Abstract
The b mating-type locus of the fungal plant pathogen Ustilago maydis encodes two multiallelic gene products, bE and bW, that control the formation and maintenance of the infectious cell type. Dimerization via the N-terminal regions of bE and bW proteins encoded by alleles of different specificities establishes a homeodomain-containing transcription factor. The bE and bW products encoded by alleles of like specificities fail to dimerize. We constructed sets of chimeric alleles for the bE1 and bE2 genes and for the bW1 and bW2 genes to identify sequences that control specificity. The mating behavior of strains carrying chimeric alleles identified three classes of specificity: b2 (class I), specificity different from either parental type (class II), and b1 (class III). Crosses between strains carrying bE and bW chimeric alleles identified two short blocks of amino acids that influence specificity and that are located in the N-terminal variable regions of the b proteins. Comparisons of pairs of chimeric alleles encoding polypeptides differing in specificity and differing at single amino acid positions identified 16 codon positions that influence the interaction between bE and bW. Fifteen of these positions lie within the blocks of amino acids identified by crosses between the strains carrying chimeric alleles. Overall, this work provides insight into the organization of the regions that control recognition.
Recognition mediated by protein-protein interactions plays a fundamental role in many biological processes. Well-characterized examples include antibody-antigen interactions (8, 9, 23), ligand-receptor binding (22, 35), and the establishment and maintenance of tissue integrity by cadherins (19). The proteins involved in sexual reproduction and incompatibility in fungi provide relatively simple examples of determinants of self versus nonself recognition. In this paper, we describe a molecular genetic approach to identify the determinants of recognition for the proteins encoded by the b mating-type locus of the fungal corn pathogen Ustilago maydis.
U. maydis is commonly found in nature as black diploid teliospores on infected corn plants (6). The teliospores germinate, and meiosis occurs to produce haploid, yeast-like progeny. Nonself recognition between compatible haploid mating partners is a prerequisite to the establishment of an infectious, dikaryotic cell type, and the genes at the a and b mating-type loci are considered pathogenicity factors (reviewed in references 2 and 18). The a locus, with alternate specificities a1 and a2, encodes pheromones and pheromone receptors and controls recognition of mating partners at the level of cell fusion (3, 11, 31). The b locus controls the formation and maintenance of the infectious cell type after cell fusion has occurred. If the cells participating in mating have different specificities (nonself) at the b locus, a vigorous, straight dikaryotic filament is formed and this cell type will be infectious. In contrast, mating partners that carry b sequences of like specificities (self) do not form an infectious dikaryon. Interestingly, the b locus is believed to have at least 25 different naturally occurring specificities (24, 29), and all of the nonself combinations of alleles are able to promote pathogenicity and sexual development.
The b locus of U. maydis was initially cloned by transformation of a library of DNA from a strain with b1 specificity into a diploid strain with b2 specificity and by subsequent screening of transformants for filamentous growth (16). The molecular characterization of the b locus revealed the presence of two divergently transcribed genes called bE (encoding a polypeptide of 473 amino acids) and bW (encoding a polypeptide of 644 amino acids) (12, 17, 28). These genes exist in an allelic series such that each of the 25 specificities at the b locus is determined by the specific bE and bW alleles present in a haploid strain. The bE and bW gene products do not show sequence similarity to each other except that each contains a homeodomain-like region of approximately 60 amino acids that lies between a variable amino-terminal region (N-terminal region; 100 to 150 amino acids) and a conserved carboxy-terminal region (C-terminal region) (12, 28). Gene disruption experiments revealed that the b gene products are necessary to establish the filamentous dikaryon. That is, strains compatible at the a locus but carrying null mutations in both bE and bW are defective in mating (12, 17). Furthermore, deletion of the b genes revealed that the presence of one bE and one bW from each mating partner (e.g., bE1 plus bW2 or bE2 plus bW1) is sufficient to allow mating and pathogenic development in the plant (12).
It is believed that any combination of bE and bW gene products encoded by different alleles is capable of triggering dikaryon formation. In contrast, the bE and bW products from the same strain fail to initiate pathogenic development. Experiments using the two-hybrid system with Saccharomyces cerevisiae and an in vitro protein binding assay indicate that the N-terminal regions of bE and bW promote dimerization between gene products from alleles of different specificities (15). The bE and bW products from genes of the same strain (e.g., bE2 and bW2) fail to dimerize. Thus, it appears that the variable regions of bE and bW are dimerization domains and that the formation of active heterodimers requires b polypeptides from genes with different specificities. In this context, the critical question is the following: what mechanism prevents the dimerization of bE and bW gene products from the same locus?
In previous work, we defined a specificity region for the bE1 and bE2 genes by the construction and analysis of chimeric alleles (37). This work identified a 40-amino-acid sequence within the N-terminal variable region that was thought to contain the residues that control specificity. Surprisingly, chimeric alleles between bE1 and bE2 that contained recombination sites within the sequence encoding the 40-amino-acid region displayed specificities different from that of either parental bE allele. These alleles were designated class II to distinguish them from alleles that had not changed specificity (class I) or that had switched specificity from one parental type to the other (class III). Kämper et al. have shown that single-amino-acid changes within a similar portion of the variable region of bE2 allow dimerization with bW2 (15). These observations prompted the proposal that a dimerization interface mediates the attraction between bE and bW proteins from different alleles. In this context, bE and bW proteins that fail to dimerize must fail to do so because of key interfering residues that block association by preventing productive interactions or by establishing disruptive interactions, e.g., by polar or hydrophobic effects or by steric hindrance (14).
In this paper, we report the construction of additional chimeric alleles for the bE1 and bE2 genes and the construction of a large set of chimeric alleles for the bW1 and bW2 genes. Overall, these sets of alleles provided a refined view of the 40-amino-acid specificity region for bE1 and bE2 and identified an analogous 70-amino-acid region for the bW1 and bW2 alleles. In addition, crosses between all combinations of bE and bW chimeric alleles revealed that the borders of the regions defined by class II alleles contain the important determinants for recognition. For bE1 and bE2 alleles, the specificity borders lie between codons 31 and 39 and between codons 79 and 92; for the bW1 and bW2 alleles, these borders are found between codons 2 and 9 and 74 and 83. Our data suggest that the 40-amino-acid region for bE and the 70-amino-acid region for bW represent the intervals between the specificity determinants in the border regions. Key amino acid positions within the borders were identified by comparisons of chimeric alleles that differed at a single codon and had different specificities when tested against strains carrying either wild-type or chimeric alleles. Additional chimeric alleles, constructed between bW1 and bW3, indicated that a single border region can be sufficient to control the interaction for certain allele pairs.
(This work fulfills part of the requirements for a Ph.D. in plant science from the University of British Columbia for A. R. Yee.)
MATERIALS AND METHODS
Strains and growth conditions.
Escherichia coli DH5α [F− endA1 hsdR17 (rK− mK+) supE44 Thi−1 recA1 φ80dlac ZM15] (Bethesda Research Laboratories) was used for DNA manipulations and was grown in Luria-Bertani medium (26). U. maydis wild-type prototrophic strains were 518 (a2 b2), 521 (a1 b1), 031 (a1 b2), and 032 (a2 b1) (16). The genotypes of strains carrying chimeric b alleles (for bE1 and bE2 or bW1 and bW2) were designated bEx and bWx where x is followed by a number indicating the amino acid position corresponding to the codon at which the sequence changes from E1 to E2 or W1 to W2. The genotypes for chimeric b alleles for bW1 and bW3 were distinguished from those for the bW1 and bW2 alleles by the designation bW1/3x followed by the codon number at which the sequence changes from bW1 to bW3. It should be noted that the procedure for constructing chimeric alleles disrupts the wild-type allele of bE or bW in each strain due to the insertion of a hygromycin resistance cassette and leaves only one functional b gene (the chimeric gene) (37). U. maydis cultures were grown in potato dextrose medium (Difco Laboratories) or complete medium (13), and mating reactions were carried out on solid double-complete medium containing 1% activated charcoal (13).
Construction of chimeric alleles.
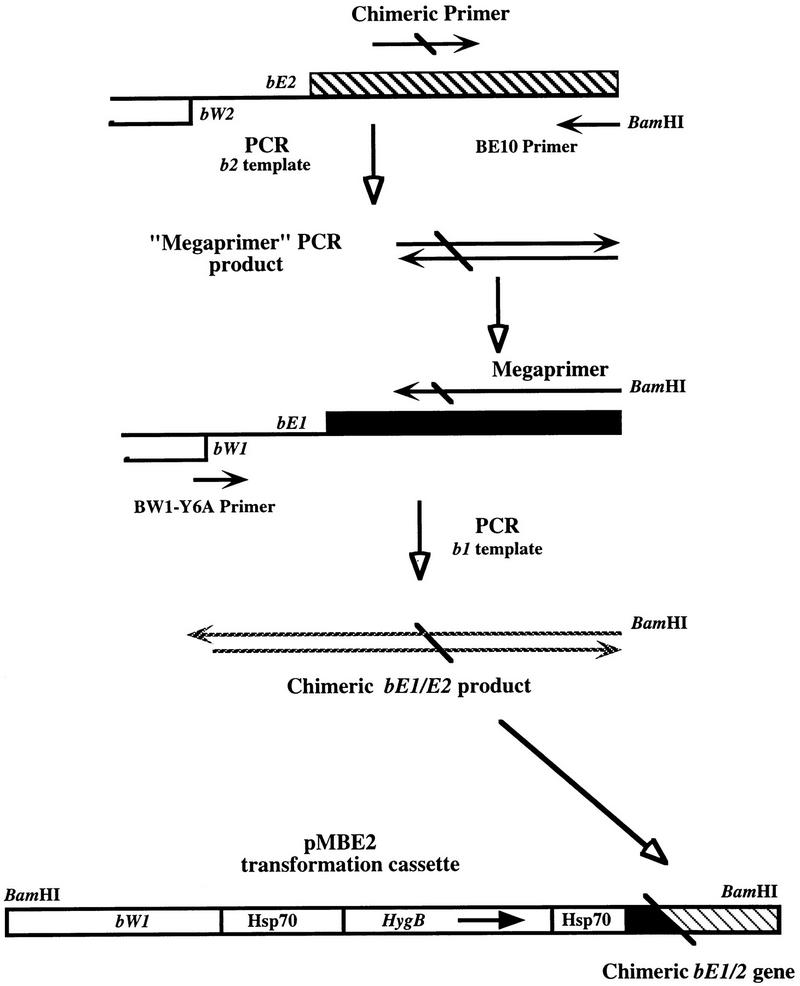
The strategy for the in vivo construction of chimeric alleles by transformation and homologous integration was described previously (37). This approach involved the transformation of deletion derivatives of bE and bW genes into strains of opposite b specificities and the generation of chimeric alleles by homologous integration at b. Additional chimeric alleles were constructed in vitro by a PCR approach and subsequently used to replace wild-type alleles by transformation. The PCR approach was based on the Megaprimer PCR procedure (27) and is diagrammed in Fig. 1. This technique requires two rounds of PCR amplification and a “chimeric” primer designed to overlap the junction between the bE1 and bE2 or bW1 and bW2 sequences. The first round involves PCR with the chimeric primer and a second primer to produce a PCR product called the “megaprimer.” This round of PCR employed bE2 or bW2 sequences as templates. The megaprimer was then used in a second round of PCR with a third primer and bE1 or bW1 template DNA to produce the final chimeric product. The primers employed for constructing chimeric alleles are shown in Table 1, and the chimeric alleles are listed in Table 2.
