Abstract
MicroRNAs have emerged as critical regulators of gene expression. Originally shown to regulate developmental timing, microRNAs have since been implicated in a wide range of cellular functions including cell identity, migration and signaling. miRNA-430, the earliest expressed microRNA during zebrafish embryogenesis, is required to undergo morphogenesis and has previously been shown to regulate maternal mRNA clearance, Nodal signaling, and germ cell migration. The functions of miR-430 in brain morphogenesis, however, remain unclear. Herein we find that miR-430 instructs oriented cell divisons in the neural rod required for neural midline formation. Loss of miR-430 function results in mitotic spindle misorientation in the neural rod, failed neuroepithelial integration after cell division, and ectopic cell accumulation in the dorsal neural tube. We propose that miR-430, independently of canonical apicobasal and planar cell polarity (PCP) pathways, coordinates the stereotypical cell divisons that instruct neural tube morphogenesis.
Keywords: MicroRNAs, Zebrafish, Neural tube
1. Introduction
MicroRNAs are evolutionarily conserved small noncoding RNAs that induce the translational repression and degradation of target mRNAs (Fabian et al., 2010; Huntzinger and Izaurralde, 2011). Animal genomes contain hundreds of microRNA genes, and each microRNA, in turn, can directly regulate hundreds of target mRNAs (Alvarez-Garcia and Miska, 2005; Ambros, 2004; Bartel, 2009; Takacs and Giraldez, 2010). Given this widespread reach, an individual microRNA has the potential to both regulate multiple components within particular cellular pathways as well as impact several distinct processes simultaneously within the cell (Flynt and Lai, 2008). The global impact of microRNA function can be assessed though removal of dicer, a RNAse III ribonuclease required for microRNA biogenesis (Bernstein et al., 2001; Denli et al., 2004; Harfe et al., 2005; Ketting et al., 2001; Lee et al., 2004; Murchison et al., 2007; Wienholds et al., 2003). Zebrafish deficient for dicer activity develop normal A-P and D-V body axes, as well as undergo cell fate specification and tissue differentiation (Giraldez et al., 2005). MZdicer mutant embryos display morphogenetic defects during gastrulation and brain ventricle development. Reintroduction of a single microRNA, miR-430, rescues these defects, indicating that loss of miR-430 alone is primarily responsible for these early dicer phenotypes (Giraldez et al., 2005). Endogenous miR-430 is expressed ubiquitously in the early embryo and regulates the expression and stability of hundreds of maternal mRNAs (Chen et al., 2005; Giraldez et al., 2006; Lund et al., 2009). Distinct phenotypes can be attributed to the misregulation of specific cellular pathways and target mRNAs. For instance, miR-430 directly represses squint and lefty mRNA expression to dampen and balance Nodal signaling during mesendoderm induction (Choi et al., 2007; Rosa et al., 2009), buffers chemokine signaling effectors sdf1a and cxcr7 to ensure robust germ cell migration (Staton et al., 2011) and regulates the expression of germline-specific genes in the soma (Mishima et al., 2006). Perhaps the most striking MZdicer phenotype is severely reduced brain ventricle formation and subsequent morphogenesis defects (Giraldez et al., 2005). However, the cellular basis underlying neural defects remains unclear.
Brain morphogenesis in zebrafish initiates with the convergence of neural progenitors to the dorsal midline (Hong and Brewster, 2006; Papan and Camposortega, 1994). Mutants in the planar cell polarity (PCP) pathway display delayed convergence movements and neurulation defects (Quesada-Hernandez et al., 2010; Tawk et al., 2007). As convergence proceeds, neural progenitors ingress ventrally to generate a solid neural rod primordium (Lowery and Sive, 2004; Papan and Camposortega, 1994). Teleost neurulation is distinctive in that neural progenitors do not undergo apicobasal polarization or organize into distinct neuroepithelia until after the neural rod forms (Geldmacher-Voss et al., 2003; Hong and Brewster, 2006). Neural midline formation is mediated by a stereotypic series of oriented cell divisions in which daughter pairs segregate across the presumptive midline and establish mirror-symmetric apicobasal polarity (Ciruna et al., 2006; Clarke, 2009; Quesada-Hernandez et al., 2010; Tawk et al., 2007). The localization, timing, and mitotic orientation of these divisions are critical to ensure the faithful distribution of daughter cells and the establishment of a coherent neural midline. Defects in any of these steps leads to ectopic neural tube formation and/or disorganization of the midline (Ciruna et al., 2006; Quesada-Hernandez et al., 2010; Tawk et al., 2007; Zigman et al., 2014; Zigman et al., 2011).
In this study, we investigate how microRNAs regulate neural tube formation in zebrafish. We find that loss of miR-430 activity causes the dorsal accumulation of ectopic neural progenitors that fail to incorporate into the nascent neuroepithelium. In the absence of miR-430 function, apicobasal and planar cell polarity are established. In contrast, we show that miR-430 is required for proper mitotic spindle orientation of neural rod cell divisions during neural midline formation. Finally, we identify a set of neuroectodermal miR-430 target genes, providing an entry point for understanding how miR-430 coordinates oriented cell divisions during neural tube development and tissue morphogenesis.
2. Methods
2.1. Zebrafish lines
MZdicer fish were generated as previously described (Giraldez et al., 2005). Embryos used were dicerhu896/hu896, dicerhu896/hu715 or dicerhu715/hu715.
2.2. mRNA reporter constructs
Capped mRNA was transcribed from reporter constructs using mMessage mMachine kit (Ambion). 1 nl of mRNA was injected at one-cell stage at following concentrations: pCS2 + GFP (50 pg). GFP-Pk ([10]; 75 ng/ul), histone2B-mCherry (100 ng/ul) and pCS2-Kaede (100 ng/ul). Scatter labeling was performed by injecting a subset of blastomeres in16-cell stage embryos. For chimaeric analysis of miR-430 function, wildtype and MZdicer embryos were injected with GFP mRNA at one-cell stage. Equivalently staged donor and host embryos were grown under similar environmental conditions until mid-blastula stages, at which point donor cells were transferred to wildtype host embryos (20–50 cells/host).
2.3. Embryo sectioning and labeling
Embryos were fixed in 4% paraformaldehyde O/N at 4C, mounted in 2% low-melting point agarose and sectioned at 200–250 um intervals using a vibratome (1500 System, Vibratome). Transverse sections through the hindbrain were identified based on the presence of otic vesicles and size/presence of notochord. Immunohistochemistry was performed on floating sections with: rabbit-aPKC (C-20; Santa Cruz Biotechnology 1:200) and rabbit-GFP (Molecular Probes; 1:1000). Secondary antibodies conjugated to Alexa-488 (Invitrogen) were used at a 1:200 dilution. Nuclei and F-actin were stained with Topro-3 (Life Technologies; 1:2000) and rhodamine-phalloidin (Molecular probes; 1:75), respectively. Imaging was performed on a Zeiss 510 Meta confocal microscope.
