Abstract
We have compared the ability of two mammalian Notch homologs, mouse Notch1 and Notch2, to inhibit the granulocytic differentiation of 32D myeloid progenitor cells. 32D cells undergo granulocytic differentiation when stimulated with either granulocyte colony-stimulating factor (G-CSF) or granulocyte-macrophage colony-stimulating factor (GM-CSF). Expression of the activated intracellular domain of Notch1 inhibits the differentiation induced by G-CSF but not by GM-CSF; conversely, the corresponding domain of Notch2 inhibits differentiation in response to GM-CSF but not to G-CSF. The region immediately C-terminal to the cdc10 domain of Notch confers cytokine specificity on the cdc10 domain. The cytokine response patterns of Notch1 and Notch2 are transferred with this region, which we have termed the Notch cytokine response (NCR) region. The NCR region is also associated with differences in posttranslational modification and subcellular localization of the different Notch molecules. These findings suggest that the multiple forms of Notch found in mammals have structural differences that allow their function to be modulated by specific differentiation signals.
Hematopoiesis can be considered a developmental process in which pluripotent stem cells give rise to committed progeny that undergo proliferation and differentiation, resulting in the continuous production of appropriate numbers of mature blood cells throughout the lifetime of a vertebrate organism (for reviews, see references 33 and 38). Considerable progress has been made in understanding the regulation of hematopoiesis, including the effects of and interactions among cytokines and the interactions of progenitors with stromal elements (for reviews, see references 23 and 30). Despite these advances, many aspects of hematopoiesis remain obscure, including the mechanisms by which multipotent progenitors choose to differentiate along one of multiple pathways or to self-renew and remain multipotent. In other developmental systems, cell fate decisions by multipotent progenitors are mediated by members of the Notch family (for reviews, see references 2, 9, and 46). We have previously demonstrated the expression of Notch genes in hematopoietic progenitors (27) and the functional activity of Notch1 in 32D myeloid progenitors (20, 26) and have proposed that members of this receptor family play a similar role in the determination of hematopoietic cell fates.
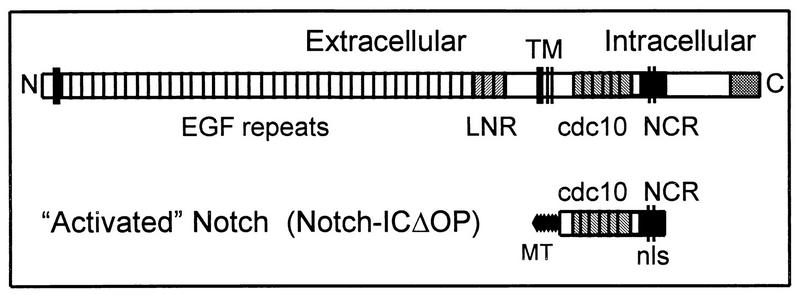
The general function of Notch as a mediator of cell fate decisions has been highly conserved throughout evolution (for a review, see reference 1), and this is reflected in the conservation of its molecular structure (Figure 1). The extracellular domain of Notch, which contains 33 to 36 tandem epidermal growth factor repeats and three lin-12/Notch repeats (LNR), functions as a receptor in cell-cell interactions. There is a single transmembrane domain. The intracellular domain contains six cdc10/SWI6/ankyrin repeats, putative nuclear localization signals (NLS), and a C-terminal OPA/PEST region. The cdc10 region is the most highly conserved portion of the molecule and is crucial for intracellular signal transduction (6, 10, 15, 21, 32, 34, 36, 40). Activation of the Notch molecule by ligand (DSL proteins, such as Delta, Serrate, and Lag-2) binding to the extracellular domain inhibits differentiation along a specific cell fate pathway in response to inductive signals (5, 20, 22). Thus, among a group of cells having equivalent cell fate potentials, a limited number of cells will adopt the specific cell fate while others (those expressing higher levels of Notch) will remain multipotent and competent to subsequently adopt an alternate cell fate (for reviews, see references 1 and 13).
FIG. 1.
Diagram of the full-length Notch molecule and the activated intracellular Notch construct. Both Notch1 and Notch2 consist of an extracellular domain containing 36 epidermal growth factor (EGF) repeats and 3 LNR; there is a single transmembrane domain (TM). The intracellular domain contains six cdc10/ankyrin repeats (cdc10) and the newly defined NCR region, which contains a putative bipartite NLS. The activated Notch constructs (Notch-ICΔOP) consist of the region of the intracellular domain including the cdc10 repeats and the NCR region. Constructs also encode N-terminal myc epitope tags (MT) to facilitate detection of protein expression.
Evaluation of the phenotypic effects of mutant Notch molecules in several different systems has helped elucidate the functions of different parts of the Notch molecule (21, 34, 40). These studies have demonstrated that truncated Notch molecules lacking most or all of the extracellular domain behave as constitutively activated forms of Notch (10, 21, 40). Thus, expression of only the intracellular domain (or a portion of the intracellular domain) results in effects comparable to those observed or expected from unregulated or continuous Notch activation through ligand binding. We have previously demonstrated that expression of a truncated intracellular form of mNotch1 (as illustrated in Fig. 1) in 32D myeloid cells inhibits granulocyte colony-stimulating factor (G-CSF)-induced granulocytic differentiation and permits the expansion of undifferentiated progenitors, effects consistent with Notch activity in other systems (26). More recently, we have demonstrated that activation of full-length mNotch1 by the Notch ligand, Jagged1, results in the same functional effects on 32D differentiation (20). These findings suggest that signaling through the Notch pathway may function in hematopoiesis to regulate cell fate decisions and to maintain progenitor populations.
In contrast to Drosophila, in which a single Notch molecule mediates a variety of cell fate decisions during the development of different tissues (2, 4, 7, 31, 37), mammals express at least four distinct Notch genes (11, 18, 41, 44, 45). These individual Notch molecules have both overlapping and distinct patterns of expression, but differences in function, if any, have not been characterized. In Caenorhabditis elegans, two Notch homologs, lin-12 and glp-1, are expressed in different types of progenitors and mediate the cell fate determination of vulval or germ line cells, respectively (3, 39, 46); however, these two Notch homologs appear to be functionally interchangeable (8, 36). We have found that Notch1, Notch2, and Notch3 are all expressed in hematopoietic progenitors, including the mouse myeloid progenitor cell lines 32D and FDCP mixA4 (reference 26 and unpublished observations). In preliminary studies to compare Notch1 and Notch2 function in hematopoietic cells, we found that the activated intracellular form of Notch1, but not Notch2, inhibited G-CSF-induced differentiation of 32D myeloid cells (26). This observation suggested that Notch1 and Notch2 might have distinct functions in hematopoietic differentiation.
In the studies presented here, we show that activated forms of Notch1 and Notch2, when constitutively expressed in 32D myeloid progenitors, have effects that depend on the specific cytokine inductive signal. Activated Notch1 specifically inhibits differentiation in response to G-CSF, whereas Notch2 inhibits differentiation only in response to granulocyte-macrophage colony-stimulating factor (GM-CSF). In addition, we provide evidence that a previously uncharacterized region, termed the Notch cytokine response (NCR) region, modulates these specific effects. We also show that the NCR region is associated with different posttranslational modifications and subcellular localizations of Notch1 and Notch2 molecules, supporting the conclusion that structural differences between the Notch1 and Notch2 NCR regions contribute to functional specificity in this system. We propose a model through which the NCR region could modulate the activity of Notch1 and Notch2 in response to different cytokines.
MATERIALS AND METHODS
Preparation of retroviral vectors containing Notch constructs.
The N1-ICΔOP and N2-ICΔOP fragments were cloned into the pLXSN retroviral vector as described elsewhere (26). Briefly, the intracellular Notch1 and Notch2 regions containing the cdc10 and the NCR region (starting 55 amino acids N-terminal to the cdc10 region and ending 23 amino acids C-terminal to the putative NLS) were cloned into the pCS2+6MT vector in frame with the six myc epitope tags (6MT) in the vector. A fragment containing the Notch-ICΔOP and the MT (ClaI-XhoI fragment) was subcloned into a pLXSN (25) vector modified to contain a ClaI site.
The N1-CDC/N2-NCR hybrid molecule contains the Notch1 cdc10 region (TKKFRF to LLDEYN) and the Notch2 NCR region (VTPSPP to PVDSLE). The Notch2 NCR fragment was PCR amplified and cloned in the EcoRV-XhoI sites of N1-ICΔOP/pCS2+6MT, replacing the Notch1 NCR region. A similar strategy was used to make the N2-CDC/N1-NCR hybrid molecule. In this case, the Notch1 NCR fragment (corresponding to LVRSPQ to PVDSLE) was PCR amplified and cloned in the XmaI-XhoI sites of N2ICΔOP/pCS2+6MT, replacing N2-NCR. Nucleotide changes were made in the 5′ primer of the Notch1 NCR fragment, so that both hybrid molecules exchange the NCR fragment at the equivalent amino acid. N1cdcIR/N2NLR, N1ΔIR, and N1ΔNLR were generated by a similar PCR strategy. The constructs all contained the myc epitope tag (6MT) from the pCS2+6MT vector and were subcloned into the ClaI-modified pLXSN retroviral vector. The correct nucleotide sequences of all constructs were verified by sequencing.
