Abstract
Terminal differentiation of many cell types involves permanent withdrawal from the cell division cycle. The p18INK4c protein, a member of the p16/INK4 cyclin-dependent kinase (CDK) inhibitor family, is induced more than 50-fold during myogenic differentiation of mouse C2C12 myoblasts to become the predominant CDK inhibitor complexed with CDK4 and CDK6 in terminally differentiated myotubes. We have found that the p18INK4c gene expresses two mRNA transcripts—a 2.4-kb transcript, p18(L), and a 1.2-kb transcript, p18(S). In proliferating C2C12 myoblasts, only the larger p18(L) transcript is expressed from an upstream promoter. As C2C12 cells are induced to differentiate into permanently arrested myotubes, the abundance of the p18(L) transcript decreases. The smaller p18(S) transcript expressed from a downstream promoter becomes detectable by 12 h postinduction and is the predominant transcript expressed in terminally differentiated myotubes. Both transcripts contain coding exons 2 and 3, but p18(L) uniquely contains an additional noncoding 1.2-kb exon, exon 1, corresponding exclusively to the 5′ untranslated region (5′ UTR). The expression pattern of the shorter p18(S) transcript, but not that of the longer p18(L) transcript, correlates with terminal differentiation of muscle, lung, liver, thymus, and eye lens cells during mouse embryo development. The presence of the long 5′ UTR in exon 1 attenuated the translation of p18(L) transcript, while its absence from the shorter p18(S) transcript resulted in significantly more efficient translation of the p18 protein. Our results demonstrate that during terminal muscle cell differentiation, induction of the p18 protein is regulated by promoter switching coupled with translational control.
Work over the past decade with several model systems has identified a number of transcription factors that play a critical role in initiating a cascade of events leading to activation of lineage-specific genes and, ultimately, to conversion of precursor cells into functionally specialized cells. Coupled with this process is withdrawal of proliferating undifferentiated cells from the mitotic cell cycle at a specific point in G1 phase to become permanently arrested, terminally differentiated cells. Myogenesis is a complex, multistep process in which determined muscle precursor cells first enter the differentiation pathway and then undergo phenotypic differentiation which is characterized by the expression of muscle structural genes before fusing to form multinucleated myotubes (reviewed in references 23, 32, and 35). Irreversible withdrawal from the cell cycle occurs after myogenin induction, and establishment of the postmitotic state is required for the expression of muscle-specific contractile proteins. The MyoD family of basic helix-loop-helix transcription factors regulates the determination and differentiation of muscle precursor cells and, in conjunction with the MEF2 family of MADS box transcription factors, also activates muscle structural genes. In contrast to the progress in understanding the mechanisms that regulate the initiation of differentiation and the subsequent expression of lineage-specific genes, little is known about how cell cycle arrest is initiated and maintained during terminal differentiation.
Primary control of the eukaryotic cell cycle is provided by the activity of a family of serine/threonine protein kinases, CDKs (cyclin-dependent kinases; see two recent reviews in references 15 and 30). The enzymatic activity of a CDK is regulated by several mechanisms, including positively by the binding of a cyclin and negatively by the binding of a CDK inhibitor. In mammalian cells, there exist at least two distinct families of CDK inhibitors, represented by the two prototype CDK inhibitors p21 and p16 (31, 34). p21 (also variously known as CIP1, WAF1, SDI1, and CDKN1), first identified in normal human fibroblasts as a component of quaternary cyclin D-CDK complexes that also contain proliferating cell nuclear antigen, is a potent inhibitor of multiple cyclin-CDK enzymes. The p21 family contains two other related CDK inhibitor genes, p27Kip1 and p57Kip2. Expression of the p21 gene can be induced by a wide range of cell growth-regulatory signals, including DNA damage, tumor suppressor p53, cellular senescence, and antiproliferative transforming growth factor β. Induction of p21 mRNA has also been correlated temporally with terminal differentiation of cultured hematopoietic and myoblast cell lines and during mouse embryo development (10, 11, 27). These observations led to the suggestion that p21 may play a critical role in causing and/or maintaining cell cycle arrest during cell differentiation. p16INK4a represents the other family of CDK inhibitors that includes three additionally isolated genes: p15INK4b (also known as MTS2 and p14) (8, 12, 17), p18INK4c (8), and p19INK4d (2, 9, 14). Members of the INK4 gene family are related in sequence and evolution, all encoding proteins composed of four repeated ankyrin motifs and containing an intron interrupting the coding sequence at the same position. Unlike p21, p27, and p57, which bind to and inhibit the activity of a wide spectrum of CDK enzymes, the p16 family of inhibitors specifically regulates two closely related CDK proteins, CDK4 and CDK6. The p16 family suppresses cell growth in a pRb-dependent manner (8, 18, 22), suggesting a potential mechanism by which these members accomplish this goal; that is, inhibition of active CDK6 and CDK4 kinases prevents phosphorylation of pRb, thereby keeping pRb in its active, growth-suppressing state.
Their biochemical characteristic of inhibiting CDK activity and the induction of their expression by different cell growth-inhibitory conditions present CDK inhibitors as a group of ideal molecules to link the cell cycle machinery with a variety of cell growth-regulatory pathways, including terminal cell differentiation. Supporting this hypothesis is the observation that the expression of several CDK inhibitor genes exhibits distinct tissue specificity (8, 9, 36). To explore the function of CDK inhibitors in cell differentiation, we have recently analyzed the expression of CDK inhibitors and their interaction with target CDK proteins during terminal differentiation by using two model systems, muscle (7) and adipocyte differentiation (28a). We found that during G1 exit in both cell lineages, the p18INK4c protein is markedly induced, more than 50-fold during myogenesis and 12-fold during adipogenesis. Nearly all of CDK6 and a major fraction of CDK4 were complexed with p18 in terminally differentiated C2C12 myotubes and 3T3-L1 adipocytes. Abundant p18 protein and p18-CDK4/6 complexes were also found in adult mouse muscle and adipose tissues. These observations suggest an important role for p18 in causing and/or maintaining the permanent cell cycle arrest associated with terminal cell differentiation. The mechanisms regulating p18 protein induction, however, are unknown. The present study was directed toward this issue.
MATERIALS AND METHODS
Cell culture and transfection.
Culture and myogenic induction of murine C2C12 myoblasts (ATCC CRL 1772; American Type Culture Collection, Rockville, Md.) were described previously (7) and were assessed morphologically, with myotubes becoming visible between 4 and 5 days postinduction, and monitored by Northern analysis of the myogenin gene, a genetic marker for myogenesis (Fig. 1A). For transfection of C2C12 cells, cells were plated at a low density in p100 dishes and allowed to grow to 50% confluence (24 to 48 h after seeding), at which time they were transfected with 12 μg of DNA premixed with 36 μl of lipofectamine (Gibco-BRL, Gaithersburg, Md.) in a total volume of 6 ml of Optimem-1 medium (Gibco-BRL) in accordance with the manufacturer’s instructions. The cells were transfected with 12 μg of pcDNA3 or 8 μg of the indicated p18 expression plasmid (see Fig. 6A). The total amount of DNA transfected was adjusted to 12 μg with pcDNA3. Following a 5-h incubation with the DNA-lipid mixture, the cells were washed with phosphate-buffered saline (PBS) before replenishment with growth medium. The cells were harvested 24 h posttransfection by trypsinization and then divided into two equal fractions for Northern and Western blot analyses.
FIG. 1.