In situ hybridization using dlx3 riboprobe (plasmid obtained from Rachel Brewster, University of Maryland) was performed as described (Thisse and Thisse, 2008)
2.4. Time-lapse confocal microscopy
Live embryos were mounted in 1.2% low-melting point agarose prior to imaging. For lineage tracing, embryos injected with kaede mRNA were mounted at approximately 3-somite stage, photoconverted by scanning ROI with a 405uM-diode laser on a Leica TCS SP5 Spectral Confocal Microscope, then imaged immediately and approximately 10 h later.
2.5. Cell division inhibition
To block cell divisons at neural keel/rod stages, embryos were placed in medium containing 100 uM aphidicolin (Sigma) and 20 mM hydroxyurea (Sigma) in 4% dimethylsulfoxide (DMSO) at 90% epiboly stage. Control embryos were incubated for same time period in media containing 4% DMSO.
2.6. Target site analysis
Fold enrichment for 135 microRNA target sites (7mers) in the UTRs of upregulated genes (> 1.5 fold) was determined:
Fold enrichment for single miRNAs
Gene ontology clusters were defined using DAVID (http://david.abcc.ncifcrf.gov) Gene Functional Annotation Clustering on GO ‘FAT’ annotations and ‘high’ stringency. Clusters are annotated with representative GO terms and corresponding Benjamini–Hochberg FDR corrected P values.
3. Results
3.1. Loss of miR-430 results in neural midline defects
Neurocoel formation is severely impaired in MZdicer embryos (Giraldez et al., 2005). MiR-430 is the earliest expressed miRNA in the early embryo (Chen et al., 2005; Giraldez et al., 2005) and, reintroduction of the miR-430 duplex (which does not require Dicer processing) into MZdicer embryos rescues neurulation defects (Giraldez et al., 2005), indicating that early neurulation defects in MZdicer embryos are exclusively due to loss of miR-430 activity. To further investigate neural tube defects in the absence of miR-430, we first compared neural rod morphology in the hindbrain of wildtype and MZdicer embryos at the ~20 somite stage (Fig. 1). At this stage, neural plate ingression has occurred and neural progenitors are organized within coherent epithelia that display mirror-symmetric apicobasal polarity. Localized enrichment of cell polarity and junctional proteins within apical membranes defines the nascent neural midline from which lumen inflation soon follows. We analyzed apicobasal polarity (aPKC; Fig. 1A–C; (Tabuse et al., 1998)), cell morphology (scatter labeling; Fig. 1G–J) and cytoskeletal organization (F-actin; Fig. 1D–F). In contrast to wildtype progenitors, MZdicer neural cells display defects in epithelial organization. Specifically, we observe the accumulation of multiple rounded cells within medial regions of the dorsal neural rod. Scatter labeling with membrane GFP reveals that these cells often fail to elongate and establish contact with the basal membrane (Fig. 1I), and display abnormal F-actin organization (Fig. 1E), reminiscent of neural rod defects associated with PCP/Wnt pathway impairment (Ciruna et al., 2006). The presence of ectopic cells prevents the formation of a continuous apical membrane along the neural midline as assessed by the apical marker aPKC, precluding proper lumen inflation and neurocoel formation (Fig. 1B). In most cases (77%, n=30 embryos), we observed ectopic neural midline formation within dorsal regions and/or a branching or bifurcation of the neural midline. Variability in phenotypic severity was observed both among different embryos as well as within different hindbrain regions of the same embryo (Fig. S1). Importantly, reintroduction of mature miR-430 rescued neural midline defects in MZdicer embryos displaying a single coherent midline (Fig. 1C, F and J). Taken together, these data indicate that miR-430 function is required for proper neuroepithelial organization in the zebrafish dorsal neural rod.
Fig. 1.
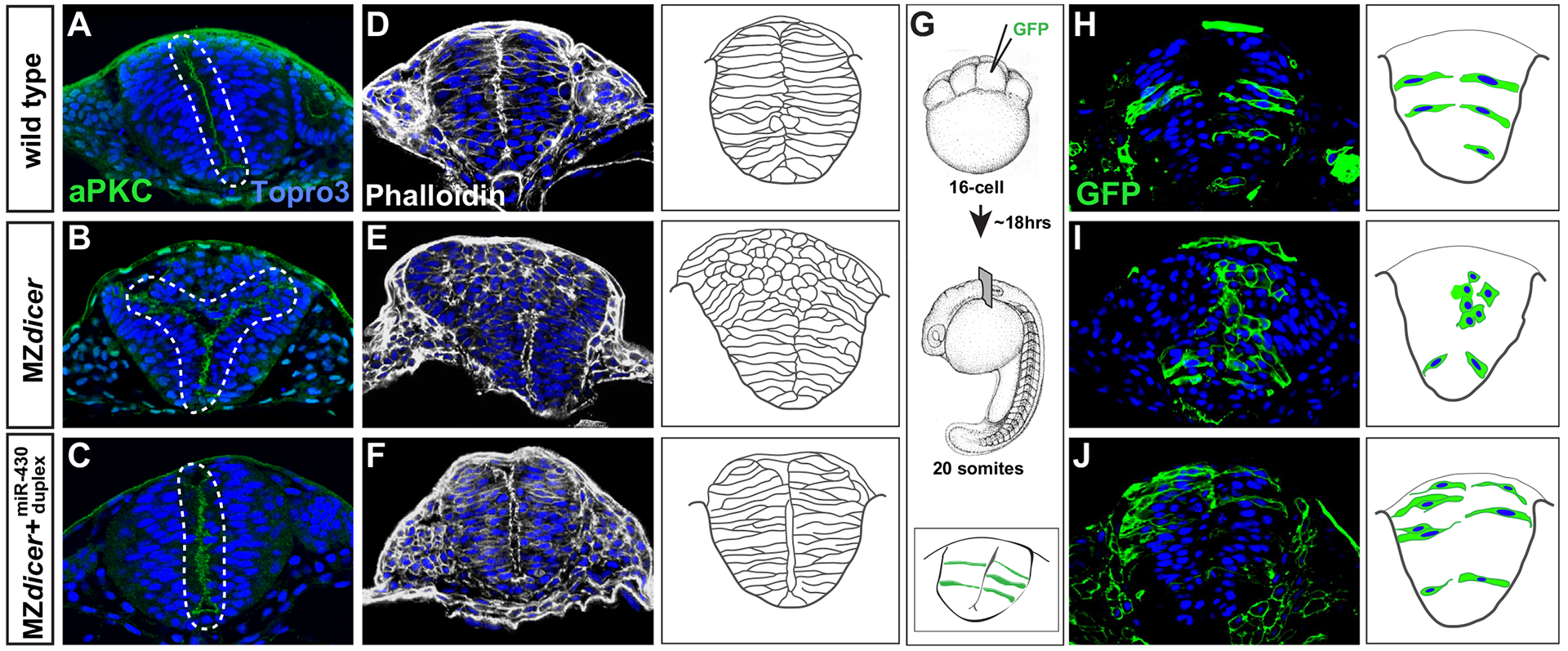
miR-430 is required for neural tube morphogenesis. Transverse sections through the hindbrain region of 20-somite stage embryos. Topro3, nuclear stain. Wildtype neural progenitors (A, D and H) display apical localization of aPKC and bilateral organization along the presumptive neural midline (A), display normal subcellular localization of F-actin (D), and acquire elongated cell morphology (H). In contrast, MZdicer neural progenitors (B, E and I) form ectopic apical membranes (B), display abnormal F-action organization (E), and fail to establish contact with lateral membrane and acquire epithelial character in dorsal regions (I). (C, F and J) Neural progenitors from MZdicer embryos injected at the 1-cell stage with miR-430 RNA duplex display rescued cell morphology and organization. (G) Scatter labeling strategy. 1–2 blastomeres of 16-cell embryos were injected with GFP mRNA.