Retroviral transductions.
Retroviral producer cell lines were established as previously described (25). Briefly, retroviral vectors were transfected into the ecotropic viral packaging cell line, PE501, by calcium phosphate precipitation, and the supernatant containing the transiently expressed virus was used to infect the amphotropic viral packaging cell line, PA317. G418-resistant clones were assayed by reverse transcriptase PCR (RT-PCR) and/or Western blotting for expression of the constructs. 32D cells were transduced by a 24-h cocultivation with PA317 cells in the transwell system or by direct incubation with the transfected PE501 cell supernatant. In both cases, 4 μg of Polybrene per ml was added to the media. After 24 to 48 h, the cells were plated in 1% methylcellulose with 10% fetal bovine serum (FBS), 10% WEHI 3B conditioned medium (WCM), and 1 mg of G418 per ml. Resistant colonies were expanded and screened for construct expression by RT-PCR and/or Western blotting. The Notch1 deletion mutants (N1DIR and N1DNLR) and N1cdcIR/N2NLR constructs were electroporated into 32D cells (260 V and 960 μF) rather than retrovirally transduced. G418-resistant cells were selected, expanded, and screened as above.
Cell cultures.
32D cells were maintained in Iscove’s modified Dulbecco’s medium with 10% fetal bovine serum and 10% WCM as a source of interleukin-3 (IL-3). The cells were induced to differentiate as described previously (26), with minor modifications. Briefly, the day before the experiment, 32D cell cultures were split to constant density and fed with fresh medium to ensure similar log-phase growth for all clones. On day −1, the cells were washed and replated at constant density (3 × 105 cells/ml) in Iscove’s modified Dulbecco’s medium containing 10% fetal bovine serum and 10 ng of recombinant human G-CSF (Amgen, Thousand Oaks, Calif.) per ml. Priming the cells in G-CSF for 17 to 20 h upregulates GM-CSF receptors (17) and improves the survival of 32D clones when cultured in GM-CSF. The cells were then washed, recounted, and plated at 2 × 105/ml (six-well plates; 4 ml/well) in differentiation media containing 10 ng of G-CSF or GM-CSF (Pharmingen, San Diego, Calif.) per ml; this point was considered day 0. The cultures were evaluated daily for the total number of viable cells and the relative percentages of undifferentiated cells and mature granulocytes. In all cultures, 10% of the medium was replaced every day. Viable cells were counted, and Wright-stained cytospin preparations were evaluated for granulocytic differentiation. The criteria for differentiation included nuclear segmentation, an increased cytoplasm/nucleus ratio, and increased eosinophilia and granularity of the cytoplasm. Considerable care was taken to validate accurate differential counts. All differential counts were done on 100 to 200 cells on several occasions by the same individual (A.B.) to ensure consistency; the results were confirmed by two other independent observers in a blinded fashion.
Immunofluorescent staining and confocal imaging.
Immunofluorescent staining of 32D cells was performed in 96-well plates as follows: the cells were harvested, washed in phosphate-buffered saline (PBS), fixed with 3% paraformaldehyde for 30 min on ice, washed three times with cold PBS–5% normal goat serum (NGS), permeabilized with 0.1% Triton X-100–PBS–NGS, washed three times, and blocked with FC Block (10 μg/ml; Pharmingen) for 30 min before the primary antibody (anti-myc tag 9e10 or isotype control; 2 μg/ml) was added; the cells were incubated overnight at 4°C, washed three times with PBS–2% NGS, and incubated with the secondary antibody (fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G; 10 μg/ml) on ice in the dark for 30 min; propidium iodide was added to 2 μg/ml for 5 min; and the cells were washed three times with PBS–2% NGS and once with PBS and mounted on slides with Vectashield (Vector Labs). The cells were visualized with a Bio-Rad MRC 600 laser scanning confocal microscope with COSMOS software (Bio-Rad) installed for digital analysis. The images were combined for illustration with Adobe Photoshop and Windows Powerpoint software.
Western blot analysis.
Whole-cell lysates prepared from 32D cells were electrophoresed through 4 to 12% gradient polyacrylamide gels (Novex) in the presence of 10% sodium dodocyl sulfate (SDS) under reducing conditions (β-mercaptoethanol). The total amount of protein loaded was adjusted to give bands of comparable intensity (1 to 160 μg). The proteins were electrotransferred from the gels to nitrocellulose membranes, and the membranes were immunoblotted with the anti-myc antibody (9e10; 2 μg/ml) and visualized by chemiluminescence with ECL reagents as previously described (26).
RESULTS
32D myeloid progenitor cells differentiate in response to either G-CSF or GM-CSF.
The myeloid progenitor cell line, 32D Cl 3, is an IL-3-dependent cell line derived from mouse bone marrow cultures (12, 42). When maintained in IL-3, 32D cells proliferate as undifferentiated blasts with an approximate doubling time of 17 h. However, they can also be induced to undergo myeloid differentiation and thus have been widely used for the study of hematopoietic differentiation (14, 17). Although 32D cells are used primarily to study G-CSF-induced differentiation, these cells, or subclones of these cells, also have the capacity to differentiate in response to other cytokines (17, 24).
We have used 32D cells as a model system to study the effects of Notch expression on myeloid cell differentiation. In our previous studies, we found that expression of an activated form of Notch1 in 32D cells inhibited granulocytic differentiation in response to G-CSF (26). In those studies, we also noted that expression of the comparable form of Notch2 did not have the same inhibitory effect. To further explore this apparent difference in function between Notch1 and Notch2, we asked whether the expression of activated forms of Notch1 and Notch2 would have the same or different effects on the differentiation of 32D cells in response to other cytokines.
Since not all 32D cells have the capacity to respond to cytokines other than G-CSF, we first evaluated differentiation of the parental 32D cells in the presence of various cytokines. The growth and differentiation characteristics of 32D cells cultured in the presence of G-CSF or GM-CSF are shown in Fig. 2. Differentiation was induced and assessed essentially as previously described (26) (see Materials and Methods). Cells were plated at constant density in media containing 10 ng of G-CSF or GM-CSF per ml and evaluated daily for the total number of viable cells and characteristics of granulocytic differentiation. To permit adequate cell survival for evaluation of the effects of GM-CSF, the cells were primed in G-CSF before being replated in media containing GM-CSF (see Materials and Methods); priming in G-CSF has previously been shown to upregulate GM-CSF receptors and improve cell survival in the presence of GM-CSF (17).
FIG. 2.
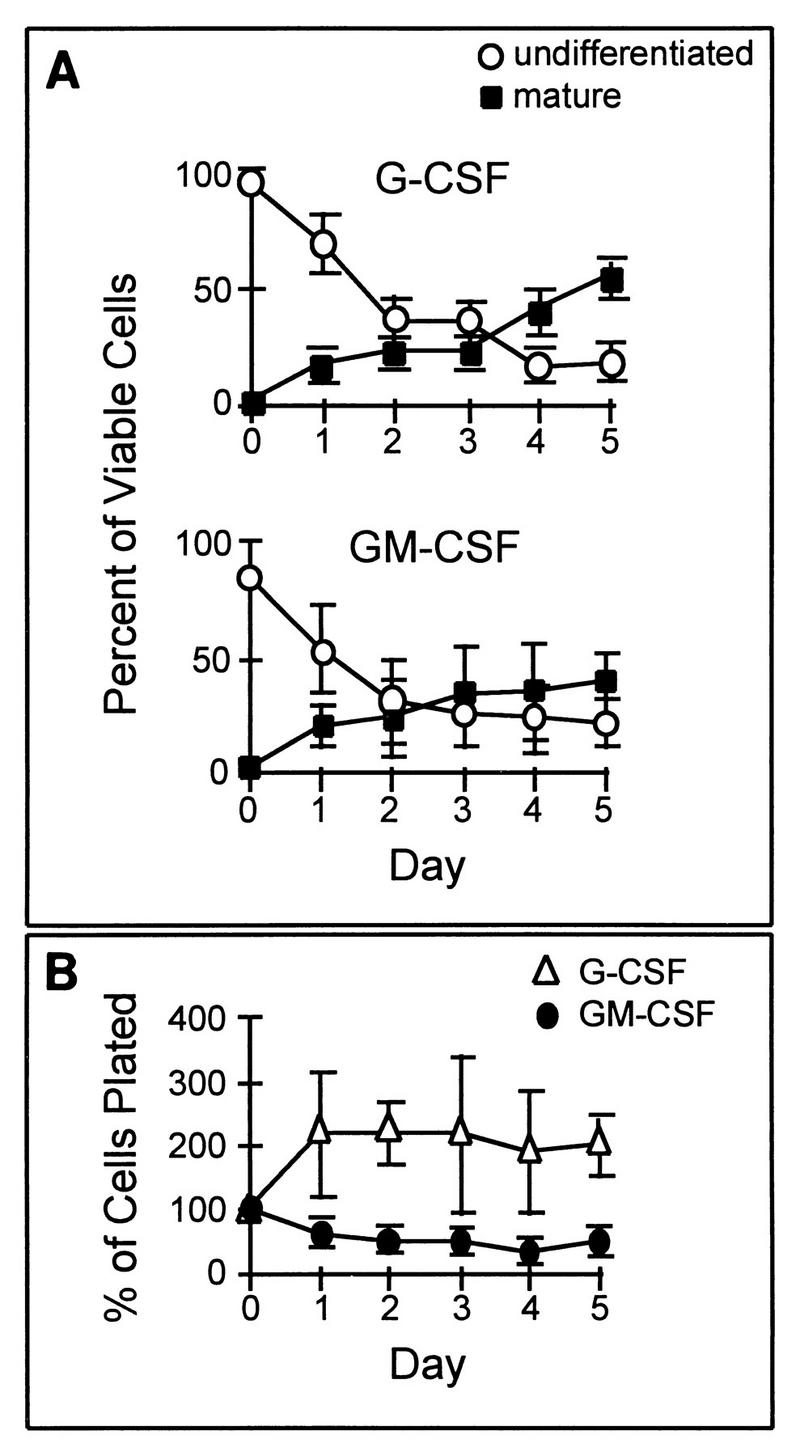
Growth and differentiation characteristics of parental 32D Cl3 cells in the presence of G-CSF or GM-CSF. (A) Granulocytic differentiation in response to G-CSF (upper graph) and to GM-CSF (lower graph) is illustrated by plotting the relative percentage of viable cells maintaining an undifferentiated blast morphology or having attained a terminally differentiated (bands and segmented neutrophils) mature phenotype after successive days in culture. In the presence of either cytokine, there is a continuous fall in the proportion of undifferentiated cells and a concomitant rise in the proportion of differentiated cells. (B) The total number of cells, relative to the original number of cells plated, present after successive days of culture in G-CSF or GM-CSF is shown. There is a slight increase in the cell number in G-CSF and a slight decline in GM-CSF. In all three graphs, the values shown are the averages of three independent experiments; error bars denote standard errors of the mean (SEM).
