots were removed from each culture at the indicated times. Total cell proteins were extracted (78), and equal amounts of proteins were analyzed by SDS-PAGE followed by immunoblotting with antibodies to F1β, Tim23p, or the HA epitope on Tim23C-HA. Lanes: 1 to 4, protein from strain RJ500; 5 to 8, protein from strain KRR131.
We found that tim23-1 cells overproducing Tim23Cp were defective in protein import (Fig. 5). tim23-1 cells containing the GAL1-tim23C construct were pregrown on raffinose-containing medium and split into two aliquots. In one sample, the expression of Tim23C-HA was induced by adding galactose; the other sample remained in raffinose medium. After 4 h, the cells were pulse-labeled for 5 min with [35S]methionine and total proteins were then immediately isolated. The F1β protein was immunoprecipitated from both samples and analyzed by SDS-PAGE and fluorography. As shown in Fig. 5, a large amount of the precursor form of F1β accumulated in cells overproducing Tim23Cp. High levels of Tim23Cp thus block the import of F1β into the mitochondrial matrix and prevent its processing. In contrast, cells not expressing Tim23p (those grown on raffinose medium) were not significantly defective in import and contained mostly the mature-sized F1β protein. Similarly, we found that the import of the cytochrome oxidase subunit IV precursor was defective in cells overproducing Tim23Cp (data not shown).
FIG. 5.
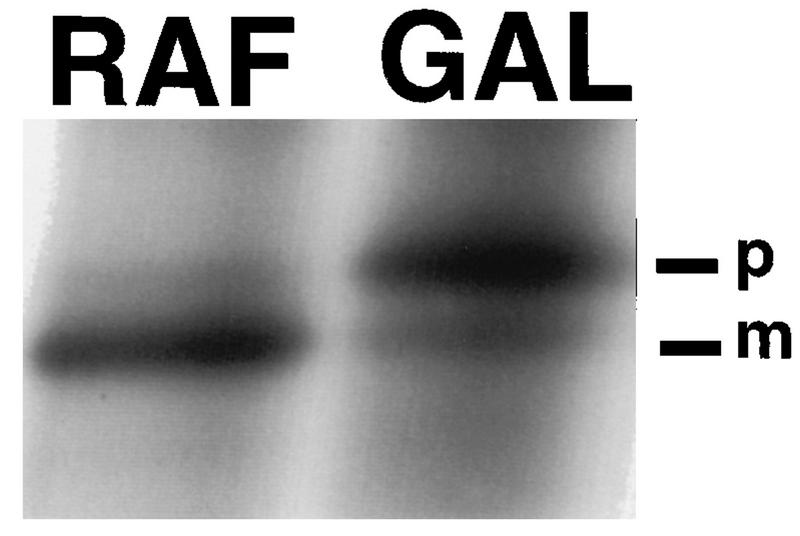
Overproduction of Tim23p causes a defect in protein import. The tim23-1 strain KRR131 carrying the GAL1-tim23C plasmid pKR17 was grown to an OD600 of 0.5 at the permissive temperature, 23°C, in selective medium containing 2% raffinose and then split into two aliquots. Galactose (GAL) was added to one aliquot to a final concentration of 4%, and the other sample remained in raffinose (RAF) medium. After 4 h, an aliquot of each culture was removed, the cells were pulse-labeled for 5 min with [35S]methionine (13), and further labeling was stopped by the addition of 1 mM sodium azide. Total proteins were extracted from cells and immunoprecipitated with antiserum to F1β, the β subunit of F1-ATPase (13, 78). Precipitates were analyzed by SDS-PAGE followed by fluorography. The precursor (p) and mature (m) forms of F1β are indicated.
Overexpression of Tim17p relieves the toxicity of Tim23Cp and prevents the loss of the Tim23-1 protein.
A possible explanation for the deleterious effect of the Tim23C protein in tim23-1 mutants is that Tim23Cp competes with Tim23-1p for binding to another component of the mitochondrial protein import machinery. This competition may remove Tim23-1p from its normal environment and enhance its degradation. Since genetic and biochemical studies indicate that Tim17p and Tim23p physically interact, we tested the possibility that Tim23Cp is toxic because it titrates Tim17p. We found that an increased amount of Tim17p from a 2μm plasmid overcomes the toxicity of Tim23Cp. tim23-1 and TIM23 (wild-type) cells that carry GAL1-tim23C, GAL1-TIM23, or a GAL1 vector alone were transformed with a 2μm-TIM17 plasmid. Transformants were grown on raffinose-containing medium and then replica plated onto galactose medium to induce the expression of the GAL1-driven constructs. As shown in Fig. 6A, tim23-1 cells with normal levels of Tim17p failed to grow after induction of Tim23Cp. However, increased levels of Tim17p from 2μm-TIM17 cells allowed the tim23-1 strain to grow in the presence of Tim23Cp. 2μm-TIM17 thus relieves the toxicity of Tim23Cp. We found, however, that excess Tim17p reduced but did not completely eliminate the deleterious effect of Tim23Cp. In particular, we examined the growth of single colonies on galactose medium (Fig. 6B). In these experiments, we placed TIM17 under the control of the GAL1 promoter (GAL1-TIM17) (57) and introduced this construct into tim23-1 cells that also carry GAL1-tim23C. While tim23-1 cells expressing only Tim23Cp failed to grow into single colonies, cells containing both GAL1-tim23C and GAL1-TIM17 were able to grow into small colonies on galactose-containing medium. These colonies, however, were markedly smaller than colonies from tim23-1 cells that contained only empty GAL1 vectors (Fig. 6B, control). Since excess Tim17p did not fully reverse the toxic effects of Tim23Cp, other import components in addition to Tim17p may be titrated by high levels of Tim23Cp.
FIG. 6.
