Abstract
In order to isolate novel estrogen-responsive genes, we utilized a CpG island library in which the regulatory regions of genes are enriched. CpG islands were screened for the ability to bind to a recombinant estrogen receptor protein with a genomic binding site (GBS) cloning method. Six CpG islands were selected, and they contained perfect, imperfect, and/or multiple half-palindromic estrogen-responsive elements (EREs). Northern blot analysis of various human cells showed that all these genomic fragments hybridized to specific mRNAs, suggesting that the genes associated with these EREs might be transcribed in human cells. Then cDNAs associated with two of them, EB1 and EB9, were isolated from libraries of human placenta and MCF-7 cells derived from a human breast cancer, respectively. Both transcripts were increased by estrogen in MCF-7 cells. The increase is inhibited by actinomycin D but not by cycloheximide, indicating that no protein synthesis is required for the up-regulation. The cDNA associated with EB1 encodes a 114-amino-acid protein similar to the cytochrome c oxidase subunit VIIa, named COX7RP (cytochrome c oxidase subunit VII-related protein). The cDNA associated with EB9 is homologous only to an express sequence tag and was named EBAG9 (estrogen receptor-binding fragment-associated gene 9). The palindromic ERE of EB1 is located in an intron of COX7RP, and that of EB9 is in the 5′ upstream region of the cDNA. Both EREs had significant estrogen-dependent enhancer activities in a chloramphenicol acetyltransferase assay, when they were inserted into the 5′ upstream region of the chicken β-globin promoter. We therefore propose that the CpG-GBS method described here for isolation of the DNA binding site from the CpG island library would be useful for identification of novel target genes of certain transcription factors.
Estrogen is essential for the development of female organs and for the regulation of the growth, differentiation, and function of target cells. Estrogen receptor (ER), a member of the steroid-thyroid hormone receptor superfamily, mediates its action by binding ligand dependently to the estrogen-responsive element (ERE) that exists in the enhancer region of target genes, regulating their transcription directly (13, 16). ER has been found in female organs and also in nonreproductive systems of both sexes, such as the central nervous system (6, 39, 47), the skeletal system (12, 25), and the cardiovascular system (37, 49). However, despite the wide variety of estrogen actions, relatively few genes that are directly responsive to this hormone have been identified (2, 33, 53, 55). In order to isolate more estrogen-responsive genes, a method called genomic binding site (GBS) cloning, by which ER-binding fragments from human genomic DNA were isolated with recombinant ER protein (18), identifying a novel estrogen-responsive gene (efp, encoding estrogen-responsive finger protein) at an adjacent region (19), was developed previously.
Human CpG islands selected by the methyl-CpG binding domain of MeCP2, which binds DNA methylated at CpG (29), were inserted into plasmid vector for construction of a human CpG island library (8). CpG islands are short stretches of DNA containing a high density of nonmethylated CpG dinucleotides and distributed throughout the genome in 45,000 short regions of about 1 kb (5, 7). About 60% of human genes are found to be associated with CpG islands, including most of the housekeeping genes and 40% of the tissue-specific genes. These regions often include the promoter region and one or more exons of associated genes (1, 14, 28).
In this study, we have utilized a human CpG island library as the source of genomic DNA for GBS cloning and named the technique the CpG-GBS method. It is shown that ER-binding fragments isolated from CpG islands contain exons of associated genes at a high frequency. Among these associated genes, two transcripts are characterized in detail and shown to be actually regulated by estrogen in MCF-7 cells.
MATERIALS AND METHODS
Isolation of ER-binding fragments.
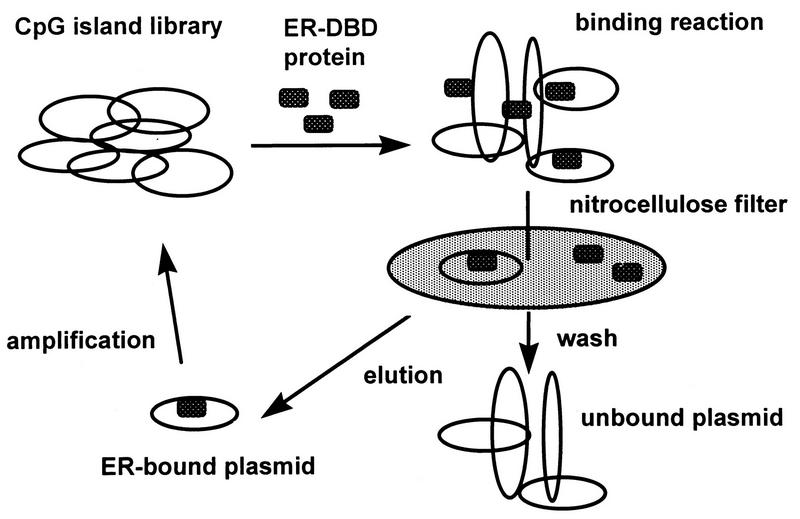
A human CpG island library (8) was kindly provided by the HGMP Resource Centre, Clinical Research Centre, Harrow, United Kingdom. Filter binding selection was performed as described previously (18). Briefly, the plasmid DNA (10 μg) was prepared from the human CpG island library and incubated with the recombinant DNA binding domain of the ER (ER-DBD) (10 pmol). The binding reaction was carried out also as described previously (18). The resulting solution was then passed slowly through a presoaked nitrocellulose filter and washed. The trapped plasmid DNA was eluted from the filter and transformed into DH5α competent cells (18). The cells were cultured on a Luria broth-ampicillin plate for 24 h, and the plasmid DNA was prepared by alkaline treatment followed by centrifugation in a cesium chloride-ethidium bromide gradient (31). The DNA (10 μg) was subsequently incubated with the recombinant protein again, and the selection cycle was repeated five times.
Plasmid construction.
An expression vector of pGEX/ER-DBD was prepared in the following manner. The DNA binding domain (amino acids 177 to 281) of rat ER cDNA was amplified by using primers 5′-ACTGGATCCATGATCATGGAGTCTGCC-3′ and 5′-TGCGAATTCCATTTCGGCCTTCCAAGTC-3′. The resulting fragment was digested with BamHI and EcoRI and inserted into these sites of pGEX-2T (Pharmacia). The plasmid vitERE/pBluescript SK(−) was constructed as follows. Oligonucleotides containing the wild-type ERE of the Xenopus vitellogenin gene A2 enhancer [vitERE (−338/−310)] (23) were synthesized, annealed, and inserted into the SacI site of pBluescript SK(−) (Stratagene). Chloramphenicol acetyltransferase (CAT) reporter plasmids of EB1 and EB9 were constructed as follows. Oligonucleotides containing the EREs of EB1 and EB9 were synthesized as 5′-GGTGCAGGGGTCAAGGTGACCCCCGGGGTCAC-3′ and 5′-GGGTGACCCCGGGGGTCACCTTGACCCCTGCA-3′ for EB1-ERE and 5′-GGTACTGTTTCCGGGTCAGGGTGACCTCTGGG-3′ and 5′-GGCCCAGAGGTCACCCTGACCCGGAAACAGTA-3′ (underlining indicates the palindromic core of ERE) for EB9-ERE. These oligonucleotides were annealed and blunted. The resulting fragments were inserted into blunting BamHI sites for pGCAT (18).
Preparation of glutathione S-transferase/ER-DBD fusion protein.