3.2. Neural daughter cells fail to segregate across nascent neural midline
Neural midline formation is instructed by a series of crossing divisions that result in the bilateral segregation of daughter cells across the axial midline with mirror-symmetric apicobasal polarity (Clarke, 2009). To determine whether midline defects in the MZdicer mutant arise from aberrant crossing divisions, we employed the photoconvertible fluorophore Kaede (Hatta et al., 2006) to follow the fate of neural progenitors and their progeny during the neural keel/rod transition (Fig. 2A). We assessed the degree of midline crossing by photoconverting progenitors exclusively on one side of the midline before the onset of crossing divisions and then recording the position of daughter cells ~10 h later after midline formation. A successful crossing division requires that a daughter cell originating from the ipsilateral side contacts, and integrates into, the contralateral epithelium (Ciruna et al., 2006; Tawk et al., 2007). The majority, if not all, of cell divisions during the neural keel/rod stage result in midline crossing (Concha and Adams, 1998; Geldmacher-Voss et al., 2003). In wildtype embryos the progeny of converted cells become bilaterally distributed between opposing epithelia (Fig. 2C). Consistent with proper midline formation in the ventral neural rod of MZdicer embryos, MZdicer progenitors in more ventral regions displayed a heterogeneous bilateral distribution. In contrast MZdicer progenitors remained segregated within dorsal regions (Fig. 2E). Failure to cross was associated with an accumulation of cells at the presumptive midline. Previous studies have shown that midline defects arising from ectopic or misoriented crossing divisions can be rescued through chemical inhibition of mitosis (Ciruna et al., 2006; Quesada-Hernandez et al., 2010; Tawk et al., 2007; Zigman et al., 2014; Zigman et al., 2011). To determine whether crossing divisions contribute to MZdicer neural midline defects, we incubated MZdicer embryos with cell cycle inhibitors before the onset of crossing divisions. We find that cell cycle inhibition rescues ectopic cell accumulation in the MZdicer mutant (Fig. 2F–H). Taken together, these results suggest that ectopic MZdicer neural progenitors arise from defective crossing division mitoses in which daughter cells fail to successfully navigate midline crossing and contralateral integration.
Fig. 2.
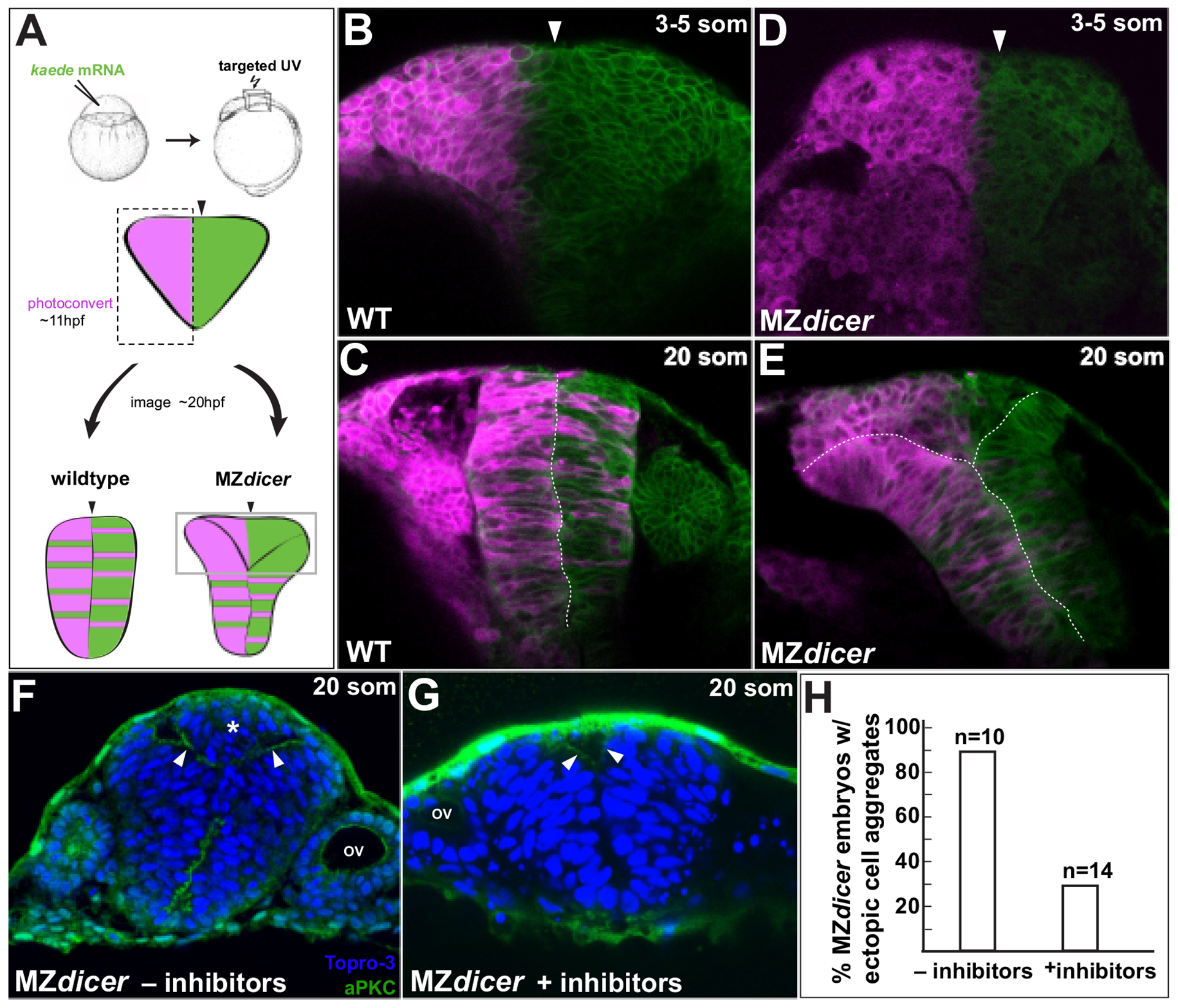
MZdicer neural progenitors in dorsal regions fail to cross midline. (A) Lineage tracing after unilateral Kaede photoconversion at 11 hpf. (B–E) Kaede lineage tracing after photoconversion (3–5 somites; B and D) and ~8 h later (20 somites; C and E). (B–C) Wildtype neural progenitors undergo bilateral distribution. (D–E) In contrast, MZdicer neural progenitors retain unilateral distribution in dorsal regions of the neural anlage. Arrowhead demarcates the presumptive neural midline. (F–H) Cell cycle inhibition reduces MZdicer ectopic neural cell accumulation. Asterisk indicates presence of ectopic cell aggregate. Arrowheads indicate apical membrane. OV, otic vesicle.
