Abstract
Adf-1 is an essential Drosophila melanogaster sequence-specific transactivator that binds the promoters of a diverse group of genes. We have performed a comprehensive mapping of the functional domains of Adf-1 to study the role of transactivators in the process of gene activation. Using a series of clustered point mutations and small deletions we have identified regions of Adf-1 required for DNA binding, dimerization, and activation. In contrast to most enhancer-binding factors, the Adf-1 activation regions are nonmodular and depend on an intact protein, including the Adf-1 DNA-binding domain, for activity. Like many transcriptional activators, Adf-1 contains a TFIID-binding domain that can interact with specific TAF subunits. Although TAFs are required for Adf-1-directed activation, TAF binding is not sufficient, suggesting that Adf-1 may direct multiple essential steps during activation. Interestingly, both the TAF-binding domain and the DNA-binding domain contain sequences homologous to those of the Myb family of DNA-binding domains. Thus, Adf-1 has evolved an unusual structure containing two versions of the Myb motif, one that binds DNA and one that binds proteins.
A major form of cellular regulation occurs at the level of transcriptional activation. The control of transactivation is, in turn, largely due to the modulation of site-specific transcription factor activities in the nuclei of eukaryotic cells. A significant advance was the realization that many promoter- and enhancer-specific transcriptional activators are modular, with separable DNA-binding and activation domains (15, 25). The modularity of transcription factors has been useful in elucidating the structure and function of DNA-binding and activation domains. For example, many DNA-binding domains (DBDs) have been well characterized and fall within one of less than a dozen major structural families (e.g., helix-turn-helix, zinc finger, basic helix-loop-helix) (29). In contrast, the characterization of the transcription activation domains of these proteins has proven more challenging.
Numerous mechanisms have been proposed for transcriptional activation including DNA-bending, recruitment of various basal transcription factors such as TFIID and RNA polymerase, alterations of chromatin structure, and the targeting of cofactors (16, 35). The complexity of most transcription factors suggests that multiple mechanisms are likely to be utilized for efficient activation by any given activator and at any given promoter. In the context of the paradigm of modularity, transactivating regions of proteins have often been studied by fusing them to heterologous DBDs. Various types of activation domains have been mapped by this approach, revealing, in many cases, protein sequences with a preponderance of certain residues (e.g., acidic, glutamine, proline) (22, 36) but little information concerning three-dimensional structure. Despite these studies, discrete activation domain boundaries have often been difficult to define. Rather, increasingly large deletions incrementally compromise the potency of activation regions (20). Alternatively, the expedient construction of artificial “hybrid” activators has been useful for mapping minimal functional activators in many cases. However, the use of heterologous protein fusions eliminates potentially important intramolecular interactions that may occur between activation domains and other regions of the native protein. Likewise, other factors that may play a role in the potency of these activation domains, such as their natural oligomerization states and their positioning with respect to DNA, cannot be studied when activation domains are analyzed out of their native context. Consequently, our understanding of the mechanisms by which these activation domains function remains superficial or incomplete in most cases.
In an attempt to circumvent some of these inherent difficulties in dissecting the activities of a transcriptional activator, we have chosen to analyze the Drosophila melanogaster transcription factor Adf-1. Although first identified as a factor that bound the distal promoter of the gene for alcohol dehydrogenase (Adh) (18), Adf-1 recognizes specific sites in a diverse group of promoters including antennapedia P1 and dopa decarboxylase (14). Its ubiquitous expression (13) and its requirement for viability (10) establish the important role of Adf-1 in Drosophila biology. The sequence of Adf-1 revealed that its presumptive DBD is a distantly related member of the Myb helix-turn-helix family (13, 24), whereas no significant similarities to the sequences associated with any activation domain categories were observed. This suggested that Adf-1 may function through a novel type of transactivation domain. Adf-1 is also one of the smallest eukaryotic transcription factors thus far identified, with a calculated mass of less than 30 kDa. Its small size presented an opportunity to define a concise activation domain that could be studied in its native context. This, taken together with the essential role that Adf-1 plays in Drosophila and the opportunity to dissect a novel activation domain, prompted us to undertake a detailed functional analysis of this enhancer-binding protein.
Here, we report a detailed functional mapping of Adf-1. We generated a series of clustered point mutations and small deletions throughout the protein to probe functional regions of Adf-1. These alterations were relatively small so as to minimize potential structural perturbations, although such perturbations can never be completely avoided. The mutant proteins were assayed for transactivation, DNA binding, and oligomerization through a variety of assays. By measuring reporter activity in Drosophila cells cotransfected with mutant Adf-1 genes as well as by testing purified recombinant proteins by in vitro transcription, we have discovered that Adf-1 differs dramatically from typical modular transcription factors. Activation by Adf-1 critically depends on its DBD. The nonmodular activation regions contain a TFIID interaction domain that binds specifically to TAFII110 and TAFII250. Although we demonstrate that TAFs are required for Adf-1 activity, they are not sufficient. Our data suggest that regions of Adf-1 besides the TAF-binding domain play additional essential roles in transactivation. Interestingly, the Adf-1 dimerization and TAF-binding region also appears to be a Myb-like domain, suggesting a novel use for the Myb domain in mediating protein-protein interactions rather than binding DNA. Additionally, this domain has similarity to a putative protein-binding region of the transcriptional adapter ADA2, implying that there may be a wider family of proteins containing Myb-like protein interaction domains.
MATERIALS AND METHODS
Construction of Adf-1 mutants.
PCR primers matching the sequences immediately downstream and upstream of an Adf-1 cDNA cloned into pBlueScript KS+ were generated (wt left 5′ primer and wt right 3′ primer). The wt left 5′ primer included an EcoRI restriction site, and the wt right 3′ primer included an XbaI site for later subcloning. At the site of each of the alanine-scanning mutations, two mutagenizing PCR primers were generated. The mutagenizing left 3′ primer (m left 3′ primer) consisted of approximately 12 nucleotides of wild-type sequence followed by a sequence coding for five alanines and containing a NotI site, followed by a stop codon and an XbaI site. The mutagenizing right 5′ primer (m right 5′ primer) contained an EcoRI site followed by a sequence coding for five alanines, with PstI and NotI sites, followed by roughly 12 nucleotides of wild-type sequence. PCR was performed with Taq polymerase, with the pBlueScript KS+ Adf-1 plasmid as the template and either wt left 5′ and m left 3′ primers (left PCR) or m right 5′ and wt right 3′ primers (right PCR). The products of each pair of PCRs were digested with EcoRI and XbaI and cloned into pBlueScript KS+. These were sequenced, and the correct clones were digested with either EcoRI and NotI (left product) or NotI and XbaI (right product). The two DNA fragments were ligated together into EcoRI- and XbaI-digested pBlueScript KS+. Most of the deletions were generated by ligating left and right halves from PCRs for adjoining pairs of mutants. ΔN5, ΔN12, and 5A245 were generated with only one mutagenizing PCR primer and one wild-type PCR primer. Mutant Adf-1 clones were subcloned from pBlueScript KS+ and ligated into pPAC Nde for Schneider cell transfections or into pET-3a or pET-21a for Escherichia coli protein expression.
Transfections and CAT assays.
Transfection of Schneider SL2 cells was done by a CaPO4 protocol previously described (30). Into each well of a six-well plate, 2.5 ml of cells at a density of 2.5 × 106 cells per ml was added. These cells were incubated with 0.4 ml of a CaPO4 mixture including 0.5 μg of reporter plasmid, 1.25 μg of expression plasmid, and salmon sperm DNA to a final total of 5.5 μg of DNA. The reporter plasmid was a G5BCAT construct (23) containing nucleotides −71 to −47 of the alcohol dehydrogenase distal promoter cloned into the SalI site. The expression plasmid was either pPAC Nde (3) alone or pPAC Nde containing the gene for wild-type or mutant Adf-1 cloned into the NdeI and XbaI sites.
Two days posttransfection, cells were harvested, washed with phosphate-buffered saline, and resuspended in 40 μl of 0.25 M Tris (pH 7.9). These samples were subjected to three freeze-thaw cycles and spun briefly to eliminate cellular debris. From them 20 μl was removed, heated for 5 min at 65°C, and used for chloramphenicol acetyltransferase (CAT) assays as described previously (9). Data from at least two sets of transfections were quantitated on a Fuji phosphorimager and averaged. The protein concentrations of the extracts used for CAT assays, measured by a Bradford assay, were used to adjust the CAT assay data. Additionally, 10 μl of the frozen and thawed extracts was run on sodium dodecyl sulfate (SDS)–10% polyacrylamide gels, blotted to nitrocellulose, and probed with anti-Adf-1 polyclonal antibodies to qualitatively measure protein levels. For electrophoretic mobility shift assay (EMSA) experiments, 2.5 μl of the frozen and thawed extracts was used.
Nuclear localization was assayed by using nuclear and cytoplasmic extracts from transfected cells (1) for Western blots, which were probed with anti-Adf-1 antibodies.
Protein expression and purification.
Mutant and wild-type genes for Adf-1 cloned into pET-3a (13) or pET-21a were transformed into E. coli BL21 (DE3) containing the pLysS plasmid. These were grown in Luria broth to an optical density at 600 nm of 0.5 and induced for 1 h with 0.8 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were lysed with a French press in buffer containing 25 mM HEPES (pH 7.6), 0.4 M NaCl, 0.5 mM EDTA, 0.01% Nonidet P-40 (NP-40), 5 mM dithiothreitol (DTT), 5 mM β-mercaptoethanol, 0.2 mM phenylmethylsulfonyl fluoride, and 1 mM sodium metabisulfate. This extract was centrifuged for 1 h at 35,000 rpm in a Ti45 rotor. The supernatant was passed over a 10 ml DEAE-Sepharose column. The flowthrough was diluted with an equal volume of 0 M NaCl buffer. This was passed over a 5-ml SP-Sepharose column. The column was washed with 0.2 M NaCl buffer and eluted with 0.5 M NaCl buffer. Fractions containing Adf-1 protein were pooled and diluted with an equal volume of 0 M NaCl buffer. This protein was passed over a 0.5-ml DNA affinity column containing streptavidin-Sepharose beads bound to biotinylated DNA corresponding to nucleotides −89 to −47 of the Adh distal promoter. The column was washed with 0.25 M NaCl buffer and eluted with 0.5 M NaCl buffer. Adf-1 eluted from the DNA affinity column was nearly homogenous as determined by Coomassie staining. During storage, Adf-1 is converted to a slower-electrophoretic-mobility form. This form of Adf-1 cannot be separated from the primary form chromatographically and may be the result of a proline isomerization.