Granulocytic differentiation of parental 32D cells in response to G-CSF and GM-CSF stimulation is compared in Fig. 2A. Differential cell counts on Wright-stained cytospin preparations were used to separate cells into three general categories: (i) undifferentiated (blasts), (ii) mature (bands and segmented granulocytes), and (iii) intermediate (myelocytes, metamyelocytes, and undetermined). Cells in the intermediate group were excluded from the analysis presented in Fig. 2. Undifferentiated 32D cells generally have a single large, relatively round nucleus and scant dark blue cytoplasm containing few granules. Criteria for granulocytic differentiation included nuclear segmentation, an increased cytoplasm/nucleus ratio, and increased eosinophilia and granularity of the cytoplasm. As illustrated in Fig. 2A, parental 32D cells differentiate in a similar temporal pattern and to a comparable extent in response to G-CSF and GM-CSF. After 5 days, 39 ± 13% of the 32D cells in GM-CSF had attained a mature granulocytic morphology compared to 54 ± 18% of the cells in G-CSF. Less than 25% of the cells remained undifferentiated in the presence of either cytokine.
The effects of G-CSF and GM-CSF on 32D cell proliferation are compared in Fig. 2B. In GM-CSF, 32D cells did not proliferate, and by day 5, cultures contained approximately half of the original number of cells plated. Cells cultured in G-CSF showed an initial proliferation and then stabilized, so that after 5 days in culture they had approximately twice the original number of cells plated. Proliferation was minimal compared to that of cells maintained in IL-3, which generally results in approximately a 30-fold increase in cell number after 5 days (26). We conclude that parental 32D cells normally differentiate to a comparable degree in response to stimulation with G-CSF or GM-CSF and that neither cytokine has a significant proliferative effect.
Expression of the activated intracellular domain of Notch2, but not Notch1, inhibits GM-CSF-induced differentiation.
Truncated intracellular Notch molecules containing the cdc10 repeats and NLS have been shown to behave as constitutively activated forms of Notch in a number of different systems (6, 15, 32, 40). We previously demonstrated that expression of a truncated intracellular form of Notch1 in the 32D myeloid progenitor cell line inhibits G-CSF-induced differentiation but permits the continued proliferation of undifferentiated cells (26), effects consistent with those of Notch activation in other systems. However, in those studies, the corresponding region of the Notch2 molecule did not have any inhibitory effect on differentiation induced by G-CSF, nor did it permit continued proliferation. This result suggested that the Notch1 and Notch2 molecules might have different functions in hematopoietic cells. Since 32D cells will undergo granulocytic differentiation in response to GM-CSF, we compared the effects of expression of the activated Notch1 and Notch2 molecules on differentiation in response to G-CSF and GM-CSF.
We evaluated G-CSF and GM-CSF-induced differentiation of parental 32D cells (32D WT), 32D cells expressing activated forms of mNotch1 (N1-ICΔOP) and mNotch2 (N2-ICΔOP), and cells containing control retroviral constructs expressing only the myc epitope tag (LXSN-MT). Addition of 10 ng of GM-CSF per ml induced differentiation of the cells in the 32D WT cultures (Fig. 2) and the LXSN-MT and N1-ICΔOP cultures (Fig. 3). By day 5, 40 to 60% of the cells in these cultures had attained a mature granulocytic phenotype and less than 10% remained undifferentiated. In contrast, 50 to 60% of the cells expressing Notch2 retained an undifferentiated blast morphology and less than 10% had attained a mature granulocytic phenotype after 5 days (Fig. 3). As we described previously (26) and as shown in Fig. 3, the same clones showed the opposite phenotypes when stimulated with G-CSF. After 5 days of culture in 10 ng of G-CSF per ml, 50 to 60% of the cells present in 32D WT cultures (Fig. 2) and LXSN-MT and N2-ICΔOP cultures (Fig. 3) showed a differentiated granulocytic morphology, while the N1-ICΔOP clones predominantly maintained an undifferentiated blast morphology (50 to 60%) and very few (<5%) of the cells were terminally differentiated. Thus, while parental 32D cells and control clones demonstrate the capacity to differentiate in response to either G-CSF or GM-CSF, 32D cells expressing activated Notch1 differentiate in response to GM-CSF but not to G-CSF whereas 32D cells expressing activated Notch2 differentiate in response to G-CSF but not to GM-CSF. Therefore, we conclude that the expression of Notch1 specifically inhibits G-CSF-induced differentiation and the expression of Notch2 specifically inhibits GM-CSF-induced granulocytic differentiation of 32D cells.
FIG. 3.
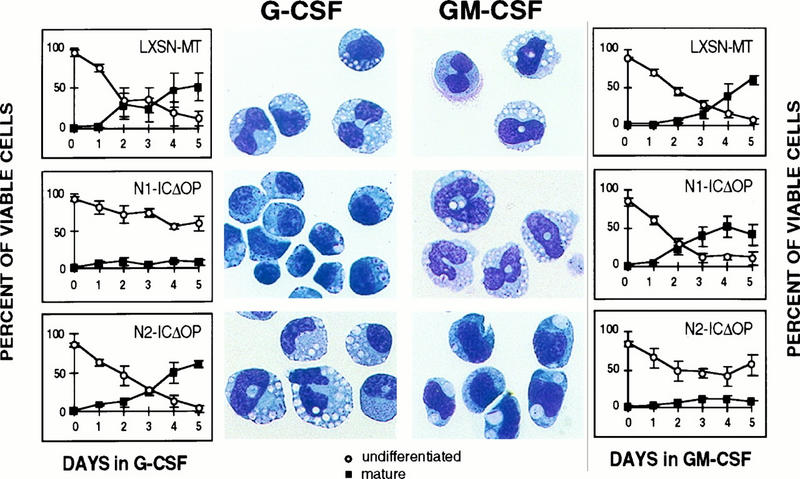
Differentiation of 32D cells expressing activated Notch1 (N1-ICΔOP) or Notch2 (N2-ICΔOP) molecules in response to either G-CSF or GM-CSF stimulation. The percentages of viable cells that are either undifferentiated or differentiated after successive days in culture in G-CSF (left) or GM-CSF (right) are shown in the graphs, and photomicrographs of Wright-stained cells from cultures on the final day are shown beside the corresponding graph. Control clones (LXSN-MT) are shown for comparison. As previously demonstrated (26), Notch1, but not Notch2, inhibits the differentiation induced by G-CSF. The converse effect is noted when the cells are induced with GM-CSF: Notch2 inhibits differentiation, but Notch1 does not. A representative experiment is shown; graphs represent the averages of results obtained with two (LXSN-MT) or three (N1-ICΔOP and N2-ICΔOP) clones, with error bars representing SEM. The same clones were used for the G-CSF and GM-CSF cultures. In this experiment, 1% WCM was included in the medium for G-CSF-induced differentiation to improve the uniformity of cell survival; we have previously reported that the addition of 1% WCM does not interfere with G-CSF-induced differentiation (26).