Overexpression of TIM17 relieves tim23C toxicity and prevents the loss of Tim23-1p. (A) The tim23-1 strain KRR131 was transformed with either 2μm-TIM17 (pKR7) (57) or a vector control (pRS426) (67). These cells were then cotransformed with one of the following GAL1 plasmids: pRS314GU (GAL1 vector control) (47), GAL1-TIM23 (13), or pKR17 (GAL1-tim23C). The cells were pregrown on medium containing 2% raffinose and then replica plated onto medium containing 2% galactose. All the cells were grown at 24°C, the permissive temperature for tim23-1. (B) The tim23-1 strain KRR131 was transformed with the plasmids listed below as described in Materials and Methods. Transformants were streaked onto selective medium containing either 2% raffinose or 2% galactose. The cells were grown at 24°C for 5 days to obtain single colonies. Cells labeled control contain the empty vectors pRS314GU and pRS316GU (47). Cells labeled tim23C contain plasmids pRS316GU and GAL1-tim23C (pKR17). Cells labeled tim23C + TIM17 contain plasmids GAL1-tim23C (pKR17) and GAL1-TIM17 (57). (C) The tim23-1 strain KRR131 was transformed with the plasmids listed below and grown to an OD600 of 0.5 on selective medium containing 2% raffinose. Galactose was added to a final concentration of 2%, and aliquots were removed from each culture at the indicated times. Total cell proteins were extracted, and equal amounts of proteins were analyzed by SDS-PAGE and immunoblotting with antibodies to Tim23p. The left-hand lanes contain empty GAL1 vectors pRS314GU plus pRS316GU; the middle lanes contain pRS316GU plus GAL1-tim23C (pKR17); the right-hand lanes contain GAL1-tim23C (pKR17) plus GAL1-TIM17.
We found that an increased amount of Tim17p prevented the loss of the Tim23-1 protein caused by overproduction of Tim23Cp. tim23-1 cells carrying either GAL1-tim23C or both GAL1-tim23C and GAL1-TIM17 were shifted to galactose-containing medium (0 h). We removed aliquots of cells at the indicated times and extracted proteins from the cells. Immunoblotting showed that induction of Tim23Cp in the absence of GAL1-TIM17 caused the loss of the Tim23-1 protein (Fig. 6C, middle four lanes). At 5 h after the addition of galactose, little or no Tim23-1 protein was detected (seventh lane). In contrast, when both Tim23Cp and Tim17p were induced, the amount of Tim23-1p remained relatively constant throughout the induction (last four lanes). We detected similar amounts of Tim23-1p in cells overexpressing both Tim23Cp and Tim17p and in tim23-1 cells (compare first four lanes with last four lanes). Our results raise the possibilities that excess Tim23Cp enhances the degradation of Tim23-1p by displacing it from its normal interaction with Tim17p and that increasing the level of Tim17p overcomes the titration by Tim23Cp.
Tim23Cp interacts with Tim17p but not with full-length Tim23p.
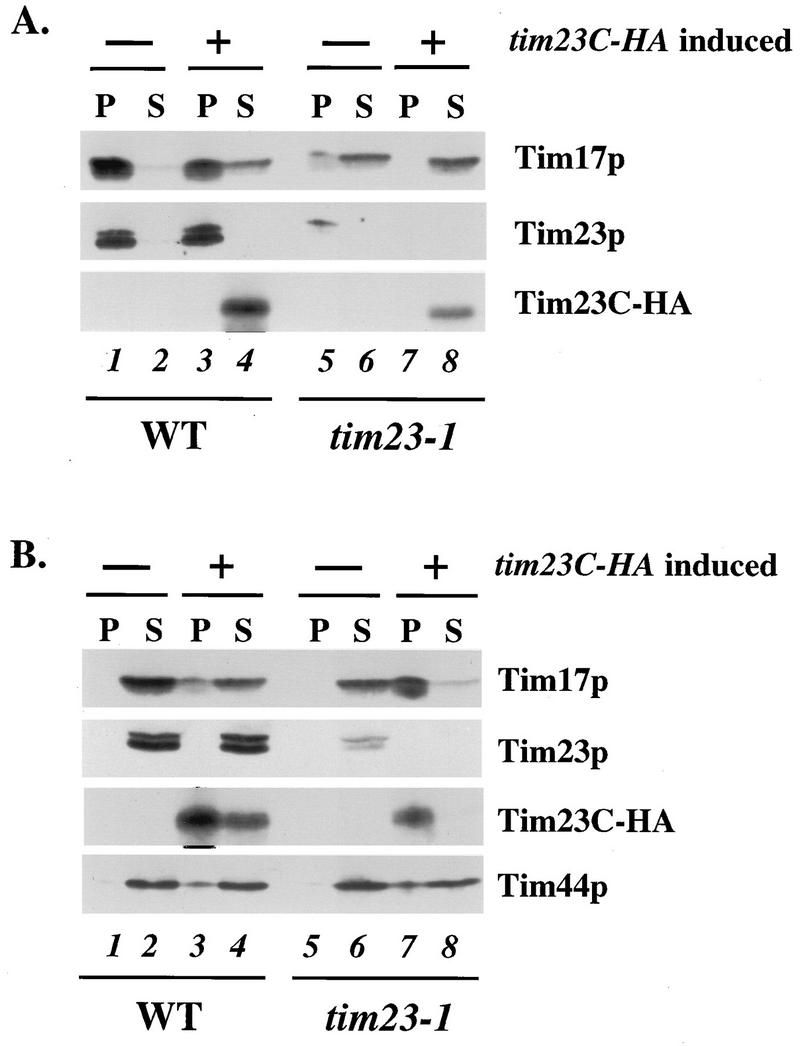
To determine if the hydrophobic domain of Tim23p physically interacts with Tim17p, we examined whether Tim23Cp and Tim17p could be coimmunoprecipitated from detergent-solubilized mitochondria. Wild-type and tim23-1 cells, each carrying either GAL1-tim23C-HA or an empty GAL1 vector, were pregrown in raffinose medium. After 6 h of galactose induction, we collected the cells and prepared mitochondria from each of the four cultures. We then solubilized the mitochondria in a buffer containing 0.5% digitonin and immunoprecipitated the proteins with antiserum to Tim23p (Fig. 7A) or antibodies to the HA epitope on Tim23C-HA (Fig. 7B). Immunoprecipitates and supernatants were then analyzed by immunoblotting.
FIG. 7.
Tim23Cp physically interacts with Tim17p in the mitochondrial inner membrane. Mitochondria were isolated from the tim23-1 strain KRR131 and the wild-type (WT) strain RJ500, each carrying either pRS314GU or GAL1-tim23C-HA (pKR18). The mitochondria were solubilized in a buffer containing 0.5% digitonin and immunoprecipitated with antibodies against Tim23p (A) or against the HA epitope on Tim23C-HA (B). Immunoprecipitates and supernatants were analyzed by SDS-PAGE and immunoblotting with antibodies to Tim23p, Tim17p, Tim44p, and the HA epitope. Lanes: 1 and 2, mitochondrial protein from wild-type cells; 3 and 4, mitochondrial protein from wild-type cells expressing Tim23C-HA; 5 and 6, mitochondrial protein from tim23-1 cells; 7 and 8, mitochondrial protein from tim23-1 cells expressing Tim23C-HA. Lanes 1, 3, 5, and 7 contain immunoprecipitate from 60 μg of solubilized mitochondrial protein, and lanes 2, 4, 6, and 8 contain supernatant from the immunoprecipitation, containing 60 μg of mitochondrial protein.