EMSAs.
For EMSA experiments using Schneider cell extracts, 2.5 μl of transfected cell extract was incubated for 10 min at 20°C in a 16-μl reaction mixture containing approximately 70 fmol of labeled Adf-1 binding site probe, 0.9 μg of salmon sperm DNA as a nonspecific competitor (a ratio of approximately 2,000:1 relative to the specific probe), and 1× EMSA buffer (10 mM Tris [pH 7.9], 4.5% Ficoll 400, 4 mM MgCl2, 0.1 mM EDTA, 50 μg of bovine serum albumin per ml, 0.2% NP-40, 1 mM DTT). Samples were electrophoresed over a 5% acrylamide gel (0.5× Tris-borate-EDTA (TBE), 0.75 mM EDTA, 0.05% NP-40) in 0.5× TBE buffer at 250 V. The specific probe was prepared by end labeling oligonucleotides corresponding to nucleotides −71 to −47 of the alcohol dehydrogenase distal promoter with 32P. EMSA experiments with purified protein were essentially identical except that 50 mM NaCl was added to the binding-reaction mixtures.
Glutaraldehyde cross-linking.
To a 40-μl reaction mixture containing 50 mM HEPES (pH 7.6) and 100 mM NaCl, 80 ng of purified Adf-1 protein was added. To this, 1.25 μl of 0.08% glutaraldehyde was added, and the mixture was incubated at 20°C for 2.5 min. Ten-microliter samples were removed and added to 5 μl of SDS-polyacrylamide gel electrophoresis (PAGE) loading dye to quench the reactions. These samples were then run on SDS–10% acrylamide gels, blotted to nitrocellulose, and probed with anti-Adf-1 antibodies.
Protein-binding assays.
Full length TAFs and portions of TAFII110 and TAFII250 were transcribed and translated in vitro in 25 μl of rabbit reticulocyte lysates from the TNT kit (Promega) containing [35S]methionine programmed with the appropriate ptβStop vectors (21). To this, 200 μl of 0.1 M NaCl HEMG buffer (25 mM HEPES [pH 7.6], 0.1 mM EDTA, 12.5 mM MgCl, 10% glycerol) containing 0.01% NP-40 was added. After 10 min of high-speed centrifugation, 50 μl of lysate was added to 5 μl of the appropriate glutathione beads. The beads had been prepared by incubating them with E. coli extract containing either glutathione S-transferase (GST) protein or GST fused to the carboxy-terminal 92 amino acids of Adf-1 (C92 fragment) and then washing them extensively with 0.5 M NaCl HEMG buffer. After binding for 4 h at 4°C, the beads were washed four times with 0.1 M NaCl HEMG containing 0.01% NP-40. Bound protein was eluted by boiling the beads in SDS-PAGE loading buffer and was subjected to electrophoresis followed by autoradiography.
For binding with full-length wild-type and mutant Adf-1 proteins, 500 ng of E. coli purified Adf-1 proteins was incubated for 5 h at 4°C with 2.5 to 5 μl of the appropriate beads, followed by five washes with 0.2 M NaCl HEMG containing 0.1% NP-40. Bound protein was eluted by boiling the beads in SDS-PAGE loading buffer and then was subjected to electrophoresis, followed by Western blotting and probing with anti-Adf-1 antibodies. Western blots were also probed with anti-TAF antibodies to determine that equal amounts of target protein were used. Beads were created by incubating whole-cell extracts from Sf9 cells infected with baculovirus containing expression constructs for HA-TAFII110, HA-hTAFII250, or GST-hNTAFII250 (amino acids 1 to 414) or by incubating E. coli extracts containing GST-NTAFII110 (amino acids 75 to 208) or GST alone with either anti-HA-Sepharose (HA monoclonal antibodies covalently linked to protein A-Sepharose beads) or glutathione-Sepharose beads (Pharmacia), as appropriate. TFIID beads were created by incubating a purified Drosophila TFIID fraction (Mono-Q) (17) with protein A-Sepharose beads (Pharmacia) bound to anti-TAFII250 monoclonal antibodies. All beads were then washed extensively with 0.5 or 1.0 M NaCl HEMG–0.1% NP-40–0.1% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}.
The yeast two-hybrid experiments were performed as described previously (19). DNA coding for the carboxy-terminal 92 amino acids of Adf-1 and for the Sp1 B domain was cloned into the GAL4 activation domain containing plasmid pAS1 (11), and DNA coding for fragments TAFII110 and hTAFII250 was cloned into the GAL4 DBD (amino acids 1 to 147) plasmid, pGAD (25). These were cotransformed into Saccharomyces cerevisiae Y153 containing LacZ under the control of a GAL4-responsive promoter. After several days of growth on selective media containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), yeast colonies were scored for the presence or absence of blue color. Controls containing either the GAL4 DBD alone cotransformed with the C92-GAL4 acidic activation domain (AAD) or the GAL4 AAD alone cotransformed with each of the TAF-GAL4 DBD fusions scored negative.
All the TAF sequences used were from Drosophila except for human hTAFII250 in some cases, as indicated in the figure legends.
In vitro transcription.
In vitro transcription and primer extension experiments were performed as described previously (18). Mutant and wild-type Adf-1 transcription was assayed in 20-μl reaction volumes containing 15 ng of activator, 1.5 μl of H.4 Drosophila embryo extract (12, 38) (that had been immunodepleted of endogenous Adf-1; see below) 40 ng of nucleotide −86 template (18), 40 ng of nucleotide −46 template (similar to nucleotide −86 template except for containing nucleotides −46 to +13 of the alcohol dehydrogenase distal promoter and producing a shorter transcript), 0.5× HEMG buffer, 50 mM KCl, 8 U of RNasin, 1 mM DTT, and 0.625 mM ribonucleoside triphosphates. The activator was preincubated with buffer for 15 min at 20°C. To this, depleted H.4 extract was added, and the mixture was incubated for an additional 5 min, followed by the addition of nucleotides and transcription for 10 min at 20°C. In vitro transcription products were assayed by primer extension (23). H.4 extract was immunodepleted of Adf-1 by incubating the extract with protein A-Sepharose beads bound to affinity-purified anti-Adf-1 antibodies overnight at 4°C.
To determine TFIID requirements, transcription was performed with a mixture containing 10 ng of activator, the H.4 fraction that had been immunodepleted against Adf-1, TAFII250, and TATA-binding protein (TBP); the mixture was supplemented with 25 ng of E. coli-produced TFIIA, 6 ng of purified Drosophila TFIIE, and 0.84 μl of a Drosophila S-300 fraction containing TFIIF, TFIIH, and PolII. To this, either 1.2 μl of a Drosophila Mono-Q fraction containing TFIID or 5 ng of E. coli-produced TBP was added. The basal transcription factors were purified as described previously (17). All protein fractions were preincubated together for 15 min at 20°C, then buffer and nucleotides were added, and the mixture was incubated for 15 min at 20°C to allow transcription to proceed. The reaction mixtures were reverse transcribed and electrophoresed to assay in vitro transcription products.
RESULTS
Mutational analysis of Adf-1 maps DNA-binding, dimerization, and activation regions.
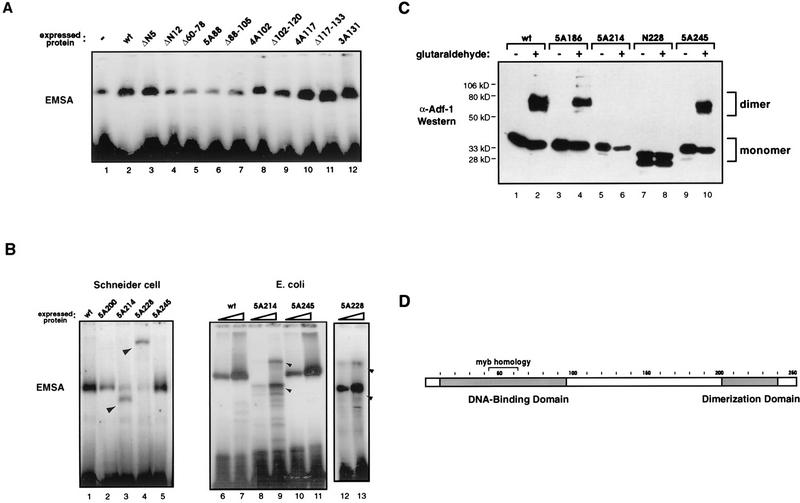
To probe the functions of various parts of Adf-1, we performed an alanine-scanning mutagenesis analysis by substituting clusters of three to five amino acids at a time. These mutations were spaced approximately 10 amino acids apart except in the amino-terminal 80 amino acids, which has homology to the Myb family of helix-turn-helix DBDs (13). Twelve groups of alanine-scanning mutations directed towards the carboxy-terminal two-thirds of Adf-1 were sufficient to probe the activities of the protein (Fig. 1). Additionally, small deletions were made by deleting the amino acids between adjacent pairs of alanine-scanning mutations. Two small deletions at the amino terminus and one at the carboxy terminus were also constructed (Fig. 1). The number of alanines and the position at which they start are indicated in the name of each of the substitution mutants, and the range of amino acids removed is indicated for each of the deletions.
