Abstract
Transcription by RNA polymerase II is highly regulated at the level of initiation and elongation. Well-documented transcription activation mechanisms, such as the recruitment of TFIID and TFIIB, control the early phases of preinitiation complex formation. The heat shock genes provide an example for transcriptional regulation at a later step: in nuclei TFIID can be detected at the TATA box prior to heat induction. Using cell-free systems for chromatin reconstitution and transcription, we have analyzed the mechanisms by which heat shock factor (HSF) increases transcription of heat shock genes in chromatin. HSF affected transcription of naked DNA templates in multiple ways: (i) by speeding up the rate of preinitiation complex formation, (ii) by increasing the number of productive templates, and (iii) by increasing the reinitiation rate. Under the more physiological conditions of potentiated chromatin templates, HSF affected only the reinitiation rate. Activator-dependent reinitiation of transcription, obviating the slow assembly of the TFIID-TFIIA complex on a promoter, may be especially crucial for genes requiring a fast response to inducers.
Transcription by RNA polymerase II requires the remodeling of chromatin to allow the binding of transcription factors to their recognition elements, the coordinated assembly of a preinitiation complex (PIC), the initiation of transcription, promoter clearance, and efficient elongation. Each of these processes can be subdivided mechanistically into steps with distinct factor requirements (for reviews, see references 6, 17, 28, and 41). The first event during PIC formation is the binding of TFIID to the TATA box. TFIIA associates with the DNA-bound TFIID, adding stability to the complex. Interaction of TFIIB with the TATA box-TFIID complex provides the docking site for the subsequent association of further general transcription factors (GTFs) and RNA polymerase II, perhaps in a preformed holopolymerase complex (2, 27, 30). In principle, PIC formation can be speeded up, and hence transcription can be activated, by active recruitment of any component of the PIC. So far, the recruitment of the GTFs TFIID and TFIIB (11, 29, 40) during PIC formation and the recruitment of the entire holoenzyme machinery (27, 30) are well-documented activation mechanisms. The PIC complex is disassembled again upon transcription initiation when the polymerase clears the promoter. While TFIIF travels with the polymerase, all other GTFs dissociate, with the exception of TFIID, which remains bound at the TATA box (59). The presence of TFIID and/or the topological changes at the promoter DNA as a result of the first cycle of transcription create a different setting for any subsequent reinitiation event.
Therefore, GTFs alone or in a holoenzyme complex with polymerase need to be recruited anew for every cycle of transcription (40, 59). The fact that many activators have been shown to interact with GTFs other than TFIID in vitro, as well as the recent demonstration of the requirement of activators for ongoing transcription in vivo (22), raises the question of whether transcription initiation is also regulated after the formation of the first PIC. Although it has previously been noted that transcriptional activation may lead to the synthesis of multiple transcripts per template (10, 12, 21, 46, 57), its contribution to transcriptional regulation at the level of reinitiation remains largely elusive.
Heat shock genes serve as models for an expanding class of genes whose activity is regulated at the postinitiation level (33, 55). RNA polymerase II initiates transcription of heat shock genes in the absence of heat shock factor (HSF), the activator of heat shock genes, but pauses 20 to 40 nucleotides downstream of the start site (39, 42, 43). Genomic footprinting suggests the constitutive occupancy of the TATA box, presumably by TFIID (reviewed in reference 33). Therefore, in contrast to that by other activators studied to date, activation by HSF is unlikely to be limited to the induction of PIC formation but may affect later steps towards productive transcription, including the release of the “paused polymerase” (9).
In order to understand the mechanism of transcriptional activation by HSF, we have reconstituted transcriptional regulation of the Drosophila melanogaster hsp26 promoter in a cell-free system derived from Drosophila embryos. Using a transcription extract that supports multiple reinitiations of transcription (25) and a chromatin assembly extract that reconstitutes important aspects of the chromatin architecture of the hsp26 promoter (54), we studied the influence of HSF on transcription from both nucleosome-free and chromatin templates. On naked templates HSF activates transcription rates in multiple ways, including a modest stimulation of PIC formation kinetics, a considerable effect on the number of productive templates, and a profound effect on the rate of reinitiation from a potentiated promoter that is constitutively loaded with TFIID and TFIIA. In chromatin, activation of reinitiation prevailed. We will also discuss the importance of regulation at the level of reinitiation.
MATERIALS AND METHODS
DNA templates.
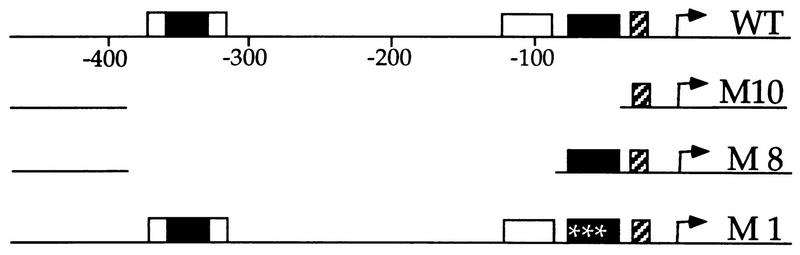
phsp26M (a plasmid containing the Drosophila hsp26 minigene with wild-type flanking sequences) and derivatives M10 (the minimal promoter, with promoter sequences upstream of the TATA box deleted), M8 (containing the proximal heat shock elements [HSEs] upstream of the TATA box), and M1 (with the proximal HSEs mutated) have been described previously (45). The minigenes give rise to a shortened hsp26 transcript which could be distinguished readily from the native hsp26 RNA endogenous to the extracts from heat-shocked embryos. In order to facilitate immobilization, we introduced a NotI restriction site and an AflII restriction site into their polylinker between XhoI and ApaI. Digestion with NotI and SpeI generated one short (13-bp) and two long fragments (vector and insert). Biotinylation at the SpeI site by incorporation of biotin-21-dUTP selectively labeled the hsp26 insert so that the vector fragment was not immobilized. The short biotinylated fragment and free deoxynucleoside triphosphates were removed by gel filtration, and DNA was quantitated by the measurement of optical density at 260 nm. Immobilization to Dynabeads M280 (Dynal, Oslo, Norway) was as described previously (44). Agarose gel analysis of the DNA fragments remaining in the supernatant after the coupling reaction and comparison to standards and to the nonbiotinylated, free fragment allowed precise quantitation of immobilized template DNA.
The plasmid pHSE was used to compete for binding of HSF as before (3). pHSE is a pUC derivative containing 14 tandem repeats of consensus HSEs.
In vitro transcription.