Escherichia coli BL21 cells transformed with pGEX/ER-DBD expression vector were grown to an optical density of 0.4 to 0.8 in 200 ml of 2× YT-ampicillin medium, and the culture was induced by 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 8 h. The induced cells were harvested and resuspended in phosphate-buffered saline containing 1% Triton X-100, 0.2% sarcosyl, and 10 mM dithiothreitol (DTT) and disrupted by sonication. The lysate was centrifuged at 10,000 × g for 30 min and then affinity purified with glutathione Sepharose 4B.
Filter binding assay.
The binding ability of isolated fragments was measured by filter binding assay (18). The fragments were amplified by PCR using primers 5′-CGGCCCCTGCAGGTCGACCTTAA-3′ and 5′-AACGCGTTGGGAGCTCTCCCTTAA-3′. PCRs were carried out in buffer III (41) supplemented with 10% dimethyl sulfoxide. Thirty cycles of amplification (1 min at 94°C, 1 min at 55°C, 3 min at 72°C) were performed. Plasmids pBluescript SK(−) and vitERE/pBluescript SK(−) were digested with HindIII and dephosphorylated. These plasmids and fragments were end labeled and incubated with the ER-DBD protein. The DNA trapped by the filter was analyzed by liquid scintillation counting to determine the recovery (18).
Cell culture.
COS-7, MCF-7, HEC-1, and HOS-TE85 cells were grown to a subconfluent state in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS). One day prior to estrogen treatment, the medium was changed to phenol red-free DMEM containing 10% FBS treated with dextran-coated charcoal.
Gel mobility shift assays.
Briefly, 106 COS-7 cells were plated in 60-mm-diameter petri dishes and maintained in DMEM containing 10% FBS for 24 h. One hour prior to transfection, the medium was changed to phenol red-free DMEM containing 10% FBS treated with dextran-coated charcoal. Cells were transfected with 10 μg of human ER expression vector (HEG0 [52]). After 12 h of incubation, the cells were cultured further in the absence of hormone for 24 h. One hour prior to harvest of the cells, 10 nM 17β-estradiol was added to the medium. The cells were washed with phosphate-buffered saline, and whole-cell extract (WCE) was prepared by freezing and thawing the cells in buffer containing 20 mM Tris-HCl (pH 7.5), 2 mM DTT, 20% glycerol, and 0.4 M KCl. As a negative control, oligonucleotides for the glucocorticoid response element (GRE) were synthesized as reported previously (26). EB1-ERE, EB9-ERE, vitERE (−338/−310), and GRE were 32P end labeled. An appropriate volume (8 to 12 μg of protein) of WCE was incubated with 2 μg of poly(dI-dC) (Pharmacia) at 0°C for 15 min. The binding reaction was started by addition of 32P-end-labeled probes (2 × 104 cpm) and incubation at 20°C for 15 min in binding buffer (10 mM Tris-HCl [pH 7.5], 1 mM DTT, 100 mM KCl, and 10% glycerol). The receptor-DNA complexes were separated on a 5% polyacrylamide gel. For the competition and supershift assays, different concentrations of unlabeled competitor probes or purified polyclonal ER antibody were mixed with the labeled probe before being added to the binding reaction mixture.
cDNA screening and DNA sequencing.
An MCF-7 cDNA library was prepared from poly(A)+ RNA of MCF-7 cells with a λZAPII cDNA synthesis kit (Stratagene), and a human placental cDNA library was purchased (Stratagene). A total of 5 × 105 phages from human placental library were screened with a 32P-labeled EB1 probe, and phages from the MCF-7 library was screened with an EB9 probe. Each cDNA was isolated according to the manufacturer’s instructions. These clones were sequenced by the dideoxy chain termination method using a BcaBest sequencing kit (Takara). The products of the sequencing reactions were analyzed on an automatic DNA sequencer (Pharmacia).
Northern blot analysis.
Total RNA was isolated from various cell lines by using ISOGEN (Nippon Gene) according to the manufacturer’s instructions. Poly(A)+ RNA was prepared by using Oligotex dT30 super (Takara). Poly(A)+ RNA (2 μg) was electrophoretically separated on a 1% agarose gel containing 2.2 M formaldehyde and blotted onto Biodyne nylon membranes. The filters were hybridized in 0.35 M Na2HPO4 (pH 7.2)–7% sodium dodecyl sulfate (SDS)–30% formamide–1 mM EDTA (pH 8.0)–1% bovine serum albumin at 60°C. The filters were then washed at 60°C with 0.4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% SDS. All probes were labeled with [α-32P]dCTP by using rediprime (Amersham) according to the manufacturer’s instructions. The filters were hybridized, washed, and then exposed to X-ray film for autoradiography or to an imaging plate for the BAS2000 bio-image analyzer (Fuji Film).
CAT analysis.
CAT analysis was performed as described elsewhere (18). Briefly, 106 MCF-7 cells were used instead of COS-7 cells. The cells were transfected with 2 μg of reporter plasmids and 2 μg of PCH110 β-galactosidase expression vector (Pharmacia). After 12 h of incubation, the cells were cultured further in the absence or presence of 10 nM 17β-estradiol for 24 h. Cell extracts were prepared and assayed for β-galactosidase and CAT activity.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases with the following accession numbers: AB007618 for COX7RP, AB007619 for EBAG9, AB007620 for EB1, and AB007621 for EB11.
RESULTS
Isolation and characterization of the ER-binding fragments in the human CpG island library.