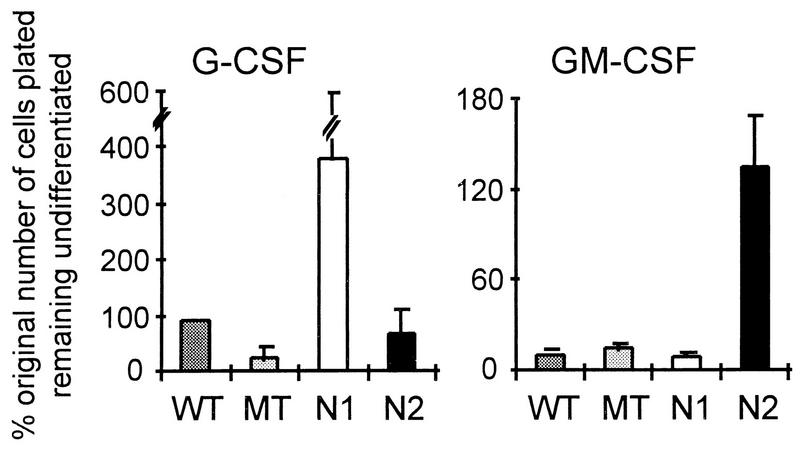
In addition to determining the relative proportion of differentiated and undifferentiated cells, we evaluated the total number of undifferentiated cells remaining after culture for successive days in G-CSF or GM-CSF. Figure 4 compares the total number of undifferentiated cells (expressed as a percentage of original number of cells plated) present after 3 to 5 days of stimulation with G-CSF or with GM-CSF. In cultures stimulated with G-CSF, significantly greater numbers of undifferentiated cells were present in the cultures containing activated Notch1-expressing 32D cells than in cultures of parental, control MT-expressing, or activated Notch2-expressing cells. Parental 32D cultures showed somewhat greater proliferation than did control (MT) clones, probably because of the heterogeneous nature of this population; individual 32D subclones have growth characteristics comparable to those of the control MT transduced clones (data not shown). While the N2 clones showed slightly greater proliferation than the MT clones, this difference was not statistically significant. In contrast to the G-CSF cultures, for the cultures stimulated with GM-CSF, those containing activated Notch2-expressing 32D cells contained significantly more undifferentiated cells than did any of the other cultures. Furthermore, only the Notch1 G-CSF and Notch2 GM-CSF cultures contained more undifferentiated cells than were present in the original cultures. When individual clones were evaluated for baseline proliferation rates in IL-3 (10% WCM), we found no significant differences among control MT, N1-ICΔOP, and N2-ICΔOP groups (although there was some clonal variation); each of the individual clones showed greater proliferation when cultured in the presence of IL-3 than in the presence of either G-CSF or GM-CSF (data not shown). Thus, expression of activated Notch1 or Notch2 does not stimulate proliferation in response to G-CSF or GM-CSF, respectively, but, rather, permits survival and continued cell division in the absence of differentiation.
FIG. 4.
Effects of Notch1 and Notch2 activity on the maintenance of undifferentiated 32D cells in the presence of G-CSF or GM-CSF stimulation. Parental 32D cells (WT) and individual clones transduced with a control myc tag (MT) retroviral construct or activated forms of Notch1 (N1; mNotch1-ICΔOP) or Notch2 (N2; mNotch2-ICΔOP) were evaluated for proliferation and differentiation in the presence of 10 ng of G-CSF or GM-CSF per ml. The total number of cells remaining undifferentiated on day 5 (or the last day when at least 40% of the original number of cells were still viable) is expressed relative to the original number of cells plated. The G-CSF graph represents a single experiment involving three independent clones for each construct; the results were comparable to those reported previously (26). The GM-CSF graph represents combined data from four experiments involving the same three independent clones for each construct. Error bars represent the SEM. Note that different scales are used in the two graphs, because of the lower overall proliferation of 32D cells in GM-CSF (Fig. 2).
A region C-terminal to the cdc10 repeats is responsible for the different effects of Notch1 and Notch2 on 32D myeloid cell differentiation.
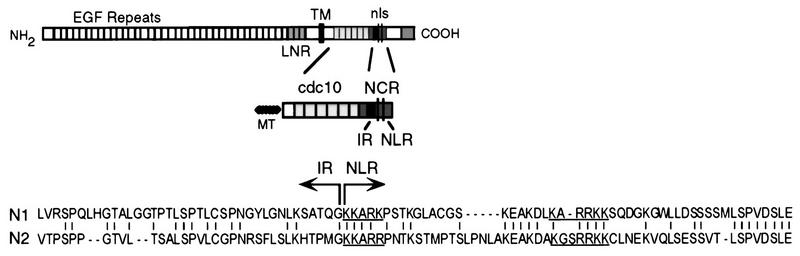
We next asked which part of the truncated Notch molecule is responsible for conferring the cytokine-specific effects of Notch1 and Notch2. The activated Notch1 and Notch2 molecules used in the studies described above contain, in addition to the cdc10 repeats, the adjacent C-terminal region (the NCR region [Fig. 1 and 5]). The cdc10 repeats of the two Notch molecules have a high degree of overall similarity (70% amino acid identity). However, there is a variable degree of similarity among the individual repeats, ranging from 45% identity for repeat 1 to 85 to 88% for repeats 3, 4, 5, and 6. The NCR regions of the two molecules have approximately 50% identity in amino acid sequence. Figure 5 shows the amino acid sequence alignment of the Notch1 and Notch2 NCR regions. To determine if the cdc10 domain or the NCR region or both were responsible for the specific effects of Notch1 and Notch2, we generated hybrid Notch1/Notch2 molecules in which these two regions were exchanged. These reciprocal Notch hybrid molecules, referred to as N1CDC/N2NCR and N2CDC/N1NCR, are represented in Fig. 6. We derived 32D clones expressing each of the hybrid molecules, confirmed their expression by Western blotting, and then evaluated their differentiation compared to that of clones expressing the activated Notch1 and Notch2 (N1-ICΔOP and N2-ICΔOP) molecules.
FIG. 5.
NCR region. The NCR region is shown as part of the full-length (top) and activated (middle) Notch molecules. At the bottom, the amino acid sequences of the Notch1 and Notch2 (N1 and N2) molecules are compared and the demarcation of the IR and NLR of the NCR is denoted. The putative NLS are underlined.
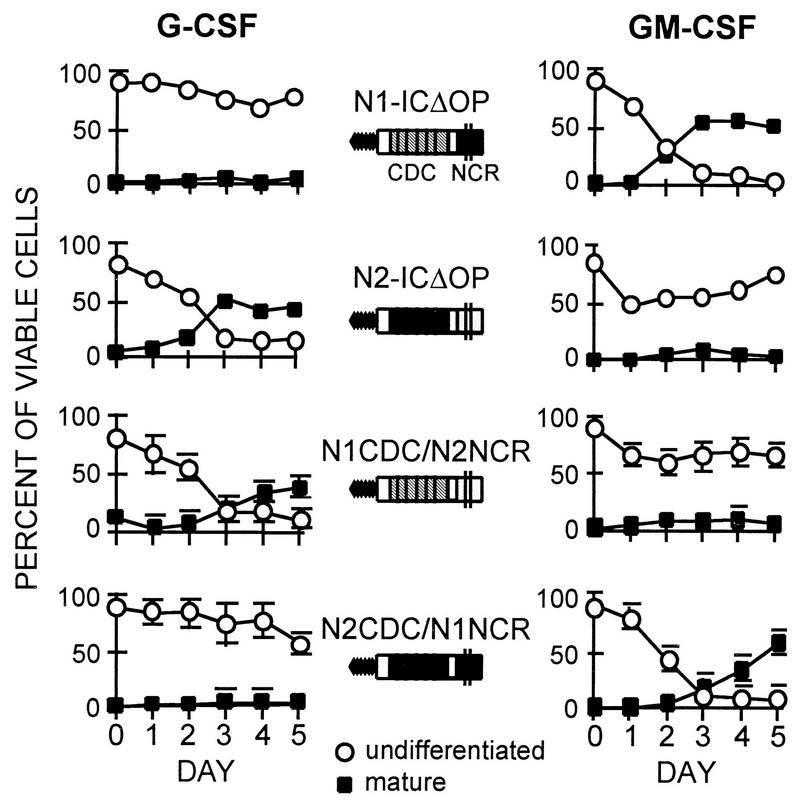
FIG. 6.
