Abstract
Yeast two-hybrid screening has led to the identification of a family of proteins that interact with the repetitive C-terminal repeat domain (CTD) of RNA polymerase II (A. Yuryev et al., Proc. Natl. Acad. Sci. USA 93:6975–6980, 1996). In addition to serine/arginine-rich SR motifs, the SCAFs (SR-like CTD-associated factors) contain discrete CTD-interacting domains. In this paper, we show that the CTD-interacting domain of SCAF8 specifically binds CTD molecules phosphorylated on serines 2 and 5 of the consensus sequence Tyr1Ser2Pro3Thr4Ser5Pro6Ser7. In addition, we demonstrate that SCAF8 associates with hyperphosphorylated but not with hypophosphorylated RNA polymerase II in vitro and in vivo. This result suggests that SCAF8 is not present in preinitiation complexes but rather associates with elongating RNA polymerase II. Immunolocalization studies show that SCAF8 is present in granular nuclear foci which correspond to sites of active transcription. We also provide evidence that SCAF8 foci are associated with the nuclear matrix. A fraction of these sites overlap with a subset of larger nuclear speckles containing phosphorylated polymerase II. Taken together, our results indicate a possible role for SCAF8 in linking transcription and pre-mRNA processing.
The carboxy-terminal domain (CTD) of the largest subunit of RNA polymerase II (pol II) consists of multiple repeats of the heptapeptide sequence YSPTSPS (2, 24). The CTD is found in pol II from a variety of species but is not present on pol I or pol III, indicating a role in mRNA biogenesis (22, 23). Deletion studies have demonstrated that the CTD is essential for cell viability in yeast, Drosophila, and mammalian cells (3, 4, 55, 75). While roles in transcription activation, enhancer function, and pre-mRNA processing have been proposed, the essential role of the CTD remains elusive.
The CTD contains an uncommonly high density of potential phosphorylation sites (26, 27). Both hyper- and hypophosphorylated pol II can be detected in vivo, but the distribution of phosphates among the more than 100 potential sites has not been determined. Substitution of alanine for serines in either position 2 or 5 of each repeat is lethal in yeast, suggesting that phosphorylation of both of these sites is essential (70).
In vitro transcription experiments have led to the hypothesis that the CTD is reversibly phosphorylated with each transcription cycle (26, 27). One role of phosphorylation may be to disrupt contacts between the unphosphorylated CTD and components of the holoenzyme. In addition, phosphorylation may prepare the CTD for a role subsequent to initiation. Indeed, transcription elongation is CTD dependent (1, 44) and is temporally linked to CTD phosphorylation (1, 31, 44, 56, 69).
Yeast two-hybrid screens have led to the identification of a family of CTD-binding proteins containing serine/arginine-rich motifs (17, 64, 74). The SCAFs (SR-like CTD-associated factors) contain conserved N-terminal or C-terminal CTD-interacting domains (25). We have previously shown that the C-terminal CTD-interacting domain of SCAF1 (formerly rA1 [74]) binds either highly or partially phosphorylated pol II.
In this paper, we demonstrate that SCAF8 (formerly rA8 [74]) interacts specifically with a highly phosphorylated form of the CTD. We also show that a protein recognized by an antibody directed against the CTD-binding domain of SCAF8 coimmunoprecipitates with pol II0. Consistent with these biochemical findings, a fraction of SCAF8 colocalizes with sites of active transcription and partially colocalizes with phosphorylated pol II in nuclei of whole cells as well as in cells extracted for nuclear matrix. Finally, we show that SCAF8 is enriched in a nuclear matrix fraction known to contain proteins involved in pre-mRNA processing (13, 54).
MATERIALS AND METHODS
Plasmids and strains.
Plasmids for expression of glutathione S-transferase (GST) consensus CTDs and serine-substituted CTDs are derived from a set of hemagglutinin-tagged yeast CTD mutants constructed earlier (70) and are described in detail elsewhere (57). The CTD-interacting domains of SCAF8 (residues 2 to 200) and SCAF4 (residues 2 to 200) were expressed as a histidine-tagged proteins with the pTrcHis(B) expression vector (Invitrogen).
Purification of SCAF8 CTD-interacting domain.
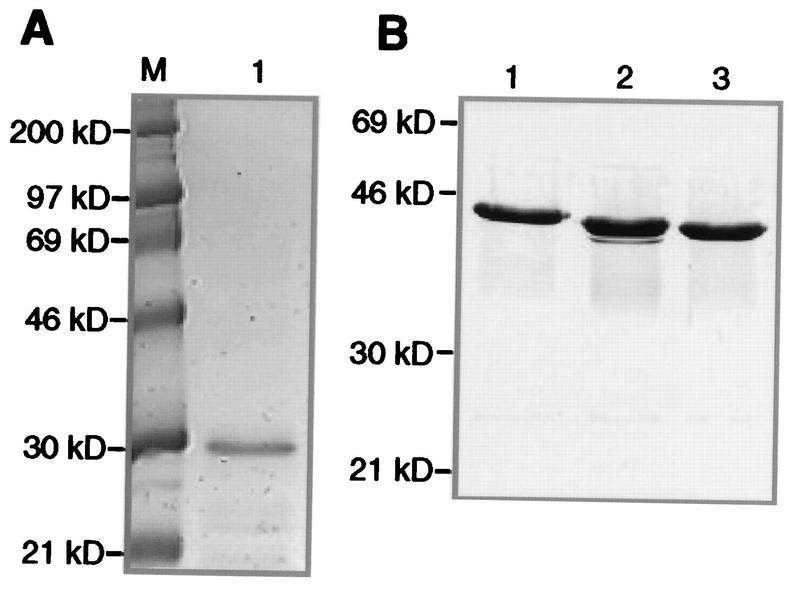
His-tagged SCAF8 and SCAF4 CTD-interacting domain (CID) fusion proteins were purified by metal affinity chromatography with Ni2+-agarose (Pharmacia) or Co2+-agarose (Clontech). Escherichia coli BL21 expressing His-tagged fusion protein was grown to an optical density at 600 nm of 0.8, induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to 0.1 mM, and grown for an additional 4 h. Cells were harvested by centrifugation, resuspended in 200 ml of buffer A (50 mM Tris-HCl [pH 8], 150 mM NaCl, 2 μM pepstatin A, 0.6 μM leupeptin, 2 mM benzamidine HCl, and 1 mM phenylmethylsulfonyl fluoride [PMSF]). After addition of Triton X-100 to 0.1%, the cells were lysed by freezing overnight at −20°C and thawing at room temperature. Thawed lysate was sonicated for 10 min and centrifuged at 4°C for 1 h at 12,000 × g. The supernatant was incubated with Ni2+-agarose or Co2+-agarose for 1 h at 4°C with shaking. Beads were collected by centrifugation at 1,500 × g for 10 min and washed with two column volumes of buffer A containing 10 mM imidazole, resuspended in the same buffer, packed onto a column, and washed with 10 column volumes of buffer A containing 10 mM imidazole. Bound protein was eluted with two column volumes of buffer A containing 100 mM imidazole. The affinity-purified SCAF8 fusion protein was greater than 90% pure as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE [10% polyacrylamide]) (Fig. 1A).
FIG. 1.
Fusion proteins used in binding studies. (A) Coomassie blue staining of recombinant SCAF8 CID. Five micrograms of purified SCAF8 CID was run on SDS-PAGE (10% polyacrylamide) and stained with Coomassie blue. Lane 1, marker proteins; lane 2, SCAF8 CID. (B) Coomassie blue staining of GST-CTD fusion proteins. Five micrograms of the affinity-purified proteins was run on SDS-PAGE (10% polyacrylamide) and stained with Coomassie blue. Lane 1, wild-type CTD (16 repeats); lane 2, A5 mutant CTD (15 repeats); lane 3, A2 mutant CTD (14 repeats).
Purification and phosphorylation of CTD fusion proteins.
E. coli (DH5α) strains expressing GST-CTD fusion proteins were grown overnight to saturation. Cultures were diluted 10-fold in fresh L-broth containing 100 μg of ampicillin per ml and grown for 1 h before addition of IPTG to 0.1 mM. After 4 h of induced expression, cells were collected by centrifugation and sonicated, and the fusion protein was purified by glutathione-agarose (Pharmacia) affinity chromatography as described previously (57). Purified GST fusions containing 16 copies of the wild-type heptapeptide YSPTSPS, 15 copies of YSPTAPS (A5), or 14 copies of YAPTSPS (A2) (mutated residues are underlined) were separated by SDS-PAGE (10% polyacrylamide) and stained with Coomassie blue dye (Fig. 1B).
