Abstract
The U1 snRNP functions to nucleate spliceosome assembly on newly transcribed pre-mRNA. Saccharomyces cerevisiae is unusual among eukaryotes in the greatly extended length of its U1 snRNA and the apparent increased polypeptide complexity of the corresponding U1 snRNP. In this paper, we report the identification of a novel U1 snRNP protein, Prp42p, with unexpected properties. Prp42p was identified by its surprising structural similarity to the essential U1 snRNP protein, Prp39p. Both Prp39p and Prp42p possess multiple copies of a variant tetratricopeptide repeat, an element implicated in a wide range of protein assembly events. Yeast strains depleted of Prp42p by transcriptional repression of a GAL1::PRP42 fusion gene arrest for splicing prior to pre-mRNA 5′ splice site cleavage. Prp42p was not observed in a recent biochemical analysis of purified U1 snRNPs from S. cerevisiae (28). Nevertheless, antibodies directed against an epitope-tagged version of Prp42p specifically precipitate U1 snRNA from yeast extracts. Furthermore, Prp42p is required for U1 snRNP biogenesis, because yeast strains depleted of Prp42p formed incomplete U1 snRNPs that failed to produce stable complexes with pre-mRNA in vitro. The evidence shows that Prp39p and Prp42p are both required to configure the atypical yeast U1 snRNP into a structure compatible with its evolutionarily conserved role in pre-mRNA splicing.
The basic steps in spliceosome assembly are well conserved between Saccharomyces cerevisiae and humans (reviewed in references 17 and 27). An initiating event in intron selection is recognition of the pre-mRNA by the U1 snRNP. Proteins bound to the pre-mRNA and associated with the U1 snRNP (i) stabilize the highly specific 4- to 7-bp interaction between the 5′ splice site and the 5′ end of the U1 snRNA and (ii) mediate branch point recognition by the U1 snRNP (reviewed in references 12 and 31). The resulting structure, the commitment complex, serves as a substrate for prespliceosome formation through the ATP-dependent addition of the U2 snRNP. The subsequent addition of the U4-U6.U5 snRNP–tri-snRNP complex and an unknown number of non-snRNP proteins completes spliceosome assembly and promotes intron removal.
Yeast uses an atypical U1 snRNP in commitment complex formation. The yeast U1 snRNA is 3.5 times larger than its metazoan equivalent (19, 40), and associated with it are at least six proteins without obvious counterparts in the mammalian U1 snRNP (28). The relevance of this increased yeast U1 snRNP complexity to splice site identification and spliceosome assembly is poorly understood. Early experiments revealed that much of the yeast-specific U1 snRNA could be deleted without significant detriment to cell viability (24, 41). The remaining U1 snRNA appears to fold into a structure similar to that found in the mammalian snRNP (18). Thus, the bulk of the yeast-specific U1 snRNA, while possibly contributing to the efficiency of the splicing event, does not provide an essential function in yeast.
Genetic and biochemical studies have confirmed the presence of the ubiquitous U1-specific proteins in the yeast U1 snRNP. The genes for yeast strains U1-70k (Snp1p [44]) and U1-C (47) were identified from genomic DNA sequence data based on the phylogenetic conservation of the encoded proteins. Each proved essential for U1 snRNP function. A synthetic lethal screen for mutations that exacerbate a mutant U1 snRNA phenotype led to the identification of the yeast U1-A counterpart (Mud1p [25]). Curiously, Mud1p is not required for pre-mRNA splicing or cellular viability.
In addition to the conserved U1 snRNP protein genes, two genes have been found that encode proteins defined to date only in yeast. PRP39 was discovered in a screen for temperature-sensitive mutants defective in pre-mRNA splicing (26). In vitro studies revealed that Prp39p is a U1 snRNP protein and is required to assemble a productive U1 snRNP–pre-mRNA complex. Prp40p was initially identified as a suppressor of a cold-sensitive C-to-U nucleotide change at position 4 of the U1 snRNA (16). Consistent with a primary role in intron removal, Prp40p is required for pre-mRNA splicing in vivo and in vitro. A recent investigation of commitment complex assembly revealed that Prp40p interacts with an evolutionarily conserved protein, BBP, which binds to the pre-mRNA branch point sequence (2, 4, 5). Yeast BBP also binds the yeast U2AF counterpart, Mud2p (1, 2). A Prp40p-BBP-Mud2p association may underpin the U1 snRNP-mediated branch point recognition that occurs in commitment complex formation (see references 2, 5, and 27 and references within). A recent yeast two-hybrid study (8) suggests that Prp39p may also interact with Mud2p. If so, Prp39p would provide an additional tether between the U1 snRNP and the branch point region of the intron. Thus, in contrast to the dispensable nature of the excess yeast U1 snRNA sequence, at least two of the yeast-specific proteins are critical for U1 snRNP function.
A number of proteins with probable common ancestry function in pre-mRNA splicing. Examples of such proteins include the Sm core snRNP proteins (14, 35), the snRNP-specific mammalian proteins U1A (42) and U2B" (13), the serine- and arginine-rich (SR) splicing factors (9), and the family of DEAD/H-box proteins (see references 17 and 27). Here we report the identification of a novel yeast protein, Prp42p, with extensive similarity to the U1 snRNP protein, Prp39p. Embedded within the Prp39p and Prp42p proteins are multiple copies of a 34-amino-acid repeat (tetratricopeptide repeat [TPR]) similar to the TPR elements found in the crooked neck (crn) protein of Drosophila melanogaster (52). Prp39p and Prp42p are not functionally redundant, because each is needed for cellular pre-mRNA splicing. The data show that like Prp39p, Prp42p is required to assemble a stable U1 snRNP capable of productive interaction with cellular pre-mRNA.
MATERIALS AND METHODS
Identification of Prp42p.
A BLASTP comparison (3) of Prp39p amino acids 1 to 629 with the S. cerevisiae nonredundant protein database was performed at site www-genome.stanford.edu/ with the Blosum 62 matrix. The best match for a novel protein, P[N] = 2.5e−10, was obtained with the 544-amino-acid hypothetical protein YDR235W. Pairwise sequence comparisons of Prp39p and Prp42p were performed with the Gap and BestFit programs (University of Wisconsin GCG package). TPR structures were initially selected on the basis of the presence of five or more conserved tryptophan and hydrophobic residues at positions 4, 7, 8, 11, 21, 24, and 27. The GCG Helical Wheel program and the Protein Sequence Analysis System (45, 50) were then used to screen for amphipathic helical sequences within the putative TPRs. Consensus sequences were derived from the Prp39p and Prp42p TPRs with the GCG Pretty program.
Cloning and characterization of PRP42.
The PRP42 open reading frame (ORF) was amplified from yeast genomic DNA by PCR (upstream primer, 5′ AGA GGA TCC ATG GAT AAA TAT ACT GCT TTG ATT CAC 3′; downstream primer, 5′ CCA GGA TCC AAT AAA TGA CAA TGC CTT TTG GCT AAG G 3′). The primer-encoded BamHI sites (underlined) were used to fuse the PRP42 ORF to the GAL1 promoter of plasmid pBM150 (15) to create GAL1::PRP42. GAL1::PRP42HA was created in a similar manner, except that the downstream PCR primer contained codons for the nine-amino-acid hemagglutinin (HA) epitope (5′ TTT GGA TCC CTA AGC GTA GTC TGG AAC GTC GTA TGG GTA AGG TTC TTC AGT AAA CAT TTC CTC 3′; BamHI site, underlined; HA codons, boldface).