Differentiation induced by G-CSF (left) and GM-CSF (right) in 32D cell lines expressing N1CDC/N2NCR and N2CDC/N1NCR hybrid molecules, compared to 32D cell lines expressing the activated Notch1 (N1-ICΔOP) or Notch2 (N2-ICΔOP) proteins. Graphs show the relative percentages of viable differentiated and undifferentiated cells present in the cultures on successive days. Inhibition of differentiation in G-CSF occurs when the expressed Notch molecule contains the NCR region from Notch1. In GM-CSF, differentiation is inhibited when the NCR region is derived from Notch2. The same clones were used in the G-CSF and GM-CSF experiments. Values in the G-CSF graphs each represent the average for three different clones expressing N1-CDC/N2-NCR or N2-CDC/N1-NCR, with error bars denoting SEM. Values in the GM-CSF graphs represent the average for two of the three clones for each construct; the third clones did not survive in GM-CSF. Representative clones expressing N1-ICΔOP and N2-ICΔOP were used in this experiment (see Fig. 3 for more extensive data on N1-ICΔOP and N2-ICΔOP).
Figure 6 shows the effects of expression of the Notch1/Notch2 hybrid constructs on differentiation. As shown above (Fig. 3), 32D cells expressing control constructs differentiate in response to either G-CSF or GM-CSF; after 5 days, these cultures contained less than 10% undifferentiated cells and had 40 to 50% mature cells. The expression of activated Notch1 or Notch2 selectively inhibited differentiation in response to G-CSF or GM-CSF, respectively, as also described above (Fig. 3). 32D clones expressing the N2CDC/N1NCR hybrid molecule displayed a differentiation pattern comparable to that of cells expressing activated Notch1 (N1-ICΔOP): inhibition of differentiation in response to G-CSF but not to GM-CSF. Expression of the reciprocal hybrid molecule, N1CDC/N2NCR, produced the converse effects: differentiation was inhibited in the presence of GM-CSF but not G-CSF, the same effects as were observed with expression of the activated Notch2 molecule (N2-ICΔOP). These results suggest that the NCR region modulates the specific functional effects of Notch1 and Notch2 in cells stimulated with different cytokines. In this system, the cdc10 domain, while required for Notch activity, does not confer specificity to the effects of Notch1 and Notch2.
To further define the functional effects associated with the NCR region, we have derived additional hybrid and mutant Notch molecules. As shown in Fig. 5, the NCR region can be subdivided into an intermediate region (IR) and nuclear localization region (NLR), the latter of which contains the putative bipartite NLS. Analysis of the effects of NCR deletion mutants of Notch1 on G-CSF-induced differentiation, compared to the effects of Notch1, Notch2, and the N1/N2 CDC/NCR hybrid molecules, is shown in Fig. 7. Notch1 molecules containing a deletion of either the IR (N1ΔIR) or the NLR (N1ΔNLR) portion of the NCR were inactive: 32D cells expressing these molecules differentiated normally in response to G-CSF (Fig. 7, bottom row, panels 5 and 6). However, replacement of the NLR with the Notch2 NLR restored activity, as demonstrated by the inhibition of differentiation of 32D cells expressing this construct (N1cdcIR/N2NLR; Fig. 7, right-hand panel). These findings confirm that the NCR region of Notch1 is required for functional activity in this system. They further suggest that the presence of both the IR and NLR is necessary for function but that it is the IR that confers cytokine specificity.
FIG. 7.
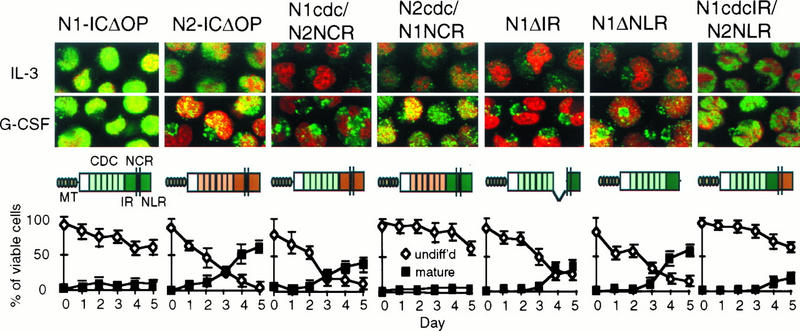
Correlation of subcellular localization and activity of Notch molecules in 32D cells stimulated with G-CSF. 32D cells transduced with the Notch1/Notch2 constructs indicated were evaluated by immunofluorescent staining and confocal microscopy for subcellular distribution of the Notch construct when the cells were grown in IL-3 or after 48 h in G-CSF. The cells were doubly stained with the nuclear stain propidium iodide (PI, red) and fluorescein isothiocyanate (green) to detect the myc epitope tags. Immunostained cells were visualized by confocal microscopy, and digital images (magnification, ×60) were reproduced and combined with Adobe Photoshop software. Below each set of micrographs is the corresponding graph of Notch activity, showing the percentages of cells remaining undifferentiated or differentiating into mature granulocytes over successive days in culture with G-CSF. Abbreviations for the Notch constructs are described in the text.
The NCR region modulates subcellular localization and electrophoretic mobility of Notch1 and Notch2 molecules.
In our previous studies evaluating expression of the activated Notch1 construct in 32D cells, we observed primary localization of the construct to the nucleus. While the corresponding Notch2 construct also showed some nuclear localization, the staining pattern was consistently more diffuse throughout the cells and was less intensely nuclear, despite comparable protein expression by Western blotting (26). We therefore asked whether there was any difference in the subcellular localization of the Notch1/Notch2 hybrid molecules, whether the subcellular localization changed with cytokine induction, and whether there was any correlation between subcellular localization and activity. We used confocal microscopy to visualize 32D cells doubly stained with the nuclear stain propidium iodide and a myc tag antibody (9e10) to detect construct expression. The four left-hand panels of Fig. 7 show 32D cells expressing the activated Notch1, Notch2, N1cdc/N2NCR, and N2cdc/N1NCR hybrid molecules cultured in IL-3 and after 48 h in G-CSF. Graphs illustrating functional activity in the context of G-CSF stimulation are shown below each set of images. The native activated Notch1 construct (N1-ICΔOP) showed intense nuclear staining, and the corresponding Notch2 construct showed mixed nuclear and cytoplasmic staining as noted previously. The N1cdc/N2NCR construct, which was inactive in G-CSF, showed only cytoplasmic staining. However, the N2cdc/N1NCR construct, which was active (inhibited differentiation) in the context of G-CSF, showed significant nuclear staining, which increased with G-CSF stimulation.
To further explore the potential correlation between the Notch1 NCR region, nuclear localization, and functional activity in G-CSF, we evaluated the subcellular expression and activity of three additional Notch constructs, as shown in the three right-hand panels of Fig. 7. Notch1 molecules lacking either the IR (N1ΔIR) or the NLR (N1ΔNLR) of the NCR (Fig. 5) were inactive and showed little or no nuclear localization. In contrast, a Notch molecule containing the Notch1 cdc10 and IR but the Notch2 NLR (N1cdcIR/N2NLR) was active and localized predominantly to the nucleus. Together, these findings suggest that Notch1 inhibition of G-CSF-induced differentiation is associated with nuclear localization, that both the IR and NLR portions of the NCR are required for functional activity, and that both the IR and the NLR are associated with subcellular trafficking in this system.
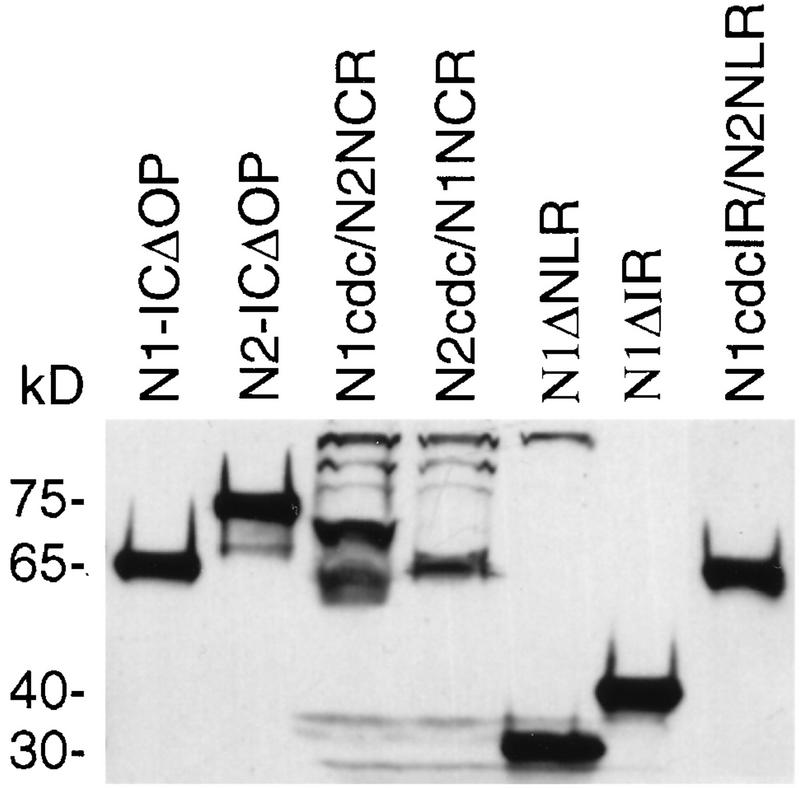
When 32D cells transduced with the various Notch1 and Notch2 constructs were evaluated for construct expression by Western blotting, the Notch1 and Notch2 molecules appeared to be of different sizes, despite having a nearly identical number of amino acid residues (385 for N1; 389 for N2). The results of Western blot analysis of representative 32D clones expressing different Notch1, Notch2, hybrid, and mutant Notch molecules are shown in Fig. 8. The N1-ICΔOP molecule runs as a single band corresponding to approximately 65 kDa. The corresponding N2-ICΔOP molecule has a slower mobility, showing a prominent band at approximately 75 kDa. The N2-ICΔOP molecule frequently also shows a minor, faster-migrating band at about 55 kDa, as previously reported (26). The N1cdc/N2NCR hybrid construct has a mobility comparable to that of Notch2, and the N2cdc/N1NCR construct has the same mobility as Notch1, indicating that electrophoretic mobility is associated with the NCR region. The N1cdcIR/N2NLR molecule also has a mobility comparable to that of Notch1, suggesting that the IR portion of the NCR is important in determining electrophoretic mobility. The Notch1 deletion molecules, N1ΔNLR and N1ΔIR, have mobilities corresponding to their smaller sizes, as expected. The differences in electrophoretic mobility through SDS-polyacrylamide gels indicate that Notch1 and Notch2 undergo different posttranslational modification processes in 32D cells. Specifically, the mobility patterns suggest that the Notch2 NCR region is associated with a covalent modification that results in delayed mobility.