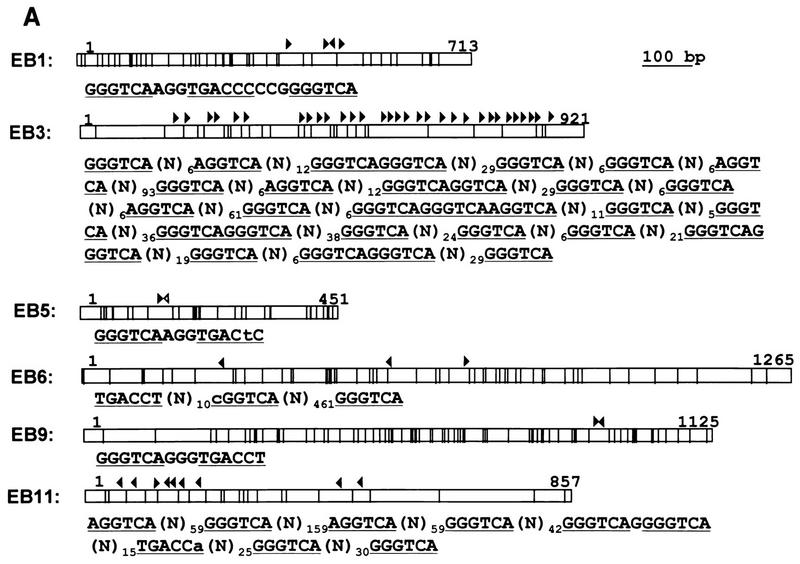
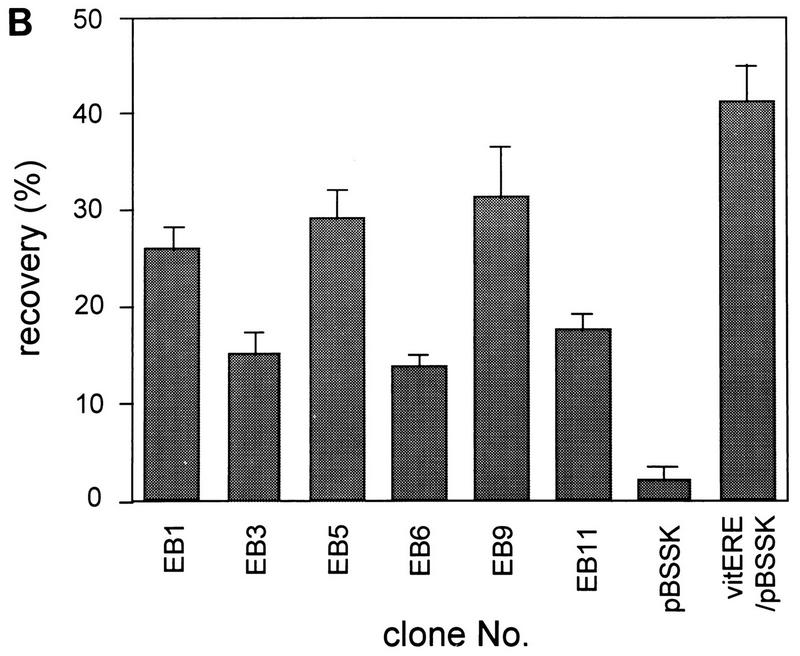
ER-binding fragments were isolated from the human CpG island library as described in Materials and Methods. DNA from the human CpG island library was bound with ER-DBD and selected with a nitrocellulose filter. The ER-binding plasmids were eluted and amplified in a bacterial culture. These plasmids were then concentrated by repeating five selection cycles (Fig. 1). Twelve colonies were picked, and the recovered plasmids were named EB1 to EB12. Cross-hybridization and restriction analysis revealed that seven clones were mutually independent. The sequences of these independent clones were analyzed to scan for CpG islands and EREs (Table 1 and Fig. 2A). Six contained many CpG dinucleotides, having over 50% GC content. However, EB12, containing only seven CpG dinucleotides, with a 48% GC content, was not analyzed further, because it seemed not to be derived from the CpG island. Of the six, the sequences of EB1 and EB9 contained a classical consensus ERE, that of EB5 contained one base mismatch to the consensus ERE, and those of EB3, EB6, and EB11 contained 27, 3, and 8 consensus ERE half-sites, respectively (Fig. 2A). The binding of these six fragments to ER-DBD was reconfirmed. The fragments were end labeled with [γ-32P]ATP, and the binding capacities were analyzed by filter binding assay. In this assay, EB1, EB5, and EB9 showed strong binding activities, as high as that of the vitellogenin ERE, used as a positive control. EB3, EB6, and EB11 also showed binding activities about 10-fold greater than that of the negative control (Fig. 2B).
FIG. 1.
Strategy of GBS cloning with a CpG island library. A CpG island library was bound with ER-DBD and passed through a nitrocellulose filter. ER-bound plasmids were eluted with Tris-HCl buffer containing 0.1% SDS and amplified in the bacterial culture. The ER-bound plasmids were concentrated by repeating five cycles.
TABLE 1.
Analysis of ER-binding human CpG islands
| Clonea | Length (bp) | No. of CGs | GC content (%) | Type of ERE (no.)b | Binding activity (%) |
|---|---|---|---|---|---|
| EB1 | 713 | 45 | 61.1 | pERE (1), hERE (3) | 26.0 |
| EB3 | 921 | 32 | 60.6 | hERE (32) | 15.3 |
| EB5 | 451 | 31 | 57.7 | ipERE (1) | 29.3 |
| EB6 | 1,265 | 54 | 53.3 | hERE (3) | 14.1 |
| EB9 | 1,125 | 75 | 63.1 | pERE (1) | 31.6 |
| EB11 | 867 | 25 | 55.0 | hERE (8) | 17.8 |
| EB12 | 483 | 7 | 48.0 | ipERE (1), hERE (1) | 23.2 |
Seven independent fragments were isolated from the human CpG island library as ER-binding fragments.
pERE, perfect palindromic ERE; ipERE, imperfect palindromic ERE; and hERE, half-palindromic ERE.
FIG. 2.
(A) Positions of CpG dinucleotides and ERE-like sequences in ER-binding CpG islands. Each fragment (open box) and its size (number at right end of open box), the positions of CpG dinucleotides in each fragment (vertical lines), the positions of ERE-like sequences (consensus ERE half-sites [GGTCA] [closed arrowheads] and imperfect EREs [open arrowheads]) are shown. The orientation of the ERE is also indicated. Only ERE-like sequences in each fragment are shown. (B) Binding capacities of the ER-binding CpG islands. The labeled fragments were incubated with ER-DBD protein, and the filter binding assay was performed. pBluescript SK(−) (pBSSK) was used as a negative control and vitERE/pBSSK was used as a positive control for ER binding. The recovery ratio is presented as means ± standard deviations of triplicate determinations.
Northern blots and DNA homology searches.
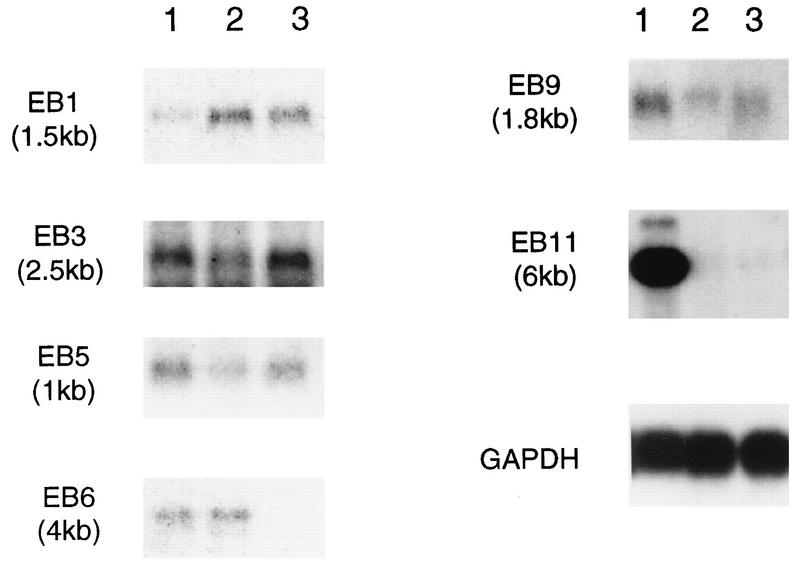
To find the estrogen-responsive genes associated with these EREs, Northern blotting and DNA database searches were performed. Some ER-positive human cell lines, namely, MCF-7, derived from a human breast cancer (48), HOS-TE85, derived from a human osteosarcoma (34), and HEC-1, derived from a human endometrial carcinoma (36), were used as the sources of mRNA. All six clones used as probes hybridized to each complementary mRNA in Northern analysis (Fig. 3). On the other hand, the EB1 and EB11 sequences showed homology to sequences in the GenBank DNA database as determined by BLAST analysis. The sequence of EB1 (GenBank accession no. AB007620) shows partial homology to a mouse cDNA (SIG81 [GenBank accession no. X80899]) (44) for approximately 100 bp near the N-terminal region. There are only 15 mismatches in the overlap region, indicating a high degree of conservation within this sequence. In addition, SIG81 had been reported to be similar to the cytochrome oxidase c subunit VII (cytox VII) gene, although EB1 had no homology with cytox VII. The sequence of EB11 (GenBank accession no. AB007621) shows partial homology to the rat N-methyl-d-aspartate (NMDA) receptor 2D subunit (NR2D [GenBank accession no. U08260]) for 490 bp of the 3′ untranslated region. In this region, the homology between EB11 and rat NR2D is 66% and the position of the polyadenylation signal (AATAAA) is also conserved. Interestingly, NR2D includes the same number of ERE half-sites as EB11, and four of eight EREs which are conserved in their positions were spaced every 50 to 100 bp.
