Abstract
Uptake of uracil by the yeast Saccharomyces cerevisiae is mediated by a specific permease encoded by the FUR4 gene. Uracil permease located at the cell surface is subject to two covalent modifications: phosphorylation and ubiquitination. The ubiquitination step is necessary prior to permease endocytosis and subsequent vacuolar degradation. Here, we demonstrate that a PEST-like sequence located within the cytoplasmic N terminus of the protein is essential for uracil permease turnover. Internalization of the transporter was reduced when some of the serines within the region were converted to alanines and severely impaired when all five serines within the region were mutated or when this region was absent. The phosphorylation and degree of ubiquitination of variant permeases were inversely correlated with the number of serines replaced by alanines. A serine-free version of this sequence was very poorly phosphorylated, and elimination of this sequence prevented ubiquitination. Thus, it appears that the serine residues in the PEST-like sequence are required for phosphorylation and ubiquitination of uracil permease. A PEST-like sequence in which the serines were replaced by glutamic acids allowed efficient permease turnover, suggesting that the PEST serines are phosphoacceptors.
Plasma membrane proteins contribute to communication between the interior and the exterior of the cell. Consequently, their expression at the cell surface must be tightly regulated to respond to changes in the extracellular environment. Endocytosis is a way of controlling the levels of many plasma membrane proteins, which are subsequently routed to the lysosome (or vacuole, in Saccharomyces cerevisiae) where they are destroyed by specific proteases. In higher cells, various types of signals, including tyrosine- and di-leucine-based signals, mediate internalization from the plasma membrane (36). Ubiquitination of several yeast and mammalian membrane proteins at the plasma membrane signals their endocytosis and subsequent vacuolar degradation (19). Examples include the two pheromone receptors Ste2p and Ste3p (18, 35); the ABC transporters Pdr5p and Ste6p (7, 23); the transporters Fur4p, Gal2p, and Gap1p (11, 21, 41) in S. cerevisiae; and the mammalian growth hormone receptor (43). Many other proteins probably undergo ubiquitin-mediated endocytosis, since a number of receptors in higher cells are ubiquitinated at the plasma membrane (5).
The covalent linkage of ubiquitin to lysine residues of substrate proteins is a common means used by eucaryotic cells to signal their degradation by the 26S proteasome, a multiprotease complex located in the cytoplasm and the nucleus (5). Conjugation of ubiquitin to proteins proceeds in three steps involving ubiquitin-activating enzymes (i.e., E1), ubiquitin-conjugating enzymes (i.e., ubc or E2), and ubiquitin ligases (i.e., E3). The component of the ubiquitin conjugation system that is thought to be the most directly involved in substrate recognition is E3. The signals that lead to ubiquitination of most naturally occurring substrates are largely unknown. The N-end rule established a relationship between the N-terminal amino acid of certain proteins and their susceptibility to ubiquitination (29). Mitotic cyclins are targeted to the ubiquitin proteolytic pathway by a consensus sequence called the destruction box (15). Rogers et al. reported that short-lived proteins generally contain regions enriched with Pro, Glu, Ser, and Thr. These so-called PEST regions were identified by statistical means as signals for protein instability (34). It was then demonstrated that PEST sequences indeed control the ubiquitination of regulatory short-lived proteins, such as transactivator Gcn4 (25) and G1 cyclins in yeast and mammalian cells. Phosphorylation of particular Ser or Thr residues in the PEST regions of these G1 cyclins specifies their recognition and processing by the ubiquitin-proteasome pathway (26, 48–50).
Regions involved in the control of ubiquitination of plasma membrane proteins have only partially been characterized (18, 24). Ligand binding induces hyperphosphorylation, rapid ubiquitination, and endocytosis of the yeast Ste2p receptor. The amino acid sequence SINNDAKSS of a truncated form of Ste2p was found to control its ubiquitination. Mutations replacing the serines within this signal abolishes ubiquitination of the receptor. Hicke and Riezman suggested that ligand-induced phosphorylation of these residues may precede and be required for ubiquitination of the receptor (18). Other cell surface proteins responding to ubiquitination have been shown to be phosphorylated. However, the role of this posttranslational modification is poorly understood (21, 35, 42, 46). Dephosphorylation of the general amino acid permease Gap1p was observed when the protein was inactivated upon transfer of the cells to repressing conditions (42). The inactivation and/or degradation of some yeast sugar transporters has been suggested to be accelerated by phosphorylation of potential PEST regions (3). However, it is unknown whether these transporters possess destabilizing regions and whether phosphorylation affects their stability.
The yeast uracil permease appears to be a good model for investigating the link, if any, between phosphorylation and ubiquitination in the turnover of a membrane protein. It is a multispanning membrane protein encoded by the FUR4 gene (4, 22). Garnier et al. have developed a two-dimensional model of its structure in which the cytoplasmic orientations of both termini and the orientations of most connecting loops with respect to the membrane were determined (13). The FUR4 gene belongs to the FUR family of homologous yeast genes (1, 31) comprising three other S. cerevisiae genes, the DAL4 gene, which encodes the allantoin permease (51), the THI10 gene, which encodes the thiamine permease (8), the open reading frame YBL042c, which encodes the uridine permease, Upl1p (20), and the gene encoding the uracil permease of Schizosaccharomyces pombe (GenBank accession no. X98696). Their deduced amino acid sequences are all very similar to that of Fur4p, especially for the hydrophobic cores of these proteins, which are probably involved in the transport function (45). The region of high similarity excludes both part of the N terminus and the entire C terminus of Fur4p. It is possible that these divergent regions are involved in the control of the stability of the proteins. Newly synthesized uracil permease is delivered to the plasma membrane via the secretory pathway, and several serine residues are phosphorylated in a post-Golgi compartment on its way to or at the plasma membrane (46). The turnover of uracil permease is constitutive and accelerated under stress conditions such as nutrient starvation and inhibition of protein synthesis (47). In contrast to receptors that display hyperphosphorylation upon ligand binding, triggering their subsequent internalization (18, 35), no variation in the degree of phosphorylation of the permease has been observed under conditions of accelerated turnover. Uracil permease undergoes cell surface ubiquitination, a process required for subsequent internalization (11). The endocytosed permease is then targeted to the vacuole for proteolysis (11, 47). Permease ubiquitination is mediated by the essential Npi1p (also known as Rsp5p) ubiquitin-protein ligase, which is also required for the degradation of two other yeast transporters, Gap1p and the maltose permease (17, 27). We investigated the relationship between the phosphorylation status of the permease and its ubiquitin-dependent turnover. We analyzed by mutagenesis the role, if any, of serines located in the two extremities of the protein which are rich in PEST residues.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The S. cerevisiae strains used were NC122sp6 (MATa leu2 fur4Δ) (22); 27061b (MATa ura3 trp1), derived from the wild-type strain Σ1278b (2); W303-1B/D (MATα ade2-1 ura3-1 his3-11 leu2-3,112 trp1) (44); NY279 (MATa ura3-52 act1-3) (carrying a single point mutation in the ACT1 gene identical to that present in act1-1) (39); and isogenic parental strain NY13 (MATa ura3-52). The chromosome-encoded uracil permease is produced in very small amounts, and cells that produce the permease from multicopy plasmids were used for accurate measurement of the permease activity and for the immunodetection of the protein (46). The multicopy plasmid pfF (2μm LEU2 FUR4) carries the FUR4 gene (22) under the control of its own promoter. The multicopy plasmid p195gF (2μm URA3 GAL-FUR4) (47) carries the FUR4 gene under the control of the GAL10 promoter. Yeast strains were transformed according to the method of Gietz et al. (14). Cells were grown at 30°C (or 24°C for thermosensitive strains) in minimal medium that contained 0.67% yeast nitrogen base without amino acids (Difco) and was supplemented with appropriate nutrients. Yeast cells to be labeled with [32P]orthophosphate were grown in a low-phosphate medium, as described in reference 40. The carbon source was 2% glucose or 4% galactose–0.05% glucose. One A600 unit corresponds to approximately 2.107 cells per ml.