Preparation of transcription extract from heat-shocked Drosophila embryos and of the standard reaction mixture were as described previously (45). Small amounts of hsp26 promoters (30 ng of immobilized DNA [9.5 fmol] or 30 ng of plasmid [6 fmol]) were transcribed with an excess of extract (12.5 μl) to ensure that no extract component became limiting. RNA accumulated linearly during the standard 30-min transcription reaction. Nonspecific DNA binding inhibitors were titrated with 1 to 3 μg of pUC for 5 min at room temperature prior to addition of the template (15). Under these conditions efficient transcription relies on the binding of endogenous HSF to the proximal HSEs upstream of the TATA box (45). For HSF titration, pUC competitor DNA was replaced by pHSE containing tandem arrays of consensus HSEs (3). To limit transcription to one cycle, PICs were assembled in a transcription reaction lacking ribonucleoside-5′-triphosphates (rNTPs) for 40 min. PIC-containing templates were purified magnetically, washed with 50 μl of HEMG 50 (45), and transcribed for 30 min in 25 μl of a transcription mix lacking extract.
Quantitation of in vitro-transcribed RNA.
In vitro-transcribed RNA was quantified by comparison to known amounts of a reference RNA (3). Reference RNA of defined length was obtained by transcribing a promoterless, truncated hsp26 gene with T7 RNA polymerase (35, 44). After repeated phenol-chloroform extractions, ethanol precipitation, and washings of the pellet, the RNA was resuspended in water and its concentration was determined by measurement of optical density at 260 nm. The reference RNA was diluted in diethyl pyrocarbonate-H2O containing 100 μg of Saccharomyces cerevisiae total RNA/ml. Typically, 1 to 5 fmol of reference RNA was added to each transcription reaction along with the stop mix. Reference RNA is longer at the 5′ end than hsp26 RNA prepared by in vitro transcription and can easily be distinguished from the newly synthesized transcripts when the RNA mixture is analyzed by the extension of a primer annealing at positions +91 and +120 relative to the hsp26 start site. Several independent preparations of reference RNA were used to evaluate the transcription efficiency.
Reconstitution and analysis of chromatin.
Preparation of chromatin assembly extract from Drosophila embryos (0 to 90 min after eggs were laid) and chromatin assembly reactions were as described previously (4, 5). Chromatin assembly was monitored by micrococcal nuclease analysis (44). PIC formation did not compromise subsequent chromatin assembly, as judged by the visualization of oligonucleosomal arrays after digestion with micrococcal nuclease and Southern blotting (data not shown). The immobilized chromatin was transcribed after being washed with 50 μl of a transcription mix devoid of extract and rNTPs. If chromatin templates were assayed for transcription without prior PIC formation, they were incubated for 30 min in the transcription reaction mix to allow formation of RNA polymerase II preinitiation complexes before the addition of nucleotides (see Fig. 6, lanes 1 and 4). For the analysis of chromatin-associated proteins, reactions were scaled up 10-fold. Chromatin beads equivalent to 300 ng of immobilized DNA were washed twice with 100 μl of transcription buffer and suspended in 10 μl of sodium dodecyl sulfate-loading buffer. After brief denaturation, the supernatants containing the eluted proteins were analyzed by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis and Western blotting with specific polyclonal antisera and the Amersham ECL kit.
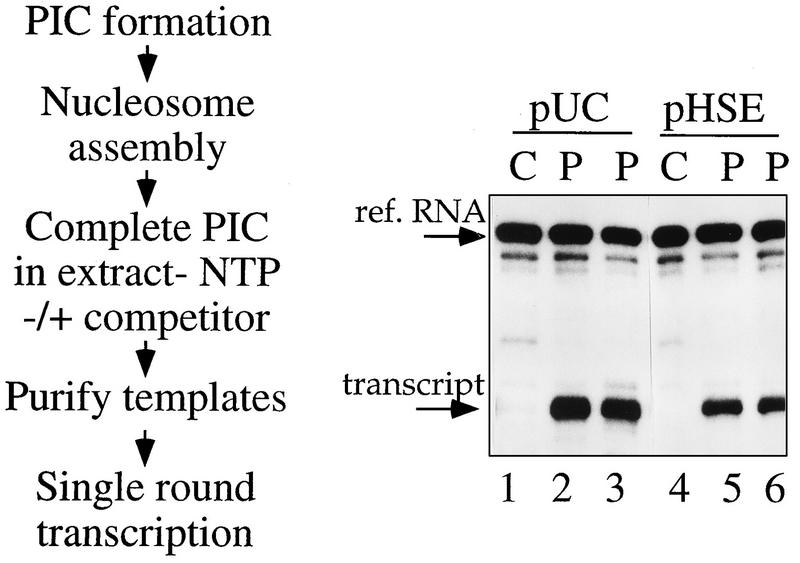
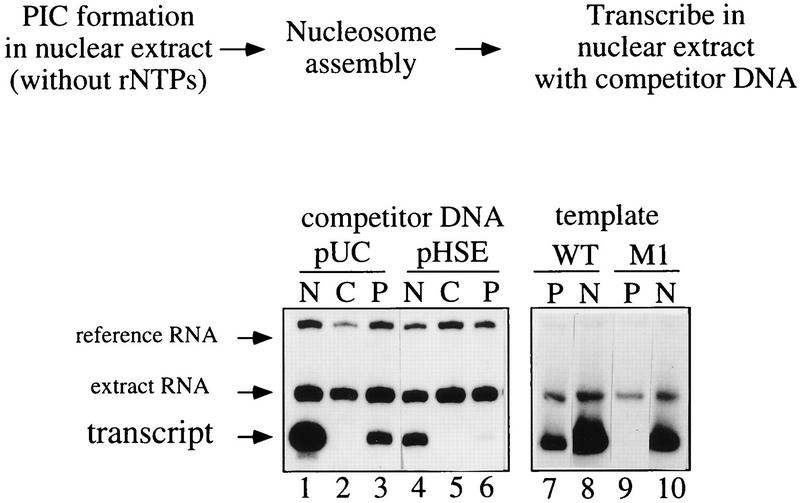
FIG. 6.
HSF does not affect the number of active chromatin templates. For single-round transcription, committed chromatin templates were incubated for 5 (lanes 2 and 5) or 15 (lanes 3 and 6) min in a transcription mix devoid of rNTPs to allow completion of PIC formation. Templates were repurified and washed, and one round of transcription was performed in rNTP-containing buffer (Fig. 2a). PIC completion after chromatin assembly occurred in the presence of 3 μg of competitor DNA, as indicated (pHSE, HSF titration; pUC, control). In the absence of prior PIC formation, chromatin assembly strongly inhibited transcription of the hsp26 promoter, whether or not HSF was present in the transcription reaction (lanes 1 and 4). Reactions were spiked with 3 fmol of reference RNA (ref. RNA). The experimental strategy is shown on the left.
RESULTS
Efficient reinitiation of heat shock gene transcription in a cell-free system.
For our analysis we optimized a cell-free transcription system derived from nuclei of heat-shocked Drosophila embryos (45) for maximal usage of the hsp26 promoter (Fig. 1 shows a summary of the promoter structure and the mutant templates used in this study). To avoid inhibition of transcription by nonspecific DNA binding proteins, such as histone H1 (15, 26), the extract was incubated with an excess of nonspecific competitor DNA prior to the addition of a small amount of template. Under these conditions antirepression by GAGA factor is not observed and transcription levels are only influenced by HSF (45). Titration of the nuclear extract/template ratio ensured that all extract components were in excess and that RNA accumulated linearly during the standard 30-min reaction (data not shown).