The PRP42 ORF described above was blunt end ligated into the SmaI site of pTZ19U (United States Biologicals). An internal deletion of approximately half of the PRP42 coding sequence was then created by excision of an internal 0.81-kbp BstBI fragment. This deletion derivative was then modified for genetic selection by blunt end insertion of a 2.2-kbp DNA fragment containing the LEU2 gene of yeast. The resultant prp42::LEU2 gene was released from the vector DNA by digestion with BamHI, followed by treatment with mung bean nuclease. Gel-purified prp42::LEU2 DNA was then used to replace one of the two wild-type copies of PRP42 in the diploid yeast strain MGD407 (33). The site of insertion was confirmed by PCR analysis of the genomic DNA. Standard techniques (37) were used to induce sporulation and dissect the meiotic progeny of this strain. To create the GAL1::PRP42 and GAL1::PRP42HA strains, the respective plasmids were transformed into the MGD407 heterozygous disruptant prior to sporulation. The asci were dissected on yeast extract-peptone medium containing 2% galactose to permit expression of the plasmid-borne fusion genes. The haploid offspring were then phenotypically scored on selective medium to screen for the presence of the prp42::LEU2 gene and for galactose-dependent cell growth.
Yeast exact preparation and immune precipitations.
Yeast extracts were prepared by grinding liquid nitrogen-frozen cell pellets with a mortar and pestle and processing the lysate as previously described (49). For immune precipitation, 1 μl of the antibody was bound to 10 μl of protein G agarose as previously described (26). The antibodies used were the HA-specific HA.11 antibody (Babco) and a nonspecific monoclonal antibody (MAb63) (gift of M. Mendenhall). For typical reactions, yeast extract containing approximately 250 μg of protein was mixed with 10 μl of antibody-bound beads in a total volume of 100 μl of HNT buffer (20 mM HEPES [pH 7.9], 100 mM NaCl, 0.05% Triton X-100) at room temperature for 30 min. The NaCl concentration of the HNT buffer was adjusted between 50 and 300 mM as needed to assay the tightness of the Prp42HAp-U1 snRNP association. The unbound extract was separated from the antibody-bound material by centrifugation in a Marathon 16 KM microcentrifuge (Fisher Scientific) at 3,500 rpm for 1 min. The beads were washed six times with 300 μl of HNT. To release the antibody-associated snRNAs, 100 μl of PK buffer (100 mM Tris-HCl [pH 7.5], 12.5 mM EDTA, 150 mM NaCl, 1% sodium dodecyl sulfate, 400 μg of proteinase K per ml) was added, and the mixture was incubated at 37°C for 10 min. After phenol extraction, the samples were concentrated by ethanol precipitation and assayed by Northern blotting with the previously described snRNA probes (6). Unbound and total snRNA preparations were recovered and assayed in parallel by analogous procedures.
Splicing analysis, spliceosome assembly, and snRNP complexes.
The analysis of in vivo pre-mRNA splicing was performed as previously described (6). RNA was extracted from yeast cultures grown on galactose and for various times after the shift to a glucose-based medium. Approximately 25 μg of total RNA was fractionated on a 1% agarose–formaldehyde gel. A transfer on Nytran-plus membrane was then hybridized with probes specific for the RP51A and ADE3 genes (6).
In vitro splicing reactions were assembled on RP51A transcripts prepared by in vitro transcription of the pSPrp51A construct (29) with SP6 RNA polymerase. The splicing reaction was fractionated on a 5% polyacrylamide–7 M urea denaturing gel to assay splicing and on a 3% polyacrylamide–0.5% agarose gel to assay the assembly of splicing complexes as previously described (26). To visualize the commitment complex bands, the U2 snRNA was cleaved with the extract’s endogenous RNase H activity (34). This was accomplished by a 10-min preincubation with 0.1 μg of the anti-U2 oligonucleotide 5′ CAG ATA CTA CAC TTG 3′ prior to pre-mRNA addition. Yeast snRNP complexes were resolved on 3 or 4% polyacrylamide–0.5% agarose gels as previously described (26), except that the heparin (Sigma) concentration was increased to 1 mg/ml. The samples were incubated on ice for 10 min prior to electrophoresis. Where antibodies were used for gel shift analysis, 1 μl of the HA.11 antibody or MAb63 was preincubated with the yeast extract at room temperature for 10 min prior to the addition of R buffer (50 mM HEPES [pH 7.5], 2.0 mM magnesium acetate, 20 mM EDTA, 1 mg of heparin per ml).
Glycerol gradient fractionation.
Prior to fractionation of the yeast snRNPs, 1 volume of yeast extract was mixed with 2 volumes of buffer A (50 mM Tris-HCl [pH 7.4], 25 mM NaCl, 5 mM MgCl2). The samples were applied to the top of a 10 to 30% linear glycerol gradient made in buffer A and centrifuged for 14 h at 37,000 rpm in a Beckman SW41 rotor. The positions of the various snRNP complexes were established by recovery of RNA from sequential 500-μl samples from the top of the gradient. Each gradient fraction was phenol extracted, and the RNA was precipitated with ethanol. The recovered RNAs and control samples were fractionated on a 5% polyacrylamide–7 M urea denaturing gel and assayed by Northern blotting for snRNAs as described previously (6). The relative intensities of the snRNA bands were determined with an LKB model 2222-010 Ultroscan XL laser densitometer.
RESULTS
Identification of a novel protein, Prp42, with sequence similarity to Prp39p.
A BLASTP search (3) of the S. cerevisiae nonredundant protein database revealed a previously uncharacterized protein of 544 amino acids, YDR235w (henceforth called Prp42p), that shares 50% sequence similarity with Prp39p (Fig. 1). The protein sequence alignment shows that the regions of Prp39p-Prp42p similarity are scattered throughout the protein coding sequences, with somewhat lower levels of sequence identity at the extreme amino and carboxyl termini. Many of the Prp39p-Prp42p sequence identities are clustered in contexts reminiscent of the TPRs of the Drosophila crn protein (52) (Fig. 2). A global BLAST search of the public protein databases highlighted this fact, because both Prp39p and Prp42p produced matches to the Drosophila crn protein, and, in each case, the similarities were restricted to the TPR elements (unpublished observations). Yeast repeats 42.1 to 42.4 and 39.1 to 39.5 show the greatest similarities to the Drosophila crn TPRs; each aligned with one or more of the crn repeats 3, 4, 7, 8, 10, 11, 12, 15, and 16 (see reference 52 for sequence definitions of the numbered repeats). The Prp39p-Prp42p TPR elements match best when arranged in register (e.g., 39.1 with 42.1 and 39.2 with 42.2). This pattern is most obvious for the first three TPR elements because the alignment based on maximum similarity places the TPRs in phase with one another. Elsewhere the Prp39p and Prp42p proteins appear to have evolved to such a degree that the TPR elements no longer align.
FIG. 1.
Comparison of the Prp39p and Prp42p protein sequences. An amino acid alignment between Prp39p and ORF YDR235w (i.e., Prp42p) was performed with the BestFit program (GCG, Inc.) to show the locations of sequence identities (vertical lines) and conservative amino acid substitutions (colons, strong similarity; single dots, moderate similarity). The positions of the putative TPRs are overlined (Prp39p) and underlined (Prp42p).
FIG. 2.
Alignment of 11 proposed TPRs within Prp39p and Prp42p. The repeats are numbered consecutively from the amino termini of the Prp39p (39.1 to 39.6) and Prp42p (42.1 to 42.5) proteins presented in Fig. 1. Columns with greater than 40% (light shading) or 70% (dark shading) amino acid sequence similarity among the Prp39p-Prp42p repeats are boxed. The amino acid residues were grouped as polar charged (D, E, H, K, and R), polar uncharged (N, Q, S, and T), nonpolar hydrophobic (L, I, V, M, F, Y, and W), small (A and G), or other (C and P). The Prp39p-Prp42p consensus sequence was assembled by BestFit and Pretty analyses (University of Wisconsin, GCG program). In many but not all instances, the calculated consensus residue was the most common amino acid within a boxed group. The asterisk indicates the position of the conserved hydrophobic residue at position 24. The vertical lines indicate sequence identity, and the colons and single dots represent degrees of conservative substitutions. Underlined in the crn sequence (52) are the box A and box B elements. Below the crn sequence is a more generalized repeat present in a diverse set of TPR proteins that consists of seven hydrophobic residues and a proline (20).