In mitochondria isolated from wild-type cells, virtually all of the Tim17 protein could be coimmunoprecipitated with the full-length Tim23 protein (Fig. 7A, lane 1). When Tim23C-HA was present in wild-type mitochondria, however, a small amount of the Tim17 protein remained in the supernatant and was no longer associated with the full-length Tim23p (lanes 3 and 4). This small amount of Tim17p was instead associated with Tim23C-HA and was found in the pellet fraction in precipitations with antibodies to the HA epitope (Fig. 7B, lane 3). Our result suggests that Tim23Cp competes with the wild-type Tim23p for interaction with Tim17p. We also noted that Tim23Cp and the wild-type Tim23 protein did not stably interact. Little or no Tim23C-HA was coimmunoprecipitated with full-length Tim23p (Fig. 7A, lanes 3 and 4), and antibodies to Tim23C-HA failed to coprecipitate Tim23p (Fig. 7B, lanes 3 and 4). Since Tim23p has recently been shown to dimerize (2), our results raise the possibility that the hydrophobic domain of Tim23 does not by itself mediate the association with other Tim23 proteins.
When we examined mitochondria isolated from tim23-1 cells, we found two striking differences from our results with wild-type mitochondria. First, tim23-1 mitochondria had only about 10% as much Tim23-1 protein as wild-type mitochondria had Tim23p (Fig. 7; compare lanes 5 and 1). Since the levels of Tim23p and Tim23-1p were similar when proteins were isolated from whole cells by rapid lysis (Fig. 4; compare lanes 1 and 5), the lack of Tim23-1p in isolated mitochondria indicated that Tim23-1p is unstable and is probably degraded during cell fractionation. However, we found that the small amount of Tim23-1p present in the isolated mitochondria was able to associate with Tim17p; a detectable amount of Tim17p was coimmunoprecipitated with Tim23-1p when Tim23 antiserum was used (Fig. 7A, lane 5).
A second difference we observed between tim23-1 and wild-type mitochondria is that in tim23-1 mitochondria, virtually all of the Tim17 protein associated with Tim23Cp. When Tim23C-HA was expressed in tim23-1 cells, most or all of Tim23-1p was lost (Fig. 7, lanes 7 and 8) and Tim17p was present in the supernatant from immunoprecipitations with Tim23 antiserum (Fig. 7A, lane 8). In contrast, virtually all of the Tim17p was precipitated along with Tim23C-HA (Fig. 7B, lane 7). Our results indicate that Tim17p and Tim23p interact via their hydrophobic domains.
To determine if Tim23C-HA interacts with other inner membrane import components, we examined our fractions with antibodies against Tim44p. In both wild-type and tim23-1 mitochondria that contained Tim23C-HA, a significant fraction of the Tim44p coprecipitated with antibodies to Tim23C-HA (Fig. 7B, lanes 3 and 7). The Tim23p hydrophobic domain can therefore interact with both Tim17p and Tim44p.
Tim23p and Tim17p can be chemically cross-linked in intact mitochondria.
To provide evidence that Tim17p and Tim23p bind each other directly, we used chemical cross-linkers to examine the interaction between the two proteins. Mitochondria were isolated from a strain containing an HA-tagged version of Tim17p (Tim17-HA) and were treated with the membrane-permeative cross-linker SMPB. Mitochondria treated with SMPB and the untreated control were examined by immunoblotting with antibodies to the Tim17-HA protein (Fig. 8, lanes 1 and 2). We observed five major cross-linked products of 30, 43, 50, 52, and 75 kDa (lane 2). Since each of these bands contains the 20-kDa Tim17-HA protein, it appears that Tim17p is in close proximity to five proteins of approximately 10, 23, 30, 32, and 55 kDa. When we solubilized SMPB-treated mitochondria and immunoprecipitated them with antibodies to Tim23p, we found a single band of 43 kDa (lane 3). Hence, this cross-linked product contained both Tim17-HA and Tim23p. Probing the immune blots with antibodies to Tim44 indicated that none of the cross-linked products contains Tim44p (data not shown). Our results suggest that Tim17p directly interacts with at least five mitochondrial proteins, one of which is Tim23p.
FIG. 8.
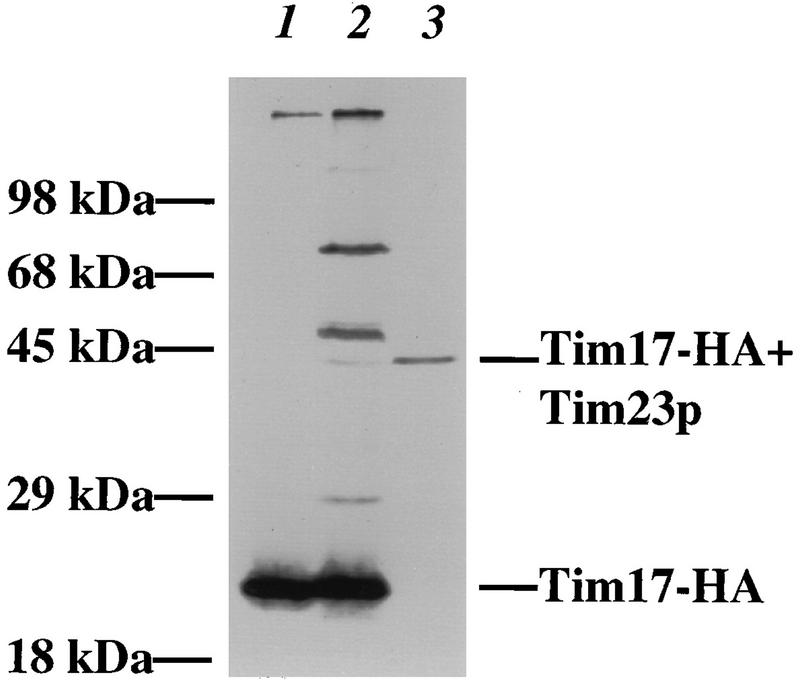
Tim17p can be cross-linked to Tim23p and four other proteins in intact mitochondria. Mitochondria were isolated from a tim17::URA3 strain carrying pKR11, which expresses Tim17p tagged with the HA epitope. Mitochondria were isolated, treated with SMPB, and split into two aliquots. One aliquot was analyzed directly, while the mitochondrial proteins in the other aliquot were solubilized and precipitated with antibodies to the full-length Tim23 protein. All samples were analyzed by SDS-PAGE and immunoblotting with antibodies to the HA epitope to detect cross-linked products containing Tim17-HA. Lane 1, 50 μg of solubilized mitochondrial protein; lane 2, 100 μg of mitochondrial protein treated with SMPB; lane 3, anti-Tim23p immunoprecipitate from 250 μg of SMPB-treated mitochondrial protein.