FIG. 1.
hsp26-derived templates used in this study. WT, wild-type promoter. The hatched boxes represent the TATA box, black boxes represent proximal and distal elements, and white boxes represent GAGA factor binding sites. A triple-point mutation that inactivates the proximal HSEs is indicated with asterisks.
To facilitate the transcriptional analysis, we used a solid-phase approach that allows the isolation of stable template-bound transcription factor complexes from the nuclear extract (13, 44, 60). Linear hsp26 templates attached to paramagnetic beads were incubated for increasing times in a transcription reaction mix without rNTPs to allow PIC formation and then purified magnetically. Upon addition of rNTPs in a buffer, transcription occurred efficiently (Fig. 2a, lane 4). Quantitation of this RNA with known amounts of a reference RNA (see Materials and Methods) established that roughly one RNA per template was synthesized. To determine whether these transcripts were due to a single transcription from a large proportion of the templates or to multiple transcriptions from a minor fraction, we employed the detergent Sarkosyl. In the presence of 0.25% Sarkosyl PICs cannot assemble, but DNA-bound PICs remain stable and active to synthesize a single transcript (21, 25). Addition of 0.25% Sarkosyl during template incubation in the transcription extract efficiently prevented PIC formation (Fig. 2a, lanes 2 and 3). By contrast, the signal did not diminish upon addition of up to 0.25% (vol/vol) Sarkosyl to the immobilized PICs (Fig. 2a, lanes 4 to 6), indicating that a major fraction of the template was active to be transcribed once. Evidently, at least the minimal set of transcription factors required for the transcription of linear templates (TFIID, TFIIB, RNA polymerase II, TFIIE, TFIIH, and TFIIF) were recruited to the template as a PIC (21) but could not be reused after the first round of transcription, presumably because of loss of GTFs and/or polymerase from the template. Indeed, Western blot analysis of factors bound to the template revealed that the PICs assembled on a promoter contained all the factors that were probed for (Fig. 2b, lane 1): TFIID (TATA-binding protein [TBP] and at least TAF42 and TAF150), TFIIA, TFIIB, TFIIF (RAP30), polymerase (revealed by an antiserum against the polymerase carboxy-terminal domain [CTD]), and HSF. The promoter specificity of this complex was illustrated by comparison to a parallel reaction in which the template DNA was substituted for by promoterless DNA (Fig. 2b, lane 3). While some nonspecific binding of TBP and polymerase was observed, no other factor associated with promoterless DNA. The selectivity of TAF42 and TAF150 for promoter-containing DNA (Fig. 2b, lanes 1 and 3) documents the specific association of TFIID with the hsp26 promoter. PIC formation occurred with a half-life of less than 5 min and reached a plateau at around 15 min (reference 25 and data not shown).
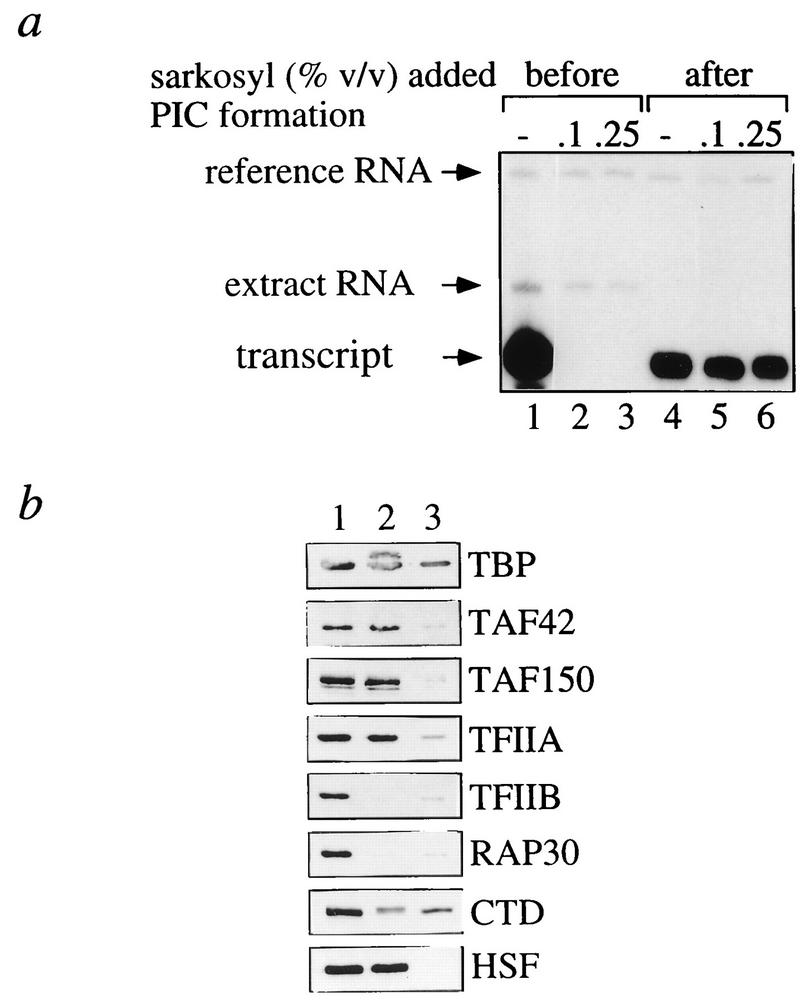
FIG. 2.
Reinitiation of transcription at the committed hsp26 promoter. (a) PICs were assembled by incubating immobilized templates in transcription reaction mixtures lacking rNTPs for 15 min. PICs were purified magnetically, washed mildly, and assayed for transcription in rNTP-containing buffer (lanes 4 to 6) or left in the extract (lanes 1 to 3). Transcriptions were done for 30 min in the presence of the indicated amounts of Sarkosyl added directly to the reaction (lanes 1 to 3) or after the purification of PICs (lanes 4 to 6). The signal in lane 4 corresponds to a single round of transcription, and that in lane 1 corresponds to multiple rounds. Samples were spiked with 2 fmol of reference RNA. The hsp26 RNA endogenous to the transcription extracts derived from heat-shocked embryos is labeled “extract RNA.” (b) Template-associated transcription factors were visualized by Western blotting with specific polyclonal antisera. Lane 1, proteins present in isolated PICs assembled on promoter-containing DNA; lane 2, committed factors that remain associated with the template after one round of transcription; lane 3, background levels of nonspecific factor binding to promoterless DNA.
