Abstract
Germ line transformation of white− Drosophila embryos with P-element vectors containing white expression cassettes results in flies with different eye color phenotypes due to position effects at the sites of transgene insertion. These position effects can be cured by specific DNA elements, such as the Drosophila scs and scs′ elements, that have insulator activity in vivo. We have used this system to determine whether human matrix attachment regions (MARs) can function as insulator elements in vivo. Two different human MARs, from the apolipoprotein B and α1-antitrypsin loci, insulated white transgene expression from position effects in Drosophila melanogaster. Both elements reduced variability in transgene expression without enhancing levels of white gene expression. In contrast, expression of white transgenes containing human DNA segments without matrix-binding activity was highly variable in Drosophila transformants. These data indicate that human MARs can function as insulator elements in vivo.
Matrix attachment regions (MARs) are DNA elements that are identified and defined by their ability to bind to DNA- and histone-depleted nuclei, which are generally termed nuclear matrices (9, 33). MARs are typically AT-rich elements that contain consensus cleavage sites for topoisomerase II, and they may contain one or more loosely defined short sequence motifs, but, in general, their structures are not highly homologous. MARs are dispersed throughout eukaryotic genomes, having been found in centromeric DNA (47), within genes (9, 10, 20, 22, 40), and in intergenic regions (4, 11, 14, 20, 29, 33, 35). The matrix-binding activities of MARs have been conserved throughout eukaryotic evolution (9, 19). The functions of MARs in vivo are largely unknown, but one commonly held view is that MARs anchor individual chromatin loops to a proteinaceous matrix or scaffold in both interphase nuclei (14, 33, 47) and mitotic chromosomes (46).
An increasing body of evidence suggests that MARs may play a direct role in the regulation of gene expression. For example, the intronic MAR of the immunoglobulin κ gene is adjacent to a tissue-specific enhancer, and both elements are required for the proper regulation of the immunoglobulin κ gene during development (24, 30). Deletion of either the MAR or the enhancer resulted in constitutive hypermethylation of the gene in all cell types and in permanent repression of the locus (24). Moreover, replacement of the κ intronic MAR with MARs from other locations in the genome or from other species restored the normal pattern of both methylation and gene expression. These results indicate that MARs can be directly involved in the regulation of gene expression, and they also suggest that MAR function may be neither tissue nor species specific.
Another putative function of MAR elements, particularly those that flank individual genes or gene clusters, is to act as insulator elements. This is an attractive hypothesis because it equates the structural boundaries of a chromatin loop, the flanking MAR elements, with the functional boundaries of the domain, the putative chromosomal insulator elements. According to this hypothesis, MAR elements, or other elements at chromatin domain boundaries, may act as insulators, shielding genes within the domain from the regulatory elements of adjacent domains. However, this hypothesis has been difficult to test experimentally. Studies designed to test the ability of MARs to insulate transgenes from position effects have been reported in both plant (1, 6, 43, 51, 52) and animal systems (28, 31, 32, 34, 38, 45, 48, 49). While many of these studies have shown that transgenes flanked by MARs are more highly expressed than similar transgenes without MAR elements, conflicting views have been expressed as to whether MAR elements can render transgene expression position-independent. For example, different groups have reported that transgene expression from concatemeric arrays was silenced (1, 21), expressed in a copy number-dependent fashion (5, 15, 31, 36, 37, 45), or neither (38). These conflicting views are due, at least in part, to the inherent limitations of these transfection assays, because the numbers and arrangements of transgene sequences within the typically multimeric arrays are difficult to determine and could differ in a number of ways. First, the number and arrangement of transgenes within a single concatemeric array could affect transgene silencing versus activation. Second, some of the transfectant clones that have been studied contained multiple transgene insertions, with different transgene arrays integrated at different chromosomal sites. Patterns of gene expression among such genotypically complex transfectants might be difficult to discern. Finally, rearrangement of transgene sequences was a common event in some experiments, although transgene expression could still be detected (1). Therefore, meaningful genotype-phenotype correlations in such transfectant clones would be difficult to establish.
One reasonable, if inefficient, means to circumvent the limitations inherent in analyses of transfectants containing multiple transgene insertions would be to study only those transfectant clones that contain single, intact transgenes integrated in the recipient cell genome. We used this approach previously to study the functions of MAR elements from the human apolipoprotein B-100 (apoB) locus (21). The human apoB gene is thought to be the sole resident of a 48-kb DNase I-sensitive domain that is flanked by MAR elements (29). When single-copy transfectants containing lacZ reporters with or without flanking apoB MARs were analyzed, a significant increase in transgene expression and a reduction in variability of expression among apoB MAR-containing clones were observed (21). This was consistent with the suggestion that the apoB MARs were insulating transgene expression from chromosomal position effects, although the possibility that the MARs resulted in integration-dependent enhancement could not be excluded by these data.
To more critically test the possibility that the apoB MARs were functional boundaries of the apoB domain, we studied their insulating activities in a position effect assay in Drosophila melanogaster, as first described by Kellum and Schedl (23). In this assay, germ line transformation of white− (w−) Drosophila embryos with P-element vectors containing white transgenes results in transgenic flies with different eye color phenotypes, as each white transgene is expressed at levels dictated by regulatory elements at the site of insertion (17, 27). In contrast, P elements containing white transgenes that are flanked by insulating elements, such as the specialized chromatin structures (scs and scs′) from the hsp70 locus of Drosophila (50), are expressed in a position-independent manner, and all of the transgenic lines display similar eye color phenotypes (23). A vertebrate regulatory element, hypersensitive site 4 (HS4) of the chicken β-globin locus control region (LCR), also functions as an insulator in this assay (8). In this study, we used the Drosophila assay to assess the insulating properties of human MAR elements. MARs from the human apoB and α1-antitrypsin (α1AT) loci displayed insulating activities much like those of scs itself. In contrast, human DNA segments without matrix-binding activity had no insulating activity in this assay. These results indicate that at least some human MAR elements can function as chromosomal insulators in vivo.
MATERIALS AND METHODS
Construction of transformation vectors.
The P-element transformation vector RW+, herein designated EmwS′, was kindly provided by Paul Schedl; the construction of this plasmid has been described (53). apoB3′MEmwS′ was generated by ligating a 786-bp XhoI fragment containing the apoB 3′ MAR (29) into the unique XhoI site upstream of white in EmwS′. The orientation of the apoB 3′ MAR fragment in the various subclones was determined by restriction analysis. The P-element vector with scs upstream of white (SEmwS′) was made by directional ligation of a gel-purified 1.7-kb KpnI/SalI fragment containing scs (23, 50) into the unique KpnI/XhoI site upstream of white in EmwS′. SmwS′ was generated by digesting EmwS′ with KpnI and SpeI, which removed the white upstream regulatory region, and inserting a gel-purified 1.7-kb KpnI/SpeI fragment containing scs into the linearized plasmid at this site. This resulted in the replacement of the white upstream regulatory region with scs. All of the other constructs in which the white upstream regulatory region was deleted were derivatives of ΔXS, herein designated mwS′, which was provided by Paul Schedl. mwS′ was prepared from EmwS′ by removing an XhoI/SpeI fragment from the white upstream regulatory region. apoB3′MmwS′, ATRMmwS′, apoBmwS′, and apoB5′MmwS′ were generated by blunt-end ligation of the appropriate restriction fragments into the unique XbaI site upstream of white in mwS′. The inserts used in these constructions were a 786-bp XhoI fragment containing the apoB 3′ MAR (29) for apoB3′MmwS′; a 4.1-kb XhoI/SalI fragment containing the ATR MAR (39a) for ATRMmwS′; an 800-bp EcoRI/NcoI fragment containing part of intron 5, exon 6, and part of intron 6 of the apoB gene (3) for apoBmwS′; and a 1.0-kb XbaI/XhoI fragment containing the apoB 5′ MAR (29) for apoB5′MmwS′. The orientations of the inserted DNA fragments were determined by restriction analysis. The constructs without the scs′ element, apoB3′Mmw and apoB3′MEmw, were prepared by deleting an ∼400-bp PvuII/SalI fragment from mwS′.
