Abstract
Eukaryotic telomeres are variable at several levels, from the length of the simple sequence telomeric repeat tract in different cell types to the presence or number of telomere-adjacent DNA sequence elements in different strains or individuals. We have investigated the sequence organization of Xenopus laevis telomeres by use of the vertebrate telomeric repeat (TTAGGG)n and blot hybridization analysis. The (TTAGGG)n-hybridizing fragments, which ranged from less than 10 to over 50 kb with frequently cutting enzymes, defined a pattern that was polymorphic between individuals. BAL 31 exonuclease treatment confirmed that these fragments were telomeric. The polymorphic fragments analyzed did not hybridize to 5S RNA sequences, which are telomeric according to in situ hybridization. When telomeric fragments from offspring (whole embryos) were compared to those from the spleens of the parents, the inheritance pattern of some bands was found to be unusual. Furthermore, in one cross, the telomeres of the embryo were shorter than the telomeres of the parents’ spleen, and in another, the male’s testis telomeres were shorter than those of the male’s spleen. Our data are consistent with a model for chromosome behavior that involves a significant amount of DNA rearrangement at telomeres and suggest that length regulation of Xenopus telomeres is different from that observed for Mus spretus and human telomeres.
Chromosomes in a wide range of eukaryotes terminate in a nucleoprotein complex, including a tract of simple sequence DNA that generally contains clusters of G’s on the strand having the 3′ end (3). These telomeric repeats serve as a substrate for the replenishment of terminal DNA by the enzyme telomerase. The adjacent DNA often harbors families of more-complex repeated sequences referred to as telomere-associated sequences (44). While these sequences have not been ascribed a particular required function, their existence in most organisms studied suggests that they play an as-yet-unknown role or that they are the result of a common telomere behavior.
In contrast to the classically described function of providing a stable end to the chromosome, telomeres at the molecular level are remarkably fluid structures. At the most basic level, telomeric restriction fragments usually appear in gels as broad or fuzzy bands, due to heterogeneity in the number of telomeric repeats at a given chromosome end that results from the activities that lengthen and shorten telomeres. Furthermore, the average length of the telomere repeat tract is under the influence of a number of factors. Different strains of Saccharomyces cerevisiae maintain different average lengths of the telomere repeat region (26, 41); in humans, the (TTAGGG)n tract length varies for different individuals, is heritable to some degree (38), and is shorter in somatic tissue than in gametes (15, 17). Telomere-binding proteins (12, 40), including those involved in gene silencing (19, 27, 36), also influence telomere length. In the wild mouse Mus spretus, the lengths of the terminal restriction fragments are reproducibly different in different tissues of the same mouse and these length differences are generated over the course of development (34).
At another level, there are wide-ranging examples of polymorphism in telomere-adjacent sequences that may reflect active recombinational behaviors for chromosome ends. The telomere-adjacent sequence families and genes of yeast are differently distributed among the chromosomes of different strains (9, 10, 31, 43), are known to be involved in frequent homologous recombination (22, 24), and are composed of a mosaic of smaller sequence families (25). Significant chromosome size polymorphisms in species of Plasmodium can be explained by rearrangements within subtelomeric repetitive elements (13, 32) which consist of a number of different sequence families (16). Mus musculus telomere fragments are quite long and define a pattern that varies between individuals of the same strain, and they frequently undergo rearrangements to generate new band sizes among siblings of a single cross (23, 39). A similar polymorphism of telomeric arrays has been demonstrated in plants (5, 35). Human telomere-adjacent DNA sequences exhibit variation in their maps (11, 17) and/or presence on specific chromosomes in different individuals (6, 14), sometimes producing large size differences at a particular terminus (42). Together, these examples point to a fluid, dynamic nature of telomere regions.
Extensive data on vertebrate telomere variation are still lacking. An appropriate system requires large numbers of quickly generated progeny that can be examined for meiotic telomere rearrangements and easily accessible developmental stages to assess telomere length variation and regulation. The frog Xenopus laevis readily meets these criteria, since numerous embryos develop aquatically and rapidly from controlled in vitro fertilizations. Xenopus was the subject of some of the earliest studies that demonstrated the existence of particular sequences at telomeres and their specific association in the pachytene “bouquet” stage (33). As for all vertebrates examined to date, the telomeres of Xenopus metaphase chromosomes hybridize to tandem repeats of TTAGGG (29); a TTAGGG-binding protein from Xenopus eggs has been characterized (8), and Mantell and Greider (28) have identified telomerase activity in Xenopus that is expressed not only in germ cells but throughout early development. We have characterized the DNA of telomere regions in Xenopus, and our studies show extensive polymorphism in telomere-adjacent DNA as well as developmental effects on telomere length that differ from those in humans and M. spretus.
MATERIALS AND METHODS
X. laevis frogs were obtained from Steve Black or Laurens Ruben. The frogs were anesthetized with 0.04% benzocaine in water and sacrificed by cervical cleavage. Organs for DNA preparation were kept in ice-cold phosphate-buffered saline (PBS) and processed as soon as possible; testes for in vitro fertilizations were stored at 4°C in 1× modified Ringer’s solution (100 mM NaCl, 1.5 mM KCl, 0.18 mM MgCl2, 0.75 mM CaCl2, 10 μM ZnCl2, 5 mM Na HEPES [pH 7.4]). For controlled crosses, females were given a subcutaneous injection of human chorionic gonadotropin (600 to 800 U; Sigma), kept at 18°C for ∼16 h, and manually squeezed to produce eggs; the females were then kept isolated. The eggs were fertilized in 33% modified Ringer’s solution with 1/10 of one testis, and once sufficient numbers of developing embryos were obtained, the female was sacrificed for organ harvest.
DNA preparations.
Agarose block DNA preparations followed the recipes of Barlow (2). Tissues were rinsed thoroughly in ice-cold PBS and then homogenized in a small volume of PBS. Spleens from mature frogs were further diluted to a total volume of 750 to 1,000 μl, but immature spleens (from individual frogs IV, V, and VI) were diluted to smaller volumes, typically 300 μl per 10 mg of tissue. Liver samples were diluted to 500 μl for each 100 mg of tissue; testis tissue (∼75 mg) was suspended in 500 μl of PBS. Some preparations were made twice as dilute. Large chunks of tissue or membranes were then removed. An equal volume of 1% low-melting-point agarose (SeaPlaque GTG) in PBS at 50°C was added to the cold suspension, which was mixed thoroughly and kept at 37°C while 40-μl droplets were pipetted onto Parafilm (80-μl droplets for more-dilute samples). Once the droplets were solid, they were transferred to cell lysis buffer (1% Sarkosyl, 0.5 M EDTA, 2 mg of proteinase K per ml) and incubated at 50 to 55°C for 2 days. The plugs were then rinsed four times with sterile Tris-EDTA (TE), the proteinase K was inactivated with two 15-min washes with TE plus 40 μg of phenylmethylsulfonyl fluoride at 55°C, and the plugs were rinsed again and stored in TE at 4°C. Individual embryos (stage 22 neurulae) were rinsed and then suspended in 20 μl of PBS. The solution was aspirated with a micropipette tip and supplemented with an equal volume of agarose, and a single droplet was prepared and processed as described above.