When the immobilized template was transcribed in the presence of extract, multiple transcription rounds occurred (Fig. 2a, compare lanes 1 and 4), consistent with the earlier findings of Kadonaga that on average six transcripts per template were synthesized in these extracts (25). These multiple transcripts could, in principle, originate from successive independent initiation events or from true reinitiation. Successive initiation events can be discriminated from bona fide reinitiation by the GTFs that remain bound after the first round of transcription: during reinitiation, TFIID remains associated with the TATA box (21, 51, 59), whereas a dissociation of TFIID is diagnostic for independent initiation events. To distinguish between these possibilities, PICs were allowed to form on immobilized templates and were isolated from the reaction mixture. Transcription was then initiated by the addition of rNTPs, templates were repurified, and template-associated factors were visualized by Western analysis (Fig. 2b, lane 2). TFIIB, TFIIF, and the polymerase were released concomitantly with the first round of transcription, in contrast to TFIID (judged by the presence of TAF42 and TAF150), TFIIA, and HSF, which remained stably bound to the promoter (Fig. 2b, lane 2). These results agree with those of a previous study (60) and indicate the existence of a “committed complex” after promoter clearance. They imply that the multiple transcription rounds observed represent true reinitiation events. This committed complex was not defined by TFIID alone but also contained TFIIA, a factor not accounted for previously (60).
HSF affects the rate of PIC formation and the number of productive templates.
Efficient transcription of the hsp26 promoter under these conditions depends on activated HSF in an extract from heat-shocked embryos. HSF can be neutralized efficiently by titration of excess HSE competitor DNA (3, 45). We tested for an effect of HSF on the kinetics of PIC assembly by monitoring the formation of single PICs (Fig. 2) on immobilized templates in the presence and absence of HSF. In the presence of HSF the time required to assemble a transcription-competent PIC on 50% of the templates (T50) was less than 5 min, in agreement with previous estimations (25). When HSF was titrated by specific competitor DNA, the T50 increased to about 10 min (data not shown). In order to assess the effect of HSF on the fraction of active templates, we eliminated the kinetic effects of PIC formation. PICs were allowed to form for 40 min on various hsp26-derived templates (Fig. 1) before transcription was initiated with rNTPs. The addition of Sarkosyl along with the rNTPs limits transcription to one round (Fig. 3, lanes S). Deletion of all sequences upstream of the TATA box (template M10) resulted in a drastic diminution of transcription, which was recovered again by fusing the proximal HSEs (template M8). As has been observed for other activators, HSF ensured maximal template usage during the assembly of the first PIC, perhaps by shifting an equilibrium between the formation of nonproductive and productive PICs in favor of the latter (24).
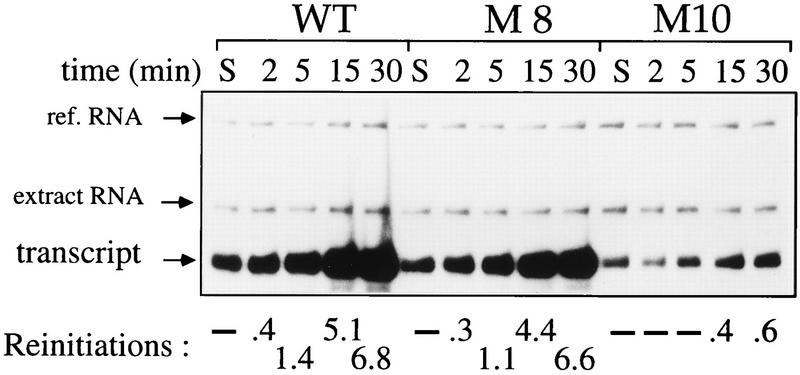
FIG. 3.
HSEs affect template usage and are necessary and sufficient to promote maximal transcription reinitiation. PICs were formed on different hsp26 minigene templates (wild type [WT], M8, and M10 [Fig. 1]) in a reaction mix lacking rNTPs for 40 min. Then rNTPs were added, and reactions were further incubated for the indicated times. Samples in lanes S received 0.05% (vol/vol) Sarkosyl (final concentration) along with rNTPs and were incubated for a further 30 min. The calculated reinitiation events are given below the lanes. WT, wild-type promoter; ref. RNA, reference RNA.
HSEs are necessary and sufficient to promote maximal reinitiation.
For the experiment shown in Fig. 3, the templates were not removed from the extract, which enabled the detection of transcript accumulation during a time course of transcription. In the absence of Sarkosyl, about eight times more transcripts were synthesized during 30 min from the wild-type promoter, reflecting approximately seven rounds of reinitiation (Fig. 3, WT). By contrast, the M10 templates that gave rise to one transcript (Fig. 3, lanes S) essentially failed to reinitiate during this time (∼0.5 reinitiations). Addition of the proximal HSEs to the TATA box (template M8) also restored the original reinitiation levels. Similar results were obtained if HSF was titrated by competing HSEs in trans. We conclude that HSF is not only able to affect the first PIC but is also necessary to induce subsequent reinitiation events.
Potentiation and activation of the hsp26 promoter reconstituted in chromatin.
The requirement for HSF for triggering reinitiation events is in agreement with its supposed role in vivo, where the first initiation occurs prior to heat induction. To study the effect of HSF under the more physiological conditions of a chromatin environment, we assembled the templates into chromatin with a cell-free system derived from Drosophila embryos (5), which recreates important aspects of native hsp26 promoter architecture (54). Nucleosome assembly strongly repressed heat shock gene transcription (Fig. 4, lanes 2 and 5) (44). The addition of recombinant HSF, GAGA factor, or TBP (or combinations thereof) before chromatin assembly was ineffective in keeping the template activatable by HSF (data not shown). Since potentiated chromatin in vivo contains a transcriptionally engaged RNA polymerase, we investigated whether the formation of a PIC prior to nucleosome assembly would keep the promoter active (Fig. 4). Preincubation of the immobilized template in the transcription extract under conditions that lead to efficient PIC formation did not inhibit subsequent chromatin assembly, as judged by the visualization of oligonucleosomal arrays after digestion with micrococcal nuclease and Southern blotting (data not shown).
FIG. 4.
HSF-dependent transcription of committed chromatin templates. The experimental strategy is outlined at the top of the figure. Lanes C, chromatin templates without prior PIC formation are tightly repressed; lanes P, PIC formation prior to chromatin assembly prevents inhibition by chromatin. PICs were formed on the wild-type (WT; lanes 1 to 8) or mutant (M1; lanes 9 and 10) template. Where indicated, transcription reactions contained 3 μg of specific (pHSE) or nonspecific (pUC) competitor DNA. Template beads were mildly washed and assembled into chromatin. Chromatin was retrieved, washed, and transcribed in a complete transcription reaction. Transcription efficiencies were compared to those of a standard transcription reaction of chromatin-free templates (N; lanes 1, 4, 8, and 10). Reactions were spiked with 3 fmol of reference RNA.
The assembly of PICs on the immobilized template prior to chromatin reconstitution effectively prevented chromatin repression such that reproducible transcriptional activity, corresponding to 20 to 30% of chromatin-free templates, was observed (Fig. 4, compare lanes 1 and 3). However, PIC formation did not suffice to overcome nucleosomal inhibition and transcription relied entirely on the action of HSF. If HSF was titrated by inclusion of HSE competitor DNA (pHSE) or if the template carried a triple-point mutation (template M1 [Fig. 1]) that impairs the binding of HSF to the proximal HSEs (45), transcription from the chromatin template was strongly diminished (Fig. 4, compare lanes 3 and 6 and lanes 7 and 9). As observed for other hsp genes (3, 9), HSF was crucial for transcription of hsp26 promoter in chromatin.