FIG. 8.
Western blot analysis of 32D cells expressing Notch1, Notch2, hybrid, and mutant Notch molecules. Whole-cell lysates from 32D cells expressing the indicated Notch constructs were subjected to SDS-polyacrylamide gel electrophoresis, and construct expression was detected by immunoblotting with an anti-myc tag antibody. Molecular masses (in kilodaltons) are shown on the left.
DISCUSSION
Members of the Notch gene family mediate cell fate decisions by multipotent progenitors in several invertebrate and vertebrate systems, and considerable evidence in support of a conserved role for Notch in hematopoietic cell fate determination is emerging. Notch1 is expressed in normal immature hematopoietic progenitors (27), and ligands for Notch are expressed by a subset of fetal liver cells (29) and bone marrow stromal cells (20). We have recently demonstrated that activation of a full-length Notch1 molecule on 32D myeloid progenitors by the Notch ligand Jagged1 on stromal cells results in an inhibition of G-CSF-induced granulocytic differentiation and expansion of undifferentiated cells (20), results comparable to those previously reported with constitutive expression of an activated intracellular form of Notch1 (26). In addition, transgenic expression of an activated intracellular Notch1 molecule influences the CD4/CD8 (35) and αβ/γδ (43) cell fate decisions in T lymphocytes. Because multiple different Notch molecules are expressed in hematopoietic progenitors, we have addressed whether they have distinct functions. In the studies presented here, we evaluated the effects of Notch1 and Notch2 activity on the capacity of 32D myeloid progenitors to differentiate in response to G-CSF and to GM-CSF. We found that while both Notch1 and Notch2 were capable of inhibiting myeloid differentiation, Notch1 did so only in response to G-CSF and Notch2 did so only in response to GM-CSF. This cytokine specificity can be attributed to a previously uncharacterized region, which we have termed the NCR region. In addition, we provide evidence that differences in subcellular localization and posttranslational modification associated with the NCR region also correlate with functional activity. Together, the results presented here suggest that structural differences between the NCR regions of the Notch1 and Notch2 molecules confer functional specificity and contribute to subcellular trafficking in this system.
Different functions for Notch1 and Notch2 in myeloid cell differentiation.
In Drosophila, Notch influences cell fate decisions in numerous different tissues during development, including the nervous system, eye, mesoderm, and oocyte (2, 4, 7, 31, 37). In addition, the product of this single gene functions in different cell types within the same tissue to mediate the specification of different cell fates at specific stages during the formation of these tissues. This is particularly notable in the formation of the compound eye, during which precise temporal and spatial expression of Notch is required for the appropriate specification of photoreceptors, cone cells, and pigment cells that comprise the ommatidia (4, 10). In C. elegans, two different Notch homologs, lin-12 and glp-1, control distinct cell fates during embryonic development (3, 39, 46). lin-12 functions specifically in vulval progenitors, whereas glp-1 functions in germ line cells. Studies with C. elegans have demonstrated that the glp-1 cdc10/ankyrin repeats can compensate for the function of lin-12 if expressed in the appropriate cell (vulval progenitors) (36) and that the glp-1 protein, when expressed under the control of lin-12 regulatory sequences, is capable of rescuing a lin-12 null mutant phenotype (8). These findings have led to the conclusion that the distinct functions of lin-12 and glp-1 in the intact organism are due to differential expression, rather than differences in their molecular structure.
Four different Notch molecules (Notch1 to Notch4) have now been identified in mammals (11, 18, 41, 44, 45). One question raised by the presence of multiple closely related forms of Notch is whether they have structural differences that make them function differently or whether they are simply used in the same way by different cell types. The mammalian Notch homologs have different temporal and spatial patterns of expression in many embryonic and adult tissues (19, 28, 45), and Notch4 expression appears to be restricted to endothelial cells (41), suggesting that differential expression may be an important determinant of Notch function in vertebrate systems. However, there is also considerable overlap in expression in some tissues, suggesting that the molecules may perform distinct functions in the same cell. We have found that multiple different Notch molecules are expressed in multipotent hematopoietic cells, including normal human bone marrow progenitors and the mouse myeloid progenitor cell lines, FDCP mix A4 and 32D (references 26 and 27 and unpublished observations). In the present studies, we show that constitutive expression of an activated form of either of two different Notch molecules, Notch1 and Notch2, influences the differentiation of the same hematopoietic cell type but in response to different cytokines. While both Notch1 and Notch2 have the same general effect (inhibition of differentiation), they are active only in the context of G-CSF and GM-CSF stimulation, respectively. These observations support the hypothesis that different Notch molecules have distinct functions in hematopoietic differentiation and elucidate a potential link between Notch and cytokine signaling. If these findings translate to hematopoietic progenitors in vivo, they raise the intriguing possibility that signaling through Notch pathways directly influences the response of hematopoietic progenitors to the diverse cytokine stimuli encountered in the hematopoietic microenvironment.
The NCR region mediates the cytokine specificity of Notch1 and Notch2.
Our observation that Notch1 and Notch2 were active in the same cell type, but in the context of stimulation by different cytokines, suggested that structural differences between the Notch molecules might be responsible for distinct molecular interactions that influence activity in 32D cells. Several additional lines of evidence support this conclusion and suggest that structural differences in the region adjacent to the cdc10 repeats, which we term the NCR region, are responsible for functional specificity. The activities of hybrid Notch1/Notch2 molecules in the context of G-CSF and GM-CSF stimulation demonstrate that the cytokine-associated specificity of the Notch1 and Notch2 molecules can be transferred with the NCR region. In addition, the activated Notch1 and Notch2 molecules have different electrophoretic mobilities (indicating different posttranslational modification) and different subcellular localization patterns; these characteristics are also transferred with exchange of the NCR regions. Thus, Notch molecules containing the Notch1 NCR region (Notch1 and the N2CDC/N1NCR hybrid molecule) are active in G-CSF, electrophorese as a single band at 65 kDa through SDS-polyacrylamide gels, and localize predominantly to the nucleus, whereas Notch molecules containing the Notch2 NCR region (Notch2 and the N1CDC/N2NCR hybrid) are active in GM-CSF, show two forms on SDS-polyacrylamide gel electrophoresis (a prominent form with slower electrophoretic mobility and a minor form with faster mobility than Notch1), and show more diffuse subcellular localization.
The NCR region consists of 88 to 89 amino acids starting 31 amino acids C-terminal to the cdc10 repeat region. We have further subdivided the NCR region into the IR and the NLR, the latter of which contains the putative NLS (Fig. 5). Mutant Notch1 molecules lacking either the IR or NLR portion of the NCR are inactive in G-CSF, indicating that the presence of both of these regions is required for Notch1 activity. However, a Notch1/Notch2 hybrid molecule containing the cdc10 and IR of Notch1 and the NLR of Notch2 is active in the context of G-CSF stimulation, suggesting that the NLR portion is necessary for function but does not mediate the specificity of Notch1 activity. Thus, it is possible that only the IR portion of the NCR region is required for cytokine specificity and that the NLR portion contributes to subcellular localization. However, since the N1ΔIR mutant also showed a lack of nuclear localization (despite the presence of the NLR and thus the NLS), it appears that the IR also participates in subcellular trafficking. Our findings suggest that nuclear localization is required for Notch1-mediated inhibition of G-CSF-induced differentiation of 32D cells. However, it appears that Notch2 activity may not require nuclear localization; further studies of Notch2 mutant molecules as well as Notch1 and Notch2 mutant molecules in the context of GM-CSF stimulation are in progress to address this question.