DISCUSSION
Tim23p, along with several other proteins of the mitochondrial inner membrane, do not carry amino-terminal presequences. Since Tim23Cp, but not Tim23Np, is correctly targeted to mitochondria, we conclude that the carboxyl-terminal domain of Tim23p carries the targeting information. Experiments to determine whether the import signal is located in a membrane-spanning segment or in one of the positively charged, intervening loops are in progress (10a). We found that the amino-terminal domain of Tim23p, Tim23Np, does not bind to the mitochondrial surface and is not imported. This observation is not surprising, since Tim17p is also targeted to mitochondria. Tim17p is homologous to the Tim23p carboxyl-terminal domain and lacks an appreciable hydrophilic amino-terminal segment.
Although the amino terminus of Tim23p does not carry targeting information, this domain is essential. In particular, the growth defect of yeast cells carrying the tim23::URA3 disruption was not rescued by the expression of Tim23Cp. It has been shown that the 9-kDa amino-terminal domain of Tim23p faces the intermembrane space (14, 36). Thus, there are several possible roles for this part of Tim23p in import. For example, the mitochondrial outer and inner membranes carry independent import machinery (26, 43, 45, 49, 66). These two import machines interact (presumably at contact sites between the two membranes) during the translocation of proteins into the mitochondrial matrix (22). The Tim23p amino terminus may mediate the interaction between the inner and outer membrane import machinery. Alternatively, the domain of Tim23p facing the intermembrane space may function as a receptor for mitochondrial proteins as they pass through the outer membrane. The Tim23p amino terminus contains several clusters of negatively charged amino acids, which would be ideal to interact with the cationic presequence (13). Supporting this possibility, we previously found that antibodies to the Tim23p amino terminus blocked both the binding and the import of precursors into mitochondria whose outer membranes were disrupted (13). Recently, Bauer et al. (2) have shown that the amino terminus of Tim23p is essential and that this domain appears to bind directly to mitochondrial presequences. Regardless of the role played by the hydrophilic domain of Tim23p, it appears that the N-terminal and C-terminal segments of Tim23p need to be physically linked to provide full Tim23p function. In particular, we fused the Tim23p hydrophilic domain to the amino terminus of Tim17p, forming a Tim23N-Tim17p chimera. When this chimera was expressed in yeast cells, it provided functional Tim17p activity, but Tim23N-Tim17p could not rescue the growth defect of tim23::URA3 cells (54a). Even coexpression of Tim23N-Tim17p and Tim23Cp did not provide Tim23p function.
Recent experiments have indicated that Tim23 protein dimerizes in mitochondria and that this interaction may be mediated by a coiled-coil segment in the hydrophilic amino-terminal domain of Tim23p (2). Supporting this conclusion, we found that Tim23Cp and the wild-type Tim23 protein do not interact. Little or no Tim23C-HA was coimmunoprecipitated with full-length Tim23p (Fig. 7A, lanes 3 and 4), and antibodies to Tim23C-HA failed to coimmunoprecipitate with Tim23p (Fig. 7B, lanes 3 and 4). Thus, the hydrophobic domain of one Tim23 molecule does not appear to mediate the interaction with other Tim23 proteins. However, since Tim23Cp is a nonfunctional fragment, further work is necessary to definitively identify regions of Tim23p that mediate dimerization.
What is the role of the carboxyl-terminal domain of Tim23p? It has been suggested that Tim23p and Tim17p form part of a protein-translocating pore or channel in the inner membrane (27, 52, 57). In support of this view, we have recently shown that Tim23p is required for the normal activity of an inner membrane channel, called MCC (39). In addition, Tim23p and Tim17p can be cross-linked to a precursor arrested in transit across the inner membrane (55). Furthermore, Berthold et al. (3) showed that an arrested precursor could be cross-linked to Tim23p and Tim17p only when the two import components were present in a larger complex containing Tim44p and mt-Hsp70. The authors suggested that Tim23p and Tim17p do not bind tightly to the precursor but that Tim23p and Tim17p instead form part of a passive channel for protein translocation. The interaction between Tim17p and the hydrophobic domain of Tim23p is consistent with a model in which Tim23p and Tim17p associate in the plane of the inner membrane to form part of a channel for mitochondrial proteins. Although Tim17p and Tim23Cp are 46% homologous, have the same number of predicted membrane spans, and have similarly sized intervening loops between their membrane spans, Tim17p cannot be replaced by Tim23Cp. Our results are consistent with the model of a channel composed of Tim23p and Tim17p, since ion channels are often composed of homologous but nonidentical subunits (9).
We have used both genetic and biochemical approaches to show that the Tim23p hydrophobic domain is responsible for binding to Tim17p. In vivo, we found that a low level of Tim23Cp is tolerated by tim23-1 cells but a high level of Tim23Cp is toxic to tim23-1 cells even at the permissive temperature. Excess Tim17p partially relieved the toxicity caused by Tim23Cp, implying that Tim23Cp kills tim23-1 cells by binding to Tim17p and displacing Tim23-1p. Since overexpressing Tim17p along with Tim23Cp did not fully restore tim23-1 to its normal rate of growth (Fig. 6B), Tim23Cp may be titrating other proteins away from Tim23-1p in addition to Tim17p. Another import component, such as Tim44p or a new member of the TIM complex (see below), may be limiting for growth under these conditions. We showed that the Tim23p hydrophobic domain associates with Tim17p in vitro by using coimmunoprecipitations from detergent-solubilized mitochondria. When Tim23C-HA is overexpressed in tim23-1 cells, the Tim23-1 protein is lost (see below) and the mitochondria contain Tim23C-HA along with an approximately wild-type amount of Tim17p. When we solubilized these mitochondria in digitonin, all of the Tim17p coprecipitated along with Tim23C-HA. Thus, Tim17p can be isolated in a complex with either the full-length Tim23p or the Tim23p hydrophobic domain, Tim23C-HA.