In addition to the proposed TPRs, a possible nuclear localization sequence, KKKLKK, is present at amino acids 231 to 235 of the Prp42p protein. This element may not be conserved within Prp39p, because none of the clustered basic residues of Prp39p match closely the canonical simian virus 40 T antigen or nucleoporin bipartite nuclear localization sequence motifs. No other strong matches to known functional sequence motifs were observed in Prp42p.
PRP42 encodes a protein required for pre-mRNA splicing in vivo.
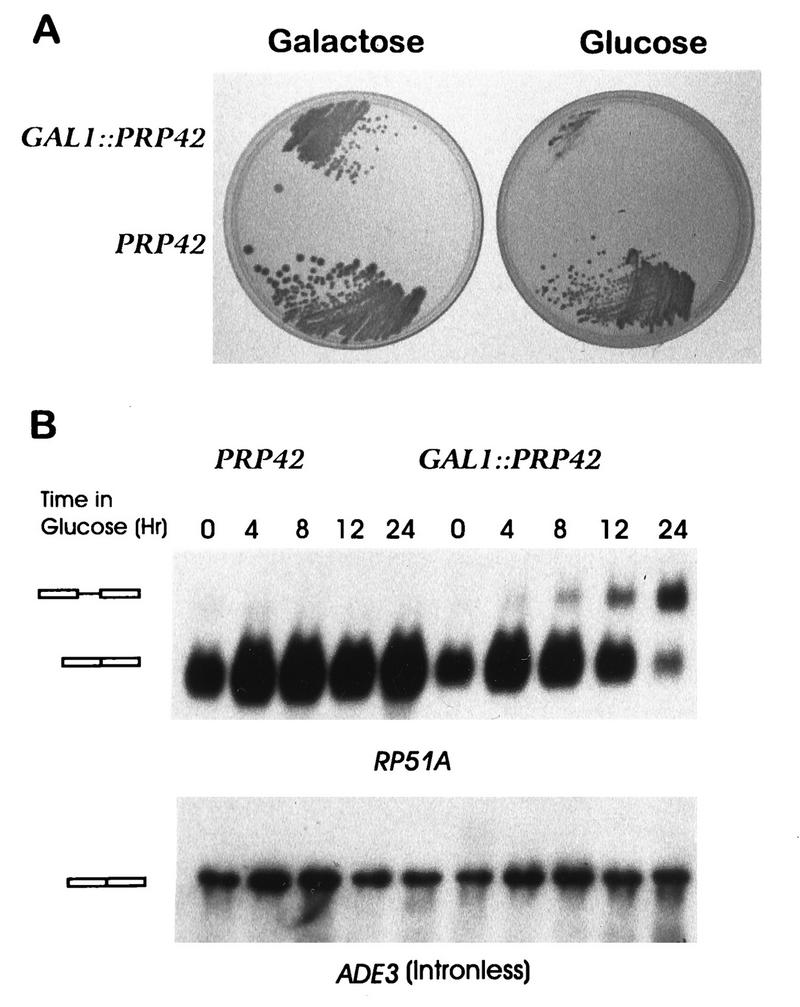
The structural similarity between Prp42p and the U1 snRNP protein Prp39p suggested that Prp42p might also function in pre-mRNA splicing. Since PRP39 is essential in yeast (26), it seemed unlikely that PRP42 acted simply as an alternate source of the Prp39p activity. Consistent with an independent cellular function, a genomic disruption of the PRP42 ORF by the yeast LEU2 gene proved lethal (see Materials and Methods). The prp42::LEU2 mutation was complemented by a plasmid-borne PRP42 allele in which the natural PRP42 promoter was replaced by the nutritionally regulated GAL1 promoter. The GAL1::PRP42 colonies were not quite as large as those from wild-type yeast when assayed on galactose medium. While the reason for this discrepancy is not known, the viability of the GAL1::PRP42 yeast clearly required expression of the fusion gene. Transfer of this strain to glucose-based medium repressed GAL1::PRP42 transcription and blocked colony formation (Fig. 3A). These experiments established that PRP42 is an essential gene and provided a valuable genetic tool with which to experimentally manipulate the intracellular levels of Prp42p.
FIG. 3.
PRP42 encodes a protein required for cellular viability and pre-mRNA splicing. (A) To assay for PRP42-dependent growth, yeast strains that expressed the endogenous wild-type copy of PRP42 or the glucose-repressible GAL1::PRP42 fusion gene were plated on galactose-based and glucose-based rich media. (B) RNA was extracted from the PRP42 and GAL1::PRP42 cultures grown continuously on galactose medium (time 0) or after a shift to glucose medium for the indicated number of hours. The Northern transfer in the upper panel was hybridized with the intron-containing RP51A gene. The identities of the mRNA and pre-mRNA (rectangles) were confirmed by primer extension analysis (data not shown). The lower panel presents the same filter after hybridization with the intronless ADE3 gene.
The intron-containing ACT1, RP51A, CYH2, and SNR17 genes were assayed to determine the impact of GAL1::PRP42 transcriptional repression on the efficiency of in vivo splicing (Fig. 3B and data not shown). RNA was extracted from wild-type (PRP42) and GAL1::PRP42 cultures grown continuously on galactose medium and from the same cultures after transfer to glucose-based medium. Yeast that expressed the GAL1::PRP42 gene showed an mRNA/pre-mRNA ratio indistinguishable from that observed with the wild-type allele. By this measure, the GAL1::PRP42 allele functioned as well as the endogenous chromosomal allele in pre-mRNA splicing. In contrast, splicing arrest resulted when GAL1::PRP42 transcription was repressed; the mRNA/pre-mRNA ratios of the interrupted genes decreased as a function of time in the glucose-based medium. Elevated pre-mRNA levels became apparent approximately 8 h after the shift of the media, while cell division slowed and then stopped some 10 to 15 h later. No differences in RNA mobilities were observed for transcripts from the intronless ADE3, SNR19, SNR20, and rDNA genes (Fig. 3B and data not shown), indicating that Prp42p is required specifically for the processing of intron bearing pre-mRNA.
Prp42p is a U1 snRNP-associated protein.