3.3. MZdicer neural progenitors establish apicobasal and PCP polarity
Both apicobasal and planar cell polarity (PCP) pathways instruct the correct partitioning daughter cells within the neural rod (Ciruna et al., 2006; Geldmacher-Voss et al., 2003; Quesada-Hernandez et al., 2010; Tawk et al., 2007; Zigman et al., 2011). We next tested whether miR-430 function is required for proper establishment of cell polarity in the neural rod. To address whether ectopic MZdicer cells retain an ability to adopt apicobasal polarity, we assessed the localization of the polarity marker aPKC (Tabuse et al., 1998) later in development (~27 hpf; Fig. 3). aPKC normally localizes to apical membrane within the neuroepithelium (Hong and Brewster, 2006). We find that, at later stages, ectopic progenitors, despite failed integration, adopt epithelial character and establish apical polarity as assessed by apical enrichment of aPKC (Fig. 3E). Moreover, we find that ectopic cells often generate bifurcated lumens indicating that they retain the ability to self organize into epithelial structures.
Fig. 3.
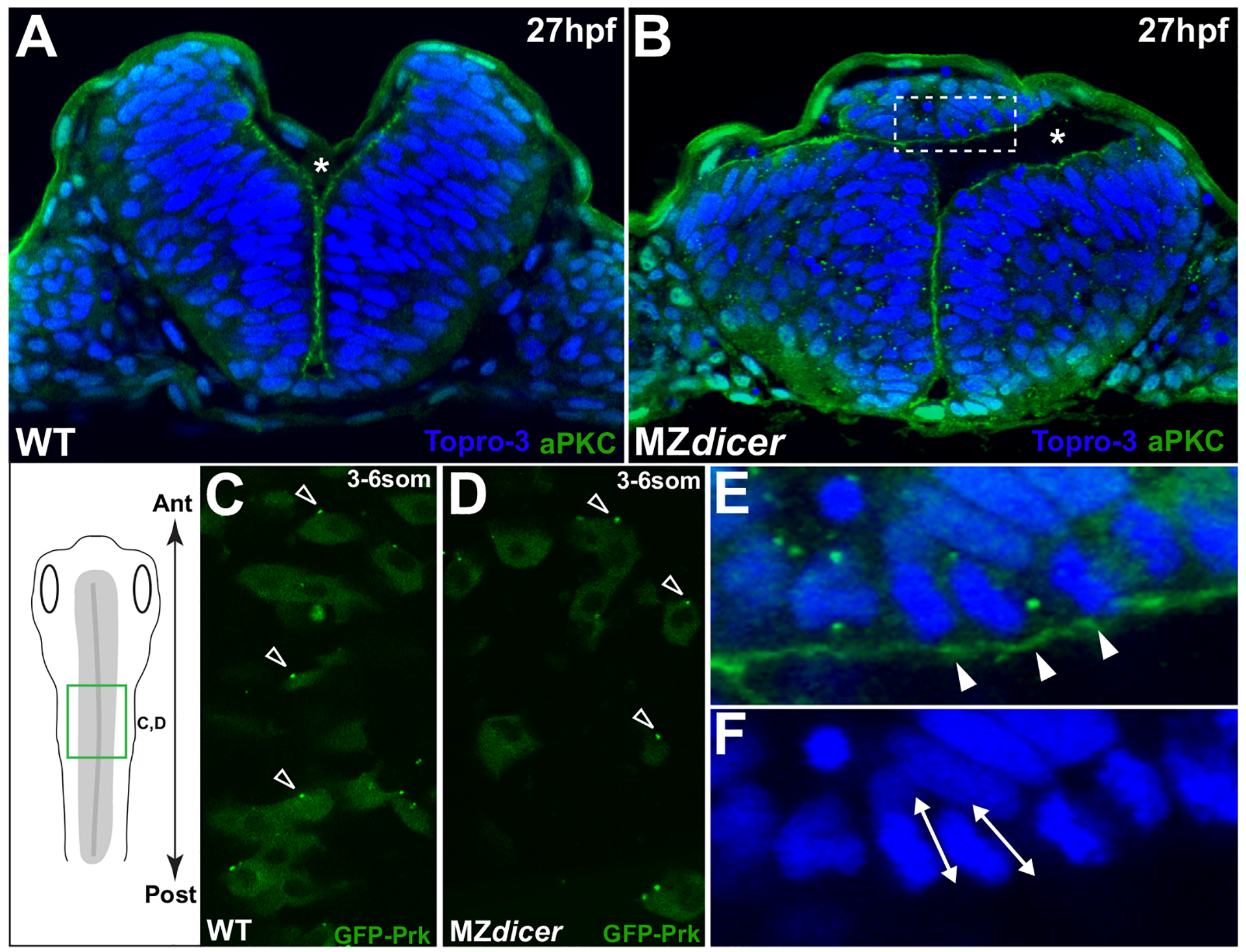
Ectopic MZdicer neural progenitors retain apicobasal and PCP polarity. (A and B) Transverse sections at 27hpf. Topro3, nuclear stain. At this stage, lumen inflation has initiated. Asterisk indicates luminal space. (B) MZdicer embryo with a dorsal ectopic cell aggregate. (E) Inset of panel B; arrowheads indicate aPKC (green) localization to apical membrane. (F) Inset of panel B; double arrows indicate elongated nuclear shape characteristic of polarized epithelial cells. (C and D) Dorsal view (green box in schematic) of GFP-Prickle scatter labeling. GFP-Prk localizes to the anterior edge (arrowheads) of (C) wildtype neural cells (71/94; 76%) and (D) MZdicer mutant cells (39/49; 80%). Anterior to top.
Loss-of-function mutations in the PCP pathway result in delayed neural convergence, failed planar polarization within neural progenitors, and disruption of C-division positioning and midline crossing (Ciruna et al., 2006; Quesada-Hernandez et al., 2010; Tawk et al., 2007; Zigman et al., 2011). To determine whether planar polarity is affected by loss of miR-430 function, we assessed the localization of the PCP component Prickle (Jenny et al., 2003). In wildtype embryos, the PCP reporter GFP-Prickle (GFP-Prk) is the earliest known marker of planar polarity during neurulation and localizes to discrete foci at the anterior edge (71/94; 76%) of neural keel stage progenitors. (Fig. 3C and D; (Ciruna et al., 2006)). We find that GFP-Prk correctly displays anterior localization in MZdicer neural cells (39/49; 80%), suggesting that the PCP pathway is not disrupted upstream of Prickle localization in MZdicer neural progenitors. Taken together, these data suggest that MZdicer cells maintain an ability to polarize in both planar and apicobasal orientations, and are both capable of, and predisposed toward, epithelial organization.