CTD fusion proteins were phosphorylated in vitro with baculovirus-expressed epitope-tagged Cdc2 kinase (29) as previously described for RNA pol II (35), but with minor modifications. For each phosphorylation reaction, 10 μl of Cdc2 kinase-bound monoclonal antibody (MAb) 12CA5 Affi-Gel beads (∼5 μg of Cdc2 kinase) was incubated with 2.5 μg of GST-CTD fusion protein in 60 mM KCl–50 mM Tris-HCl (pH 7.8)–10 mM MgCl2–0.5 mM dithiothreitol, 1 mM ATP for 20 min at 30°C. For labeling, the reaction was pulsed with 20 μCi of [γ-32P]ATP for 5 min and then chased with 1 mM ATP for 20 min at 30°C. Phosphorylated GST-CTD fusion protein was removed from the beads by spinning the supernatant through a bovine serum albumin-treated filter (UFC3 OHV 00; Millipore). The level of phosphorylation was assessed by electrophoresis in an SDS–5% polyacrylamide gel, followed by direct autoradiography of the dried gel. We estimate that about eight phosphates have been added to the wild-type GST-CTD fusion protein and that about half that number have been added to the mutant CTDs. Phosphorylation of the mouse CTD fusion protein with c-Abl kinase was essentially performed as described previously (5).
CTD binding assay.
About 2 μg of the GST fusion protein or human pol II was applied to a 15-μl Ni-agarose column containing 8 μg of bound SCAF8-CID. The column was washed with six sequential 50-μl aliquots of binding buffer (100 mM NaCl, 10 mM Tris-HCl [pH 7.9], 10 mM imidazole, 0.1% Nonidet P-40, 2 μM pepstatin A, 0.6 μM leupeptin, 2 mM benzamidine HCl, and 1 mM PMSF), and bound protein was eluted with sequential 50-μl aliquots of SDS-PAGE sample buffer. All samples were separated by SDS-PAGE (10% polyacrylamide) and subjected to Western blotting. The blots were probed with MAb 12CA5, which recognizes both phosphorylated and unphosphorylated GST-CTD fusion proteins, or with antiphosphotyrosine antibody to detect c-Abl-phosphorylated CTD. Gels containing 32P-labeled proteins were directly subjected to autoradiography.
MAbs.
MAb 8WG16 is an anti-CTD immunoglobulin G (IgG) described by Thompson et al (65). MAb H14 is an IgM directed against phosphoepitopes on the CTD (18, 19) and was a gift of Steve Warren (Nexstar Pharmaceuticals, Boulder, Colo.), while MAb B3 (49) is a mouse IgM that recognizes a partially overlapping set of CTD phosphoepitopes on the hyperphosphorylated CTD (57). MAb 230.F1E1 (F1E1) is an IgG raised in mice against the His-tagged SCAF8 CID with the help of Dennis Wilson (Johns Hopkins).
Immunoprecipitation of SCAF8 complexes.
Tissue culture supernatant containing MAb F1E1 (about 25 μg) was allowed to bind protein A/G agarose beads (50 μl; Pierce) overnight and washed three times with 500 μl of IP100 buffer (100 mM NaCl, 50 mM Tris-HCl [pH 7.6], 0.05% Nonidet P-40, 0.5 mM dithiothreitol, 5 mM MgCl2, 5 mM NaF, 5 mM β-glycerol phosphate). Beads (20 μl, containing approximately 10 μg of antibody) were incubated with 25 μl of HeLa cell nuclear extract for 1 h at 4°C with shaking. The beads were washed four times with 500 μl of IP100 buffer, and bound proteins were eluted with 50 μl of 2× SDS-PAGE loading buffer, and subjected to SDS-PAGE followed by Western blot analysis (57).
Immunostaining and laser scanning confocal microscopy.
Mouse 3T3 fibroblasts were grown on coverslips for 48 h. Cells were then fixed in 3% paraformaldehyde on ice for 2 min, permeabilized with 0.5% Triton X-100 in TBS buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM MgCl2) for 1 min on ice, blocked with 5% goat serum, and then incubated consecutively with MAbs B3 and F1E1 at room temperature for 1 h. Cells were then treated with biotin-conjugated goat anti-mouse IgG Fc fragments (Jackson ImmunoResearch Laboratories, Inc.) at 1:50 dilution at room temperature for 30 min, washed three times with TBS-Tween buffer, and incubated with a mixture of fluorescein isothiocyanate (FITC)-conjugated goat anti mouse IgM (μ chain specific, Sigma) at 1:50 and Texas red-conjugated streptavidin (GIBCO) at 1:50 at room temperature for 30 min. After a final wash with TBS-Tween buffer, coverslips were mounted on slides, and images from 0.5-μm optical sections were collected with a Bio-Rad MRC-1024 confocal microscope. A krypton argon laser was used to excite FITC and Texas red simultaneously at 488 and 568 nm, respectively. The emitted light was collected with filter 522-DF32 for FITC and filter HQ598-40 for Texas red.
For in situ nuclear matrix preparation, BALB/c 3T3 cells were grown on coverslips for 48 h and permeabilized with 0.5% Triton X-100 in TBS buffer containing 0.5 mM PMSF on ice for 1 min. Cells were then washed with TBS buffer, incubated in 500 ml of TBS buffer with 5 U of DNase I/ml (Worthington) at room temperature for 10 min, and extracted with salt extraction buffer [0.2 M (NH4)2SO4, 10 mM Tris-HCl (pH 7.4), 0.2 mM MgCl2, 0.5 mM PMSF] for 2 min on ice. The extracted cells were washed with TBS-Tween buffer and fixed with 3% paraformaldehyde for 3 min. Procedures for immunostaining of the in situ-prepared nuclear matrix with B3 and FIEI and laser scanning confocal microscopy were performed as described above for the whole-cell samples.
Isolation of nuclear matrix.
Rat liver nuclei and nuclear matrix were isolated based on the procedures of Berezney and Coffey (11) and Basler et al. (6). Briefly, rat livers were excised and homogenized with a Potter-Elvehjem apparatus and centrifuged to isolate the nuclei. To obtain the rat liver nuclear matrix, the rat liver nuclei were digested with DNase I (5 U/mg of DNA), extracted with 2 M NaCl three times, and then further extracted with 0.4% Triton X-100. The final rat liver nuclear matrix pellet was resuspended in 10 mM Tris (pH 7.4)–0.2 mM MgCl2–50% glycerol and stored in liquid nitrogen.
HeLa nuclei and nuclear matrix were isolated according to the procedure of Belgrader et al. (9). Briefly, nuclei were isolated by a syringe technique for cell disruption. To isolate nuclear matrix, the nuclei were incubated with DNase I (30 U/mg of DNA) on ice for 30 min and extracted two times with 0.6 M (NH4)2SO4, followed by centrifugation of the nuclear matrix pellet. The final matrix fraction was suspended in TM-2 buffer with 50% glycerol and stored at −20°C until use. The supernatants obtained after 0.6 M (NH4)2SO4 extraction were combined as the high-salt extraction fraction. Protein concentrations in the various fractions were determined by the bicinchoninic acid assay (Pierce).
Proteins from cytoplasmic, nuclear, matrix, and high-salt extracts (10 μg each) were loaded on SDS–12.5% polyacrylamide gels as previously described (49). The gels were stained with Coomassie blue to visualize the protein profile. For Western blots, proteins in the SDS gels were transferred to nitrocellulose membranes, blocked with 5% nonfat milk in phosphate-buffered saline (PBS) with 1% Tween 20 for 15 min, and incubated with MAb B3 or F1E1, or anti-lamin B antibody in blocking buffer at 4°C overnight. After three washes, the blots were incubated with alkaline phosphatase-conjugated secondary antibody at room temperature for 2 h, washed four more times, and developed in nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate toluidinium (BCIP) solution (Sigma).
Double labeling of transcription sites and SCAF8 in permeabilized cells.
Labeling of RNA synthesis sites was done according to published methods (41, 50, 68) with modifications. Briefly, Swiss 3T3 mouse fibroblast cells were grown on a coverslip in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum in 5% CO2 for 24 to 48 h. Cells were then washed with ice-cold TBS buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM MgCl2) and further washed with glycerol buffer (20 mM Tris-HCl [pH 7.4], 25% glycerol, 5 mM MgCl2, 0.5 mM EGTA, 0.5 mM PMSF) for 10 min on ice. Washed cells were permeabilized with 0.025% Triton X-100 in glycerol buffer with 25 U of RNasin (Promega) per ml on ice for 3 min and immediately incubated with RNA synthesis buffer (50 mM Tris-HCl [pH 7.4]; 10 mM MgCl2; 150 mM NaCl; 25% glycerol; 0.5 mM PMSF; 25 U of RNasin per ml; 1.8 mM ATP; 0.5 mM CTP, GTP, and bromo-UTP; and 25 U of RNasin per ml) to label transcription sites. After incubation for 30 min at room temperature, the cells were washed twice with ice-cold 0.5% Triton X-100 in TBS buffer and immediately fixed in 100% methanol at −20°C for 20 min and further fixed with 3% freshly made formaldehyde in PBS on ice for 5 min. Fixed cells were washed twice with ice cold TBS-Tween buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.2 mM MgCl2, 0.2% Tween 20), and blocked with 5% goat serum on ice for 5 min before incubation with primary antibodies at room temperature for 1 h. For labeling RNA sites, Sera-Lab rat anti-bromodeoxyuridine (BrdU) MAb was used at a 1:5 dilution. Tissue culture supernatant containing MAb F1E1 was used to detect SCAF8. After incubation with the primary antibody, cells were washed three times and incubated with FITC-conjugated goat anti-rat IgG (Jackson ImmunoResearch Laboratories, Inc.) at a 1:50 dilution for 40 min at room temperature to visualize transcription sites. MAb F1E1 was detected as described in the previous section. After a final wash with TBS-Tween buffer, coverslips were mount on slides in Slow Fade (Molecular Probe) and examined under a laser scanning confocal microscope as described above.