Mutagenesis.
Point mutations and the deletion of residues 42 to 60 in the FUR4 gene were performed by site-directed mutagenesis with a Stratagene Chameleon double-stranded-DNA site-directed mutagenesis kit as described by the supplier. Mutated constructs were identified by testing for restriction site polymorphisms introduced by the mutagenic primers. Mutations were confirmed by sequencing with double-strand DNA (37) and a Sequenase version 2.0 kit (U.S. Biochemicals). For each mutagenesis, two independent mutant plasmids were used to transform yeast and two yeast transformants were analyzed for each mutant plasmid.
Measurement of uracil uptake.
Uracil uptake was measured in exponentially growing cells as previously described (46). One milliliter of yeast culture was incubated with 5 μM [14C]uracil (ICN Pharmaceuticals) for 20 s at 30°C and then quickly filtered through Whatman GF/C filters, which were washed twice with ice-cold water and subjected to counting for radioactivity.
Yeast cell extracts and Western immunoblotting.
Cell extracts were prepared, and proteins were analyzed by immunoblotting as previously described with an antiserum to the last 10 residues of uracil permease (47). Primary antibodies were detected with a horseradish peroxidase-conjugated anti-rabbit immunoglobulin G secondary antibody revealed by enhanced chemiluminescence (ECL; Amersham).
Membrane preparation.
Yeast cells (80 A600 units) in exponential growth phase were harvested by centrifugation in the presence of 10 mM sodium azide and used to prepare plasma membrane-enriched fractions as previously described (11). Membrane-bound proteins were analyzed by Western blotting as described above.
Pulse-chase labeling and immunoprecipitation.
W303-1B/D cells grown with galactose as a carbon source to an appropriate A600 were labeled by adding 50 μCi of [35S]methionine (Amersham) or 80 μCi of [32P]orthophosphate (Amersham) per ml directly in the growth medium. For [35S]methionine labeling, cells were labeled for 10 min and then chased with 10 mM cold methionine for 120 min. Aliquots of the culture (0.8 ml) were removed at various times during the chase, and proteins were processed for immunoprecipitation as described previously (46). For [32P]orthophosphate labeling, cells were labeled for 1 h and then 10 mM pyrophosphate was added and the assay mixture (0.8 ml) was treated as described above. The rabbit antiserum used for the immunoprecipitation experiments, kindly provided by M. O. Blondel (our laboratory), was raised against the hydrophilic N terminus of uracil permease (amino acids 1 to 141) fused to the maltose-binding protein (MBP) with the pMAL-c2 vector from Biolabs. Immunoprecipitated proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), dried gels were read with a PhosphorImager (Molecular Dynamics), and bands were quantified with Imagequant software.
Alkaline phosphatase treatment.
Cell extracts were prepared as described above, except that EDTA was omitted. Protein extracts were diluted 1:10 with phosphatase buffer and incubated for 15 min at 37°C with or without molecular-biology-grade calf intestinal alkaline phosphatase as recommended by the supplier (Boehringer). Proteins were then precipitated with 10% trichloroacetic acid, resuspended in a twofold-concentrated sample buffer, and analyzed by immunoblotting.
RESULTS
Serines in a PEST-like sequence are required for uracil permease turnover.
Uracil permease is phosphorylated on several unidentified serine residues (46). We used point mutation analysis to investigate the role of serines at potential phosphorylation sites in the cytoplasmic extremities of Fur4p and to identify regions required for the protein turnover. Serines in these extremities were substituted for alanine (Fig. 1A). The Fur4p variants were produced in a strain in which the chromosomal copy of the FUR4 gene had been disrupted. Permease activity, corresponding to protein located in the plasma membrane, was assayed in cells expressing either wild-type or mutated permease genes after inhibition of protein synthesis. Addition of cycloheximide to cells producing the wild-type permease caused a drop in permease activity (Fig. 1B). Similar results were obtained with the Fur4p variants in which Ser12, -13, and -18; Ser24 and -25; or Ser622 (the only serine in the C-terminal domain) was replaced. In contrast, the uptake of uracil was maintained in cells harboring Fur4p variants with replacements of Ser43 or Ser55 and -56. The relative protection (1.6- to 2-fold) against loss of activity suggested that the corresponding mutations stabilized the transporter at the plasma membrane.
FIG. 1.
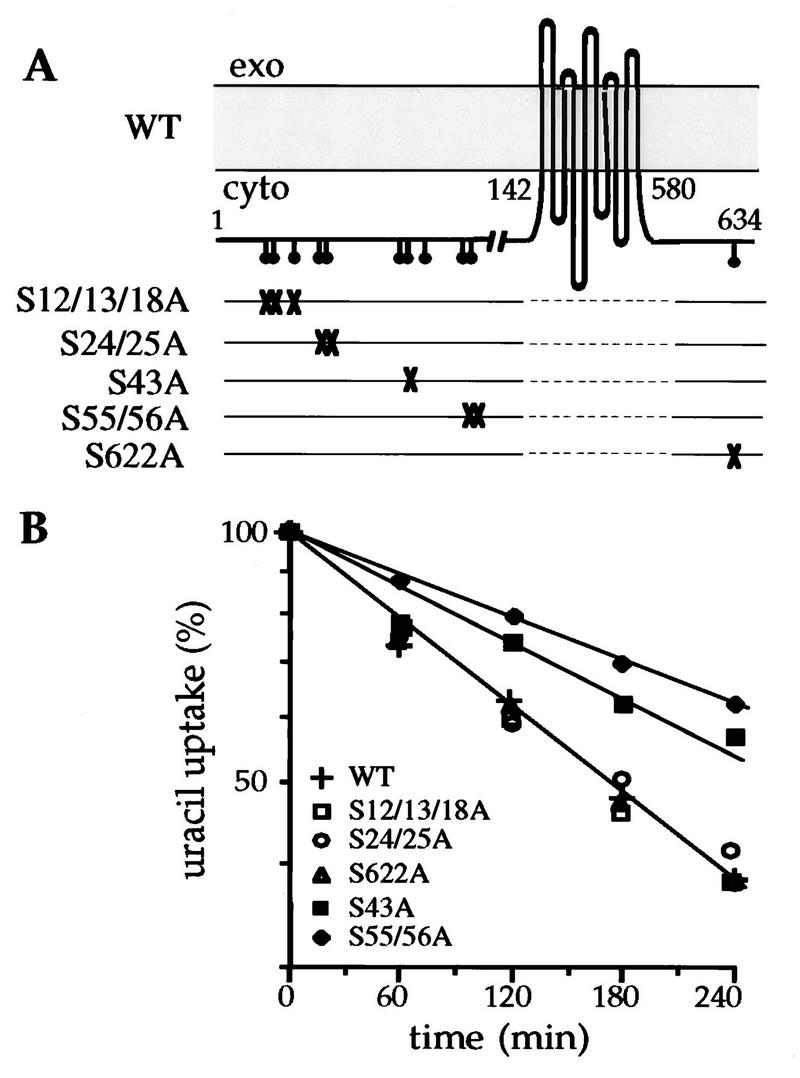
Effects of serine substitutions on the loss of uracil uptake after inhibition of protein synthesis. (A) Topological model of uracil permease in the membrane (13), showing the serines (•) in potential phosphorylation sites in the N and C termini. For each mutant represented on a separate line, the serines replaced by alanines are indicated with an X. (B) NC122sp6 cells transformed with plasmid pfF or derivatives carrying the variant permease genes were grown to logarithmic phase on glucose medium. Cycloheximide (50 μg/ml) was then added to the medium, and uracil uptake was measured at the times indicated. Results are percentages of initial activities. WT, wild type; exo, extracellular medium; cyto, cytoplasm.