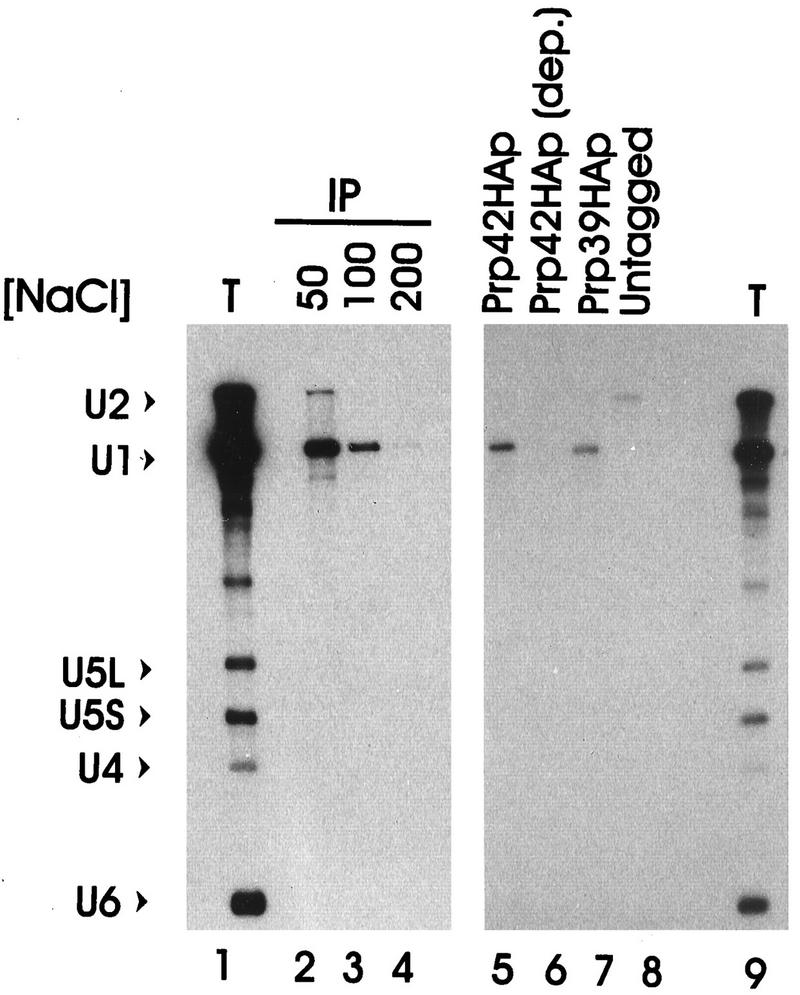
To assist in the characterization of Prp42p, the nine-codon HA epitope sequence was added to the 3′ end of the GAL1::PRP42 ORF. This derivative, GAL1::PRP42HA, retained biological activity, because it complemented the growth and splicing defects of the chromosomal prp42::LEU2 null allele. Extracts prepared from the GAL1::PRP42HA culture and from control cultures were assayed for coprecipitation of spliceosomal snRNAs with the HA-tagged proteins (Fig. 4). U1 snRNA was specifically recovered from the anti-HA (HA.11) immune pellets of the epitope-tagged Prp42HAp and Prp39HAp extracts (Fig. 4, lanes 5 and 7) (26). The coprecipitation of U1 snRNA with Prp42HAp was rather salt sensitive; peak U1 snRNA recovery was achieved below 100 mM NaCl (Fig. 4, lanes 2 to 4). The U1 snRNA recovery was epitope specific, however, because no detectable U1 snRNA coprecipitated at 100 mM NaCl from the extract metabolically depleted of Prp42HAp or from an extract that lacked an HA-tagged protein (Fig. 4, lanes 6 and 8). Coprecipitation was also antibody specific, because only background levels of U1 snRNA were recovered from the GAL1::PRP42HA extract when the irrelevant MAb63 was substituted for HA.11 (reference 26 and unpublished observations). Because the U1 snRNA levels varied less than twofold in all extracts tested (see below), the failure to recover U1 snRNA with the HA.11 antibody after Prp42HAp depletion was not a consequence of extensive U1 snRNA degradation. Rather, the recovery of U1 snRNA from GAL1::PRP42HA extracts correlated directly with the presence or absence of the Prp42HAp protein.
FIG. 4.
Prp42HAp specifically associates with the U1 snRNP. A splicing-competent yeast extract prepared from the GAL1::PRP42HA yeast strain was immune precipitated with the HA-specific antibody HA.11 under conditions of increasing NaCl concentration (lanes 2 to 4). RNA present in the immune pellets (IP) or the unfractionated total extract (T) was analyzed by Northern blotting with probes specific for the spliceosomal snRNAs (U1, U2, U4, U5, and U6). In lanes 5 to 9, the immune precipitation was repeated at 100 mM NaCl with the splicing-competent GAL1::PRP42HA extract (lane 5), an extract prepared from GAL1::PRP42HA cultures metabolically depleted of Prp42HAp (Prp42HAp dep.), an extract with an HA-tagged PRP39 allele (Prp39HAp), and an extract without an HA-tagged gene (Untagged).
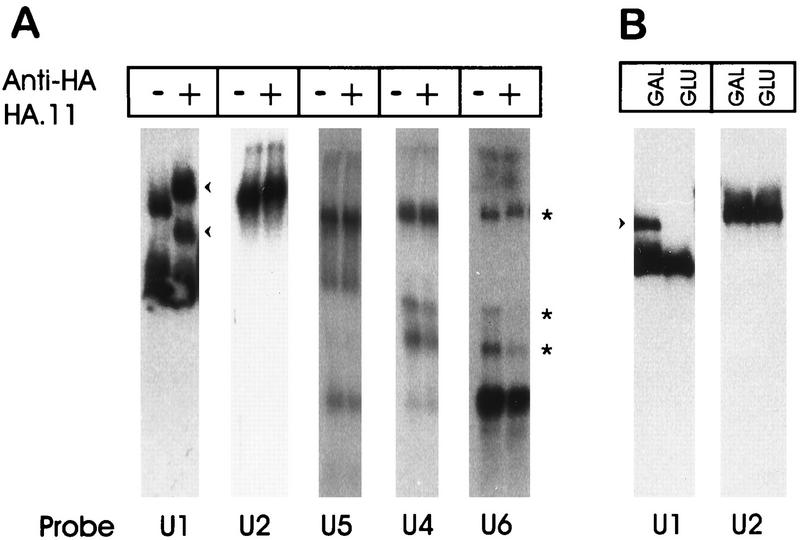
As a more direct means of viewing the Prp42HAp-U1 snRNP association, the GAL1::PRP42HA extract was resolved by native gel electrophoresis with or without prior preincubation with the HA.11 antibody (Fig. 5A). In the absence of HA.11, we routinely observed two distinct U1 snRNP bands upon hybridization (Fig. 5A) (26). The upper band was previously deduced to be the mature form of the U1 snRNP (26). In contrast, the lower band appeared to be an incomplete U1 snRNP, since it was formerly shown to migrate very near the position of naked U1 snRNA and it lacked the U1 snRNP-specific protein, Prp39p (26). The incomplete U1 snRNP band was also detected in wild-type yeast extracts, but the abundance of the incomplete form increased in the epitope-tagged Prp39HAp and Prp42HAp extracts. This observation suggests that the HA epitope reduces the stability or inhibits the function of the tagged proteins. However, the negative impact of epitope addition is clearly limited, because the Prp39HAp and Prp42HAp extracts both contain fully assembled U1 snRNPs and are splicing competent. When the HA.11 antibody was added to the GAL1::PRP42HA extract, 100% of the upper U1 band was shifted (Fig. 5A), consistent with Prp42HAp being present in each of the fully assembled U1 snRNPs. Approximately 50% of the lower U1 band was shifted by the HA.11 antibody. We interpret this to signify heterogeneity within the incompletely assembled U1 particles (i.e., some possessed the Prp42HAp protein and therefore shifted, while others either lacked Prp42HAp or had it sequestered in an antibody-inaccessible location. The U1 snRNP complexes did not change mobility when the HA.11 antibody was added to an untagged extract or when the irrelevant MAb63 was substituted for HA.11 in the GAL1::PRP42HA extract (reference 26 and unpublished observations).
FIG. 5.
Prp42p contributes directly to U1 snRNP structure. (A) Splicing-competent GAL1::PRP42HA extracts were incubated with (+) or without (−) the HA.11 antibody and then resolved on a 3% polyacrylamide–0.5% agarose gel. After electrophoresis, parallel lanes of the Northern transfer were probed for the U1, U2, U4, U5, and U6 snRNA-bearing snRNP complexes. The arrowheads to the right of the U1 snRNA hybridization show the positions of the novel bands present after incubation with the HA.11 antibody. The asterisks to the right of the U6 hybridization identify the location of the U4-U6.U5 tri-snRNP (top band) and two distinct U4-U6-hybridizing bands (based on comigration of the U4, U5, and U6 snRNA signals). (B) GAL1::PRP42HA extracts prepared before (GAL) and after (GLU) depletion of Prp42HAp were resolved by native gel electrophoresis and probed for the U1 and U2 snRNA-bearing complexes. The arrowhead to the left of the U1 hybridization indicates the position of the fully assembled U1 snRNP.
In contrast to the U1 hybridization results, none of the U2, U4, U5, or U6 snRNP complexes showed decreased electrophoretic mobility after HA.11 antibody addition. Occasionally, changes in the relative levels of the U4-U6 versus U4-U6.U5 snRNP complexes were noted after antibody addition (Fig. 5A, asterisks, compare U4 and U6 hybridizations). This appeared to be a nonspecific consequence of the antibody preparation and was independent of the presence of an epitope-tagged protein in the extract (26a).