FIG. 1.
In vitro construction of chimeric alleles. The megaprimer PCR method (27) was employed to construct chimeric alleles of bE1 and bE2 and bW1 and bW2. The procedure is diagrammed for the construction of bE1 and bE2 chimeric alleles; the same approach was used for bW1 and bW2 chimeric alleles except that the final chimeric amplification product was cloned into plasmid pAR69 (see Materials and Methods). The chimeric primer is designed to overlap the anticipated junction point of the chimeric allele. The megaprimer approach makes use of two additional, flanking primers. For bE chimeric alleles, primers BE10 and bW1-Y6A were used to produce the megaprimer and to isolate a fragment containing 5′ promoter sequences, respectively. For bW chimeric alleles, the equivalent primers were BW6 and BE1-Y31A (Materials and Methods). Note that the final transformation cassette disrupts bE or bW upon homologous integration and leaves the chimeric allele as the only functional b gene in the transformant.
TABLE 1.
Oligonucleotide primers used for the construction of chimeric alleles
| Primer name | Sequencea |
|---|---|
| bEx31 | ACGGGGTAATTTTCCCCTTTATCTCGC |
| bEx42 | GTCTTTCGTCGCAGCTCTCGGAG |
| bEx45 | AACGTTGTTGGGTGTCTTTTGTTGCAGCTCTCG |
| bWx4 | AAACATTCAAGATCTTTCATGTTGGG |
| bWx6 | ATCTCGGAGAAACATTCAAAATCTTTCATGTTG |
| bWx68 | GTCTGTCGAGATACACTCATCAAGCTT |
| bWx72 | CTCGAGGAGATCTTCTTGGGACATTTG |
| bWx73 | CTCGAGGAGATCTTCATAGGACATTTGAACGAA |
| bWx74 | GAGGAGATCTTCATAGAGCATTTGAACGAACTA |
| bWx76 | ATCTTCATAGAGTATCTGAACGAACTACACATA |
| bWx77 | TTCATAGAGTATCTGAGGGAACTACACATAGGG |
| bWx79 | GAGTATCTGAGGAAGCTACACATAGGG |
| bWx80 | TATCTGAGGAAGCTACGAATAGGGTGCCAAGCT |
| bWx81 | CTGAGGAAGCTACGACGAGGGTGCCAAGCTCAG |
| bWx82 | AGGAAGCTACGACGAGTGTGCCAAGCTCAGTAC |
| bWx83 | AAGCTACGACGAGTGTATCAAGCTCAGTACGAG |
| bWx88 | CGACGAGTGTATGAGGCTCAGTACGAG |
| bWx91 | CAATACGAAAATGCGTTCGCGATATGG |
| bWx140 | CAGGGGGAGAAGAATCAGTTCG |
| BE10 | AAGGATCCATAGCGTGAGCTGATGA |
| BW1-Y6A | CAAAATCTTGGAGAAAGCTTCAAAATCTTTCAT |
| BW6 | TTGGATCCAGTGACCTCTGAAAG |
| BE1-Y31A | ACGCACGAGAACGGGGGCATTCTCCCC |
The primers are shown in the 5′-to-3′ orientation.
TABLE 2.
Results of mating tests between strains carrying wild-type and chimeric alleles
| Allele | Mating test resulta for allele:
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| bE2 | bEx28 | bEx31 | bEx39 | bEx45 | bEx48 | bEx49 | bEx51 | bEx57 | bEx60 | bEx70 | bEx79 | bEx82 | bEx87 | bEx89 | bEx90 | bEx92 | bEx107 | bEx128 | bEx156 | bE1 | |
| bW2 | − | − | − | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| bWx4 | − | − | − | − | − | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ | +++ | ++ | ++ | +++ | +++ | +++ | +++ |
| bWx6 | − | − | − | − | − | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ |
| bWx9 | +++ | +++ | +++ | − | − | +++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | ++ | +++ | ++ | ++ | +++ | +++ | +++ | +++ |
| bWx12 | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − | − | +++ | ++ | ++ | +++ | +++ | +++ | +++ |
| bWx19 | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − | − | +++ | ++ | ++ | +++ | +++ | +++ | +++ |
| bWx36 | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − | − | +++ | ++ | ++ | +++ | +++ | +++ | +++ |
| bWx38 | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − | − | ++ | ++ | +++ | +++ | +++ | +++ | +++ |
| bWx48 | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − | − | ++ | ++ | ++ | +++ | +++ | +++ | +++ |
| bWx49 | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − | − | ++ | ++ | ++ | +++ | +++ | +++ | +++ |
| bWx52 | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − | − | + | ++ | +++ | +++ | +++ | +++ | +++ |
| bWx68 | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − | − | − | − | +++ | +++ | +++ | +++ | +++ |
| bWx72 | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − | − | − | − | +++ | +++ | +++ | +++ | +++ |
| bWx73 | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − | − | − | − | +++ | +++ | +++ | +++ | +++ |
| bWx74 | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − | − | − | − | ++ | +++ | +++ | +++ | +++ |
| bWx76 | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − | − | + | ++ | − | ++ | ++ | ++ | − |
| bWx77 | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ++ | ++ | ++ | − |
| bWx79 | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − | − | +++ | +++ | − | +++ | +++ | +++ | ++ |
| bWx80 | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − | − | +++ | ++ | − | +++ | +++ | +++ | − |
| bWx81 | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − | − | − | ++ | − | − | − | − | − |
| bWx82 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | − | − | +++ | ++ | ++ | +++ | +++ | +++ | ++ |
| bWx83 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − |
| bWx88 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − |
| bWx91 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − | − |
| bWx109 | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ | +++ | +++ | ++ | − | − | − | − | − | − | − | − | − | − |
| bWx140 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − |
| bWx171 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − |
| bW1 | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | +++ | − | − | − | − | − | − | − | − | − |
A minus sign indicates that the mating reaction between b gene products was incompatible and did not result in the formation of white aerial hyphae. The strength of a compatible interaction was judged visually and assigned one to three plus signs to indicate the density of aerial hyphae present on the colonies. Representative positive mating reactions are shown in Fig. 3 and 5.
The protocol of Sarkar and Sommer (27) was employed for PCR except that Vent polymerase (New England BioLabs, Beverly, Mass.) was used instead of Taq polymerase. The reaction mixture (100 μl) for PCR with Vent polymerase, as recommended by Cease et al. (5), contained 10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris-HCl (pH 8.8 at 25°C), 3 mM MgSO4, a 500 μM concentration of each dNTP, 0.1% Triton X-100, 1 μM concentrations of primers, 1 fmol of DNA template, and 1 U of Vent polymerase. Thermal cycling was done with a Perkin-Elmer 480 with an initial 3-min time delay at 94°C and a step cycle of 1 min at 94°C, 1 min at 52°C, and 1 min at 72°C; the samples were held for 10 min at 72°C at the end of the cycles.
The chimeric PCR products were first cloned as blunt-end fragments into pBluescript KS+ (Stratagene) which had been linearized with EcoRV. The cloned chimeric or mutant product was then subcloned from pBluescript KS+ into a Ustilago transformation construct. For bE, this transformation construct was pMBE2 (Fig. 1) which had been linearized with SacI, made blunt with T4 polymerase, and dephosphorylated. Plasmid pMBE2 contains a 1.8-kb BglII-SalI fragment carrying the hygromycin resistance cassette in pUC9 (34). The plasmid also contains a 1.4-kb fragment encoding the N-terminal portion of the bW1 polypeptide. For bW chimeras, the transformation construct was pAR69 which had been linearized with HindIII, made blunt with T4 polymerase, and dephosphorylated. Plasmid pAR69 is based on pBluescript KS+ and contains a 1.9-kb BglII-XbaI fragment encoding the hygromycin resistance cassette (34) and a 5.8-kb XbaI-BamHI fragment encoding the bE1 gene and 3′ flanking sequences. Subclones containing the correct orientation of insert were chosen for transformation into U. maydis. The bE and bW chimeric allele constructs were linearized with BamHI, extracted once with phenol-chloroform-isoamyl alcohol (24:24:1) and once with chloroform-isoamyl alcohol (24:1), ethanol precipitated, and dissolved in Tris-EDTA. This DNA was then used to transform U. maydis protoplasts. Transformation of U. maydis was accomplished by a protoplast-polyethylene glycol-CaCl2 procedure modified from Wang et al. (34) and Specht et al. (30). Homologous integration and replacement of sequences at the b locus were confirmed by DNA hybridization (data not shown).
