Abstract
In Drosophila melanogaster, the fruitless (fru) gene controls essentially all aspects of male courtship behavior. It does this through sex-specific alternative splicing of the fru pre-mRNA, leading to the production of male-specific fru mRNAs capable of expressing male-specific fru proteins. Sex-specific fru splicing involves the choice between alternative 5′ splice sites, one used exclusively in males and the other used only in females. Here we report that the Drosophila sex determination genes transformer (tra) and transformer-2 (tra-2) switch fru splicing from the male-specific pattern to the female-specific pattern through activation of the female-specific fru 5′ splice site. Activation of female-specific fru splicing requires cis-acting tra and tra-2 repeat elements that are part of an exonic splicing enhancer located immediately upstream of the female-specific fru 5′ splice site and are recognized by the TRA and TRA-2 proteins in vitro. This fru splicing enhancer is sufficient to promote the activation by tra and tra-2 of both a 5′ splice site and the female-specific doublesex (dsx) 3′ splice site, suggesting that the mechanisms of 5′ splice site activation and 3′ splice site activation may be similar.
Alternative pre-mRNA processing is widely used as a regulatory mechanism for the control of gene expression in higher eukaryotes. Developmentally regulated pre-mRNA processing can give rise to gene expression patterns that are specific with regard to tissue, developmental stage, and sex.
In Drosophila melanogaster, the fruitless (fru) gene regulates essentially all aspects of male courtship behavior, including sexual orientation (33, 54). It does this by functioning in about 0.5% of the neurons in the central nervous system (CNS). fru is part of the hierarchy of Drosophila sex determination genes (5), where it controls one of the two branches identified downstream of the genes transformer (tra) and transformer-2 (tra-2) (63). The other branch controls all known aspects of somatic sexual differentiation outside the CNS, as well as certain aspects of sexual differentiation within the CNS, and is regulated by the doublesex (dsx) gene (13). Fundamental to the sex-specific functions of fru, transcripts from the distal fru promoter undergo sex-specific alternative splicing (54). In males, a male-specific 5′ splice site (5′ SS) is used that is located 1,590 nucleotides (nt) upstream of the female-specific 5′ SS used in females. Both 5′ SS are spliced to a common 3′ splice site (3′ SS) over 70 kb downstream. Male-specific and female-specific fru cDNAs contain a common coding region downstream of the common 3′ SS. In males, usage of the distal male-specific 5′ SS extends the open reading frame in the 5′ direction such that an additional 101 amino acids are present at the N terminus.
Females mutant for either tra or tra-2 exhibit male-specific fru splicing and male courtship behavior (54), indicating that tra and tra-2 are required either directly or indirectly for female-specific fru splicing. tra and tra-2 are known to activate directly the female-specific dsx 3′ SS by binding to cis-acting tra and tra-2 (tra/tra-2) repeat elements, (T/A)C(T/A)(T/A)C(A/G)ATCAACA (27, 30, 53). While tra-2 is expressed in both sexes (2, 25), tra, which is expressed only in females (11), is one of the few cell-specific splicing regulators known to date. In dsx, the tra/tra-2 repeat elements are present in six copies which are part of an exonic splicing enhancer located 300 nt downstream of the female-specific dsx 3′ SS (13, 48). Three copies of the tra/tra-2 repeat elements are also present upstream of the female-specific 5′ SS in fru (33, 54), suggesting that regulation of fru splicing by tra/tra-2 may occur directly. However, it is not known whether the tra/tra-2 repeat elements in fru are required for the regulation of fru sex-specific splicing.
tra and tra-2 contain protein domains known to be involved in regulated and general splicing. tra-2 contains a RRM-type RNA binding domain flanked by two SR domains, regions rich in serine and arginine residues, while tra contains a single SR domain. RRM-type RNA binding domains have been shown to be essential for the recognition of pre-mRNAs by a variety of RNA processing factors (12). SR domains have been implicated in protein-protein interactions between splicing factors (37, 68) and also in the intracellular localization of splicing factors to nuclear speckles (41), regions of the nucleus enriched in splicing factors. Other RNA processing factors containing RRM-type RNA binding domains and/or SR domains include the splicing factors of the SR protein family, general splicing factors which can also affect splice site choices (4, 15, 16, 18, 21, 23, 36, 39, 52, 55, 67, 72, 73), and the splicing factors U1-70K (64) and U2AF (74) involved in the general recognition of splice sites.