FIG. 3.
Results of Northern blot analysis using ER-binding CpG islands as probes. Poly(A)+ RNAs (2 μg each) were prepared from HEC-1 (lanes 1), HOS (lanes 2), and MCF-7 (lanes 3) cells, and Northern blot analysis was performed. The probes and the sizes of the detectable transcripts are indicated. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Cloning of a cDNA similar to cytox VII from the human placental cDNA library.
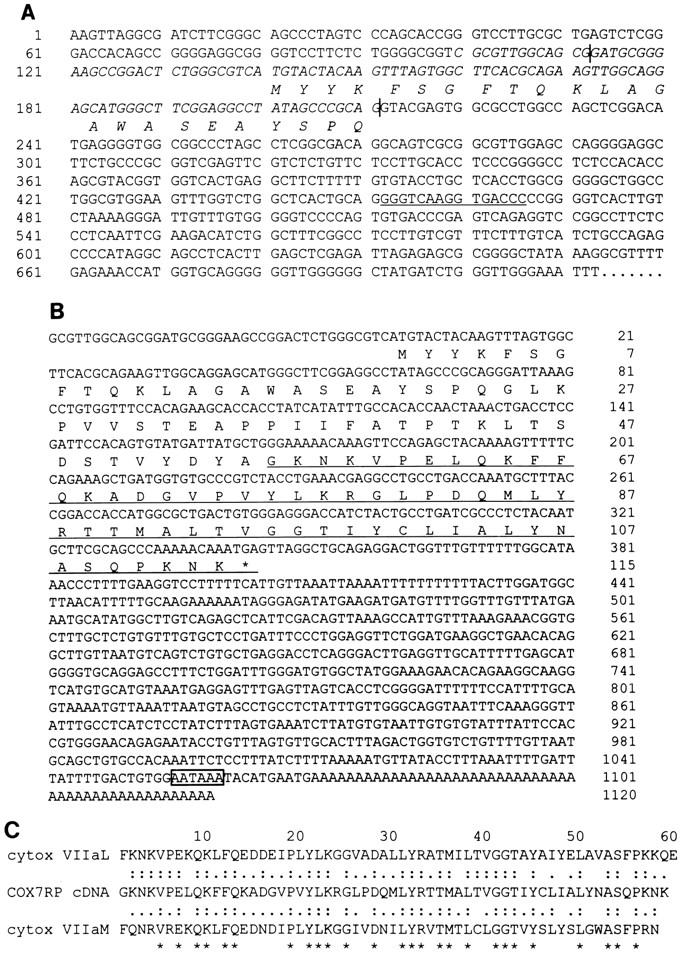
Using EB1 as a probe, we screened human placental cDNA library and isolated a cDNA named COX7RP (cytochrome c oxidase subunit VII [cytox VII]-related protein). The 5′ untranslated and further upstream region of COX7RP was contained in the EB1 sequence. The perfect palindromic ERE was present in the putative first intron of the gene (Fig. 4A). The nucleotide sequence and predicted amino acid sequence of this gene are shown in Fig. 4B. The entire gene is homologous to SIG81 at the nucleotide as well as the amino acid level, and the translational initiation site of SIG81 is also the same as for this cDNA (data not shown). Hence, SIG81 seems to be the mouse homolog of COX7RP. Only the C-terminal 30 amino acids of COX7RP are homologous with the mature protein regions of cytox VIIa isoforms. The levels of C-terminal homology between COX7RP and the mature regions of cytox VIIa liver- and muscle-type isoforms are 58.6 and 50.9%, respectively, at the amino acid level. On the other hand, the homology between the liver- and muscle-type isoforms is 62% (Fig. 4C). These facts suggest some functional relationship between COX7RP and cytox VIIa.
FIG. 4.
(A) Putative first exon of COX7RP and ERE in EB1 fragment. The complement sequence with COX7RP cDNA is indicated (italics), and the amino acid sequence is indicated below the DNA sequence. The 5′ and 3′ termini of the exon of COX7RP (vertical lines) and the perfect palindromic ERE (underlined) are also shown. (B) Nucleotide and predicted amino acid sequences of COX7RP. A putative predicted polyadenylation signal (open box) and the conserved mature cytox VIIa protein region (underlined) are indicated. (C) Multiple-sequence alignment of COX7RP and cytox VIIaL and VIIaM. Identical amino acid sequences (two dots) and similar amino acid sequences (single dot) and three consensus sequences (asterisks) are indicated.
Cloning of a novel cDNA associated with EB9 from the MCF-7 cDNA library.
EB9 had no homologous sequences in the DNA database but hybridized to a complement RNA, suggesting the existence of a novel ERE-associated gene. Using EB9 as a probe, we screened the MCF-7 cDNA library and isolated a novel cDNA named EBAG9 (ER-binding fragment-associated gene 9). The nucleotide sequence and predicted amino acid sequence of this gene are shown in Fig. 5. EB9 spanned the 5′-terminal and far-upstream region of the cDNA, and the consensus ERE was located in a 5′ upstream position of the cDNA. The in-frame stop codon was observed in this cDNA, indicating that this cDNA includes the full-length open reading frame. The amino acid sequence of the EBAG9 cDNA also had no homology with protein data bank sequences. Protein localization analysis (PSORT) showed the possibility that the product would be localized in the endoplasmic reticulum.
FIG. 5.
Nucleotide and predicted amino acid sequences of EBAG9. The amino acid sequence of EBAG9 is indicated below the nucleotide sequence. A putative Kozak consensus sequence (CCACCATGG) is indicated (underlined).