Metabolic depletion of Prp42HAp caused a profound, yet specific, increase in the electrophoretic mobility of the U1 snRNP and the loss of splicing competence (Fig. 5B and see below). Although incomplete, the Prp42HAp-depleted extract was not otherwise denatured and could be functionally reconstituted by the addition of a micrococcal nuclease-treated (i.e., RNA free) wild-type extract (unpublished observations). No shift in electrophoretic mobility was observed with the U2 snRNP or any other snRNP complex after Prp42HAp depletion (Fig. 5B and data not shown).
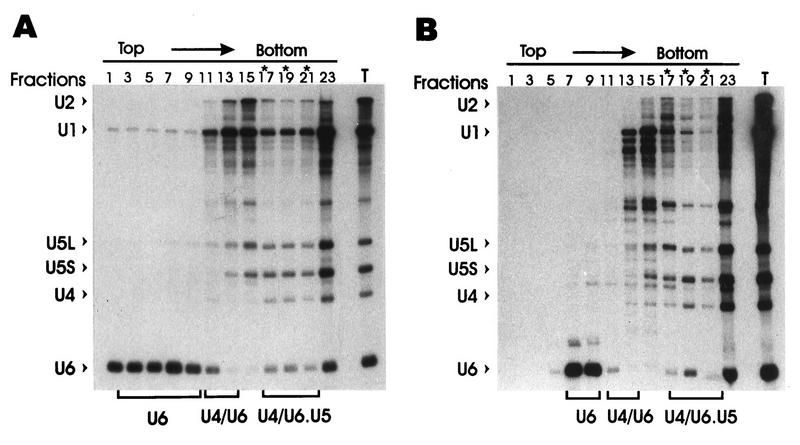
Since the magnitudes of electrophoretic shifts after subunit removal were not necessarily predictable, glycerol gradient sedimentation was performed as an independent assay for Prp42HAp-dependent changes in snRNP structure (Fig. 6). A reproducible reduction in the U1 snRNA signal was observed for the fastest sedimenting and presumably most complete U1 snRNP particles after Prp42HAp depletion (compare lanes 13 through 21, Fig. 6A and B). In contrast, the relative amounts and positions of the U4 and U5 snRNAs did not vary significantly between these two parallel gradient preparations. Densitometric scans showed an approximate 70% reduction in the ratio of the U1 to U5L plus U5S snRNA signals in the heaviest gradient fractions after Prp42HAp depletion (averaged ratio of fractions 19 to 21 = 1.1 in Fig. 6A and 0.3 in Fig. 6B). (Note that fraction 23 contains nonspecific aggregates from the tube bottom.) The U1/U5L plus U5S ratio increased roughly twofold for the bulk of the remaining U1 snRNP fractions (averaged ratio of fractions 13 to 17 = 2.0 in Fig. 6A and 3.9 in Fig. 6B), which is consistent with the recruitment of the Prp42HAp-deficient particles into the lighter U1 snRNP fractions.
FIG. 6.
Sedimentation of the U1 snRNP is altered after Prp42HAp depletion. GAL1::PRP42HA extracts prepared before (A) and after (B) metabolic depletion of Prp42HAp were fractionated on a 10 to 30% linear glycerol gradient. The snRNAs present in alternate gradient fractions were assayed by Northern blotting with a hybridization cocktail which probed for each of the spliceosomal snRNAs. The U1 snRNP fractions most sensitive to Prp42HAp depletion are indicated by asterisks. The location of the free U6 snRNP, the U4-U6 snRNP, and the U4-U6.U5 tri-snRNP are indicated below the figure. In both panels, sample 23 comes from the bottom of the gradient tube and contains all of the spliceosomal snRNAs, presumably, from native spliceosomes or nonspecific aggregates. T, total, unfractionated RNA.
The U1 snRNA decreased by ∼35% and the U6 snRNA decreased by ∼50% after Prp42HAp depletion (compare Fig. 6A and B). SnRNA reductions have been reported after the inactivation or removal of certain other spliceosomal proteins (for examples, see references 6, 32, and 33) and appear to be caused by enhanced snRNA degradation upon snRNP perturbation or splicing arrest. The decrease in U1 snRNA could be accounted for by modestly decreased U1 snRNA stability in the absence of Prp42HAp. The reduction in U6 snRNA is less easily explained but likely occurs as an indirect consequence of the splicing arrest. The free U6 snRNP pool (i.e., the lightest gradient fractions) decreased most dramatically after Prp42HAp depletion, suggesting that the higher-order U4-U6 and U4-U6.U5 snRNP complexes stabilize the U6 snRNA.
Pre-mRNA–U1 snRNP complex formation is impaired by Prp42p depletion.
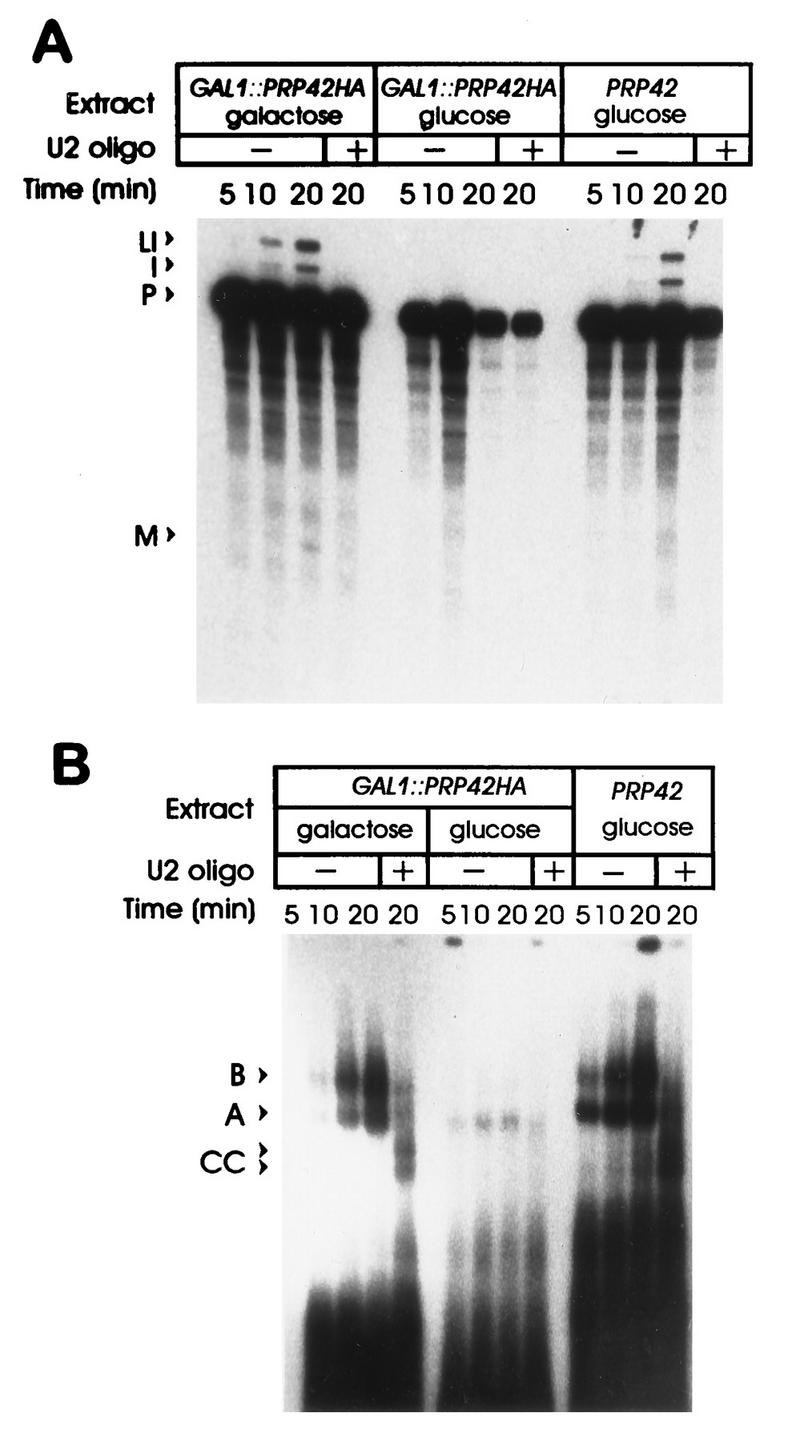
The molecular contacts that secure the 5′ splice site-U1 snRNP interaction in the commitment complex are poorly understood. Since the Prp42HAp-depleted U1 snRNP particles were stable, the formation of a pre-mRNA–U1 snRNP interaction was credible, despite possible defects in other aspects of U1 snRNP function. In vitro spliceosome assembly and splicing were monitored in the Prp42HAp-complete and Prp42HAp-depleted extracts (Fig. 7). Consistent with the in vivo results, RP51A pre-mRNA assembled into prespliceosome and spliceosomal complexes in the Prp42HAp-complete extract (Fig. 7B, GAL1::PRP42HA galactose lanes), and the levels of the splicing intermediates and products in the reaction increased with time of incubation (Fig. 7A). When an oligonucleotide directed against the 5′ end of the U2 snRNA was preincubated with the Prp42HAp complete extract prior to substrate addition, splicing was blocked and spliceosome assembly was arrested at the commitment complex stage. Roughly equivalent amounts of the 5′ splice site-dependent (CC1 [Fig. 7B, upper arrowhead]) and the 5′ splice site and branch point-dependent (CC2 [Fig. 7B, lower arrowhead]) commitment complex bands were observed (Fig. 7) (36). Analogous results were obtained with a wild-type extract prepared from glucose-grown cultures (PRP42 glucose). In contrast, when pre-mRNA was added to the Prp42HAp-depleted extract (GAL1::PRP42HA glucose), a greatly reduced level of spliceosome assembly and no splicing were observed. The low level of splicing complex (mostly prespliceosome) observed was likely due to trace amounts of Prp42HAp remaining in the extract, although inefficient spliceosome assembly in the absence of Prp42HAp cannot be ruled out.