FIG. 1.
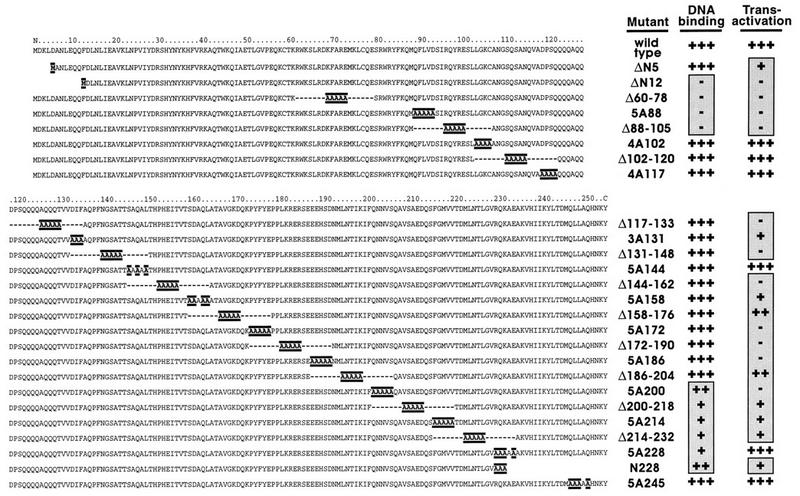
The mutagenesis of Adf-1 allowed the mapping of DNA-binding and transactivation activities by using Schneider cell transfections. Twenty-six mutant forms of Adf-1 were constructed and assayed following Schneider cell transfections. The amino and carboxy halves of Adf-1 are shown with nonnative amino acids highlighted. Internally deleted amino acids are indicated with dashes. Data obtained from quantitating mobility shift experiments using extracts from Schneider cells transfected with wild-type and mutant expression vectors and Adf-1 binding site probes are summarized in the “DNA binding” column. Data obtained from quantitating CAT assays using extracts of cells cotransfected with Adf-1 expression vectors and an Adf-1-responsive CAT vector are summarized in the “Transactivation” column. Symbols (both columns): +++, activity at least 70% of that of the wild type; ++, activity between 30 and 69% of that of the wild type; +, activity between 10 and 29% of that of the wild type; −, activity less than 10% of that of the wild type. Background activity was standardized to 0% of that of the wild type.
To assay the activities of these proteins, expression vectors containing the wild-type or mutant genes under the control of a constitutive actin promoter were transfected into Drosophila Schneider L2 cells. A minimal promoter containing an Adf-1 binding site from the alcohol dehydrogenase distal promoter (13, 14, 17, 18) (data not shown) and the adenovirus E1B TATA sequence were used to drive the expression of a CAT reporter gene. Schneider cells constitutively express a low level of Adf-1, so careful titration of expression vector, reporter vector, and carrier DNA was performed to minimize the background signal. Following transfection, extracts were made from the Schneider cells and assayed for CAT activity. These cell extracts were also used in EMSAs. A summary of the data obtained from these DNA-binding and transactivation assays for all the mutants is shown in Fig. 1. Additionally, Western blots were made with whole-cell and nuclear extracts of transfected cells and probed with anti-Adf-1 antibodies to confirm that all the transfected genes were expressed at similar levels and were localized in the nucleus (data not shown).
The EMSAs identified two regions in Adf-1 that contribute to DNA binding (Fig. 1). The most dramatic effects on DNA binding were seen with four amino-terminal mutants (ΔN12, Δ60-78, 5A88, and Δ88-105) (Fig. 2A). This region contains the Myb motif (13), and a truncated protein containing only the amino-terminal 130 amino acids retains sequence-specific DNA-binding activity (data not shown). These data suggest that this amino-terminal region is a bona fide DBD. A second group of mutants lying near the carboxy terminus of Adf-1 also compromised DNA binding (Fig. 1). Interestingly, two of these, mutants 5A214 and 5A228, gave rise to Adf-1–DNA complexes that migrated aberrantly in EMSA experiments (Fig. 2B, lanes 3 and 4). Mutant 5A214 gave rise to a weak shift with mobility higher than that of the other full-length forms of Adf-1. In contrast, the 5A228 protein produced a shifted species that migrated more slowly than the other complexes. A mobility shift product at the position of the wild-type protein-DNA complex in each of these cases is attributed to DNA binding by wild-type endogenous Adf-1 (such as in Fig. 2A, lane 1).
FIG. 2.
Mutations in two regions of Adf-1 decrease DNA-binding affinity. (A) Wild-type and amino-terminal mutant forms of Adf-1 were tested for their abilities to bind an Adf-1 binding site in mobility shift experiments. Lane 1, background DNA binding due to endogenous Adf-1 in extracts from cells transfected with a negative-control expression vector; lanes 2 to 12, mobility-shifts for cell extracts transfected with the indicated expression vectors. (B) Wild-type and carboxy-terminal mutant forms of Adf-1 were tested for their abilities to bind an Adf-1 binding site-containing probe. Lanes 1 to 5, mobility shifts obtained with extracts from cells transfected with wild-type and mutant Adf-1 as described for panel A; lanes 6 to 11, mobility shifts obtained with 5 (lanes 6, 8, and 10) and 50 ng (lanes 7, 9, and 11) of purified recombinant wild-type and mutant Adf-1 proteins, respectively; lanes 12 and 13, mobility shifts obtained with 0.5 and 1 μl, respectively, of partially purified recombinant 5A228 protein. The positions of aberrant mobility shifts are indicated by arrowheads. (C) Chemical cross-linking of wild-type and mutant forms of Adf-1. Wild-type and mutant forms of Adf-1 were incubated in the presence or absence of 0.0025% glutaraldehyde and then subjected to SDS-PAGE followed by Western blotting and probing with anti-Adf-1 antibodies. The positions of prestained molecular weight standards and the presumed oligomeric states of the Adf-1 species are indicated. The lower band in the N228 samples (lanes 7 and 8) indicates a degradation product. kD, kilodaltons. (D) Schematic of Adf-1 in which the regions that affect DNA binding and the region of Myb homology are indicated. The amino-terminal region is the bona fide DBD, while the carboxy-terminal region is required for dimerization.
To further characterize the behavior of these mutants, wild-type Adf-1 and several carboxy-terminal mutant proteins were expressed in E. coli and purified to homogeneity for EMSA characterization. Mutant 5A228 protein was expressed very poorly and could not be purified easily, so this protein was only partially purified from E. coli extract. In the EMSA, purified mutant 5A214 produced a species that migrated similarly to the 5A214-DNA complex produced from transfected Schneider cells (Fig. 2B; compare lanes 3, 8, and 9). In addition a less-abundant species that migrated more slowly than the wild-type Adf-1–DNA complex was observed (Fig. 2B, lanes 8 and 9). Recombinant 5A228 protein also produced a similarly migrating pair of complexes (Fig. 2B, lanes 12 and 13). These observations prompted us to hypothesize that Adf-1 may bind DNA as a dimer and that mutations in the region between amino acids 200 and 228 may disrupt this dimerization. We predicted that mutant monomeric forms of Adf-1 bound singly or doubly to a dimer DNA site, producing the two observed shifts. Wild-type Adf-1, in contrast, would bind exclusively as a dimer, producing only one species of protein-DNA complex.
To obtain additional evidence for dimer formation by Adf-1, we performed glutaraldehyde cross-linking experiments with purified wild-type and mutant Adf-1 proteins (Fig. 2C). In the presence of 0.0025% glutaraldehyde, wild-type protein is efficiently converted from its monomeric form (migrating at 35 kDa in SDS-PAGE) to a covalently dimerized species (migrating at 70 kDa) (Fig. 2C, lanes 1 and 2). Many of the mutant forms of Adf-1, including mutant 5A186 (Fig. 2C, lanes 3 and 4) and mutant 5A245 (Fig. 2C, lanes 9 and 10), were also efficiently cross-linked by glutaraldehyde. In contrast, mutants 5A214 (Fig. 2C, lanes 5 and 6) and N228 (Fig. 2C, lanes 7 and 8) produced virtually no detectable dimer species upon glutaraldehyde treatment. Although high concentrations of the 5A214 mutant led to a complex in the EMSA with lower mobility, which may be caused by two molecules of 5A214 bound adjacently on DNA containing a dimeric Adf-1 binding site (Fig. 2B, lane 9), the addition of DNA to the glutaraldehyde cross-linking reaction mixture failed to induce the cross-linking of the 5A214 mutant protein (data not shown). The results of these chemical cross-linking experiments, taken together with the aberrant mobility shift results, suggest that mutant 5A214 is unable to form dimers in solution and that two monomeric 5A214 molecules, when bound to adjacent sites on DNA, are in a conformation different from that of a wild-type dimer bound to DNA. Mutant 5A228 protein from Drosophila transfections forms a low-mobility complex in an EMSA, which cannot be reproduced with E. coli-produced protein. Mutant 5A228 may only be partially defective in dimerization and is perhaps able to interact with endogenous wild-type Adf-1 in an unusual complex leading to its aberrant migration. Alternatively, it may be forming higher-order multimers or fortuitously interacting with other unknown Drosophila proteins. Mutant 5A200 protein from transfections formed a normally migrating complex, although to a lesser extent than did the wild-type protein (Fig. 2B, lane 2). Unfortunately, we could not test the E. coli-produced 5A200 protein for dimerization because of its toxicity to E. coli. Thus, our data suggest that wild-type Adf-1 contains an amino-terminal DBD and that DNA binding by this domain is enhanced by protein dimerization that requires a region of Adf-1 between amino acids 200 and 245 (summarized in Fig. 2D).
Transcriptional activation by Adf-1.