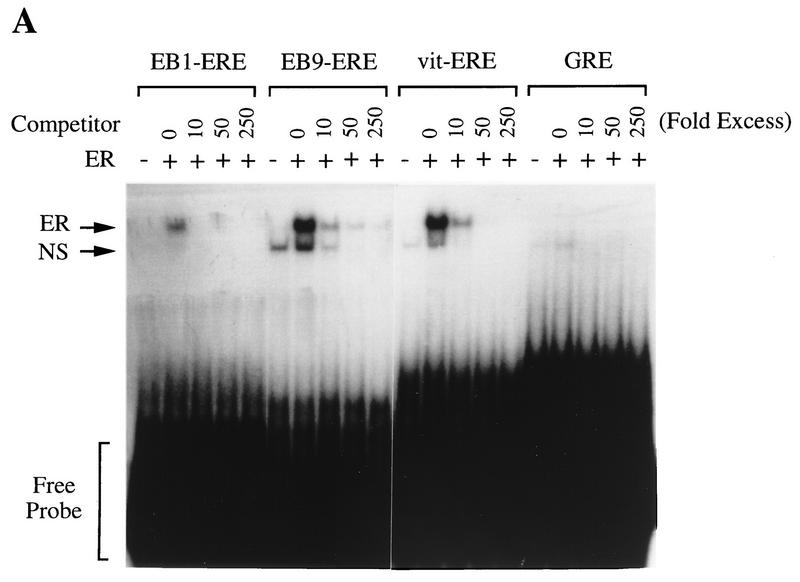
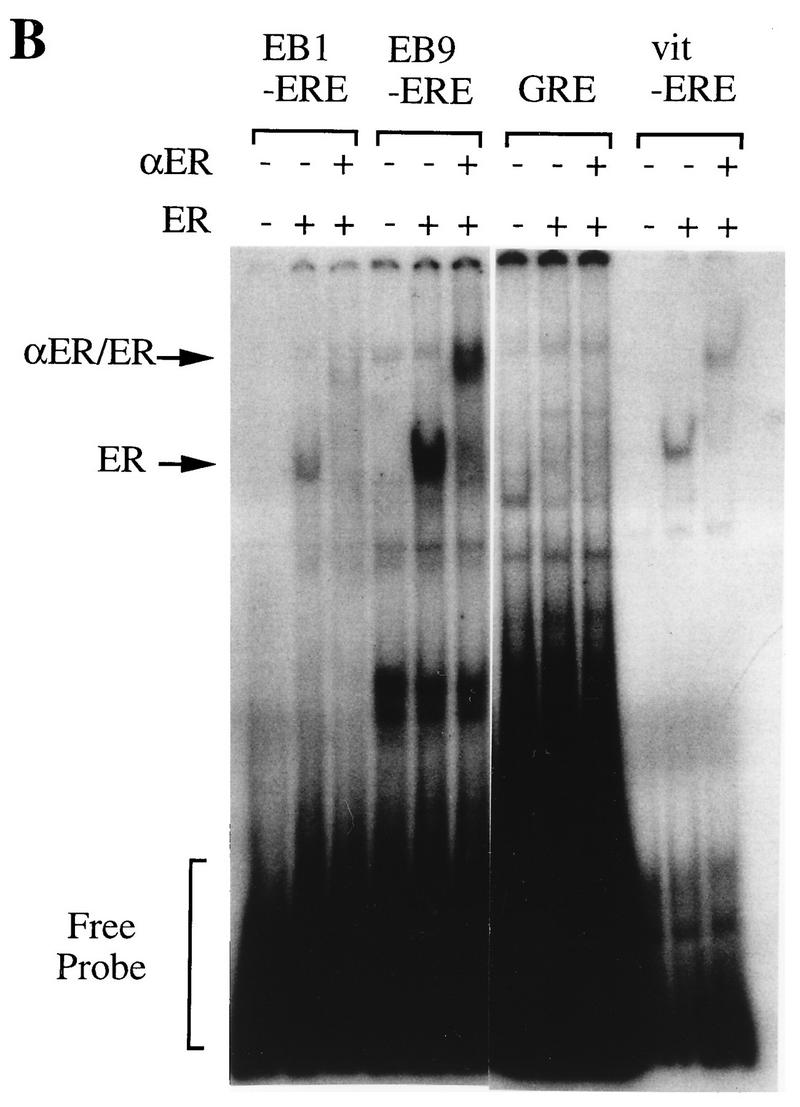
Gel mobility shift assay.
To confirm that the EREs of these fragments actually bind to full-length human ER, we have done gel mobility shift assays using these consensus EREs as probes. WCEs of COS-7 cells transiently transfected with vectors expressing wild-type human ER (HEG0) were used for this assay. While minor and more rapidly migrating protein-DNA complexes were seen when COS-7 cells were transfected with control vector pGEM3Zf(−), a major signal was detected when COS-7 cells were transfected with HEG0 using the palindromic ERE of both EB1 and EB9 (Fig. 6A). To investigate the specificity of the migrating protein-DNA complexes, competition analysis was performed. Both major and minor protein-DNA complexes were competed by addition of the cold consensus ERE probe (Fig. 6A). The specificities of these protein-DNA complexes were further confirmed by adding ER-specific polyclonal antibody. When the antibody was included in the binding reaction mixture, only the major protein-DNA complex was disrupted and supershifted (Fig. 6B). Thus, the specific binding of human ER was demonstrated by use of the EREs of both EB1 and EB9.
FIG. 6.
(A) Specific binding of perfect palindromic EREs present in EB1 and EB9 to human ER. WCEs of COS-7 cells transfected with either the control (pSG5) or human ER expression vector (HEG0) were incubated with the radiolabeled probes as indicated above the lanes and increasing amounts of unlabeled probes. ER-DNA complexes (ER) and nonspecific signal (NS) are indicated. (B) Specific binding further confirmed by including anti-ER polyclonal antibody (αER) in ER-DNA incubations as indicated above the lanes. ER-DNA complexes (ER) and supershifted signals (αER/ER) are indicated. +, addition; −, no addition.
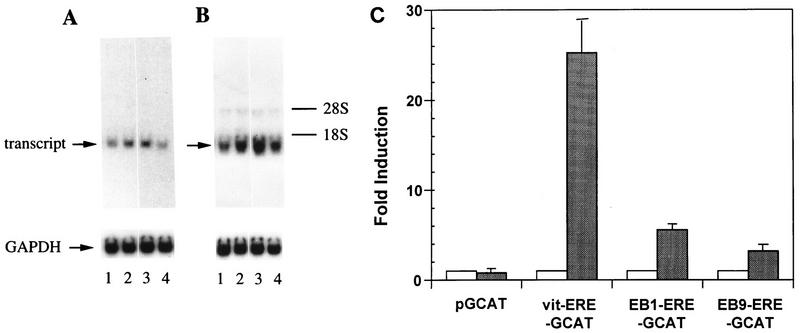
Regulation of COX7RP and EBAG9 mRNAs by estrogen in MCF-7 cells.
The signals detected by COX7RP and EBAG9 cDNA probes were identical to those detected by EB1 and EB9 probes in Northern blot analysis. These mRNAs increased by 6 h of estrogen treatment in MCF-7 cells, and this effect was inhibited by actinomycin D but not by cycloheximide (Fig. 7). Then the EREs of EB1 and EB9 were inserted into an upstream region of the β-globin promoter of pGCAT to construct EB1-ERE-GCAT and EB9-ERE-GCAT, respectively. These reporter plasmids were transfected into MCF-7 cells. As negative and positive controls, the reporter plasmids without (pGCAT) and with (vitERE-GCAT) the insertion of the ERE of the vitellogenin enhancer, respectively, were used. The CAT activities of EB1-ERE-GCAT, EB9-ERE-GCAT, and vitERE-GCAT were stimulated 5.6-, 3.2-, and 25.2-fold, respectively (Fig. 7C).
FIG. 7.