Notch molecules as mediators of hematopoietic differentiation: a model for cytokine-specific activity of Notch1 and Notch2.
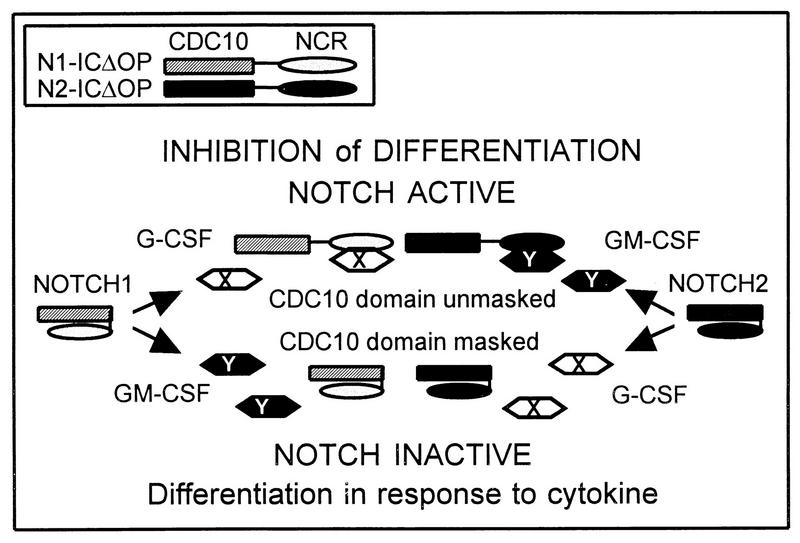
The studies presented here provide evidence that Notch1 and Notch2 have distinct functions that can be attributed to structural differences in the NCR region. We speculate that the NCR region modulates the activity of the cdc10 domain, which previously has been shown to be the effector portion of Notch (6, 10, 15, 21, 32, 34, 36, 40). Figure 9 depicts a model in which the NCR region could modulate Notch activity through posttranslational modifications or conformational changes that affect molecular interactions. For example, when Notch is in an inactive form, the NCR region itself or molecules interacting with the NCR region may mask the cdc10 domain; in the context of specific cytokine induction, the NCR region may interact with molecules involved in the cytokine signaling pathway, resulting in unmasking of the cdc10 domain and permitting Notch activity (inhibition of differentiation). Our observations suggest that the activated forms of Notch1 and Notch2 may interact with distinct molecules involved in different cytokine signal transduction pathways (as indicated by X and Y in Fig. 9). In addition, given the correlation of Notch1 activity with the presence of the Notch1 NCR and with nuclear localization, it is possible that interactions involving the NCR region affect subcellular trafficking, which could also influence functional activity. While our results are not definitive, they suggest that Notch2 activity may not require nuclear localization. Since nuclear localization and posttranslational modification are both associated with the NCR region, it is possible that posttranslational modification of Notch2 in 32D cells prevents nuclear targeting, contributing to a function distinct from that of Notch1 in these cells. A difference in the subcellular localization of Notch1 and Notch2 is particularly intriguing in light of the controversial significance of nuclear localization in other systems, with some studies demonstrating nuclear localization of activated forms of Notch (10, 15, 16, 21, 40) and others demonstrating that Notch homologs have functional activity in the absence of nuclear localization (10, 36).
FIG. 9.
Model of cytokine specificity mediated by the NCR domain. In this model, the cdc10 domain is required for inhibitory activity, but this activity is masked by the NCR region. Cytokine stimulation activates signal transduction pathways, represented by X for G-CSF and Y for GM-CSF. The product of the X pathway is able to dissociate the Notch1 NCR from the cdc10 domain, thereby unmasking its activity, but is unable to act on the Notch2 NCR. Conversely, the product of the Y pathway can dissociate the Notch2 NCR but has no effect on the Notch1 NCR. The result is an inhibition of differentiation which is conditional on both Notch activation and cytokine stimulation.
In contrast to Drosophila development, in which the numerous cell fate decisions mediated by Notch are temporally and/or spatially distinct, diverse signaling molecules coexist in the hematopoietic microenvironment, a variety of hematopoietic cell fates are continuously being determined, and the proportion of cell types produced may change in response to environmental factors. In addition, individual hematopoietic cells simultaneously express multiple cytokine receptors and thereby have the capacity to respond to different signals. Thus, a theoretical need for multiple Notch molecules and potential evolutionary pressure for diversification exists in this system. By influencing the differentiation of hematopoietic progenitors in response to distinct cytokines, the different Notch molecules could provide an important link between cytokine stimulation and cell-cell signaling in hematopoiesis. We would predict that signaling through the Notch pathway in the normal hematopoietic microenvironment is conditional on both activation of Notch by an external signal (Notch ligand on adjacent cells) and specific cytokine stimulation.
Activation of different Notch molecules could inhibit the differentiation of hematopoietic progenitors in response to specific cytokines in a number of different ways: by interacting with molecules specific to different cytokine pathways, by influencing the expression of cytokine receptors, or by regulating the expression of distinct lineage-specific genes. For example, Notch1 may interact with molecules specific to the G-CSF signaling pathway (such as specific JAK/STAT molecules), may downregulate G-CSF receptors and/or upregulate other cytokine receptors, or may inhibit the expression of genes normally induced by G-CSF stimulation. Similarly, Notch2 may interact specifically with molecules induced by GM-CSF, may influence GM-CSF or other cytokine receptors, and/or may regulate the expression of genes induced by GM-CSF. The effects of Notch1 and Notch2 may vary considerably among individual cells, since the effects in a given cell are likely to be influenced by the relative levels of Notch and Notch ligand expressed on neighboring cells as well as the maturational state and the capacity of that cell to express particular gene products. Thus, signaling through Notch receptors may provide a mechanism by which hematopoietic progenitors could communicate with adjacent cells, permitting some cells to differentiate in response to specific inductive signals while inhibiting the differentiation of others, thus regulating the number of mature cells produced while also maintaining a pool of multipotent progenitors.
ACKNOWLEDGMENTS
This work was supported by grants to L.M. from the James S. McDonnell Foundation and the University of Washington Child Health Research Center (NIH grant P30 HD28834), by NIH grant P50 HL54881, and by NIH 5RO1HL48790 to D.M. D.M. is a Scholar of the Leukemia Society of America.
We thank Michele Black and Brian Hall for technical assistance, Irv Bernstein for support of A.B., and Claire Francastel and Barbara Varnum-Finney for helpful discussions.