Transformation and line establishment.
Samples (500 μg/ml) of each P-element transformation vector were coinjected with 150 μg of helper plasmid pπ25.7wcΔ2-3 per ml into w1118 embryos as described by Spradling and Rubin (44). Survivors were crossed to each other in groups of five (three females and two males), and transformants were identified by eye pigmentation. Mixed populations of transgenic flies were separated on the basis of eye color. Chromosome assignments of the various transgene insertions were made by crossing the transformants with w− balancer stocks containing dominant markers: In(2LR)O,Cy for the second chromosome, In(3LR)TM3,Sb for the third chromosome, and In(1)FM6,B for the X chromosome. All lines were maintained as balanced stocks. Transgene copy number was determined by Southern hybridization. Genomic DNA was isolated from 20 flies of each transformed line, digested with a restriction endonuclease that cuts once within the transgene (XbaI for white enhancer-containing constructs and for mwS′; SpeI for all other constructs), separated on 1% agarose gels by field inversion gel electrophoresis, and transferred to nylon membranes in 0.2 M NaOH–0.6 M NaCl denaturing solution. The filters were hybridized with various radiolabeled DNA probes: a 786-bp XhoI fragment containing the apoB 3′ MAR for apoB3′MEmwS′ and apoB3′MmwS′, a 4.1-kb XhoI/SalI fragment containing the ATR MAR for ATRMmwS′, a 1.0-kb XbaI/XhoI fragment containing the apoB 5′ MAR for apoB5′MmwS′, and a 400-bp EcoRI/BamHI fragment containing scs′ for EmwS′, mwS′, SEmwS′, and SmwS′.
Eye pigment assay.
Five mating pairs of flies for each line were placed at 25°C. After 5 days the adults were removed to prevent overcrowding of larvae, which can result in variations in head and body size among the progeny. Heterozygous virgin females were then collected and aged for 6 days at 25°C. To quantitate eye pigment, flies from each phenotypic class were collected, frozen, and decapitated by vortexing for 10 s. Heads from each eye color category (13 for pale yellow [phenotypic class I], 10 for yellow [II], 8 for orange [III], 5 for dark orange [IV], and 4 for red [V] and dark red [VI]) were pooled, and pigment was extracted by incubating the heads in 30% ethyl alcohol, pH 2, at room temperature for 4 days. Pigment absorption was determined at 450 nm. Each group of heads was assayed in triplicate, and absorption-per-head values were determined.
Photography.
The eyes of 4-day-old heterozygous females were photographed using a Zeiss SR microscope fitted with a Nikon FX-35 WA camera. Illumination was supplied by a Nikon MK II fiber optic light source, and photographic images were prepared with Fujichrome tungsten 64T film. Transformed lines carrying P-element insertions in the X chromosome were backcrossed to w1118 stocks to remove the In(1)FM6,B balancer chromosome prior to photography. The eye color phenotypes were identical in both genetic backgrounds.
RESULTS
The human apoB 3′ MAR functions as an insulator element in Drosophila.
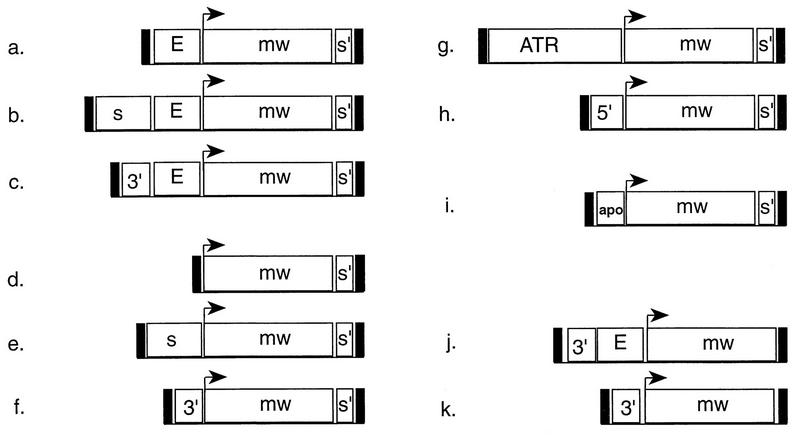
The P-element transformation vectors used in this study are shown in Fig. 1. In the first set of experiments, three different P-element transposons were employed. EmwS′ contained the white gene promoter and enhancer (E), a white cDNA coding cassette (termed mini-white [mw]), and the Drosophila scs′ (S′) element downstream of the white transcription unit (Fig. 1a). In addition, SEmwS′ (Fig. 1b) contained the Drosophila scs element (S) upstream of white, and apoB3′MEmwS′ (Fig. 1c) contained the 3′ MAR from the human apoB locus (apoB3′M) inserted upstream of white.
FIG. 1.
white P-element transformation vectors. Symbols: E, the upstream regulatory region of white that contains the eye- and testes-specific enhancers; mw, mini-white cDNA expression cassette which includes the proximal promoter, with the white transcription start site depicted by the arrow; S′, scs′; S, scs; 3′, apoB 3′ MAR; 5′, apoB 5′ MAR; ATR, ATR MAR; apo, apoB transcribed sequence; black rectangles at the ends of each transposon, 5′ and 3′ P elements. Vectors: a, EmwS′; b, SEmwS′; c, apoB3′MEmwS′; d, mwS′; e, SmwS′; f, apoB3′mwS′; g, ATRMmwS′; h, apoB5′MmwS′; i, apoBmwS′; j, apoB3′MEmw; and k, apoB3′Mmw.