RESULTS
SCAF8 interacts with serine-phosphorylated CTD.
In previous work, we showed that the SCAF1 (rA1) CID binds hypophosphorylated RNA pol IIA and the hyperphosphorylated II0 form (74). Whether phosphorylation is necessary for this interaction could not be determined from these experiments, because both forms of pol II contained phosphorylated CTD residues. Here we show that phosphorylation of the CTD is required for the CTD-SCAF8 interaction. When a mixture of phosphorylated and unphosphorylated wild-type CTD fusion proteins is passed over a column containing the SCAF8 CID, most of the unphosphorylated CTD flowed through (Fig. 2A, lanes FT and W1 to W6). Bound phosphorylated GST-CTD fusion protein was eluted with SDS-PAGE sample buffer (lanes E1, E2, and E3). The SCAF8-CTD interaction is resistant to treatment with DNase or RNase, ruling out a nucleic acid “bridge” and suggesting a direct interaction between SCAF8 and the CTD (Fig. 2B). Figure 2B also shows that the SCAF8-CTD interaction is sensitive to 0.1% Sarkosyl and 1 M NaCl. The SCAF8 CID does not interact with tyrosine-phosphorylated CTD (Fig. 2C), indicating that the interaction does not reflect a general affinity for phosphorylated proteins.
FIG. 2.
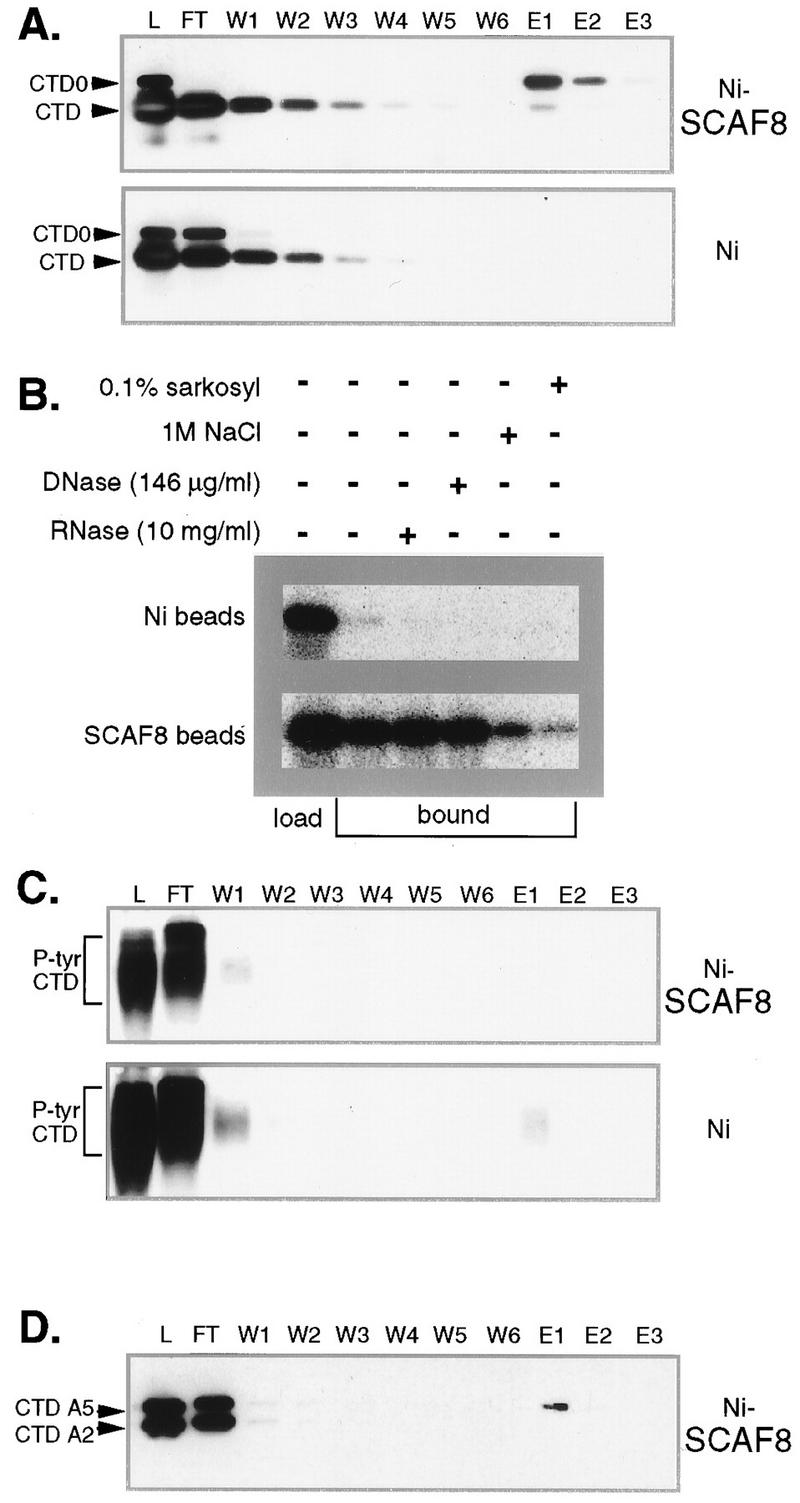
SCAF8 CTD binding assays. (A) SCAF8 interacts with phosphorylated CTD fusion proteins. The binding assay for SCAF8 CID and the different CTD fusion proteins is described in Materials and Methods. All column fractions were separated by SDS-PAGE (10% polyacrylamide) and then subjected to Western blot analysis. Proteins were detected with monoclonal antibody 12CA5, which detects both phosphorylated and unphosphorylated CTD fusion proteins (indicated at left). The phosphorylated fusion protein can be distinguished by its retarded mobility. The column matrix is indicated to the right. Lane L, protein load; lane FT, flowthrough; lanes W1 to W6, column washes; lanes E1 to E3, SDS sample buffer eluates. (B) Binding controls. A binding assay similar to that described for panel A was performed in the presence of additional components indicated above the figure. In this experiment, the wild-type CTD fusion protein was phosphorylated with Cdc2 kinase in the presence of [γ-32P]ATP, and the bound fractions eluted with SDS sample buffer were pooled. (C) SCAF8 does not bind tyrosine-phosphorylated CTD (P-tyr CTD). A binding assay similar to that described for panel A was performed with tyrosine-phosphorylated CTD (Materials and Methods). Lanes are as indicated for panel A. (D) SCAF8 does not bind mutant CTDs. A binding reaction similar to that for panel A was performed with a mixture of phosphorylated A5 and A2 mutant CTD fusion proteins. The identity of the fusion protein (indicated at left) was determined by running them individually. Lanes are as described for panel A.
SCAF8 does not interact with serine-phosphorylated mutant CTDs.
Our earlier studies indicated that serine-to-alanine substitutions in either position 2 or 5 of each consensus repeat is lethal in yeast (70). We have used the same mutant CTDs to study the specificity of the SCAF8-CTD interaction. Cdc2 kinase, which phosphorylates both positions 2 and 5 in the wild-type CTD (80), was used to phosphorylate mutant CTDs containing serine-to-alanine substitutions in serine 2 or 5 of each repeat. The resulting phosphorylated fusion proteins contain phosphates only in position 5 or 2, respectively. When a mixture of the phosphorylated mutant fusion proteins is passed over a SCAF8 CID column, we observe that most of the protein fails to bind (Fig. 2D), suggesting that SCAF8 interacts with CTD phosphorylated on serine residues in both positions 2 and 5.
SCAF8 interacts with phosphorylated pol II.
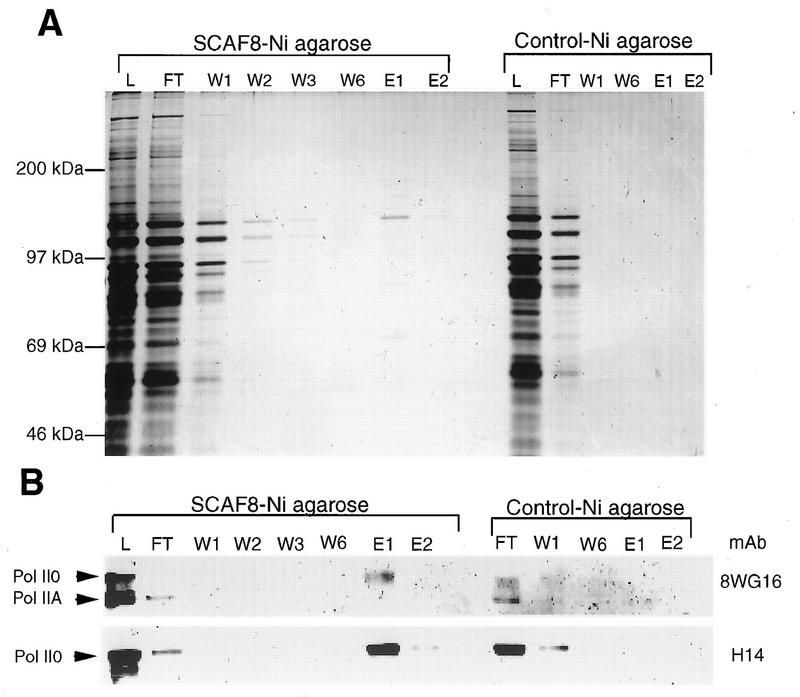
The results described so far demonstrate that SCAF8 interacts with in vitro-phosphorylated CTD. To extend our observation to full-length pol II, we passed semipurified pol II derived from HeLa cells over a SCAF8 CID-Ni2+-agarose column and monitored the fate of the unphosphorylated (IIA) and phosphorylated (II0) forms by Western blotting. As shown in Fig. 3A, most protein flows through the column. The flowthrough contains essentially all of the pol IIA, while most of the pol II0 remains bound to the column (Fig. 3B), thus suggesting that SCAF8 interacts specifically with pol II containing the phosphorylated CTD.