Molecular biology procedures.
Agarose blocks were digested with restriction enzymes (2) in 200-μl volumes with 0.1 mg of bovine serum albumin, the recommended buffer, and 20 U of enzyme for several hours to overnight; reactions were stopped by the addition of EDTA. Excess liquid was removed, and the plugs were melted at 68 to 72°C and then treated with Agarase (Calbiochem) for at least 3 h at 37°C. Samples were run on low-percentage agarose gels, usually 0.4% SeaKem GTG, at low voltage for 24 to 48 h. The gels were poured thick enough (∼7 mm) that careful handling did not break them, and the SeaKem GTG formulation provided additional tensile strength. Ethidium bromide-stained gels were photographed and blotted onto Magnagraph (MSI) nylon by standard procedures, including depurination with 0.25 N HCl for sufficient time to change the color of the tracking dyes (up to 25 min for thick gels). Digoxigenin-labelled probes were prepared and detected with colorimetric or chemiluminescent substrates by the Genius kit procedures (Boehringer Mannheim), with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate washes at 65°C for (TTAGGG)n probes and 0.5× SSC–0.1% sodium dodecyl sulfate washes at <65°C for the oocyte-type 5S spacer probe. Plasmids containing (TTAGGG)n inserts of up to 800 bp were the generous gifts of T. de Lange. Plasmid pXlOsp1, containing 312 bp of the oocyte-type 5S spacer region, was kindly provided by Sandya Narayanswami. Mixes of HindIII, XhoI, and NdeI digests of phage lambda DNA, plus uncut lambda DNA, served as size markers.
BAL 31 digestions.
Approximately 20 μg of DNA prepared by standard procedures (1) was suspended in 100 μl of the recommended buffer and supplemented with intact or HindIII-digested lambda DNA. After the solution was preheated to 30°C, a portion was removed and the remaining 75 to 80 μl was digested with 1.5 to 3 U of BAL 31 exonuclease (New England Biolabs or Boehringer Mannheim). At various times, portions were removed and reactions were terminated with EGTA and ethanol precipitation prior to digestion with restriction enzymes. For DNA in agarose blocks, the blocks were melted and treated with Agarase and then manipulated as described above, with approximately 12 μg of DNA and 3 U of BAL 31 in a volume of 300 μl.
RESULTS
Highly polymorphic terminal restriction fragments in X. laevis.
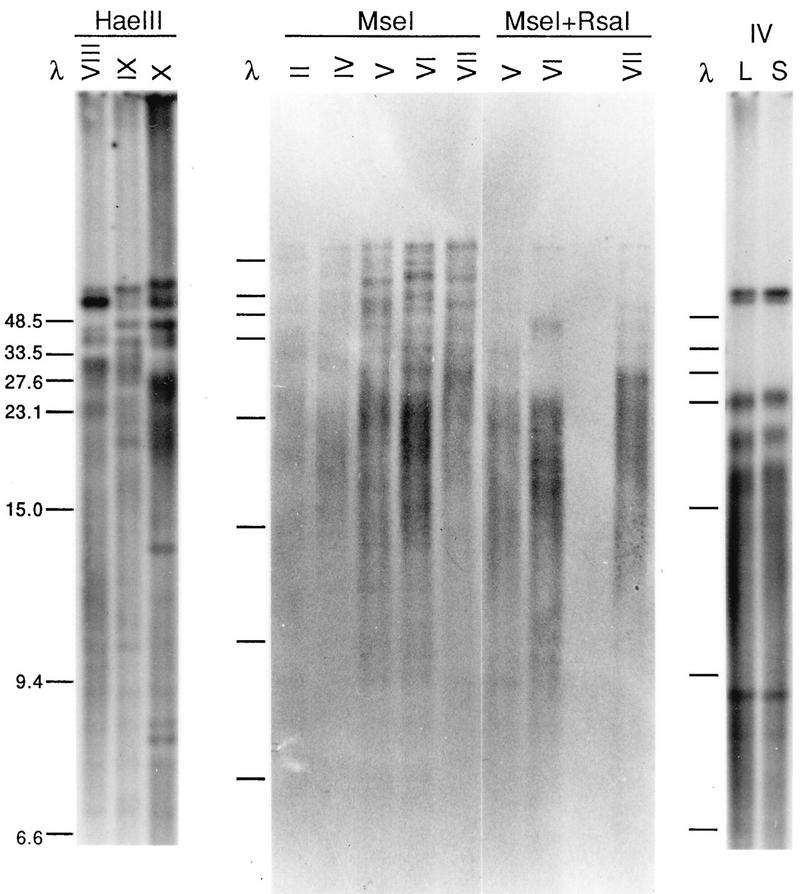
Xenopus DNA was analyzed by blot hybridization analysis with a (TTAGGG)n probe. When agarose-embedded DNA was used, with frequently cutting restriction enzymes and stringent hybridization washes, a clear pattern of fragments was obtained. Of the enzymes tested, most left the (TTAGGG)n signal exclusively in the >20-kb range, including some frequent cutters such as Sau3AI, MspI, and HinfI (data not shown). However, HaeIII, MseI, and RsaI generated broad smears below 20 kb in addition to discrete large-molecular-size bands ranging to over 48.5 kb. Some examples are shown in Fig. 1. Furthermore, this banding pattern was distinct for different individual frogs (e.g., compare individuals VIII, IX, and X or individuals II through VII in Fig. 1) and was most readily noticeable for the large-molecular-size fragments, although variation could be discerned in the smaller-molecular-size regions as well (see, for example, Fig. 4). Bands below approximately 25 kb were more likely to have the typical broad, fuzzy appearance of telomeric fragments, with a length heterogeneity of about ±1 kb; occasional sharp (presumedly internal) bands appeared and were more dependent on the stringency of washing and the probe concentration.
FIG. 1.