The coimmunoprecipitation of Tim23p and Tim17p suggests that they bind each other directly but does not rule out the possibility that they interact via an intermediary subunit of the inner membrane complex. Chemical cross-linking has been used to identify nearest neighbors within multisubunit complexes (29). We cross-linked Tim23p to Tim17-HA in intact mitochondria by using SMPB, a heterobifunctional cross-linker with a spacer-arm length of 14.5 Å. Thus, Tim23p and Tim17p appear to bind each other directly.
By using myc-tagged versions of Tim17p and Tim23p, Blom et al. (4) found that Tim17 and Tim23 are expressed at approximately equal levels in the cell. Here, we found that Tim17p is quantitatively coimmunoprecipitated along with Tim23p (Fig. 7A, lane 1). In other experiments, when we immunoprecipitated Tim23p with 40% efficiency from solubilized mitochondria, we also coimmunoprecipitated 40% of Tim17p (data not shown). If cells contain equal amounts of Tim17p and Tim23p and the two proteins coimmunoprecipitate with similar efficiencies, it is likely that Tim23p and Tim17p bind each other with 1:1 stoichiometry. Recently, Bömer et al. (7) have detected two subcomplexes containing Tim23p molecules in the mitochondrial inner membrane. One complex appears to contain Tim23p, Tim17p, and mt-Hsp70, whereas the other complex contains Tim23p, Tim44p, and mt-Hsp70. Additional experiments are required to determine if the stoichiometry of Tim23p and Tim17p is actually 2:1 and if only 50% of the total Tim23 protein coimmunoprecipitates along with Tim17p.
We also determined by coimmunoprecipitation that Tim23C-HA is capable of interacting with Tim44p, in addition to Tim17p. Tim44 is the third mitochondrial inner membrane protein known to be required for import. In the absence of cross-linking or functional data, however, we cannot determine if Tim44p binds directly to the Tim23p hydrophobic domain or interacts via another subunit of the inner membrane complex. Tim44p is not quantitatively coimmunoprecipitated along with Tim23C-HA, even when Tim17p is (Fig. 7B, lanes 7 and 8). This result is consistent with prior reports suggesting either that Tim44p is loosely bound to the complex containing Tim23p and Tim17p or that Tim44p may cycle on and off the inner membrane import complex during protein translocation (3, 4).
If Tim23Cp is toxic because it competes for binding to Tim17p, why does Tim23Cp kill only tim23-1 cells and not wild-type cells as well? It is possible that the level of Tim23Cp needs to be higher in wild-type cells than we have expressed in tim23-1 cells to see an effect. Recently, we have expressed increased levels of Tim23Cp from multicopy vectors and from a strong constitutive promoter, but we have not seen any growth defect in wild-type cells carrying these constructs (38a). Even when wild-type cells are grown on nonfermentable medium, which requires full mitochondrial function, overproduction of Tim23Cp has no observable effect. We suggest an alternative possibility. In particular, Tim23Cp in wild-type and mutant cells binds to Tim17p and displaces other full-length Tim23p or Tim23-1p molecules. However, we suggest that Tim23-1p is selectively degraded at an accelerated rate when it is displaced from its normal binding partners. First, when Tim23Cp is induced, the Tim23-1 protein disappears while the wild-type Tim23p remains at normal levels (Fig. 4). Second, Tim23-1p has increased susceptibility to proteolysis because it is selectively degraded during mitochondrial isolation. Finally, in wild-type mitochondria containing both Tim23p and Tim23C-HA, part of the Tim17 protein coprecipitates with Tim23C-HA (Fig. 7B, lanes 3 and 4), indicating that the hydrophobic domain of Tim23p binds to Tim17p even in the presence of wild-type Tim23p. Taken together, these data strongly suggest that Tim23Cp can displace both wild-type Tim23p and Tim23-1p from Tim17p but only Tim23-1p is degraded when it is removed from its normal environment in the inner membrane. Recently, two protease complexes have been identified which degrade nonnative proteins in the mitochondrial inner membrane (1, 38). It will be of interest to determine if either the Yme1p or the YTA10-12 complex is responsible for degrading Tim23-1 molecules that are displaced from the inner membrane import complex.
In this report, we have focused on the interaction between Tim23p and Tim17p, but preliminary data indicate that other members of the inner membrane import complex have yet to be identified and characterized. In one previous study, the authors used coimmunoprecipitation to isolate a complex containing Tim23p, Tim17p, and two proteins of 14 and 33 kDa (3). In another study, the authors identified a different complex containing Tim23p, Tim17p, and two proteins of 55 and 20 kDa (4). In this study, we showed that Tim17-HA can be cross-linked to Tim23p. In addition, Tim17-HA is present in three cross-linked products of approximately 29, 45, and 75 kDa. It has recently been shown that Tim23p can dimerize with itself and that the inner membrane potential is required for this dimerization (2). In contrast, we found similar amounts of all five cross-links to Tim17-HA in the presence or absence of membrane potential (38a). We did find, however, that both the Tim17p-Tim23p cross-link and the 75-kDa cross-link were significantly decreased in mitochondria isolated from cells that overproduced Tim23Cp (54a). Our cross-linking results thus may indicate that there are new members of the inner membrane import complex that interact directly with Tim17p. The 45- and 75-kDa cross-linked products appear to contain Tim17-HA along with proteins of approximately 25 and 55 kDa, respectively. Further experiments are needed to determine if these cross-linked proteins represent new members of the TIM complex.
ACKNOWLEDGMENTS
We thank Jennifer Emtage and Jennifer Kalish for TIM23-containing plasmids, Ken Saville for the pKS1 construct, Jeff Schatz for antiserum to Tom44, and Casey Kinnally for peptides to Tim17. We also thank Alison Davis, Carolyn Machamer, Dan Isaac, Mike Maceyka, and Steve Doberstein for critical comments on the manuscript.
This work was supported by grant R01-GM46803 from the U.S. Public Health Service to R.E.J. and by Medical Scientist Training Program grant GM07309 to K.R.R.