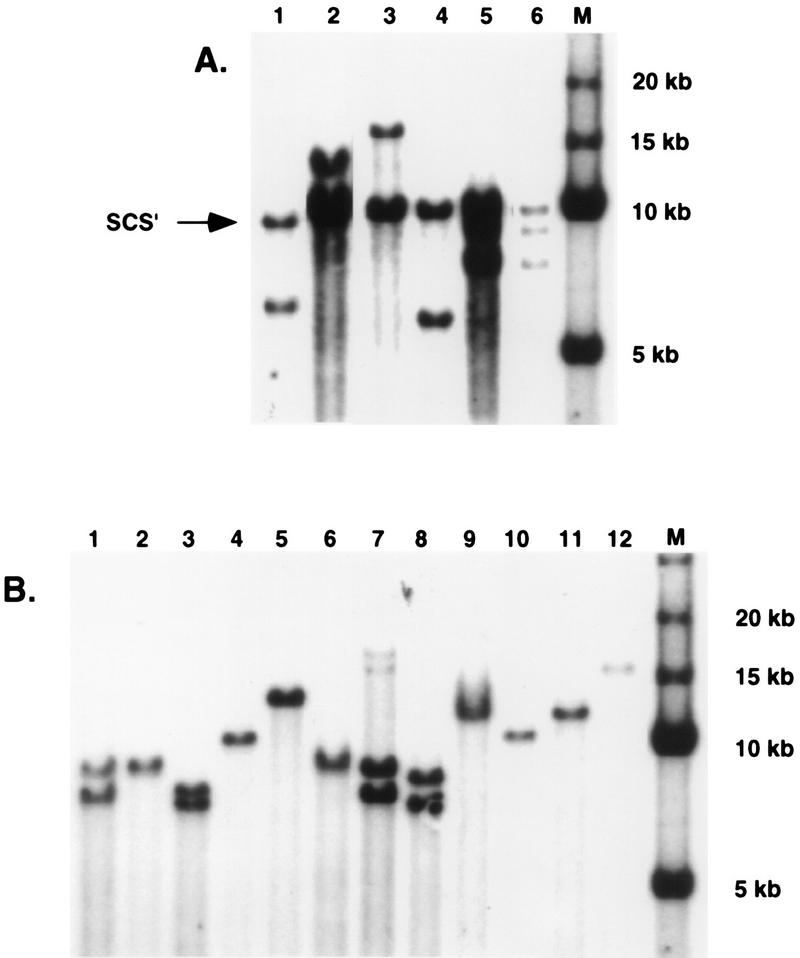
Each P-element transposon was injected into w− Drosophila embryos, and transformed lines were established from flies with any detectable eye pigment. Transgene copy number was determined in each transformed line by Southern hybridization (Fig. 2), and the eye color phenotypes of age-matched females heterozygous for the various white insertions were compared. Each transformant was assigned to one of six phenotypic classes: I (light yellow), II (yellow), III (light orange), IV (orange), V (red), and VI (dark red), as shown in Fig. 3, 4, and 5. Seven transformed lines were obtained using the control construct EmwS′, and all seven lines contained single transgene insertions (data not shown). As shown in Fig. 3a, three of these lines had light orange eyes (phenotypic class III), three had red eyes (V), and one had dark red eyes (VI). Thus, transgenes without an insulating element upstream of white were expressed at different levels in different transformed lines. This observation suggested that the white transgenes in these lines were sensitive to position effects at the sites of insertion. These results are in accord with those of Kellum and Schedl (23), who showed that transformants containing white transgenes without insulating elements had eye color phenotypes that ranged from orange to dark red. In contrast, all three single-copy lines that contained SEmwS′, in which the white transcription unit is flanked by scs and scs′, had dark red (VI) eyes (Fig. 3b). This suggested that expression of white transgenes containing scs and scs′ was largely position independent, as shown previously by Kellum and Schedl (23).
FIG. 2.
Copy number determinations for the P-element transformants. (A) mwS′ transformants. Genomic DNA was isolated, digested with XbaI, separated on an agarose gel, and probed with a labeled, ∼400-bp EcoRI/BamHI DNA fragment containing scs′. The endogenous scs′ DNA fragment of ∼ 10 kb is indicated by the arrow. Each transformant contained, in addition, one (lanes 1 to 4) or two (lanes 5 and 6) additional scs′ fragments, corresponding to single or double transgene insertions. The marker (M) lane contains DNA fragments of 5, 10, 15, and 20 kb. (B) apoB5′MmwS′ transformants. SpeI-digested genomic DNAs were probed with a labeled, ∼1.0-kb XbaI/XhoI DNA fragment containing the apoB 5′ MAR. Single (lanes 2, 4, 5, 6, 9, 10, 11, and 12) and double (lanes 1, 3, 7, and 8) transgene insertions were obtained.
FIG. 3.
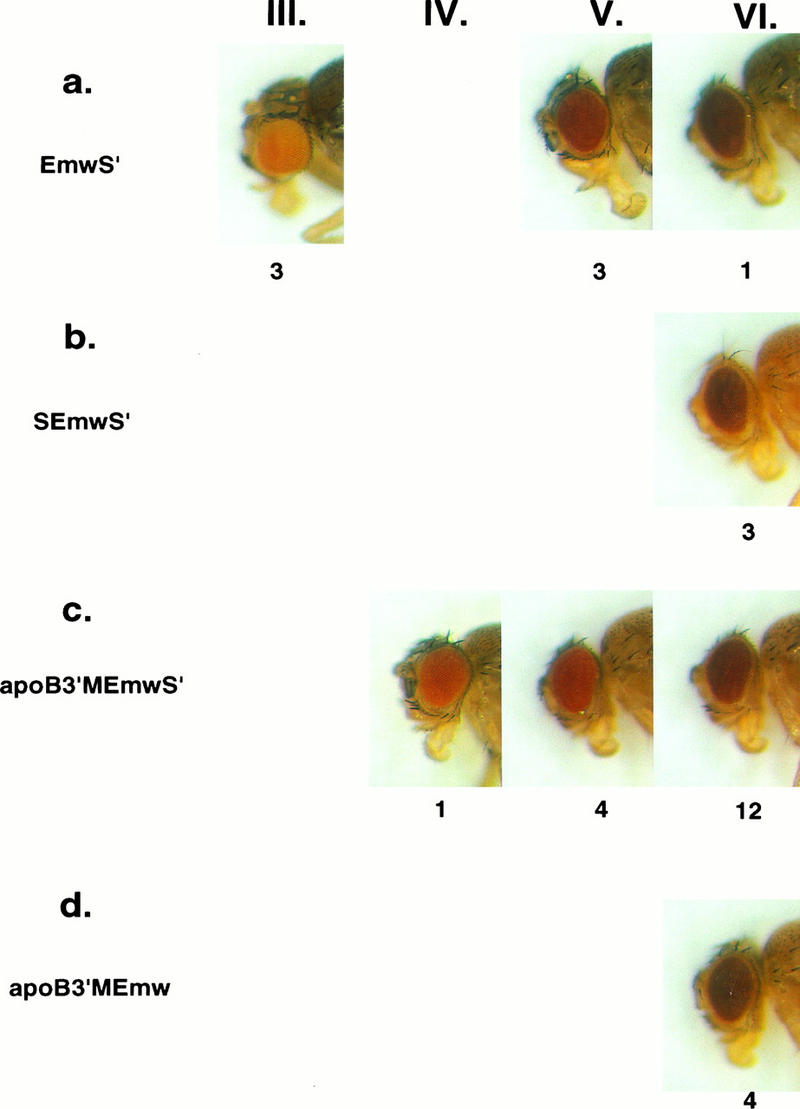
Eye color phenotypes of Drosophila transformants expressing mini-white from white enhancer-containing vectors. Eyes of 4-day-old females heterozygous for each of the different, single-copy P-element insertions were classified as being light orange (phenotypic class III), orange (IV), red (V), or dark red (VI). A representative of each of the phenotypes obtained with the different white vectors is shown, and the number of independent transformed lines with that phenotype is indicated below each picture. (a) EmwS′ transformants had eye color phenotypes that varied widely. (b) SEmwS′ transformants had dark red (VI) eyes. (c) apoB3′MEmwS′ transformants had primarily red (V) or dark red (VI) eyes. (d) apoB3′MEmw transformants had red (VI) eyes.
FIG. 4.