HSF induces transcription reinitiation of a chromatin template.
We next assessed which components of the transcription machinery maintained the promoter in an active configuration during chromatin assembly. The efficient formation of complete PICs on the hsp26 promoter was visualized as before by Western blotting (Fig. 5, lane 4). The PIC-containing template was then assembled into chromatin, repurified, and analyzed for associated transcription factors. From the complete PICs formed on the template prior to chromatin assembly, only TFIID and TFIIA remained on the template after chromatin reconstitution while TFIIB, TFIIF (RAP30 subunit), the RNA polymerase, and the vast majority of HSF were released from the template during nucleosome assembly (Fig. 5, lane 2). These factors might either be displaced during chromatin reconstitution or, more likely, released as a result of uncontrolled transcription fed by rNTPs endogenous to the chromatin assembly extract (unpublished observations). The important conclusion of the experiment is that the presence of this TFIID-TFIIA complex, which also characterizes the reinitiation-competent template, sufficed to potentiate the template during chromatin reconstitution.
FIG. 5.
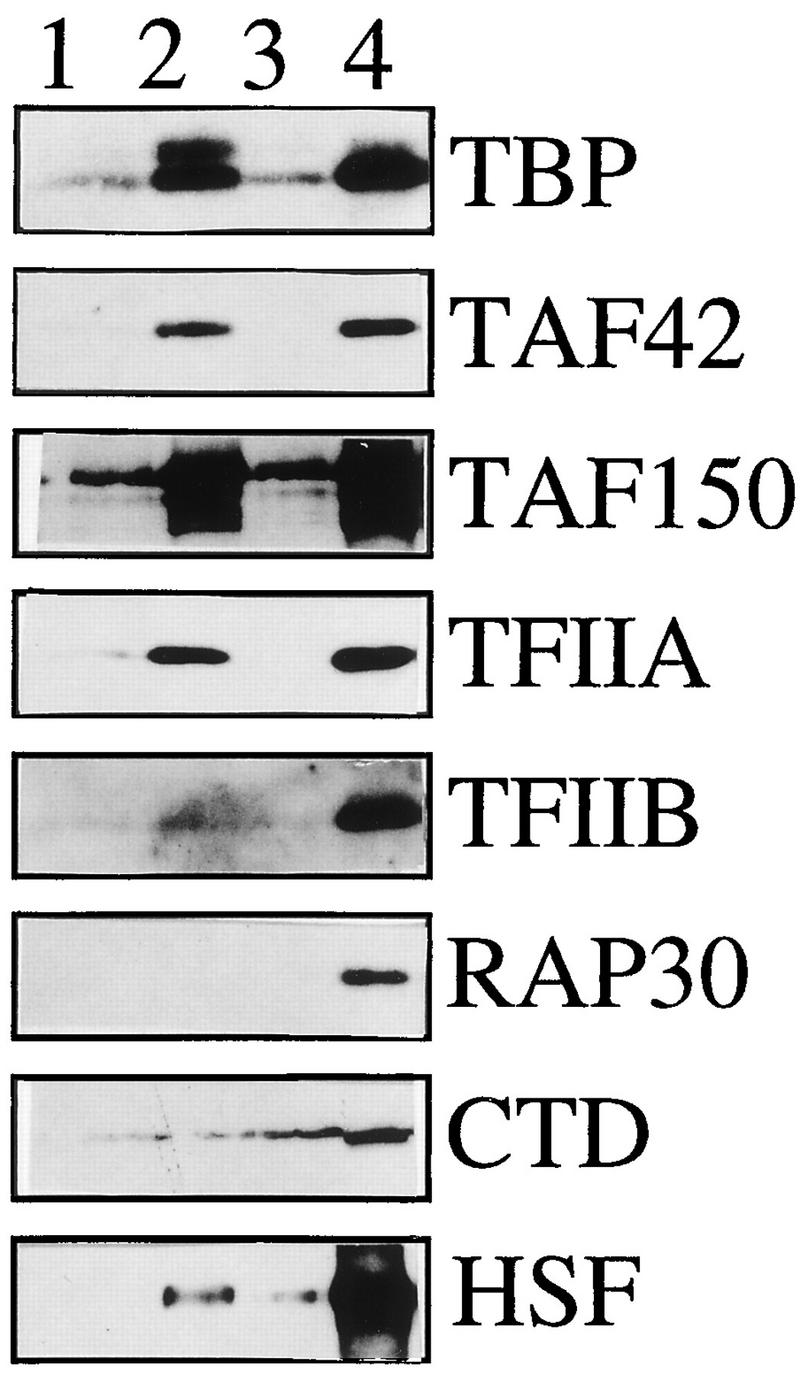
Characterization of the committed complex. Template-associated components of the transcription machinery were visualized by Western blotting. Lane 1, background of factors associated with nonspecific DNA; lane 2, PIC-containing templates were assembled into chromatin, reisolated, and analyzed; lane 3, templates without prior PIC formation were reconstituted into chromatin, revealing the background of transcription factors in the chromatin assembly reaction; lane 4, complete PICs prior to chromatin assembly.
The experiment shown in Fig. 4 demonstrated that HSF can activate transcription in chromatin under conditions that allow multiple rounds of transcription. The finding that a committed complex consisting of TFIID and TFIIA allows activation by HSF in chromatin suggests that HSF activates the reinitiation of transcription from a chromatin template. By a modification of the experimental protocol, we directly tested whether HSF would also affect a single round of transcription in chromatin. We formed and purified committed TFIID-TFIIA complexes in chromatin as before. During chromatin assembly, HSF was released from the template, perhaps as a result of its inactivation in the environment of an extract derived from nonshocked embryos, which resembles recovery after heat shock (Fig. 5, lanes 2 and 4). Therefore, the effect of HSF on transcription from the committed chromatin template could be dissected. The committed chromatin templates were then incubated in transcription extract for 30 min in the presence or absence of HSF (titrated as before by specific competitor DNA) without rNTPs to allow completion of the PICs. If HSF affected the fraction of active templates at this stage, we would expect to see a large difference in transcription efficiency in the presence or absence of HSF. The templates were repurified again, and transcription was triggered by the addition of rNTPs (Fig. 6). As shown in Fig. 1, the addition of rNTPs in the absence of extract supports only a single round of transcription. The presence or absence of HSF did not affect the transcription signal significantly in this experiment (Fig. 6, compare lanes 5 and 6 to lanes 2 and 3; note that the gel was exposed much longer than the one shown in Fig. 4 and that the signal in Fig. 6, lanes 5 and 6, corresponds to the faint signal in Fig. 4, lane 6). We conclude that the critical mode of activation of a chromatin template by HSF is the induction of efficient reinitiation.