3.4. Neural midline defects are not due to mislocalized crossing divisions
The failure of neural progenitors to cross the midline could be a function of their distance from the midline at the time of crossing division. For example, in the PCP mutant vang2, convergent extension defects result in delayed medial convergence of neural progenitors to the axial midline (Quesada-Hernandez et al., 2010; Tawk et al., 2007). Mislocalized crossing divisions on either side of the axial midline give rise to ectopic neural midlines in the vang2 mutant. To address whether MZdicer neural defects arise from convergence defects, we first assessed the temporal requirements for miR-430 in neural tube development. Embryos that are maternally deficient for dicer but express Dicer zygotically (Mdicer) do not generate significant levels of mature miR-430 until 8hpf (vs. ~4–5 hpf in wildtype embryos) (Fig. 4A). However, in contrast to MZdicer embryos, Mdicer embryos undergo brain morphogenesis and form a single contiguous lumen through the hindbrain region (Fig. 4C). This observation suggests that MZdicer neurulation defects are due to relatively late function(s) of miR-430 in neurulation as opposed to an earlier role in convergent extension. Indeed, analysis of dlx3, a marker of the lateral edges of the neural anlage, does not reveal differences in the width of the neural keel, indicating that early defects in epiboly and convergence do not explain the observed neural midline defects in MZdicer embryos. (Fig. 4D and E).
Fig. 4.

miR-430 activity is required post-gastrulation within proximal neural progenitors (A) Northern blot analysis of miR-430 expression: lane 1, embryos derived from dcr/dcr female germline (maternally deficient) X dcr/+ male; lane 2, embryos derived from dcr/dcr male germline X dcr/+ female (maternally sufficient). Embryos deficient for maternal dicer function do not generate significant levels of mature miR-430 until after 8hpf. (B) MZdicer embryo (maternally and zygotically deficient) fails to generate contiguous neural lumen and ventricular organization. (C) Mdicer embryo (maternally deficient) undergoes ventricle formation, suggesting that zygotic Dicer contribution (and delayed miR-430 production) is sufficient for neural tube generation. (D and E) In situ analysis of dlx3 expression, marking the lateral edges of neural keel. (F–I) Kaede lineage tracing after photoconversion at dorsal midline (tailbud-1somite stage; F, G) and ~10 hours later (20 somite stage; H, I). Arrowheads demarcate dorsal midline. As ingression proceeds, proximal progenitors adopt ventral positions in neural rod. In MZdicer embryos, a subset of proximal progenitors fails to ingress ventrally (arrowheads in panel I).
We reasoned that if the distance from the axial midline is not a determining factor in whether a neural progenitor undergoes a defective crossing division, then neural progenitors positioned close to the midline in the neural keel should also give rise to ectopic daughter cells that fail to cross and integrate into the contralateral epithelium. To follow the fate of medial progenitors, we photoconverted cells located close to the midline in the early neural keel (Fig. 4F–G). In a wildtype embryo, medial neural cells adopt ventral positions in the neural rod due to ingression as neurulation proceeds (Fig. 4H; Fig S2); (Lowery and Sive, 2004; Papan and Camposortega, 1994). Accordingly, neural progenitors at the lateral edges of the neural keel are destined to the dorsal neural rod. This dorsoventral profile is maintained through the crossing division phase since opposing daughter pairs maintain similar dorsoventral positions as ingression proceeds. In contrast to wildtype progenitors, we observed labeled midline MZdicer cells that failed to co-migrate ventrally during epithelial ingression. Labeled cells separated from their ventral neighbors and remained “left behind” within MZdicer dorsal cell aggregates (6/6 embryos displayed > 1 labeled cell within dorsal aggregate: Fig. 4I). These results provide multiple insights. First, a subset of progenitors proximal to the midline give rise to ectopic cells, indicating that distance from the midline is not a principal determining factor in failed MZdicer epithelial integration. Second, MZdicer ectopic cells fail to migrate ventrally with the surrounding neuroepithelium and, as a result, are relegated to the dorsal midline. Taken together, these results suggest that miR-430 activity facilitates neuroepithelial integration after crossing division within progenitors positioned at the axial midline
3.5. Transplanted MZdicer neural progenitors display defects in neuroepithelial integration
The above results suggest that defects underlying MZdicer neurulation originate during the neural keel-to-rod transition. To assess whether Dicer function is required within individual neural progenitors, we transplanted labeled dicer cells into wildtype hosts (Fig. 5A). A failure of mutant daughter cells to cross after division would suggest that Dicer function is required cell-autonomously. This could result in either the deposition of a daughter cell outside of the nascent epithelium or, alternatively, its reintegration within the same side leading to a predominantly unilateral distribution. Labeled wildtype cells transplanted into wildtype hosts generated bilaterally organized clones suggestive of successful crossing events (15/17 (88%) clones). Transplanted mutant MZdicer clones (or miR430 KD cells; Fig. S3A) of similar size (< 6 cells) into wildtype hosts also displayed bilateral organization in wildtype hosts (Fig. 5B and F) albeit to a lesser extent (20/28 (71%) clones; pval=0.2759). However, in a subset of clones that failed to establish bilateral organization (2/8 clones), we observed the exclusion of mutant cells from the neuroepithelium (Fig. 5G–I; Fig. S4). Larger clones (> 6 cells) revealed additional qualitative differences between wildtype and mutant MZdicer clones. Specifically, larger mutant clones formed unilateral clusters that remained segregated from the neuroepithelium (Fig. 5B). In one instance, we observed the formation of ectopic apical membrane through a larger clone (Fig. 5C). These behaviors were never observed for wildtype cell clones of similar size. Taken together, these results indicate that MZdicer cell morphology and organization can be largely rescued by neighboring wildtype cells and is consistent with cell nonautonomous functions.
Fig. 5.

Transplanted MZdicer cells display reduced ability to integrate into wildtype neuroepithelium. (A) At midblastula stage, ~50 cells (WT or MZdicer) were transplanted into equivalently staged WT embryos. (B) MZdicer clones that display unilateral distribution (asterisk) and/or segregation away from wildtype epithelium (arrowhead). Arrows demarcate a MZdicer clone that displays normal cell shape and bilateral distribution. (C and D) Unilateral MZdicer clone that is organized around ectopic apical membrane (localized aPKC signal; arrowhead in panel D). (E and F) Transverse views of wildtype clone (E) and MZdicer clone (F) displaying epithelial integration and bilateral distribution. (G) Transverse view of MZdicer progeny cells excluded from the neuroepithelium. (H and I) Inset of panel F.