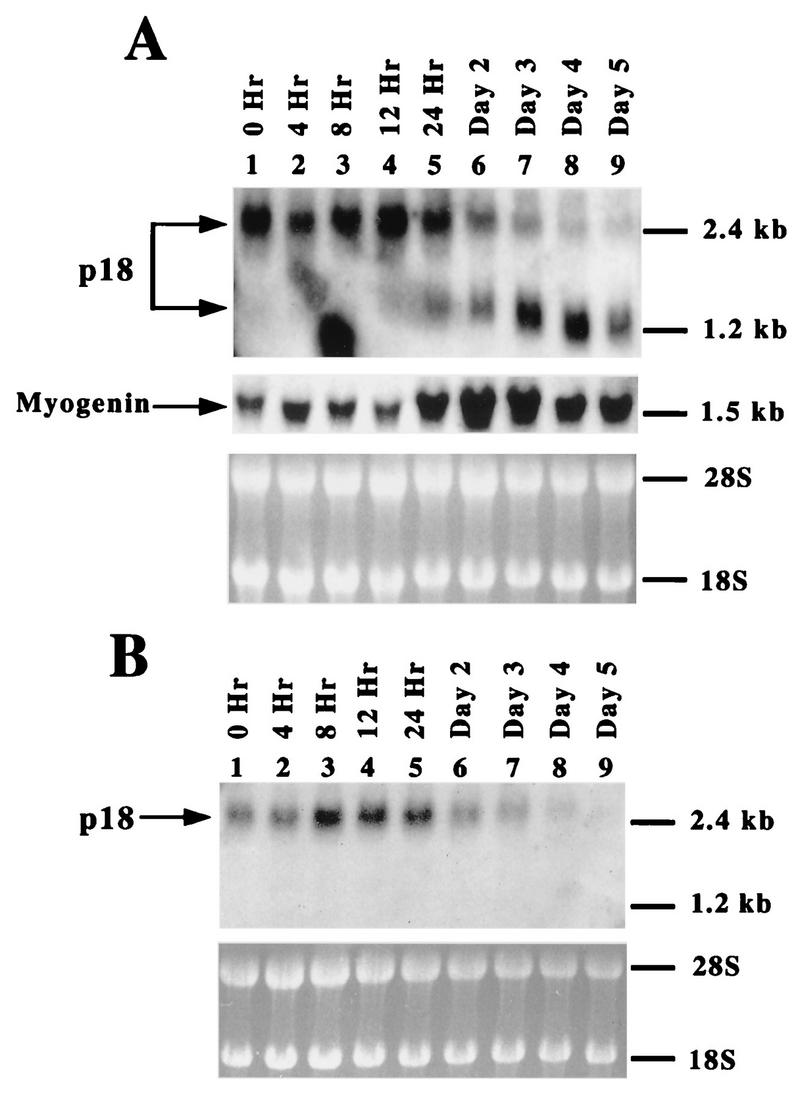
Differential expression of p18 mRNA during C2C12 cell differentiation. (A) Total RNA was prepared from proliferating C2C12 cells cultured in growth medium (lane 1) or in differentiation medium for the time indicated above each lane (lanes 2 to 9). Ten micrograms of each RNA sample was resolved on a 1% agarose–formaldehyde–MOPS gel, transferred to a nitrocellulose filter, and hybridized with a probe derived from the coding region of mouse p18 cDNA (top). The same blot was stripped and rehybridized with a probe derived from the mouse myogenin gene to monitor the progression of myogenesis (middle). Approximately equal amounts of RNA were loaded, as determined by ethidium bromide staining (bottom). (B) Expression of p18 exon 1 during C2C12 cell differentiation. Ten micrograms of total RNA prepared from C2C12 cells cultured in growth medium (lane 1) or in differentiation medium for the time indicated above each lane (lanes 2 to 9) were resolved on a 1% agarose–formaldehyde–MOPS gel, transferred to a nitrocellulose filter, and hybridized with a probe derived from exon 1 of NIH 3T3 cDNA clone T9 (top). Approximately equal amounts of RNA were loaded, as determined by ethidium bromide staining (bottom).
FIG. 6.
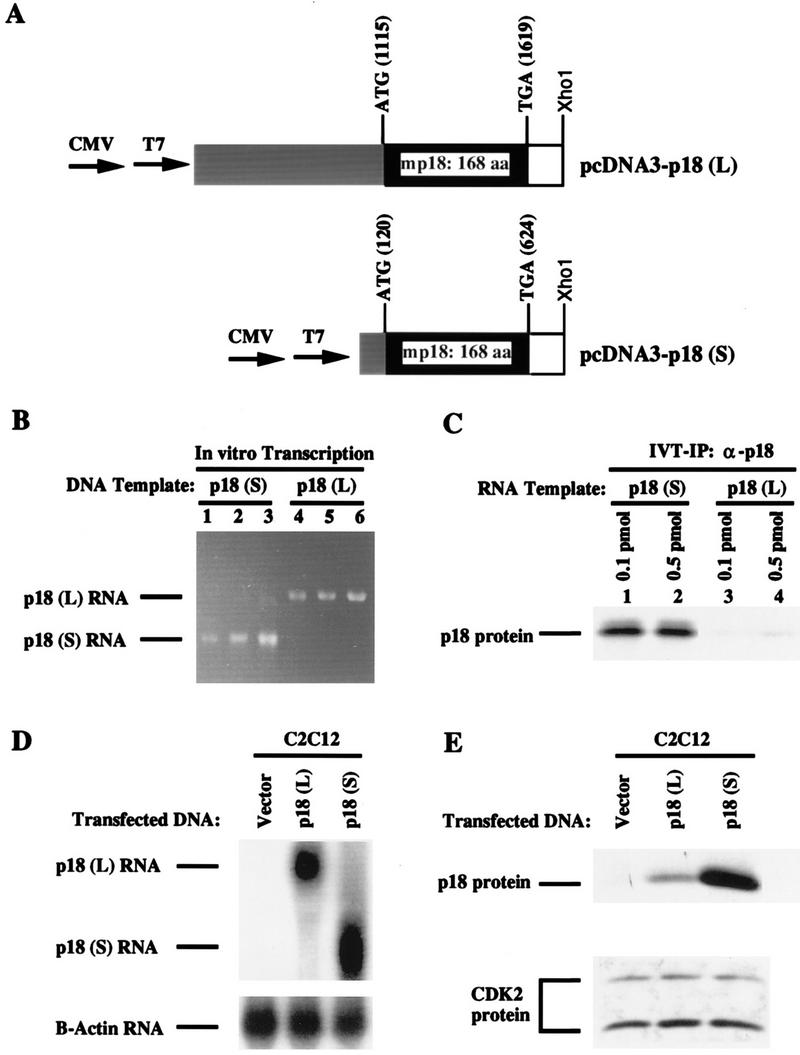
Translational regulation of p18 mRNA by the 5′ UTR. (A) Diagram of the relevant regions of the pcDNA3-p18(L) and pcDNA3-p18(S) plasmids. A 1,840-bp DNA fragment corresponding to the p18(L) cDNA isolated from NIH 3T3 cells and an 845-bp DNA fragment corresponding to the p18(S) cDNA isolated from mouse skeletal muscle tissue were inserted into the pcDNA3 expression vector downstream of the heterologous CMV and phage T7 promoters. The 5′ UTRs are indicated by gray boxes, the 3′ UTRs are indicated by white boxes, and the p18 coding sequences are indicated by black boxes. The positions of the translation initiation codons (ATG) and stop codons (TGA) are indicated. (B) In vitro transcription of the two p18 cDNAs. Both pcDNA3-p18(L) and pcDNA3-p18(S) (0.1 pmol of each) were used as templates for in vitro transcription of 5′ capped mRNA using phage T7 RNA polymerase. The RNA products were verified by loading 0.1 (lanes 1 and 4), 0.25 (lanes 2 and 5), or 0.5 (lanes 3 and 6) μl of each reaction mixture onto a 1% agarose–formaldehyde–MOPS gel and quantitated by spectrophotometry (lanes 1 to 6, respectively, contained 180, 450, 900, 160, 400, and 800 ng). A discrete RNA transcript consistent with the size of the appropriate cloned cDNA fragment was produced from each reaction, as indicated to the left of the gel. (C) In vitro translation of the two p18 cDNAs. The in vitro-transcribed p18 mRNAs (0.1 or 0.5 pmol of each) were in vitro translated in the presence of [35S]methionine. Ten microliters of each translation reaction mixture was immunoprecipitated with antiserum against p18 and resolved by SDS-PAGE. (D and E) C2C12 cells were transiently transfected with the indicated p18 expression plasmid or the parental pcDNA3 plasmid. Cells were harvested 24 h posttransfection and divided into two equal fractions for RNA (D) and protein (E) analyses. Ten micrograms of total RNA from the transfected C2C12 cells was resolved on a 1% agarose–formaldehyde–MOPS gel, transferred to a nylon membrane, and hybridized with a probe derived from the coding region of mouse p18 cDNA (top). Approximately equal amounts of RNA were loaded, as determined by stripping and reprobing of the same blot with a human β-actin probe (bottom). (E) Fifty micrograms of total protein lysate from the transfected C2C12 cells was resolved by SDS-PAGE, transferred to a nitrocellulose filter, and blotted with an antibody specific for the p18 (top) or CDK2 (bottom) protein to verify equal protein loading.
NIH 3T3 cells (ATCC CRL 1658) were plated onto six-well plates at a density of 170,000/well 24 h prior to transfection. For each transfection, 1 μg of the indicated p18 luciferase plasmid or the corresponding promoterless luciferase plasmid pGL2-Basic (Promega, Madison, Wis.) and 1 μg of CMV-LacZ, an internal control for transfection efficiency, were premixed with 6 μl of lipofectamine in a total volume of 1 ml of Optimem-1. After incubation with the DNA-lipid mixture for 5 h, the cells were washed with PBS, replenished with fresh medium, and then harvested at 24 h posttransfection for luciferase and β-galactosidase assays. All transfections were performed in triplicate.
Luciferase assays.
Twenty-four hours posttransfection, cells were washed in PBS and harvested by scraping in 250 μl of 1× Reporter lysis buffer (Promega). The cells were lysed for 10 min at 4°C with rotation and clarified by centrifugation for 5 min at 4°C in a microcentrifuge at maximum speed. Ten microliters of the clarified cell lysate was assayed for luciferase activity as previously described (20). For β-galactosidase assays, 75 μl of the clarified lysate was incubated at 37°C with 500 μl of Z buffer (21 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 40 mM β-mercaptoethanol [pH 7.0]) containing chlorophenol red–β-d-galactopyranoside (Boehringer Mannheim, Indianapolis, Ind.) at a final concentration of 0.1 mg/ml. The reaction was stopped by adding Na2CO3 to 260 mM, and the optical density at 595 nm was determined. The luciferase activity for each sample was normalized to β-galactosidase activity to control for transfection efficiency. The normalized luciferase activity of the pGL2-Basic plasmid was set to 1.
Genomic and cDNA structure analysis.