(TTAGGG)n-hybridizing fragments in Xenopus are polymorphic. Spleen DNA samples from different individuals (indicated by roman numerals), suspended in agarose blocks, were digested with the indicated enzymes, separated in 0.4% agarose gels, blotted, and hybridized to a (TTAGGG)n probe. In the rightmost lanes, DNA from liver (L) and spleen (S) tissues of individual IV was digested with MseI-RsaI. The sizes of the marker fragments (lanes λ) are indicated on the left in kilobases.
FIG. 4.
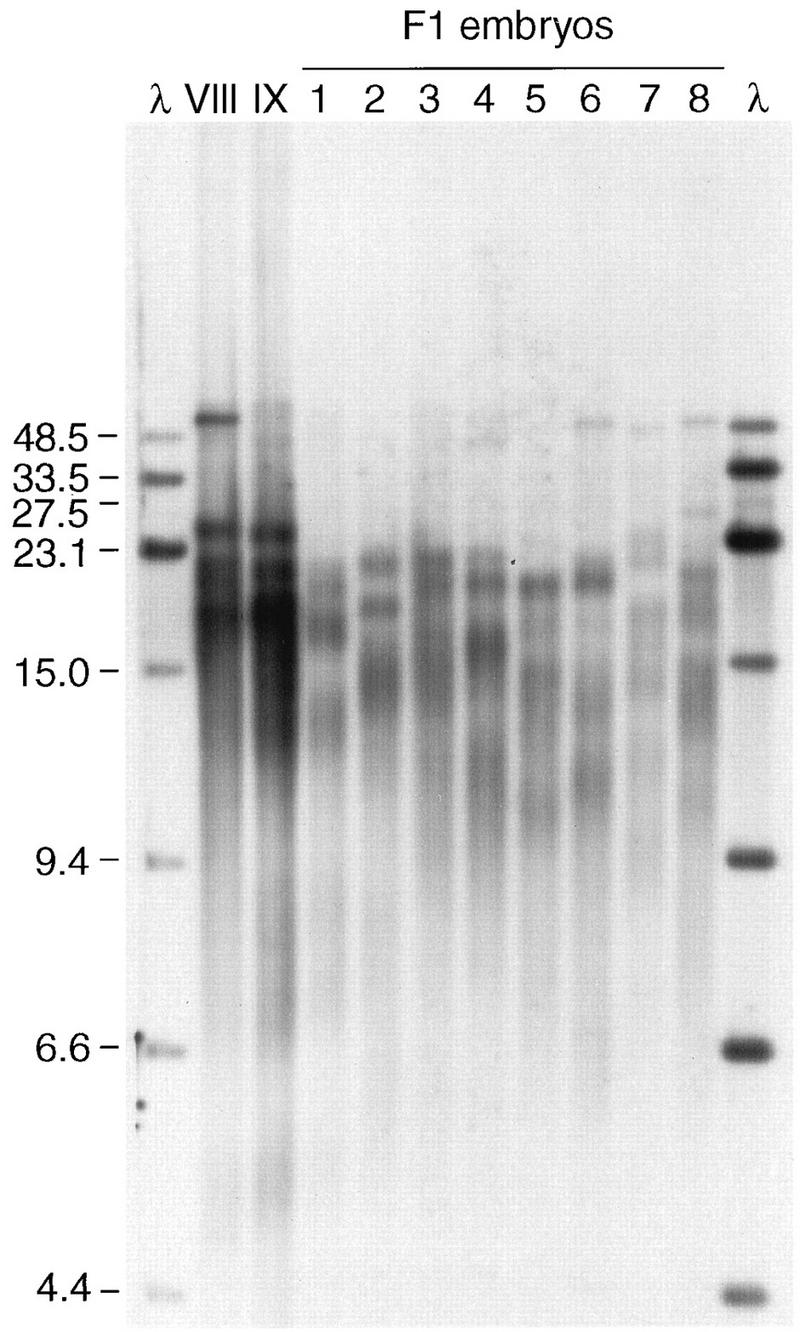
Terminal-fragment variation in parents and offspring. Spleen DNA samples from the father (individual VIII) and the mother (individual IX) and whole-embryo DNA samples from eight offspring of the in vitro fertilization (F1) were all digested with MseI-RsaI and then processed for blot hybridization with a mix of telomere repeat probe and lambda probe, the latter to provide reliable size comparisons on the autoradiogram. The sizes of the marker fragments (lanes λ) are indicated on the left in kilobases. (The 27.5-kb fragment is provided by the 23.1- and 4.4-kb λ fragments that have annealed at the cos sites.)
The overall kind of pattern for a particular enzyme was always the same; for example, EcoRI always left only large-molecular-size fragments, and the MseI-RsaI double digest always left a very small number of >20-kb fragments plus the smaller-molecular-size broad smears, but the position of each band (or presence or absence of a particular band) varied independently for different individuals. Furthermore, the variation was not simply that the DNA from some frogs had generally longer telomeres as might be expected from longer average (TTAGGG)n tract lengths, since in that case all the DNA fragments from one frog would simply be shifted up or down relative to those of another. For example, as shown in Fig. 1, the DNA from individual VII has three MseI-RsaI bands in the 20- to 33-kb range, while that from individual VI has only one. The data are consistent with a high degree of polymorphism in the most-terminal restriction sites relative to the ends of the individual chromosomes, that is, within the telomere-associated sequences internal to the telomeric repeats themselves.
In contrast, the overall banding pattern of TTAGGG-hybridizing fragments was the same in different tissues from the same frog. Figure 1 shows an example of MseI-RsaI digests of liver and spleen samples from individual IV. The pattern was clearly the same in the different tissues, although the liver DNA bands were slightly shorter (∼0.5 to 1 kb for the three broad bands between 15 and 23 kb) than the corresponding spleen DNA bands. Other experiments using MseI alone on DNA from other individual frogs demonstrated the same basic pattern for the lung, liver, and spleen (data not shown). An analysis of testis samples is presented below. Overall, since the pattern is maintained in different tissues of the same frog, the wide variation in these long fragments is likely to be meiotic in origin.
To confirm that these polymorphic, TTAGGG-hybridizing fragments were telomeric, DNA from agarose blocks was digested for increasing lengths of time with the exonuclease BAL 31 and then subjected to restriction enzyme digestion and blot analysis. An example is shown in Fig. 2. By the first time point of exonuclease treatment (15 min), the MseI fragments were already significantly affected, whereas ethidium bromide-stained bulk genomic DNA in the high-molecular-weight range was not detectably degraded throughout the experiment (data not shown). Lambda DNA fragments in the same reaction tube as a control for nonspecific degradation were shortened by 850 bp per end in 30 min. In other experiments, using standard phenol-chloroform preparations of DNA and longer BAL 31 digestion times, followed by cleavage with EcoRI, the (TTAGGG)n signal was completely removed when <3 kb of DNA had been removed from the lambda fragments (data not shown). It appeared that the (TTAGGG)n-bearing fragments were more sensitive than the lambda DNA or were digested more asynchronously. The length of simple-sequence DNA at the telomeres is likely to be much longer than 3 kb since combinations of two or more enzymes (e.g., MseI-RsaI and others not shown), while cleaving some of the long fragments left by only one enzyme, do not significantly reduce the size of the smaller-molecular-size broad smear to less than approximately 9 to 15 kb. Whether the fragments ranging to over 50 kb contain mostly telomeric repeats is unknown.