FIG. 3.
SCAF8 interacts with phosphorylated pol II. The binding experiment using partially purified pol II is described in Materials and Methods. Proteins were separated by SDS-PAGE (5% polyacrylamide) and then subjected to either silver staining (A) or Western blotting (B). Lane L, column load; lane FT, column flowthrough; lanes W1 to W6, column washes; lanes E1 and E2, SDS sample buffer eluates.
SCAF8 binds phosphorylated pol II in vivo.
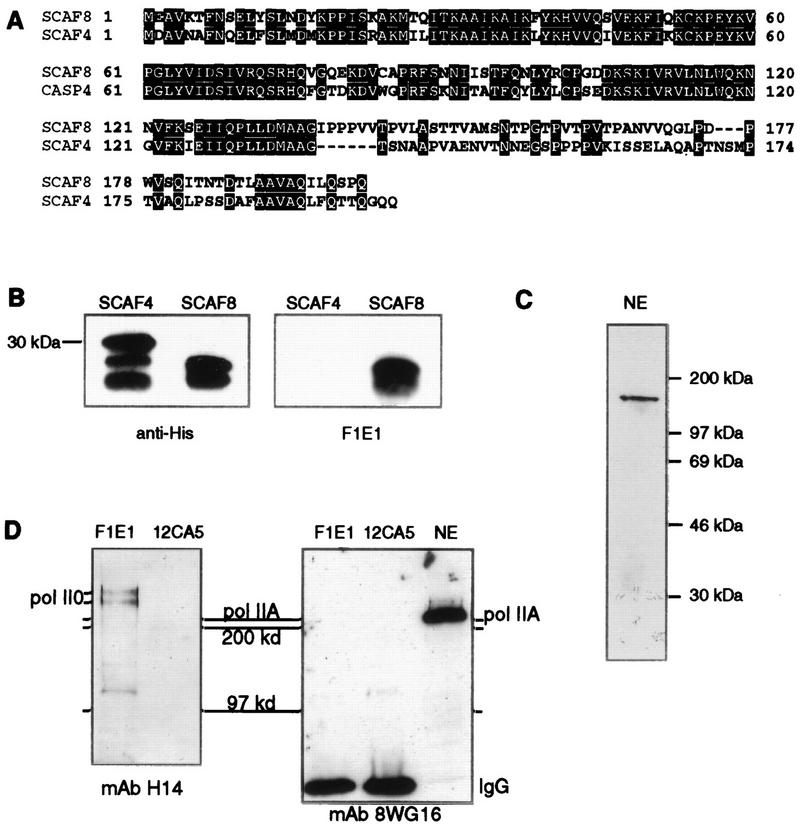
To determine if SCAF8 interacts with pol II in vivo, we performed immunoprecipitation experiments with a MAb directed against the SCAF8-CID. MAb F1E1 was raised against a fusion protein containing the first 200 amino acids of SCAF8. Because the CIDs of SCAF4 and SCAF8 are similar (Fig. 4A), it was important to determine whether F1E1 cross-reacts with SCAF4. Figure 4B shows that F1E1 reacts with a SCAF8 CID fusion protein but not with a similar SCAF4 fusion protein. F1E1 recognizes a protein of around 170 kDa in Hela nuclear extract (Fig. 4C) and in total rat liver nuclear proteins (see Fig. 6B). A similar-sized protein is observed in insect cells infected with a baculovirus expressing SCAF8 but not in control cells (not shown). While the apparent molecular mass is slightly larger than the 140 kDa predicted from the cDNA sequence (74), we note that the protein is rich in proline residues (10%) and therefore may display aberrant mobility in SDS gel electrophoresis. In addition, phosphorylation of the SCAF8 Arg-Ser repeats may retard mobility in SDS gel electrophoresis.
FIG. 4.
SCAF8 interacts with phosphorylated pol II in vivo. (A) Alignment of SCAF8 and SCAF4 CTD interacting domains. These sequences were expressed as 6His-tagged fusion proteins as described in Materials and Methods. (B) MAb F1E1 detects the SCAF8 but not the SCAF4 CID. Partially purified 6His-tagged SCAF CID fusion proteins were separated by SDS gel electrophoresis and immunoblotted with anti-His tag antibody (BABCO) or with MAb F1E1. (C) Western blot analysis of HeLa nuclear extract (NE). SCAF8 was detected with MAb F1E1. (D) Immunoprecipitation of SCAF8 complexes. Immunoprecipitation of the SCAF8 complexes was carried out as described in Materials and Methods. The same blot was probed with MAb H14 (left panel) or with 8WG16 (right panel). The immunoprecipitating antibody is indicated above the lane, while the blotting antibody is indicated below the panel. Markers are indicated to both sides and between the blots. The right lane in the right panel contains nuclear extract to indicate the position of the unphosphorylated RNA pol II largest subunit (IIA).
FIG. 6.
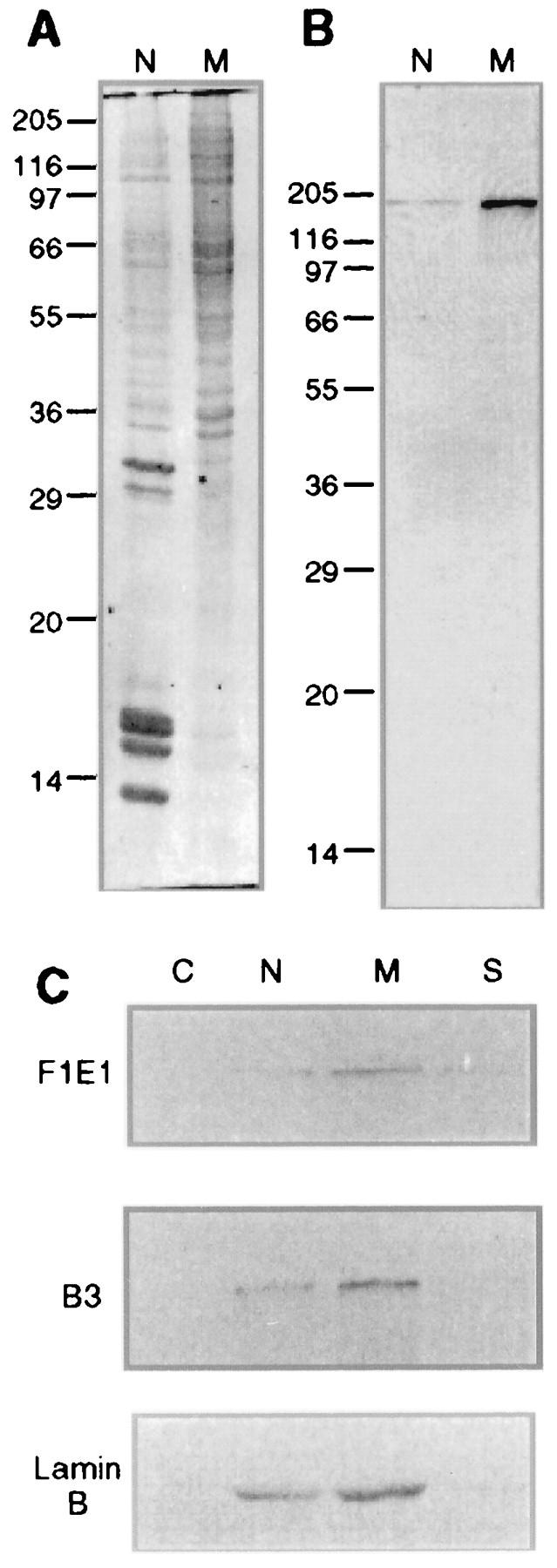
SCAF8 fractionates with nuclear matrix proteins. (A) Coomassie blue staining of rat liver nuclear (N) and nuclear matrix (M) proteins. Ten micrograms of protein from each sample was run on SDS-PAGE (12.5% polyacrylamide). The molecular mass markers (in kilodaltons) are displayed on the left side. (B) Western blot of rat liver nuclear and nuclear matrix proteins with FIEI antibody. (C) The distribution of the SCAF8 protein, RNA pol II0, and lamin B in the HeLa cytoplasmic fraction (C), purified nuclear fraction (N), nuclear matrix fraction (M), and salt-extracted nuclear proteins (S) was determined by Western blot analysis.