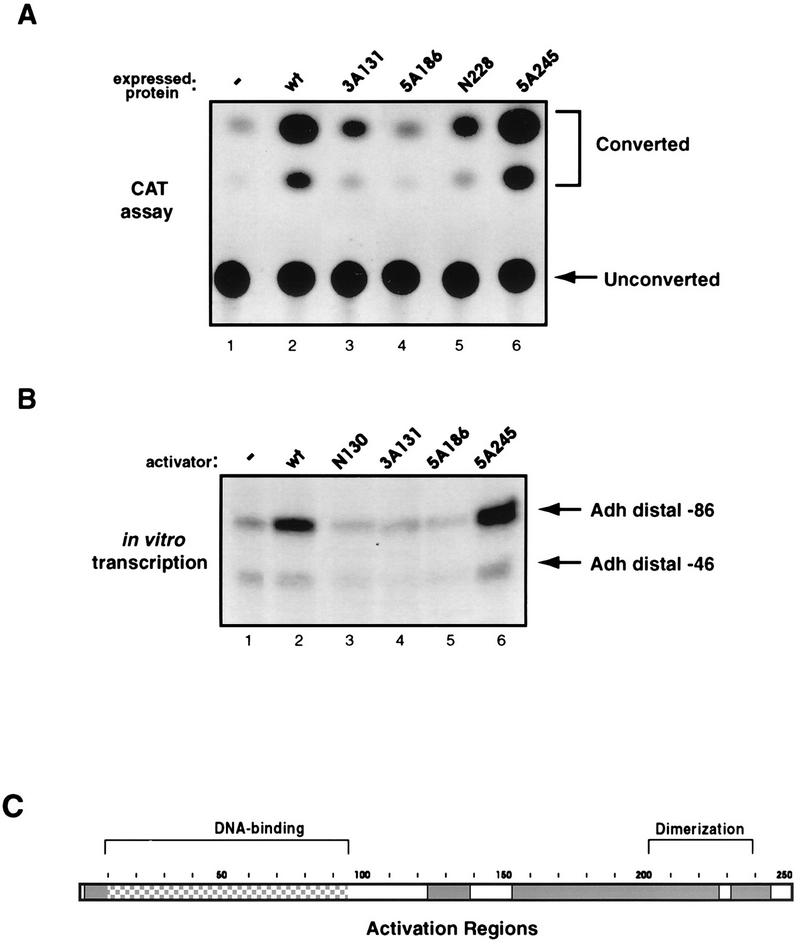
To probe the activation properties of Adf-1, CAT assays were performed with extracts from Schneider cells transfected with wild-type and mutant Adf-1. Wild-type protein directs activation of the CAT reporter at levels significantly above background, while many of the mutants gave rise to little or no CAT activity above endogenous background levels (Fig. 3A; summarized in Fig. 1). Surprisingly, few alanine-scanning or deletion mutants retained transcriptional activity. The only region clearly dispensable for activation lies between amino acids 102 and 120 (Fig. 1), immediately adjacent to the DBD. This region is followed by a short glutamine-rich region, although the importance of this region in activation is not clear from the data. Contrary to expectations, the Δ158-176 and Δ186-204 deletion mutants are more transcriptionally active than the alanine-scanning mutants which they incorporate, and the reasons for this are not clear. Interestingly, removal of just the amino-terminal five amino acids (ΔN5) produced a protein that can bind DNA but that has very little transcriptional activity (Fig. 1). These results provide the first evidence that transactivation by Adf-1 may require an integrated rather than a modular activation domain.
FIG. 3.
Regions of Adf-1 that are required for transcriptional activation. (A) Extracts from transfected cells were used for CAT assays to measure the transcriptional activities of wild-type and mutant forms of Adf-1. Lane 1, background CAT activity from extracts of cells transfected with a negative-control expression vector; lanes 2 to 6, results of CAT assays using extracts of cells transfected with Adf-1 expression vectors. Converted and unconverted forms of [14C]chloramphenicol are indicated. (B) Purified wild-type and mutant forms of Adf-1 behave similarly in in vitro and in vivo transcription. Lane 1, in vitro transcription signal from a reaction mixture containing a partially purified Drosophila embryo extract; the Adh distal position −86 template contains Adf-1 binding sites, while the Adh distal position −46 template does not; lanes 2 to 6, the same transcription system as in lane 1 with 15 ng of purified wild-type or mutant Adf-1 added. (C) A schematic of Adf-1 in which the regions that affect activation are indicated. The DBD is partially filled since it is not possible to discriminate between direct and indirect effects on activation by mutations in this region.
To further characterize the transcriptional activation properties of wild-type and mutant forms of Adf-1, several of these proteins were tested in an in vitro transcription system composed of partially purified Drosophila embryo extracts that had been immunodepleted of endogenous Adf-1. A template containing the alcohol dehydrogenase distal promoter extending to position −86 (containing the natural Adf-1 binding sites) (18) and a control template containing a truncated enhancer extending to position −46 (lacking the Adf-1 binding sites and giving rise to a shorter transcript) were added to our transcription system along with purified recombinant wild-type or mutant forms of Adf-1. The addition of wild-type Adf-1 produced a high level of transcriptional activation on the template containing the Adf-1 binding site (Fig. 3B, lane 2). The addition of no activator or the Adf-1 minimal DBD alone (N130) produced no significant activation (lanes 1 and 3). Two other mutants, 3A131 and 5A186, that displayed little transcriptional activity in vivo were also largely inactive in this in vitro transcription system (lanes 4 and 5). Conversely, an alanine variant that is active in vivo, 5A245, is also fully active in vitro (lane 6). A slight activation of the negative-control template with active proteins (lanes 2 and 6) and a slight inhibition with inactive proteins (lanes 3 to 5) can be seen, presumably due to low levels of Adf-1 binding to cryptic binding sites and dominant-negative effects of the inactive mutants.
The mapping of sequences required for transactivation is summarized in Fig. 3C. Interestingly, no region of Adf-1 is competent for transactivation when separated from its natural DBD domain and fused to the heterologous DBDs of GAL4 or Sp1 (data not shown). These results, taken together, strongly suggest that competency for activation by Adf-1 requires a structure that includes an intact DBD; this structure deviates substantially from the typical modular construction of most other enhancer-binding proteins.
Adf-1 binds TAFs to activate transcription.
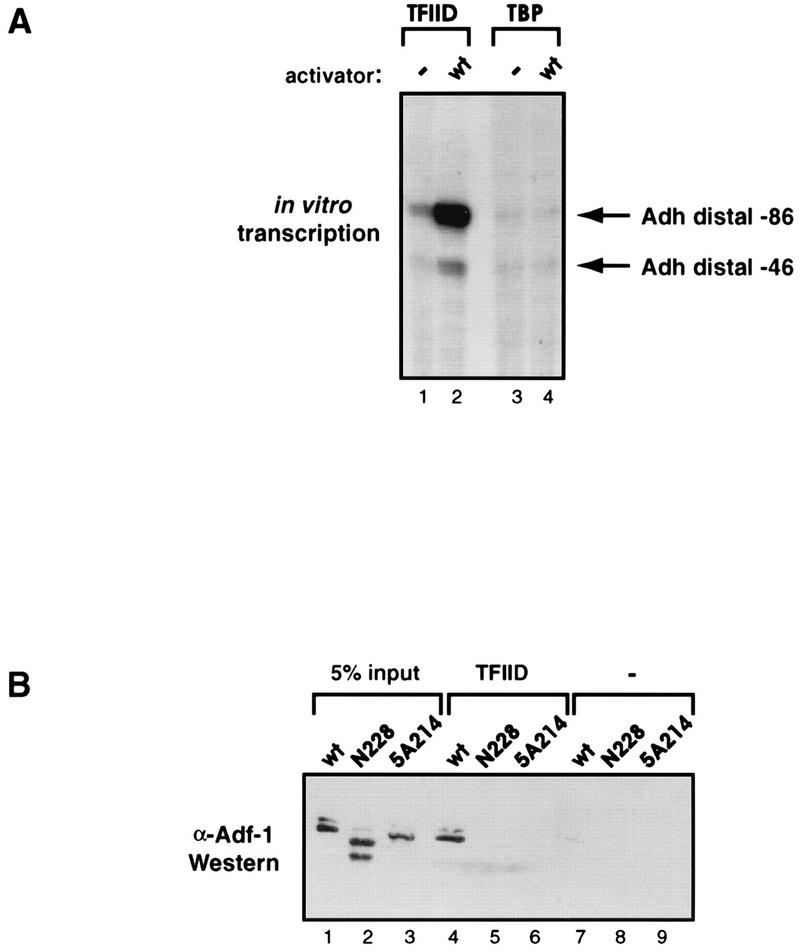
Many transcriptional activators have been shown to interact with the TAF subunits of the TFIID complex, and this interaction has been correlated with transactivation in several cases (6, 7, 31, 33, 34). We next tested whether TAFs were important in Adf-1-directed transcription and, if so, whether there was any correlation between activation and potential Adf-1–TAF interactions. Our Adf-1-dependent in vitro transcription system was depleted of TFIID by using antibodies against both TAFII250 and TBP. In the absence of an exogenous source of TFIID, this transcription system was inactive (data not shown). The addition of either purified TFIID or purified recombinant TBP to the depleted transcription system restored basal transcription levels (Fig. 4A, lanes 1 and 3; the slight activation seen in lane 1 is due to residual endogenous Adf-1). The addition of purified Adf-1 to these reaction mixtures led to a high level of activation when TFIID was present but had no effect on the reaction mixture containing only TBP (Fig. 4A, lanes 2 and 4). These results suggest that Adf-1, like many Drosophila activators, requires the presence of TAFs in order to activate transcription in vitro.
FIG. 4.