Mating tests.
Routine mating tests employed a “drop-on-drop” procedure. Overnight cultures of U. maydis cells were grown at 30°C and 225 rpm for 16 to 20 h until late log or early stationary phase (optical density at 600 nm, 1.8 to 2.2). Then, 10 to 30 μl of culture was dropped on a charcoal mating plate containing 1% glucose and allowed to dry. A second drop of the tester culture was placed on top of the first and allowed to dry. The plates were then taped with a double layer of Parafilm, incubated in the dark at room temperature for 24 to 48 h, and scored for mycelial growth.
RESULTS
Chimeric alleles of bE1 and bE2.
Previously, we reported the construction of 16 chimeric alleles of the bE1 and bE2 genes (37). The strategy for constructing chimeric alleles at the b locus involves transformation of U. maydis cells with truncated bE or bW genes such that chimeric alleles are generated as a result of homologous recombination within the variable 5′ portion of the gene (37). In addition, it was also possible to construct chimeric alleles in vitro and to replace wild-type alleles by transformation and homologous recombination. In each case, the chimeric alleles were constructed such that deletions or insertions did not occur at the point of recombination between sequences from different alleles, as confirmed by sequence analysis.
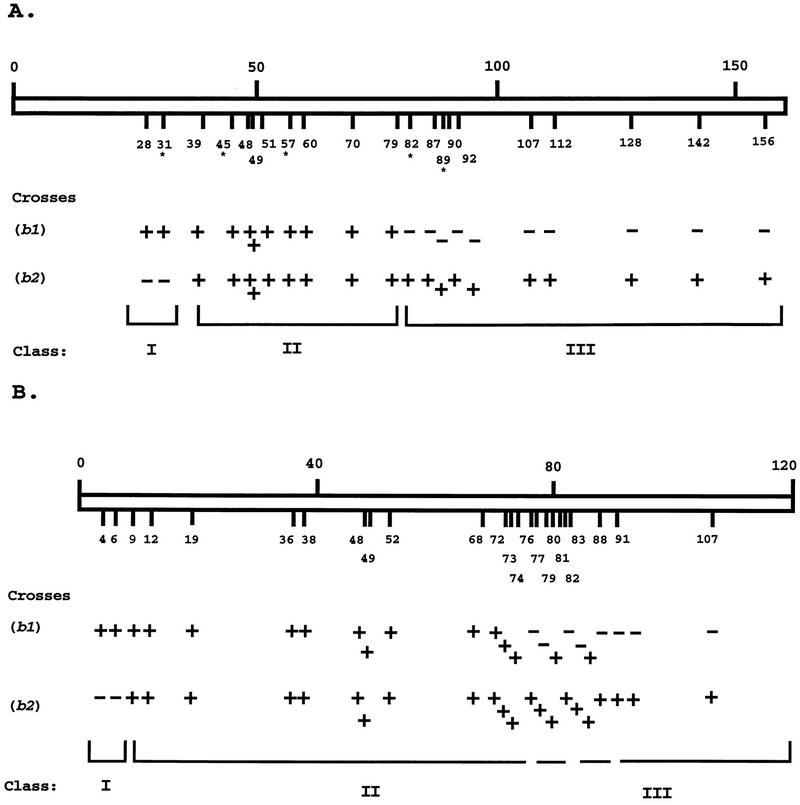
Our earlier work on chimeric bE alleles identified three classes based on the mating activity of the host strains; those that had a bE2 specificity (class I), those with a bE1 specificity (class III) and those with a specificity different from bE1 or bE2 (class II). The determination of the positions of the recombination sites for these classes identified a region involved in specificity between codons 39 and 87 which encodes a portion of the N-terminal variable region. We have now obtained five new chimeric alleles for bE1 and bE2 (bEx31, bEx45, bEx57, bEx82, bEx89) to develop a more detailed map of the three specificity classes for the chimeric bE alleles. The new chimeric alleles are shown with the previously constructed alleles on a map of the bE1 and bE2 variable region in Fig. 2A. It should be noted that allele bEx39 was previously scored as having a specificity like that of bE2 (37); the subsequent isolation of additional strains carrying this allele and further incompatibility tests revealed mating activity with both bW1 and bW2 tester strains. With the additional chimeric alleles for bE1 and bE2, we have now obtained chimeras for 17 of the 36 potential positions in the variable region between codons 1 and 107. These chimeras include 15 of the 27 potential chimeras in the first 92 codons that encode the N- and C-terminal borders of the region identified by the analysis of the class II alleles.
FIG. 2.
Map of the specificity regions for chimeric alleles of bE1 and bE2 and chimeric alleles of bW1 and bW2. (A) The positions of recombination sites for the sequence encoding the N-terminal variable region (positions 1 to 160 of the 473-amino-acid polypeptide) are shown for 21 chimeric alleles. The numbers on the map indicate the codons at which each recombination event changed the coding sequence from bE1 (N terminal) to bE2 (C terminal). The mating reactions of strains carrying the chimeric alleles are indicated below the map. The strains carrying chimeric alleles have a2 specificity and were tested against strains with a1 b1 and a1 b2 specificity. A plus sign indicates a compatible mating reaction that results in the formation of white aerial hyphae on mating colonies. A minus sign indicates a failure of the mating mixture to form aerial hyphae. Three classes of mating behavior are exhibited by the strains carrying chimeric alleles: class I (bE2 specificity), class II (novel specificity), and class III (bE1 specificity). The newly constructed chimeric alleles are marked with an asterisk to distinguish them from the alleles described previously (37). (B) The positions of recombination sites for the sequence encoding the N-terminal variable region (positions 1 to 120 of the 644-amino-acid polypeptide) are shown for 24 chimeric alleles. The numbers on the map indicate the codons at which each recombination event changed the coding sequence from bW1 (N terminal) to bW2 (C terminal). The mating reactions of strains carrying the chimeric alleles are indicated below the map. The strains carrying chimeric alleles have a1 specificity and were tested against strains with a2 b1 and a2 b2 specificities (see Fig. 4). As with the strains carrying the bE chimeric alleles shown in panel A, three classes of mating behavior were found: class I (bW2 specificity), class II (novel specificity), and class III (bW1 specificity).
Chimeric alleles of bW1 and bW2.
Gillissen et al. (12) presented genetic evidence that the specificity of recognition determined by b is mediated by interactions between the bE and bW gene products from strains with different b specificities rather than by bE-bE or bW-bW interactions. In addition, our previous analysis of bE chimeric alleles indicated that each allele with novel specificity (class II alleles) was found to have identical mating behavior when tested against a set of strains carrying naturally occurring bW alleles (37). Therefore, we constructed a set of chimeric alleles for bW1 and bW2 to further explore the interaction between bE and bW and to attempt to collect bW chimeric alleles that might identify differences between class II bE chimeric alleles. As shown in Fig. 2B, chimeric alleles were obtained for 24 of the 43 potential positions (56%) between codons 1 and 109 of bW1 and bW2. The potential positions for the formation of chimeric alleles represent the codons that specify different amino acids in bW1 and bW2. Mating tests with strains carrying wild-type bE1 and bE2 alleles revealed that the transformants carrying the chimeric bW alleles represented three classes: bW2 (class I), specificity different from bW1 and bW2 (class II), and bW1 (class III) (Fig. 3). A sequence analysis of the bW chimeric alleles from each class allowed the identification of a region involved in determining specificity comparable to that found for bE1 and bE2 and located between codons 6 and 83 (the N-terminal variable region for bW is encoded by codons 1 to 150). Interestingly, the region between codons 76 and 83 did not show a distinct transition between chimeric alleles that had a novel specificity (class II) and those that had a bW1 (class III) specificity. This feature suggests that amino acids involved in determining specificity may be clustered in the part of the variable region specified by these codons. The C-terminal border of the region defined by the class II alleles of bE did not show a similar complexity (Fig. 2A) (37).
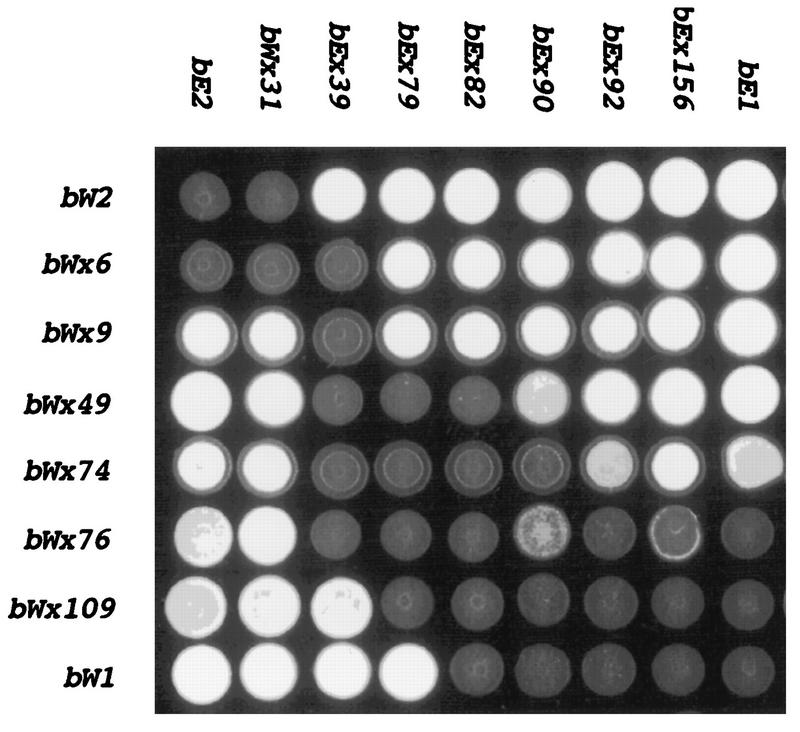
FIG. 3.