To determine the structure of the 1.2-kb p18 mRNA expressed in terminally differentiated mouse myotubes, a mouse skeletal muscle cDNA library (Clontech Laboratories, Inc., Palo Alto, Calif.) was screened by using the coding region of human p18 cDNA as a probe (8) at reduced stringency (final wash in 0.1% sodium dodecyl sulfate [SDS]–1× SSC [0.15 M NaCl plus 0.015 M sodium citrate] at 52°C). To determine the structure of the 2.4-kb p18 mRNA expressed in proliferating cells, a lambda cDNA library derived from proliferating NIH 3T3 cells was screened by using the 1.1-kb mouse skeletal muscle p18 cDNA clone as a probe at high stringency (final wash in 0.1% SDS–1× SSC at 68°C). To isolate genomic clones containing the mouse p18 gene, the same 1.1-kb cDNA fragment was used as a probe to screen a λFIXII library constructed from 129/SV mouse genomic DNA (Stratagene, La Jolla, Calif.) at high stringency. The complete sequences of the 1.1-kb cDNA clone isolated from a skeletal muscle library, M1 (1,040 bp); the cDNA clone isolated from the NIH 3T3 library, T9 (1,840 bp); and the genomic region (encompassing all three exons, the exon-intron junctions, and portions of the introns) were determined by the University of North Carolina DNA Sequencing Facility by using an automated sequencing system (ABI-373A; Perkin-Elmer). Two single-nucleotide polymorphisms between the genomic and cDNA sequences of mouse skeletal muscle clone M1 were detected (indicated by asterisks in Fig. 2A).
FIG. 2.
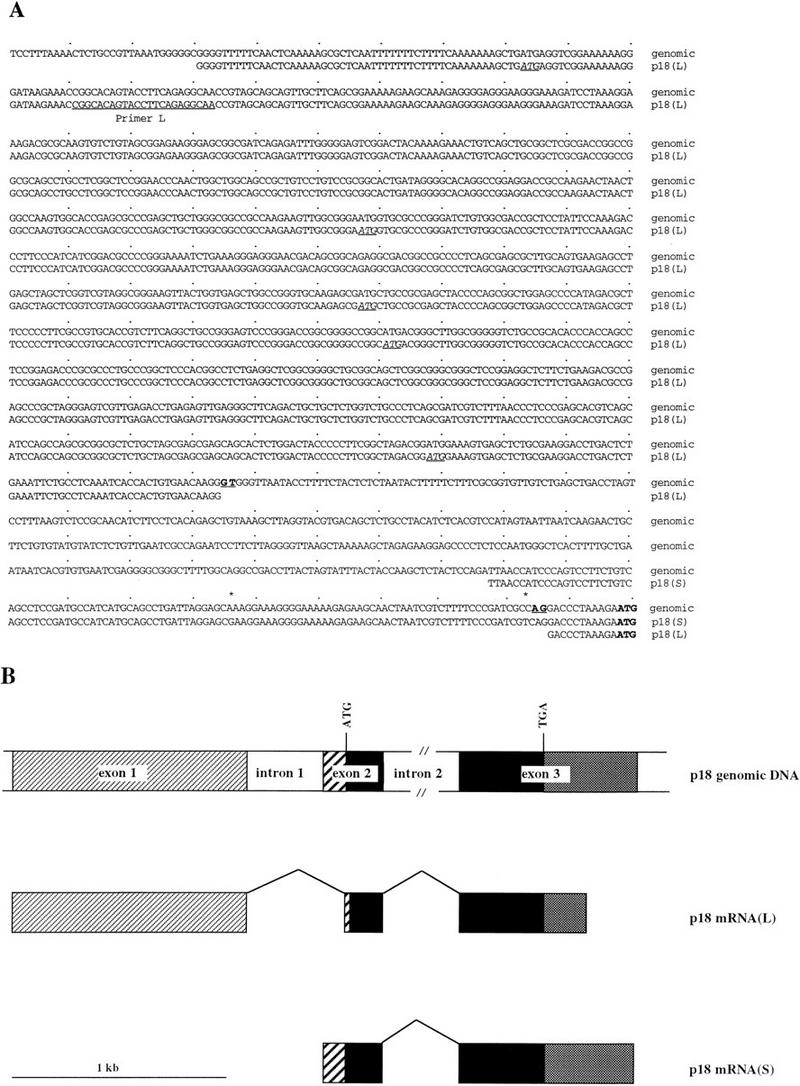
Genomic and cDNA structures of the mouse p18 gene. (A) Comparison of p18 cDNA 5′ UTRs with the p18 genomic sequence. Nucleotide sequences of the 5′ UTR of a mouse p18 genomic fragment and the p18 cDNAs isolated from proliferating NIH 3T3 cells [cDNA T9, p18(L)] and mature skeletal muscle cells [cDNA M1, p18(S)] are aligned. The splice donor and acceptor sites are boldfaced and underlined. The ATG translation initiation codon for p18 is boldfaced, and other ATG codons encoding short ORFs present in the 5′ UTR (uORFs) of cDNA T9 are italicized and underlined. Two single-nucleotide polymorphisms between the genomic and skeletal muscle sequences are indicated with asterisks. The antisense oligonucleotide primer, L, used for the primer extension experiments is underlined on the coding strands. (B) Schematic representation of the mouse p18 genomic and cDNA structures. The 5′ UTRs of the p18(L) and p18(S) transcripts are indicated by thin- and thick-striped boxes, respectively, the 3′UTR is indicated by grey boxes, introns are indicated by white boxes, and coding regions are indicated by black boxes. Splicing events are indicated by bridged lines.
Plasmids.
The pcDNA3-p18(L) expression plasmid was generated by subcloning a 1,840-bp EcoRI restriction fragment of p18 cDNA clone T9, isolated from NIH 3T3 cells, into expression plasmid pcDNA3 (Invitrogen, San Diego, Calif.) downstream of the phage T7 and cytomegalovirus (CMV) promoters. A DNA fragment corresponding to skeletal muscle p18 cDNA clone M1 was generated by PCR (sense primer; 5′-GTAATACGACTCACTATAGGGC-3′; antisense primer, 5′-ATGGGCCCAACCATCCCAGTCCTTCTG-3′). After digestion with ApaI and AflIII and filling in with T7 DNA polymerase, the blunt-ended DNA fragment was subcloned into the EcoRV site of pcDNA3 to generate pcDNA3-p18 (S). The two p18 clones contain identical coding sequences and 3′ untranslated regions (UTRs), but their 5′ UTRs differ in length (see Fig. 6A).
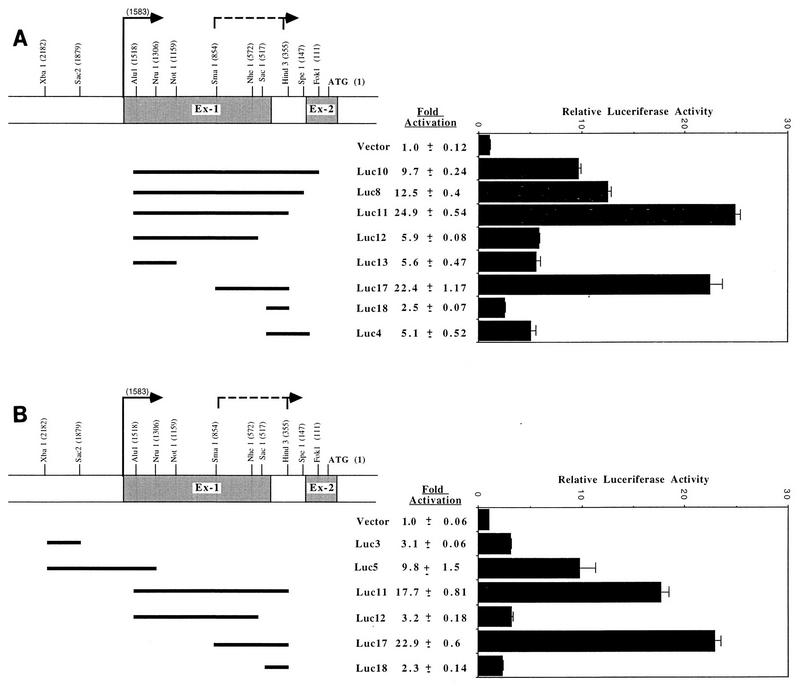
The luciferase constructs were generated by subcloning mouse p18 genomic fragments into the pGL2-Basic luciferase reporter plasmid, which lacks eukaryotic promoter and enhancer sequences. For the luciferase constructs (see the schematic depiction in Fig. 4), the A nucleotide located within the ATG codon was arbitrarily defined as +1, such that numbering for these constructs represents the distance 5′ from this nucleotide. Luc3 is a 303-bp SacII-XbaI fragment spanning nucleotides (nt) 1879 to 2182, Luc4 is a 370-bp SpeI-SacI fragment spanning nt 147 to 517, Luc5 is an 876-bp NruI-XbaI fragment spanning nt 1306 to 2182, Luc8 is a 1,371-bp SpeI-AluI fragment spanning nt 147 to 1518, Luc10 is a 1,407-bp FokI-AluI fragment spanning nt 111 to 1518, Luc11 is a 1,163-bp HindIII-AluI fragment spanning nt 355 to 1518, Luc12 is a 1,001-bp SacI-AluI fragment spanning nt 517 to 1518, Luc13 is a 359-bp NotI-AluI fragment spanning nt 1159 to 1518, Luc17 is a 499-bp HindIII-SmaI fragment spanning nt 355 to 854, and Luc18 is a 162-bp HindIII-SacI fragment spanning nt 355 to 517.