Figure 4D shows that several pol II0 phosphoisoforms coimmunoprecipitate with the F1E1 reactive protein (left panel). Hypophosphorylated pol IIA does not coimmunoprecipitate with SCAF8 (right panel). This result indicates that SCAF8 interacts with pol II0 in vivo. Since pol II0 has been shown to be involved in transcription elongation, this result indicates that SCAF8 may be present in elongation complexes.
SCAF8 colocalizes with transcription sites.
Recent studies have shown that pol II transcription is distributed among several thousand nuclear sites which are revealed as small “dots” when cells pulse-labeled with BrUTP are stained with anti-BrdU antibody (36, 39, 41, 52, 68, 78). To determine whether SCAF8 localizes to these sites of transcription we labeled both transcription sites and SCAF8 in the same nuclei. Figure 5 shows that both transcription sites and SCAF8 are localized to numerous punctate extranucleolar staining regions. The merged images indicate a significant level of overlap at the light microscopy level of resolution. The expanded regions in the lower panels indicate that most, but not all, punctate extranucleolar staining regions overlap. Colocalization of SCAF8 with transcription sites is also detected over the nucleolar regions (left side of Fig. 5F). The SCAF8 decoration of nucleolar transcription sites is very weak, however, making the significance of this observation unclear.
FIG. 5.
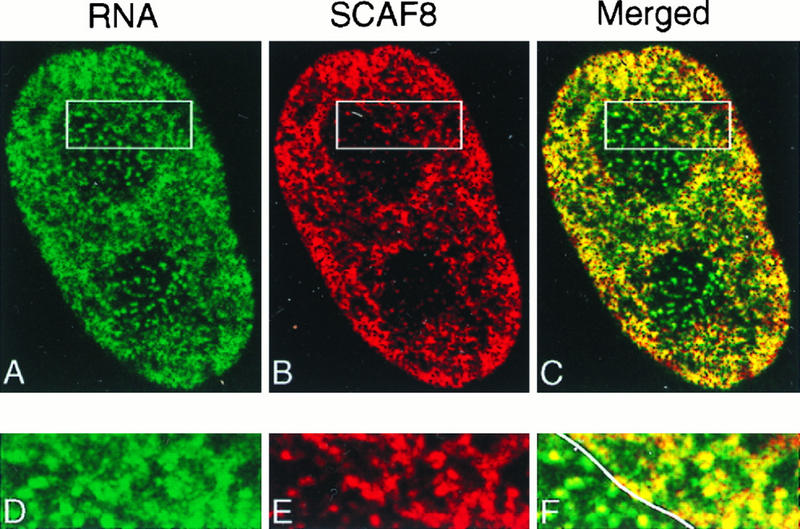
SCAF8 is associated with transcription sites. Mouse Swiss 3T3 cells were permeabilized and incubated with BrUTP to label transcription sites in situ (Materials and Methods). Transcription sites were detected with rat anti-BrUTP antibody (Sera-Lab) and visualized with green (A). SCAF8 was detected with MAb F1E1 and then visualized with red (B), and the merged images are displayed in panel C. The boxed area, including both extranucleolar and nucleolar regions, is further enlarged and displayed in panels D (transcription sites), E (SCAF8), and F (merged images). The line through F indicates the boundary between the nucleolus (left) and the nucleus (right).
Association of SCAF8 with the nuclear matrix.
Previous studies have shown that hyperphosphorylated pol II is associated with the nuclear matrix (49). Since purified SCAF8 binds to the phosphorylated CTD, it was of further interest to determine whether SCAF8 is also a nuclear matrix-associated protein. As a first step, we ran Western blots of total rat liver nuclei and nuclear matrix proteins prepared by classical procedures (6, 11) and probed with the F1E1 antibody. Figure 6A shows the total protein profiles stained with Coomassie blue. Approximately 25% of the total nuclear protein was recovered in the isolated nuclear matrix fraction, which was devoid of histones and enriched in higher-molecular-mass nuclear proteins. Immunoblot analysis of the total rat liver nuclear and nuclear matrix protein fractions revealed a single band with an apparent molecular mass of 170 kDa (Fig. 6B). The four- to fivefold enrichment of SCAF8 in the rat liver nuclear matrix versus the total nuclear fraction indicates a nearly quantitative association of SCAF8 with the nuclear matrix.
We next studied the subnuclear distribution of the SCAF8 protein in HeLa cells fractionated into cytoplasmic, nuclear, nuclear matrix, and high-salt nuclear extract fractions (9). For comparison, we also blotted with antibodies to pol II0 and lamin B, since both of these proteins are known to be constitutively associated with the isolated nuclear matrix fraction (9, 49, 51). The results (Fig. 6C) show a virtually identical distribution pattern of SCAF8 protein compared to those of pol II0 and lamin B. In all cases, the proteins are present in the nuclear fraction, but are enriched in the nuclear matrix. The trace amounts of SCAF8 detected in the high-salt nuclear extract also parallel the findings with pol II0 and lamin B (Fig. 6C), supporting the conclusion that SCAF8 is nearly quantitatively associated with the HeLa nuclear matrix.
It is important to note that the procedure used to isolate the HeLa nuclear matrix is both rapid (<1 h) and occurs at very low temperature (<4°C) to minimize potential degradation and preparative alterations in the isolated nuclear matrix (9). So-called “stabilization steps” used by some investigators to demonstrate nuclear matrix associations (9), including incubations at 30 to 37°C or addition of stabilizing agents such as sodium tetrathionate, had no effect on the subnuclear distribution of the SCAF8 protein (results not shown).
Spatial relationships between SCAF8 and pol II0.
The distribution of SCAF8 with respect to a particular pol II phosphoisoform was studied by fluorescence laser scanning confocal microscopy. Consistent with the results shown in Fig. 5 and the immunoblot studies, the SCAF8 protein was detected exclusively in the interphase cell nucleus of BALB/c 3T3 mouse fibroblast cells, with no detectable signal in the cytoplasm (Fig. 7). Numerous granule-like foci were present in the extranucleolar regions of the nucleus, with far fewer distributed over the nucleoli. Extraction of the coverslip-grown cells preserving nuclear matrix (50) resulted in a strikingly similar pattern of decoration and staining intensity. This is consistent with the immunoblotting experiments, which indicated a nearly quantitative association of the SCAF8 protein with the isolated nuclear matrix, and further suggests that certain key features of the structural organization of SCAF8 observed in whole cells are also maintained in the cells extracted for nuclear matrix.
FIG. 7.
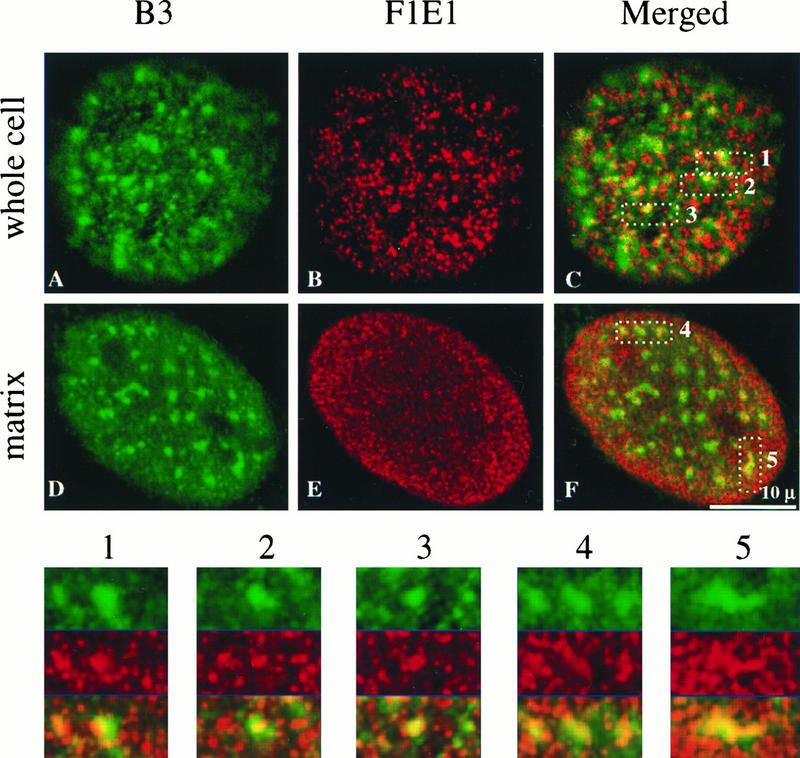
Double labeling of 3T3 cells and in situ-prepared nuclear matrix with B3 and FIEI antibodies. The hyperphosphorylated RNA pol II large subunit was recognized with the B3 antibody (green). SCAF8 antigen was recognized with the FIEI antibody (red). The merged image is displayed on the right. Five speckle regions indicated by the B3 staining were magnified and displayed underneath the whole images. The single red and green channel images for the magnified areas of the nucleus in the whole cell are also displayed above the merged magnified regional images. The 0.5-μm optical sections are displayed.