DISCUSSION
Activation of transcription by HSF.
HSF affected transcription in the cell-free system in multiple ways. It stimulated the kinetics of the first PIC assembled, increased template usage, and was absolutely required for reinitiation from committed promoters. These different effects could be due to a single function of HSF that affects several steps leading to efficient transcription, or alternatively, HSF may be able to induce transcription in multiple independent ways. The fact that multiple, nonredundant activator domains have been identified in yeast HSF and human HSF1 (8, 19) supports the idea of multiple roles for HSF in transcriptional control.
In vivo, the first initiation of hsp26 transcription occurs in the absence of heat shock under conditions where HSF is unable to bind promoters with high specificity. Under those conditions productive transcription from the hsp26 promoter requires the efficient release of the pausing polymerase as well as the recruitment of the transcription machinery to the TFIID-TFIIA complex for reinitiation. Conceivably, HSF may contact component(s) of the transcription machinery other than TFIID-TFIIA (13, 40, 57) or a putative polymerase holo-enzyme complex (2, 30). Such a recruitment would speed up the formation of the first PIC and subsequent reinitiation events and might also increase the template usage. In crude transcription systems an equilibrium between the formation of productive and nonproductive preinitiation complexes exists, brought about by competition between negative and positive regulators. HSF might increase the fraction of active templates by shifting this equilibrium towards the formation of productive complexes, similar to the action of UBX (24). It is, however, also possible that HSF increases the affinity of the committed complex for TFIIB by altering the conformation of a ternary TFIIA-TFIID-TATA complex. Such an activator-dependent “isomerization” of a bound TFIID-TFIIA complex has recently been described (12).
Reinitiation by a newly recruited machinery is only possible if the previous polymerase has cleared the promoter. In heat shock genes promoter clearance is a limiting step which can be overcome by activated HSF (33). Promoter clearance involves the phosphorylation of the polymerase CTD by the TFIIH kinase (23, 34), a process that is integrated by TFIIE: TFIIE associates selectively with the hypophosphorylated form of the polymerase and stimulates the TFIIH kinase at a late step in PIC assembly (34, 37). In vivo the paused polymerase at the hsp26 promoter should be in the elongation phase (59), but it has an unphosphorylated CTD (36). Since HSF induction leads to the release of this polymerase, it will be interesting to investigate whether HSF rerecruits TFIIE and TFIIH (18, 23, 37), as has been suggested (34), or an as yet unidentified heat shock-specific CTD kinase (16).
A comparison of our results with the recent findings of Brown and colleagues (9), who used a model system to demonstrate that HSF is able to stimulate a pausing polymerase to elongate efficiently in chromatin, may be instructive. The pausing of the polymerase at heat shock promoters occurs naturally in the absence of chromatin (32) and is strongly enhanced by the presence of a nucleosome downstream of the transcription start site (9). In the experiments of Brown et al. (9) release of the polymerase pause in chromatin required, in addition to HSF, a protein fraction enriched in the chromatin modulator SWI/SNF, suggesting that nucleosomes contribute to the stability of the polymerase pause. It will be interesting to investigate whether nucleosome remodeling factors, such as SWI/SNF complex (38), NURF (50), and CHRAC (53), present in our transcription extracts are essential for efficient transcription in the context of chromatin. While it is clear that the release of the pause is required for efficient reinitiation, the stimulation of reinitiation appears not to be due simply to the release of the nucleosome-stimulated pause. While neither HSF nor the SWI/SNF fraction affected promoter clearance of naked DNA in the study of Brown et al. (9), HSF was required for reinitiation on chromatin-free templates in our experiments. While promoter-proximal pausing, which appears to be a rather general phenomenon (31, 55), presumably affects the reinitiation potential of a promoter, a direct mechanistic link between efficient reinitiation and the relief of the polymerase pause has not yet been established.
Since our primer extension assay detects transcripts that are at least 100 nucleotides long, an effect of HSF on elongation processivity (7) cannot be excluded. Szentirmay and Sawadogo have shown that if elongation factor SII was limiting, the reinitiation rate of a promoter was affected, presumably due to slow promoter clearance (47). Recombinant Dm-SII facilitates elongation in vitro (20, 47) but did not affect reinitiation in our system, either from nucleosome-free or from chromatin template (data not shown).
Transcriptional regulation at the level of reinitiation.
In potentiated chromatin, which reflects the in vivo situation best, HSF affected only the reinitiation. While the importance of the presence of transcription activators for ongoing transcription in vivo was recently suggested by an elegant study (22), our data provide a direct biochemical demonstration of a role for an activator for reinitiation on chromatin templates. We attempted to define the mechanism of activator-dependent reinitiation by utilizing purified GTFs from Drosophila (1) and recombinant HSF that is constitutively active (14). However, HSF did not activate transcription, suggesting that the purified system lacked a cofactor for either HSF-mediated activation or reinitiation (unpublished results). Interestingly, recombinant HSF enhanced the number of active templates but did not affect reinitiation on chromatin-free templates when added to an extract from unshocked embryos (data not shown). The deficiency with respect to the reinitiation function could be due to the lack of posttranslational modifications, known to regulate HSF activity in vivo (reviewed in reference 58), or to the absence of a heat shock-specific cofactor in extracts from unshocked embryos (e.g., a heat shock-specific CTD kinase [16]).
Previous reports on multiple rounds of initiation in vitro (10, 57) suggested that activators may promote reinitiation by stabilizing committed TFIID-TFIIA complexes. In our assays TFIID-TFIIA remained stable on the template during a 6-h incubation under chromatin assembly conditions whereas HSF dissociated, which argues against a stabilizing role for HSF.
Regulation of heat shock genes in chromatin.
The stable association of TFIIA and TFIID with the chromatin template sufficed to potentiate the promoter for subsequent activation by HSF. Since the prebinding of these factors rendered the promoter active during nucleosome reconstitution, we conclude that the failure of TFIID to interact with chromatin limits transcription initiation despite the known dynamic features of reconstituted chromatin (49, 52–54). The finding that HSF did not increase the absolute number of active templates but activated the reinitiation is in accord with the “preset” promoter structure observed in vivo (55). Most of the many steps towards productive transcription have already been taken under noninducing circumstances: binding sites for transcription factors are permanently kept accessible in chromatin, and GAGA elements are always occupied by GAGA factor, which is thought to act at an early stage of the assembly of the preset promoter by keeping the HSE clear of repressive nucleosomes (33, 48, 55). Occupation of the TATA box, presumably by TFIID, is also constitutive (48, 56). Transcription has already been initiated, but the polymerase pauses some 20 nucleotides downstream of the transcription start site (39, 42, 43). Clearly, the time-consuming processes leading to the first initiation have already been completed under nonshock conditions but productive transcription has been halted at the latest possible step, ready for the quickest possible induction by HSF in an emergency. Therefore, the mechanism by which HSF activates heat shock promoters is clearly distinct from the presently known activation strategies.