FIG. 2.
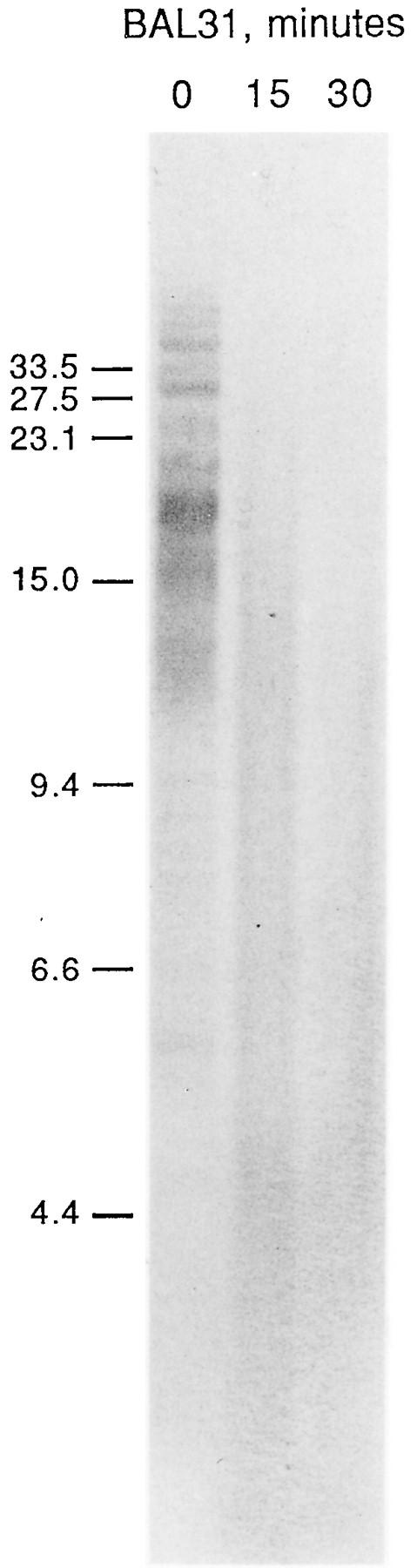
Polymorphic fragments are BAL 31 sensitive. Agarose blocks of spleen DNA from individual VII were treated with Agarase, mixed with lambda DNA, and treated with BAL 31 for the indicated numbers of minutes. Samples were then digested with MseI, run on a 0.4% gel, blotted, and hybridized to a (TTAGGG)n probe. A portion of the BAL 31 reaction mixture was digested with HindIII and hybridized to a lambda probe (data not shown). The sizes of the marker fragments are indicated on the left in kilobases.
Together, these data show that the terminal restriction fragments of Xenopus chromosomes are highly polymorphic. Furthermore, it appears that most enzymes do not cut closer than 9 to 15 kb from the chromosome ends; a subset of chromosome ends bears even longer stretches of telomeric or telomere-adjacent DNA lacking recognition sites for some frequently cutting restriction enzymes.
Telomere-adjacent DNA of long polymorphic fragments does not contain 5S RNA genes.
One explanation for a large tract of DNA that is not cleaved with even frequently cutting restriction enzymes is that it consists of tandemly repeated DNA of relatively low complexity, such as telomeric repeats themselves, or longer repeats of up to a few hundred base pairs. If there are no sites for a particular enzyme in the consensus repeat unit, then only occasional cuts will be made depending on the degree of divergence from the consensus. There is long-standing cytogenetic evidence that the tandemly repeated 5S RNA genes are found at many chromosome ends in Xenopus (7, 30, 33). More specifically, the 5S class that is expressed exclusively in oocytes maps to the long-arm telomeres of most chromosomes; the somatic 5S RNA genes map to one locus at the telomere of chromosome 9 (21).
We tested the hypothesis that these very long telomeric fragments carry 5S genes by hybridizing digested DNA to a 312-bp fragment of the oocyte-type 5S spacer region. The 5S probe detects high-molecular-weight fragments in some digests, but for MseI, RsaI, and HaeIII, which give the clearest spread of discrete high-molecular-weight telomeric bands, there is no signal in this region. An example is shown in Fig. 3; a blot of MseI and RsaI digests was hybridized to 5S oocyte-type spacer and then to (TTAGGG)n. The 5S signal is collapsed almost entirely into very small fragments; this is consistent with the published sequence of a cloned copy of the 650-bp 5S repeat unit (18) which contains sites for each of these enzymes. Similar results were found with other individual frogs and even longer exposures (data not shown). Longer-range mapping with less frequently cutting enzymes and pulsed-field gels produced some bands that appeared to hybridize to both 5S and (TTAGGG)n probes (17a). Thus, the 5S repeats may extend to a position close to some telomeres, but the bulk are removed from telomeric fragments with digests such as MseI.
FIG. 3.
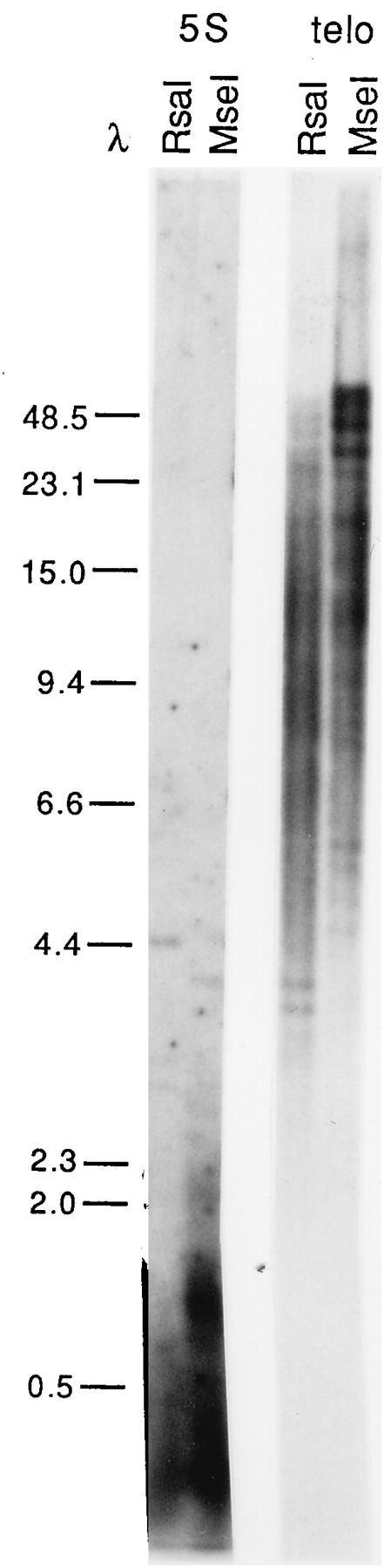
Hybridization of oocyte-type 5S sequences to Xenopus DNA digests. DNA samples from individual VII were digested with MseI or RsaI, run on a gel, and blotted; the filter was first hybridized to the oocyte 5S spacer probe and then stripped and hybridized to (TTAGGG)n (telo) under appropriate conditions for each. The sizes of the marker fragments are indicated on the left in kilobases.