REFERENCES
- 1.Arlt H, Tauer R, Feldmann H, Neupert W, Langer T. The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 1996;85:875–885. doi: 10.1016/s0092-8674(00)81271-4. [DOI] [PubMed] [Google Scholar]
- 2.Bauer M, Sirrenberg C, Neupert W, Brunner M. Role of Tim23 as voltage sensor and presequence receptor in protein import into mitochondria. Cell. 1996;87:33–41. doi: 10.1016/s0092-8674(00)81320-3. [DOI] [PubMed] [Google Scholar]
- 3.Berthold J, Bauer M F, Schneider H C, Klaus C, Dietmeier K, Neupert W, Brunner M. The MIM complex mediates preprotein translocation across the mitochondrial inner membrane and couples it to the mt-Hsp70/ATP driving system. Cell. 1995;81:1085–1093. doi: 10.1016/s0092-8674(05)80013-3. [DOI] [PubMed] [Google Scholar]
- 4.Blom J, Dekker P J T, Meijer M. Functional and physical interactions of components of the mitochondrial inner-membrane import machinery (MIM) Eur J Biochem. 1995;232:309–314. doi: 10.1111/j.1432-1033.1995.tb20813.x. [DOI] [PubMed] [Google Scholar]
- 5.Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 6.Bolliger L, Junne T, Schatz G, Lithgow T. Acidic receptor domains on both sides of the outer membrane mediate translocation of precursor proteins into yeast mitochondria. EMBO J. 1995;14:6318–6326. doi: 10.1002/j.1460-2075.1995.tb00322.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bömer U, Meijer M, Maarse A C, Hönlinger A, Dekker P J T, Pfanner N, Rassow J. Multiple interactions of components mediating preprotein translocation across the inner mitochondrial membrane. EMBO J. 1997;16:2205–2216. doi: 10.1093/emboj/16.9.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonner W M, Laskey R A. A film detection method for tritium-labeled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974;46:83–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- 9.Canessa C M, Schild L, Buell G, Thorens B, Gautschi I, Horisberger J-D, Rossier B C. Amiloride-sensitive epithelial Na+ channel is made of three homologous subunits. Nature. 1994;367:463–476. doi: 10.1038/367463a0. [DOI] [PubMed] [Google Scholar]
- 10.Daum G, Böhni P C, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J Cell Biol. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 10a.Davis, A., K. Ryan, and R. Jensen. Unpublished data.
- 11.Dekker P, Keil P, Rassow J, Maarse A C, Pfanner N, Meijer M. Identification of MIM23, a putative component of the protein import machinery of the mitochondrial inner membrane. FEBS Lett. 1993;330:66–70. doi: 10.1016/0014-5793(93)80921-g. [DOI] [PubMed] [Google Scholar]
- 12.Doolittle R F. Of URFS and ORFS. Mill Valley, Calif: University Science Books; 1986. [Google Scholar]
- 12a.Emtage, J. Unpublished data.
- 13.Emtage J L T, Jensen R E. MAS6 encodes an essential inner membrane component of the yeast mitochondrial import pathway. J Cell Biol. 1993;122:1003–1012. doi: 10.1083/jcb.122.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emtage, J. L. T., and R. E. Jensen. Submitted for publication.
- 15.Glick B S. Can Hsp70 proteins act as force-generating motors? Cell. 1995;80:11–14. doi: 10.1016/0092-8674(95)90444-1. [DOI] [PubMed] [Google Scholar]
- 16.Gratzer S, Lithgow T, Bauer R E, Lamping E, Paltauf F, Kohlwein S D, Haucke V, Junne T, Schatz G, Horst M. Mas37p, a novel receptor subunit for protein import into mitochondria. J Cell Biol. 1995;129:25–34. doi: 10.1083/jcb.129.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hachiya N, Komiya T, Alam R, Iwahashi J, Sakaguchi M, Omura T, Mihara K. MSF, a novel cytoplasmic chaperone which functions in precursor targeting to mitochondria. EMBO J. 1994;13:5146–5154. doi: 10.1002/j.1460-2075.1994.tb06844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hachiya N, Mihara K, Suda K, Horst M, Schatz G, Lithgow T. Reconstitution of the initial steps of mitochondrial protein import. Nature. 1995;376:705–709. doi: 10.1038/376705a0. [DOI] [PubMed] [Google Scholar]
- 19.Haid A, Suissa M. Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 1983;96:192–205. doi: 10.1016/s0076-6879(83)96017-2. [DOI] [PubMed] [Google Scholar]
- 20.Hawlitschek G, Schneider H, Schmidt B, Tropschug M, Hartl F U, Neupert W. Mitochondrial protein import: identification of processing peptidase and of PEP, a processing enhancing protein. Cell. 1988;53:795–806. doi: 10.1016/0092-8674(88)90096-7. [DOI] [PubMed] [Google Scholar]
- 21.Hines V, Brandt A, Griffiths G, Horstmann H, Brutsch H, Schatz G. Protein import into yeast mitochondria is accelerated by the outer membrane protein MAS70. EMBO J. 1990;9:3191–3200. doi: 10.1002/j.1460-2075.1990.tb07517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horst M, Hilfiker-Rothenfluh S, Oppliger W, Schatz G. Dynamic interaction of the protein translocation systems in the inner and outer membranes of yeast mitochondria. EMBO J. 1995;14:2293–2297. doi: 10.1002/j.1460-2075.1995.tb07223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horwich A L, Kalousek F, Mellman I, Rosenberg L E. A leader peptide is sufficient to direct mitochondrial import of a chimeric protein. EMBO J. 1985;4:1129–1135. doi: 10.1002/j.1460-2075.1985.tb03750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hurt E C, Pesold-Hurt B, Schatz G. The amino-terminal region of an imported mitochondrial precursor polypeptide can direct cytoplasmic dihydrofolate reductase into the mitochondrial matrix. EMBO J. 1984;3:3149–3156. doi: 10.1002/j.1460-2075.1984.tb02272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurt E C, Pesold-Hurt B, Schatz G. The cleavable prepiece of an imported mitochondrial protein is sufficient to direct cytosolic dihydrofolate reductase into the mitochondrial matrix. FEBS Lett. 1984;178:306–310. doi: 10.1016/0014-5793(84)80622-5. [DOI] [PubMed] [Google Scholar]
- 26.Hwang S, Jascur T, Vestweber D, Pon L, Schatz G. Disrupted yeast mitochondria can import precursor proteins directly through their inner membrane. J Cell Biol. 1989;109:487–493. doi: 10.1083/jcb.109.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen R E, Kinnally K W. The mitochondrial import pathway: are precursors imported through membrane channels? J Bioenerg Biomembr. 1997;29:3–10. doi: 10.1023/a:1022470303365. [DOI] [PubMed] [Google Scholar]
- 28.Jensen R E, Yaffe M P. Import of proteins into yeast mitochondria: the nuclear MAS2 gene encodes a component of the processing protease that is homologous to the MAS1-encoded subunit. EMBO J. 1988;7:3863–3871. doi: 10.1002/j.1460-2075.1988.tb03272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Joshi S, Burrows R. ATP synthase complex from bovine heart mitochondria: subunit arrangement as revealed by nearest neighbor analysis and susceptibility to trypsin. J Biol Chem. 1990;265:14518–14525. [PubMed] [Google Scholar]
- 30.Kaiser C, Michaelis S, Mitchell A. Methods in yeast genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 30a.Kalish, J. Unpublished data.