Ser43 and Ser55 and -56 are located within the longest sequence of contiguous PEST residues in the protein. This sequence extends from amino acids 42 to 59. PEST-rich sequences are potential degradation signals. It has been reported that phosphorylation of serines or threonines can activate latent PEST sequences (33). To determine whether other serines in the PEST-like sequence of Fur4p are required for Fur4p’s turnover, we replaced each of the five serines within region 42 to 59 with alanines and tested the effects on Fur4p stability after inhibition of protein synthesis. We also deleted the entire PEST-like sequence (Fig. 2A). First, we monitored cell surface delivery of the variant uracil permeases. Wild-type and mutant permease were produced from the inducible GAL10 promoter by addition of galactose to cells grown on lactate. Uracil uptake was monitored for 2 h. Permease activity appeared at the same level and with the same kinetics in all cells, whatever the permease produced (data not shown). Thus, the mutations did not delay delivery of uracil permease to the plasma membrane. We then compared levels of internalization of wild-type and variant permeases. Addition of cycloheximide caused a sharp decrease in uracil uptake by cells grown on galactose (rather than glucose) and producing wild-type permease (Fig. 2B). Extracts from cells withdrawn at different times after addition of cycloheximide were analyzed by immunoblotting. Permease immunoreactivity declined in parallel to the drop in uracil uptake (Fig. 2C). Uracil uptake decreased less rapidly in cells containing the double mutation S55A-S56A (2SA). The relative protection was 1.4-fold. The 2SA mutation also protected permease against degradation. Similar results were obtained with the single mutation S43A (data not shown). When the three serines S43, S55, and S56 were replaced by alanine (3SA), the loss of uracil uptake was further slowed (relative protection, 2.7-fold) and the permease was more resistant to degradation. When all five serines in the sequence were replaced (5SA) or when the entire sequence was deleted (ΔPEST), almost no loss in uracil uptake could be detected and the amount of immunodetected permease was maintained throughout the 2 h of the experiment. Therefore, replacement of serine residues within the PEST-like sequence by alanines or deletion of the sequence prevented the degradation of the permease observed upon inhibition of protein synthesis. Moreover, the mutant proteins were stabilized at the cell surface as judged by the maintenance of uracil uptake. Therefore, stress-induced degradation of the permease is dependent of the PEST-like region.
FIG. 2.
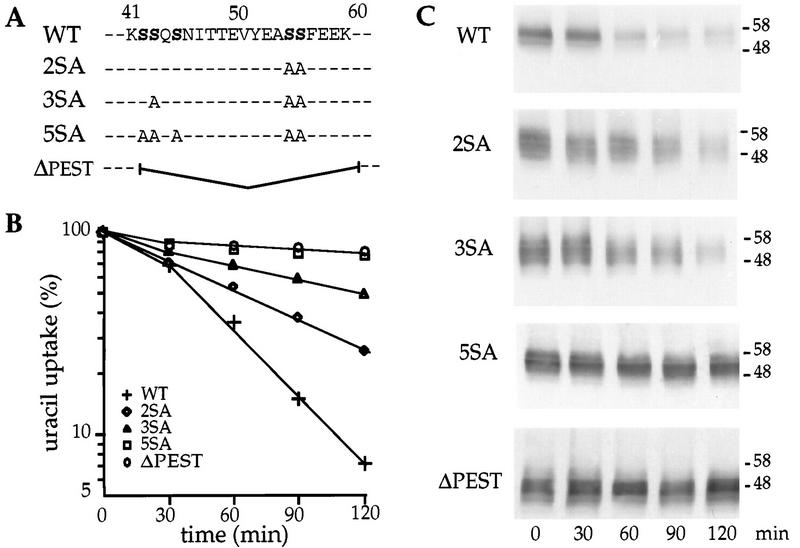
Defective internalization of mutant versions of uracil permease in cells in which protein synthesis has been inhibited. (A) The PEST-like sequence of the permease is indicated by the one-letter code. For each mutant, the serines replaced by alanines or the 19-residue deletion is indicated. (B) 27061b cells transformed with plasmid p195gF or derivatives carrying the variant permease genes were grown to logarithmic phase on galactose medium. The turnover of uracil permease is significantly faster when cells are supplied with galactose rather than glucose (11). Cycloheximide (50 μg/ml) was then added to the medium, and uracil uptake was measured at the times indicated. Results are percentages of initial activities. (C) Protein extracts were prepared at the same times and analyzed for uracil permease by Western immunoblotting. The molecular masses of the markers are given at the right in kilodaltons. WT, wild type.
We then tested whether the constitutive turnover is also affected in the mutant permeases. The abundance of the mutant permeases was 1.5- to 4.5-fold higher, depending of the number of introduced mutations, than that of the native permease as estimated by immunoblotting serial dilutions of cell extracts (data not shown). This result suggested that the protection against degradation is also effective in growing cells. To firmly establish that the constitutive turnover of Fur4p in exponentially growing cells depends on the PEST-like region, we investigated the life spans of the permeases by pulse-chase labeling. Exponentially growing cells were labeled for 10 min with [35S]methionine and then chased with cold methionine for appropriate times. Fur4p was immunoprecipitated and analyzed by SDS-PAGE (Fig. 3A). Native and variant permeases were synthesized at the same rate: the same amount of permease was immunoprecipitated after a 10-min labeling of cells producing any of these proteins (Fig. 3B). A slight decrease in electrophoretic mobility appeared after a 30-min chase for the wild-type permease, which resulted from the phosphorylation of the permease at the plasma membrane (46). A similar mobility shift was observed only for the 2SA variant. The decay of each permease was determined by quantitative analysis by PhosphorImaging (Fig. 3B). Serine-to-alanine substitutions slowed down permease turnover. The half-life of the 2SA permease was 3.6-fold longer than that of the wild-type permease. The variants with three to five serine-to-alanine replacements were almost not degraded during the 2-h chase period. Similarly, permease lacking the entire PEST-like sequence was stable. These results demonstrate that the constitutive turnover of uracil permease is dependent on the PEST-like region and in particular on its serine residues.