Telomere length variation in offspring and parents.
The significant pattern differences observed for DNA from individual frogs suggested that new fragment lengths might be generated rather frequently. To test this hypothesis, we performed controlled in vitro fertilizations and compared the telomere banding patterns of DNA from available parent frog tissue samples to that from whole F1 embryos (neurulae) at stage 22. It was anticipated that the fragment bands from parents would be transmitted to some of the offspring in a Mendelian fashion, with occasional new bands not evident in the parents’ DNA fragments occurring in the offspring. Our experiments suggested that DNA from embryo samples could be distinctly different from parental-spleen DNA samples in telomere length as well.
MseI-RsaI digests of the DNA from parents and offspring from two separate crosses are shown in Fig. 4 and 5. In both experiments, a small amount of labelled lambda DNA (0.5 ng/ml) was included in the hybridization for unambiguous size marker identification; earlier blots with the two probes separately showed no hybridization of genomic Xenopus DNA to the lambda probe (data not shown). In Fig. 4, showing results for a cross between individuals VIII and IX, the DNAs from both parents had most of their hybridization to three to four very strong bands, the centers of which were in the 17- to 26-kb range. The DNAs from the offspring also all had strong hybridization to three to five broad bands, but the bands ranged from 11 to 23 kb in size; with the exception of DNA from embryo 8, none of the F1 embryo DNAs had a band at 26 kb, whereas the DNAs from both parents had strong bands at this position. Thus, the predominant, broad DNA bands of the offspring embryos appear to be 3 to 6 kb shorter than those of the parental spleen. In this interpretation, the strong band at >48.5 kb for DNA from the father is the likely source of the longest DNA fragments visible for embryos 6, 7, and 8, which are closer in size to the 48.5-kb size marker than the DNA fragment from the father.
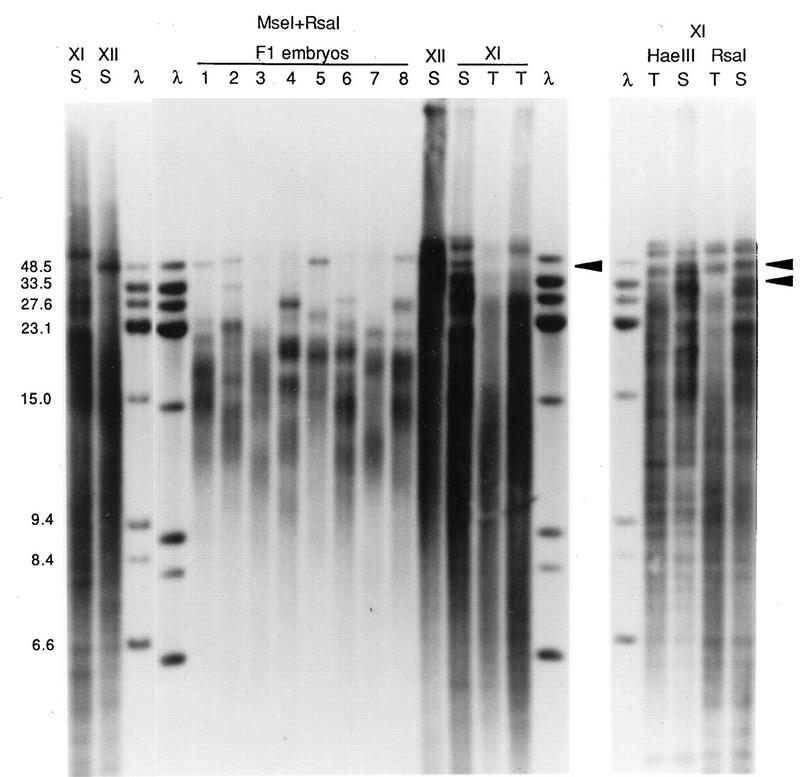
FIG. 5.
Terminal-fragment variation in offspring and in testis DNA samples. In an experiment with different parents, DNA from samples of spleen (S) or testis (T; two different concentrations) from the father (individual XI) or the spleen (S) of the mother (individual XII) and of whole embryos (numbered 1 to 8) were digested with MseI-RsaI and hybridized to a mix of telomere repeat probe and lambda probe. Results of duplicate experiments with the parent DNA digests are shown; the mother’s DNA sample next to embryo 8 lane was only partially digested, as indicated by staining with ethidium bromide. The right panel shows HaeIII or RsaI digests of spleen and the more concentrated testis DNA sample from individual XI. The sizes of marker fragments (lanes λ) are indicated on the left in kilobases. Arrowheads indicate examples of the bands from the father’s spleen DNA that are clearly different in size from the testis sample DNA bands.
Whole neurulae typically contained approximately 1/4 to 1/6 as much DNA as a spleen DNA agarose block. The spleen samples themselves contained only about 2.5 μg; with the double digest, there was no visible DNA on the agarose gel and thus overloading was not likely to affect DNA migration seriously. Nevertheless, the remaining parental and neurula samples were used to determine if the amounts of TTAGGG-hybridizing material were quantitatively different, as might be expected for the apparent size differences. Similar amounts of each DNA sample were run on a minigel, which was blotted and probed with TTAGGG repeats. Under these conditions, the TTAGGG-hybridizing bands in each lane were not resolved but appeared as a tight smear that was darker in the parental samples than in those of the neurula; the neurula sample had at least the same amount of DNA as the parental sample, as measured by ethidium bromide staining or hybridization of (TTAGGG)n to a low-molecular-weight, presumably internal band (data not shown). In addition, dilutions containing similar amounts of each DNA sample were used to make dot blots, probed with TTAGGG and/or total Xenopus DNA, and analyzed densitometrically. The expected trends were evident; however, the variation between replicate dots was occasionally great enough (>10%) that differences between samples were sometimes not significant, which is consistent with a large amount of (TTAGGG)n overall (data not shown).