Activation by Adf-1 requires the presence of TAFs, and Adf-1 is able to interact directly with the TFIID complex. (A) The TFIID complex is required by Adf-1 to activate transcription. The transcription system used for Fig. 3B was depleted of TFIID activity. Purified TFIID (lanes 1 and 2) or purified recombinant TBP (lanes 3 and 4) was added back either in the absence of added activator (lanes 1 and 3) or with 10 ng of purified wild-type Adf-1 (lanes 2 and 4). (B) Adf-1 can interact directly with purified TFIID, while some mutants cannot. Purified TFIID was immunoprecipitated with beads containing anti-TAFII250 antibodies. These beads (lanes 4 to 6) or negative-control GST beads (lanes 7 to 9) were incubated with wild-type or mutant forms of Adf-1, washed extensively, and subjected to SDS-PAGE followed by Western blotting and probing with anti-Adf-1 (α-Adf-1) antibodies. Results for 5% of the input wild-type and mutant Adf-1 are also shown (lanes 1 to 3). The upper bands seen in lanes 1 and 4 are believed to result from an isomeric form of Adf-1 (see Materials and Methods). The lower band seen in lane 2 results from a degradation product.
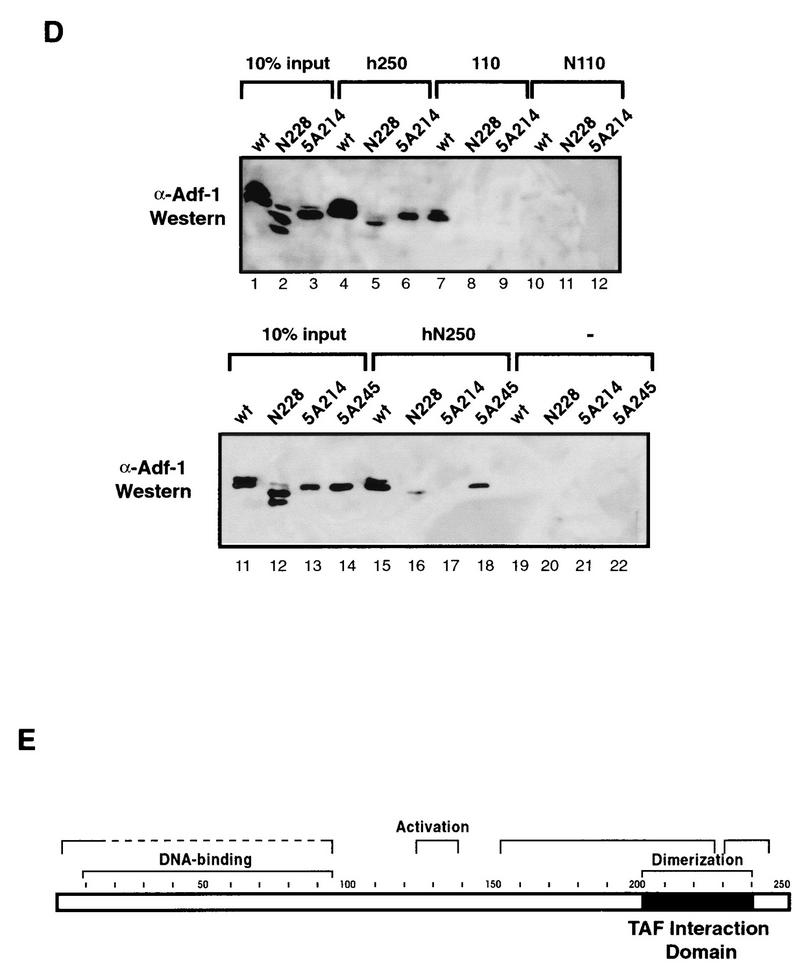
To determine if the transcriptional requirement for TAFs was due to direct interactions between one or more of these proteins and Adf-1, in vitro binding experiments were conducted. Beads containing anti-TAFII250 monoclonal antibodies were used to immunoprecipitate TFIID from partially purified TFIID fractions. Silver staining and Western blotting using anti-TAFII80 antibodies confirmed that an intact holo-TFIID complex was immunoprecipitated (data not shown). Purified wild-type or mutant Adf-1 protein was incubated with these TFIID beads or with negative-control beads. Anti-Adf-1 antibodies were used to visualize the amounts of bound and input Adf-1 proteins on Western blots (Fig. 4B). We could not detect any of the Adf-1 proteins binding to the negative-control beads (lanes 7 to 9), whereas wild-type Adf-1 bound strongly to beads containing TFIID (lane 4). In contrast, the two carboxy-terminal transcriptionally inactive mutants, 5A214 and N228, failed to bind the TFIID beads (lanes 5 and 6), suggesting that the carboxy terminus of Adf-1 is necessary for TFIID binding.
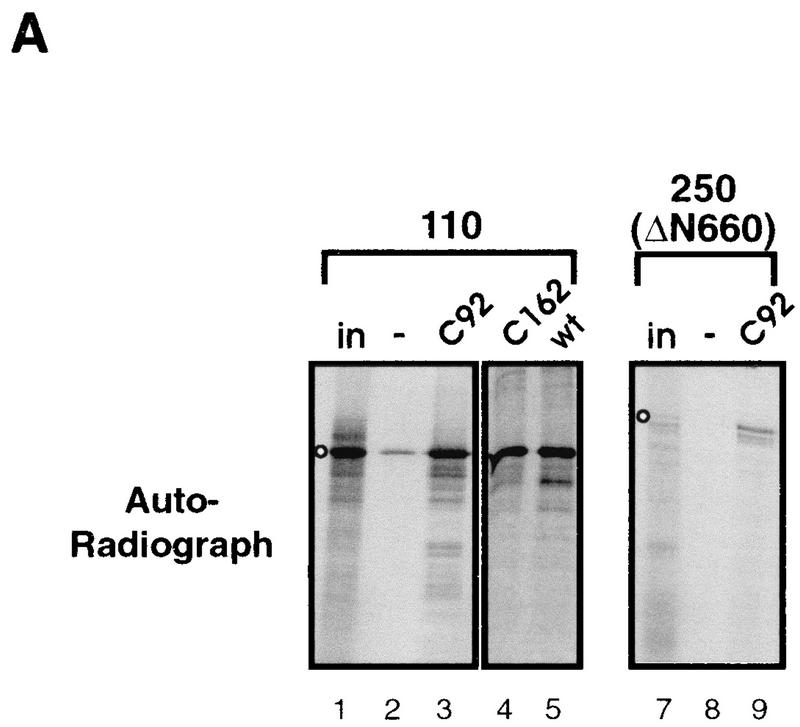
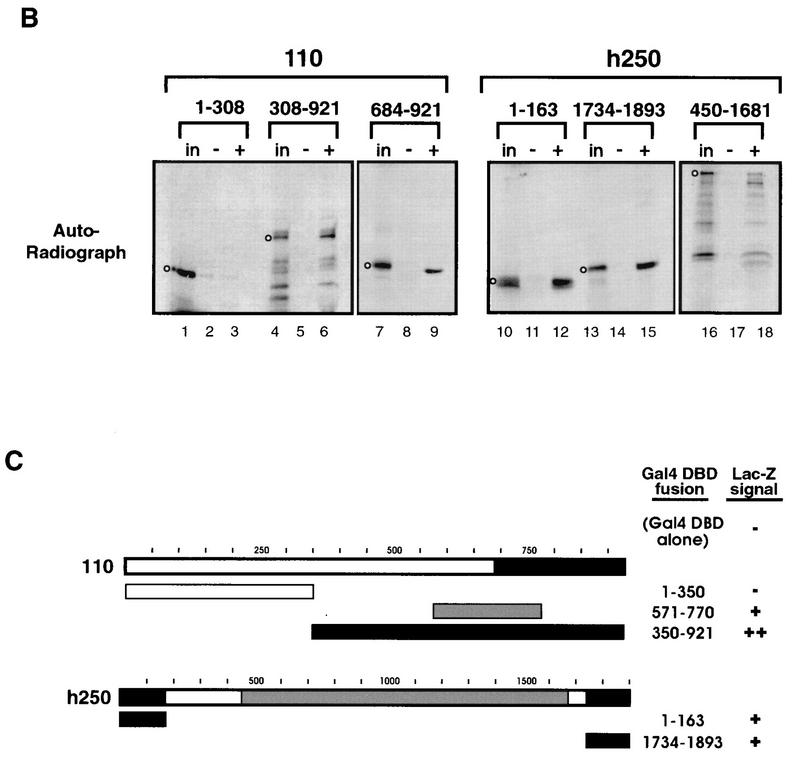
The Drosophila TFIID complex includes TBP and approximately eight major TAFs (37). To determine which of these were bound by Adf-1, each of the eight TAFs and TBP were produced by in vitro transcription and translation in the presence of 35S-labeled methionine. The lysates were then incubated with glutathione beads bound to either GST fused to the carboxy terminus of Adf-1 (C92; amino acids 162 to 253) or to GST alone. The C92 fragment was used since the TFIID-binding data suggested that TAF-binding activity may reside in the carboxy terminus of Adf-1. After extensive washing, bound and input TAFs were visualized by autoradiography. Of the nine proteins, only TAFII110 and TAFII250 bound at significant levels to Adf-1 (Fig. 5A and data not shown). The similar amounts of TAF binding observed with full-length Adf-1 and with proteins having amino-terminal deletions of Adf-1 further confirm that the carboxy terminus of Adf-1 is the major TAF-binding determinant in the protein (Fig. 5A; compare lanes 3 to 5). Smaller fragments of TAFII110 and hTAFII250 were also tested for their ability to bind Adf-1 (Fig. 5B). We found that the carboxy-terminal 240 amino acids of TAFII110 were sufficient for binding Adf-1, while binding with regions at both the amino and carboxy termini of hTAFII250 was seen. The central portion of hTAFII250 also bound Adf-1, though to a lesser extent. This binding also demonstrated that the carboxy-terminal region of Adf-1 is sufficient for TAF binding.