FIG. 3.
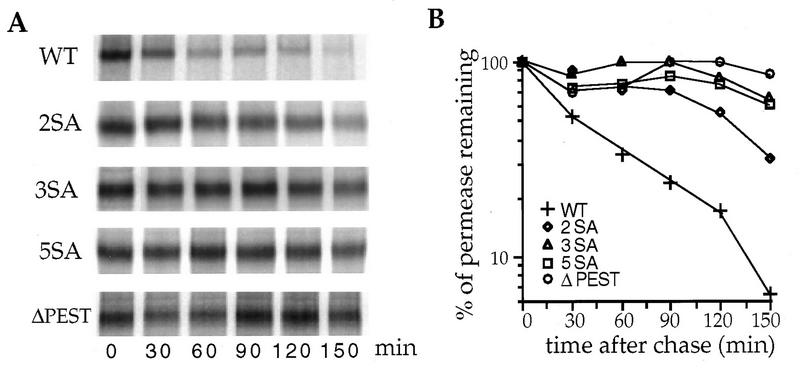
Stabilization of mutant versions of uracil permease during cell growth. Strain W303-1B/D was used because uracil permease is particularly unstable in this genetic background. Cells transformed with plasmid p195gF or derivatives carrying the variant permease genes were grown to mid-log phase on galactose medium. Cells were labeled by incubation for 10 min with [35S]methionine in the growth medium and chased for the indicated times. Uracil permease was immunoprecipitated and resolved by SDS-PAGE. (A) PhosphorImager readout. (B) For each variant, the intensities of the bands were measured with Imagequant software. Results are percentages of the time zero bands. WT, wild type.
Replacement of serines in the PEST-like sequence affects the phosphorylation status of the permease.
Under steady-state conditions, phosphorylation of the permease causes it to give several bands on immunoblots. The slower-running bands correspond to higher levels of phosphorylation, as revealed by alkaline phosphatase treatment (46). We compared the electrophoretic mobilities of the variant permeases (Fig. 4A). Serine-to-alanine substitutions in the PEST-like region altered the banding pattern of the permease. The progressive loss of slower-migrating bands and appearance of faster-migrating bands correlated with the number of substitutions. The ΔPEST variant migrated faster than the other variant proteins due to the 19-residue deletion. Treatment of extracts containing wild-type permease with alkaline phosphatase resulted in a change in the electrophoretic pattern with the appearance of faster-running bands (Fig. 4B). Alkaline phosphatase treatment only slightly affected the banding pattern of the 5SA variant. Moreover, the banding pattern of the 5SA variant was roughly similar to that of the wild-type permease when the latter extract was treated with alkaline phosphatase. This result suggests that the 5SA variant may be poorly phosphorylated. However, some phosphorylation events may be insensitive to alkaline phosphatase or may not affect electrophoretic mobility.
FIG. 4.
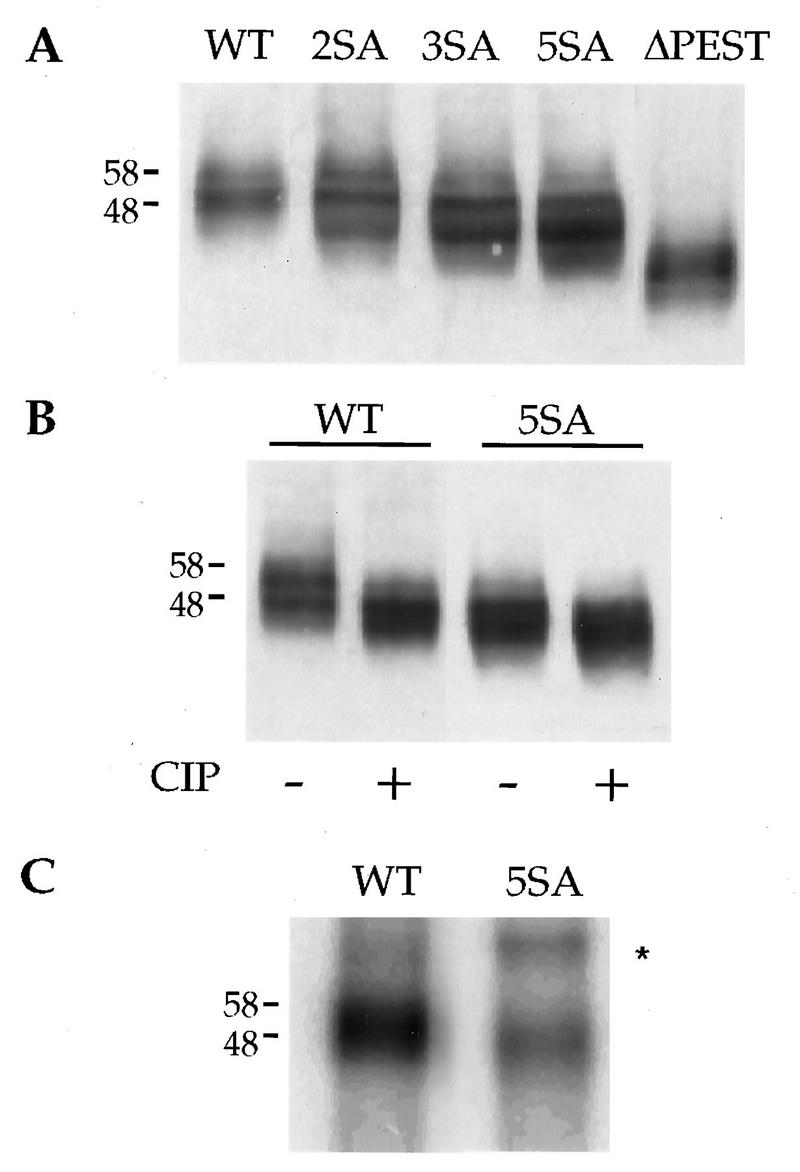
Mutant versions of uracil permease are less phosphorylated than wild-type permease. Cells transformed with plasmid p195gF or derivatives carrying the variant permease genes were grown to logarithmic phase on galactose medium. (A) Protein extracts were prepared, resolved by SDS-PAGE, and analyzed for uracil permease by Western immunoblotting. The concentrations of variant permeases were 1.5 to 4.5 times higher than that of the wild-type permease as estimated by Western blot analysis of serial dilutions of protein extracts (data not shown). Sample volumes were adjusted to have roughly the same amount of permease in each. (B) Protein extracts prepared from cells expressing the wild-type permease or the 5SA mutant permease were incubated at 37°C for 15 min in the presence or absence of calf intestinal phosphatase (CIP) prior to separation by SDS-PAGE and analysis for uracil permease by Western immunoblotting. (C) Cells transformed with plasmid p195gF or derivatives carrying the variant permease genes were grown in low-phosphate medium to an A600 of 1. Cells were then labeled by incubation for 60 min with [32P]orthophosphate in the growth medium. Uracil permease was immunoprecipitated and resolved by SDS-PAGE. A PhosphorImager readout is shown. A contaminating band is marked by an ∗. The molecular masses of the markers are given at the left in kilodaltons. WT, wild type.