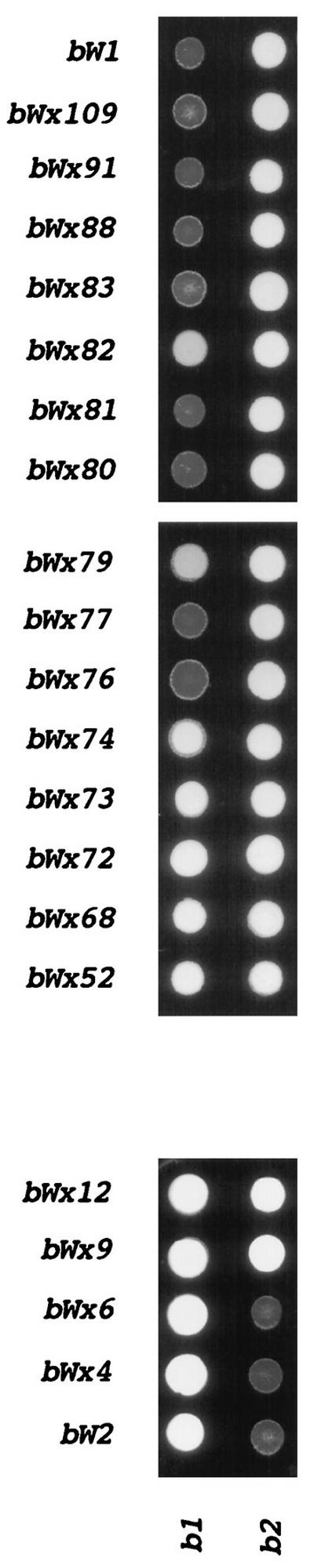
Mating reactions between strains carrying wild-type alleles and bW chimeric alleles. Representative mating reactions between strains with chimeric alleles and strains with wild-type alleles (032 [a2 b1] and 518 [a2 b2]) to demonstrate the activity of the chimeric alleles shown in Fig. 2. Each colony develops from the coinoculation of the two strains to be tested on medium containing activated charcoal (to enhance the reaction). Positive controls (white aerial hyphae) for mating are shown for the interactions of wild-type strains 032 (a2 b1) and 031 (a1 b2) and 521 (a1 b1) and 518 (a2 b2). Negative controls (flat gray colonies) include the interaction of 031 (a1 b2) with 518 (a2 b2) and 032 (a2 b1) with 521 (a1 b1); these strains, although capable of fusion, have the same specificity at b. Note that certain allele combinations give weak mating reactions: bWx79 and bWx82 with bE1 (032).
During the construction of the bW chimeric alleles, a specific attempt was made to obtain a large number of recombinant alleles in the borders of the region defined by the class II alleles. As a result, chimeric alleles were obtained for 10 of the 12 potential positions (83%) between codons 1 and 52 (encoding the N-terminal border region) and for 14 of the 18 potential positions (77%) between codons 68 and 109 (encoding the C-terminal border region). As described above, the potential positions for generating chimeric alleles are the codons for bW1 that differ from those for bW2. These chimeric alleles were initially obtained to provide a detailed analysis of the regions of transition between classes with different specificities. As described below, however, these chimeric alleles have also allowed the identification of specific amino acid positions that control recognition between bE and bW gene products.
Chimeric alleles of bW1 and bW3.
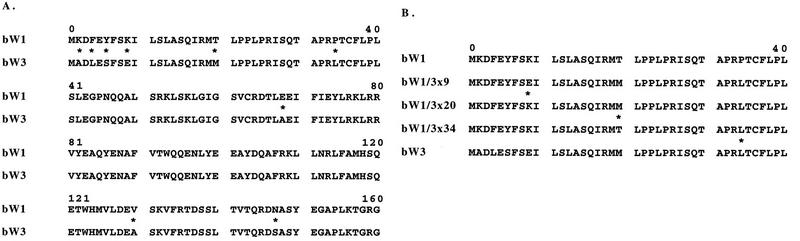
To date, our chimeric allele analysis has focused on bE and bW genes of b1 and b2 specificities. We were interested in expanding the analysis to include additional b specificities to determine whether similar sequences in the N-terminal variable regions were important in different allele combinations. The b3 locus provided a straightforward starting point for this analysis because alignments of the predicted amino acid sequences encoded by bW1 and bW3 revealed that the variability between these alleles shows up primarily in the first 20 amino acids at the corresponding N termini (Fig. 4A). Therefore, this region potentially contains all of the specificity determinants for this allele pair. Interestingly, differences exist at only 5 of the first 20 positions and at only 4 of the remaining 140 amino acids in the N-terminal region upstream of the homeodomain. The sequence similarity suggests that bW1 and bW3 are evolutionarily close, relative to other allele pairs, and that only a few differences in one region are sufficient to change specificity.
FIG. 4.
Construction of chimeric alleles for bW1 and bW3. (A) An alignment of the sequences at the N-terminal regions of bW1 and bW3 shows a high degree of identity; amino acid differences are found at nine positions in the first 160 positions. Alignments of additional bW sequences have been described previously (12). (B) Three chimeric alleles (bW1/3x9, -20, and -34) were constructed between bW1 and bW3; the number following the x indicates the first position of the bW3 sequence. The amino acid sequences encoded by the first 40 codons of the chimeric alleles are shown aligned with the sequences of the comparable regions from bW1 and bW3. For the chimeric alleles, the sequences before the asterisk are from the bW1 protein and all three have bW1 specificity.
Three chimeric alleles were obtained between bW1 and bW3 to confirm the position of the specificity determinants for this allele pair (Fig. 4B). Each of the chimeric alleles (bW1/3x9, bW1/3x20, and bW1/3x34) had class III (bW1) specificity, and alleles with class I and II specificities were not found. The relatively small region of variable sequence between bW1 and bW3 probably accounts for the absence of chimeric alleles of the class I and II types. The region for recombination to generate these alleles would be relatively small, and it would be necessary to construct this type of allele in vitro. The class III specificity of the alleles between bW1 and bW3 indicates that the specificity region for this allele pair lies upstream of codon 9, as predicted by sequence inspection. Overall, these results indicate that a quite different map of the bW N-terminal variable region can be obtained depending on the bW and bE alleles under consideration. However, in terms of specificity determinants, the bW1/bW3 combination revealed that a short N-terminal region is important; this region may play an identical role to that of the N-terminal region identified for the bW1/bW2 combination. In particular, codon 6, which plays an important role in specificity (described below), encodes an amino acid in this region and was found to encode different amino acids when the bW1 and bW3 polypeptide sequences were compared.
Crosses between strains carrying bE and bW chimeric alleles.
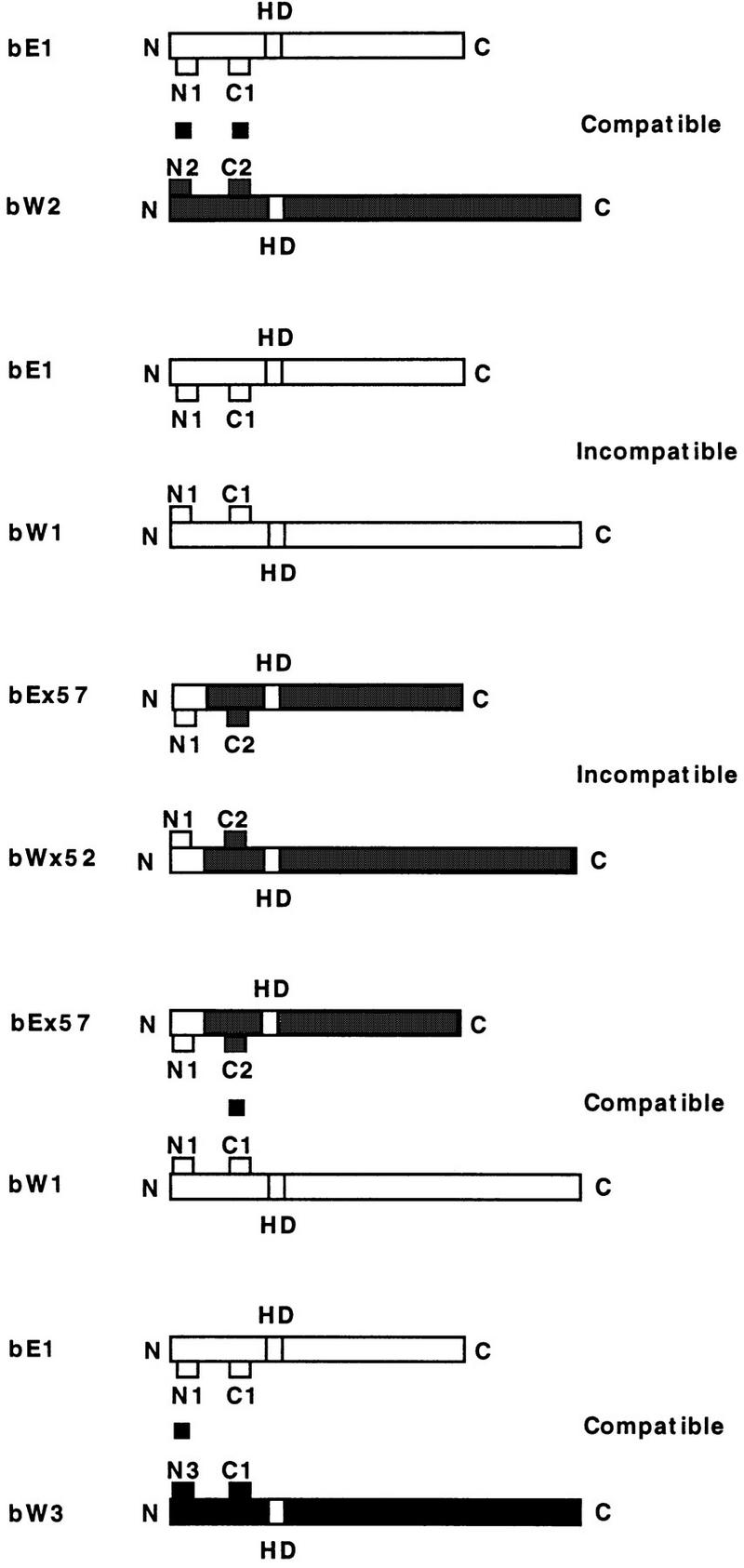
The availability of sets of bE1/bE2 and bW1/bW2 chimeric alleles presented an opportunity to further investigate the roles of the specificity regions through incompatibility tests between strains carrying different chimeric bE and bW genes. As described above, these sets of chimeras each defined three specificity classes: class I, wild-type b2 specificity; class II, novel specificity (different from b1 or b2); and class III, wild-type b1 specificity. Mating tests on culture medium were performed for all combinations between strains carrying each of 19 different bE1/bE2 chimeras and strains carrying each of the 26 different bW1/bW2 chimeras (494 combinations). The results of these crosses are summarized in Table 2, and representative mating tests are shown in Fig. 5. These tests provided an interesting insight into the organization of the specificity regions of the bE and bW proteins. Specifically, the striking general result was that the majority of strains carrying chimeric alleles from class II failed to give a positive mating reaction, suggesting that the borders of the regions defined by these alleles contain the important residues for recognition and dimerization between bE and bW. As diagrammed in Fig. 6, these border sequences have been designated N1 and C1 for the b1 specificity genes and N2 and C2 for the b2 specificity genes. The bE and bW chimeric alleles of class II specificity would therefore be associated with the N1 and C2 border combination. Note that our strategy would not yield chimeric alleles encoding the N2 and C1 border combination.