Both in vitro and in vivo studies indicate that SCAF8 binds to the CTD of pol II0 and suggest that these proteins colocalize in the nucleus. To determine the extent of colocalization, we differentially labeled SCAF8 and pol II0 in whole cells and used laser scanning confocal microscopy to examine the distribution of these proteins. pol II0 was decorated with MAb B3 at a limited number of relatively large nuclear foci (Fig. 7, green labeling), which previous studies have shown to colocalize with the splicing factor-rich nuclear speckles (19, 30, 42, 49, 78) as well at numerous other small granule-like foci throughout the extranucleolar nuclear region (49, 78). Merging of the green pol II labeling with the red SCAF8 labeling revealed an association of some (but not all) SCAF8 foci with nuclear pol II0 speckles. Indeed, virtually every pol II-decorated nuclear speckle contained or was in close association with one or more SCAF8 foci. This can be more readily observed at a higher magnification of regions of the nucleus containing one or more pol II-decorated speckles (see inserts in Fig. 7). Moreover, an identical pattern of colocalization at nuclear speckles was observed following extraction of cells for nuclear matrix (Fig. 7).
DISCUSSION
Recent studies have shown that the pol II CTD plays a role in coupling transcription and pre-mRNA processing (reviewed in references 25, 53 and 63), but the molecular details of the interface between the transcription and processing machinery have not been established. In this paper, we demonstrate that a recently discovered CTD-binding protein, SCAF8, interacts specifically with a phosphorylated form of pol II. We further show that SCAF8 colocalizes with sites of active transcription. These transcription sites are enriched in nuclear matrix preparations that have previously been shown to contain components of the processing machinery (reviewed in references 12, 13, and 54). The ability of SCAF8 to interact with both transcription and processing structures indicates that it could play a direct role in coupling these two processes.
The CTD is involved in pre-mRNA processing.
Although in vitro transcription and pre-mRNA processing can occur in isolation, in vivo these processes occur in close proximity. Electron micrographs of active transcription units first demonstrated that intron removal can occur on elongating transcripts (14). Subsequent studies have supported a model in which nascent transcripts are cotranscriptionally processed (7, 8, 43, 79).
Cotranscriptional processing could be a simple matter of mass action with excess processing components initiating processing as soon as the nascent pre-mRNA emerges from the elongating polymerase. Alternatively, cotranscriptional processing may be facilitated by unique features of the transcription and processing machines. In support of the second model, processing occurs inefficiently on transcripts synthesized by pol I or pol III (37, 60, 61), indicating that pol II is specialized for coupled transcription and processing.
Recent evidence has indicated that the distinctive feature of pol II responsible for coupling transcription and processing is the CTD (25, 53, 63). This highly repetitive domain is not present on pol I or pol III, indicating it plays a role in mRNA biogenesis. In vitro studies first showed that antibodies against the CTD or CTD peptides can inhibit splicing (74). Subsequent in vivo studies have shown that pol II containing only 5 of the normal 52 repeats is capable of transcription, but capping (46), splicing (47), and cleavage and polyadenylation (47) are inhibited. Overexpression of a detached CTD also inhibits splicing (42), presumably by competing with pol II for essential CTD-binding factors. Taken together, these results suggest that the CTD plays a direct role in pre-mRNA processing.
Pre-mRNA processing and the CTD phosphorylation cycle.
The CTD is rich in potential phosphorylation sites and is reversibly phosphorylated during the transcription cycle in vivo, thus giving rise to at least two distinct forms of pol II (27). Pol IIA has an unphosphorylated or hypophosphorylated CTD and is involved in the formation of preinitiation complexes (27). CTD phosphorylation in the context of the preinitiation complex converts pol IIA to pol II0, coinciding with the elongation phase of the transcription cycle. The involvement of pol II0 in transcription elongation suggests that processing components will interact with the phosphorylated CTD. Indeed, capping activities have been shown to interact with the phosphorylated CTD (21, 46, 58, 73). Cleavage and polyadenylation factors also bind to the CTD, but here the role of phosphorylation is not clear (47). Loading the 3′-end-forming factors onto the transcription complex also requires the general transcription factor TFIID (28). Splicing small nuclear ribonucleoprotein particles (snRNPs) and members of the SR family of non-snRNP pre-mRNA splicing factors associate with hyperphosphorylated forms of pol II (20, 42, 49, 66), but in most of these cases, it is not known whether these splicing components interact directly with the CTD.
SCAF8 binds to the phosphorylated CTD.
Yeast two-hybrid interaction screening has led to the identification of proteins that interact with the pol II CTD (17, 64, 74). These proteins resemble the SR proteins previously implicated in splicing (33) and have been designated SCAFs.
Three classes of SCAFs have been identified. SCAF1, -5, and -9 (rA1, rA5, and rA9, respectively, in reference 74 and later CASP1, -5, and -9, respectively, in reference 25), and SCAF11 and 12 (SRrp129 and C10, respectively in reference 64) contain conserved ∼80-amino-acid C-terminal CTD-interacting domains. With the exception of their Ser/Arg-rich motifs, these proteins are unrelated. SCAF4 and SCAF8 (rA4 and rA8, respectively, in reference 74, and CASP4 and CASP8, respectively, in reference 25) contain conserved ∼120-amino-acid N-terminal CIDs that are unrelated to the SCAF1-type sequence. SCAF4 and SCAF8 also contain atypical RNA recognition motifs, but with the exception of these and the Ser/Arg-rich motifs, they are unrelated. SCAF10 (SRcyp) is a unique CTD-interacting protein containing a peptidyl-prolyl cis-trans isomerase domain (17). SCAF10 differs from the other SCAFs by interacting with the CTD through ill-defined sequences in the vicinity of its Ser/Arg-rich domain (17). The ability of SCAFs to interact with pol II and their similarity to splicing factors make them excellent candidates for coupling transcription and splicing.
We have investigated the role of CTD phosphorylation on the interaction between SCAFs and the CTD. In a previous paper, we demonstrated that SCAF1 binds both pol IIA and pol II0 (74), but because the pol IIA preparation had been substoichiometrically phosphorylated, it was impossible to determine whether phosphorylation influenced the interaction. In the present study, we show that CTD phosphorylation is absolutely required for SCAF8 interaction. This interaction is direct and specific, requiring phosphorylation of both serine 2 and serine 5 in the consensus repeat. Interestingly, preliminary studies indicate that SCAF1 also requires CTD phosphorylation for interaction but will bind to either serine 2- or serine 5-phosphorylated CTDs (56a). This difference in specificity is not unexpected given the difference in sequence of the SCAF8 and SCAF1 CIDs (74).
SCAF8 also associates with the phosphorylated CTD in vivo. Hyperphosphorylated pol II0 coimmunoprecipitates with SCAF8, while the hypophosphorylated pol IIA does not (Fig. 4D). This result is consistent with previous work indicating that pol II0 forms a complex with snRNPs and SR proteins (20, 42, 49, 66, 74). Because SCAFs are the only splicing-related proteins shown to directly interact with pol II they may play a crucial role in establishing links with processing components. SCAFS are well designed for interfacing the transcription and processing machines. As shown here, the SCAF8 CID interacts specifically with the form of the CTD present in elongation complexes. The SCAF serine/arginine-rich motifs would then be available to interact with other SR proteins (71), leading to the assembly of spliceosomes.
Subnuclear localization of transcription and pre-mRNA processing.
Antibodies directed against splicing components stain mammalian nuclei in a nonuniform manner, suggesting the presence of subnuclear structures associated with pre-mRNA processing (62). Typical mammalian cells contain 20 to 50 such “speckles,” which were first detected with an antibody directed against the splicing factor SC35 (34). The size and number of these structures can vary according to the staining conditions (52), with more dilute staining leading to smaller, more abundant speckles. The method of signal processing (32, 59) can also influence the perception of speckles. Careful quantification of SC35 distribution in the nucleus indicates that the local concentration varies by as little as twofold. Together, these factors make structural characterization of speckles difficult. The localization of green fluorescent protein-tagged proteins to speckle-like structures supports the nonuniform distribution of splicing factors in vivo (48).
Another controversial question is whether speckles represent processing or storage sites (45, 59). Staining of active transcription sites by uridine incorporation does not detect speckles but rather has revealed more numerous, smaller punctate sites (32, 36, 41, 52, 68, 78). Up to several thousand of these sites may exist in each nucleus. Modified staining procedures have shown that pol II (36, 78) and some pre-mRNA processing components (52, 78) colocalize to transcription sites. These results are consistent with coupled transcription and processing but at the limited resolution of light microscopy.
The disunion of transcription sites and speckles has led to the suggestion that speckles are storage or recycling sites. Consistent with this model, inactivation of transcription or splicing leads to an enlargement of speckles (48, 78). Furthermore, time-lapse fluorescence microscopy of GFP-tagged splicing factor SF2/ASF indicates that splicing factors can leave speckles in peripheral extensions and accumulate at nearby transcription sites (48). Thus, a large portion of pol II-mediated transcription appears to occur in the more diffuse nucleoplasm between speckles. None of these studies, however, rule out the possibility that transcription also occurs on or within the speckle itself. Indeed, this is suggested by the findings of Xing et al. (72).