- 31.Kiebler M, Becker K, Pfanner N, Neupert W. Mitochondrial protein import—specific recognition and translocation of preproteins. J Membr Biol. 1993;135:191–207. doi: 10.1007/BF00211091. [DOI] [PubMed] [Google Scholar]
- 32.Kiebler M, Keil P, Schneider H, Vanderklei I J, Pfanner N, Neupert W. The mitochondrial receptor complex—a central role of MOM22 in mediating preprotein transfer from receptors to the general insertion pore. Cell. 1993;74:483–492. doi: 10.1016/0092-8674(93)80050-o. [DOI] [PubMed] [Google Scholar]
- 33.Kiebler M, Pfaller R, Söllner T, Griffiths G, Horstmann H, Pfanner N, Neupert W. Identification of a mitochondrial receptor complex required for recognition and membrane insertion of precursor proteins. Nature. 1990;348:610–616. doi: 10.1038/348610a0. [DOI] [PubMed] [Google Scholar]
- 34.Komiya T, Sakaguchi M, Mihara K. Cytoplasmic chaperones determine the targeting pathway of precursor proteins to mitochondria. EMBO J. 1996;15:399–407. [PMC free article] [PubMed] [Google Scholar]
- 35.Kronidou N G, Oppliger W, Bolliger L, Hannavy K, Glick B S, Schatz G, Horst M. Dynamic interaction between Isp45 and mitochondrial hsp70 in the protein import system of the yeast mitochondrial inner membrane. Proc Natl Acad Sci USA. 1994;91:12818–12822. doi: 10.1073/pnas.91.26.12818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kubrich M, Keil P, Rassow J, Dekker P J, Blom J, Meijer M, Pfanner N. The polytopic mitochondrial inner membrane proteins MIM17 and MIM23 operate at the same preprotein import site. FEBS Lett. 1994;349:222–228. doi: 10.1016/0014-5793(94)00670-9. [DOI] [PubMed] [Google Scholar]
- 37.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 38.Leonhard K, Herrmann J M, Stuart R A, Mannhaupt G, Neupert W, Langer T. AAA proteases with catalytic sites on opposite membrane surfaces comprise a proteolytic system for the ATP-dependent degradation of inner membrane proteins in mitochondria. EMBO J. 1996;15:4218–4229. [PMC free article] [PubMed] [Google Scholar]
- 38a.Leung, R., and R. Jensen. Unpublished observations.
- 39.Lohret T A, Jensen R E, Kinnally K W. Tim23, a protein import component of the mitochondrial inner membrane, is required for normal activity of the multiple conductance channel, MCC. J Cell Biol. 1997;137:377–386. doi: 10.1083/jcb.137.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maarse A C, Blom J, Grivell L A, Meijer M. MPI1, an essential gene encoding a mitochondrial membrane protein, is possibly involved in protein import into yeast mitochondria. EMBO J. 1992;11:3619–3628. doi: 10.1002/j.1460-2075.1992.tb05446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maarse A C, Blom J, Keil P, Pfanner N, Meijer M. Identification of the essential yeast protein MIM17, an integral mitochondrial inner membrane protein involved in protein import. FEBS Lett. 1994;349:215–221. doi: 10.1016/0014-5793(94)00669-5. [DOI] [PubMed] [Google Scholar]
- 42.Martin J. Molecular chaperones and mitochondrial protein folding. J Bioenerg Biomembr. 1997;29:35–43. doi: 10.1023/a:1022407705182. [DOI] [PubMed] [Google Scholar]
- 43.Mayer A, Lill R, Neupert W. Translocation and insertion of precursor proteins into isolated outer membranes of mitochondria. J Cell Biol. 1993;121:1233–1243. doi: 10.1083/jcb.121.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayer A, Nargang F E, Neupert W, Lill R. MOM22 is a receptor for mitochondrial targeting sequences and cooperates with MOM19. EMBO J. 1995;14:4204–4211. doi: 10.1002/j.1460-2075.1995.tb00094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mayer A, Neupert W, Lill R. Mitochondrial protein import: reversible binding of the presequence at the trans side of the outer membrane drives partial translocation and unfolding. Cell. 1995;80:127–137. doi: 10.1016/0092-8674(95)90457-3. [DOI] [PubMed] [Google Scholar]
- 46.Moczko M, Dietmeier K, Söllner T, Segui B, Steger H F, Neupert W, Pfanner N. Identification of the mitochondrial receptor complex in Saccharomyces cerevisiae. FEBS Lett. 1992;310:265–268. doi: 10.1016/0014-5793(92)81345-m. [DOI] [PubMed] [Google Scholar]
- 47.Nigro J M, Sikorski R, Reed S I, Vogelstein B. Human p53 and CDC2Hs genes combine to inhibit the proliferation of Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:1357–1365. doi: 10.1128/mcb.12.3.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Niman H L, Houghten R A, Walker L E, Reisfeld R A, Wilson I A, Hogle J M, Lerner R A. Generation of protein-reactive antibodies by short peptides is an event of high frequency: implications for the structural basis of immune recognition. Proc Natl Acad Sci USA. 1983;80:4949–4953. doi: 10.1073/pnas.80.16.4949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohba M, Schatz G. Disruption of the outer membrane restores protein import to trypsin-treated yeast mitochondria. EMBO J. 1987;6:2117–2122. doi: 10.1002/j.1460-2075.1987.tb02478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pfanner N, Craig E A, Meijer M. The protein import machinery of the mitochondrial inner membrane. Trends Biochem Sci. 1994;19:368–372. doi: 10.1016/0968-0004(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 51.Pfanner N, Douglas M G, Endo T, Hoogenraad N J, Jensen R E, Meijer M, Neupert W, Schatz G, Schmitz U K, Shore G. Uniform nomenclature for the protein transport machinery of the mitochondrial membranes. Trends Biochem Sci. 1996;21:51–52. [PubMed] [Google Scholar]
- 52.Pfanner N, Hartl F U, Guiard B, Neupert W. Mitochondrial precursor proteins are imported through a hydrophilic membrane environment. Eur J Biochem. 1987;169:289–293. doi: 10.1111/j.1432-1033.1987.tb13610.x. [DOI] [PubMed] [Google Scholar]
- 53.Rassow J, Maarse A C, Krainer E, Kubrich M, Muller H, Meijer M, Craig E A, Pfanner N. Mitochondrial protein import—biochemical and genetic evidence for interaction of matrix Hsp70 and the inner membrane protein Mim44. J Cell Biol. 1994;127:1547–1556. doi: 10.1083/jcb.127.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53a.Rine J. Gene overexpression in studies of Saccharomyces cerevisiae. Methods Enzymol. 1991;194:239–251. doi: 10.1016/0076-6879(91)94019-9. [DOI] [PubMed] [Google Scholar]
- 54.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 54a.Ryan, K. Unpublished data.