To measure the relative loss of phosphorylation of the 5SA variant, exponentially growing cells producing the wild-type or the 5SA permease were labeled for 1 h with [32P]orthophosphate. The permease was immunoprecipitated from equal aliquots of cells that had incorporated identical 32P counts and analyzed by SDS-PAGE (Fig. 4C). Band intensity was measured by PhosphorImaging. The level of incorporation of [32P]orthophosphate into the mutant protein was 25% of that into the wild-type protein. As cells producing the 5SA variant protein contain up to 4.5 times more permease than those producing the wild-type permease, the value of 25% is a large overestimation. The true value may be as low as 5 to 6% if all the phosphates on the permease are replaced by radiolabeled species within the 1-h period of labeling. The immunoprecipitated 5SA variant protein migrated faster by SDS-PAGE than the wild-type permease (Fig. 4C), in agreement with its lower level of phosphorylation.
To investigate further whether serine residues within the PEST-like region are phosphoacceptors, we progressively replaced the alanines of the 5SA variant with glutamic acids (AE variants) (Fig. 5A) and tested the effects of these substitutions on permease turnover after inhibition of protein synthesis (Fig. 5B). Addition of cycloheximide caused almost no loss in uracil uptake by cells producing the 5SA variant. A significant decrease in permease activity was observed for cells producing the 2AE and 3AE variants. Replacement of all five alanines in the sequence with glutamic acids caused a sharp decrease in uracil uptake after inhibition of protein synthesis and thus almost reversion to the wild-type phenotype. The loss in the amount of immunodetected permease paralleled the loss in uracil uptake (data not shown). These results indicate that glutamic acids partially substituted for serines to mediate permease degradation, which suggests that several of the serine residues within the PEST-like sequence are probably targets for phosphorylation and that this region is quantitatively the most important site for phosphorylation of the protein.
FIG. 5.

Replacing alanines with glutamic acids reverses the 5SA phenotype. (A) Substitutions in the PEST-like sequence of the permease are indicated by the one-letter code. For each mutant, the alanines replaced by glutamates are indicated. (B) 27061b cells transformed with plasmid p195gF or derivatives carrying the variant permease genes were grown to logarithmic phase on galactose medium. Cycloheximide (50 μg/ml) was then added to the medium, and uracil uptake was measured at the times indicated. Results are percentages of initial activities. WT, wild type.
Mutations in the PEST-like sequence dramatically reduce ubiquitination of the permease.
The absence of ubiquitination of uracil permease due either to defective ubiquitin ligase Npi1p/Rsp5p or to the absence of ubiquitin-isopeptidase Doa4p stabilizes uracil permease at the plasma membrane (10, 11). On the other hand, permease conjugates accumulate in cells deficient for the internalization step of endocytosis (11, 30). These data strongly suggest that ubiquitination of the permease is a cell surface event that is required prior to internalization. We investigated whether the accumulation of the S-to-A mutant proteins at the plasma membrane may be explained by reduced ubiquitination. Ubiquitinated forms of the permease were better evidenced in plasma membrane-enriched fractions. Membrane samples containing similar amounts of the permease were probed with anti-uracil permease antibodies by immunoblotting (Fig. 6). The ubiquitin-conjugated permease corresponding to the wild-type Fur4p appeared as a high-molecular-mass smear and a discrete ladder-like pattern with four faint bands with lower mobilities than that of the main permease band (Fig. 6A). It has previously been immunologically demonstrated that these bands correspond to ubiquitin-permease conjugates (10, 11). Little or no ubiquitin-conjugated permease was detected in membrane extracts from cells producing the mutant permeases, suggesting that the PEST-like region and/or specific serine residues within this element are required for ubiquitination of the permease. However, the amount of ubiquitinated wild-type permease was relatively small, as was inferred from the distribution of immune reactive material (11). Ubiquitinated forms of the permease accumulate in thermosensitive act1 cells that are conditionally deficient in the internalization step of endocytosis because of the lack of a functional actin network (11). In order to ascertain whether ubiquitin-conjugated variant permeases could be detected, plasma membrane-enriched fractions were prepared from act1 cells grown at 24°C and then incubated at 37°C for 10 min (Fig. 6B). act1 cells producing wild-type Fur4p were enriched in ubiquitin-permease conjugates compared to wild-type cells incubated under the same conditions. act1 cells producing the Fur4p variants with S-to-A substitutions contained some ubiquitin-permease conjugates but in smaller amounts, indicating that addition of ubiquitin is not entirely precluded by decreased phosphorylation of the protein. No ubiquitin-permease conjugates were evidenced in the ΔPEST mutant. The ubiquitin-permease conjugates had faster-migrating bands in the variants with more serine substitutions (Fig. 6B), indicating that ubiquitin conjugates of the wild-type permease are probably phosphorylated. These results show that variants with S-to-A substitutions in the PEST-like region are able to ligate ubiquitin but that they do so less efficiently than the wild-type permease; in contrast, the ΔPEST variant was not ubiquitinated under our experimental conditions.
FIG. 6.