3.6. Abnormal mitotic spindle orientation during neural midline divisions
In contrast to prior divisions that occur longitudinal to the anteroposterior axis, crossing divisions (usually the 16th cell division) orient perpendicularly to the anteroposterior axis through a 90° rotation of the mitotic spindle (Concha and Adams, 1998; Geldmacher-Voss et al., 2003; Kimmel et al., 1994). It has been reported that proper mitotic spindle orientation during the crossing division phase is critical for both the establishment of an organized neural midline as well as for the faithful bilateral segregation of daughter cells (Quesada-Hernandez et al., 2010; Zigman et al., 2014; Zigman et al., 2011). To assess cell division orientation, we conducted time-lapse recording of neural keel/rod stages (8som-14som stages) in wildtype and MZdicer embryos injected with an mRNA encoding the fusion protein histone2A-Cherry. We limited our analysis to cell mitoses within 30 um on either side of the axial midline. Consistent with previous reports, cell mitoses during this stage predominantly oriented orthogonal to the anteroposterior axis in wildtype embryos (median 81.6° ± 11.4°; Fig. 6A and B) In contrast, cell divisions in MZdicer embryos displayed a heterogeneous distribution without final orientation preference (median 52.7° ± 25.0°; Mann–Whitney Test (p < 0.0001); Fig. 6D and E). Misoriented cell divisions could be due to abnormal cell orientation at start of mitosis and/or due to abnormal spindle rotation as mitosis proceeds (Zigman et al., 2011). To distinguish between these two possibilities, we measured the initial orientation of neural progenitors during prophase and compared to orientation during anaphase (Fig. 6C and F). Consistent with previous work, wildtype cells predominantly oriented along the anteroposterior axis upon entry into mitosis (median 7° ± 17°; n=12 cells, 2 embryos) and rotated an average of 80° ± 28° (Fig. 6C). In contrast, MZdicer progenitors were less consistently oriented in planar orientation (median 30° ± 27°; n=12 cells, 2 embryos) and additionally, displayed reduced rotation (average of 29° ± 24°; Fig. 6F). Taken together, these results suggest that miR-430 function is required to both establish initial spindle orientation and facilitate spindle rotation as mitosis proceeds during neural rod cell divisions.
Fig. 6.

MZdicer neural progenitors undergo misoriented neural rod cell divisions. (A and D) Time-lapse recording from dorsal view (green box in schematic). Histone2A-cherry localizes to chromosomes and enables measurement of mitotic orientation (red lines) at anaphase. White line indicates orientation at initiation of mitosis. (B and E) Quantification of mitotic orientation at anaphase relative to anteroposterior axis (0 degrees) in (B) wildtype (96 cells; 4 embryos) and (E) MZdicer embryos (72 cells; 5 embryos). (C and F) Representation of mitotic spinde rotation for randomly chosen (C) wildtype (n=12) and (F) MZdicer (n=12) progenitors. Inferred spindle orientation (relative to anteroposterior axis) was measured at prophase (start of line) and then again at anaphase (red). (G) miRNA target site (7-mer sites) enrichment in 3’UTRs of upregulated genes (> 1.5 fold) in MZdicer neuroectodermal cells. The enrichment of 135 target sites compared to a control set is plotted. The p-value cutoff after multiple test correction is shown as a dashed line. (H) Selected candidate list of upregulated transcripts in MZdicer mutant. Fold-change upregulation and p-value derived from published microarray data (Shkumatava et al., 2009).
3.7. Upregulated neural mRNAs are enriched for miR-430 target sites
Our phenotypic analysis has uncovered abnormal cellular behaviors that underlie the MZdicer neural phenotype. We reasoned that this phenotype arises from the derepression of miR-430 targets in neuroectoderm. To identify misregulated miR-430 target genes involved in neural tube morphogenesis, we analyzed RNA-seq datasets derived from whole embryos (shield stage) (Lee et al., 2013), as well as microarray datasets derived exclusively from neuroectoderm (18hpf; (Shkumatava et al., 2009)). In both cases, we compared wildtype and MZdicer mutant expression profiles to identify transcripts upregulated in the absence of microRNA function. This analysis revealed 166 transcripts that were significantly upregulated (> 1.5 fold; pval < .05) in MZdicer neuroectoderm. Examination of 3’UTRs for the presence of microRNA target sites (pairing to seed region) revealed significant enrichment of miR-430 target sequences within 3’UTRs of upregulated genes (Fig. 6G), with 85 (51%) transcripts possessing at least one miR-430 target site (8mer sites, n=22; 7mer-m8 sites, n=34; 7mer-A1, n=11; 6mer, n=53), suggesting that miR-430 activity directly regulates mRNA transcript stability within neuroectoderm. To identify affected cellular processes, we conducted gene ontology enrichment analysis for misregulated genes. Representative GO terms for the top five enriched functional clusters were lysosome (GO:0005764), cell migration (GO:0016477), peptidase inhibitor activity (GO:0030414), extracellular matrix (GO:0031012), and cytoskeletal organization (GO:0007010). Closer inspection revealed genes involved in cell–cell adhesion (e.g. cdh1, mif1), ECM-cell interactions (e.g. mmp2, timp2), and cytoskeletal dynamics (e.g. tuba2, arpc5b, dynll2a) (Fig. 6H). Taken together, these results suggest that miR-430 provides an essential and pervasive layer of regulation across a range of cellular processes to coordinate mitotic spindle orientation during neural tube morphogenesis.
4. Discussion
A fundamental challenge in developmental biology is to understand how molecular information influences cellular behaviors that shape tissue morphogenesis. We find that miR-430 coordinates a specialized cell division that directly instructs neural tube development. Photolabeling experiments indicate that a subset of neural progenitors that lack miR-430 activity fail to incorporate into the nascent neural epithelium. We propose that spindle misorientation results in the failed integration and dorsal accumulation of ‘orphan’ daughter cells that self-organize to form ectopic neural structures (Fig. 7). Our data suggest that MZdicer neural progenitors are in the right location to engage in effective crossing divisions. However, we cannot rule out the possibility that the timing of crossing divisions is askew in the MZdicer mutant. MicroRNAs have been shown to regulate developmental timing in multiple contexts (Ambros, 2011; Decembrini et al., 2009; La Torre et al., 2013), and in turn, miR-430 might function to ensure that crossing divisions are temporally coordinated within a defined developmental window. Such coordination would be especially important given that crossing divisions deviate dramatically in their orientation compared to both precedent and subsequent cell divisions (which orient parallel to the anteroposterior axis). Recently published transplantation experiments suggest that crossing division timing is regulated by the neural progenitor’s intrinsic schedule and not from surrounding environmental cues (Girdler et al., 2013). Interestingly, larger clones of transplanted MZdicer cells resemble heterochronically-displaced cell transplants in that they tend to cluster rather than integrate into the host neuroepithelium, suggesting that MZdicer cells might possess incompatible cellular properties (e.g. cell adhesive properties) due to an asynchrony in developmental time (Girdler et al., 2013).
Fig. 7.