The regulation of transcription at the level of reinitiation is of great importance for a cell: transcription reinitiations occur much more rapidly than successive de novo initiation events, since the slow step of TFIID-TFIIA binding to the promoter is avoided (21, 23). Therefore, activation of reinitiation allows the most rapid transcriptional response in an emergency, such as heat shock or other stresses. Conversely, it accounts for the rapid down-regulation of transcription once the activator is released from its binding site.
ACKNOWLEDGMENTS
R.S. acknowledges the receipt of an EMBL predoctoral fellowship.
We thank the following researchers for providing antibodies against transcription factors: Y. Nakatani (CTD, TAF42, TBP, and RAP30), A. Greenleaf (CTD), R. Tjian (TFIIA and TAF150), and J. Kadonaga (TFIIB). We thank M. Biggin for providing purified GTFs, D. Price for recombinant Dm-SII, and I. W. Mattaj, H. Stunnenberg, and C. Wu for valuable comments on the manuscript.
Footnotes
R.S. dedicates this work to his father, Mattheos.
REFERENCES
- 1.Austin R J, Biggin M D. Purification of the Drosophila RNA polymerase II general transcription factors. Proc Natl Acad Sci USA. 1996;93:5788–5792. doi: 10.1073/pnas.93.12.5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barberis A, Pearlberg J, Simkovich N, Farrell S, Reinagel P, Bamdad C, Sigal G, Ptashne M. Contact with a component of the polymerase II holoenzyme suffices for gene activation. Cell. 1995;81:359–368. doi: 10.1016/0092-8674(95)90389-5. [DOI] [PubMed] [Google Scholar]
- 3.Becker P B, Rabindran S, Wu C. Heat shock-regulated transcription in vitro from a chromatin template. Proc Natl Acad Sci USA. 1991;88:4109–4113. doi: 10.1073/pnas.88.10.4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becker P B, Tsukiyama T, Wu C. Chromatin assembly extracts from Drosophila embryos. Methods Cell Biol. 1994;44:207–223. doi: 10.1016/s0091-679x(08)60915-2. [DOI] [PubMed] [Google Scholar]
- 5.Becker P B, Wu C. Cell-free system for assembly of transcriptionally repressed chromatin from Drosophila embryos. Mol Cell Biol. 1992;12:2241–2249. doi: 10.1128/mcb.12.5.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentley D L. Regulation of transcriptional elongation by RNA polymerase II. Curr Opin Genet Dev. 1995;5:210–216. doi: 10.1016/0959-437x(95)80010-7. [DOI] [PubMed] [Google Scholar]
- 7.Blau J, Xiao H, McCracken S, O’Hare P, Greenblatt J, Bentley D. Three functional classes of transcriptional activation domains. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonner J J, Heyward S, Fackenthal D L. Temperature-dependent regulation of a heterologous transcriptional activation domain fused to yeast heat shock transcription factor. Mol Cell Biol. 1992;12:1021–1030. doi: 10.1128/mcb.12.3.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown S A, Imbalzano A N, Kingston R E. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- 10.Carcamo J, Lobos S, Merino A, Buckbinder L, Weinmann R, Natarajan V, Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II. Role of factors IID and MLTF in transcription from the adenovirus major late and IVa2 promoters. J Biol Chem. 1989;264:7704–7714. [PubMed] [Google Scholar]
- 11.Chatterjee S, Struhl K. Connecting a promoter-bound protein to TBP bypasses the need for a transcriptional activation domain. Nature. 1995;374:820–822. doi: 10.1038/374820a0. [DOI] [PubMed] [Google Scholar]
- 12.Chi T, Carey M. Assembly of the isomerized TFIIA-TFIID-TATA ternary complex is necessary and sufficient for gene activation. Genes Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- 13.Choy B, Green M R. Eukaryotic activators function during multiple steps of preinitiation complex assembly. Nature. 1993;366:531–536. doi: 10.1038/366531a0. [DOI] [PubMed] [Google Scholar]
- 14.Clos J, Westwood T, Becker P B, Wilson S, Lambert K, Wu C. Molecular cloning and expression of a hexameric Drosophila heat shock factor subject to negative regulation. Cell. 1990;63:1085–1097. doi: 10.1016/0092-8674(90)90511-c. [DOI] [PubMed] [Google Scholar]
- 15.Croston G E, Kerrigan L A, Lira L M, Marshak D R, Kadonaga J T. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription. Science. 1991;251:643–649. doi: 10.1126/science.1899487. [DOI] [PubMed] [Google Scholar]
- 16.Dubois M-F, Vincent M, Vigneron M, Adamczewski J, Egly J-M, Bensaude O. Heat shock inactivation of the TFIIH-associated kinase and change in the phosphorylation sites on the C-terminal domain of RNA polymerase II. Nucleic Acids Res. 1997;25:694–700. doi: 10.1093/nar/25.4.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodrich J A, Cutler G, Tjian R. Contacts in context: promoter specificity and macromolecular interactions in transcription. Cell. 1996;84:825–830. doi: 10.1016/s0092-8674(00)81061-2. [DOI] [PubMed] [Google Scholar]
- 18.Goodrich J A, Tjian R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- 19.Green M, Schuetz T J, Sullivan E K, Kingston R E. A heat shock responsive domain of human HSF1 that regulates transcription activation domain functions. Mol Cell Biol. 1995;15:3354–3362. doi: 10.1128/mcb.15.6.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo H, Price D H. Mechanism of DmS-II-mediated pause suppression by Drosophila RNA polymerase II. J Biol Chem. 1993;268:18762–18770. [PubMed] [Google Scholar]
- 21.Hawley D K, Roeder R G. Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J Biol Chem. 1987;262:3452–3461. [PubMed] [Google Scholar]
- 22.Ho S N, Biggar S R, Spencer D M, Schreiber S L, Crabtree G R. Dimeric ligands define a role for transcriptional activation domains in reinitiation. Nature. 1996;382:822–826. doi: 10.1038/382822a0. [DOI] [PubMed] [Google Scholar]
- 23.Jiang Y, Yan M, Gralla J D. A three-step pathway of transcription initiation leading to promoter clearance at an activated RNA polymerase II promoter. Mol Cell Biol. 1996;16:1614–1621. doi: 10.1128/mcb.16.4.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson F B, Krasnow M A. Differential regulation of transcription preinitiation complex assembly by activator and repressor homeo domain proteins. Genes Dev. 1992;6:2177–2189. doi: 10.1101/gad.6.11.2177. [DOI] [PubMed] [Google Scholar]
- 25.Kadonaga J T. Assembly and disassembly of the Drosophila RNA polymerase II complex during transcription. J Biol Chem. 1990;265:2624–2631. [PubMed] [Google Scholar]
- 26.Kerrigan L A, Croston G E, Lira L M, Kadonaga J T. Sequence-specific transcriptional antirepression of the Drosophila Krüppel gene by the GAGA factor. J Biol Chem. 1991;266:574–582. [PubMed] [Google Scholar]