FIG. 4.
The mouse p18 gene contains two promoters. DNA fragments containing mouse p18 genomic sequences were subcloned into the pGL2-Basic luciferase reporter plasmid, which lacks eukaryotic promoter and enhancer sequences. The restriction sites used in the construction of these plasmids are indicated. The A nucleotide in the ATG codon has been arbitrarily set to 1. The promoter activity of each construct was determined by measuring the luciferase activity of each construct 24 h after transient transfection into proliferating NIH 3T3 cells. Transfection efficiency was normalized by including a CMV-LacZ expression plasmid. Relative luciferase activity was normalized to β-galactosidase activity, and the normalized luciferase activity of the promoterless pGL2-Basic plasmid was set to 1. The data are averages of three independent experiments. The transcription initiation site of the p18(S) transcript, localized by promoter activity assays between the SmaI (nt 854) and HindIII (nt 355) restriction sites (highlighted with a dashed line), has not been precisely determined. (A) Localization of a second mouse p18 promoter; (B) comparison of two promoter strengths.
RNA analysis and primer extension.
Total RNA was prepared by using TRI-REAGENT in accordance with the manufacturer’s (Molecular Research Center, Inc., Cincinnati, Ohio) instructions. Ten micrograms of each RNA sample was separated on a 1% agarose–formaldehyde–morpholinepropanesulfonic acid (MOPS) gel and transferred to a nitrocellulose or nylon filter. For nitrocellulose filters, hybridizations were carried out at 60°C for 16 h in a solution containing 5× Denhardt’s solution, 2× SSC, 0.1% SDS, 100 μg of denatured salmon sperm DNA per ml, 25 μM sodium phosphate buffer (pH 7.0), and 10% dextran sulfate. For nylon membranes, hybridizations were carried out at 68°C for 2 h in QuiKHyb (Stratagene) followed by two washes in 2× SSC–0.1% SDS and then two washes in 0.2× SSC–0.1% SDS for 15 min each at 50°C. The p18 probe was either a 0.6-kb cDNA fragment derived from the mouse p18 coding region (Fig. 1A; see Fig. 6D) or a 1.1-kb cDNA fragment corresponding to p18 exon 1 (Fig. 1B). The myogenin probe was a 1.4-kb mouse myogenin cDNA fragment kindly provided by W. Wright (33). Ethidium bromide staining was used to demonstrate even RNA loading (Fig. 1A and B), or the membranes were stripped and reprobed with a human β-actin cDNA probe (see Fig. 6D). p18 and β-actin RNA levels were quantitated with a densitometer (see Fig. 6D).
Primer extension was performed with 30 μg of total RNA from either proliferating or 4-day differentiated mouse C2C12 cells or yeast tRNA (Ambion, Austin, Tex.) as a negative control, by using an antisense oligonucleotide specific for the 2.4-kb p18(L) transcript, primer L, which corresponds to nt 102 to 80 in exon 1 of the NIH 3T3 cDNA clone (5′-TTGCCTCTGAAGGTACTGTGCCG-3′; underlined in Fig. 2A). Ten picomoles of primer L was 5′ end labeled with T4 polynucleotide kinase (New England Biolabs, Beverly, Mass.) and purified through a G-25 Sephadex column. The radiolabeled primer was mixed with total RNA and then precipitated with ethanol. The pelleted samples were resuspended in hybridization buffer consisting of 40 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 6.4), 1 mM EDTA, 400 mM NaCl, and 80% formamide; denatured at 100°C for 5 min; and allowed to hybridize at 42°C for at least 10 h. Following hybridization, the samples were precipitated with ethanol and then reverse transcribed at 37°C for 2 h in a 20-μl volume containing 50 mM Tris-HCl (pH 8.3), 8 mM MgCl2, 10 mM dithiothreitol, 40 U of RNase inhibitor (Boehringer Mannheim), 1 mM deoxynucleoside triphosphates, and 50 U of Moloney murine leukemia virus reverse transcriptase (New England Biolabs). The reactions were stopped by adding 1 μl of 0.5 M EDTA, and then 1 μl of 10-mg/ml RNase A was added and the mixture was incubated for 10 min at 37°C to digest the RNA templates. After addition of 80 μl of water and 50 μl of 7.5 M ammonium acetate; the samples were extracted once with an equal volume of phenol-chloroform, precipitated with ethanol, and resuspended in 8 μl of sequencing loading dye buffer. The reverse-transcribed products were denatured for 3 min at 100°C and resolved by polyacrylamide gel electrophoresis (PAGE) on a 7% sequencing gel. The gel was dried and exposed to film.
In situ hybridization.
Paraffin sections of mouse embryos were prepared as previously described (19). In situ hybridization was performed as previously described (4; also see reference 19 for details). Single-stranded antisense mouse p18 RNA probes were generated by in vitro transcription using [33P]UTP. The p18 probe corresponding to the coding sequence was a 0.6-kb DNA fragment derived from the p18 cDNA clone isolated from a mouse skeletal muscle cDNA library and was prepared by in vitro transcription using a T7 promoter. The p18 probe corresponding to the 5′ UTR of exon 1 was a 1.1-kb cDNA fragment derived from the p18 cDNA clone isolated from a proliferating mouse NIH 3T3 cDNA library [p18(L); Fig. 2] and was prepared by in vitro transcription using an Sp6 promoter.
Coupled in vitro transcription and translation.
pcDNA3-p18(L) and pcDNA3-p18(S) (0.1 pmol of each) were linearized with XhoI and used as templates to in vitro transcribe the two species of 5′ capped p18 mRNAs using T7 RNA polymerase in a Cap-Scribe buffer (Boehringer Mannheim). After digestion of the DNA templates with RNase-free DNase I, the RNAs were precipitated with ethanol, resuspended in water, quantitated by spectrophotometry, and visualized on a 1% agarose–formaldehyde–MOPS gel stained with ethidium bromide. Capped RNA templates (0.1 and 0.5 pmol) were translated in vitro in a rabbit reticulocyte lysate system containing [35S]methionine (Promega). Ten microliters of each in vitro translation reaction mixture was immunoprecipitated with an antibody specific to p18 (7) and resolved by SDS-PAGE. The gel was dried, and radiolabeled proteins were quantitated by using a PhosphorImager and ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.).
Western blot analysis.
Cells were washed in PBS, lysed in Nonidet P-40 lysis buffer and immunoblotted as previously described (16). The protein concentration of cell lysates was determined by the Bradford assay (Bio-Rad, Hercules, Calif.), and 50 μg of total cell lysate was resolved by SDS-PAGE. Equal loading was verified by Ponceau staining and blotting the top half of the membrane with a 1:2,500 dilution of an antibody specific to the CDK2 protein (7). The bottom half of the membrane was blotted with a 1:2,500 dilution of an antibody specific to p18 (7). Antibodies were visualized by using an ECL detection kit (Amersham) and quantitated by using a densitometer.
RESULTS
Differential expression of p18 mRNA during terminal muscle differentiation.
To determine the mechanisms that regulate p18 protein expression during terminal cell differentiation, we analyzed p18 mRNA during myogenesis of cultured C2C12 myoblasts. Total RNA was isolated from C2C12 cells over a time course of induced differentiation, and the steady-state level of p18 mRNA was determined by Northern blot analysis using the mouse p18 cDNA coding sequence as a probe (Fig. 1A). Two p18 transcripts, referred to hereafter as p18(L) for the longer form and p18(S) for the shorter form, were detected by using this probe. Differentiation of C2C12 myoblasts was monitored by stripping and reprobing the same blot with myogenin, a genetic marker for myogenesis (Fig. 1A) (33), as well as by microscopic examination of morphological changes (data not shown). In proliferating C2C12 myoblasts (lane 1), a single 2.4-kb transcript [p18(L)] was detected. Immediately following the induction of myogenesis, there was a transient increase in p18(L) (lanes 3 and 4). Twenty-four hours after induction, the steady-state level of p18(L) mRNA began to decline (lane 5) and was reduced to a nearly undetectable level by 5 days postinduction (lane 9). In contrast, a shorter 1.2-kb transcript [p18(S)] not present in proliferating myoblasts (lane 1) became detectable between 12 and 24 h postinduction and steadily increased during differentiation (lanes 4 to 9). The shorter p18 transcript isolated from the early stage of C2C12 cell differentiation (24 to 48 h) exhibited slower mobility than at the later stages (3 to 5 days; compare lanes 5 and 6 to lanes 7 to 9), suggesting the possibility that poly(A) tail lengthening associated with the short p18 transcript may facilitate more active synthesis of p18 protein in cells initially exiting the cell cycle. In proliferating murine macrophages, p18 mRNA is periodically expressed, with its lowest level in G1 and its highest level during S phase (14). Thus, while the enrichment of the G1 cell population caused by the differentiation could explain the observed decline in the p18(L) level, it argues that p18(S) transcript accumulation is a regulatory event. The data also show that the steady-state level of the p18(S) transcript in differentiated cells (lane 9) is similar to or slightly lower than that of the p18(L) transcript in proliferating cells (lane 1), despite the more-than-50-fold induction of p18 protein during C2C12 cell differentiation (7). Together, these results indicate that there is a preferential switch from the longer p18(L) to the shorter p18(S) transcript during terminal muscle differentiation and that the induction of p18 protein during myogenesis may involve both transcriptional and posttranscriptional regulation.