The telomere bands might have been shorter in embryo DNA because of early development or because germ line telomeres were shorter than spleen telomeres. In several comparisons of testis and spleen telomeres for individual XI, the father of the cross depicted in Fig. 5, length differences were observed. Results for three different enzyme combinations are shown in the right portion of Fig. 5. For the highest-molecular-weight bands, migration rates were only slightly different, but these are near the limit of mobility of the gel and differences of several kilobases will translate to only a fraction of a millimeter. Other bands were measurably shorter in testis DNA than in spleen DNA. An RsaI band that was 46.5 kb in spleen DNA appeared to be ∼42.5 kb in testis DNA; the spleen RsaI sample yielded a band at 33.5 kb, but the most similar band in testis DNA was ∼28.5 kb. The general intensity of the hybridization smears appeared to be shifted to bands of smaller molecular sizes in both the HaeIII and RsaI testis DNA digests. Similarly, for the MseI-RsaI digests, a spleen DNA band at 45 kb and two very intense bands just below it appeared to be shifted to 38 kb and lower in testis DNA and the largest-molecular-size bands in spleen DNA also appeared shorter in the testis DNA samples. Sharp bands that presumably represent internal fragments did not migrate differently in the two samples. This suggests a difference of 4 to 7 kb in at least some of the terminal restriction fragments of testis and spleen DNA for this individual frog. Analysis of a small number of additional individual frogs indicated that TTAGGG-hybridizing fragments could be similar in size in testis and spleen DNAs, with occasional bands that were apparently shorter or longer in testis DNA, even in the same individual (22a). However, unambiguous identification of the bands will require more-specific probes.
The embryonic samples appeared to have a more distinct series of broad bands in the lower-molecular-weight region than the adult samples. This may be due to the facts that the embryo samples have undergone only 17 to 18 cell divisions (as estimated by the amount of DNA), the differentiated tissues have undergone more, and, in yeast, more rounds of replication generate more length variability (37); in addition, any effects on length regulation directly related to the differentiated state itself have not yet come into play.
Telomere pattern variation in offspring and parents.
Differences in DNA fragment lengths in offspring and parents might reflect mechanisms that generate polymorphism. While a telomere-by-telomere comparison of DNA bands in offspring with those in parents is problematic, given the number of bands and apparent length difference in embryo-germ line and spleen samples, some observations can be made. Specifically, for both crosses used in this study (Fig. 4 and 5), there were bands that appeared in the DNA from a single F1 embryo and not in that from the other seven. As shown in Fig. 4, a 26.5-kb band is present only in embryo 8 DNA, and, possibly, a 45-kb band is found only in embryo 4 DNA. As shown in Fig. 5, the second largest telomere fragment from embryo 2 is 33.5 kb; the closest-sized DNA bands in any other embryos were of 48.5 and 27.6 kb. Although the DNA from the father appears to have a band which occurs at 33.5 kb in the germ line, the inheritance of such a band in only one of eight offspring is not expected for unmodified Mendelian inheritance (see Discussion).
These results show that the polymorphic telomeric fragments are not always transmitted to offspring embryos in a simple fashion. At least in male frogs, there is an effect of germ line status on telomere length; terminal fragments can be shorter in testis DNA than in spleen DNA. Whether all bands are similarly shifted cannot be discerned from these blots. Nevertheless, the degree of apparent shortening of telomeres in testis DNA (or greater lengthening in spleen DNA) cannot entirely account for the band sizes and frequencies observed in the offspring DNA.
DISCUSSION
Xenopus terminal DNA fragments are wide-ranging in size, define a pattern that is polymorphic between individuals, and can be shorter in the germ line and embryos than in the adult spleen. The latter is supported by the testis-spleen comparison of a male used for a controlled cross (Fig. 5), in which at least some terminal fragments of testis DNA were shorter by 4 to 7 kb than the spleen telomeres. (We did not attempt to do a similar analysis with the yolk-rich oocytes.) The simplest way to accomplish this is through a shortening of (TTAGGG)n tracts in the germ line or a relative lengthening of them in the spleen. This difference is not necessarily universal in Xenopus since not every terminal fragment can be distinctly visualized and we have observed other patterns of testis-spleen differences in other males. The results of the cross depicted in Fig. 4 support the conclusion that male and female germ line telomeres can be shorter than spleen telomeres, since both parents fortuitously had fairly simple and similar spleen telomere patterns, and the embryo offspring had similar telomere patterns but the fragments were consistently shorter. Early, rapid embryonic cleavage events could have an effect as well.
These results are in contrast to those obtained with M. spretus and human telomeres, which in published examples are longer in sperm or testis than in other adult tissues (15, 17, 34). The mammalian observations have been important in models suggesting that telomere length is generated in the germ line but degraded somatically, coupled with a shutdown of telomerase and implications for cellular senescence and carcinogenesis (for an example, see reference 20), although the somatic shutdown of telomerase may not be as universal as originally thought (4). Mantell and Greider (28) found Xenopus telomerase in oocytes and testis and readily detected activity in embryos through neurulation. The presence of telomerase in embryos may be necessary to generate, in concert with other regulatory factors, the lengths ultimately found in the differentiated tissues; in M. spretus, different telomere lengths in different tissues are generated slowly over time, although in this system, the testis telomeres do become relatively long (34). While it is self-evident that, in a telomerase-dependent system, cell doublings in the absence of telomerase will lead to telomere shortening, our results show that the reported presence of telomerase does not guarantee the increase, or even the maintenance, of telomere length in Xenopus. A variety of mechanisms can act to modify telomere length and/or telomerase activity (45), and the abundance and nature of telomere-binding proteins have a clear effect on telomere length (12, 40). The identification of additional components of the telomere complex, such as the 3′-overhang-specific binding protein stockpiled in Xenopus oocytes (8) and other genetic factors, will be necessary to sort out this complex phenomenon.
The long restriction fragments, taken together, define a pattern that is distinct for different individuals. A relatively simple way to generate large differences in fragment lengths is to change the extent of some tandemly repeated sequence in the telomere-adjacent region via unequal crossover between homologous chromosomes or between homologous sequences on different chromosomes. The repetitive element rep20 found in the terminal regions of Plasmodium falciparum chromosomes is responsible for polymorphism (13); a subtelomeric macrosatellite is found at hypervariable tomato telomeres (5). Since the number of distinct bands we observed is fewer than the 72 telomeres in a diploid Xenopus cell, some bands may represent several telomeres with a conserved restriction enzyme site position, arguing for a family or families of telomere-adjacent sequences, but confirmation will require the cloning and sequence analysis of specific telomeres.