Regulation of COX7RP (A) and EB9 (B) mRNA by estrogen. MCF-7 cells were treated without (lane 1) or with (lanes 2 to 4) 10−7 M 17β-estradiol for 6 h in the presence or absence of cycloheximide (lane 3) or actinomycin D (lane 4). Poly(A)+ RNA (2 μg) was prepared from the cells, and Northern blot analysis was performed using COX7RP, EB9 cDNA, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as probes. The signals of transcripts and GAPDH are indicated. Migration positions of rRNA are shown on the right. (C) Estrogen-dependent enhancer activity of the ERE present in the first intron of the COX7RP gene and in the 5′ upstream region of the EBAG9 gene. Reporter plasmids containing pGCAT, vitERE-GCAT, EB1-ERE-GCAT, and EB9-ERE-GCAT were transfected into MCF-7 cells. After culture in the absence or presence of 10−7 M 17β-estradiol, a CAT assay was performed. The intensities of the signals were analyzed with a BAS2000 image analyzer. The results are presented as means ± standard deviations of triplicate determinations.
DISCUSSION
In this study, we have isolated ER-binding fragments by the CpG-GBS method. These ER-binding CpG islands contained perfect, imperfect, and/or half-palindromic EREs and had the capacity to bind to the ER in vitro. The transcript could be detected in estrogen target cells when the ER-binding CpG islands were used as probes. This high frequency of detection of the associated transcripts is remarkable and may be fortified by the use of CpG islands for the GBS method in combination with the use of ER-positive cell lines MCF-7, HOS-TE85, and HEC-1 for the Northern blot analysis. While we found estrogen-responsive genes associated with perfect palindromic EREs of EB1 and EB9, it has been reported that an imperfect or multiple half-palindromic ERE could be functional as an estrogen-dependent enhancer in vivo (2, 33, 53, 55). Therefore, imperfect or multiple half-palindromic EREs of these fragments might also be functional in vivo.
COX7RP cDNA, related to cytox VIIa, was isolated from a human placental cDNA library. The perfect palindromic ERE found in the putative first intron of the gene was shown to have an estrogen-dependent enhancer activity in a CAT assay when it was inserted at the 5′ upstream position. A number of enhancers are known to exist in introns (22, 27), including an example of an ERE in mouse hepatocyte growth factor gene (30). Recently, Segade et al. reported a new subunit of cytox VIIa, SIG81, from the mouse (45). They indicated that SIG81 localized in the mitochondrial fraction and that the fraction contained some cytochrome oxidase c activity. They also deduced the human SIG81 sequence by connecting the available expressed sequence tag sequences. The C-terminal portion (56 to 113 amino acids) of the assembled sequence described in that report is identical to the corresponding portion (57 to 114 amino acids) of the COX7RP sequence described here. The cytox VIIa protein is composed of an N-terminal mitochondrial presequence (17) and a C-terminal mature cytox VII protein region (43). COX7RP cDNA also had a mature-protein sequence in its C-terminal region that is well conserved with cytox VIIa, but its N-terminal region was different from cytox VIIa.
Cytochrome c oxidase is a complex enzyme composed of 13 subunits in mammals, of which three (cytox I, II, and III) are encoded by the mitochondrial genome and the others are encoded by the nuclear genome (21, 35). Interestingly, it has been reported that type II subunit in GH4C1 rat pituitary tumor cells (54) and type III subunit in the rat hippocampus (3) are regulated by estrogen. However, no ERE-like sequence was found in the mitochondrial genome, and no estradiol binding activity was detected in the mitochondria (20). cytox VII is a nuclear subunit, but little is known about the coordinate synthesis of the various subunits. It has also been postulated that the mitochondrial mRNA levels of cytox I, II, and III could be affected by increased levels of the nuclear subunits (3). Thus, estrogen might modulate the transcription of some nuclear subunits and then affect the synthesis or stabilization of the mitochondrial subunit mRNAs. Indeed, the nuclear subunits appear to modulate the entire enzymatic activity by regulating the holoenzyme assembly in yeast (11, 42). We have shown here that COX7RP mRNA was up-regulated by estrogen in MCF-7 cells. It is possible that this COX7RP represents a regulatory subunit of cytochrome c oxidase and mediates the higher level of energy production in target cells by estrogen.
Recently, mutations in the mitochondrial cytox I and II genes were found to segregate at a higher frequency with Alzheimer’s disease (AD) than with other neurodegenerative and metabolic diseases (9). These mutations cause a decrease of cytochrome c oxidase activity and increased production of reactive oxygen species. It has also been suggested that estrogen plays an important role in memory and learning and even in the treatment of AD (50). If the decrease of cytochrome c oxidase activity underlies the pathogenesis of AD, it is possible that the mechanism of estrogen action in the treatment of AD is associated with the modulation of cytox I and II and/or COX7RP by estrogen.
A novel cDNA, EBAG9, was isolated from the MCF-7 cDNA library by using EB9 as a probe. A perfect palindromic ERE of EB9 was not included in the 5′ untranslated region of EBAG9 but was located in the far-upstream region of the genomic EBAG9 fragment. Thus, this ERE may be located in the 5′-flanking region of this gene, just like in the Xenopus vitellogenin A2 gene (23) and the rat prolactin gene (33). To confirm this, the transcription initiation site of EBAG9 must be identified. This perfect palindromic ERE was shown to have an estrogen-dependent enhancer activity when it was placed in the upstream region of the β-globin promoter. The mRNA was up-regulated by estrogen in MCF-7 cells, and the up-regulation was dependent on the RNA synthesis but not on new protein synthesis, indicating that the transcriptional activation could take place via the perfect palindromic ERE. The tissue distribution of this mRNA was almost ubiquitous (data not shown), but significant homology was not found in the DNA or protein database. Although the functional association between estrogen and this gene remains unclear, it seems to be a novel estrogen target gene in vivo.
Several methods for isolation of the target gene of transcription factors have been reported. Differential or subtractive hybridization methods (32, 38) are useful for isolation of any regulatory gene, including direct as well as indirect target genes. DNA binding sites of transcription factors can be isolated from native chromatin by immunoprecipitation (15, 40, 51). Among these methods, the GBS method (18, 19) can be applicable if the recombinant protein of the transcription factor is available in certain amounts. In this method, target elements could be isolated, depending on their binding activities but not on the expression levels of the associated genes. However, the GBS cloning method would not be suitable when the binding of the transcription factor is weak or the binding requires heterodimers or cofactors. The use of CpG islands for isolation of the target elements has a significant advantage over the use of whole genomic fragments, because CpG islands are enriched with active genes (8). Furthermore, most of CpG islands exist as single copies in the genome. Thus, when the CpG islands were isolated as ER-binding fragments, they could be used efficiently as probes for Northern and Southern blotting for detection of their associated genes. Of course, it must be noted that CpG islands include a maximum of 60% of the human gene, so the target elements which are not associated with the CpG islands cannot be isolated by this strategy.
In this report, we have shown that the efficiency of cDNA screening can be increased when the target elements are isolated from a CpG island library by the GBS method. A random appearance of the 10-bp sequence of one perfect palindromic ERE may be 1 in about every 1,000 kb. Therefore, by calculation, the human genome would contain a few thousand ERE sequences. If the recognition sequence of the transcription factor is 6 to 8 bp long, the human genome could contain more than 100,000 recognition sequences. Most of these sequences would be nonspecific, which makes the application of the GBS method difficult. The human CpG island library contains 45,000 copies of the gene, and the average insert size is 1 kb. Thus, the CpG island library would contain only 50 ERE sequences, and these ERE sequences could be suitable for further analysis.