Structure of the mouse p18 gene.
We sought to determine the structures of both p18 transcripts by isolating their corresponding cDNA clones. Screening of a mouse skeletal muscle library yielded a 1.1-kb cDNA clone (M1) that, given its size and origin, most likely corresponds to the 1.2-kb p18(S) transcript that accumulated in differentiated C2C12 cells (Fig. 1A). To identify a cDNA clone corresponding to the 2.4-kb p18(L) transcript, we screened a mouse cDNA library derived from proliferating NIH 3T3 fibroblasts, based on the observation that this longer transcript is preferentially expressed in proliferating, undifferentiated myoblasts (Fig. 1A). Several independent p18 cDNA clones were isolated from the 3T3 library. The longest of the NIH 3T3 clones, T9 (1,840 bp), was completely sequenced. The sequence of T9 contains a 504-bp open reading frame and an apparently truncated 221-bp 3′ UTR that are identical to those encoded by the shorter 1.1-kb p18(S) M1 cDNA (data not shown). However, T9 contains a 1,115-bp 5′ UTR that is completely different from the 5′ UTR of the skeletal muscle cDNA clone, with the exception of an 11-bp sequence immediately upstream from the ATG initiation codon (Fig. 2A).
To determine the origin of these two different transcripts, we isolated a mouse genomic clone encompassing the p18 gene. The complete sequences of both p18 cDNAs were identified in the genomic clone. Comparison of the cDNA and genomic sequences revealed that the coding sequence of the p18 gene contains only one intron which is located in a position corresponding to intron 1 in the p16INK4a gene (in p18, between amino acid residues 43 and 44). This is unlike the p16 gene, whose coding sequence contains a second intron that interrupts the coding sequence prior to the last five amino acid residues (17). In addition to the two coding exons (exons 2 and 3) that are shared by both p18 cDNAs, the longer p18(L) cDNA contains a noncoding exon (exon 1) that is separated from exon 2 by a 343-bp intron (Fig. 2A and B). Another mouse p18 cDNA clone, previously isolated from proliferating T-lymphoma cells (14), contains a 78-bp 5′ UTR which includes the 11-bp sequence common to both p18(S) and p18(L) transcripts, as well as 53 bp of a sequence corresponding to that found in the 5′ UTR of cDNA clone T9 after removing the same 343-bp intron.
To confirm that the cDNA clones isolated from NIH 3T3 cells correspond to the 2.4-kb p18(L) transcript, whose abundance diminishes during C2C12 cell differentiation, exon 1 of clone T9 was used as a probe for Northern analysis of p18 mRNA steady-state levels during myogenesis. This probe only detected the 2.4-kb p18(L) transcript (Fig. 1B). The pattern of the p18(L) transcript’s steady-state level detected by the exon 1-specific probe is the same as that detected by the coding probe (Fig. 1A). These results indicate that the two p18 transcripts differ mainly, if not only, in the 5′ UTR and that the two transcripts, one containing noncoding exon 1 and one lacking exon 1, are differentially expressed during myogenic differentiation.
The p18 gene contains two distinct promoters.
The two p18 transcripts could be generated by either alternative splicing or promoter switching. The possibility that the 1.2-kb p18(S) transcript was initiated at the same site as the 2.4-kb p18(L) transcript, followed by an alternative splicing event that added a small exon, seems less likely than initiation from a separate promoter, given that the exon 1 probe did not hybridize with the p18(S) transcript (Fig. 1B) and that to generate two transcripts by alternative splicing would require two separate splice donor and acceptor sites on the same transcript. Comparison of the p18(L) cDNA with the p18 genomic sequence identified perfect splice donor and acceptor sites (Fig. 2). For p18(S), the size of the p18 cDNA clone isolated from the skeletal muscle library [1,040 bp with no poly(A) tail] is very close to the size of the p18 transcript detected by Northern hybridization, estimated as 1.2 kb by us (Fig. 1) and 1.1 kb by others (14). Hence, the skeletal muscle cDNA clone we isolated may be missing only a very small sequence at its 5′ end. We have examined the genomic sequences and found that no sequence sufficiently resembles a splice acceptor site within the 300-bp intron region upstream of the 5′ end of the skeletal muscle p18(S) cDNA clone. To determine the mechanism generating the two p18 transcripts, we mapped the p18 transcription start site(s) in both proliferating and differentiated C2C12 cells by primer extension. A 23-mer antisense oligonucleotide primer specific for the longer p18(L) transcript (primer L [underlined sequence in Fig. 2A]), which corresponds to nucleotides 102 to 80 of NIH 3T3 p18 cDNA clone T9, generated a primer extension product approximately 100 nt long (Fig. 3). This indicates that clone T9 appears to be full length at its 5′ end. The level of the p18 extension product is considerably higher in proliferating C2C12 cells than in differentiated C2C12 cells, consistent with its preferential expression in proliferating myoblasts (Fig. 1A and 3). To confirm the presence of promoter sequences within this region of the mouse p18 gene, an 876-bp XbaI-NruI genomic DNA fragment spanning this start site was inserted into the pGL2-Basic luciferase reporter plasmid, which lacks eukaryotic promoter and enhancer sequences (Fig. 4B, Luc5). The resulting plasmid, when transiently transfected into proliferating NIH 3T3 cells, exhibited 9.8-fold induction of luciferase activity compared to the parental pGL2-Basic plasmid (Fig. 4B). Deletion of the 573-bp SacII-NruI region, which includes the p18(L) start site (Luc3), abolished promoter activity, thus confirming the presence of promoter sequences within this region (Fig. 4B).
FIG. 3.
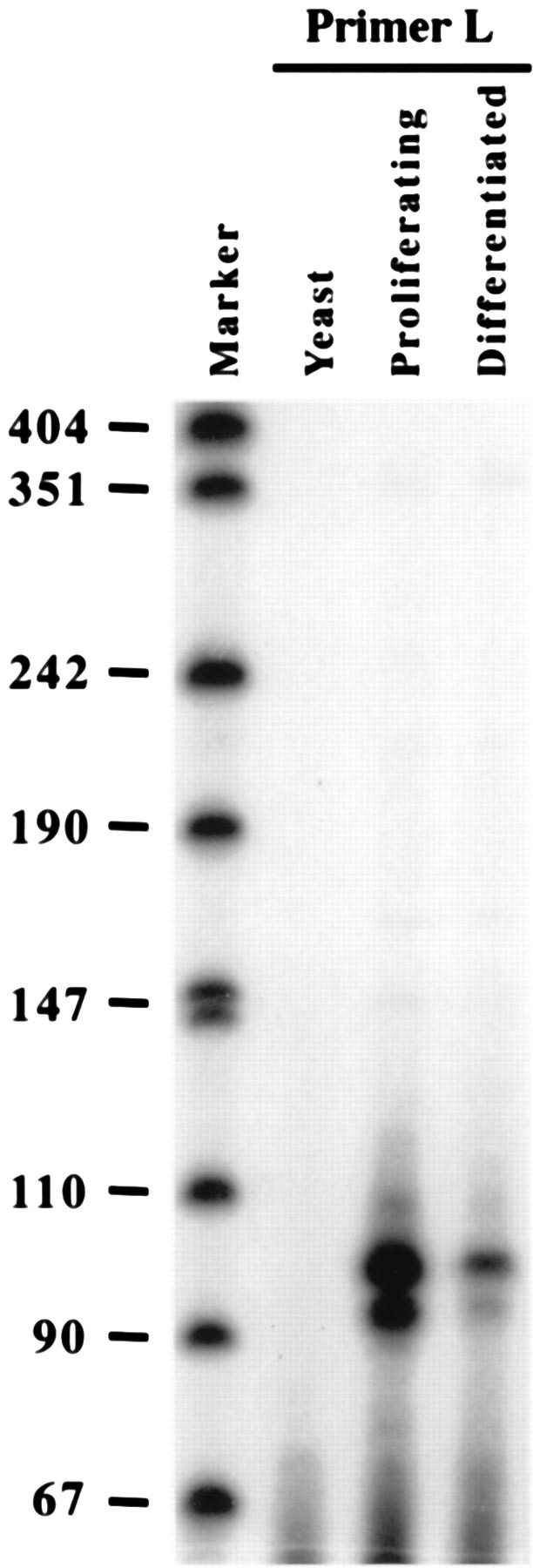
Primer extension of the p18(L) transcript. Total RNA was prepared from C2C12 cells cultured in growth medium (Proliferating) and C2C12 cells cultured in differentiation medium for 4 days (Differentiated). Yeast tRNA was used as a negative control. Thirty micrograms of each RNA sample was hybridized with an antisense primer specific to the p18(L) transcript, primer L, and extended with reverse transcriptase. The extension products were resolved on a 7% urea denaturing gel. The marker lane is pUC18 digested with HpaII and 5′ end labeled. The fragment lengths (in nucleotides) are indicated on the left.