FIG. 5.
Mating reactions between strains carrying wild-type or chimeric alleles of bE1 and bE2 or bW1 and bW2. Representative mating reactions are shown to illustrate the data summarized in Table 2. The appearance of vigorous white aerial hyphae indicates a strongly compatible interaction between bE and bW polypeptides; this type of reaction (e.g., bW2 with bE1) is assigned three pluses in Table 2; weaker compatible reactions are assigned one or two pluses (e.g., bEx90 with bWx76).
FIG. 6.
Model for the interactions of the borders of the specificity regions of bE and bW. The borders are designated N and C followed by a number (1 or 2) to indicate the specificity of the parental allele. Compatible interactions (dimerization) would align N and C borders of different specificities (top), and incompatibility would result from the interaction of borders of like specificity (second from top). An interaction leading to dimerization is indicated by the black boxes between borders of different specificities. Chimeric alleles with recombination points between the N and C borders (e.g., class II alleles such as bEx57 and bWx52) give an incompatible reaction (Table 2) because borders of like specificity are aligned. In contrast, chimeric alleles of class II specificity (e.g., bEx57) give compatible reactions with wild-type alleles (e.g., bW1). In this case, it is sufficient for either the N or the C border to be recognized as non-self. A similar situation can be found for some naturally occurring alleles such as the combination of bW1 and bW3 where the determinants of specificity are found only in the N-terminal border region.
As shown in Fig. 6, the recombination events between the border regions that generate class II alleles would result in b gene products that are capable of interacting with products from either parental allele. For example, the product of class II allele bEx57 would allow a positive mating interaction with strains carrying bW1 and bW2 alleles. This suggests that dimerization is not prevented when sequences of like specificities are present at just one of the borders (e.g., a bE N1/C2 interaction with a bW N1/C1). Conversely, recognition of nonself at one border is sufficient to allow dimerization. We propose that the products of the class II alleles fail to interact with each other because self combinations are present for both of the borders (e.g., N1/C2 with N1/C2). An example of this situation is depicted in Fig. 6 for class II alleles bWx52 and bEx57. These combinations would be similar, in terms of border combinations, to the wild-type self combinations of N1/C1 with N1/C1, or N2/C2 with N2/C2. Overall, the mating tests between strains with chimeric alleles indicated that the borders of the specificity intervals contain amino acid residues that are important for recognition; this is consistent with the border locations of residues that influence specificity (see below) and the location of a single short N-terminal border for the bW1/bW3 combination described earlier.
The crosses presented in Table 2 also revealed that the bEx107, bEx128, and bEx156 alleles were found to apparently have specificities different from that of the wild-type bE1 allele when tested against bW chimeras bWx76, bWx77, bWx80, and bWx81. Surprisingly, the bEx156 allele contains all of the variable region of the bE1 gene (encoding amino acids 1 to 156) fused to a portion of the bE2 gene encoding part of the C-terminal region (amino acids 157 to 473). A comparison of the predicted amino acid sequences of the products of the bEx156 and bE1 alleles revealed three differences in the homeodomain and one difference in the C-terminal region. It is possible that these residues in the homeodomain contribute to the specificities of interactions in other allele combinations because variability was found in this region when the sequences of six bE alleles were compared (17). This result suggests a possible role for the homeodomain in specificity that is only revealed through test crosses with specific bW chimeric alleles. It is possible that sequences in the homeodomain could directly influence dimerization or that the amino acid differences in the homeodomain might have a long-range influence on the conformation of the specificity region resulting in different interactions with some of the chimeric bW alleles.
Identification of single amino acid positions that influence specificity.
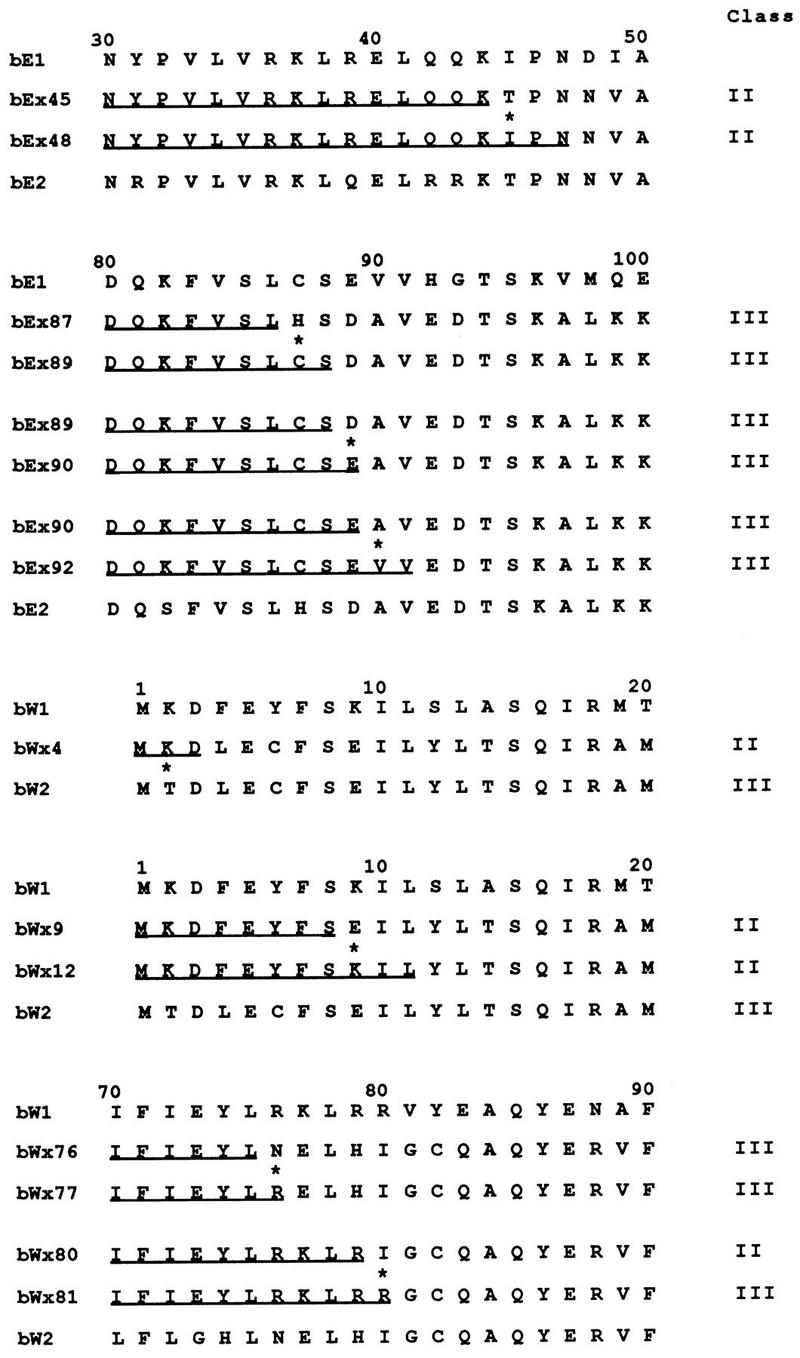
In earlier work, we noted that sequence comparisons of chimeric alleles with different specificities allowed the identification of amino acid residues that were important for specificity (37). Our expanded collection of chimeric alleles for bE1/bE2 and the new collection for the bW1/bW2 alleles provided an opportunity to compile a list of amino acid positions that are involved in the specificity of interaction. In particular, crosses between strains carrying chimeric alleles and strains carrying wild-type alleles identified several pairs of alleles (e.g., bWx6 and bWx9) whose products differ in sequence at only one amino acid position but which are found to confer a difference in specificity when tested against strains carrying wild-type alleles. The mating behavior of the strains carrying the bW chimeric alleles, which is believed to reflect the specificity of the interaction between bE and bW gene products, is shown in Fig. 3. The sequence alignments for the amino acids encoded by some of those chimeric alleles and for those encoded by other chimeras that were found to differ in specificity when tested with wild-type alleles are shown in Fig. 7. These alignments focus attention on key positions within the border sequences of the regions defined by the class II bE and bW alleles and identify eight amino acid positions that influence specificity. These positions include those encoded by codons 31 and 79 of bE and by codons 6, 74, 77, 79, 81, and 82 of bW.