We show here that SCAF8 colocalizes primarily with sites of transcription (Fig. 5). In this sense, SCAF8 localization is similar to pol II0 detected by MAbs H5 and H14 (36, 78) and the SR protein SRp20 (52). In addition, SCAF8 partially overlaps with nuclear speckles detected by MAb B3 (Fig. 7). This result could indicate that some transcription sites are located on or within speckles. The observation of large B3-reactive speckles suggests that a significant fraction of the pol II0 phosphoisoform detected by this antibody is not active in transcription. In a similar fashion, only a fraction of SC35 staining colocalizes with sites of transcription (32, 78). Apparently some transcription and processing components are present in excess and accumulate in speckles, while other factors, like SCAF8 and SRp20, are less abundant and primarily associate with sites of active transcription. While further research is needed to clarify the role of nuclear speckles in transcription and processing events, our findings support the emerging concept that they may be dynamic structures involved in the assembly of active transcription/processing sites.
SCAF8 and pol II0 are nuclear matrix proteins.
The nuclear structure remaining after removal of soluble nuclear proteins and chromatin has been termed the nuclear matrix (10–13, 54). The nuclear matrix contains the majority of the heterogeneous nuclear RNA (38), and pulse-labeled transcripts also associate with the nuclear matrix (40), indicating that this structure contains transcription components. Nascent, unprocessed pre-mRNAs are apparently held in a functional state, because addition of soluble nuclear factors to matrix preparations leads to rapid processing of bound mRNA precursors (76, 77). In addition to RNA, the nuclear matrix contains proteins, including spliceosome components (67), a novel set of high-molecular-mass SR-like proteins (15, 16), phosphorylated pol II0 (20, 49, 66), and, as we show here, SCAF8.
The presence of SCAF8 in the nuclear matrix and its interaction with pol II0 suggest a possible role in targeting some pol II0 phosphoisoforms to the nuclear matrix. The B3 antibody detects a pol II0 phosphoisoform that is distributed both in larger speckles and in smaller foci resembling transcription sites. While not all of the pol II0 detected with MAb B3 colocalizes with SCAF8, the larger pol II0 foci often overlap with several smaller SCAF8 foci.
We have recently demonstrated that the anti-phospho-CTD antibodies H5, H14, and B3 recognize different CTD phosphoepitopes (57). In yeast, CTD phosphorylation generating these different epitopes is regulated independently in response to extracellular signals (57). While the function of differential CTD phosphorylation is not known, it seems likely that differential phosphorylation modifies the transcriptional capacity and localization of different pol II0 phosphoisoforms. Indeed, H5 preferentially stains pol II0 in speckles, while H14 preferentially stains the diffuse nucleoplasmic pol II0 (19). SCAFs could play a role in the localization of different pol II0 phosphoisomers by interacting with different CTD phosphorylation patterns.
Summary.
The results presented here support two main conclusions. First, SCAF8 interacts directly with a highly phosphorylated form of pol II. This observation indicates that SCAF8 is not involved in transcription initiation but rather associates with pol II during elongation. The colocalization of SCAF8 with sites of active transcription supports this conclusion. Second, SCAF8 is present in the nuclear matrix. The second conclusion is consistent with the presence of large nuclear structures containing both transcription and pre-mRNA processing components. The role of SCAF8 could be to bind newly initiated pol II and recruit processing factors to the elongation complex. In this case, SCAF8 (and possibly other SCAFs) would act as coupling factors, responding to CTD phosphorylation by linking transcription elongation complexes to an appropriate processing machine. The details of this mechanism and its potential regulatory function await further investigation.
ACKNOWLEDGMENTS
We thank R. Baskaran for c-Abl kinase and Danny Reinberg for RNA pol II. Robert G. Summers, Jr. provided valuable advice and assistance with the dual-labeling confocal microscopy. Geraldine Seydoux and Nick Conrad provided helpful comments on the manuscript.
This work was supported by grants from the NIH GM53600 (J.L.C.) and GM23922 (R.B.).
M.P. and X.W. contributed equally to this work.
REFERENCES
- 1.Akhtar A, Faye G, Bentley D L. Distinct activated and non-activated RNA polymerase II complexes in yeast. EMBO J. 1996;15:4654–4664. [PMC free article] [PubMed] [Google Scholar]
- 2.Allison L A, Moyle M, Shales M, Ingles C J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985;42:599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- 3.Allison L A, Wong J K-C, Fitzpatrick V D, Moyle M, Ingles C J. The C-terminal domain of the largest subunit of RNA polymerase II of Saccharomyces cerevisiae, Drosophila melanogaster, and mammals: a conserved structure with an essential function. Mol Cell Biol. 1988;8:321–329. doi: 10.1128/mcb.8.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartolomei M S, Halden N F, Cullen C R, Corden J L. Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II. Mol Cell Biol. 1988;8:330–339. doi: 10.1128/mcb.8.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baskaran R, Dahmus M E, Wang J Y. Tyrosine phosphorylation of mammalian RNA polymerase II carboxyl-terminal domain. Proc Natl Acad Sci USA. 1993;90:11167–11171. doi: 10.1073/pnas.90.23.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Basler J, Hastie N D, Pietras D, Matsui S I, Sandberg A A, Berezney R. Hybridization of nuclear matrix attached deoxyribonucleic acid fragments. Biochemistry. 1981;20:6921–6929. doi: 10.1021/bi00527a027. [DOI] [PubMed] [Google Scholar]
- 7.Bauren G, Jiang W Q, Bernholm K, Gu F, Wieslander L. Demonstration of a dynamic, transcription-dependent organization of pre-mRNA splicing factors in polytene nuclei. J Cell Biol. 1996;133:929–941. doi: 10.1083/jcb.133.5.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- 9.Belgrader P, Siegel A J, Berezney R. A comprehensive study on the isolation and characterization of the HeLa S3 nuclear matrix. J Cell Sci. 1991;98:281–291. doi: 10.1242/jcs.98.3.281. [DOI] [PubMed] [Google Scholar]
- 10.Berezney R, Coffey D S. Identification of a nuclear protein matrix. Biochem Biophys Res Commun. 1974;60:1410–1417. doi: 10.1016/0006-291x(74)90355-6. [DOI] [PubMed] [Google Scholar]
- 11.Berezney R, Coffey D S. Nuclear matrix. Isolation and characterization of a framework structure from rat liver nuclei. J Cell Biol. 1977;73:616–637. doi: 10.1083/jcb.73.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berezney R, Mortillaro M, Ma H, Meng C, Samarabandu J, Wei X, Somanathan S, Liou W S, Pan S J, Cheng P C. Connecting nuclear architecture and genomic function. J Cell Biochem. 1996;62:223–226. doi: 10.1002/(SICI)1097-4644(199608)62:2%3C223::AID-JCB10%3E3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 13.Berezney R, Mortillaro M J, Ma H, Wei X, Samarabandu J. The nuclear matrix: a structural milieu for genomic function. Int Rev Cytol. 1995;162A:1–65. doi: 10.1016/s0074-7696(08)61228-0. [DOI] [PubMed] [Google Scholar]
- 14.Beyer A L, Osheim Y N. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- 15.Blencowe B J, Issner R, Kim J, McCaw P, Sharp P A. New proteins related to the Ser-Arg family of splicing factors. RNA. 1995;1:852–865. [PMC free article] [PubMed] [Google Scholar]
- 16.Blencowe B J, Nickerson J A, Issner R, Penman S, Sharp P A. Association of nuclear matrix antigens with exon-containing splicing complexes. J Cell Biol. 1994;127:593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bourquin J P, Stagljar I, Meier P, Moosmann P, Silke J, Baechi T, Georgiev O, Schaffner W. A serine/arginine-rich nuclear matrix cyclophilin interacts with the C-terminal domain of RNA polymerase II. Nucleic Acids Res. 1997;25:2055–2061. doi: 10.1093/nar/25.11.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bregman D B, Du L, Li Y, Ribisi S, Warren S L. Cytostellin distributes to nuclear regions enriched with splicing factors. J Cell Sci. 1994;107:387–396. doi: 10.1242/jcs.107.3.387. [DOI] [PubMed] [Google Scholar]
- 19.Bregman D B, Du L, Vanderzee S, Warren S L. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chabot B, Bisotto S, Vincent M. The nuclear matrix phosphoprotein p255 associates with splicing complexes as part of the [U4/U6.U5] tri-snRNP particle. Nucleic Acids Res. 1995;23:3206–3213. doi: 10.1093/nar/23.16.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho E J, Takagi T, Moore C R, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Corden J L, Ingles C J. Carboxy-terminal domain of the largest subunit of eukaryotic RNA polymerase II. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Vol. 1. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 81–108. [Google Scholar]
- 23.Corden J L. Tails of RNA polymerase II. Trends Biochem Sci. 1990;15:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 24.Corden J L, Cadena D L, Ahearn J, Jr, Dahmus M E. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci USA. 1985;82:7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corden J L, Patturajan M. A CTD function linking transcription to splicing. Trends Biochem Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- 26.Dahmus M E. Phosphorylation of the C-terminal domain of RNA polymerase II. Biochim Biophys Acta. 1995;1261:171–182. doi: 10.1016/0167-4781(94)00233-s. [DOI] [PubMed] [Google Scholar]
- 27.Dahmus M E. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- 28.Dantonel J C, Murthy K G, Manley J L, Tora L. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature. 1997;389:399–402. doi: 10.1038/38763. [DOI] [PubMed] [Google Scholar]