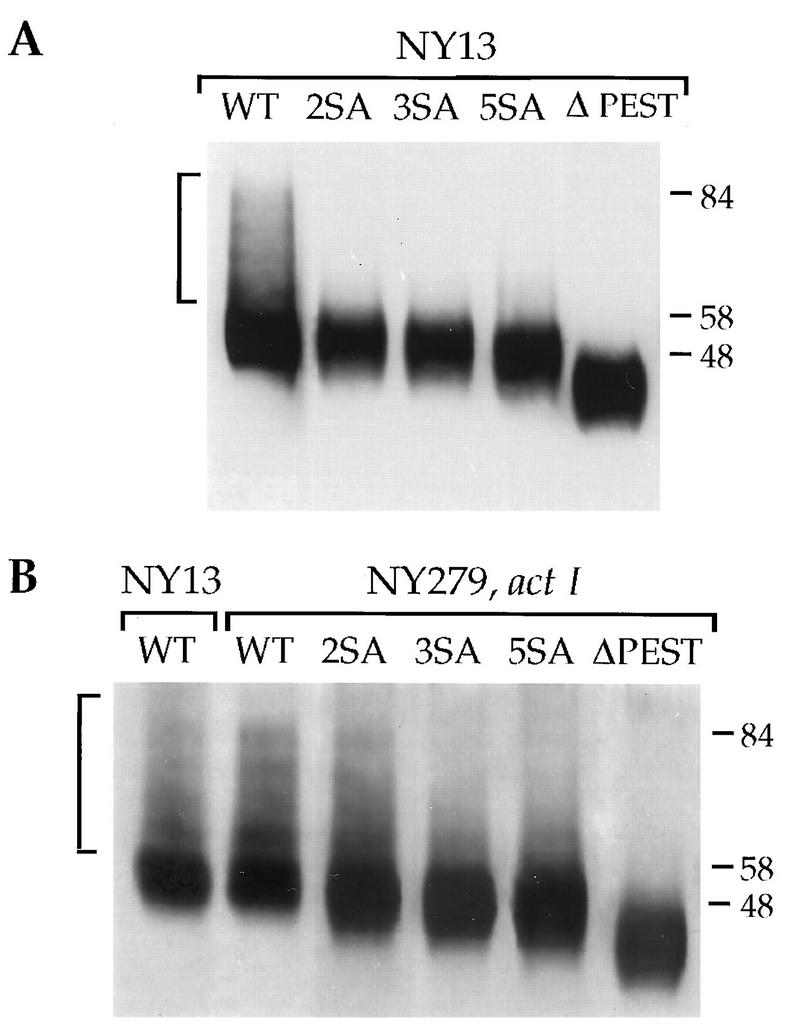
SA variants of the uracil permease are poorly ubiquitinated. NY279 (act1) and NY13 (parental strain) were transformed with plasmid p195gf or derivatives carrying the variant permeases genes. Cells were grown at 30°C (parental cells) or 24°C (act1 cells) to logarithmic phase with galactose as the carbon source and were collected before (A) or after (B) incubation for 10 min at 37°C. Plasma membrane-enriched fractions were prepared, and volumes, chosen to have roughly the same amount of permease signal, were resolved by SDS-PAGE and analyzed for uracil permease by Western immunoblotting. Brackets indicate ubiquitin-permease conjugates. The molecular masses of the markers are given at the right in kilodaltons. WT, wild type.
DISCUSSION
This study shows that a PEST-like region extending from positions 42 to 59 in the hydrophilic N terminus of uracil permease is required for phosphorylation and ubiquitination of the protein. The Fur4p variants lacking the PEST-like region or containing substitutions of some or all of the serines within this domain were stabilized at the plasma membrane. Uracil permease is to our knowledge the first example of a plasma membrane protein, the turnover of which is PEST sequence dependent.
Protein instability has been correlated to so-called PEST sequences. As defined, PEST sequences are sequences rich in proline, glutamic acid, serine, and threonine in any order or combination. The only other constraint is that they have to be flanked by basic amino acids. The region shown in this report to be required for efficient turnover of uracil permease has in fact no proline and, by these criteria, cannot appear as a PEST sequence by the algorithm used to rate PEST sequences (33). However, sequence analysis of Fur4p and two closely related permeases of the FUR family, Dal4p and Upl1p, shows that the extremities of the three N termini appear to share a repetitive structural organization consisting of two consecutive acidic regions enriched in PEST residues and flanked by basic amino acids. For each protein, the second acidic region, including the 42-to-59 region in Fur4p, is the most similar to a PEST sequence. The best score by an algorithm used to rate PEST sequences (33) was for the second region of the uridine permease (Upl1p), with a value of +13.6, which is considered high. This fact, together with the fact that the concerned region in Fur4p also possesses proline residues in the vinicity of the 42-to-59 acidic sequence, led us to suggest that the region involved in the instability of Fur4p is a true PEST sequence. Various secondary structure programs predict that the regions including the PEST-like sequences in Fur4p, Dal4p, and Upl1p are surface-exposed loops. The two other proteins of the FUR family, the thiamine permease and the uracil permease of S. pombe, have shorter N-terminal hydrophilic regions and possess equivalents of these PEST-like sequences in their C-terminal hydrophilic tails.
We tested the importance of the five serine residues within the PEST element of the uracil permease by progressively converting each of them to alanine. The extent of phosphorylation of the permease appeared to correlate with the number of remaining serines. Simultaneous mutation of the five serine residues strongly decreases phosphorylation of the permease. Although we identify several serines or combinations of serines whose replacement by nonphosphorylable alanine prevented basal and stress-induced turnover of the permease and resulted in underphosphorylated species, we provide no direct evidence that these residues are indeed the phosphoacceptor sites. However, the substitution of alanine residues for glutamic acid residues at positions 55 56 and/or 42, 43, and 45 by mutating the alanines of the 5SA variant partially restored the wild-type phenotype. The rate of degradation of these AE variants correlated with the number of alanine residues replaced with glutamic acid. This finding indicates that addition of negative charges improves permease internalization, suggesting that phosphorylation rather than the mere presence of serines is necessary for activating this PEST sequence. To check whether the PEST serines are indeed phosphoacceptors, we also replaced all of them with threonines (5ST variants) and tested for phosphothreonine residues in the resulting protein. However, we failed to detect any phosphothreonine in a phosphoamino acid analysis of the 32P-labeled Fur4p 5ST variant. In agreement with this apparent absence of phosphorylation, the 5ST mutant permease was not degraded with wild-type kinetics, having a low turnover rate similar to that of the 3SA variant (data not shown). A similar failure of threonine substitutions to mimic the reactivities of their serine counterparts was also reported for IκBα, the inhibitor of the transcriptional regulator NFκB (6). DiDonato et al. suggested that the kinase involved may have a very strong preference for serine residues (6). The same may be true of the kinase(s) involved in Fur4p phosphorylation, as most known serine kinases can also use threonine but do so somewhat less efficiently.
A relevant aspect of PEST sequences may be their richness in kinase target sites rather than their PEST amino acids per se. Casein kinase II phosphorylates some short half-life proteins containing PEST sequences (28, 33, 38). In vivo phosphorylation by the cyclin-dependent kinase partner of the yeast G1 cyclins, Cdc28, of specific sites in the PEST sequences of Cln2 and Cln3 has been implicated in their instability (26, 50). Phosphorylation of cyclin E on Thr380 in its PEST-like region is important in the regulation of cyclin E destruction (49). The PEST sequence identified in the hydrophilic NH2 part of Fur4p contains several potential phosphorylation sites conforming to the consensus sequences for casein kinase I, casein kinase II, and protein kinase C. The two yeast homologs of mammalian casein kinase I, Yck1p and Yck2p, are good candidates for possible involvement in permease phosphorylation as they are plasma membrane-bound proteins involved in the phosphorylation of the plasma membrane H+-ATPase, (reference 9 and references therein). Preliminary results indicate that mutants defective in the yeast casein kinase I proteins, Yck1p and Yck2p, are impaired in the internalization of Fur4p (unpublished results), and recently CK1 activity was also shown to be required for internalization of Ste3p (32).