Recent studies have implicated additional proteins in splicing regulation by tra/tra-2. The TRA-2 protein has been reported to interact with the TRA protein (3, 32), the Drosophila SR protein RBP1 (29), human SR proteins (68), and the human U2AF35 protein (68). The Drosophila SR protein RBP1 participates in the activation of the female-specific dsx 3′ SS together with tra/tra-2 by specifically binding to the dsx pre-mRNA in the proximity of the female-specific dsx 3′ SS (28, 43). Mutations in the gene of another Drosophila SR protein, B52, inhibit female-specific dsx splicing, suggesting indirect or direct involvement of B52 in dsx splicing regulation (50). Unlike TRA, SR proteins appear to be ubiquitous, although there are differences in the relative abundance of members of this family in various tissues (71). Two other tissue-specific splicing factors, the soma-specific PSI protein in Drosophila (57) and a human neuron-specific 75-kDa protein (46), have been reported to regulate splicing of their target genes together with particular hnRNP proteins. These findings indicate that in several systems, tissue-specific splicing regulation involves both tissue-specific regulators and target-specific contributions from ubiquitous factors.
Recently, several mammalian tra-2 homologs have been identified (6, 17, 44, 56). At least one human tra-2 homolog can functionally replace tra-2 in Drosophila (17), suggesting that the mechanisms governing splice site selection by tra/tra-2 in Drosophila may also be significant in humans. The mechanism by which sex-specific fru splicing is regulated, is currently unknown. A priori, tra/tra-2 could induce female-specific fru splicing by either blocking the male-specific 5′ SS or activating the female-specific 5′ SS. In fru, the tra/tra-2 repeat elements are located between the two sex-specific 5′ SSs, 38 nt upstream of the female-specific 5′ SS and 1,352 nt downstream of the male-specific 5′ SS (54). Thus, the close proximity of the tra/tra-2 repeat elements to the female-specific fru 5′ SS would suggest that the likely mechanism of fru regulation is 5′ SS activation. In the case of dsx, instead of affecting 5′ SS selection, tra and tra-2 activate the female-specific dsx 3′ SS (27, 30, 53). Furthermore, the activation model would predict that in fru the tra/tra-2 repeat elements function as an upstream splicing enhancer, while in dsx their role is that of a downstream splicing enhancer. Unlike transcriptional enhancers, splicing enhancers can be position dependent. For example, shortening the distance of the tra/tra-2 repeat elements to the female-specific dsx 3′ SS inhibits dsx splicing regulation by tra/tra-2 (65). Indeed, while other downstream splicing enhancers, generally containing multiple repeats of the sequence motif GAR, where R is a purine, have been identified in exons of vertebrate genes (14, 19, 24, 31, 40, 51, 62, 66, 69, 70), and several of these purine-rich downstream splicing enhancers are recognized by SR proteins (40, 51, 60, 61), to our knowledge no upstream splicing enhancer has been reported to date. Hence, an involvement of tra/tra-2 and of the tra/tra-2 repeat elements in the selection of fru 5′ SSs would indicate a novel regulatory mechanism.
In order to determine whether tra/tra-2 regulate fru sex-specific splicing by acting through the tra/tra-2 repeat elements and, if they do, to determine the regulatory mechanism and its relationship to the mechanism of dsx splicing regulation, we developed a fru splicing assay system. Here we report that tra and tra-2 regulate fru sex-specific splicing in transfected Drosophila cells. Our data show that tra/tra-2 induce female-specific fru splicing by activating the female-specific fru 5′ SS.
MATERIALS AND METHODS
Plasmid constructs.