In our experiments, when the banding pattern of offspring was compared to that of the parents, unexpected results were obtained. By a conservative measure, i.e., chi-square analysis, a band present in only one of eight offspring, even if it is present in a parent and that parent is heterozygous and has only one copy of a fragment of that length, rejects the null hypothesis of unmodified Mendelian inheritance (chi-square value of 4.5 when the critical value is 3.84). In both of our experiments, a particular band was visible in one of eight offspring; in both cases, this band was not in the uppermost region of the gel, where degradation could explain its absence, nor was its size explained by the general phenomenon of telomere shortening. The apparent modification of Mendelian inheritance could take several forms. First, in the case of the 26.5-kb band in embryo 8 (Fig. 4), it could be argued that the length heterogeneity of parental telomeres (generally ±1 kb) could allow, at a low frequency, a particular gamete to acquire a long version of that telomere, with the majority being ∼22 kb; these initial differences would still be evident over a small number of cell doublings. Second, an unexpected fragment size could result from the combination of a specific terminal fragment and a genetic effect that creates generally longer or shorter (TTAGGG)n tract lengths in a particular offspring relative to those in the parents. Finally, a rearrangement in telomere-adjacent sequences, such as those documented for other systems, would change the position of the terminal restriction site; this could either generate a novel-size fragment or remove some expected-size fragments from those observed.
The data for Xenopus are consistent with some degree of meiotic rearrangement, as has been observed for yeast, mice, and some plants (22, 23, 35, 39). The variable distribution of telomere-adjacent sequences in several organisms suggests that an interaction between nonhomologous chromosomes could be a relatively common occurrence. Mitotic recombination between telomeres of nonhomologous chromosomes is observed in yeast cells selected for Y′ copy number change (24). The presence of tandemly repeated 5S genes at many Xenopus telomeres suggested to us a possible mechanism for such an interaction, and these genes are prone to translocation (30), but the 5S gene sequences or closely related variants do not appear to be present on the long polymorphic fragments we have examined. The 5S sequences could lie close to the chromosome ends that have short (<20-kb) terminal fragments, since the enzymes that cut in the array (MseI, RsaI, and HaeIII) all generate a subset of short terminal fragments. The cloning of one or more specific telomere-adjacent regions from Xenopus will provide probes to visualize one or a few chromosome ends; this will allow the unambiguous identification of meiotic products and sources, reveal the nature of telomere polymorphism, and permit a full description of variation within the terminal telomeric repeats themselves.
ACKNOWLEDGMENTS
We thank Titia de Lange and Sandya Narayanswami for providing probes, the Ruben and Black labs for frogs, members of the Black laboratory for assistance and advice with frog procedures, and Keith Karoly, Steve Black, and Liz Blackburn for helpful discussions or comments.
This work was supported by a grant from the Oregon Health Sciences Foundation (Medical Research Foundation of Oregon) to J.S. and by a grant to Reed College under the Howard Hughes Medical Institute Undergraduate Biological Sciences Initiative.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons, Inc.; 1994. [Google Scholar]
- 2.Barlow D P. Preparation, restriction, and hybridization analysis of mammalian genomic DNA for pulsed field gel electrophoresis. Methods Mol Biol. 1992;12:107–128. doi: 10.1385/0-89603-229-9:107. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn E H. Telomeres: no end in sight. Cell. 1994;77:621–623. doi: 10.1016/0092-8674(94)90046-9. [DOI] [PubMed] [Google Scholar]
- 4.Broccoli D, Young J W, de Lange T. Telomerase activity in normal and malignant hematopoietic cells. Proc Natl Acad Sci USA. 1995;92:9082–9086. doi: 10.1073/pnas.92.20.9082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Broun P, Ganal M W, Tanksley S D. Telomeric arrays display high levels of heritable polymorphism among closely related plant varieties. Proc Natl Acad Sci USA. 1992;89:1354–1357. doi: 10.1073/pnas.89.4.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown W R A, MacKinnon P J, Villasanté A, Spurr N, Buckle V J, Dobson M J. Structure and polymorphism of human telomere-associated DNA. Cell. 1990;63:119–132. doi: 10.1016/0092-8674(90)90293-n. [DOI] [PubMed] [Google Scholar]
- 7.Callan H G, Gall J G, Berg C A. The lampbrush chromosomes of Xenopus laevis: preparation, identification, and distribution of 5S DNA sequences. Chromosoma (Berlin) 1987;95:236–250. doi: 10.1007/BF00294780. [DOI] [PubMed] [Google Scholar]
- 8.Cardenas M E, Bianchi A, de Lange T. A Xenopus egg factor with DNA-binding properties characteristic of terminus-specific telomeric proteins. Genes Dev. 1993;7:883–894. doi: 10.1101/gad.7.5.883. [DOI] [PubMed] [Google Scholar]
- 9.Carlson M, Celenza J L, Eng F J. Evolution of the dispersed SUC gene family of Saccharomyces by rearrangements of chromosome telomeres. Mol Cell Biol. 1985;5:2894–2902. doi: 10.1128/mcb.5.11.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charron M J, Read E, Haut S R, Michels C A. Molecular evolution of the telomere-associated MAL loci of Saccharomyces. Genetics. 1989;122:307–316. doi: 10.1093/genetics/122.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooke H J, Brown W R A, Rappold G A. Hypervariable telomeric sequences from the human sex chromosomes are pseudoautosomal. Nature. 1985;317:687–692. doi: 10.1038/317687a0. [DOI] [PubMed] [Google Scholar]
- 12.Cooper J P, Nimmo E R, Allshire R C, Cech T R. Regulation of telomere length and function by a Myb-domain protein in fission yeast. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- 13.Corcoran L M, Thompson J K, Walliker D, Kemp D J. Homologous recombination within subtelomeric repeat sequences generates chromosome size polymorphisms in P. falciparum. Cell. 1988;53:807–813. doi: 10.1016/0092-8674(88)90097-9. [DOI] [PubMed] [Google Scholar]
- 14.Cross S, Lindsey J, Fantes J, McKay S, McGill N, Cooke H. The structure of a subterminal repeated sequence present on many human chromosomes. Nucleic Acids Res. 1990;18:6649–6657. doi: 10.1093/nar/18.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cross S H, Allshire R C, McKay S J, McGill N I, Cooke H J. Cloning of human telomeres by complementation in yeast. Nature. 1989;338:771–774. doi: 10.1038/338771a0. [DOI] [PubMed] [Google Scholar]
- 16.de Bruin D, Lanzer M, Ravetch J V. The polymorphic subtelomeric regions of Plasmodium falciparum chromosomes contain arrays of repetitive sequence elements. Proc Natl Acad Sci USA. 1994;91:619–623. doi: 10.1073/pnas.91.2.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Lange T, Shiue L, Myers R M, Cox D R, Naylor S L, Killery A M, Varmus H E. Structure and variability of human chromosome ends. Mol Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Emond, M., and J. Shampay. Unpublished observations.