For many years, the most thoroughly characterized function for protein ubiquitination has been that of a signal for degradation of proteins by the proteasome (19). Ubiquitination of plasma membrane proteins targets them for endocytosis. As the phosphorylation-defective mutants in the PEST element of Fur4p failed to undergo efficient ubiquitination, phosphorylation of the permease may be required for its ubiquitination. In other words, we propose that phosphorylation converts the protein into an efficient substrate for the ubiquitin machinery. Uracil permease also carries a sequence similar to that of the destruction box, a motif required for ubiquitin-dependent proteolysis of mitotic cyclins. A mutation within this sequence partially protects Fur4p against stress-induced degradation (12). However, further analysis of the role of this sequence was not feasible because extensive modification of this destruction box led to misfolded proteins that did not reach the plasma membrane (9a). Other regions involved in degradation of yeast plasma membrane proteins have only partially been characterized. An SINNDAKSS sequence was described as being essential for the ubiquitin-dependent endocytosis of a truncated form of Ste2p (18). A linker region containing a DAKSS-like sequence within an acidic box has been recently described as being essential for ubiquitination and fast turnover of the Ste6p (24). A di-leucine and a DAKSS-like motif together with 11 amino acids in its tail extremity were found to be required for degradation of Gap1p (16).
How a PEST element might influence recognition of uracil permease or other PEST-containing proteins by the ubiquitin machinery is unclear. The E3 ubiquitin-protein ligases contribute to control substrate specificity. Only a few ubiquitin ligases have been characterized so far. One of them, the yeast-essential Npi1p/Rsp5p, which is similar to human E6-AP and related proteins (17), is implicated in the degradation of three transporters, including uracil permease (11, 17, 27). Uracil permease lacks regions rich in proline residues which may be recognized by the WW(P) motifs of Npi1p/Rsp5p (17), but auxiliary factors might mediate the E3-permease interaction. It remains to be established whether the acidic PEST region is an important recognition determinant or whether phosphorylation of the PEST element induces a conformational change in the permease, thus unmasking a region recognized by the Npi1p/Rsp5p ubiquitin ligase.
ACKNOWLEDGMENTS
We are grateful to M. O. Blondel for providing antiserum against the N-terminal hydrophilic part of uracil permease. Special thanks are also due to C. Volland for stimulating discussions, advice, and critical reading of the manuscript. We thank K. Seron and J. M. Galan for helpful discussions. We thank A. Edelman for editorial assistance.
This work was supported by a special grant of the CNRS (program, Biologie Cellulaire; project 96105).
REFERENCES
- 1.André A. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast. 1995;11:1575–1611. doi: 10.1002/yea.320111605. [DOI] [PubMed] [Google Scholar]
- 2.Béchet J, Grenson M, Wiame J M. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. Eur J Biochem. 1970;12:31–39. doi: 10.1111/j.1432-1033.1970.tb00817.x. [DOI] [PubMed] [Google Scholar]
- 3.Bisson L F, Coons D M, Kruckeberg A L, Lewis D A. Yeast sugar transporters. Crit Rev Biochem Mol Biol. 1993;28:259–308. doi: 10.3109/10409239309078437. [DOI] [PubMed] [Google Scholar]
- 4.Chevallier M R. Cloning and transcriptional control of a eucaryotic permease gene. Mol Cell Biol. 1982;2:977–984. doi: 10.1128/mcb.2.8.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ciechanover A. The ubiquitin-proteasome proteolytic pathway. Cell. 1994;79:13–21. doi: 10.1016/0092-8674(94)90396-4. [DOI] [PubMed] [Google Scholar]
- 6.DiDonato J, Mercurio F, Rosette C, Wu-Li J, Suyang H, Ghosh S, Karin M. Mapping of the inducible IκB phosphorylation sites that signal its ubiquitination and degradation. Mol Cell Biol. 1996;16:1295–1304. doi: 10.1128/mcb.16.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Egner R, Kuchler K. The yeast multidrug transporter Pdr5 of the plasma membrane is ubiquitinated prior to endocytosis and degradation in the vacuole. FEBS Lett. 1996;378:177–181. doi: 10.1016/0014-5793(95)01450-0. [DOI] [PubMed] [Google Scholar]
- 8.Enjo F, Nosaka K, Ogata M, Iwashima A, Nishimura H. Isolation and characterization of a thiamin transport gene, THI10, from Saccharomyces cerevisiae. J Biol Chem. 1997;272:19165–19170. doi: 10.1074/jbc.272.31.19165. [DOI] [PubMed] [Google Scholar]
- 9.Estrada E, Agostinis P, Vandenheede J R, Goris J, Merlevede W, François J, Goffeau A, Ghislain M. Phosphorylation of yeast plasma membrane H+ATPase by casein kinase I. J Biol Chem. 1996;271:32064–32072. doi: 10.1074/jbc.271.50.32064. [DOI] [PubMed] [Google Scholar]
- 9a.Galan, J.-M. Personal communication.
- 10.Galan J-M, Haguenauer-Tsapis R. Ubiquitin Lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J. 1997;16:5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galan J M, Moreau V, André B, Volland C, Haguenauer-Tsapis R. Ubiquitination mediated by the Npi1p/Rsp5p ubiquitin-protein ligase is required for endocytosis of the yeast uracil permease. J Biol Chem. 1996;271:10946–10952. doi: 10.1074/jbc.271.18.10946. [DOI] [PubMed] [Google Scholar]
- 12.Galan J M, Volland C, Urban-Grimal D, Haguenauer-Tsapis R. The yeast plasma membrane uracil permease is stabilized against stress induced degradation by a point mutation in a cyclin-like “destruction box.”. Biochem Biophys Res Commun. 1994;201:769–775. doi: 10.1006/bbrc.1994.1767. [DOI] [PubMed] [Google Scholar]
- 13.Garnier C, Blondel M O, Hagenauer-Tsapis R. Membrane topology of the yeast uracil permease. Mol Microbiol. 1996;21:1061–1073. doi: 10.1046/j.1365-2958.1996.621430.x. [DOI] [PubMed] [Google Scholar]
- 14.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glotzer M, Murray A W, Kirschner M W. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- 16.Hein C, André B. A C-terminal di-leucine motif and nearby sequences are required for NH4+-induced inactivation and degradation of the general amino acid permease, Gap1p, of Saccharomyces cerevisiae. Mol Microbiol. 1997;24:607–616. doi: 10.1046/j.1365-2958.1997.3771735.x. [DOI] [PubMed] [Google Scholar]
- 17.Hein C, Springael J Y, Volland C, Haguenauer-Tsapis R, André B. NPI1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- 18.Hicke L, Riezman H. Ubiquitination of a yeast plasma membrane receptor signals its ligand-stimulated endocytosis. Cell. 1996;84:277–287. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 19.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 20.Horak J. Yeast nutrients transporters. Biochim Biophys Acta. 1997;1331:41–79. doi: 10.1016/s0304-4157(96)00015-9. [DOI] [PubMed] [Google Scholar]