Eye color phenotypes of Drosophila transformants expressing mini-white from white enhancerless vectors, series 1. Four-day-old females heterozygous for each of the different, single-copy P-element insertions were classified as having light yellow (phenotypic class I), yellow (II), light orange (III), orange (IV), or red (V) eyes. A representative of each of the phenotypes obtained with the different white vectors is shown, and the number of transformed lines with that phenotype is indicated below each picture. (a) mwS′ transformants were widely distributed in all five phenotypic classes. (b) SmwS′ transformants all had light orange (III) eyes. (c) apoB3′MmwS′ transformants had primarily yellow (II) or light orange (III) eyes. (d) Most apoB3′Mmw transformants had yellow (II) eyes, but flies with light orange (III), orange (IV) and red (V) eyes were also obtained.
FIG. 5.
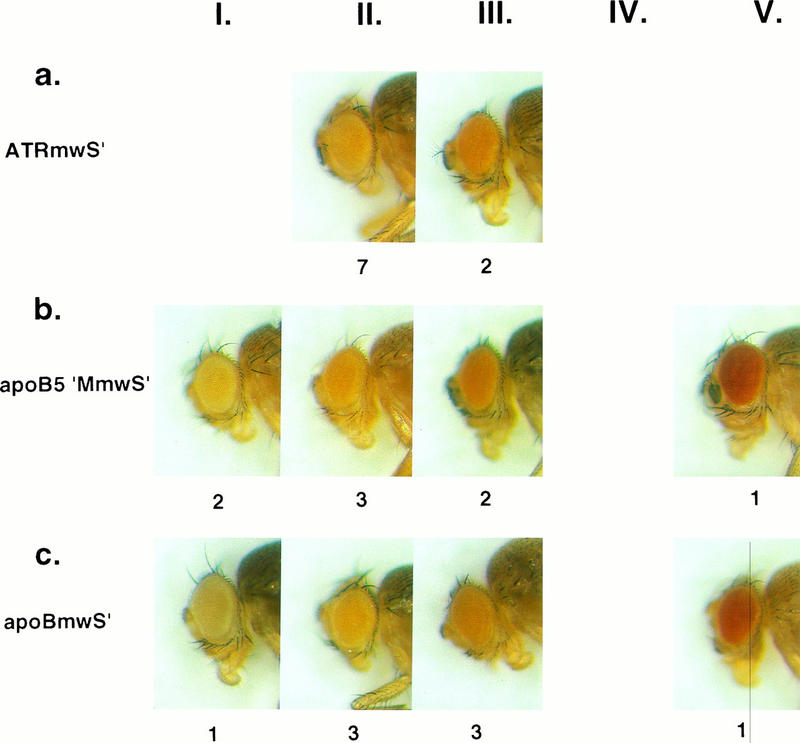
Eye color phenotypes of Drosophila transformants expressing mini-white from white enhancerless vectors, series 2. Four-day-old females heterozygous for each of the different, single-copy P-element insertions were classified as having light yellow (phenotypic class I), yellow (II), light orange (III), orange (IV), or red (V) eyes. A representative of each of the phenotypes obtained with the different white vectors is shown, and the number of transformed lines with that phenotype is indicated below each picture. (a) ATRmwS′ transformants had yellow (II) or light orange (III) eyes. (b) apoB5′MmwS′ transformants and (c) apoBmwS′ transformants were widely distributed in the light yellow (I), yellow (II), light orange (III), and red (V) phenotypic classes.
To determine whether the MAR from the 3′ boundary of the human apoB domain could function as an insular element in Drosophila, transformants containing apoB3′MEmwS′ (Fig. 1c) were prepared. This P-element vector was derived from EmwS′ by inserting a 786-bp XhoI fragment containing the apoB 3′ MAR (29) upstream of the white transcription unit. Seventeen transformed lines were established, all of which contained single P-element insertions (data not shown). Among the 17 single-copy lines, 16 had red (V) or dark red (VI) eyes (Fig. 3c). One transformant had orange (IV) eyes (Fig. 3c). Thus, inserting the apoB 3′ MAR upstream of white in a vector that contained scs′ downstream largely eliminated the phenotypic variation in white gene expression that was observed with a similar vector containing scs′ alone. Furthermore, the predominant eye color phenotype of the apoB3′MEmwS′ transformants, dark red (VI), was the same as that observed in SEmwS′ transformants in which transgene expression was shielded from chromosomal position effects by the Drosophila insulators scs and scs′. These observations suggest that the apoB 3′ MAR can function as an insulator element in Drosophila. An alternate interpretation of these results would be that the apoB 3′ MAR was functioning as a strong enhancer in these experiments, stimulating but not insulating white gene expression. To address this possibility, experiments using enhancer-sensitive vectors were performed.
The human apoB 3′ MAR insulates but does not enhance white gene expression.
To distinguish between the possibilities that the human apoB 3′ MAR was acting as an insulator versus an enhancer, we prepared P-element transformation vectors in which putative enhancement of white gene expression by the apoB MAR would be readily apparent. To do this, constructs similar to those employed in the experiments described above but lacking the white gene enhancers were prepared. The vectors mwS′, SmwS′, and apoB3′MmwS′ (Fig. 1d, e, and f) were similar to EmwS′, SEmwS′, and apoB3′MEmwS′ (Fig. 1a, b, and c), respectively, but they lacked an ∼1.6-kb XhoI/SpeI DNA fragment upstream of mini-white that contains the white gene enhancers (53). white gene expression from such P-element vectors is reduced compared to that from enhancer-containing vectors, so that enhancement of white gene expression can be readily distinguished from insulation (23).
Nineteen mwS′ transformants were isolated, and the genotypes of the transformed lines were determined by Southern hybridization. DNA from each transformant was digested with XbaI, which cuts once within the P-element vector, resolved by electrophoresis, and hybridized with a Drosophila scs′ probe. Figure 2a shows results for six of the mwS′ transformants. As shown in the figure, each line contained an ∼10-kb fragment that corresponded to the endogenous scs′ element at hsp70. In addition, each transformant contained one (Fig. 2a, lanes 1 to 4) or two (lanes 5 to 6) additional scs′ fragments, indicating single or double transgene insertions. In total, 16 of the 19 mwS′ transformants contained single P-element insertions.
As expected, white gene expression in the mwS′ lines, as judged by eye pigmentation, was generally less than that of transformants expressing EmwS′, which includes the white enhancers (compare Fig. 3a and 4a). Nonetheless, the eye color phenotypes of the mwS′ transformants varied considerably, ranging from light yellow (I) to red (V) (Fig. 4a). The 16 mwS′ transformants were widely distributed in five phenotypic classes (I, 1; II, 5; III, 6; IV, 2; and V, 2). Thus, mwS′ transposons, like EmwS′ vectors, were sensitive to chromosomal position effects when integrated in the Drosophila genome. In contrast, all five lines derived from SmwS′-injected embryos fell into a single phenotypic class, light orange (III) (Fig. 4b), indicating that white expression from this P-element vector, which contains scs and scs′, was position-independent.
Twenty-four of 27 lines transformed with apoB3′MmwS′ (Fig. 1f) contained single P-element insertions (data not shown). Twenty of these 24 lines had yellow (II) or light orange (III) eyes (Fig. 4c). These results were similar to those obtained with apoB3′MEmwS′, in that >80% of the transformed lines in each experiment fell into two similar phenotypic classes. Furthermore, only 3 of the 24 apoB3′MmwS′ transformants had orange (IV) eyes, and none of them had red (V) eyes, so the apoB 3′ MAR was clearly not acting as an enhancer in this system (Fig. 4a versus 4c). Thus, inserting the apoB 3′ MAR upstream of the white transcription unit of mwS′ enriched for yellow-light orange transformants and eliminated red transformants without enhancing white gene expression. These data indicate that the apoB 3′ MAR can function as an insulator element in Drosophila. However, the insulating activity of the apoB MAR appeared to be less than that of scs itself, because variation in white gene expression was reduced but not eliminated.