- 18.Fedoroff N V, Brown D D. The nucleotide sequence of oocyte 5S DNA in Xenopus laevis. I. The AT-rich spacer. Cell. 1978;13:701–716. doi: 10.1016/0092-8674(78)90220-9. [DOI] [PubMed] [Google Scholar]
- 19.Hardy C F J, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 20.Harley C B, Villeponteau B. Telomeres and telomerase in aging and cancer. Curr Opin Genet Dev. 1995;5:249–255. doi: 10.1016/0959-437x(95)80016-6. [DOI] [PubMed] [Google Scholar]
- 21.Harper M E, Price J, Korn L J. Chromosomal mapping of Xenopus 5S genes: somatic-type vs. oocyte type. Nucleic Acids Res. 1983;11:2313–2323. doi: 10.1093/nar/11.8.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horowitz H, Thorburn P, Haber J E. Rearrangements of highly polymorphic regions near telomeres of Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:2509–2517. doi: 10.1128/mcb.4.11.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Hummasti, S., and J. Shampay. Unpublished observations.
- 23.Kipling D, Cooke H J. Hypervariable ultra-long telomeres in mice. Nature. 1990;347:400–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- 24.Louis E J, Haber J E. Mitotic recombination among subtelomeric Y′ repeats in Saccharomyces cerevisiae. Genetics. 1990;124:547–559. doi: 10.1093/genetics/124.3.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Louis E J, Naumova E S, Lee A, Naumov G, Haber J E. The chromosome end in yeast: its mosaic nature and influence on recombinational dynamics. Genetics. 1994;136:789–802. doi: 10.1093/genetics/136.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lustig A J, Petes T D. Identification of yeast mutants with altered telomere structure. Proc Natl Acad Sci USA. 1986;83:1398–1402. doi: 10.1073/pnas.83.5.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lustig A J, Kurtz S, Shore D. Involvement of the silencer and UAS binding protein RAP1 in regulation of telomere length. Science. 1990;250:549–553. doi: 10.1126/science.2237406. [DOI] [PubMed] [Google Scholar]
- 28.Mantell L L, Greider C W. Telomerase activity in germline and embryonic cells of Xenopus. EMBO J. 1994;13:3211–3217. doi: 10.1002/j.1460-2075.1994.tb06620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyne J, Ratliff R L, Moyzis R K. Conservation of the human telomere sequence (TTAGGG)n among vertebrates. Proc Natl Acad Sci USA. 1989;89:7049–7053. doi: 10.1073/pnas.86.18.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narayanswami S, Hamkalo B A. High resolution mapping of Xenopus laevis 5S and ribosomal RNA genes by EM in situ hybridization. Cytometry. 1990;11:144–152. doi: 10.1002/cyto.990110117. [DOI] [PubMed] [Google Scholar]
- 31.Ness F, Aigle M. RTM1: a member of a new family of telomeric repeated genes in yeast. Genetics. 1995;140:945–956. doi: 10.1093/genetics/140.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pace T, Ponzi M, Dore E, Janse C, Mons B, Frontali C. Long insertions within telomeres contribute to chromosome size polymorphism in Plasmodium berghei. Mol Cell Biol. 1990;10:6759–6764. doi: 10.1128/mcb.10.12.6759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pardue M L, Brown D D, Birnstiel M L. Location of the genes for 5S ribosomal RNA in Xenopus laevis. Chromosoma (Berlin) 1973;42:191–203. doi: 10.1007/BF00320940. [DOI] [PubMed] [Google Scholar]
- 34.Prowse K R, Greider C W. Developmental and tissue-specific regulation of mouse telomerase and telomere length. Proc Natl Acad Sci USA. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Röder M S, Lapitan N L V, Sorrells M E, Tanksley S D. Genetic and physical mapping of barley telomeres. Mol Gen Genet. 1993;238:294–303. doi: 10.1007/BF00279558. [DOI] [PubMed] [Google Scholar]
- 36.Runge K W, Zakian V A. TEL2, an essential gene required for telomere length regulation and telomere position effect in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3094–3105. doi: 10.1128/mcb.16.6.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shampay J, Blackburn E H. Generation of telomere-length heterogeneity in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1988;85:534–538. doi: 10.1073/pnas.85.2.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slagboom P E, Droog S, Boomsma D I. Genetic determination of telomere size in humans: a twin study of three age groups. Am J Hum Genet. 1994;55:876–882. [PMC free article] [PubMed] [Google Scholar]
- 39.Starling J A, Maule J, Hastie N D, Allshire R C. Extensive telomere repeat arrays in mouse are hypervariable. Nucleic Acids Res. 1990;18:6881–6888. doi: 10.1093/nar/18.23.6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Steensel B, de Lange T. Control of telomere length by the human telomeric protein TRF1. Nature. 1997;385:740–742. doi: 10.1038/385740a0. [DOI] [PubMed] [Google Scholar]
- 41.Walmsley R M, Petes T D. Genetic control of chromosome length in yeast. Proc Natl Acad Sci USA. 1985;82:506–510. doi: 10.1073/pnas.82.2.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilkie A O M, Higgs D R, Rack K A, Buckle V J, Spurr N K, Fischel-Ghodsian N, Ceccherini I, Brown W R A, Harris P C. Stable length polymorphism of up to 260 kb at the tip of the short arm of human chromosome 16. Cell. 1991;64:595–606. doi: 10.1016/0092-8674(91)90243-r. [DOI] [PubMed] [Google Scholar]
- 43.Zakian V A, Blanton H M. Distribution of telomere-associated sequences on natural chromosomes in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:2257–2260. doi: 10.1128/mcb.8.5.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zakian V A. Structure and function of telomeres. Annu Rev Genet. 1989;23:579–604. doi: 10.1146/annurev.ge.23.120189.003051. [DOI] [PubMed] [Google Scholar]
- 45.Zakian V A. Telomeres: beginning to understand the end. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]