FIG. 7.
Spliceosome assembly is blocked by Prp42HAp removal. (A) 32P-labeled RP51A pre-mRNA was incubated for the indicated times in wild-type extracts (PRP42 glucose) and in extracts prepared before (GAL1::PRP42HA galactose) and after (GAL1::PRP42HA glucose) Prp42HAp depletion. The pre-mRNA (P) and spliced RNA products (lariat intermediate [LI], excised intron [I], and mRNA [M]) were then resolved on a 5% polyacrylamide denaturing gel. In some cases (+), an oligonucleotide directed against the 5′ end of the U2 snRNA was included to block splicing and arrest spliceosome assembly at the commitment complex stage. (B) A portion of each splicing reaction was resolved in parallel on a 3% polyacrylamide–0.5% agarose gel to monitor spliceosome assembly. The commitment complexes (CC1, upper band; CC2, lower band) and assignments of the prespliceosome (A) and spliceosome (B) forms indicated by the arrowheads were based on our (26, 30) and others’ (36) previous characterizations of these complexes. The lane descriptions are the same as those indicated for panel A.
Prominent commitment complex bands were not observed in the Prp42HAp-depleted extracts in either the presence or absence of the U2 oligonucleotide. Since the mobility of the U1 snRNP is changed upon Prp42HAp depletion, it is possible that functional commitment complex 1-commitment complex 2-like complexes formed but did not resolve from the bulk uncomplexed pre-mRNA. This appears unlikely, however, since only a minor amount of spliced pre-mRNA (presumably derived from the residual prespliceosomal complexes) was observed when commitment complex formation was assayed by the substrate challenge method (23) rather than by gel electrophoresis (unpublished observations). These results are consistent with a spliceosome assembly defect prior to stable U1 snRNP addition but do not rule out the possibility that specific, yet weak or transient, U1 snRNP–pre-mRNA interactions occur in the absence of Prp42p.
DISCUSSION
Approximately 108 years ago, S. cerevisiae is believed to have undergone a whole genome duplication (51). This ancient duplication and subsequent genetic events (e.g., deletions and single-gene amplifications) left the modern yeast with approximately 800 pairs of duplicated genes. In many instances, members of individual gene pairs have evolved distinct yet related biological functions. With this in mind, we used the amino acid sequence of the Prp39p protein to search for and identify a novel component of the yeast U1 snRNP, Prp42p. Prp42p shares 50% amino acid sequence similarity with Prp39p and, like Prp39p, contains multiple copies of the TPR protein recognition domain. The results of this study show that the PRP39 and PRP42 genes are not functionally redundant; each protein coded for by these genes is required at an early stage of splicing to assemble a splicing-competent U1 snRNP. The identification of structurally related, snRNP-specific proteins is unprecedented and indicates that the U1 snRNP is more complex than originally envisioned.
Several lines of evidence implicate Prp42p as a legitimate component of the U1 snRNP. First, the structure of Prp42p is quite similar to that of Prp39p, a protein shown genetically and biochemically to be part of the U1 snRNP complex (26, 28). Second, an antibody directed against an epitope-tagged version of Prp42p specifically coprecipitates U1 snRNA and supershifts the U1 snRNP in native polyacrylamide gels. Third, the U1 snRNP in extracts depleted of Prp42HAp shows increased electrophoretic mobility and a decreased sedimentation rate, indicative of a change in U1 snRNP structure or stability. Finally, certain mutations within Prp42p are synthetically lethal with a biologically active U1 snRNA deletion derivative (30a), again consistent with an intimate association between Prp42p and the U1 snRNP.
The absence of Prp42p from a biochemically purified preparation of the yeast U1 snRNP (28) can possibly be resolved by the observation that the Prp42HAp-U1 snRNP interaction is quite salt sensitive. If the native protein behaves like Prp42HAp, then the 300 mM salt washes used during affinity purification would strip the Prp42p from the U1 snRNP. This apparently weak interaction might reflect a mainly protein-based contact between Prp42p and the U1 snRNP. The snRNP association of yeast U1-C with the U1 snRNP is likely protein mediated and is nearly as salt sensitive as Prp42p (47). In contrast, the U1-snRNA-binding proteins Snp1p and Mud1p remain bound to the snRNP at NaCl concentrations of at least 200 mM (Mud1p [25]) and 500 mM (Snp1p [21]). Whatever the mode of interaction, all available data are consistent with an essential, although possibly salt-sensitive, interaction of Prp42p with the U1 snRNP.
It seems likely that Prp39p and Prp42p are present in the same particle and do not substitute for one another in alternative forms of the U1 snRNP with distinct properties (e.g., distinct substrate preferences). The transcripts of every interrupted gene assayed (i.e., ACT, CYH2, RP51A, and SNR17) required both Prp39p and Prp42p to be spliced in vivo. In addition, the gel shifts associated with anti-HA antibody addition and with Prp42HAp depletion included all of the fully assembled U1 snRNPs independent of whether Prp39HAp or Prp42HAp was being manipulated. If Prp39HAp and Prp42HAp were present in different U1 snRNP populations, these experiments should have revealed a pre-mRNA or snRNP subset insensitive to the manipulation of one or the other factor. Thus, although we have not directly assayed for Prp39p in Prp42p-bearing complexes, no precedent for functionally distinct U1 snRNP subpopulations exists in yeast, and the available data all support Prp39p-Prp42p colocalization in the U1 snRNP.