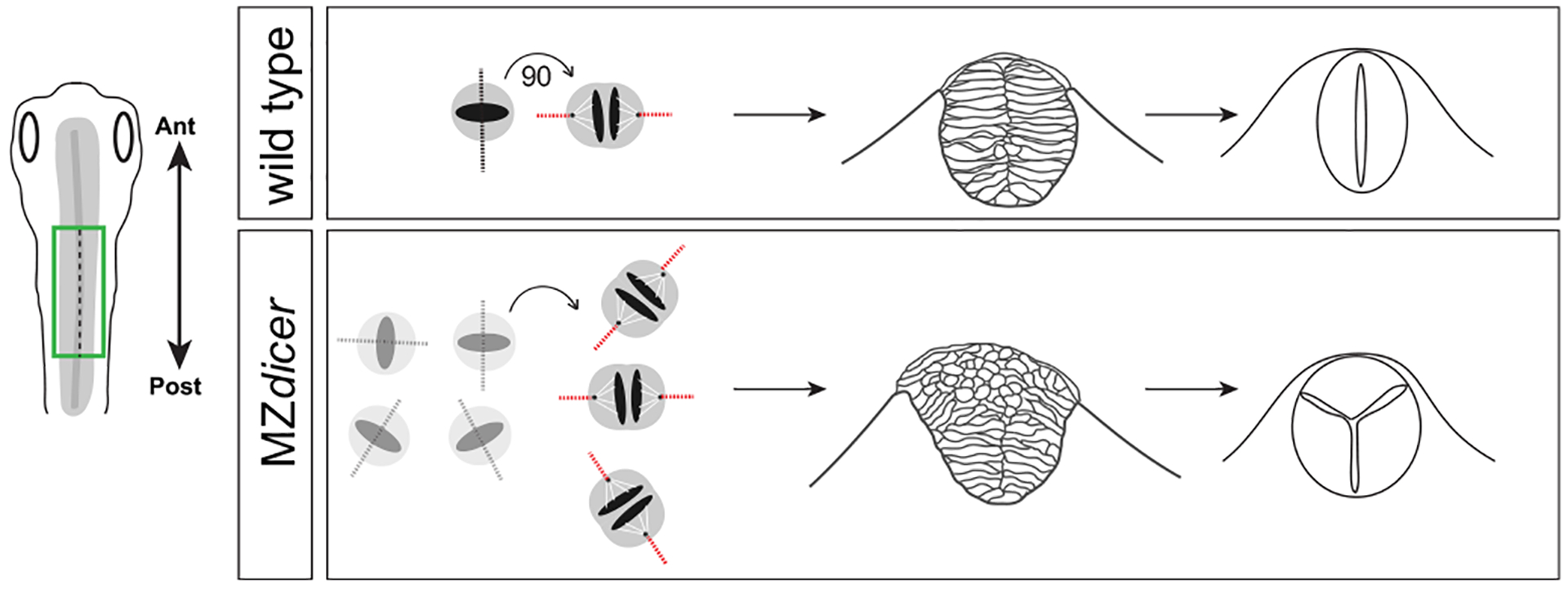
Model of neurulation defects in MZdicer embryos. Loss of miR-430 results in abnormal spindle orientation of neural keel cell mitoses and failed neuroepithelial integration. Displaced neural progenitor daughter cells within dorsal regions retain ability to organize into ectopic neural structures.
Alternatively, the crossing division per se is defective. Spindle positioning is determined by forces exerted on astral microtubules associated with the cell cortex. The localization of cortical protein complexes that pull on astral microtubules (i.e. Gi-LGN-NuMA/Dynein/Dynactin) is determined, in turn, by an interactive network of intrinsic mechanisms that sense and respond to external cues. Among these, we first assessed cell polarity cues. The apical par-3/par-6/aPKC complex coordinates spindle geometry in early C. elegans embryos (Colombo et al., 2003) and Drosophila neuroblasts (Cai et al., 2003), as well as centrosome positioning during zebrafish neurulation (Hong et al., 2010). A recent study by Buckley et al. demonstrates that broad apical localization of Pard3-GFP is initiated immediately before and independently of crossing divisions. Therefore, localized Pard3 could potentially influence the mitotic orientation of crossing divisions as apical polarization proceeds. Planar cell polarity has also been shown to instruct spindle orientation in multiple contexts (Gong et al., 2004; Segalen and Bellaiche, 2009), and loss of the PCP component frizzled7 results in misoriented crossing divisions in the zebrafish neural keel (Quesada-Hernandez et al., 2010). In contrast, we find that MZdicer neural progenitors establish apical and planar polarity and therefore, do not interpret spindle orientation defects as due to the loss of these polarizing cues.
In addition to cell polarity cues, defects in cell adhesion might also contribute to MZdicer neural tube misorganization. N-cadherin mutant embryos display neural tube defects that are similar to those observed in MZdicer embryos(Hong and Brewster, 2006; Lele et al., 2002). Specifically, dorsal cells become excluded from the neuroepithelium and often form aggregates that retain the ability to self-organize. Knockdown of zebrafish N-cadherin (cdh2) function results in spindle misorientation in the ventral neural tube (Zigman et al., 2011). Further implicating cell adhesion and spindle orientation, reduced alpha-catenin accumulation at nascent adhesive junctions is associated with misoriented neural keel divisions in MZscribble neural progenitors (Zigman et al., 2011). Interestingly, MZscribble neural progenitors display both initial misorientation and reduced spindle rotation, suggesting that similar defects observed in MZdicer embryos may be rooted in altered cell adhesion. Consistent with this hypothesis, several misregulated molecules in MZdicer neuroectoderm are involved in cell adhesion, as well as cell-extracellular matrix interactions, suggesting that miR-430 loss affects the adhesive interactions between neighboring cells or with the basal lamina
MicroRNAs can interact with hundreds of mRNAs in the cell and provide a versatile mechanism whereby multiple target mRNAs within a common cellular process can be coordinately regulated. Given this complexity, it is likely that the majority of microRNA phenotypes arise as a composite outcome of derepression across a range of targets affecting multiple pathways. We have been able to show that microRNA activity provides an essential regulatory layer to ensure the precise and coordinated elaboration of a series of oriented cell divisions involved in neural tube morphogenesis. Future work will entail identifying those target mRNAs whose functions in oriented division are most sensitive to perturbations in gene expression.
Supplementary Material
Appendix A. Supplementary material
Supplementary data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.ydbio.2015.11.016.
Acknowledgments
We thank Brian Ciruna and Rachel Brewster for reagents; Alena Shkumatava and David Bartel for microarray data; Mustafa Khoka for sharing laboratory equipment; Alison Staton and Minsun Jeong for generously providing MZdicer adult fish; Heather Patnode for fish husbandry, and members of the Giraldez lab for discussion and critical reading of the manuscript. This work was supported by NIH grants F32HD061194-03 (C.M.T) and R01GM08160206A1, R01GM10110803, R0GM10225103, March of Dimes 1-FY12-230 (A. J.G).