FIG. 5.
Regions of TAFII110 and TAFII250 bind the carboxy terminus of Adf-1. (A) The carboxy terminus of Adf-1 binds TAFII110 and TAFII250. TAFII110 and TAFII250 (ΔN660) were incubated with glutathione beads bound to GST alone (lanes 2 and 8), GST–Adf-1 amino acids 162 to 253 (C92) (lanes 3 and 9), GST–Adf-1 amino acids 92 to 253 (C162) (lane 4), or GST–Adf-1 full length (wt) (lane 5). Bound TAFs were visualized by autoradiography of SDS-PAGE gels after extensive washing. in (lanes 1 and 7), 10% lysate input; −, binding to negative-control beads. The positions of the TAF products are indicated by open circles. (B) One region of TAFII110 and multiple regions of TAFII250 are responsible for Adf-1 binding. Full-length and truncated forms of TAFII110, TAFII250, and hTAFII250 were incubated with glutathione beads bound to GST–Adf-1 C92 or to GST alone and visualized by autoradiography after extensive washing. +, binding to GST–Adf-1 C92; in and − are as defined for panel A. The positions of TAF products are indicated by open circles. (C) Adf-1 binds the same regions of TAFII110 and TAFII250 in vivo as it does in vitro. A C92 fragment of Adf-1 fused to the GAL4 AAD and portions of TAFII110 and TAFII250 fused to the GAL4 DBD or the DBD alone (as indicated) were coexpressed in S. cerevisiae containing a GAL4-responsive LacZ gene. Schematics of TAFII110 and hTAFII250 are shown with shading indicating the relative amounts of binding to Adf-1 in vitro, as shown in panel B. Below these are bars representing the portions of TAFII110 and hTAFII250 fused to the DBD. The amounts of LacZ activity seen are summarized in the “Lac-Z” column; the symbols represent the relative level of LacZ signal observed. (D) Some of the Adf-1 mutants are defective for TAF binding. Purified Adf-1 proteins were incubated with beads bound to hTAFII250 (lanes 4 to 6), TAFII110 (lanes 7 to 9), NTAFII110 (amino acids 75 to 208) (lanes 10 to 12), NTAFII250 (amino acids 1 to 414) (lanes 15 to 18), and GST (lanes 19 to 22). The beads were washed extensively and subjected to SDS-PAGE, followed by Western blotting and probing with anti-Adf-1 antibodies. Lanes 1 to 3 and 11 to 14 show results for 10% input wild-type and mutant Adf-1 proteins. The upper bands seen in some of the lanes represent isomeric forms of Adf-1 (see Materials and Methods), and the lower bands in the N228 samples (lanes 2 and 12) represent degradation products. (E) Schematic of Adf-1, with the region required for binding to TAFII110 and TAFII250 indicated.
To verify that the in vitro binding we observed reflected events that can occur inside of a cell, we also tested the interaction between Adf-1 and TAFII110 and hTAFII250 in S. cerevisiae. Several portions of both TAFs were fused to the GAL4 DBD, and the carboxy terminus of Adf-1 was fused to the GAL4 AAD. These vectors were cotransformed into S. cerevisiae containing GAL4 sites upstream of the gene for β-galactosidase. As summarized in Fig. 5C, the regions of TAFII110 and hTAFII250 that bound Adf-1 in vitro also interacted efficiently in this “in vivo” two-hybrid assay.
Several of the mutations in the carboxy-terminal region of Adf-1 decreased its ability to activate transcription. As was shown in Fig. 4B, some of these carboxy-terminal Adf-1 mutants are also deficient for binding to the holo-TFIID complex. Similar experiments using individual TAFs and other Adf-1 mutants were performed to examine these results in more detail. Paralleling the TFIID-binding results, mutants 5A214 and N228 bound with low efficiency to full-length hTAFII250 and TAFII110 as well as to an amino-terminal fragment of hTAFII250 (amino acids 1 to 414) (Fig. 5D, lanes 5 to 9 and 15 to 17). As expected, none of these proteins bound to GST alone or to GST fused to the amino terminus of TAFII110 (amino acids 75 to 208) (Fig. 5D, lanes 10 to 12 and 19 to 21). In contrast, another carboxy-terminal mutant, 5A245, that is fully active for transactivation retains the ability to bind hTAFII250 efficiently (Fig. 5D, lane 18). Mutants 3A131 and 5A186, bearing alterations in the central portion of Adf-1, also bound TAFs at levels similar to that of the wild-type protein (data not shown). These results provide a good correlation between TAF binding and transcriptional activation by the carboxy-terminal region of Adf-1. However, because activation by Adf-1 requires sequences outside of the carboxy-terminal region, we suspect that binding may be necessary but not sufficient to stimulate transcriptional activation by Adf-1. Instead, our results suggest that sequences outside of the TAF interaction region contribute additional activities essential for transactivation.
The carboxy-terminal TAF-binding domain of Adf-1 bears homologies to ADA2 and Myb.
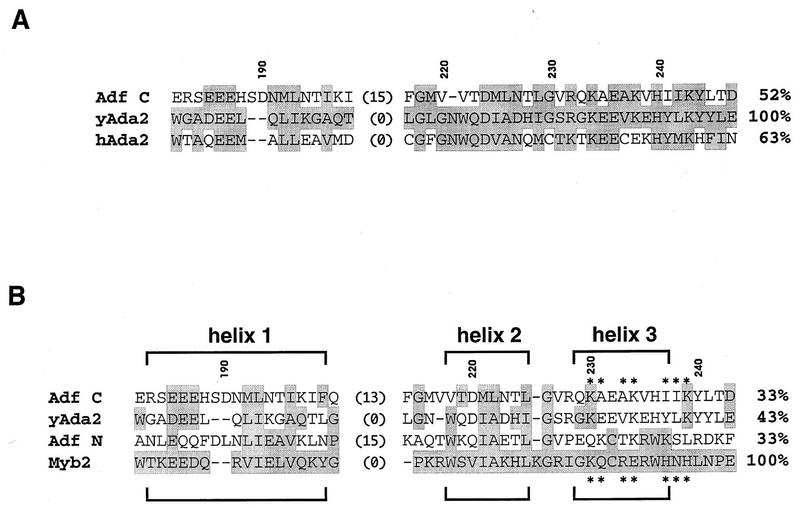
A BLAST search for sequences similar to that of the TAF-binding domain of Adf-1 revealed a sequence similarity with a region of the S. cerevisiae transcriptional adapter protein ADA2 (Fig. 6A). In fact, the number of residues at the carboxy terminus of Adf-1 (AdfC) that are identical or homologous to those of the yeast ADA2 is almost as high as that between the human and yeast ADA2 proteins over this region. The region of highest homology is in the region between amino acids 223 and 247 of Adf-1. The corresponding region of ADA2 has been shown to be essential for its biological function and is required for binding another transcriptional adapter protein, GCN5 (4). Examination of the sequence of Adf-1 by eye also suggested that this same region has homology with the Adf-1 amino-terminal Myb domain (AdfN). Interestingly, it has been reported that the corresponding region of ADA2 also contains a Myb domain homology (2, 5). ADA2 is more similar to Adf-1 than to Myb (52% similarity between Adf-1 and yADA2 versus 43% similarity between mouse Myb repeat 2 and yADA2) (Fig. 6B), suggesting that Adf-1 and ADA2 may contain domains belonging to a novel subfamily of Myb sequences.
FIG. 6.
The carboxy terminus of Adf-1 is homologous to ADA2 and to Myb DBDs. (A) The carboxy terminus of Adf-1 is homologous to a region of the S. cerevisiae and human ADA2 proteins. An alignment between amino acids 182 and 247 of Adf-1 (Adf C), amino acids 65 and 110 of yeast ADA2 (yADA2), and amino acids 75 and 120 of human ADA2 (hADA2) is shown. The position of a 15-residue insertion in Adf-1 is indicated. All residues that are highly similar or identical to those of yADA2 are indicated by shading. The percent similarities between all sequences and that of yADA2 are indicated. Similar amino acids are PG, ST, DE, NQ, RHK, AVLIM, and FTW. (B) The carboxy terminus of Adf-1 may be a Myb-like domain. AdfC and yADA2 are aligned to two DNA-binding Myb domains, the amino terminus of Adf-1 (AdfN; amino acids 6 to 69) and M. musculus Myb repeat 2 (Myb2; amino acids 95 to 141). Amino acid positions in AdfC are indicated, as are the positions of sequence insertions in AdfC and AdfN. Residues that are identical or highly similar to those of Myb2 are indicated as in panel A. Percent similarities between all sequences and that of Myb2 are indicated. The positions of helices 1, 2, and 3 in Myb2 are indicated by brackets. Seven residues whose side chains project from the exposed face of mMyb helix 3 are indicated by asterisks.
Unlike the AdfC and ADA2 sequences, previously characterized Myb domains have only been shown to bind DNA. The high-resolution structures of several Myb domains have been solved (26, 28), revealing that they are variants of the helix-turn-helix motif. An alignment of AdfC to yADA2 and two DNA-binding Myb domains, AdfN and the second Myb repeat of the mouse Myb protein, is shown in Fig. 6B. The level of similarity between AdfC and Myb is similar to that between Myb and the two other sequences which are already recognized as being Myb homologs, AdfN and yADA2 (Fig. 6B) (2, 5, 13, 24). Interestingly, the region of greatest similarity between AdfC and yADA2 is also the region of least similarity between those two proteins and Myb and AdfN. This region corresponds to Myb helix 3, the DNA-recognition helix of DNA-binding Myb domains. The mouse Myb structure reveals that seven residues (Fig. 6B) in helix 3, including those that specifically recognize DNA (26–28), project out from the protein. While all seven of these residues are hydrophilic in the mouse Myb sequence and six are hydrophilic in the Adf-1 DBD, only three are hydrophilic in the Adf-1 carboxy-terminal region. Thus, this region of Adf-1, which may correspond to the DNA-recognition surface of typical Myb domains, may be much more hydrophobic than that of any of the DBDs. Fewer hydrophobic substitutions are seen in this region with ADA2, but there are several large hydrophobic residues in both ADA2 and AdfC immediately carboxy terminal to the helix 3 region that may further contribute to a potential helical hydrophobic surface. This structural arrangement of the Adf-1 carboxy-terminal Myb-like domain is consistent with a role for this domain in protein-protein rather than protein-DNA interactions.