We have also attempted to map the transcription initiation site of the p18(S) transcript. Despite our repeated efforts, however, we failed to conclusively determine the precise transcription start site for the p18(S) transcript by nuclease protection, 5′ rapid amplification of cDNA ends by PCR, and primer extension. We do not know the exact cause for such difficulties, and we can only attribute them to a high GC content surrounding the 5′ end of the shorter transcript and/or possible multiple initiation sites, as suggested by the heterogeneous nature of the short transcript (Fig. 1A). We therefore turned our effort to determining whether a separate downstream promoter exists by assaying for p18 promoter activity by using luciferase reporter promoter assays. A series of partially overlapping mouse p18 genomic DNA fragments spanning exon 1 and intron 1 were subcloned into the pGL2-Basic luciferase reporter plasmid (Fig. 4A). The resulting plasmids were transiently transfected into proliferating NIH 3T3 cells, and the expression of luciferase activity was measured (Fig. 4A). A 1,407-bp genomic fragment, Luc10, that starts 65 bp downstream of the initiation site of the first p18 promoter displayed a 9.7-fold induction of luciferase activity (Fig. 4A), indicating the presence of a second mouse p18 promoter. Serial deletions from both directions were generated from the Luc10 construct to confirm the activity of, and further localize, the second promoter. Two reporter constructs, Luc8 and Luc11, containing deletions from the 3′ end of Luc10 both exhibited increased luciferase activity, with Luc11 displaying the highest luciferase activity of the constructs examined, a 24.9-fold increase compared to the parental pGL2-Basic plasmid. The 3′ end of the mouse p18 genomic sequence present within Luc11 is 239 bp upstream of the 5′ end of the longest p18(S) cDNA, p18M1 (Fig. 2A), that we have isolated, suggesting that p18M1 is truncated at its 5′ end. A further 162-bp (Luc12) or 804-bp (Luc13) deletion from the 3′ end of Luc11 resulted in a similar, drastic decrease of luciferase activity. These results indicate that a 162-bp sequence (nucleotides 355 to 517) within exon 1 is required for full activity of the downstream promoter. However, this 162-bp fragment (Luc18) by itself exhibited only weak promoter activity compared to Luc11. An extension of this 162-bp fragment to include an additional 337 nucleotides of the 5′ sequence (Luc17), but not a shorter 55 bp fragment (to the NheI site; data not shown), restored the promoter activity close to that of Luc11. Notably, the 5′ end of the Luc17 plasmid is 729 bp downstream from the initiation site of p18(L), ruling out a possible contribution by the upstream promoter and localizing the initiation site for the p18(S) transcript within the 499-bp sequence between SmaI and HindIII.
To further confirm the presence of two distinct promoters within the mouse p18 gene, we compared in the same set of experiments the luciferase activity expressed from the constructs containing the upstream promoter (Luc5) and the downstream promoter (Luc11 and Luc17). All three constructs exhibited significant luciferase activity compared to the pGL2-Basic plasmid (Fig. 4B). Especially Luc5 and Luc17, which do not share any overlapping sequences, each exhibited promoter activity, consistent with the presence of two distinct and separable regions within the p18 gene that contain promoter sequences.
Expression of p18 mRNA during mouse embryo development.
To confirm the differential expression of the p18 gene in vivo, we determined the expression of both species of p18 transcripts in developing mouse embryos by in situ hybridization. By using the mouse p18 antisense RNA probe encompassing the coding sequence, abundant expression of p18 mRNA was detected in differentiated muscle cells of the cervix (Fig. 5A), diaphragm (Fig. 5C), and heart (Fig. 5E) in day 15 embryos. In addition to the muscle cells, there was also abundant p18 mRNA in the lung (Fig. 5C), the liver (Fig. 5E), the thymus (Fig. 5E), and the lens of the eye (data not shown). No signal was detected with a sense control probe (data not shown). p18 mRNA was not detected in several tissues, including the neural tube and retina (data not shown). To determine which form of the p18 transcript is highly expressed in these differentiated cells, we conducted in situ hybridization in the same tissues of day 15 mouse embryos by using a p18 antisense RNA probe encompassing the 5′ UTR (noncoding exon 1) uniquely present in the longer p18(L) transcript. No or extremely low levels of p18 mRNA were detected by this probe in differentiated skeletal muscle cells of the cervix (Fig. 5G), heart (Fig. 5I), and diaphragm (Fig. 5I). In addition, the exon 1 probe also failed to detect any p18 mRNA in the lungs (Fig. 5I) and liver (Fig. 5I). In contrast, the p18(L)-specific probe detected abundant p18 mRNA in adult mouse testis tissue, which contains largely undifferentiated, mitotically active cells (Fig. 5K). This is consistent with our previous Northern analysis showing a high level of expression of the longer p18(L) transcript in spermatogonia of the adult mouse testis (7, 36), substantially higher than that of the p18(S) form. These observations provide in vivo evidence correlating expression of the shorter p18(S) transcript with terminal differentiation of several cell types, including the muscle cell lineage.
FIG. 5.
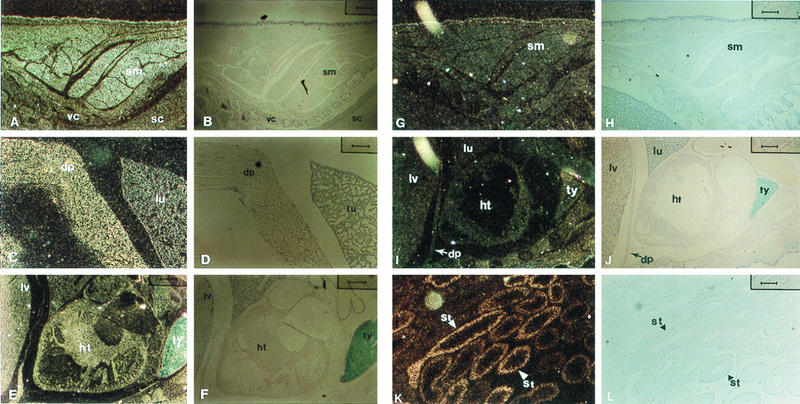
Expression of p18 mRNA in mouse embryos. Sagittal sections from E15.0 embryos were hybridized with the mouse p18 antisense RNA probe corresponding to the coding sequences common to both long and short transcripts (A, C, and E) or the 5′ UTR uniquely present in the long transcript (G, I, and K). Both dark-field (left) and corresponding bright-field (right) photographs are shown. Testis tissue was from 2-month-old mice (K and L). dp, diaphragm; ht, heart; lv, liver; lu, lung; sc, spinal cord; sm, skeletal muscle; ST, seminiferous tubule; ty, thymus; vc, vertebral cartilage.
Translational regulation of p18 mRNA by the 5′ UTR.
While switching of the p18 transcripts occurs during C2C12 cell differentiation, the steady-state levels of the p18(L) transcript in actively proliferating cells and the p18(S) transcript in differentiated cells are similar (Fig. 1A). Therefore, induction of p18 protein, from a nearly undetectable level in proliferating C2C12 cells to a 50-fold increase in mature C2C12 myotubes (7), is not regulated by a change in the steady-state level of p18 mRNA. To elucidate the mechanism underlying p18 protein induction, we examined the possibility of a difference in the translational regulation of the two p18 transcripts both in vitro and in vivo. The cDNA sequences encoding the p18(S) and p18(L) transcripts were placed under the control of the heterologous CMV and phage T7 promoters located within the pcDNA3 expression plasmid. The two cDNA sequences contain the same coding and 3′ UTR sequences but different 5′ UTRs. The p18(L) cDNA contains the 1,115-bp 5′ UTR derived from 2.4-kb cDNA clone T9, and the p18(S) cDNA contains the 120-bp 5′ UTR derived from 1.2-kb cDNA clone M1 (Fig. 6A). 5′ capped p18 RNA transcripts were generated in vitro by using T7 RNA polymerase and quantitated by spectrophotometry and gel electrophoresis (Fig. 6B). Both DNA templates generated a single RNA species of the expected size. The in vitro-transcribed p18 RNA products (0.1 and 0.5 pmol of each) were used as templates for in vitro translation in rabbit reticulocyte lysate. Equal volumes of the translation reactions were immunoprecipitated with antiserum raised against p18, and the samples were resolved by SDS-PAGE (Fig. 6C). The only translation product generated from either form of p18 RNA is an 18-kDa protein which was recognized by an antibody specific to p18, confirming that both transcripts encode the same protein species (Fig. 6C). Strikingly, the shorter p18(S) transcript is 50-fold more efficiently translated than the longer p18(L) transcript (Fig. 6C), providing a plausible molecular basis for p18 protein induction during myogenesis.
The same p18 expression plasmids were transiently transfected into proliferating C2C12 cells to determine if the translational attenuation by the 5′ UTR in p18(L) observed in vitro was recapitulated in vivo. Twenty-four hours posttransfection, cells were harvested and divided into two fractions for RNA and protein analyses, respectively. The same results were obtained from two independent experiments. Northern blot analysis of total RNA using a probe corresponding to the mouse p18 coding region detected RNA species of the expected sizes (Fig. 6D, top). The relative amounts of the p18(L) and p18(S) messages were nearly equal, as determined by densitometer scanning of the autoradiograph. Western blot analysis of total cell lysate using an antibody specific for the p18 protein detected 12-fold more p18 protein in cells transfected with the p18(S) plasmid than in cells transfected with the p18(L) plasmid (Fig. 6E, top). The level of p18 protein in cells transfected with the empty pcDNA3 expression plasmid was undetectable within this exposure time. These results demonstrate that the short p18 transcript lacking 1.1-kb 5′ UTR exon 1 is significantly more efficient for translation.