- 29.Desai D, Gu Y, Morgan D O. Activation of human cyclin-dependent kinases in vitro. Mol Biol Cell. 1992;3:571–582. doi: 10.1091/mbc.3.5.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Du L, Warren S L. A functional interaction between the carboxy-terminal domain of RNA polymerase II and pre-mRNA splicing. J Cell Biol. 1997;136:5–18. doi: 10.1083/jcb.136.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egyhazi E, Ossoinak A, Pigon A, Holmgren C, Lee J M, Greenleaf A L. Phosphorylation dependence of the initiation of productive transcription of Balbiani ring 2 genes in living cells. Chromosoma. 1996;104:422–433. doi: 10.1007/BF00352266. [DOI] [PubMed] [Google Scholar]
- 32.Fay F S, Taneja K L, Shenoy S, Lifshitz L, Singer R H. Quantitative digital analysis of diffuse and concentrated nuclear distributions of nascent transcripts, SC35 and poly(A) Exp Cell Res. 1997;231:27–37. doi: 10.1006/excr.1996.3460. [DOI] [PubMed] [Google Scholar]
- 33.Fu X D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 34.Fu X D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 35.Gebara M M, Sayre M H, Corden J L. Phosphorylation of the carboxy-terminal repeat domain in RNA polymerase II by cyclin-dependent kinases is sufficient to inhibit transcription. J Cell Biochem. 1997;64:390–402. [PubMed] [Google Scholar]
- 36.Grande M A, Vanderkraan I, Dejong L, Vandriel R. Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J Cell Sci. 1997;110:1781–1791. doi: 10.1242/jcs.110.15.1781. [DOI] [PubMed] [Google Scholar]
- 37.Gunnery S, Mathews M B. Functional mRNA can be generated by RNA polymerase III. Mol Cell Biol. 1995;15:3597–3607. doi: 10.1128/mcb.15.7.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He D C, Martin T, Penman S. Localization of heterogeneous nuclear ribonucleoprotein in the interphase nuclear matrix core filaments and on perichromosomal filaments at mitosis. Proc Natl Acad Sci USA. 1991;88:7469–7473. doi: 10.1073/pnas.88.17.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iborra F J, Pombo A, Jackson D A, Cook P R. Active RNA polymerases are localized within discrete transcription “factories” in human nuclei. J Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- 40.Jackson D A, Cook P R. Transcription occurs at a nucleoskeleton. EMBO J. 1985;4:919–925. doi: 10.1002/j.1460-2075.1985.tb03719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jackson D A, Hassan A B, Errington R J, Cook P R. Visualization of focal sites of transcription within human nuclei. EMBO J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim E, Du L, Bregman D B, Warren S L. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.LeMaire M F, Thummel C S. Splicing precedes polyadenylation during Drosophila E74A transcription. Mol Cell Biol. 1990;10:6059–6063. doi: 10.1128/mcb.10.11.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marshall N F, Peng J M, Xie Z, Price D H. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 45.Mattaj I W. RNA processing—splicing in space. Nature. 1994;372:727–728. doi: 10.1038/372727a0. [DOI] [PubMed] [Google Scholar]
- 46.McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Program A E, Shuman S, Bentley D L. 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson S D, Wickens M, Bentley D L. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 48.Misteli T, Caceres J F, Spector D L. The dynamics of a pre-mRNA splicing factor in living cells. Nature. 1997;387:523–527. doi: 10.1038/387523a0. [DOI] [PubMed] [Google Scholar]
- 49.Mortillaro M J, Blencowe B J, Wei X Y, Nakayasu H, Du L, Warren S L, Sharp P A, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakayasu H, Berezney R. Mapping replicational sites in the eucaryotic cell nucleus. J Cell Biol. 1989;108:1–11. doi: 10.1083/jcb.108.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nakayasu H, Berezney R. Nuclear matrins: identification of the major nuclear matrix proteins. Proc Natl Acad Sci USA. 1991;88:10312–10316. doi: 10.1073/pnas.88.22.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Neugebauer K M, Roth M B. Distribution of pre-mRNA splicing factors at sites of RNA polymerase II transcription. Genes Dev. 1997;11:1148–1159. doi: 10.1101/gad.11.9.1148. [DOI] [PubMed] [Google Scholar]
- 53.Neugebauer K M, Roth M B. Transcription units as RNA processing units. Genes Dev. 1997;11:3279–3285. doi: 10.1101/gad.11.24.3279. [DOI] [PubMed] [Google Scholar]
- 54.Nickerson J A, Blencowe B J, Penman S. The architectural organization of nuclear metabolism. Int Rev Cytol. 1995;162A:67–123. doi: 10.1016/s0074-7696(08)61229-2. [DOI] [PubMed] [Google Scholar]
- 55.Nonet M, Sweetser D, Young R A. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987;50:909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- 56.O’Brien T, Hardin S, Greenleaf A, Lis J T. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 56a.Patturajan, M., and J. L. Corden. Unpublished observations.
- 57.Patturajan M, Schulte R J, Sefton B M, Berezney R, Vincent M, Bensaude O, Warren S L, Corden J L. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1997;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- 58.Shuman S. Origins of mRNA identity-capping enzymes bind to the phosphorylated C-terminal domain of RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:12758–12760. doi: 10.1073/pnas.94.24.12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singer R H, Green M R. Compartmentalization of eukaryotic gene expression—causes and effects. Cell. 1997;91:291–294. doi: 10.1016/s0092-8674(00)80411-0. [DOI] [PubMed] [Google Scholar]
- 60.Sisodia S S, Sollner-Webb B, Cleveland D W. Specificity of RNA maturation pathways: RNAs transcribed by RNA polymerase III are not substrates for splicing or polyadenylation. Mol Cell Biol. 1987;7:3602–3612. doi: 10.1128/mcb.7.10.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smale S T, Tjian R. Transcription of herpes simplex virus tk sequences under the control of wild-type and mutant human RNA polymerase I promoters. Mol Cell Biol. 1985;5:352–362. doi: 10.1128/mcb.5.2.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Spector D L. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- 63.Steinmetz E J. Pre-mRNA processing and the CTD of RNA polymerase II: the tail that wags the dog? Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 64.Tanner S, Stagljar I, Georgiev O, Schaffner W, Bourquin J P. A novel SR-related protein specifically interacts with the carboxyl-terminal domain (CTD) of RNA polymerase II through a conserved interaction domain. Biol Chem. 1997;378:565–571. doi: 10.1515/bchm.1997.378.6.565. [DOI] [PubMed] [Google Scholar]
- 65.Thompson N E, Steinberg T H, Aronson D B, Burgess R R. Inhibition of in vivo and in vitro transcription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J Biol Chem. 1989;264:11511–11520. [PubMed] [Google Scholar]
- 66.Vincent M, Lauriault P, Dubois M F, Lavoie S, Bensaude O, Chabot B. The nuclear matrix protein p255 is a highly phosphorylated form of RNA polymerase II largest subunit which associates with spliceosomes. Nucleic Acids Res. 1996;24:4649–4652. doi: 10.1093/nar/24.23.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vogelstein B, Hunt B F. A subset of small nuclear ribonucleoprotein particle antigens is a component of the nuclear matrix. Biochem Biophys Res Commun. 1982;105:1224–1232. doi: 10.1016/0006-291x(82)91099-3. [DOI] [PubMed] [Google Scholar]
- 68.Wansink D G, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weeks J R, Hardin S E, Shen J, Lee J M, Greenleaf A L. Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: correlations with gene activity and transcript processing. Genes Dev. 1993;7:2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- 70.West M L, Corden J L. Construction and analysis of yeast RNA polymerase II CTD deletion and substitution mutants. Genetics. 1995;140:1223–1233. doi: 10.1093/genetics/140.4.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 72.Xing Y, Johnson C V, Moen P T, Jr, McNeil J A, Lawrence J. Nonrandom gene organization: structural arrangements of specific pre-mRNA transcription and splicing with SC-35 domains. J Cell Biol. 1995;131:1635–1647. doi: 10.1083/jcb.131.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yue Z Y, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin A J. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci USA. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yuryev A, Patturajan M, Litingtung Y, Joshi R V, Gentile C, Gebara M, Corden J L. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zehring W A, Lee J M, Weeks J R, Jokerst R S, Greenleaf A L. The C-terminal repeat domain of RNA polymerase II largest subunit is essential in vivo but is not required for accurate transcription initiation in vitro. Proc Natl Acad Sci USA. 1988;85:3698–3702. doi: 10.1073/pnas.85.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zeitlin S, Parent A, Silverstein S, Efstratiadis A. Pre-mRNA splicing and the nuclear matrix. Mol Cell Biol. 1987;7:111–120. doi: 10.1128/mcb.7.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zeitlin S, Wilson R C, Efstratiadis A. Autonomous splicing and complementation of in vivo-assembled spliceosomes. J Cell Biol. 1989;108:765–777. doi: 10.1083/jcb.108.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zeng C, Kim E, Warren S L, Berget S M. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J. 1997;16:1401–1412. doi: 10.1093/emboj/16.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang G, Taneja K L, Singer R H, Green M R. Localization of pre-mRNA splicing in mammalian nuclei. Nature. 1994;372:809–812. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]
- 80.Zhang J, Corden J L. Identification of phosphorylation sites in the repetitive carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J Biol Chem. 1991;266:2290–2296. [PubMed] [Google Scholar]