The human ATR MAR functions as an insulator element in Drosophila.
To determine whether other human MARs can function as insulator elements in Drosophila, we utilized a MAR that is located approximately 2 kb downstream of the α1-antitrypsin-related (ATR) sequence on human chromosome 14q32.1. This MAR is one of five matrix-binding elements that we have identified in an ∼120-kb segment of 14q32.1 that includes three related serine protease inhibitor genes, α1AT, ATR, and corticosteroid-binding globulin (CBG) (39; unpublished data). A P-element vector in which the ATR MAR was inserted upstream of the white transcription unit (ATRMmwS′ [Fig. 1g]) was prepared and used to transform w− Drosophila embryos. Nine of 10 transformed lines contained single-copy ATRMmwS′ insertions (data not shown). Seven of the nine single-copy lines had yellow (II) eyes, and two had light orange (III) eyes (Fig. 5a). This distribution of eye color phenotypes was similar to that observed in apoB3′MmwS′ transformants (Fig. 4c), suggesting that the human ATR MAR has insulating properties similar to those of the apoB 3′ MAR.
A putative apoB 5′ MAR does not function as an insulator element in Drosophila.
MARs have been mapped both upstream and downstream of the human apoB gene, and these elements have been proposed to define the limits of the apoB chromatin domain (29). In view of our finding that the apoB 3′ MAR acted as an insulator element in Drosophila, it might have been expected that the apoB 5′ MAR would have similar properties. To test this possibility, an ∼1-kb DNA fragment reported to contain the 5′ apoB MAR was inserted upstream of the white transcription unit in the white enhancerless P-element vector (Fig. 1h). Twelve apoB5′MmwS′ transformants were obtained; eight of these contained single transgene insertions and four contained double transgene insertions (Fig. 2b). The eye color phenotypes of the eight single-copy transformants varied considerably, ranging from light yellow (I) to red (V) (Fig. 5b). This range of phenotypes, without enrichment for any particular phenotypic class, was much like that seen in the mwS′ transformants (Fig. 4a), which do not contain an insulator element upstream of white. These results indicate that the putative apoB 5′ MAR does not function as an insulator element in Drosophila. This observation prompted us to reassess the matrix-binding activity of this human DNA fragment. Despite repeated attempts, we have been unable to detect matrix-binding activity of the apoB 5′ MAR DNA fragment in any of the standard assays (9, 33). Furthermore, DNA sequencing studies demonstrated that the “apoB 5′ MAR” fragment is not particularly AT-rich, nor does it contain the characteristic features of MAR elements (39a). Therefore, the status of this DNA element as a matrix-associated region is uncertain at present.
A DNA segment from within the apoB gene does not insulate in Drosophila.
The results described above suggest that at least some human MARs can function as insulator elements in Drosophila. In contrast, a DNA fragment without matrix-binding activity failed to insulate white gene expression from position effects in Drosophila. To explore this difference further, we prepared a transformation vector in which an ∼800-bp human DNA fragment from within the apoB gene was inserted upstream of the white transcription unit (apoBmwS′ [Fig. 1i]). This human DNA fragment is clearly devoid of matrix-binding activity. Eight of 11 transgenic lines containing apoBmwS′ had single-copy transgene insertions. The eye color phenotypes of these eight transformed lines varied widely, ranging from pale yellow (I) to red (V) (Fig. 5c). These results were similar to those obtained with the other position-sensitive P elements, mwS′ (Fig. 3a) and apoB5′MmwS′ (Fig. 5b). Therefore, this DNA fragment, which contains intron and exon sequences from the human apoB gene, does not function as an insulator element in Drosophila. Thus, human DNA does not have intrinsic insulating activity in this assay, nor is the insulating phenotype due to a distance effect between the white transcription units and Drosophila genomic elements upstream of the P-element insertions.
scs′ affects insulator function of the apoB 3′ MAR in Drosophila.
It has been reported that the scs′ fragment used in these and other mini-white transformation vectors is not fully functional as an insulator element in Drosophila (16, 53). To determine whether scs′ was required for insulator function of the apoB 3′ MAR to be apparent in our assays, transformation vectors containing the apoB 3′ MAR upstream of the white transcription unit, but without scs′ downstream, were prepared. Two different transformation vectors were prepared, one containing the white gene enhancers (apoB3′MEmw [Fig. 1j]) and one without the white enhancers (apoB3′Mmw [Fig. 1k]). Four of seven apoB3′M Emw transformants had single P-element insertions (data not shown), and all four of these lines had dark red (VI) eyes (Fig. 3d). This was the same phenotypic class as most of the apoB3′MEmwS′ transformants, which contained scs′ (Fig. 3c). However, the distribution of eye color phenotypes among the apoB3′Mmw transformants was more complex. Eight of 13 single-copy apoB3′Mmw transformants had yellow (II) eyes (Fig. 4d), like most of the scs′-containing apoB3′MmwS′ transformants (Fig. 4c). However, three of the apoB3′Mmw transformants had red (V) eyes, a phenotype that was not observed among 24 apoB3′MmwS′ transformants. These observations suggest that white transformation vectors containing the apoB 3′ MAR alone are more sensitive to chromosomal position effects than vectors containing both the apoB 3′ MAR and scs′.
Eye color phenotypes versus eye pigment expression—quantitative aspects.
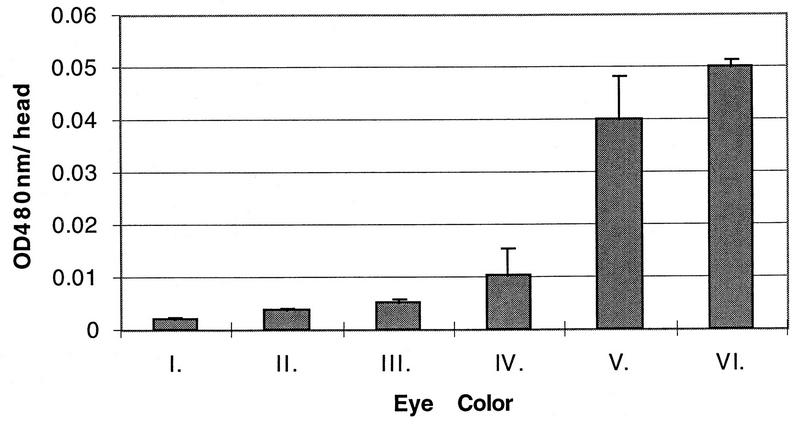
The eye color phenotypes of our transformants varied in a continuous fashion from light yellow to dark red (Fig. 3, 4, and 5). We divided these phenotypes into six classes, I through VI, which could be described as various shades of yellow, orange, and red. To determine how the different phenotypic classes corresponded to the amount of pigment in the Drosophila eyes, eye pigment was quantitated spectrophotometrically in the different phenotypic classes. Transformants from each phenotypic class were obtained, fly heads were pooled in triplicate, pigment was extracted, and absorbance at 480 nm was determined. The mean absorbance per head for each phenotypic class is shown in Fig. 6. The most common phenotypic classes among the enhancerless white transformants, light yellow (I), yellow (II), and light orange (III), differed from each other only about twofold, and the amount of pigment per head varied linearly in this range. Thus, small changes in eye pigment expression among the enhancerless transformants yielded readily discriminated eye color phenotypes. The orange (IV), red (V), and dark red (VI) phenotypes corresponded to approximately 2-, 9-, and 11-fold more pigment than light orange (III), so that eye color readout was a less sensitive indicator of white expression in this range. These data demonstrate that eye pigment expression in our transformants varied over an approximately 20-fold range, and small variations within this range could be readily resolved into the different phenotypic classes. Thus, the eye color phenotypes of white transformants are very sensitive indicators of white gene expression, which makes this system particularly useful as a position effect assay. Among the white enhancerless transformants, the apoB 3′ MAR and the ATR MAR both enriched for yellow (II) to light orange (III) transformants (Fig. 4c and 5a). These phenotypes differed only about 1.2-fold in eye pigment expression. These observations provide further support for the conclusion that these two human MARs can function as insulator elements in Drosophila.