A consensus element built from the TPR sequences observed in Prp39p and Prp42p is most closely related to that observed in the Drosophila crn protein (Fig. 2). The level of sequence match to the TPR consensus element for the individual Prp39p and Prp42p repeats is low but not unlike that observed for many other TPR elements (10, 11). In all cases, the Prp39p and Prp42p TPR elements are predicted to present amphipathic alpha-helical surfaces characteristic of the TPR. TPR proteins can be grouped into distinct subfamilies based on their distinctive primary structure features (10, 11, 39). For instance, domain A of the Drosophila crn repeats generally has charged residues at positions 6 and 9, aromatic residues at positions 7 and 10, and an aspartic acid residue at position 11. Other than a reduced prevalence of the position 11 aspartic acid, each of these amino acids is well conserved within the Prp39p and Prp42p repeats. Other TPR proteins, including the one other known TPR protein of the spliceosome, Prp6p (22), show different distributions of amino acids in these positions (10, 11, 39). The remaining positions from amino acid 1 through the end of domain A of the Prp39p-Prp42p repeats are mostly perfect matches or conservative substitutions for the crn consensus sequence. The domain B structure is less conserved in primary sequence, but individual repeats generally match the crn or an elaborated TPR consensus (52) at multiple positions and show the overall alpha-helical character of the TPR. In 7 of 11 cases, the predicted domain B alpha helix terminates with a characteristic proline residue (10, 11, 39) located with a more relaxed positioning between TPR residues 30 and 34.
A number of polypeptide targets for TPR interaction have been identified. For instance, the TPR region of the yeast Cyc8p protein binds to the homeodomain of the yeast α2 protein (43, 48), while the TPRs of yeast Cdc23p bind to a helix-loop-helix region of Sin1p (38). In addition, a mutation in the TPR region of the Cdc27p protein reduces its ability to associate with the Cdc23p TPR protein, suggesting a possible TPR-TPR interaction (20). While these target polypeptides differ considerably in primary sequence, each presents a helical surface for interaction with the corresponding TPR helices. The unique sequence characteristics of the Prp39p and Prp42p repeats (as well as those of other TPR proteins) likely reflect distinctions in the complementary surfaces of their interacting ligands. Genetic screens and direct protein assays are currently under way to identify the natural ligands of Prp39p and Prp42p interaction.
Database comparisons revealed multiple TPRs of the Prp39p-Prp42p-crn domain A sort in only one other protein set, the yeast Rna14p, Drosophila Su(f), and human CstF77 proteins, in which six to eight repeats were present (this study and reference 50a). These proteins are part of the RNA cleavage stimulation factor required for pre-mRNA 3′ end processing (see reference 46 and references within). Thus, with the possible exception of crn, all of the known proteins with this variant TPR are involved in RNA processing. Mutations in the Drosophila crn gene show a pleiotropic embryonic lethal phenotype with impaired neurological (52) and muscle (7) development. Based in part on the reduced DNA synthesis observed in mutant embryos, it was suggested that crn may be involved in the cell cycle. We have cloned the likely yeast homolog of crn and are currently investigating its activity in the yeast cell cycle and RNA processing (6a). A role for yeast crn in splicing or polyadenylation would support the view that the crn-like TPR is reserved for the assembly or intracellular trafficking of complexes involved in RNA maturation.
Prp39p and Prp42p are essential in yeast, while mammalian homologs have not been found. Either these proteins provide a truly yeast-specific function or they are present in mammals but lost in the U1 snRNP isolation procedures used to date. In the first case, the Prp39p and Prp42p proteins might substitute, for instance, for one or more of the mammalian SR proteins absent in yeast but required for early spliceosome assembly events in mammals (e.g., ASF/SF2 and SC35 [reviewed in references 9 and 17]). However, given the general high level of conservation observed between the yeast and mammalian basal spliceosomal components (17, 27), perhaps a more likely view is that the mammalian equivalents of Prp39p and Prp42p exist but are tenuously associated with the U1 snRNP. The initial failure to identify the phylogenetically conserved, yet weakly bound SF3a and SF3b proteins with the U2 snRNP offers precedent for this (see references in references 17 and 27). The identification of mammalian Prp39p and Prp42p equivalents would reveal the yeast U1 snRNP as more conserved than is suggested by its exaggerated snRNA length.
ACKNOWLEDGMENTS
We thank Charles Query, Martha Peterson, John Woolford, and our laboratory colleagues for helpful comments on the manuscript. Carol Williams and Kevin O’Hare are thanked for pointing out the TPRs within Su(f). Seyung Chung is gratefully acknowledged for assistance with the Prp39p-Prp42p TPR alignments, and Elizabeth Otte is acknowledged for help with the quantitative analysis of the snRNA levels.
This work was supported by grant GM42476 from the National Institutes of Health to B.C.R.
REFERENCES
- 1.Abovich N, Liao X C, Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- 2.Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Arning S, Gruter P, Bilbe G, Krämer A. Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA. 1996;2:794–810. [PMC free article] [PubMed] [Google Scholar]
- 5.Berglund J A, Chua K, Abovich N, Reed R, Rosbash M. The specificity factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- 6.Blanton S, Srinivasan A, Rymond B C. PRP38 encodes a yeast protein required for pre-mRNA splicing and maintenance of stable U6 small nuclear RNA levels. Mol Cell Biol. 1992;12:3939–3947. doi: 10.1128/mcb.12.9.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6a.Chung, S., and B. C. Rymond. Unpublished observations.
- 7.Drysdale R, Rushton E, Bate M. Genes for embryonic muscle development in Drosophila melanogaster. Roux’s Arch Dev Biol. 1993;202:276–295. doi: 10.1007/BF00363217. [DOI] [PubMed] [Google Scholar]
- 8.Fromont-Racine M, Rain J C, Legrain P. Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nat Genet. 1997;16:277–282. doi: 10.1038/ng0797-277. [DOI] [PubMed] [Google Scholar]
- 9.Fu X-D. The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- 10.Gindhart J G, Jr, Goldstein L S. Tetratrico peptide repeats are present in the kinesin light chain. Trends Biochem Sci. 1996;21:52–53. [PubMed] [Google Scholar]
- 11.Goebl M, Yanagida M. The TPR snap helix: a novel protein repeat motif from mitosis to transcription. Trends Biochem Sci. 1991;16:173–177. doi: 10.1016/0968-0004(91)90070-c. [DOI] [PubMed] [Google Scholar]
- 12.Guthrie C. Messenger RNA splicing in yeast: clues to why the spliceosome is a ribonucleoprotein. Science. 1991;253:157–163. doi: 10.1126/science.1853200. [DOI] [PubMed] [Google Scholar]
- 13.Habets W J, Sillekens P T, Hoet M H, Schalken J A, Roebroek A J M, Leuissen J A M, van de Ven W J M, Venrooij W J. Analysis of a cDNA clone expressing a human autoimmune antigen. Full length sequence of the U2 small nuclear RNA-associated B" antigen. Proc Natl Acad Sci USA. 1987;84:2421–2425. doi: 10.1073/pnas.84.8.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermann H, Fabrizio P, Raker V A, Fouaki K, Hornig H, Brahms H, Lührmann R. SnRNP Sm proteins share two evolutionarily conserved sequence motifs which are involved in Sm protein-protein interactions. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnston M, Davis R W. Sequences that regulate the divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:1440–1448. doi: 10.1128/mcb.4.8.1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kao H Y, Siliciano P G. Identification of Prp40, a novel essential yeast splicing factor associated with the U1 small nuclear ribonucleoprotein particle. Mol Cell Biol. 1996;16:960–967. doi: 10.1128/mcb.16.3.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krämer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 18.Kretzner L, Krol A, Rosbash M. Saccharomyces cerevisiae U1 small nuclear RNA secondary structure contains both universal and yeast-specific domains. Proc Natl Acad Sci USA. 1990;87:851–855. doi: 10.1073/pnas.87.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kretzner L, Rymond B C, Rosbash M. S. cerevisiae U1 RNA is large and has limited primary sequence homology to metazoan U1 snRNA. Cell. 1987;50:593–602. doi: 10.1016/0092-8674(87)90032-8. [DOI] [PubMed] [Google Scholar]
- 20.Lamb J R, Tugendreich S, Hieter P. Tetratrico peptide repeat interactions: to TPR or not to TPR? Trends Biochem Sci. 1995;20:257–259. doi: 10.1016/s0968-0004(00)89037-4. [DOI] [PubMed] [Google Scholar]
- 21.Lauber J, Fabrizio P, Teigelkamp S, Lane W S, Hartmann E, Lührmann R. The HeLa 200 kDa U5 snRNP-specific protein and its homolog in Saccharomyces cerevisiae are members of the DEXH-box protein family of putative RNA helicases. EMBO J. 1996;15:4001–4015. [PMC free article] [PubMed] [Google Scholar]
- 22.Legrain P, Choulika A. The molecular characterization of PRP6 and PRP9 yeast genes reveals a new cysteine/histidine motif common to several splicing factors. EMBO J. 1990;9:2775–2781. doi: 10.1002/j.1460-2075.1990.tb07465.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Legrain P, Seraphin B, Rosbash M. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol Cell Biol. 1988;8:3755–3760. doi: 10.1128/mcb.8.9.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao X, Kretzner L, Seraphin B, Rosbash M. Universally conserved and yeast-specific U1 snRNA sequences are important but not essential for U1 snRNP function. Genes Dev. 1990;4:1766–1774. doi: 10.1101/gad.4.10.1766. [DOI] [PubMed] [Google Scholar]
- 25.Liao X C, Tang J, Rosbash M. An enhancer screen identifies a gene that encodes the yeast U1 snRNP A protein: implications for snRNP protein function in pre-mRNA splicing. Genes Dev. 1993;7:419–428. doi: 10.1101/gad.7.3.419. [DOI] [PubMed] [Google Scholar]
- 26.Lockhart S R, Rymond B C. Commitment of yeast pre-mRNA to the splicing pathway requires a novel small nuclear ribonucleoprotein polypeptide, Prp39p. Mol Cell Biol. 1994;14:3623–3633. doi: 10.1128/mcb.14.6.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26a.Lockhart, S. R., and B. C. Rymond. Unpublished observations.