DISCUSSION
Regulation of the abundance of CDK inhibitor proteins has been shown to play a critical role in a cell’s response to a variety of growth signals. The elevation in the abundance of the CDK inhibitor p21 following DNA damage (6), during cell differentiation (21), and after induction by gamma interferon (3), as well as induction of the CDK inhibitor p15INK4b by the antiproliferative cytokine transforming growth factor β (12), are all mediated by transcriptional activation. The protein abundance of the CDK inhibitor p27Kip1 decreases following mitogen stimulation and is regulated posttranscriptionally, by either a ubiquitin-mediated protein degradation pathway (26) or a translational mechanism (1, 13). In this report, we present the first evidence for the coupled transcriptional and translational regulation of the abundance of a CDK inhibitor, p18INK4c, in response to the specific cell growth control signal, terminal muscle cell differentiation.
Upon myogenic induction of C2C12 cells, p18 protein is rapidly and markedly induced from negligible levels in proliferating myoblasts to clearly detectable levels within 8 h of myogenic induction, accumulating to a more than 50-fold increase in terminally differentiated myotubes. p18 is the only CDK inhibitor that was found to continuously accumulate during the differentiation process. Induction of p18 protein correlated with increased complex formation, first with CDK6 and then with CDK4, as well as decreased pRb kinase activity of CDK4. In terminally differentiated C2C12 myotubes and in adult mouse muscle tissue, p18 is complexed with nearly all of the CDK6 and half of the CDK4 (7). In this paper, we present four lines of evidence demonstrating that this rapid and sustained p18 protein induction during C2C12 cell differentiation is largely regulated by coupled transcriptional and translational control. First, p18 mRNA is differentially expressed during G1 exit of C2C12 cells. Proliferating C2C12 myoblasts preferentially express the larger 2.4-kb p18 transcript whose abundance decreases as C2C12 cells differentiate into terminally differentiated myotubes. Coinciding with this down regulation is the induction of the short 1.2-kb p18 transcript from nearly undetectable level in proliferating cells to the predominating p18 transcript in permanently arrested myotubes. Structural analysis of genomic and cDNA sequences indicates that the two p18 transcripts most likely originate from alternative transcription initiation start sites. Second, the expression of the shorter, but not the longer, p18 transcript correlates with terminal differentiation in vivo during mouse embryo development. Third, the mouse p18 gene contains two distinct promoters that are localized to two separate regions in the genomic DNA; one is upstream of the longer p18(L) transcript, while the other is upstream of the shorter p18(S) transcript. Lastly, while the steady-state level of the shorter p18 transcript in arrested C2C12 myotubes is similar to or slightly lower than that of the longer transcript in proliferating myoblasts, it is significantly more efficiently translated in vitro, as well as in vivo, than the longer p18 transcript, providing a molecular basis for the 50-fold induction and sustained expression of p18 protein during C2C12 cell differentiation. A similar switch of the two species of p18 transcripts was also observed during G1 exit of murine macrophages, where the shorter transcript is preferentially expressed in cells arrested by growth factor deprivation and the longer transcript is preferentially expressed in proliferating cells (14). Together, these results suggest that the coupled transcriptional and translational control of p18 expression may not be unique to myogenesis and could potentially be utilized in other cell types to regulate the abundance of p18 protein.
The major structural difference between the two forms of p18 transcripts is in their 5′ UTRs. Both transcripts contain coding exons 2 and 3 and encode the same protein (Fig. 6A), but the larger p18 transcript uniquely contains an additional 1.2-kb noncoding exon, exon 1. Removal of this long 5′ UTR significantly increased translational efficiency (Fig. 6), indicating an inhibitory effect of the 5′ UTR encoded by exon 1. Translational inhibition by the 5′ UTR has been documented in a number of cellular genes that function in regulating cell growth and development (see the recent review in reference 5). cis-acting elements within the 5′ UTR that may be involved in translational inhibition include high GC content, which has a predictable tendency to form stable stem-loop RNA secondary structures that block ribosomal scanning, multiple upstream AUG codons that encode small open reading frames (uORFs) which can interfere with translation initiation at the downstream AUG codon, and sequences that can be specifically bound by trans-acting factors (RNA binding proteins) to hinder translation. The 5′ UTR encoded by exon 1 of the p18(L) transcript has an unusually high GC content of 62%, predicting an increased tendency to form stable secondary structures that attenuate the translation of p18 protein. The 5′ UTR of the longer form of p18 transcript also contains five AUG codons (uORFs) which could potentially encode polypeptides ranging from 12 to 43 amino acids residues in length (Fig. 2A). These features may provide a structural basis for the low efficiency of translation of the 2.4-kb p18 transcript, accounting for the low level of accumulation of the p18 protein in proliferating cells. We have not examined possible changes in p18 protein stability that could also contribute to p18 protein accumulation. However, a similar increase in p18 translation efficiency upon removal of the 5′ UTR in p18(S) observed in vitro, as well as in vivo (Fig. 6C and E), and the long half-life of the homologous protein p16INK4a (little change within 3 h of experimental design [28]) argue against a significant contribution by a change in p18 protein stability.
Our findings raise a question concerning the function of p18 in proliferating cells. As cells exit from the G0 quiescent state upon growth factor stimulation, the pRb kinase activity of both CDK4 and CDK6 is activated during early to mid-G1 phase and remains elevated in proliferating cells (24, 25). Thus, while the high level of p18 protein induced during differentiation and sustained in terminally differentiated cells may play an important role in inhibiting both CDK4 and CDK6, which is necessary for entry into and maintenance of cell cycle arrest, the function of p18 in proliferating cells, if any, is unclear. We suggest that p18 may perform an unidentified surveillancelike function in proliferating cells that requires only a low level of p18 protein synthesized from the p18(L) transcript. The expression of the translationally attenuated longer p18 transcript may provide proliferating cells with the ability to rapidly and reversibly respond to certain growth conditions through translational control, such as a mechanism which removes translational repressor proteins bound to the 5′ UTR, similar to the iron response element (29). As cells enter a state of permanent cell cycle arrest, switching to the expression of the translationally efficient, shorter p18 transcript would result in an elevated and sustained level of p18 protein to ensure stable cell cycle arrest, most likely through a pRb-dependent pathway. To date, the only cell cycle-regulatory molecule shown to have an essential, nonredundant role during skeletal muscle development is the retinoblastoma protein (reviewed in references 23 and 32). In addition, differentiated myotubes can re-enter the cell cycle following expression of viral oncoproteins which functionally inactivate pRb, thus supporting a role of pRb in negatively regulating proliferation of these cells and suggesting that the proliferative capacity of these cells is not lost but is actively repressed by pRb. p18 suppresses cell growth in a pRb-dependent manner, suggesting a potential mechanism by which p18 plays a role in the pRb-dependent initiation and maintenance of cell cycle arrest during myogenesis: by directly inhibiting the kinase activity of CDK4 and CDK6, thereby keeping pRb in its active, growth-suppressing state (8). In support of this is the finding that the retinoblastoma protein functions downstream of the CDK complexes to regulate myogenesis (23, 32).
The results presented in this paper indicate that the p18 gene is expressed from two promoters: one used primarily in proliferating, undifferentiated myoblast cells and the other used in terminally differentiated, nondividing myotube cells. Preferential expression of the upstream promoter in dividing cells gives rise to the longer 2.4-kb transcript with a long 5′ UTR that interferes with translation. Activation of the downstream promoter during cell differentiation leads to expression of the shorter 1.2-kb transcript that is more efficiently translated and, in turn, is responsible for the accumulation of p18 protein. A key issue raised by our findings concerns the unknown mechanism that regulates p18 promoter switching during cell differentiation. The downstream promoter may be repressed in proliferating cells by transcription from the upstream promoter and then become activated by default when the upstream promoter is turned off. Although both promoters exhibited similarly high activities in proliferating cells when linked to a transiently transfected luciferase reporter gene, this cell culture system prevents us from further determining possible repressive effects of the upstream promoter on the downstream one. Alternatively, the two promoters may each be regulated by a different set of trans-acting factors. In either scenario, transcriptional regulation of the p18 gene would involve a specific trans-regulatory factor(s). It is attractive to speculate that a transcription factor(s) that regulates p18 expression may directly participate in the control of myogenesis, thus coupling terminal muscle cell differentiation with cell cycle control. The discovery and characterization of two separate p18 promoters that are differentially expressed during myogenesis should facilitate the identification of such transcription factors.
ACKNOWLEDGMENTS
D.E.P. and K.-M.H. contributed equally to this work. We thank Bill Marzluff, Jenny Ting, and Al Baldwin for discussions during the course of this work and Chris Jenkins and Jennifer Michel for critical reading of the manuscript.