- 27.Kim Y, Bjorklund S, Li Y, Sayre M H, Kornberg R D. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 28.Kingston R, Bunker C, Imbalzano A N. Repression and activation by multiprotein complexes that alter chromatin structure. Genes Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- 29.Klages N, Strubin M. Stimulation of RNA polymerase II transcription initiation by recruitment of TBP in vivo. Nature. 1995;374:822–823. doi: 10.1038/374822a0. [DOI] [PubMed] [Google Scholar]
- 30.Koleske A J, Young R A. An RNA polymerase II holoenzyme responsive to activators. Nature. 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- 31.Krumm A, Hickey L B, Groudine M. Promoter proximal pausing of RNA polymerase II defines a general rate-limiting step after transcriptional initiation. Genes Dev. 1995;9:559–572. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- 32.Li B, Weber J A, Chen Y, Greenleaf A L, Gilmour D S. Analyses of promoter-proximal pausing by RNA polymerase II on the hsp70 heat shock gene promoter in a Drosophila nuclear extract. Mol Cell Biol. 1996;16:5433–5443. doi: 10.1128/mcb.16.10.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lis J T, Wu C. Transcriptional regulation of heat shock genes. In: Conaway R C, Conaway J W, editors. Transcription: mechanisms and regulation. New York, N.Y: Raven Press; 1994. pp. 459–475. [Google Scholar]
- 34.Maxon M E, Goodrich J A, Tjian R. Transcription factor IIE binds preferentially to RNA polymerase IIa and recruits TFIIH: a model for promoter clearance. Genes Dev. 1994;8:515–524. doi: 10.1101/gad.8.5.515. [DOI] [PubMed] [Google Scholar]
- 35.Mitchelmore C. In vitro analysis of DNA-binding proteins involved in transcriptional regulation. Ph.D. thesis. London, United Kingdom: Open University; 1993. [Google Scholar]
- 36.O’Brien T, Hardin S, Greenleaf A, Lis J T. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 37.Ohkuma Y, Roeder R G. Regulation of TFIIH ATPase and kinase activities by TFIIE during active initiation complex assembly. Nature. 1994;368:160–163. doi: 10.1038/368160a0. [DOI] [PubMed] [Google Scholar]
- 38.Peterson C L, Tamkun J W. The SWI-SNF complex: a chromatin remodeling machine? Trends Biochem Sci. 1995;20:143–146. doi: 10.1016/s0968-0004(00)88990-2. [DOI] [PubMed] [Google Scholar]
- 39.Rasmussen E B, Lis J T. Short transcripts of the ternary complex provide insight into RNA polymerase II pausing. J Mol Biol. 1995;252:522–535. doi: 10.1006/jmbi.1995.0517. [DOI] [PubMed] [Google Scholar]
- 40.Roberts S G E, Choy B, Walker S S, Lin Y S, Green M R. A role for activator-mediated TFIIB recruitment in diverse aspects of transcriptional regulation. Curr Biol. 1995;5:508–516. doi: 10.1016/s0960-9822(95)00103-5. [DOI] [PubMed] [Google Scholar]
- 41.Roeder R. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 42.Rougvie A E, Lis J T. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 43.Rougvie A E, Lis J T. Postinitiation transcriptional control in Drosophila melanogaster. Mol Cell Biol. 1990;10:6041–6045. doi: 10.1128/mcb.10.11.6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sandaltzopoulos R, Blank T, Becker P B. Transcriptional repression by nucleosomes but not H1 in reconstituted preblastoderm Drosophila chromatin. EMBO J. 1994;13:373–379. doi: 10.1002/j.1460-2075.1994.tb06271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandaltzopoulos R, Mitchelmore C, Bonte E, Wall G, Becker P B. Dual regulation of the Drosophila hsp26 promoter in vitro. Nucleic Acids Res. 1995;23:2479–2487. doi: 10.1093/nar/23.13.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szentirmay M N, Sawadogo M. Transcription factor requirement for multiple rounds of initiation by human RNA polymerase II. Proc Natl Acad Sci USA. 1991;88:10691–10695. doi: 10.1073/pnas.88.23.10691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szentirmay M N, Sawadogo M. Synthesis of reinitiated transcripts by mammalian RNA polymerase II is controlled by elongation factor SII. EMBO J. 1993;12:4677–4684. doi: 10.1002/j.1460-2075.1993.tb06156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thomas G H, Elgin S C R. Protein/DNA architecture of the DNase I hypersensitive region of the Drosophila hsp26 promoter. EMBO J. 1988;7:2191–2201. doi: 10.1002/j.1460-2075.1988.tb03058.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsukiyama T, Becker P B, Wu C. ATP-dependent nucleosome disruption at a heat-shock promoter mediated by binding of GAGA transcription factor. Nature. 1994;367:525–532. doi: 10.1038/367525a0. [DOI] [PubMed] [Google Scholar]
- 50.Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- 51.Van Dyke M W, Sawadogo M, Roeder R G. Physical analysis of transcription preinitiation complex assembly on a class II gene promoter. Science. 1988;241:1335–1338. doi: 10.1126/science.3413495. [DOI] [PubMed] [Google Scholar]
- 52.Varga-Weisz P D, Blank T A, Becker P B. Energy-dependent chromatin accessibility and nucleosome mobility in a cell-free system. EMBO J. 1995;14:2209–2216. doi: 10.1002/j.1460-2075.1995.tb07215.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Varga-Weisz P D, Wilm M, Bonte E, Dumas K, Mann M, Becker P B. Chromatin remodelling factor CHRAC contains the ATPases ISWI and toposiomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 54.Wall G, Varga-Weisz P D, Sandaltzopoulos R, Becker P B. Chromatin remodeling by GAGA factor and heat shock factor at the hypersensitive Drosophila hsp26 promoter in vitro. EMBO J. 1995;14:1727–1736. doi: 10.1002/j.1460-2075.1995.tb07162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wallrath L L, Quinn L, Granok H, Elgin S C R. Architectural variations of inducible eukaryotic promoters: preset and remodeling chromatin structures. Bioessays. 1994;16:165–170. doi: 10.1002/bies.950160306. [DOI] [PubMed] [Google Scholar]
- 56.Weber J A, Gilmour D S. Genomic footprinting of the hsp70 and histone H3 promoters in Drosophila embryos reveals novel protein-DNA interactions. Nucleic Acids Res. 1995;23:3327–3334. doi: 10.1093/nar/23.16.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White J, Brou C, Wu J, Lutz Y, Moncollin V, Chambon P. The acidic transcriptional activator GAL-VP16 acts on preformed template-committed complexes. EMBO J. 1992;11:2229–2240. doi: 10.1002/j.1460-2075.1992.tb05282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 59.Zawel L, Kumar P, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 60.Zawel L, Reinberg D. Advances in RNA polymerase II transcription. Curr Opin Cell Biol. 1992;4:488–495. doi: 10.1016/0955-0674(92)90016-6. [DOI] [PubMed] [Google Scholar]