FIG. 6.
Eye pigment expression in the different phenotypic classes. Eye pigment was extracted from pools of each phenotypic class and quantitated spectrophotometrically as described in Materials and Methods. The mean optical density at 480 nm per head is indicated. The phenotypic classes are light yellow (I), yellow (II), light orange (III), orange (IV), red (V), and dark red (VI).
DISCUSSION
MARs are DNA segments that are defined by their abilities to bind to isolated nuclear matrices in vitro (9, 33). These binding properties are consistent with the view that at least some MAR elements might represent structural boundaries of individual chromatin domains, serving to tether the ends of individual chromatin loops to a proteinaceous nuclear matrix in vivo. One functional activity that is often ascribed to DNA elements that are located at or near chromatin domain boundaries is insulation, an activity by which a DNA sequence prevents interactions between neighboring regulatory elements. This suggests that specific DNA segments between neighboring chromatin domains might have both matrix-binding activity and insulator function. Although this model is consistent with currently available data, it has not been critically tested, and alternate interpretations of the data remain viable (26, 54).
The experiments reported here were designed to test whether human MAR elements can function as chromosomal insulators in vivo. To do this, we used the mini-white position effect assay of Kellum and Schedl (23) to study insulator function of human MAR elements in Drosophila. This assay was used previously to demonstrate the insulating properties of scs and scs′, which are well-characterized insulator elements from Drosophila (23). Using this approach, we have demonstrated that two different human MARs can insulate transgenes from position effects in Drosophila. w− flies transformed with P elements in which either the apoB 3′ MAR or the ATR MAR was inserted upstream of a white transcription unit had substantially less variability in white transgene expression than control transformants without human MARs. Furthermore, variability of transgene expression was reduced without increasing the levels of white transgene expression. This contrasts with conclusions drawn from transfection experiments, in which MAR elements have been suggested to increase transgene expression without eliminating variation in expression levels (1, 38). However, the interpretation of transfection studies is complicated by the complex rearrangements of transgene sequences that generally occur in transfected cells. Our experiments clearly show that neither the apoB 3′ MAR nor the ATR MAR enhanced white gene expression in Drosophila, as most transformants that expressed white from enhancerless P-element vectors had yellow or orange eyes irrespective of the presence or absence of MAR elements. This conclusion is in accord with previous studies, in which the apoB 3′ MAR did not enhance expression of transiently transfected reporter genes in mammalian cells (21). Thus, the apoB 3′ MAR and the ATR MAR do not contain associated enhancers, unlike some Drosophila MARs (14), the chicken lysozyme 5′ MAR (45), and MARs from the human immunoglobulin κ and μ (10, 30) and beta interferon (25) genes.
In contrast to results obtained with the apoB 3′ MAR and the ATR MAR, insulator activity was not observed in human DNA segments that were devoid of matrix-binding activity. For example, an ∼800-bp fragment of the apoB gene that included parts of introns 5 and 6 and all of exon 6 had no insulator function in Drosophila. Furthermore, an ∼1,000-bp DNA fragment from the upstream region of apoB also failed to function as an insulator. This result was surprising in view of the fact that this DNA fragment had been reported to have matrix-binding activity; indeed, this element is thought to be the upstream boundary of a 48-kb chromatin domain that contains the apoB gene as its sole resident (29). However, we were unable to detect matrix-binding activity of this DNA fragment in any of the standard assays (9, 33). Moreover, the DNA sequence of this fragment has none of the features that commonly occur in MAR elements, except for a single core binding site for topoisomerase II (42). Thus, we conclude that the putative apoB 5′ MAR has neither matrix-binding nor insulator activities, and we view its identification as the upstream boundary of the apoB domain with skepticism. This suggests that the chromatin domain structure of the human apoB locus may be more complex and extensive than previously envisioned.
Both the apoB 3′ MAR and the ATR MAR insulated white gene expression from position effects in Drosophila. Transformants expressing control vectors without insulator elements upstream of white generally displayed a wide range of eye color phenotypes with no apparent enrichment for any particular phenotypic class. In contrast, most of the transformed lines containing either human MAR upstream of white fell into a single phenotypic class, and variation in white gene expression was greatly reduced. However, a few lines in each collection displayed slightly different eye color phenotypes. Thus, the human apoB 3′ MAR and the ATR MAR reduced, but did not completely eliminate, variability in transgene expression. This suggests that, in some lines, insulation was not complete. This could be due to any of several factors. First, it has been shown that two Drosophila insulators, scs and suppressor-of-hairy-wing [su(HW)], can block the effects of some, but not all, enhancers and repressors in enhancer-blocking assays (7, 41). These assays are thought to mimic some aspects of insulator function. Thus, some of our P-element insertions may have occurred in the vicinity of strong enhancers or repressors, resulting in incomplete insulation. The notion that insulator elements may vary in intrinsic activity is also consistent with the observation that two copies of HS4 of the chicken β-globin locus control region were required for insulator function in Drosophila (8). Second, recent reports suggest that scs′ is a weaker insulator element than either scs or su(HW) (16, 53). The scs′ fragment used in those experiments, and in those described here, is a derivative of a larger scs′ fragment originally employed by Kellum and Schedl (23). This derivative seems to have less insulator activity than the larger scs′ fragment, which may account for some of the variation in white expression in our transformants.
The demonstration that human MARs can function as insulator elements in Drosophila suggests that this activity is evolutionarily conserved. It has already been shown that the matrix-binding activities of MARs from different species are highly conserved (9, 19). However, it is not clear at present whether matrix attachment is required for insulator function. We presume that the two activities are distinct because other insulator elements, including scs, scs′, su(HW), and HS4, are not known to have matrix-binding activity. Conversely, not all MAR elements establish boundaries between chromatin domains, as some of them map within expressed genes (9, 10, 20, 22, 40), so it seems unlikely that all MARs will function as insulators. Thus, we anticipate that insulator function and matrix-binding activities will prove to be separable activities, although they may colocalize to discrete DNA fragments in some instances. These issues will be interesting to explore.
Finally, the genomic locations of the two human MAR elements that have insulator activity are consistent with the possibility that they might represent the boundaries of individual chromatin domains. The apoB 3′ MAR is just downstream of the apoB transcription unit (29), but the chromatin configuration of sequences further downstream has not yet been explored. The ATR MAR is located ∼2 kb downstream of ATR, an antitrypsin-related sequence that may (18) or may not (2) be a pseudogene. The α1AT gene is located ∼32 kb upstream of the ATR MAR, and the CBG gene is ∼33 kb downstream (39). α1AT and CBG are highly expressed in hepatic cells but differentially expressed in macrophages and intestinal epithelium. The entire region is inactive in most other cell types. This provides an opportunity to determine whether the ATR MAR functions as an insulator between putative α1AT and CBG chromatin domains by constructing and analyzing specifically modified human chromosomes using recombination-proficient cell hybrids (12, 13).