D.E.P. and D.S.F. are recipients of National Research Service awards from the NIH/NIGMS and the NIH/NIAMS, respectively. Y.X. is a Pew Scholar in Biomedical Science and a recipient of an American Cancer Society junior faculty award. This study was supported by National Institutes of Health grant CA68377 to Y.X.
REFERENCES
- 1.Agrawal D, Hauser P, McPherson F, Dong F, Garcia A, Pledger W J. Repression of p27kip1 synthesis by platelet-derived growth factor in BALB/c 3T3 cells. Mol Cell Biol. 1996;16:4327–4336. doi: 10.1128/mcb.16.8.4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan F K M, Zhang J, Cheng L, Shapiro D N, Winoto A. Identification of human and mouse p19, a novel CDK4 and CDK6 inhibitor with homology to p16ink4. Mol Cell Biol. 1995;15:2682–2688. doi: 10.1128/mcb.15.5.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chin Y E, Kitagawa M, Su W-C S, You Z-H, Iwamoto Y, Fu X-Y. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21WAF1/CIP1 mediated by STAT1. Science. 1996;272:719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 4.Cox K H, DeLeon D V, Angerer L A, Angerer R C. Detection of mRNA in sea urchin embryos by in situ hybridization using asymmetric RNA probes. Dev Biol. 1984;101:485–502. doi: 10.1016/0012-1606(84)90162-3. [DOI] [PubMed] [Google Scholar]
- 5.Curtis D, Lehmann R, Zamore P D. Translational regulation in development. Cell. 1995;81:171–178. doi: 10.1016/0092-8674(95)90325-9. [DOI] [PubMed] [Google Scholar]
- 6.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Lin D M, Mercer W E, Kinzler K W V, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 7.Franklin D S, Xiong Y. Induction of p18INK4c and its predominant association with CDK4 and CDK6 during myogenic differentiation. Mol Biol Cell. 1996;7:1587–1599. doi: 10.1091/mbc.7.10.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guan K-L, Jenkins C W, Li Y, Nichols M A, Wu X, O’Keefe C L, Matera A G, Xiong Y. Growth suppression by p18, a p16INK4/MTS1- and p14INK4B/MTS2-related CDK6 inhibitor, correlates with wild-type pRb function. Genes Dev. 1994;8:2939–2952. doi: 10.1101/gad.8.24.2939. [DOI] [PubMed] [Google Scholar]
- 9.Guan K-L, Jenkins C W, Li Y, O’Keefe C L, Noh S, Wu X, Zariwala M, Matera A G, Xiong Y. Isolation and characterization of p19INK4dd, a p16-related inhibitor specific to CDK6 and CDK4. Mol Biol Cell. 1996;7:57–70. doi: 10.1091/mbc.7.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo K, Wang J, Andres V, Smith R C, Walsh K. MyoD-induced expression of p21 inhibits cyclin-dependent kinase activity upon myocyte terminal differentiation. Mol Cell Biol. 1995;15:3823–3829. doi: 10.1128/mcb.15.7.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Halevy O, Novitch B G, Spicer D B, Skapek S X, Rhee J, Hannon G J, Beach D, Lassar A B. Terminal cell cycle arrest of skeletal muscle correlates with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 12.Hannon G J, Beach D. p15INK4B is a potential effector of TGF-β-induced cell cycle arrest. Nature. 1994;371:257–261. doi: 10.1038/371257a0. [DOI] [PubMed] [Google Scholar]
- 13.Hengst L, Reed S I. Translational control of p27Kip1 accumulation during the cell cycle. Science. 1996;271:1861–1864. doi: 10.1126/science.271.5257.1861. [DOI] [PubMed] [Google Scholar]
- 14.Hirai H, Roussel M F, Kato J, Ashmun R A, Sherr C J. Novel INK4 proteins, p19 and p18, are specific inhibitors of cyclin D-dependent kinases CDK4 and CDK6. Mol Cell Biol. 1995;15:2672–2681. doi: 10.1128/mcb.15.5.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter T, Pines J. Cyclins and cancer II: cyclin D and CDK inhibitors come of age. Cell. 1994;79:573–582. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins C W, Xiong Y. Immunoprecipitation and immunoblotting in cell cycle studies. In: Pagano M, editor. Cell cycle: material and methods. New York, N.Y: Springer-Verlag; 1995. pp. 250–263. [Google Scholar]
- 17.Kamb A, Gruis N A, Weaver-Feldhaus J, Liu Q, Harshman K, Tavitgian S V, Stockert E, Day R S, Johnson B E, Skolnick M H. A cell cycle regulator potentially involved in genesis of many tumor types. Science. 1994;264:436–440. doi: 10.1126/science.8153634. [DOI] [PubMed] [Google Scholar]
- 18.Koh J, Enders G H, Dynlacht B D, Harlow E. Tumor-derived p16 alleles encoding proteins defective in cell-cycle inhibition. Nature. 1995;375:506–510. doi: 10.1038/375506a0. [DOI] [PubMed] [Google Scholar]
- 19.Lee E Y-H P, Hu N, Yuan S S, Cox L A, Bradley A, Lee W-H, Herrup K. Dual roles of the retinoblastoma protein in cell cycle regulation and neuron differentiation. Genes Dev. 1994;8:2008–2021. doi: 10.1101/gad.8.17.2008. [DOI] [PubMed] [Google Scholar]
- 20.Li Y, Nichols M A, Shay J W, Xiong Y. Transcriptional repression of the D-type cyclin-dependent kinase inhibitor p16 by the retinoblastoma susceptibility gene product, pRb. Cancer Res. 1994;54:6078–6082. [PubMed] [Google Scholar]
- 21.Liu M, Lee M-H, Cohen M, Bommakanti M, Freedman L P. Transcriptional activation of the Cdk inhibitor p21 by vitamin D3 leads to the induced differentiation of the myelomonocytic cell line U937. Genes Dev. 1996;10:142–153. doi: 10.1101/gad.10.2.142. [DOI] [PubMed] [Google Scholar]
- 22.Lukas J, Parry D, Aagaard L, Mann D J, Bartkova J, Strauss M, Peters G, Bartek J. Retinoblastoma-protein-dependent cell cycle inhibition by the tumor suppressor p16. Nature. 1995;375:503–506. doi: 10.1038/375503a0. [DOI] [PubMed] [Google Scholar]
- 23.Maione R, Amati P. Interdependence between muscle differentiation and cell-cycle control. Biochim Biophys Acta. 1997;1332:M19–M30. doi: 10.1016/s0304-419x(96)00036-4. [DOI] [PubMed] [Google Scholar]
- 24.Matsushime H, Quelle D E, Shurtleff S A, Shibuya M, Sherr C J, Kato J Y. D-type cyclin-dependent kinase activity in mammalian cells. Mol Cell Biol. 1994;14:2066–2076. doi: 10.1128/mcb.14.3.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyerson M, Harlow E. Identification of G1 kinase activity for cdk6, a novel cyclin D partner. Mol Cell Biol. 1994;14:2077–2086. doi: 10.1128/mcb.14.3.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagano M, Tam S W, Theodoras A M, Beer-Romero P, Sal G D, Chau V, Yew P R, Draetta G F, Rolfe M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 27.Parker S B, Eichele G, Zhang P, Rawls A, Sands A T, Bradley A, Olson E N, Harper J W, Elledge S J. p53-independent expression of p21Cip1 in muscle and other terminally differentiating cells. Science. 1995;267:1024–1027. doi: 10.1126/science.7863329. [DOI] [PubMed] [Google Scholar]
- 28.Parry D, Bates S, Mann D J, Peters G. Lack of cyclin D-Cdk complexes in Rb-negative cells correlates with high levels of p16INK4/MTS1 tumour suppressor gene product. EMBO J. 1995;14:503–511. doi: 10.1002/j.1460-2075.1995.tb07026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Phelps. D., and Y. Xiong. Unpublished data.
- 29.Rouault T A, Klausner R D, Harford J B. Translational control of ferritin. In: Hershey J W B, Mathews M B, Sonenberg N, editors. Translational control. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 335–362. [Google Scholar]
- 30.Sherr C J. G1 phase progression: cycling on cue. Cell. 1994;79:551–555. doi: 10.1016/0092-8674(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 31.Sherr C J, Roberts J M. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 1995;9:1149–1163. doi: 10.1101/gad.9.10.1149. [DOI] [PubMed] [Google Scholar]
- 32.Walsh K, Perlman H. Cell cycle exit upon myogenic differentiation. Curr Biol. 1997;7:597–602. doi: 10.1016/s0959-437x(97)80005-6. [DOI] [PubMed] [Google Scholar]
- 33.Wright W E, Sassoon D A, Lin V K. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD1. Cell. 1989;56:607–617. doi: 10.1016/0092-8674(89)90583-7. [DOI] [PubMed] [Google Scholar]
- 34.Xiong Y. Why are there so many CDK inhibitors? Biochim Biophys Acta. 1996;1288:1–5. doi: 10.1016/0304-419x(96)00012-1. [DOI] [PubMed] [Google Scholar]
- 35.Yun K, Wold B. Skeletal muscle determination and differentiation: story of a core regulatory network and its context. Curr Opin Cell Biol. 1996;8:877–889. doi: 10.1016/s0955-0674(96)80091-3. [DOI] [PubMed] [Google Scholar]
- 36.Zindy F, Quelle D E, Roussel M F, Sherr C J. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]