DISCUSSION
Our understanding of the mechanisms of transcriptional activation and the structure of activation domains remains rudimentary. Most characterized enhancer-binding proteins have revealed modular activation domains that can function when fused to heterologous DBDs. Many activators contain multiple activation regions, but studying these functional domains in isolation as chimeric fusions fails to provide us with information about how they may interact with each other and with their natural DBDs to influence activation. Additionally, structural studies of these activation domains have thus far been difficult and not highly informative. Here we report a detailed analysis of the Drosophila activator Adf-1. We have probed the activities of sequences throughout the protein through alanine-scanning and deletion mutagenesis. These small modifications of Adf-1 have allowed us to study the functional domains of Adf-1 in their native context, revealing an unusual activation region and a novel dual use of the Myb motif.
A distinct DBD is located in the amino-terminal 100 amino acids of Adf-1. This domain contains homologies to the Myb DNA-binding motif and Drosophila protein Stonewall (8). Adf-1 dimerizes in solution through a domain found near the carboxy terminus. Although dimerization increases the affinity of Adf-1 for DNA, its activity is not essential for binding, as mutants unable to dimerize retain sequence-specific DNA-binding activity. Since most Myb proteins contain two or three tandem repeats of the Myb motif, it is possible that the dimerization by Adf-1 compensates for the presence of only one DNA-binding Myb motif in the protein. Although monomeric forms of Adf-1 can still bind DNA, they do so with a lower affinity. In addition to being required for dimerization, the carboxy terminus of Adf-1 possesses TAF-binding activity. We show here that TFIID cannot be replaced by TBP for Adf-1-directed transcriptional activation, consistent with the notion that TAF binding is likely to be functionally important for the activity of Adf-1. All mutations which disrupt Adf-1 dimerization also disrupt TAF binding, while mutants of Adf-1 that are competent for dimerization are also competent for TAF binding. While these data suggest that dimerization may be required for TAF binding, our observation that a carboxy-terminal fragment of Adf-1 containing the dimerization domain is sufficient for TAF binding indicates that dimerization and TAF binding activities are likely due to closely adjacent or overlapping sequences within Adf-1.
Mutations in both regions of Adf-1, DNA-binding and dimerization/TAF-binding regions, affect transcriptional activation. Surprisingly, several other mutations that do not interfere with these activities also disrupt activation by Adf-1. For example, a deletion of the first five residues of the protein (ΔN5) severely impairs activation by Adf-1. Since DNA binding is an obvious requirement for activation and since the fusions of fragments of Adf-1 with heterologous DBDs which have been tested (data not shown) have proven to be inactive, we cannot conclusively say whether the Adf-1 DBD has activities directly required for activation. The transcriptional phenotype of the ΔN5 protein and the lack of activity of heterologous fusions, however, suggest that a direct role of the Adf-1 DBD in activation is likely. It is not uncommon for amino and carboxy termini to be near each other in the three-dimensional structures of proteins. One possibility is that there are important interactions between the amino terminus of Adf-1 containing the DBD and the carboxy-terminal domain where TAF-binding activity resides. Although these interactions are not required for TAF binding in solution, they may help to position the TAF-binding domain with respect to the rest of the protein and to the DNA such that these TAF interactions are competent for the stimulation of an active transcription complex. Alternatively, it is possible that the pathway of activation directed by Adf-1 requires at least one step in addition to TAF binding and that these other regions of Adf-1 are required for such a step.
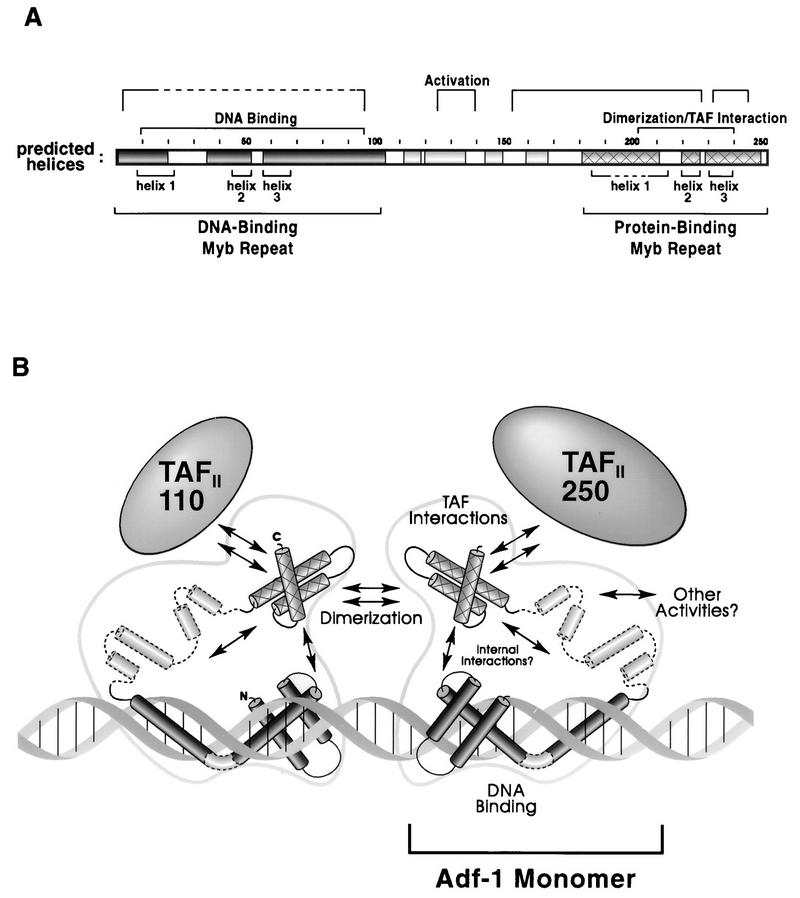
The functional mapping of Adf-1 is summarized in Fig. 7A. Indicated within the protein are regions predicted to be α-helical by the nearest-neighbor secondary structure prediction algorithm (32). Additionally, the positions of the three Myb domain-like helices predicted by the alignments of the amino and carboxy termini of Adf-1 to Myb domains are indicated. Circular dichroism spectroscopy of purified Adf-1 indicates a very high α-helical content (data not shown), supporting these sequence-based predictions. The helical predictions based on the primary structure match closely those based on the Myb homologies with one major exception. The predicted recognition helix of the DBD, helix 3, is nearly 50 amino acids long and terminates near the border of the DBD as defined by our mutagenesis. This helix is 40 amino acids longer than the corresponding helices seen in the NMR structures of Myb (26, 28). The close match with the mapped border of the DBD and the amphiphilic nature of this entire region (data not shown) support the model of a greatly extended DNA recognition helix. A linear extension of the recognition helix would, by necessity, project out of the major groove and lose contact with DNA. If contact between such an extended recognition helix and DNA is maintained, dramatic perturbations in the helix or in the DNA must exist. Alternatively, the extended recognition helix may be playing an indirect role in DNA binding.
FIG. 7.
A synthesis of structural and functional information about Adf-1. (A) Secondary structure prediction of Adf-1 correlates with mutagenesis data and protein homologies. α-Helices predicted by the nearest-neighbor secondary-structure prediction algorithm are indicated by shading. The positions of helices 1, 2, and 3 predicted by an analysis of the two Myb domain homologies (Fig. 6B) are indicated below the protein. Regions required for DNA binding, activation, TAF binding, and dimerization are indicated above the protein. (B) A model for the structure of Adf-1. The data summarized in panel A are interpreted as a possible three-dimensional structure showing the activities of Adf-1. A dimer of two Adf-1 molecules is depicted bound to DNA. A monomer of Adf-1 contains a DNA-binding helix-turn-helix domain at its amino terminus. The recognition helix that is expected to lie in the major groove of the DNA is shown as an extended helix. The relationship between this helix and DNA is unknown, and the helical kink depicted is arbitrary. The positions and orientations on DNA of the two monomers with respect to each other are also arbitrary. The activities and structures in the central portion of Adf-1 are not well understood, although the region is predicted to contain several α-helices. The carboxy terminus of Adf-1 also contains a Myb-like helix-turn-helix domain; this domain is able to dimerize as well as bind TAFII110 and TAFII250. Potential interactions are indicated by arrows.
The agreement of the nearest-neighbor analysis results and the positions of the Myb homology-predicted α-helices at the carboxy terminus of Adf-1 support the model that a second Myb-like domain exists in the protein. Unlike the case for previously characterized Myb domains, there is no evidence for direct interactions between this carboxy-terminal domain and DNA. Rather, this region contains dimerization and TAF-binding activities. The known structures of Myb domains reveal two important protein surfaces, one consisting of helices 1 and 2 and the other containing helix 3, the recognition helix. Mutations that disrupt any of these helices or the linker sequence between them, are likely to have effects on the entire structure. Thus, although both dimerization and TAF-binding activities map to the same general region, it is quite possible that these two activities reside on opposing surfaces of a Myb-like structure. This region in Adf-1 is also similar to that of the transcriptional adapter protein ADA2. The greatest similarity between Adf-1 and ADA2 is in the regions corresponding to the Myb DNA recognition helix, which, for these two domains, contains several large hydrophobic residues. This is consistent with a role for this domain in protein-protein rather than protein-DNA interactions. A deletion in ADA2 which removes the sequence corresponding to Myb helices 2 and 3 eliminates binding to another adapter protein, GCN5 (4). Thus, there is a strong likelihood that both Adf-1 and ADA2 contain a novel variant of the Myb domain adapted for protein-protein interactions rather than protein-DNA recognition.