References
- Alvarez-Garcia I, Miska EA, 2005. MicroRNA functions in animal development and human disease. Development 132, 4653–4662. [DOI] [PubMed] [Google Scholar]
- Ambros V, 2004. The functions of animal microRNAs. Nature 431, 350–355. [DOI] [PubMed] [Google Scholar]
- Ambros V, 2011. MicroRNAs and developmental timing. Curr. Opin. Genet. Dev 21, 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, 2009. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ, 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Cai Y, Yu F, Lin S, Chia W, Yang X, 2003. Apical complex genes control mitotic spindle geometry and relative size of daughter cells in Drosophila neuroblast and pI asymmetric divisions. Cell 112, 51–62. [DOI] [PubMed] [Google Scholar]
- Chen PY, Manninga H, Slanchev K, Chien M, Russo JJ, Ju J, Sheridan R, John B, Marks DS, Gaidatzis D, et al. , 2005. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes. Dev 19, 1288–1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi WY, Giraldez AJ, Schier AF, 2007. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 318, 271–274. [DOI] [PubMed] [Google Scholar]
- Ciruna B, Jenny A, Lee D, Mlodzik M, Schier AF, 2006. Planar cell polarity signalling couples cell division and morphogenesis during neurulation. Nature 439, 220–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke J, 2009. Role of polarized cell divisions in zebrafish neural tube formation. Curr. Opin. Neurobiol 19, 134–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo K, Grill SW, Kimple RJ, Willard FS, Siderovski DP, Gonczy P, 2003. Translation of polarity cues into asymmetric spindle positioning in Caenorhabditis elegans embryos. Science 300, 1957–1961. [DOI] [PubMed] [Google Scholar]
- Concha ML, Adams RJ, 1998. Oriented cell divisions and cellular morphogenesis in the zebrafish gastrula and neurula: a time-lapse analysis. Development 125, 983–994. [DOI] [PubMed] [Google Scholar]
- Decembrini S, Bressan D, Vignali R, Pitto L, Mariotti S, Rainaldi G, Wang X, Evangelista M, Barsacchi G, Cremisi F, 2009. MicroRNAs couple cell fate and developmental timing in retina. Proc. Natl. Acad. Sci. USA 106, 21179–21184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ, 2004. Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231–235. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W, 2010. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem 79, 351–379. [DOI] [PubMed] [Google Scholar]
- Flynt AS, Lai EC, 2008. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat. Rev. Genet 9, 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldmacher-Voss B, Reugels AM, Pauls S, Campos-Ortega JA, 2003. A 90-degree rotation of the mitotic spindle changes the orientation of mitoses of zebrafish neuroepithelial cells. Development 130, 3767–3780. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, Enright AJ, Thomson JM, Baskerville S, Hammond SM, Bartel DP, Schier AF, 2005. MicroRNAs regulate brain morphogenesis in zebrafish. Science 308, 833–838. [DOI] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF, 2006. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312, 75–79. [DOI] [PubMed] [Google Scholar]
- Girdler GC, Araya C, Ren X, Clarke JD, 2013. Developmental time rather than local environment regulates the schedule of epithelial polarization in the zebrafish neural rod. Neural Dev. 8, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Mo C, Fraser SE, 2004. Planar cell polarity signalling controls cell division orientation during zebrafish gastrulation. Nature 430, 689–693. [DOI] [PubMed] [Google Scholar]
- Harfe BD, McManus MT, Mansfield JH, Hornstein E, Tabin CJ, 2005. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc. Natl. Acad. Sci. USA 102, 10898–10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta K, Tsujii H, Omura T, 2006. Cell tracking using a photoconvertible fluorescent protein. Nat. Protoc 1, 960–967. [DOI] [PubMed] [Google Scholar]
- Hong E, Brewster R, 2006. N-cadherin is required for the polarized cell behaviors that drive neurulation in the zebrafish. Development 133, 3895–3905. [DOI] [PubMed] [Google Scholar]
- Hong E, Jayachandran P, Brewster R, 2010. The polarity protein Pard3 is required for centrosome positioning during neurulation. Dev. Biol 341, 335–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntzinger E, Izaurralde E, 2011. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat. Rev. Genet 12, 99–110. [DOI] [PubMed] [Google Scholar]
- Jenny A, Darken RS, Wilson PA, Mlodzik M, 2003. Prickle and Strabismus form a functional complex to generate a correct axis during planar cell polarity signaling. EMBO J. 22, 4409–4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH, 2001. Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM, Kane DA, 1994. Cell cycles and clonal strings during formation of the zebrafish central nervous system. Development 120, 265–276. [DOI] [PubMed] [Google Scholar]
- La Torre A, Georgi S, Reh TA, 2013. Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc. Natl. Acad. Sci. USA 110, E2362–E2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, Giraldez AJ, 2013. Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature 503, 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW, 2004. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell 117, 69–81. [DOI] [PubMed] [Google Scholar]
- Lele Z, Folchert A, Concha M, Rauch GJ, Geisler R, Rosa F, Wilson SW, Hammerschmidt M, Bally-Cuif L, 2002. parachute/n-cadherin is required for morphogenesis and maintained integrity of the zebrafish neural tube. Development 129, 3281–3294. [DOI] [PubMed] [Google Scholar]
- Lowery LA, Sive H, 2004. Strategies of vertebrate neurulation and a re-evaluation of teleost neural tube formation. Mech. Dev 121, 1189–1197. [DOI] [PubMed] [Google Scholar]
- Lund E, Liu M, Hartley RS, Sheets MD, Dahlberg JE, 2009. Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA 15, 2351–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima Y, Giraldez AJ, Takeda Y, Fujiwara T, Sakamoto H, Schier AF, Inoue K, 2006. Differential regulation of germline mRNAs in soma and germ cells by zebrafish miR-430. Curr. Biol.: CB 16, 2135–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ, 2007. Critical roles for Dicer in the female germline. Genes. Dev 21, 682–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papan C, Camposortega JA, 1994. On the formation of the neural keel and neuraltube in the zebrafish danio (Brachydanio) rerio. Roux Arch. Dev. Biol 203, 178–186. [DOI] [PubMed] [Google Scholar]
- Quesada-Hernandez E, Caneparo L, Schneider S, Winkler S, Liebling M, Fraser SE, Heisenberg CP, 2010. Stereotypical cell division orientation controls neural rod midline formation in zebrafish. Curr. Biol.: CB 20, 1966–1972. [DOI] [PubMed] [Google Scholar]
- Rosa A, Spagnoli FM, Brivanlou AH, 2009. The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev. Cell 16, 517–527. [DOI] [PubMed] [Google Scholar]
- Segalen M, Bellaiche Y, 2009. Cell division orientation and planar cell polarity pathways. Semin. Cell. Dev. Biol 20, 972–977. [DOI] [PubMed] [Google Scholar]
- Shkumatava A, Stark A, Sive H, Bartel DP, 2009. Coherent but overlapping expression of microRNAs and their targets during vertebrate development. Genes. Dev 23, 466–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staton AA, Knaut H, Giraldez AJ, 2011. miRNA regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat. Genet 43, 204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuse Y, Izumi Y, Piano F, Kemphues KJ, Miwa J, Ohno S, 1998. Atypical protein kinase C cooperates with PAR-3 to establish embryonic polarity in Caenorhabditis elegans. Development 125, 3607–3614. [DOI] [PubMed] [Google Scholar]
- Takacs CM, Giraldez AJ, 2010. MicroRNAs as genetic sculptors: fishing for clues. Semin. Cell. Dev. Biol 21, 760–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawk M, Araya C, Lyons DA, Reugels AM, Girdler GC, Bayley PR, Hyde DR, Tada M, Clarke JD, 2007. A mirror-symmetric cell division that orchestrates neuroepithelial morphogenesis. Nature 446, 797–800. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B, 2008. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc 3, 59–69. [DOI] [PubMed] [Google Scholar]
- Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH, 2003. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat. Genet 35, 217–218. [DOI] [PubMed] [Google Scholar]
- Zigman M, Laumann-Lipp N, Titus T, Postlethwait J, Moens CB, 2014. Hoxb1b controls oriented cell division, cell shape and microtubule dynamics in neural tube morphogenesis. Development 141, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigman M, Trinh le A, Fraser SE, Moens B,C, 2011. Zebrafish neural tube morphogenesis requires Scribble-dependent oriented cell divisions. Curr. Biol.: CB 21, 79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