REFERENCES
- 1.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 2.Artavanis-Tsakonas S, Simpson P. Choosing a cell fate: a view from the Notch locus. Trends Genet. 1991;7:403–408. doi: 10.1016/0168-9525(91)90264-q. [DOI] [PubMed] [Google Scholar]
- 3.Austin J, Kimble J. glp-1 is required in the germ line for regulation of the decision between mitosis and meiosis in C. elegans. Cell. 1987;51:589–599. doi: 10.1016/0092-8674(87)90128-0. [DOI] [PubMed] [Google Scholar]
- 4.Cagan R L, Ready D F. Notch is required for successive cell decisions in the developing Drosophila retina. Genes Dev. 1989;3:1099–1112. doi: 10.1101/gad.3.8.1099. [DOI] [PubMed] [Google Scholar]
- 5.Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- 6.Coffman C R, Skoglund P, Harris W A, Kintner C R. Expression of an extracellular deletion of Xotch diverts cell fate in Xenopus embryos. Cell. 1993;73:659–671. doi: 10.1016/0092-8674(93)90247-n. [DOI] [PubMed] [Google Scholar]
- 7.Corbin V, Michelson A M, Abmayr S M, Neel V, Alcamo E, Maniatis T, Young M W. A role for the Drosophila neurogenic genes in mesoderm differentiation. Cell. 1991;67:311–323. doi: 10.1016/0092-8674(91)90183-y. [DOI] [PubMed] [Google Scholar]
- 8.Fitzgerald K, Greenwald I. Interchangeability of Caenorhabditis elegans DSL proteins and intrinsic signalling activity of their extracellular domains in vivo. Development. 1995;121:4275–4282. doi: 10.1242/dev.121.12.4275. [DOI] [PubMed] [Google Scholar]
- 9.Fortini M E, Artavanis-Tsakonas S. Notch: neurogenesis is only part of the picture. Cell. 1993;75:1245–1247. doi: 10.1016/0092-8674(93)90611-s. [DOI] [PubMed] [Google Scholar]
- 10.Fortini M E, Rebay I, Caron L A, Artavanis-Tsakonas S. An activated Notch receptor blocks cell-fate commitment in the developing Drosophila eye. Nature. 1993;365:555–557. doi: 10.1038/365555a0. [DOI] [PubMed] [Google Scholar]
- 11.Franco del Amo F, Smith D E, Swiatek P J, Gendron-Maguire M, Greenspan R J, McMahon A P, Gridley T. Expression pattern of Motch, a mouse homolog of Drosophila Notch, suggests an important role in early postimplantation mouse development. Development. 1992;115:737–744. doi: 10.1242/dev.115.3.737. [DOI] [PubMed] [Google Scholar]
- 12.Greenberger J S, Sakakeeny M A, Humphries R K, Eaves C J, Eckner R J. Demonstration of permanent factor-dependent multipotential (erythroid/neutrophil/basophil) hematopoietic progenitor cell lines. Proc Natl Acad Sci USA. 1983;80:2931–2935. doi: 10.1073/pnas.80.10.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenwald I, Rubin G M. Making a difference: the role of cell-cell interactions in establishing separate identities for equivalent cells. Cell. 1992;68:271–281. doi: 10.1016/0092-8674(92)90470-w. [DOI] [PubMed] [Google Scholar]
- 14.Jonsson J, Wu Q, Nilsson K, Phillips R A. Use of a promoter-trap retrovirus to identify and isolate genes involved in differentiation of a myeloid progenitor cell line in vitro. Blood. 1996;87:1771–1779. [PubMed] [Google Scholar]
- 15.Kopan R, Nye J S, Weintraub H. The intracellular domain of mouse Notch: a constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of MyoD. Development. 1994;120:2385–2396. doi: 10.1242/dev.120.9.2385. [DOI] [PubMed] [Google Scholar]
- 16.Kopan R, Schroeter E H, Nye J, Weintraub H. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci USA. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kreider B L, Phillips P D, Prystowsky M B, Shirsat N, Pierce J H, Tushinski R, Rovera G. Induction of the granulocyte-macrophage colony-stimulating factor (CSF) receptor by granulocyte CSF increases the differentiative options of a murine hematopoietic progenitor cell. Mol Cell Biol. 1990;10:4846–4853. doi: 10.1128/mcb.10.9.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lardelli M, Dahlstrand J, Lendahl U. The novel Notch homologue mouse Notch 3 lacks specific epidermal growth factor-repeats and is expressed in proliferating neuroepithelium. Mech Dev. 1994;46:123–136. doi: 10.1016/0925-4773(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 19.Lardelli M, Lendahl U. Motch A and Motch B—two mouse Notch homologues coexpressed in a wide variety of tissues. Exp Cell Res. 1993;204:364–372. doi: 10.1006/excr.1993.1044. [DOI] [PubMed] [Google Scholar]
- 20.Li L, Milner L A, Deng Y, Iwata M, Banta A, Graf L, Marcovina S, Friedman C, Trask B, Hood L, Torok-Storb B. The human homolog of Rat Jagged, hJagged1, is expressed by marrow stroma and inhibits differentiation of 32D cells through interaction with Notch1. Immunity. 1998;8:43–55. doi: 10.1016/s1074-7613(00)80457-4. [DOI] [PubMed] [Google Scholar]
- 21.Lieber T, Kidd S, Alcamo E, Corbin V, Young M W. Antineurogenic phenotypes induced by truncated Notch proteins indicate a role in signal transduction and may point to a novel function for Notch in nuclei. Genes Dev. 1993;7:1949–1965. doi: 10.1101/gad.7.10.1949. [DOI] [PubMed] [Google Scholar]
- 22.Lindsell C E, Shawber C J, Boulter J, Weinmaster G. Jagged: a mammalian ligand that activates Notch1. Cell. 1995;80:909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- 23.Metcalf D. Hematopoietic regulators: redundancy or subtlety? Blood. 1993;82:3515–3523. [PubMed] [Google Scholar]
- 24.Migliaccio G, Migliaccio A R, Kreider B L, Rovera G, Adamson J W. Selection of lineage-restricted cell lines immortalized at different stages of hematopoietic differentiation from the murine cell line 32D. J Cell Biol. 1989;109:833–841. doi: 10.1083/jcb.109.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–990. [PMC free article] [PubMed] [Google Scholar]
- 26.Milner L A, Bigas A, Kopan R, Brashem-Stein C, Black M, Bernstein I D, Martin D I K. Inhibition of granulocytic differentiation by mNotch1. Proc Natl Acad Sci USA. 1996;93:13014–13019. doi: 10.1073/pnas.93.23.13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Milner L A, Kopan R, Martin D I, Bernstein I D. A human homologue of the Drosophila developmental gene, Notch, is expressed in CD34+ hematopoietic precursors. Blood. 1994;83:2057–2062. [PubMed] [Google Scholar]
- 28.Mitsiadis T A, Lardelli M, Lendahl U, Thesleff I. Expression of Notch1, 2, and 3 is regulated by epithelial-mesenchymal interactions and retinoic acid in the developing mouse tooth and associated with determination of ameloblast cell fate. J Cell Biol. 1995;130:407–418. doi: 10.1083/jcb.130.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore K A, Pytowski B, Witte L, Hicklin D, Lemischka I R. Hematopoietic activity of a stromal cell transmembrane protein containing epidermal growth factor-like repeat motifs. Proc Natl Acad Sci USA. 1997;94:4011–4016. doi: 10.1073/pnas.94.8.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison S J, Uchida N, Weissman I L. The biology of hematopoietic stem cells. Annu Rev Cell Dev Biol. 1995;11:35–71. doi: 10.1146/annurev.cb.11.110195.000343. [DOI] [PubMed] [Google Scholar]
- 31.Muskavitch M A T. Delta-Notch signaling and Drosophila cell fate choice. Dev Biol. 1994;166:415–430. doi: 10.1006/dbio.1994.1326. [DOI] [PubMed] [Google Scholar]
- 32.Nye J S, Kopan R, Axel R. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development. 1994;120:2421–2430. doi: 10.1242/dev.120.9.2421. [DOI] [PubMed] [Google Scholar]
- 33.Ogawa M. Differentiation and proliferation of hematopoietic stem cells. Blood. 1993;81:2844–2853. [PubMed] [Google Scholar]
- 34.Rebay I, Fehon R G, Artavanis-Tsakonas S. Specific truncations of Drosophila Notch define dominant activated and dominant negative forms of the receptor. Cell. 1993;74:319–329. doi: 10.1016/0092-8674(93)90423-n. [DOI] [PubMed] [Google Scholar]
- 35.Robey E, Chang D, Itano A, Cado D, Alexander H, Lans D, Weinmaster G, Salmon P. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell. 1996;87:483–492. doi: 10.1016/s0092-8674(00)81368-9. [DOI] [PubMed] [Google Scholar]
- 36.Roehl H, Kimble J. Control of cell fate in C. elegans by a GLP-1 peptide consisting primarily of ankyrin repeats. Nature. 1993;364:632–635. doi: 10.1038/364632a0. [DOI] [PubMed] [Google Scholar]
- 37.Ruohola H, Bremer K A, Baker D, Swedlow J R, Jan L Y, Jan Y N. Role of neurogenic genes in establishment of follicle cell fate and oocyte polarity during oogenesis in Drosophila. Cell. 1991;66:433–449. doi: 10.1016/0092-8674(81)90008-8. [DOI] [PubMed] [Google Scholar]
- 38.Shivdasani R A, Orkin S H. The transcriptional control of hematopoiesis. Blood. 1996;87:4025–4039. [PubMed] [Google Scholar]
- 39.Stern M J, DeVore D L. Extending and connecting signaling pathways in C. elegans. Dev Biol. 1994;166:443–459. doi: 10.1006/dbio.1994.1328. [DOI] [PubMed] [Google Scholar]
- 40.Struhl G, Fitzgerald K, Greenwald I. Intrinsic activity of the Lin-12 and Notch intracellular domains in vivo. Cell. 1993;74:331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- 41.Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996;122:2251–2259. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- 42.Valtieri M, Tweardy D J, Caracciolo D, Johnson K, Mavilio F, Altmann S, Santoli D, Rovera G. Cytokine-dependent granulocytic differentiation. Regulation of proliferative and differentiative responses in a murine progenitor cell line. J Immunol. 1987;138:3829–3835. [PubMed] [Google Scholar]
- 43.Washburn T, Schweighoffer E, Gridley T, Chang D, Fowlkes B J, Cado D, Robey E. Notch activity influences the α/β versus γ/δ T cell lineage decision. Cell. 1997;88:833–843. doi: 10.1016/s0092-8674(00)81929-7. [DOI] [PubMed] [Google Scholar]
- 44.Weinmaster G, Roberts V J, Lemke G. A homolog of Drosophila Notch expressed during mammalian development. Development. 1991;113:199–205. doi: 10.1242/dev.113.1.199. [DOI] [PubMed] [Google Scholar]
- 45.Weinmaster G, Roberts V J, Lemke G. Notch2: a second mammalian Notch gene. Development. 1992;116:931–941. doi: 10.1242/dev.116.4.931. [DOI] [PubMed] [Google Scholar]
- 46.Yochem J, Greenwald I. glp-1 and lin-12, genes implicated in distinct cell-cell interactions in C. elegans, encode similar transmembrane proteins. Cell. 1989;58:553–563. doi: 10.1016/0092-8674(89)90436-4. [DOI] [PubMed] [Google Scholar]