ACKNOWLEDGMENTS
We thank Julio Vazquez and Paul Schedl for the transformation vectors pRW+ and pΔXS. Steve Henikoff, Bob Levis, Georgette Sass, and Julio Vazquez contributed many helpful discussions. We thank Ed Giniger, Steve Henikoff, Pierre Rollini, and Steve Tapscott for their critical reviews of the manuscript and Cathy Ludlow for excellent technical assistance.
S.J.N. was the recipient of a National Research Service Award (DK09188) from the NIH. These studies were supported by grant GM26449 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Allen G C, Hall G, Jr, Michalowski S, Newman W, Spiker S, Weissinger A K, Thompson W F. High-level transgene expression in plant cells: effects of a strong scaffold attachment region from tobacco. Plant Cell. 1996;8:899–913. doi: 10.1105/tpc.8.5.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bao J J, Reed-Fourquet L, Sifers R N, Kidd V J, Woo S L. Molecular structure and sequence homology of a gene related to α1-antitrypsin in the human genome. Genomics. 1988;2:165–173. doi: 10.1016/0888-7543(88)90099-7. [DOI] [PubMed] [Google Scholar]
- 3.Blackhart B D, Ludwig E M, Pierotti V R, Caiati L, Onasch M A, Wallis S C, Powell L, Pease R, Knott T J, Chu M L, et al. Structure of the human apolipoprotein B gene. J Biol Chem. 1986;261:15364–15367. [PubMed] [Google Scholar]
- 4.Bode J, Maass K. Chromatin domain surrounding the human interferon-β gene as defined by scaffold-attached regions. Biochemistry. 1988;27:4706–4711. doi: 10.1021/bi00413a019. [DOI] [PubMed] [Google Scholar]
- 5.Bonifer C, Vidal M, Grosveld F, Sippel A E. Tissue-specific and position-independent expression of the complete gene domain for chicken lysozyme in transgenic mice. EMBO J. 1990;9:2843–2848. doi: 10.1002/j.1460-2075.1990.tb07473.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breyne P, van Montagu M, Depicker N, Gheysen G. Characterization of a plant scaffold attachment region in a DNA fragment that normalizes transgene expression in tobacco. Plant Cell. 1992;4:463–471. doi: 10.1105/tpc.4.4.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai H, Levine M. Modulation of enhancer-promoter interactions by insulators in the Drosophila embryo. Nature. 1995;376:533–536. doi: 10.1038/376533a0. [DOI] [PubMed] [Google Scholar]
- 8.Chung J H, Whiteley M, Felsenfeld G. A 5′ element of the chicken β-globin domain serves as an insulator in human erythroid cells and protects against position effect in Drosophila. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 9.Cockerill P N, Garrard W T. Chromosomal loop anchorage of the κ immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 10.Cockerill P N, Yuen M H, Garrard W T. The enhancer of the immunoglobulin heavy chain locus is flanked by presumptive chromosomal loop anchorage elements. J Biol Chem. 1987;262:5394–5397. [PubMed] [Google Scholar]
- 11.Conkling M A, Cheng C-L, Yamamoto Y T, Goodman H M. Isolation of transcriptionally regulated root-specific genes from tobacco. Plant Physiol. 1990;93:1203–1211. doi: 10.1104/pp.93.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dieken E S, Epner E M, Fiering S, Fournier R E K, Groudine M. Efficient modification of human chromosomal alleles using recombination-proficient chicken/human microcell hybrids. Nat Genet. 1996;12:174–182. doi: 10.1038/ng0296-174. [DOI] [PubMed] [Google Scholar]
- 13.Dieken E S, Fournier R E K. Homologous modification of human chromosomal genes in chicken B-cell x human microcell hybrids. Methods. 1996;9:56–63. doi: 10.1006/meth.1996.0008. [DOI] [PubMed] [Google Scholar]
- 14.Gasser S M, Laemmli U K. Cohabitation of scaffold binding regions with upstream/enhancer elements of three developmentally regulated genes of D. melanogaster. Cell. 1986;46:521–530. doi: 10.1016/0092-8674(86)90877-9. [DOI] [PubMed] [Google Scholar]
- 15.Grosveld F, van Assendelft G B, Greaves D R, Kollias G. Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 16.Hagstrom K, Muller M, Schedl P. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 1996;10:3202–3215. doi: 10.1101/gad.10.24.3202. [DOI] [PubMed] [Google Scholar]
- 17.Hazelrigg T, Levis R, Rubin G M. Transformation of white locus DNA in Drosophila: dosage compensation, zeste interaction, and position effects. Cell. 1984;36:469–481. doi: 10.1016/0092-8674(84)90240-x. [DOI] [PubMed] [Google Scholar]
- 18.Hofker M H, Nelen M, Klasen E C, Nukiwa T, Curiel D, Crystal R G, Frants R R. Cloning and characterization of an α1-antitrypsin-like gene 12 kb downstream of the genuine α1-antitrypsin gene. Biochem Biophys Res Commun. 1988;155:634–642. doi: 10.1016/s0006-291x(88)80542-4. [DOI] [PubMed] [Google Scholar]
- 19.Izaurralde E, Mirkovitch J, Laemmli U K. Interaction of DNA with nuclear scaffolds in vitro. J Mol Biol. 1988;200:111–125. doi: 10.1016/0022-2836(88)90337-3. [DOI] [PubMed] [Google Scholar]
- 20.Jarman A P, Higgs D R. Nuclear scaffold attachment sites in the human globin gene complexes. EMBO J. 1988;7:3337–3344. doi: 10.1002/j.1460-2075.1988.tb03205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kalos M, Fournier R E K. Position-independent transgene expression mediated by boundary elements from the apolipoprotein B chromatin domain. Mol Cell Biol. 1995;15:198–207. doi: 10.1128/mcb.15.1.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kas E, Chasin L A. Anchorage of the Chinese hamster dihydrofolate reductase gene to the nuclear scaffold occurs in an intragenic region. J Mol Biol. 1987;198:677–692. doi: 10.1016/0022-2836(87)90209-9. [DOI] [PubMed] [Google Scholar]
- 23.Kellum R, Schedl P. A position-effect assay for boundaries of higher order chromosomal domains. Cell. 1991;64:941–950. doi: 10.1016/0092-8674(91)90318-s. [DOI] [PubMed] [Google Scholar]
- 24.Kirillov A, Kistler B, Mostoslavsky R, Cedar H, Wirth T, Bergman Y. A role for nuclear NF-κB in B-cell-specific demethylation of the Igκ locus. Nat Genet. 1996;13:435–441. doi: 10.1038/ng0895-435. [DOI] [PubMed] [Google Scholar]
- 25.Klehr D, Maass K, Bode J. Scaffold-attached regions from the human interferon β domain can be used to enhance the stable expression of genes under the control of various promoters. Biochemistry. 1991;30:1264–1270. doi: 10.1021/bi00219a015. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K, Kas E, Poljak L, Adachi Y. Scaffold-associated regions: cis-acting determinants of chromatin structural loops and functional domains. Curr Opin Genet Dev. 1992;2:275–285. doi: 10.1016/s0959-437x(05)80285-0. [DOI] [PubMed] [Google Scholar]
- 27.Levis R, Hazelrigg T, Rubin G M. Effects of genomic position on the expression of transduced copies of the white gene of Drosophila. Science. 1985;229:558–561. doi: 10.1126/science.2992080. [DOI] [PubMed] [Google Scholar]