A model of Adf-1 based on the data we have presented is shown in Fig. 7B. Some of the features, such as the kink in the extended recognition helix and the orientation of the DBDs on DNA, are, by necessity, arbitrary. While the existence of two Myb domains in a protein is not unprecedented, the very different roles they play were unanticipated. The likely use of a Myb domain in protein-protein interactions by both Adf-1 and ADA2 suggests that a family of such proteins may exist. Another unexpected feature of Adf-1 was the nonmodularity of its transcriptional activation region. Adf-1 may be much more structurally integrated than previously characterized transcription factors. Our observation that TAF binding, while required, is not sufficient for activation suggests that Adf-1 may direct multiple steps in the pathway of activation. The typical cut-and-splice characterization of transcriptional activators is likely to obscure such multiple contributions by a protein. In the case of Adf-1, we have a good model for elucidating a more complete pathway by which an individual activator functions in the process of transcriptional activation.
ACKNOWLEDGMENTS
We thank the members of the Tjian lab, past and present, for their support of this research. We are especially grateful to P. Beaurang, S. Hansen, T. O’Brien, F. Sauer, K. Yokomori, S. Ryu, D. Azivonis, J. Zwicker, and M. Holmes for donations of critical DNA and protein reagents. We appreciate C. Day and T. Alber for their efforts with Adf-1. We additionally thank M. Rabenstein, C. Smith, L. Gandhi, L. de la Fuente, and S. Treisenberg for critical comments on the manuscript.
G.C. was supported by a Howard Hughes Predoctoral Fellowship. This work was supported in part by grant no. CA25417 from the National Institutes of Health to R.T.
REFERENCES
- 1.Attardi L D, Tjian R. Drosophila tissue-specific transcription factor NTF-1 contains a novel isoleucine-rich activation motif. Genes Dev. 1993;7:1341–1353. doi: 10.1101/gad.7.7b.1341. [DOI] [PubMed] [Google Scholar]
- 2.Berger S L, Pina B, Silverman N, Marcus G A, Agapite J, Regier J L, Triezenberg S J, Guarente L. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell. 1992;70:251–265. doi: 10.1016/0092-8674(92)90100-q. [DOI] [PubMed] [Google Scholar]
- 3.Biggin M D, Tjian R. A purified Drosophila homeodomain protein represses transcription in vitro. Cell. 1989;58:433–440. doi: 10.1016/0092-8674(89)90424-8. [DOI] [PubMed] [Google Scholar]
- 4.Candau R, Berger S L. Structural and functional analysis of yeast putative adaptors. Evidence for an adaptor complex in vivo. J Biol Chem. 1996;271:5237–5245. doi: 10.1074/jbc.271.9.5237. [DOI] [PubMed] [Google Scholar]
- 5.Candau R, Moore P A, Wang L, Barlev N, Ying C Y, Rosen C A, Berger S L. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caron C, Mengus G, Dubrowskaya V, Roisin A, Davidson I, Jalinot P. Human TAF(II)28 interacts with the human T cell leukemia virus type I Tax transactivator and promotes its transcriptional activity. Proc Natl Acad Sci USA. 1997;94:3662–3667. doi: 10.1073/pnas.94.8.3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen J L, Attardi L D, Verrijzer C P, Yokomori K, Tjian R. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell. 1994;79:93–105. doi: 10.1016/0092-8674(94)90403-0. [DOI] [PubMed] [Google Scholar]
- 8.Clark K A, McKearin D M. The Drosophila stonewall gene encodes a putative transcription factor essential for germ cell development. Development. 1996;122:937–950. doi: 10.1242/dev.122.3.937. [DOI] [PubMed] [Google Scholar]
- 9.Courey A J, Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988;55:887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- 10.Dezazzo, J. Personal communication.
- 11.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 12.Dynlacht B D, Hoey T, Tjian R. Isolation of coactivators associated with the TATA-binding protein that mediate transcriptional activation. Cell. 1991;66:563–576. doi: 10.1016/0092-8674(81)90019-2. [DOI] [PubMed] [Google Scholar]
- 13.England B P, Admon A, Tjian R. Cloning of Drosophila transcription factor Adf-1 reveals homology to Myb oncoproteins. Proc Natl Acad Sci USA. 1992;89:683–687. doi: 10.1073/pnas.89.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.England B P, Heberlein U, Tjian R. Purified Drosophila transcription factor, Adh distal factor-1 (Adf-1), binds to sites in several Drosophila promoters and activates transcription. J Biol Chem. 1990;265:5086–5094. [PubMed] [Google Scholar]
- 15.Frankel A D, Kim P S. Modular structure of transcription factors: implications for gene regulation. Cell. 1991;65:717–719. doi: 10.1016/0092-8674(91)90378-c. [DOI] [PubMed] [Google Scholar]
- 16.Goodrich J A, Cutler G, Tjian R. Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell. 1996;84:825–830. doi: 10.1016/s0092-8674(00)81061-2. [DOI] [PubMed] [Google Scholar]
- 17.Hansen S K, Tjian R. TAFs and TFIIA mediate differential utilization of the tandem Adh promoters. Cell. 1995;82:565–575. doi: 10.1016/0092-8674(95)90029-2. [DOI] [PubMed] [Google Scholar]
- 18.Heberlein U, England B, Tjian R. Characterization of Drosophila transcription factors that activate the tandem promoters of the alcohol dehydrogenase gene. Cell. 1985;41:965–977. doi: 10.1016/s0092-8674(85)80077-5. [DOI] [PubMed] [Google Scholar]
- 19.Hoey T, Weinzierl R O, Gill G, Chen J L, Dynlacht B D, Tjian R. Molecular cloning and functional analysis of Drosophila TAF110 reveal properties expected of coactivators. Cell. 1993;72:247–260. doi: 10.1016/0092-8674(93)90664-c. [DOI] [PubMed] [Google Scholar]
- 20.Hope I A, Mahadevan S, Struhl K. Structural and functional characterization of the short acidic transcriptional activation region of yeast GCN4 protein. Nature. 1988;333:635–640. doi: 10.1038/333635a0. [DOI] [PubMed] [Google Scholar]
- 21.Jantzen H M, Chow A M, King D S, Tjian R. Multiple domains of the RNA polymerase I activator hUBF interact with the TATA-binding protein complex hSL1 to mediate transcription. Genes Dev. 1992;6:1950–1963. doi: 10.1101/gad.6.10.1950. [DOI] [PubMed] [Google Scholar]
- 22.Johnson P F, Sterneck E, Williams S C. Activation domains of transcriptional regulatory proteins. J Nutr Biochem. 1993;4:386–398. [Google Scholar]
- 23.Lillie J W, Green M R. Transcription activation by the adenovirus E1a protein. Nature. 1989;338:39–44. doi: 10.1038/338039a0. [DOI] [PubMed] [Google Scholar]
- 24.Lipsick J S. One billion years of Myb. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- 25.Ma J, Ptashne M. A new class of yeast transcriptional activators. Cell. 1987;51:113–119. doi: 10.1016/0092-8674(87)90015-8. [DOI] [PubMed] [Google Scholar]
- 26.Morikawa S, Ogata K, Sekikawa A, Sarai A, Ishii S, Nishimura Y, Nakamura H. Determination of the NMR solution structure of a specific DNA complex of the Myb DNA-binding domain. J Biomol NMR. 1995;6:294–305. doi: 10.1007/BF00197810. [DOI] [PubMed] [Google Scholar]
- 27.Ogata K, Morikawa S, Nakamura H, Hojo H, Yoshimura S, Zhang R, Aimoto S, Ametani Y, Hirata Z, Sarai A, et al. Comparison of the free and DNA-complexed forms of the DNA-binding domain from c-Myb. Nat Struct Biol. 1995;2:309–320. doi: 10.1038/nsb0495-309. [DOI] [PubMed] [Google Scholar]
- 28.Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- 29.Pabo C O, Sauer R T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 30.Pascal E, Tjian R. Different activation domains of Sp1 govern formation of multimers and mediate transcriptional synergism. Genes Dev. 1991;5:1646–1656. doi: 10.1101/gad.5.9.1646. [DOI] [PubMed] [Google Scholar]
- 31.Petty K J, Krimkevich Y I, Thomas D. A TATA binding protein-associated factor functions as a coactivator for thyroid hormone receptors. Mol Endocrinol. 1996;10:1632–1645. doi: 10.1210/mend.10.12.8961272. [DOI] [PubMed] [Google Scholar]
- 32.Salamov A A, Solovyev V V. Prediction of protein secondary structure by combining nearest-neighbor algorithms and multiple sequence alignments. J Mol Biol. 1995;247:11–15. doi: 10.1006/jmbi.1994.0116. [DOI] [PubMed] [Google Scholar]
- 33.Sauer F, Wassarman D A, Rubin G M, Tjian R. TAF(II)s mediate activation of transcription in the Drosophila embryo. Cell. 1996;87:1271–1284. doi: 10.1016/s0092-8674(00)81822-x. [DOI] [PubMed] [Google Scholar]
- 34.Thut C J, Chen J L, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- 35.Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 36.Triezenberg S J. Structure and function of transcriptional activation domains. Curr Opin Genet Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 37.Verrijzer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- 38.Wampler S L, Tyree C M, Kadonaga J T. Fractionation of the general RNA polymerase II transcription factors from Drosophila embryos. J Biol Chem. 1990;265:21223–21231. [PubMed] [Google Scholar]