FIG. 7.
Sequence alignments for the products of chimeric alleles that were found to differ in specificity and in a single amino acid position when tested against the products of wild-type alleles. Eight amino acid positions that influence specificity (marked with asterisks) were identified by sequence alignments of alleles whose products were found to differ in mating reactions when tested with those of wild-type alleles. Four bE1/bE2 chimeric alleles allowed the identification of two amino acid positions that influence specificity. An additional 12 bW1/bW2 chimeric alleles identified six positions that determine specificity. The results of mating tests demonstrating the interactions of strains carrying the bW chimeric alleles are shown in Fig. 3, and the results of mating tests for all of the alleles are shown in Table 2. The specificity classes are indicated on the right, the underlined sequences represent the bE1 or bW1 portions of the products of the chimeric alleles, and the remaining sequences are from bE2 or bW2.
It is interesting to note that among the eight positions that influence specificity, four of the amino acid differences involve a Tyr residue. These positions have substitutions of Tyr for either Arg, His or, in two cases, Cys. Charged or polar amino acids are present at one or both of the positions in six of the eight examples. Only one of the positions (bE codon 79) has a substitution of two hydrophobic residues (Ile and Phe) and one (bW codon 79) has a substitution of basic residues (His for Arg). In addition, a reversal of charge (Lys or Asp) was found for one position (bW codon 77). These comparisons of the amino acids found at positions that influence specificity suggest that charge and polarity may play an important role in the interaction between the bE and bW polypeptides. Furthermore, it is striking that residues with aromatic side chain rings, i.e., His, Tyr, and Phe are prominent within the list of amino acids at the eight positions. Overall, these results indicate that it is possible to use differences between chimeric alleles to identify single-amino-acid positions important for specificity and to catalog the types of residues at those positions.
The identification of important amino acid positions within the border regions was extended by the analysis of additional chimeric alleles that were found to have different specificities when strains with chimeric alleles were used as testers. An important feature of these crosses between strains carrying chimeric alleles was the identification of interesting interactions between specific chimeras with recombination points in or near the N and C borders of the specificity regions. For example, alleles bEx87 and bEx89 show opposite specificities when tested with various bW chimeric alleles (Table 2). The behavior of these and other alleles with adjacent recombination points indicates that recombination has occurred in regions that are important for specificity, i.e., the N and C borders. These data can also be used to identify the amino acid positions that play a role in the specificity of interactions between the products of chimeric alleles. Sequence alignments of amino acids encoded by chimeric alleles with specificity differences are shown in Fig. 8. These sequence alignments reveal single-amino-acid differences at important positions in the allele products and provide an additional list of the types of residues that influence specificity. As with the eight amino acid positions identified in the analysis shown in Fig. 7, the majority of the residues are charged or polar and few are hydrophobic. In one position (bW codon 9), a clear charge difference is present (Asp versus Lys). At two other positions, the amino acid differences involve substitution of a polar or charged residue for a hydrophobic residue (e.g., bE codon 45 and bW codon 80).
FIG. 8.
Alignments of predicted sequences of the products of chimeric alleles that were found to differ at a single amino acid and have different specificities when tested against the products of other chimeric alleles. The portion of each sequence containing the single amino acid difference (asterisk) is shown, and the specificity class of each allele is indicated on the right. The patterns of interactions for each allele pair are shown in Table 2. Note that alleles bEx87, bEx89, bEx90, and bEx92 were all designated class III when assayed with wild-type b1 and b2 mating partners. Differences in specificity are revealed in mating assays with strains carrying other chimeric alleles as shown in Table 2.
The identification of bE codon 45 as a key position is interesting because this is the only position which shows an influence and which is outside of the border regions previously identified as containing the important residues. This finding suggests that the border regions that were defined by testing b1 and b2 chimeric alleles with wild-type strains may not be definitive when testing chimeric alleles against each other. That is, bE position 45 may have a residue that is important for specificity only in the context of the chimeric alleles. This finding reinforces the idea that the identification of the residues in the bE and bW N-terminal dimerization domains that are important for specificity is dependent upon the allele combinations under investigation.
Possible interactions between the N and C border regions.
Chimeric alleles bEx87 and bEx89 encode products that differ at a single amino acid position (Fig. 8) and, as shown in Table 2, have different specificities when tested against chimeric bW alleles with recombination points near the N-terminal border (e.g., bWx12, bWx19, and bWx36) and within the C-terminal border (e.g., bWx76, bWx79, bWx80, and bWx82). Although this allele pair was the only one to clearly exhibit this phenotype, this finding suggests that interactions may occur between the N- and C-terminal border regions. That is, the ability of a difference within one border (defined by bEx87 and bEx89) to alter the specificity of interactions with alleles with recombination points near or in both the N- or C-terminal borders may indicate that the borders cooperate.
DISCUSSION
Specificity determinants in the variable N-terminal portions of bE and bW.
The bE and bW genes of U. maydis each exist in a series of at least 25 alleles that are primarily distinguished by variability in the regions encoding the N-terminal 100 to 150 amino acids (12, 17, 24, 28, 29). Dimerization of bE and bW proteins encoded by alleles from mating partners of different specificities has been demonstrated (15) and is thought to establish a transcription factor that controls morphogenesis and pathogenesis. The bE and bW proteins encoded by the same strain (like specificity) fail to dimerize (15). A primary goal in the analysis of the b proteins has been the identification of regions of bE and bW that control the specificity of interaction. The N-terminal regions of bE and bW were obvious targets for this analysis because these regions contain most of the allelic variation and mediate dimerization (12, 15, 17). In addition, our previous work on chimeric bE alleles revealed that recombination within the 5′ proximal coding regions resulted in alleles with novel specificity (37).
The construction and analysis of dual sets of chimeric alleles for bE1/bE2 and bW1/bW2 that are described here provided an opportunity to further refine our view of the N-terminal specificity regions believed to control the recognition between bE and bW proteins. Previously, we found that bE1/bE2 chimeras that contained recombination points in the central portion of the variable region were of particular interest because they had specificities different from either parental allele (37). These alleles were designated class II to distinguish them from alleles that had not changed specificity (class I) or that had switched from one parental specificity to the other (class III). One major finding from the extension of our work to include chimeras of bW1 and bW2 is that the same three classes of alleles could be identified, including the class II group with novel specificity. However, in contrast to the findings for bE1 and bE2, the transition between class II and class III chimeras was not distinct for bW1 and bW2. Rather, a pattern of switching between class II and class III specificities was observed. This result served to focus attention on the borders of the region defined by the class II alleles and provided the framework for more detailed studies to identify sequences that control specificity.
Throughout our analysis of the chimeric alleles, we have made the assumption that the differences in specificity indicated by the presence or absence of filamentous cell growth in mating tests reflect differences in dimerization ability between bE and bW polypeptides. This assumption is based on the demonstrated correlation between dimerization and mating specificity reported by Kämper et al. (15) for the bE and bW proteins. That is, Kämper et al. (15) have employed in vitro and in vivo assays to show that the N-terminal variable portions of the b proteins mediate dimerization and that these same regions control mating specificity. A similar relationship has also been reported for analogous homeodomain-containing mating type proteins in Schizophyllum commune and Coprinus cinereus (1, 36, 38). It is formally possible, however, that other explanations account for the different activities of the chimeric bE and bW proteins analyzed in our study. For example, there may be differences in levels or stability of the chimeric proteins compared to the parental proteins. Furthermore, it is possible that a negative mating test may reflect a difference in the activity of a heterodimer (e.g., failure to act as a transcriptional repressor or activator) rather than a failure to dimerize. Although these other possibilities must be kept in mind, we would note that the chimeric alleles that we have constructed have been used to directly replace the parental alleles by homologous recombination. Thus, the chromosomal location is the same for the chimeric and parental alleles, and we would expect transcription of the genes to be identical. Also, the chimeric alleles were constructed such that deletions or insertions did not occur at the site of recombination. That is, the chimeric alleles represent genes with novel combinations of parental sequences rather than mutated versions. Given these considerations and the description of Kämper et al. (15) of amino acid changes in the N-terminal regions that influence both dimerization and mating specificity, we favor the interpretation that our mating tests reflect differences in the abilities of chimeric bE and bW proteins to dimerize. The discussion below is presented with this interpretation in mind.
In previous work, we explored the mating behavior of the strains carrying the class II chimeric alleles of bE1 and bE2 to gain insight into the novel specificities of these alleles (37). For example, we performed mating reactions between strains carrying class II alleles of bE1/bE2 and strains with wild-type alleles different from b1 and b2 (bD, bI, and bM) to demonstrate that the class II alleles are not simply constitutively active with any other bW protein (37). That is, recombination within the specificity region does not simply result in constitutive compatibility because the mating reactions failed with bI and bM strains. In addition, we performed the mating tests with the strains with class II alleles and strains with three additional b specificities (bH, bJ, and bL) to search for specificity differences between class II bE alleles. In these experiments, we found that all three class II alleles had the same specificity. Not surprisingly, we also found that strains of opposite a mating type that carried different class II alleles of bE1/bE2 failed to mate with each other. This was expected because of the genetic evidence indicating that specificity is determined by the interactions between bE and bW polypeptides (12). As described below, the construction of a set of bW1/bW2 chimeric alleles provided an opportunity to retest the class II alleles of bE1/bE2 for differences in specificity.
Crosses between chimeric alleles identify short regions containing specificity determinants.