- 21.Horak J, Wolf D H. Catabolite inactivation of the galactose transporter in the yeast Saccharomyces cerevisiae: ubiquitination, endocytosis, and degradation in the vacuole. J Bacteriol. 1997;179:1541–1549. doi: 10.1128/jb.179.5.1541-1549.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jund R, Weber E, Chevallier M R. Primary structure of the uracil transport protein of Saccharomyces cerevisiae. Eur J Biochem. 1988;171:417–424. doi: 10.1111/j.1432-1033.1988.tb13806.x. [DOI] [PubMed] [Google Scholar]
- 23.Kölling R, Hollenberg C P. The ABC-transporter Ste6 accumulates in the plasma membrane in a ubiquitinated form in endocytosis mutants. EMBO J. 1994;13:3261–3271. doi: 10.1002/j.1460-2075.1994.tb06627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kölling R, Losko S. The linker region of the ABC-transporter Ste6 mediates ubiquitination and fast turnover of the protein. EMBO J. 1997;16:2251–2261. doi: 10.1093/emboj/16.9.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kornitzer D, Raboy B, Kulka R G, Fink G R. Regulated degradation of the transcription factor Gcn4. EMBO J. 1994;13:6021–6030. doi: 10.1002/j.1460-2075.1994.tb06948.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanker S, Valdivieso M H, Wittenberg C. Rapid degradation of the G1 cyclin Cln2 induced by CDK-dependent phosphorylation. Science. 1996;271:1597–1600. doi: 10.1126/science.271.5255.1597. [DOI] [PubMed] [Google Scholar]
- 27.Lucero P, Lagunas R. Catabolite inactivation of the yeast maltose transporter requires ubiquitin-ligase npi1/rsp5 and ubiquitin-hydrolase npi2/doa4. FEMS Microbiol Lett. 1997;147:273–277. doi: 10.1111/j.1574-6968.1997.tb10253.x. [DOI] [PubMed] [Google Scholar]
- 28.MacElhinny J A, Trushin S A, Bren G D, Chester N, Paya C V. Casein kinase II phosphorylates IκBα at S-283, S-289, S-293, and T-291 and is required for its degradation. Mol Cell Biol. 1996;16:899–906. doi: 10.1128/mcb.16.3.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madura K, Varshavsky A. Degradation of Gα by the N-end rule pathway. Science. 1994;265:1454–1458. doi: 10.1126/science.8073290. [DOI] [PubMed] [Google Scholar]
- 30.Moreau V, Galan J-M, Devilliers G, Haguenauer-Tsapis R, Winsor B. The yeast actin-related protein Arp2p is required for the internalization step of endocytosis. Mol Biol Cell. 1997;8:1361–1375. doi: 10.1091/mbc.8.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelissen B, Mordant P, Jonniaux J-L, De Wachter R, Goffeau A. Phylogenetic classification of the major superfamily of membrane transport facilitators, as deduced from yeast genome sequencing. FEBS Lett. 1995;377:232–236. doi: 10.1016/0014-5793(95)01380-6. [DOI] [PubMed] [Google Scholar]
- 32.Panek H R, Stepp J D, Engle H M, Marks K M, Tan P K, Lemmon S K, Robinson N C. Suppressors of YCK-encoded yeast casein kinase 1 deficiency define the four subunits of a novel chlatrin AP-like complex. EMBO J. 1997;16:4194–4204. doi: 10.1093/emboj/16.14.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rechsteiner M, Rogers S W. PEST sequences and regulation by proteolysis. Trends Biochem Sci. 1996;21:267–271. [PubMed] [Google Scholar]
- 34.Rogers S, Wells R, Rechsteiner M. Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science. 1986;234:364–368. doi: 10.1126/science.2876518. [DOI] [PubMed] [Google Scholar]
- 35.Roth A F, Davis N G. Ubiquitination of the yeast a-factor receptor. J Cell Biol. 1996;134:661–674. doi: 10.1083/jcb.134.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandoval I V, Bakke O. Targeting of membrane proteins to endosomes and lysosomes. Trends Cell Biol. 1994;4:292–297. doi: 10.1016/0962-8924(94)90220-8. [DOI] [PubMed] [Google Scholar]
- 37.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwarz E M, Van Antwerp D, Verma I M. Constitutive phosphorylation of IκBα by casein kinase II occurs preferentially at serine 293: requirement for degradation of free IκBα. Mol Cell Biol. 1996;16:3554–3559. doi: 10.1128/mcb.16.7.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shortle D, Novick P, Botstein D. Construction and genetic characterization of temperature-sensitive mutant allele of the yeast actin gene. Proc Natl Acad Sci USA. 1984;81:4889–4893. doi: 10.1073/pnas.81.15.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silve S, Volland C, Garnier C, Jund R, Chevallier M R, Haguenauer-Tsapis R. Membrane insertion of uracil permease, a polytopic yeast plasma membrane protein. Mol Cell Biol. 1991;11:1114–1124. doi: 10.1128/mcb.11.2.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Springael, J.-Y., and B. André. Ammonium-induced endocytosis of Gap1 permease requires both ubiquitination and C-terminal sequences including a di-leucine peptide. Folia Microbiol., in press.
- 42.Stanbrough M, Magasanik B. Transcriptional and posttranslational regulation of the general amino acid permease of Saccharomyces cerevisiae. J Bacteriol. 1995;177:94–102. doi: 10.1128/jb.177.1.94-102.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strous G J, van Kerkhof P, Govers R, Ciechanover A, Schwarz A L. The ubiquitin conjugation system is required for ligand-induced endocytosis and degradation of the growth hormone receptor. EMBO J. 1996;15:3806–3812. [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas B J, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- 45.Urban-Grimal D, Pinson B, Chevallier J, Haguenauer-Tsapis R. Replacement of Lys by Glu in a transmembrane segment strongly impairs the function of the uracil permease from Saccharomyces cerevisiae. Biochem J. 1995;308:847–851. doi: 10.1042/bj3080847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volland C, Garnier C, Haguenauer-Tsapis R. In vivo phosphorylation of the yeast uracil permease. J Biol Chem. 1992;267:23767–23771. [PubMed] [Google Scholar]
- 47.Volland C, Urban-Grimal D, Géraud G, Haguenauer-Tsapis R. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J Biol Chem. 1994;269:9833–9841. [PubMed] [Google Scholar]
- 48.Willems A, Lanker S, Patton E E, Craig A L, Nason T F, Mathias N, Kobayashi R, Wittenberg C, Tyers M. Cdc53 targets phosphorylated G1 cyclins for degradation by the ubiquitin proteolytic pathway. Cell. 1996;86:453–463. doi: 10.1016/s0092-8674(00)80118-x. [DOI] [PubMed] [Google Scholar]
- 49.Won K A, Reed S I. Activation of cyclin E/CDK2 is coupled to site-specific autophosphorylation and ubiquitin-dependent degradation of cyclin E. EMBO J. 1996;15:4182–4193. [PMC free article] [PubMed] [Google Scholar]
- 50.Yaglom J, Linskens M H, Sadis S, Rubin D M, Futcher B, Finley D. p34Cdc28-mediated control of Cln3 cyclin degradation. Mol Cell Biol. 1995;15:731–741. doi: 10.1128/mcb.15.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoo H S, Cunningham T S, Cooper T G. The allantoin and uracil permease gene sequences of S. cerevisiae are nearly identical. Yeast. 1992;8:997–1006. doi: 10.1002/yea.320081202. [DOI] [PubMed] [Google Scholar]