- 55.Ryan K R, Jensen R E. Mas6p can be crosslinked to an arrested precursor and interacts with other proteins during mitochondrial protein import. J Biol Chem. 1993;268:23743–23746. [PubMed] [Google Scholar]
- 56.Ryan K R, Jensen R E. Protein translocation across mitochondrial membranes: what a long, strange trip it is. Cell. 1995;83:517–519. doi: 10.1016/0092-8674(95)90089-6. [DOI] [PubMed] [Google Scholar]
- 57.Ryan K R, Menold M M, Garrett S, Jensen R E. SMS1, a high-copy suppressor of the yeast mas6 mutant, encodes an essential inner membrane protein required for mitochondrial protein import. Mol Biol Cell. 1994;5:529–538. doi: 10.1091/mbc.5.5.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saiki R K, Scharf S, Faloona F, Mullis K B, Horn G T, Erlich H A, Arnheim N. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 58a.Saville, K. Unpublished data.
- 59.Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;271:31763–31766. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- 60.Schatz G, Dobberstein B. Common principles of protein translocation across membranes. Science. 1996;271:1519–1526. doi: 10.1126/science.271.5255.1519. [DOI] [PubMed] [Google Scholar]
- 61.Scherer P E, Manning-Krieg U C, Jenö P, Schatz G, Horst M. Identification of a 45-kDa protein at the protein import site of the yeast mitochondrial inner membrane. Proc Natl Acad Sci USA. 1992;89:11930–11934. doi: 10.1073/pnas.89.24.11930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schlossmann J, Dietmeier K, Pfanner N, Neupert W. Specific recognition of mitochondrial preproteins by the cytosolic domain of the import receptor MOM72. J Biol Chem. 1994;269:11893–11901. [PubMed] [Google Scholar]
- 63.Schneider H-C, Berthold J, Bauer M F, Dietmeier K, Guiard B, Brunner M, Neupert W. Mitochondrial Hsp70/MIM44 complex facilitates protein import. Nature. 1994;371:768–774. doi: 10.1038/371768a0. [DOI] [PubMed] [Google Scholar]
- 64.Sherman F. Mutants of yeast deficient in cytochrome c. Genetics. 1964;49:39–48. doi: 10.1093/genetics/49.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 66.Sikorski R, Boeke J D. In vitro mutagenesis and plasmid shuffling: from cloned gene to mutant yeast. Methods Enzymol. 1991;194:302–318. doi: 10.1016/0076-6879(91)94023-6. [DOI] [PubMed] [Google Scholar]
- 67.Sikorski R, Hieter P. A system of shuttle vectors and host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–28. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Söllner T, Griffiths G, Pfaller R, Pfanner N, Neupert W. MOM19, an import receptor for mitochondrial precursor proteins. Cell. 1989;59:1061–1070. doi: 10.1016/0092-8674(89)90762-9. [DOI] [PubMed] [Google Scholar]
- 69.Söllner T, Pfaller R, Griffiths G, Pfanner N, Neupert W. A mitochondrial import receptor for the ADP/ATP carrier. Cell. 1990;62:107–115. doi: 10.1016/0092-8674(90)90244-9. [DOI] [PubMed] [Google Scholar]
- 70.Söllner T, Rassow J, Wiedmann M, Schlossmann J, Keil P, Neupert W, Pfanner N. Mapping of the protein import machinery in the mitochondrial outer membrane by crosslinking of translocation intermediates. Nature. 1992;355:84–87. doi: 10.1038/355084a0. [DOI] [PubMed] [Google Scholar]
- 71.Spencer F, Hugerat Y, Simchen G, Hurko O, Connelly C, Hieter P. Yeast kar1 mutants provide an effective method for YAC transfer to new hosts. Genomics. 1994;22:118–126. doi: 10.1006/geno.1994.1352. [DOI] [PubMed] [Google Scholar]
- 72.Stuart R A, Gruhler A, van der Klei I, Guiard B, Koll H, Neupert W. The requirement for matrix ATP for the import of precursor proteins into the mitochondrial matrix and intermembrane space. Eur J Biochem. 1994;220:9–18. doi: 10.1111/j.1432-1033.1994.tb18593.x. [DOI] [PubMed] [Google Scholar]
- 73.Tyers M, Tokiwa G, Nash R, Futcher B. The Cln3-Cdc28 kinase complex of S. cerevisiae is regulated by proteolysis and phosphorylation. EMBO J. 1992;11:1773–1784. doi: 10.1002/j.1460-2075.1992.tb05229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van Loon A P G M, Brändli A W, Schatz G. The presequences of two imported mitochondrial proteins contain information for intracellular and intramitochondrial sorting. Cell. 1986;44:801–812. doi: 10.1016/0092-8674(86)90846-9. [DOI] [PubMed] [Google Scholar]
- 75.von Ahsen O, Voos W, Henninger H, Pfanner N. The mitochondrial protein import machinery: role of ATP in dissociation of the Hsp70-Mim44 complex. J Biol Chem. 1995;270:29848–29853. doi: 10.1074/jbc.270.50.29848. [DOI] [PubMed] [Google Scholar]
- 76.Winston F, Dollard C, Ricupero-Hovasse S L. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 77.Witte C, Jensen R E, Yaffe M P, Schatz G. MAS1, a gene essential for yeast mitochondrial assembly, encodes a subunit of the mitochondrial processing protease. EMBO J. 1988;7:1439–1447. doi: 10.1002/j.1460-2075.1988.tb02961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yaffe M P, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang M, Jensen R E, Yaffe M P, Oppliger W, Schatz G. Import of proteins into yeast mitochondria: the purified matrix processing protease contains two subunits which are encoded by the nuclear MAS1 and MAS2 genes. EMBO J. 1988;7:3857–3862. doi: 10.1002/j.1460-2075.1988.tb03271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]