The availability of dual sets of chimeric alleles of bE1/bE2 and bW1/bW2 allowed crosses to be performed between all combinations of alleles representing the three specificity classes (Table 2). The basic finding from this work was that the products of class II alleles of bE generally fail to interact in a compatible manner with the products of class II alleles of bW. Our interpretation of this result is that the borders of the region defined by class II alleles contain the important determinants of recognition and that artificial combinations of these borders (resulting from recombination in the intervening region) generate alleles with novel specificities. In this context, the 40-amino-acid region defined by the class II bE1/bE2 alleles and the analogous 70-amino-acid region for the bW1/bW2 alleles may represent the intervals between the borders that influence specificity. For bE1 and bE2, these specificity borders are encoded by codons 31 to 39 and by codons 79 to 92; for bW1 and bW2, these sequences are encoded by codons 2 to 9 and 74 to 83.
As shown in Fig. 6, we have designated the border regions (10 to 20 amino acids) defined by the analysis of class II alleles as the N and C sequences. These sequences have specificities N1 and C1 for bE1 and bW1, and N2 and C2 for bE2 and bW2. In a self interaction (e.g., bE1 with bW1), regions of like specificity (N1 with N1 and C1 with C1) would prevent formation of the heterodimer. In a nonself interaction (e.g., N1 with N2 and C1 with C2), the specificity regions would allow dimerization. Chimeric proteins encoded by class II alleles would fail to interact with each other because these products would have like specificity borders (Fig. 6). That is, N1 would interact with N1 and C2 would interact with C2, resulting in a situation similar to that occurring with the bE and bW products of wild-type self alleles. The idea that the N and C regions contain important residues for specificity is supported by the finding that the bW1 and bW2 chimeras did not show a distinct C-terminal transition between alleles with class II specificity and alleles with class III specificity. Instead, recombination events with the C sequence resulted in alleles that showed a pattern of alternating specificities (Fig. 2B). In addition, the amino acid positions that influence specificity, as identified by comparisons of chimeric alleles, are located mainly in the N and C regions (see below).
The analysis of an additional set of chimeric alleles between bW1 and bW3 (Fig. 4) supports the importance of the N and C borders defined for the b1 and b2 genes. That is, the construction of chimeric alleles for the bW1 and bW3 alleles confirmed the presence of an N-terminal specificity sequence encoded by the first 10 codons. Interestingly, the construction and analysis of chimeric alleles from bW1 and bW3 indicates that some naturally occurring alleles have products that differ at only one of the N or C regions (e.g., N1) and that sequence differences in one region are sufficient to provide a different specificity. In terms of the specificity borders, bW3 appears to be a naturally occurring chimera whose product has an N3 and C1 combination of borders (Fig. 2A). This suggests that new specificities could be generated via recombination between different alleles to reassort the N and C sequences; this type of event is demonstrated by the nonparental specificity of class II chimeric alleles.
Identification of amino acid positions important for the specificity of recognition.
The sequence comparisons of pairs of chimeric alleles that differ in specificity and that are neighbors on the specificity maps (Fig. 2) provided a means of identifying amino acid positions that influence specificity. Initially, eight of these positions were identified through crosses between strains carrying chimeric alleles and strains carrying wild-type b1 or b2 sequences (Fig. 7). In general, most of the amino acids found at the eight positions were either charged or polar and relatively few hydrophobic residues were present. An inspection of the types of residues occupying the eight positions revealed a preponderance (six positions) of aromatic amino acids (His, Phe, or Tyr). Although it is difficult to draw definite conclusions about the role of aromatic amino acids, it is interesting to note that these types of amino acids have been found to play important roles in antigen-antibody binding (8, 23, 25, 33).
An additional eight amino acid positions that influence specificity were identified from crosses between strains that each carry chimeric alleles (Fig. 8). In these amino acid positions, the majority of residues were polar or charged, but only one residue had an aromatic side chain ring (His), and Tyr and Phe were not found. We speculate that the preponderance of aromatic amino acids found in the first set of eight positions, compared with the second set, may reflect differences in the interactions of the products of chimeric alleles with wild-type products compared with interactions between chimeric proteins. Taken as a group, the 16 pairs of the alternate residues (32 amino acids) present at the key positions reflect the preponderance of polar and charged residues; that is, 24 of 32 residues were polar or charged, 7 of 32 residues were hydrophobic, and 1 was Gly.
Overall, the data from Table 2 and Fig. 7 and 8 identified six positions for bE1/bE2 (codons 31, 45, 79, 87, 89, and 90) and 10 positions for bW1/bW2 (codons 2, 6, 9, 74, 76, 77, 79, 80, 81, and 82) that influence specificity. It is noteworthy that these positions are all found in the N and C border regions except position 45 of bE. Thus, the locations of the key amino acids reinforce the idea that the failure of class II bE and bW chimeric alleles to interact results from the presence of self combinations of borders (Fig. 4A). Given that chimeric alleles were constructed for only 50 to 60% of the potential positions for bE and bW, it is possible that additional positions are important in the interactions of these allele pairs. However, for bW1 and bW2, chimeric alleles were obtained for 4 of 6 potential positions (positions 2, 6, 9, 12) in the N-terminal region (codons 1 to 15) and for 10 of 11 sites (positions 72, 73, 74, 76, 77, 79, 80, 81, 82, 83) in the C-terminal region (codons 70 to 85). For bE1 and bE2, chimeric alleles were constructed for all three potential N-terminal sites (positions 28, 31, and 39) between positions 25 and 40 and for all five potential C-terminal sites (positions 79, 80, 81, 82, and 83) between positions 75 and 90. Thus, the majority of potential chimeric alleles have been constructed for the N and C regions and the majority of the important amino acid positions have probably been identified for these allele pairs.
Kämper et al. (15) have shown that the variable N-terminal regions of bE and bW control dimerization such that heterodimers arise from polypeptides with different specificities (e.g., bE1 with bW2) but not from polypeptides with like specificities (e.g., bE1 with bW1). In addition, mutations that allowed interaction between the self polypeptide combination of bE2 and bW2 were identified. In general, these mutations were found to increase hydrophobicity, and it was suggested that the wild-type residues involved were important for the failure of self combinations to interact. Two additional mutations resulted in a change in charge, implying a contribution from polar interactions for dimerization. Combining the results from this work with the analysis of chimeric alleles leads to the general idea that a number of key amino acid positions control recognition by influencing dimerization. In general, however, the amino acid changes described by Kämper et al. that resulted in an increase in hydrophobicity promoted interaction (dimerization) between the self combination of bE2 and bW2 polypeptides (15). In contrast, the positions identified for chimeric alleles suggest a prominent role for charged or polar residues, including aromatic amino acids. These differences may reflect the fact that the substitutions identified by Kämper et al. represented changes that allowed self interaction. In the case of chimeric alleles, the interactions of novel combinations of self and nonself sequences were explored.
Chimeric alleles for other homeodomain mating proteins in fungi.
Homeodomain proteins encoded by multiallelic genes and having roles in sexual development have also been characterized for the mushroom fungi C. cinereus and S. commune. These proteins, designated HD1 and HD2 for C. cinereus (1) and Y and Z for S. commune, also contain the determinants of allelic specificity in N-terminal regions, as revealed by chimeric allele analysis. In the case of the HD1 and HD2 proteins of C. cinereus, specificity is determined by the N-terminal 160 to 170 amino acids (1). For S. commune Z proteins, seven chimeric alleles were constructed between Z4 and Z5, and these defined a specificity region between codons 19 and 60 (36). Eight chimeric alleles for Y4 and Y3 were also constructed, and a region determining specificity was found between codons 17 and 72 (38). As with the class II chimeric alleles of bE1 and bE2 (37), Y4/Y3 chimeric genes with exchange points between codons 17 and 72 had specificities different from either parental allele. This result indicates that in the case of the Y alleles of S. commune, the borders of the region defined by alleles with novel specificities carry the important determinants of recognition. A similar situation would probably be revealed by chimeric alleles with recombination in the region between positions 19 and 60 of the Z proteins. Overall, these results suggest that a common mechanism and perhaps a common structural organization may be employed to determine self versus nonself recognition for the homeodomain-containing mating-type proteins in basidiomycetes.
The use of chimeric proteins to study recognition.
A chimeric strategy for identifying specificity determinants has been employed in other systems involving recognition between polypeptides. For example, the dimerization specificity of the bacteriophage P22 repressor has been studied by making chimeras between P22 and homologous repressor protein 434 (10). In addition, an attempt to determine the basis of multiallelic self-incompatibility in plants was carried out by exchanging domains between allelic S-RNases from Nicotiana alata (39). Chimeric proteins have also been used to study protein-protein interactions during ligand-receptor recognition (7, 20, 21, 32). In fact, our observation that bE and bW class II chimeric alleles have specificities different from either parent is not unique to fungal mating-type systems; a similar phenomenon has recently been reported for chimeras of two glycoprotein hormones (4). Specifically, the chimeras of human chorionic gonadotropin (hCG) and human follitropin (hFSH) were shown to exhibit activity unique to a third family member, human thyrotropin (hTSH). This result was explained by a model stating that the specificity between ligand and receptor was mediated by “inhibitory determinants” that restricted binding to only the appropriate combinations (4, 22). The construction of chimeras was thought to disrupt the inhibitory determinants and unmask activities characteristic of other members of the protein family.
It is interesting to speculate that an inhibitory determinant model such as that described for receptor-ligand interactions may be applicable to the problem of specificity determination at the multiallelic b locus. In the case of b genes, specificity may result from interactions that prevent dimerization between bE and bW proteins derived from the same strain. That is, there may be amino acid residues positioned to interfere with dimerization between bE1 and bW1, and these inhibitory determinants may be positioned differently for each self allele combination. Thus a set of interfering residues could prevent dimerization between self allele combinations of bE and bW; presumably, the residues would not directly oppose each other for nonself allele combinations. The amino acid residues in the borders defined by chimeric allele analysis may represent inhibitory determinants. Site-directed mutagenesis and in vitro protein interaction studies, combined with access to the three-dimensional structure of the bE and bW proteins, will be needed to explore this possibility.