While this paper was being prepared, a report by Shago and Giguere (46) which described the use of CpG island-enriched DNA for the isolation of retinoic acid-responsive genes appeared. They enriched the CpG island fraction by restriction enzymes containing CpG within their recognition sequences and selected retinoic acid-responsive elements from the fraction by utilizing a combination of PCR amplification and gel mobility shift assay. Although the procedures are different from ours, the results of their study are consistent with ours in defining the importance of the use of CpG islands for isolation of the target elements of transcription factors.
ACKNOWLEDGMENTS
We thank the HGMP Resource Centre for kindly providing the human CpG island library (CGI1), P. Chambon for the kind gift of the HEGO clone, M. Maruyama for helpful discussions, and M. Hihara for expert technical assistance.
This work was supported by grants from the Ministry of Education, Science and Culture, Japan, and Maruki Memorial Foundation.
REFERENCES
- 1.Antequera P, Bird A. Number of CpG islands in human and mouse. Proc Natl Acad Sci USA. 1993;90:11995–11999. doi: 10.1073/pnas.90.24.11995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berry A, Nunez M, Chambon P. Estrogen-responsive element of the human pS2 gene is an imperfectly palindromic sequence. Proc Natl Acad Sci USA. 1989;86:1218–1222. doi: 10.1073/pnas.86.4.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bettini E, Maggi A. Estrogen induction of cytochrome c oxidase subunit III in rat hippocampus. J Neurochem. 1992;58:1923–1929. doi: 10.1111/j.1471-4159.1992.tb10070.x. [DOI] [PubMed] [Google Scholar]
- 4.Bird A. CpG islands as gene markers in the vertebrate nucleus. Trends Genet. 1987;3:342–347. [Google Scholar]
- 5.Bird A, Taggart P M, Frommer M, Miller O J, Macleod D. A fraction of the mouse genome that is derived from islands of non-methylated, CpG-rich DNA. Cell. 1985;40:91–99. doi: 10.1016/0092-8674(85)90312-5. [DOI] [PubMed] [Google Scholar]
- 6.Brown T J, Hochberg R B, Zielinski J E, Maclusky N J. Regional sex differences in cell nuclear estrogen-binding capacity in the rat hypothalamus and preoptic area. Endocrinology. 1988;123:1761–1770. doi: 10.1210/endo-123-4-1761. [DOI] [PubMed] [Google Scholar]
- 7.Cooper D, Taggart M, Bird A. Unmethylated domains in vertebrate DNA. Nucleic Acids Res. 1983;11:647–658. doi: 10.1093/nar/11.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross S, Charlton J, Nan X, Bird A. Purification of CpG islands using a methylated DNA binding column. Nat Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- 9.Davis R E, Miller S, Herrnstadt C, Ghosh S S, Fahy E, Shinobu L A, Galasko D, Thal L J, Beal M F, Howell N, Parker W D., Jr Mutations in mitochondrial cytochrome c oxidase genes segregate with late-onset Alzheimer disease. Proc Natl Acad Sci USA. 1997;94:4526–4531. doi: 10.1073/pnas.94.9.4526. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Diel P, Walter A, Fritzemeier K H, Hegele-Hartung C, Knauthe R. Identification of estrogen regulated genes in Fe33 rat hepatoma cells by differential display polymerase chain reaction and their hormonal regulation in rat liver and uterus. J Steroid Biochem Mol Biol. 1995;55:363–373. doi: 10.1016/0960-0760(95)00186-7. [DOI] [PubMed] [Google Scholar]
- 11.Dowhan W, Bibus C R, Schatz G. The cytoplasmically-made subunit IV is necessary for assembly of cytochrome c oxidase in yeast. EMBO J. 1985;4:179–184. doi: 10.1002/j.1460-2075.1985.tb02334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eriksen E F, Colvard D S, Berg N J, Graham M L, Mann K G, Spelsberg T C, Riggs B L. Evidence of estrogen receptors in normal human osteoblast-like cells. Science. 1988;241:84–86. doi: 10.1126/science.3388021. [DOI] [PubMed] [Google Scholar]
- 13.Evans R M. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 15.Gould A P, Brookman J J, Strutt D I, White R A. Targets of homeotic gene control in Drosophila. Nature. 1990;348:308–312. doi: 10.1038/348308a0. . (Erratum, 348:560.) [DOI] [PubMed] [Google Scholar]
- 16.Green S, Chambon P. Nuclear receptors enhance our understanding of transcription regulation. Trends Genet. 1988;4:309–314. doi: 10.1016/0168-9525(88)90108-4. [DOI] [PubMed] [Google Scholar]
- 17.Hendrick J P, Hodges P E, Rosenberg L E. Survey of amino-terminal proteolytic cleavage sites in mitochondrial precursor proteins: leader peptides cleaved by two matrix proteases share a three-amino acid motif. Proc Natl Acad Sci USA. 1989;86:4056–4060. doi: 10.1073/pnas.86.11.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Inoue S, Kondo S, Hashimoto M, Kondo T, Muramatsu M. Isolation of estrogen receptor-binding sites in human genomic DNA. Nucleic Acids Res. 1991;19:4091–4096. doi: 10.1093/nar/19.15.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inoue S, Orimo A, Hosoi T, Kondo S, Toyoshima H, Kondo T, Ikegami A, Ouchi Y, Orimo H, Muramatsu M. Genomic binding-site cloning reveals an estrogen responsive gene that encodes a RING finger protein. Proc Natl Acad Sci USA. 1993;90:11117–11121. doi: 10.1073/pnas.90.23.11117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ioannou I M, Tsawdaroglou N, Sekeris C E. Presence of glucocorticoid responsive elements in the mitochondrial genome. Anticancer Res. 1988;8:1405–1409. [PubMed] [Google Scholar]
- 21.Kadenbach B, Merle P. On the function of multiple subunits of cytochrome c oxidase from higher eukaryotes. FEBS Lett. 1981;135:1–11. doi: 10.1016/0014-5793(81)80932-5. [DOI] [PubMed] [Google Scholar]
- 22.Karpinski B A, Yang L H, Cacheris P, Morle G D, Leiden J M. The first intron of the 4F2 heavy-chain gene contains a transcriptional enhancer element that binds multiple nuclear proteins. Mol Cell Biol. 1989;9:2588–2597. doi: 10.1128/mcb.9.6.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klein-Hiptass L, Schorpp M, Wagner U, Ryffel G U. An estrogen-responsive element derived from the 5′ flanking region of the Xenopus vitellogenin A2 gene functions in transfected human cells. Cell. 1986;46:1053–1061. doi: 10.1016/0092-8674(86)90705-1. [DOI] [PubMed] [Google Scholar]
- 24.Koike S, Sakai M, Muaramatsu M. Molecular cloning and characterization of rat estrogen receptor cDNA. Nucleic Acids Res. 1987;15:2499–2513. doi: 10.1093/nar/15.6.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Komm B S, Christopher M T, Terpening M, Benz D J, Greme K A, Gallegos M R. Estrogen binding, receptor mRNA, and biologic response in osteoblast-like osteosarcoma cells. Science. 1988;241:81–84. doi: 10.1126/science.3164526. [DOI] [PubMed] [Google Scholar]