- 27.Moore M J, Query C C, Sharp P A. Splicing of precursors to mRNA by the spliceosome. In: Gesteland R F, Atkins J F, editors. The RNA world. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- 28.Neubauer G, Gottschalk A, Fabrizio P, Seraphin B, Lührmann R, Mann M. Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc Natl Acad Sci USA. 1997;94:385–390. doi: 10.1073/pnas.94.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pikielny C W, Rosbash M. Specific small nuclear RNAs are associated with yeast spliceosomes. Cell. 1986;45:869–877. doi: 10.1016/0092-8674(86)90561-1. [DOI] [PubMed] [Google Scholar]
- 30.Pikielny C W, Rymond B C, Rosbash M. Electrophoresis of ribonucleoproteins reveals an ordered assembly pathway of yeast splicing complexes. Nature. 1986;342:341–345. doi: 10.1038/324341a0. [DOI] [PubMed] [Google Scholar]
- 30a.Rosbash, M. Personal communication.
- 31.Rosbash M, Seraphin B. Who’s on first? The U1 snRNP-5′ splice site interaction and splicing. Trends Biochem Sci. 1991;16:187–190. doi: 10.1016/0968-0004(91)90073-5. [DOI] [PubMed] [Google Scholar]
- 32.Roy J, Zheng B, Rymond B C, Woolford J L., Jr Structurally related but functionally distinct yeast Sm D core small nuclear ribonucleoprotein particle proteins. Mol Cell Biol. 1995;15:445–455. doi: 10.1128/mcb.15.1.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rymond B C. Convergent transcripts of the yeast PRP38-SMD1 locus encode two essential splicing factors, including the D1 core polypeptide of small nuclear ribonucleoprotein particles. Proc Natl Acad Sci USA. 1993;90:848–852. doi: 10.1073/pnas.90.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rymond B C, Rosbash M. Cleavage of 5′ splice site and lariat formation are independent of 3′ splice site in yeast mRNA splicing. Nature. 1985;317:735–737. doi: 10.1038/317735a0. [DOI] [PubMed] [Google Scholar]
- 35.Seraphin B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J. 1995;14:2089–2098. doi: 10.1002/j.1460-2075.1995.tb07200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seraphin B, Rosbash M. The yeast branchpoint sequence is not required for the formation of a stable U1 snRNA-pre-mRNA complex and is recognized in the absence of U2 snRNA. EMBO J. 1991;10:1209–1216. doi: 10.1002/j.1460-2075.1991.tb08062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sherman F, Fink G P, Hicks J B. Laboratory course manual for methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- 38.Shpungin S, Liberzon A, Bangio H, Yona E, Katcoff D J. Association of yeast SIN1 with the tetratrico peptide repeats of CDC23. Proc Natl Acad Sci USA. 1996;93:8274–8277. doi: 10.1073/pnas.93.16.8274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sikorski R S, Michaud W A, Wootton J C, Buguski M S, Connelly C, Hieter P. TPR proteins as essential components of the yeast cell cycle. Cold Spring Harbor Symp Quant Biol. 1991;56:663–673. doi: 10.1101/sqb.1991.056.01.075. [DOI] [PubMed] [Google Scholar]
- 40.Siliciano P G, Jones M H, Guthrie C. Saccharomyces cerevisiae has a U1-like small nuclear RNA with unexpected properties. Science. 1987;237:1484–1487. doi: 10.1126/science.3306922. [DOI] [PubMed] [Google Scholar]
- 41.Siliciano P G, Kivens W J, Guthrie C. More than half of yeast U1 snRNA is dispensable for growth. Nucleic Acids Res. 1991;19:6367–6372. doi: 10.1093/nar/19.23.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sillekens P T, Habets W, Beijer R P, van Venrooij W J. cDNA cloning of the human U1 snRNA-associated A protein: extensive homology between U1 and U2 snRNP-specific proteins. EMBO J. 1987;6:3841–3848. doi: 10.1002/j.1460-2075.1987.tb02721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith R L, Redd M J, Johnson A D. The tetratricopeptide repeats of Ssn6 interact with the homeo domain of alpha 2. Genes Dev. 1995;9:2903–2910. doi: 10.1101/gad.9.23.2903. [DOI] [PubMed] [Google Scholar]
- 44.Smith V, Barrell B G. Cloning of a yeast U1 snRNP 70k protein homologue: functional conservation of an RNA binding domain between humans and yeast. EMBO J. 1991;10:2627–2634. doi: 10.1002/j.1460-2075.1991.tb07805.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stultz C M, White J V, Smith T F. Structural analysis based on state-space modeling. Protein Sci. 1993;2:305–314. doi: 10.1002/pro.5560020302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takagaki Y, Manley J L. A polyadenylation factor subunit is the human homologue of the Drosophila suppressor of forked protein. Nature. 1994;372:471–481. doi: 10.1038/372471a0. [DOI] [PubMed] [Google Scholar]
- 47.Tang J, Abovich N, Fleming M L, Seraphin B, Rosbash M. Identification and characterization of yeast homolog of U1 snRNP-specific protein C. EMBO J. 1997;16:4082–4091. doi: 10.1093/emboj/16.13.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tzamarias D, Struhl K. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev. 1995;9:821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- 49.Umen J G, Guthrie C. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes Dev. 1995;9:855–868. doi: 10.1101/gad.9.7.855. [DOI] [PubMed] [Google Scholar]
- 50.White J V, Stultz S M, Smith T F. Protein classification by stochastic modeling and optimal filtering of amino acid sequences. Math Biosci. 1994;119:35–75. doi: 10.1016/0025-5564(94)90004-3. [DOI] [PubMed] [Google Scholar]
- 50a.Williams, C., and K. O’Hare. Personal communication.
- 51.Wolfe K H, Shields D C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 52.Zhang K, Smouse D, Perrimon N. The crooked neck gene of Drosophila contains a motif found in a family of yeast cell cycle genes. Genes Dev. 1991;5:1080–1091. doi: 10.1101/gad.5.6.1080. [DOI] [PubMed] [Google Scholar]