- 28.Levy-Wilson B. Transcriptional control of the human apolipoprotein B gene in cell culture and in transgenic animals. Prog Nucleic Acids Res Mol Biol. 1995;50:161–190. doi: 10.1016/s0079-6603(08)60814-4. [DOI] [PubMed] [Google Scholar]
- 29.Levy-Wilson B, Fortier C. The limits of the DNase I-sensitive domain of the human apolipoprotein B gene coincide with the locations of chromosomal anchorage loops and define the 5′ and 3′ boundaries of the gene. J Biol Chem. 1989;264:21196–21204. [PubMed] [Google Scholar]
- 30.Lichtenstein M, Keini G, Cedar H, Bergman Y. B cell-specific demethylation: a novel role for the intronic κ chain enhancer sequence. Cell. 1994;76:913–923. doi: 10.1016/0092-8674(94)90365-4. [DOI] [PubMed] [Google Scholar]
- 31.McKnight R A, Shamay A, Sankaran L, Wall R J, Hennighausen L. Matrix-attachment regions can impart position-independent regulation of a tissue-specific gene in transgenic mice. Proc Natl Acad Sci USA. 1992;89:6943–6947. doi: 10.1073/pnas.89.15.6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McKnight R A, Spencer M, Wall R J, Hennighausen L. Severe position effects imposed on a 1 kb mouse whey acidic protein gene promoter are overcome by heterologous matrix attachment regions. Mol Reprod Dev. 1996;44:179–184. doi: 10.1002/(SICI)1098-2795(199606)44:2<179::AID-MRD6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 33.Mirkovitch J, Mirault M E, Laemmli U K. Organization of the higher-order chromatin loop: specific DNA attachment sites on nuclear scaffold. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- 34.Palmiter R D, Sandgren E P, Koeller D M, Brinster R L. Distal regulatory elements from the mouse metallothionein locus stimulate gene expression in transgenic mice. Mol Cell Biol. 1993;13:5266–5275. doi: 10.1128/mcb.13.9.5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phi-Van L, Stratling W H. The matrix attachment regions of the chicken lysozyme gene co-map with the boundaries of the chromatin domain. EMBO J. 1988;7:655–664. doi: 10.1002/j.1460-2075.1988.tb02860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phi-Van L, Stratling W H. Dissection of the ability of the chicken lysozyme gene 5′ matrix attachment region to stimulate transgene expression and to dampen position effects. Biochemistry. 1996;35:10735–10742. doi: 10.1021/bi9603783. [DOI] [PubMed] [Google Scholar]
- 37.Phi-Van L, von Kries J P, Ostertag W, Stratling W H. The chicken lysozyme 5′ matrix attachment region increases transcription from a heterologous promoter in heterologous cells and dampens position effects on the expression of transfected genes. Mol Cell Biol. 1990;10:2302–2307. doi: 10.1128/mcb.10.5.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poljak L, Seum C, Mattioni T, Laemmli U K. SARs stimulate but do not confer position independent gene expression. Nucleic Acids Res. 1994;22:4386–4394. doi: 10.1093/nar/22.21.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rollini P, Fournier R E K. Molecular linkage of the human α1-antitrypsin and corticosteroid-binding globulin genes on chromosome 14q32.1. Mamm Genome. 1997;8:913–916. doi: 10.1007/s003359900610. [DOI] [PubMed] [Google Scholar]
- 39a.Rollini, P., and R. E. K. Fournier. Unpublished results.
- 40.Romig H, Ruff J, Fackelmayer F O, Patil M S, Richter A. Characterisation of two intronic nuclear-matrix-attachment regions in the human DNA topoisomerase I gene. Eur J Biochem. 1994;221:411–419. doi: 10.1111/j.1432-1033.1994.tb18753.x. [DOI] [PubMed] [Google Scholar]
- 41.Roseman R R, Pirrotta V, Geyer P K. The su(Hw) protein insulates expression of the Drosophila melanogaster white gene from chromosomal position-effects. EMBO J. 1993;12:435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sander M, Hsieh T S. Drosophila topoisomerase II double-strand DNA cleavage: analysis of DNA sequence homology at the cleavage site. Nucleic Acids Res. 1985;13:1057–1072. doi: 10.1093/nar/13.4.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schoffl F, Schroder G, Kliem M, Rieping M. An SAR sequence containing 395 bp DNA fragment mediates enhanced, gene-dosage-correlated expression of a chimaeric heat shock gene in transgenic tobacco plants. Transgenic Res. 1993;2:93–100. doi: 10.1007/BF01969382. [DOI] [PubMed] [Google Scholar]
- 44.Spradling A C, Rubin G M. Transposition of cloned P-elements into Drosophila germ line chromosomes. Science. 1982;218:341–347. doi: 10.1126/science.6289435. [DOI] [PubMed] [Google Scholar]
- 45.Stief A, Winter D M, Stratling W H, Sippel A E. A nuclear DNA attachment element mediates elevated and position-independent gene activity. Nature. 1989;341:343–345. doi: 10.1038/341343a0. [DOI] [PubMed] [Google Scholar]
- 46.Strick R, Laemmli U K. SARs are cis DNA elements of chromosome dynamics: synthesis of a SAR repressor protein. Cell. 1995;83:1137–1148. doi: 10.1016/0092-8674(95)90140-x. [DOI] [PubMed] [Google Scholar]
- 47.Strissel P L, Espinosa III R, Rowley J D, Swift H. Scaffold attachment regions in centromere-associated DNA. Chromosoma. 1996;105:122–133. doi: 10.1007/BF02509522. [DOI] [PubMed] [Google Scholar]
- 48.Talbot D, Descombes P, Schibler U. The 5′ flanking region of the rat LAP (C/EBP β) gene can direct high-level, position-independent, copy number-dependent expression in multiple tissues in transgenic mice. Nucleic Acids Res. 1994;22:756–766. doi: 10.1093/nar/22.5.756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson E M, Christians E, Stinnakre M G, Renard J P. Scaffold attachment regions stimulate HSP70.1 expression in mouse preimplantation embryos but not in differentiated tissues. Mol Cell Biol. 1994;14:4694–4703. doi: 10.1128/mcb.14.7.4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Udvardy A, Maine E, Schedl P. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol. 1985;185:341–358. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- 51.van der Geest A H, Hall A H M, Spiker S, Hall T C. The β-phaseolin gene is flanked by matrix attachment regions. Plant J. 1994;6:413–423. [Google Scholar]
- 52.van der Geest A H, Hall T C. The β-phaseolin 5′ matrix attachment region acts as an enhancer facilitator. Plant Mol Biol. 1997;33:553–557. doi: 10.1023/a:1005765525436. [DOI] [PubMed] [Google Scholar]
- 53.Vazquez J, Schedl P. Sequences required for enhancer blocking activity of scs are located within two nuclease-hypersensitive regions. EMBO J. 1994;13:5984–5993. doi: 10.1002/j.1460-2075.1994.tb06944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou J, Barolo S, Szymanski P, Levine M. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev. 1996;10:3195–3201. doi: 10.1101/gad.10.24.3195. [DOI] [PubMed] [Google Scholar]