ACKNOWLEDGMENTS
This work was supported by Research and Collaborative Project grants from NSERC (to J.W.K.) and by postgraduate fellowships from NSERC and the Killam Foundation (to A.R.Y.).
We thank Jeanette Johnson-Beatty, George Athwal, and Alan Au for technical assistance and Lisa Gentile and Lawrence McIntosh for comments on the manuscript.
REFERENCES
- 1.Banham A H, Asante-Owusu R N, Gottgens B, Thompson S A J, Kingsnorth C S, Mellor E J C, Casselton L A. An N-terminal dimerization domain permits homeodomain proteins to choose compatible partners and initiate sexual development in the mushroom Coprinus cinereus. Plant Cell. 1995;7:773–783. doi: 10.1105/tpc.7.6.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banuett F. Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu Rev Genet. 1995;29:179–208. doi: 10.1146/annurev.ge.29.120195.001143. [DOI] [PubMed] [Google Scholar]
- 3.Bölker M, Urban M, Kahmann R. The a mating type locus of U. maydis specifies cell signalling components. Cell. 1992;68:441–450. doi: 10.1016/0092-8674(92)90182-c. [DOI] [PubMed] [Google Scholar]
- 4.Campbell R K, Bergert E R, Wang Y, Morris J C, Moyle W R. Chimeric proteins can exceed the sum of their parts: implications for evolution and protein design. Nat Biotechnol. 1997;15:439–443. doi: 10.1038/nbt0597-439. [DOI] [PubMed] [Google Scholar]
- 5.Cease K B, Potcova C A, Lohff C J, Zeigler M E. Optimized PCR using Vent polymerase. PCR Methods Appl. 1994;3:298–300. doi: 10.1101/gr.3.5.298. [DOI] [PubMed] [Google Scholar]
- 6.Christensen J J. Corn smut caused by Ustilago maydis. Monograph no. 2. Saint Paul, Minn: American Phytopathological Society; 1963. [Google Scholar]
- 7.Cunningham B C, Jhurani P, Ng P, Wells J A. Receptor and antibody epitopes in human growth hormone identified by homolog-scanning mutagenesis. Science. 1989;243:1330–1336. doi: 10.1126/science.2466339. [DOI] [PubMed] [Google Scholar]
- 8.Davies D R, Cohen G H. Interactions of protein antigens with antibodies. Proc Natl Acad Sci USA. 1996;93:7–12. doi: 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies D R, Padlan E A, Sheriff S. Antibody-antigen complexes. Annu Rev Biochem. 1990;59:439–473. doi: 10.1146/annurev.bi.59.070190.002255. [DOI] [PubMed] [Google Scholar]
- 10.Donner A L, Carlson P A, Koudelka G B. Dimerization specificity of P22 and 434 repressors is determined by multiple polypeptide segments. J Bacteriol. 1997;179:1253–1261. doi: 10.1128/jb.179.4.1253-1261.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Froeliger E H, Leong S A. The a mating-type alleles of Ustilago maydis are idiomorphs. Gene. 1991;100:113–122. doi: 10.1016/0378-1119(91)90356-g. [DOI] [PubMed] [Google Scholar]
- 12.Gillissen B, Bergemann J, Sandmann C, Schroer B, Bölker M, Kahmann R. A two-component system for self/non-self recognition in Ustilago maydis. Cell. 1992;68:647–657. doi: 10.1016/0092-8674(92)90141-x. [DOI] [PubMed] [Google Scholar]
- 13.Holliday R. The genetics of Ustilago maydis. In: King R C, editor. Handbook of genetics. Vol. 1. New York, N.Y: Plenum Press; 1974. pp. 575–595. [Google Scholar]
- 14.Kahmann R, Bölker M. Self/nonself recognition in fungi: old mysteries and simple solutions. Cell. 1996;85:145–148. doi: 10.1016/s0092-8674(00)81091-0. [DOI] [PubMed] [Google Scholar]
- 15.Kämper J, Reichmann M, Romeis T, Bölker M, Kahmann R. Multiallelic recognition: nonself-dependent dimerization of the bE and bW homeodomain proteins in Ustilago maydis. Cell. 1995;81:73–83. doi: 10.1016/0092-8674(95)90372-0. [DOI] [PubMed] [Google Scholar]
- 16.Kronstad J W, Leong S A. Isolation of two alleles of the b locus of Ustilago maydis. Proc Natl Acad Sci USA. 1989;86:978–982. doi: 10.1073/pnas.86.3.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kronstad J W, Leong S A. The b mating-type locus of Ustilago maydis contains variable and constant regions. Genes Dev. 1990;4:1384–1395. doi: 10.1101/gad.4.8.1384. [DOI] [PubMed] [Google Scholar]
- 18.Kronstad J W. Pathogenesis and sexual development of the smut fungi. In: Stacey G, Keen N T, editors. Plant-microbe interactions. Vol. 1. New York, N.Y: Chapman and Hall; 1996. pp. 141–186. [Google Scholar]
- 19.Lasky L A. From sticky zippers to morphology. Nat Struct Biol. 1995;2:258–261. doi: 10.1038/nsb0495-258. [DOI] [PubMed] [Google Scholar]
- 20.Leeb T, Mathis S A, Leeb-Lundberg L M F. The sixth transmembrane domains of the human B1 and B2 bradykinin receptors are structurally compatible and involved in discriminating between subtype-selective agonists. J Biol Chem. 1997;272:311–317. doi: 10.1074/jbc.272.1.311. [DOI] [PubMed] [Google Scholar]
- 21.Maden M. Retinoids in patterning: chimeras win by a knockout. Curr Biol. 1996;6:790–793. doi: 10.1016/s0960-9822(02)00596-1. [DOI] [PubMed] [Google Scholar]
- 22.Moyle W R, Campbell R K, Myers R V, Bernard M P, Han Y, Wang X. Co-evolution of ligand-receptor pairs. Nature. 1994;368:251–255. doi: 10.1038/368251a0. [DOI] [PubMed] [Google Scholar]
- 23.Padlan E A. On the nature of antibody combining sites: unusual structural features that may confer on these sites an enhanced capacity for binding ligands. Proteins. 1990;7:112–124. doi: 10.1002/prot.340070203. [DOI] [PubMed] [Google Scholar]
- 24.Puhalla J E. Genetic studies of the b incompatibility locus of Ustilago maydis. Genet Res. 1970;16:229–232. [Google Scholar]
- 25.Rini J M, Schulze-Gahmen U, Wilson I A. Structural evidence for induced fit as a mechanism for antibody-antigen recognition. Science. 1992;255:959–965. doi: 10.1126/science.1546293. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Sarkar G, Sommer S S. The megaprimer method of site-directed mutagenesis. Biotechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 28.Schulz B, Banuett F, Dahl M, Schlesinger R, Schäfer W, Martin T, Herskowitz I, Kahmann R. The b alleles of U. maydis, whose combinations program pathogenic development, code for polypeptides containing a homeodomain-related motif. Cell. 1990;60:295–306. doi: 10.1016/0092-8674(90)90744-y. [DOI] [PubMed] [Google Scholar]
- 29.Silva J. Alleles at the b incompatibility locus in Polish and North American populations of Ustilago maydis (DC) Corda. Physiol Plant Pathol. 1972;2:333–337. [Google Scholar]
- 30.Specht C A, Munoz-Rivas A M, Novotny C P, Ullrich R C. Transformation of Schizophyllum commune: an analysis of specific properties. Exp Mycol. 1991;15:326–335. [Google Scholar]
- 31.Spellig T, Bölker M, Lottspeich F, Frank R W, Kahmann R. Pheromones trigger filamentous growth in Ustilago maydis. EMBO J. 1994;13:1620–1627. doi: 10.1002/j.1460-2075.1994.tb06425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian Y, Wu L, Oxender D L, Chung F. The unpredicted high affinities of a large number of naturally occurring tachykinins for chimeric NK1/NK3 receptors suggest a role for an inhibitory domain in determining receptor specificity. J Biol Chem. 1996;271:20250–20257. doi: 10.1074/jbc.271.34.20250. [DOI] [PubMed] [Google Scholar]
- 33.Tsumoto K, Ogasahara K, Ueda Y, Watanabe K, Yutani K, Kumagai I. Role of Tyr residues in the contact region of anti-lysozyme monoclonal antibody HyHEL10 for antigen binding. J Biol Chem. 1995;270:18551–18557. doi: 10.1074/jbc.270.31.18551. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Holden D W, Leong S A. Gene transfer system for the phytopathogenic fungus Ustilago maydis. Proc Natl Acad Sci USA. 1988;85:865–869. doi: 10.1073/pnas.85.3.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wells J A. Binding in the growth hormone receptor complex. Proc Natl Acad Sci USA. 1996;93:1–6. doi: 10.1073/pnas.93.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu J, Ullrich R C, Novotny C P. Regions in the Z5 mating gene of Schizophyllum commune involved in Y-Z binding and recognition. Mol Gen Genet. 1996;252:739–745. doi: 10.1007/BF02173981. [DOI] [PubMed] [Google Scholar]
- 37.Yee A R, Kronstad J W. Construction of chimeric alleles with altered specificity at the b incompatibility locus of Ustilago maydis. Proc Natl Acad Sci USA. 1993;90:664–668. doi: 10.1073/pnas.90.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yue C, Osier M, Novotny C P, Ullrich R C. The specificity determinant of the Y mating-type proteins of Schizophyllum commune is also essential for Y-Z protein binding. Genetics. 1997;145:253–260. doi: 10.1093/genetics/145.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zurek D M, Mou B, Beecher B, McClure B. Exchanging sequence domains between S-RNases from Nicotiana alata disrupts pollen recognition. Plant J. 1997;11:797–808. doi: 10.1046/j.1365-313x.1997.11040797.x. [DOI] [PubMed] [Google Scholar]