- 26.Kumar V, Chambon P. The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell. 1988;55:145–156. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- 27.Lacy J, Rudnick H. Transcriptional regulation of the human IgE receptor (Fc epsilon RII/CD23) by EBV. Identification of EBV-responsive regulatory elements in intron. J Immunol. 1992;148:1554–1560. [PubMed] [Google Scholar]
- 28.Larsen F, Gundersen G, Lopez R, Prydz H. CpG islands as gene markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- 29.Lewis J D, Meehan R R, Henzel W J, Maurer-Fogy I, Jeppesen P, Klein F, Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, Lin L, Zarnegar R. Modulation of hepatocyte growth factor gene expression by estrogen in mouse ovary. Mol Cell Endocrinol. 1994;104:173–181. doi: 10.1016/0303-7207(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 31.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 32.Manning D L, Archibald L H, Ow K T. Cloning of estrogen-responsive messenger RNAs in the T-47D human breast cancer cell line. Cancer Res. 1990;50:4098–4104. [PubMed] [Google Scholar]
- 33.Maurer R M, Notides A C. Identification of an estrogen-responsive element from the 5′-flanking region of the rat prolactin gene. Mol Cell Biol. 1987;7:4247–4254. doi: 10.1128/mcb.7.12.4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McAllister R M, Gardiner M B, Greene A E, Bradt C, Nichols W W, Landing B H. Cultivation in vitro of cells derived from a human osteosarcoma. Cancer. 1971;27:397–402. doi: 10.1002/1097-0142(197102)27:2<397::aid-cncr2820270224>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 35.Merle P, Kadenbach B. Kinetic and structural differences between cytochrome c oxidases from beef liver and heart. Eur J Biochem. 1982;125:239–244. doi: 10.1111/j.1432-1033.1982.tb06674.x. [DOI] [PubMed] [Google Scholar]
- 36.Morinaga M, Yonehara S, Tomita Y, Kuwata T. Insensitivity to interferon of two subclones of human endometrial carcinoma cell line, HEC-1. Int J Cancer. 1983;31:21–28. doi: 10.1002/ijc.2910310105. [DOI] [PubMed] [Google Scholar]
- 37.Orimo A, Inoue S, Ikegami A, Hosoi T, Akishita M, Ouchi Y, Muramatsu M, Orimo H. Vascular smooth muscle cells as target for estrogen. Biochem Biophys Res Commun. 1993;195:730–736. doi: 10.1006/bbrc.1993.2106. [DOI] [PubMed] [Google Scholar]
- 38.Pellerin I, Vuillermoz C, Jouvenot M, Ordener C, Royez M, Adessi G L. Identification and characterization of an early estrogen-regulated RNA in cultured guinea-pig endometrial cells. Mol Cell Endocrinol. 1993;90:R17–R21. doi: 10.1016/0303-7207(93)90161-c. [DOI] [PubMed] [Google Scholar]
- 39.Pfaff D W, Keiner M. Atlas of estradiol concentrating cells in the central nervous system of the adult female rat. J Comp Neurol. 1973;151:121–158. doi: 10.1002/cne.901510204. [DOI] [PubMed] [Google Scholar]
- 40.Phelps D E, Dressler G R. Identification of novel Pax-2 binding sites by chromatin precipitation. J Biol Chem. 1996;271:7978–7985. doi: 10.1074/jbc.271.14.7978. [DOI] [PubMed] [Google Scholar]
- 41.Ponce M R, Micol J L. PCR amplification of long DNA fragments. Nucleic Acids Res. 1992;20:623. doi: 10.1093/nar/20.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poyton R O, Trueblood C E, Wright R M, Farrell L E. Expression and function of cytochrome c oxidase subunit isologues. Modulators of cellular energy production? Ann N Y Acad Sci. 1988;550:289–307. doi: 10.1111/j.1749-6632.1988.tb35344.x. [DOI] [PubMed] [Google Scholar]
- 43.Seelan R S, Grossman L I. Cytochrome c oxidase subunit VIIa isoforms. Characterization and expression of bovine cDNAs. J Biol Chem. 1991;266:19752–19757. [PubMed] [Google Scholar]
- 44.Segade F, Claudio E, Wrobel K, Ramos S, Lazo P S. Isolation of nine gene sequences induced by silica in murine macrophages. J Immunol. 1995;154:2384–2392. [PubMed] [Google Scholar]
- 45.Segade F, Hurle B, Claudio E, Ramos S, Lazo P S. Identification of an additional member of the cytochrome c oxidase subunit VIIa family of proteins. J Biol Chem. 1996;271:12343–12349. doi: 10.1074/jbc.271.21.12343. [DOI] [PubMed] [Google Scholar]
- 46.Shago M, Giguere V. Isolation of a novel retionic acid-responsive gene by selection of genomic fragments derived from CpG island-enriched DNA. Mol Cell Biol. 1996;16:4337–4348. doi: 10.1128/mcb.16.8.4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simerly R B, Chang C, Muramatsu M, Swanson L W. Distribution of androgen and estrogen receptor mRNA-containing cells in the rat brain: an in situ hybridization study. J Comp Neurol. 1990;294:76–95. doi: 10.1002/cne.902940107. [DOI] [PubMed] [Google Scholar]
- 48.Soul H D, Vazquez J, Long A, Albert S, Brennan M. A human cell line from a pleural effusion derived from a breast carcinoma. J Natl Cancer Inst. 1973;51:1409–1416. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- 49.Stumpf W E, Sar M, Aumuller G. The heart: a target organ for estradiol. Science. 1977;196:319–321. doi: 10.1126/science.847474. [DOI] [PubMed] [Google Scholar]
- 50.Tang M X, Jacobs D, Stern Y, Marder K, Schofield P, Gurland B, Andrews H, Mayeux R. Effect of oestrogen during menopause on risk and age at onset of Alzheimer’s disease. Lancet. 1996;348:429–432. doi: 10.1016/S0140-6736(96)03356-9. [DOI] [PubMed] [Google Scholar]
- 51.Tomotsune D, Shoji H, Wakamatsu Y, Kondoh H, Takahashi N. A mouse homologue of the Drosophila tumor-suppressor gene 1(2)g1 controlled by Hox-C8 in vitro. Nature. 1993;365:69–72. doi: 10.1038/365069a0. [DOI] [PubMed] [Google Scholar]
- 52.Tora L, Mullick A, Metzger D, Ponglikitmongkol M, Park I, Chambon P. The cloned human oestrogen receptor contains a mutation which alters its hormone binding properties. EMBO J. 1989;8:1981–1986. doi: 10.1002/j.1460-2075.1989.tb03604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tora M, Gaub P, Mader S, Dierich A, Bellard M, Chambon P. Cell-specific activity of a GGTCA half-palindromic oestrogen-responsive element in the chicken ovalbumin gene promoter. EMBO J. 1988;7:3771–3778. doi: 10.1002/j.1460-2075.1988.tb03261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Itallie C M, Dannies P S. Estrogen induces accumulation of the mitochondrial ribonucleic acid for subunit II of cytochrome oxidase in pituitary tumor cells. Mol Endocrinol. 1988;2:332–337. doi: 10.1210/mend-2-4-332. [DOI] [PubMed] [Google Scholar]
- 55.Walker P, Germond J E, Brown-Luedi M, Givel F, Wahli W. Sequence homologies in the region preceding the transcription initiation site of the liver estrogen-responsive vitellogenin and apo-VLDLII genes. Nucleic Acids Res. 1984;12:8611–8626. doi: 10.1093/nar/12.22.8611. [DOI] [PMC free article] [PubMed] [Google Scholar]