To make the fru minigene construct fruM+Fwt, a 1.5-kb NotI-EcoRI fragment from the class 1 fru cDNA (54), including the male-specific 5′ SS, was ligated to a 0.645-kb genomic EcoRI fragment from phage 1H (54) spanning the tra/tra-2 repeat elements and the female-specific 5′ SS and cloned into pSK+ (Stratagene). A 1.2-kb genomic PstI fragment from cosmid HXI (54) including the common 3′ SS was placed downstream. A 2.5-kb BamHI fragment including the actin 5C promoter (53) was placed upstream and a 0.5-kB NotI fragment encompassing the tra-2 polyadenylation site (53) was placed downstream of the fru sequences, respectively. Constructs fruM+Fmut, fruM+FΔB-M, and fruFwt are derivatives of construct fruM+Fwt. In construct fruM+Fmut, the sequence of the tra/tra-2 repeat elements was changed from TCAATCAACA to GGCAGCTTAC. In construct fruM+FΔB-M, a 1-kb BbsI-MscI fragment located between the male-specific fru 5′ SS and the repeat elements was deleted. Construct fruFwt lacks the NotI-EcoRI fragment of construct fruM+Fwt, which contains the male-specific fru 5′ SS. fruM+REwt and fruM+REmut are derivatives of construct fruM+FΔB-M. Both the 0.645-kb genomic EcoRI fragment and all fru sequences 4 bp upstream of the male-specific 5′ SS were deleted, and PCR fragments encompassing the wild-type (wt) fru repeat elements and the mutated fru repeat elements, respectively, were inserted upstream of the male-specific 5′ SS instead. The fru PCR primers used were 5′-TCCCCCGGGGAATTCGAGGACGTGTGA-3′ and 5′-TAACCCGGGCGCCAGTTGGTGGGGAT-3′. Construct fruM+REds is a derivative of construct fruM+Fwt lacking the female-specific 5′ SS in which the 0.645-kb genomic EcoRI fragment was replaced by a PCR fragment containing the wt fru repeat elements. Construct dsxF+dsxRE contains genomic dsx sequences including the 114-bp intron between nt 2066 and 3048f (13). Construct dsxF+fruRE is a derivative of construct dsxF+dsxRE in which the dsx repeat region between nt 2741f and 3048f (13) was deleted and a PCR fragment containing the wt fru repeat region was inserted instead. The tra, tra-2, rbp1 and B52 expression constructs are as described previously (28, 53).
The following probes were used. For constructs fruM+Fwt, fruM+FREmut, and fruM+FΔB-M, a 179-bp BsaHI-BbsI fragment spanning the male-specific fru 5′ SS was placed upstream of the 0.645-kb genomic EcoRI fragment. For construct fruFwt, the 0.645-kb genomic EcoRI fragment was used. For construct fruM+REds, the 179-bp BsaHI-BbsI fragment was used. For constructs fruM+REwt and fruM+REmut, EcoRI fragments spanning the inserted repeat regions and the male-specific fru 5′ SS were used. For constructs dsxF+dsxRE and dsxF+fruRE, a dsx genomic fragment spanning nt 2066 to 2711f, including the 114-bp intron (13), was used as a probe.
Transfections and RNase protections.
Transfections and RNase protections were performed essentially as described previously (28, 53). If not indicated otherwise, 10 μg of the minigene constructs and 30 to 100 ng of the cDNA expression constructs were transfected. Quantitations of RNase protection products were done by densitometry and were corrected for the different numbers of labeled residues in the protection products.
RT-PCR experiments.
Reverse transcription (RT)-PCR experiments on RNA isolated from transfected SL2 cells were carried out using the primers m and c as described previously (54). Male-specific fru splicing gave rise to a 164-bp PCR product as expected. Upon cotransfection with tra/tra-2, a PCR product 1,754 bp long was detected, as expected for female-specific fru splicing. Several PCR products 350 to 1,600 bp long were also detected, possibly indicating cryptic splicing.
RESULTS
Regulation of fru sex-specific splicing by tra and tra-2 in transfected Drosophila cells.
To study the regulation of fru sex-specific splicing, we began by establishing a fru splicing assay system in transfected Drosophila Schneider (SL2) cells. As is characteristic for male somatic tissue, Schneider cells express endogenously a functional tra-2 transcript, but no functional tra transcript (53). We constructed a fru minigene fruM+Fwt, driven by an actin promoter, which contains the fru genomic region spanning the male-specific 5′ SS, the tra/tra-2 repeat elements, and the female-specific 5′ SS, fused to the downstream common 3′ SS (see Fig. 2C). In this construct, the fru intron flanked by the female-specific 5′ SS and the common 3′ SS, over 70 kb long (Fig. 1), was shortened to 1.3 kb. Construct fruM+Fwt includes 300 bp of intron sequence downstream of the female-specific 5′ SS and 1,000 bp of intron sequence upstream of the common 3′ SS, respectively. Transient transfections into Drosophila Schneider (SL2) cells were carried out, and fru splicing products were detected by an RNase protection assay.
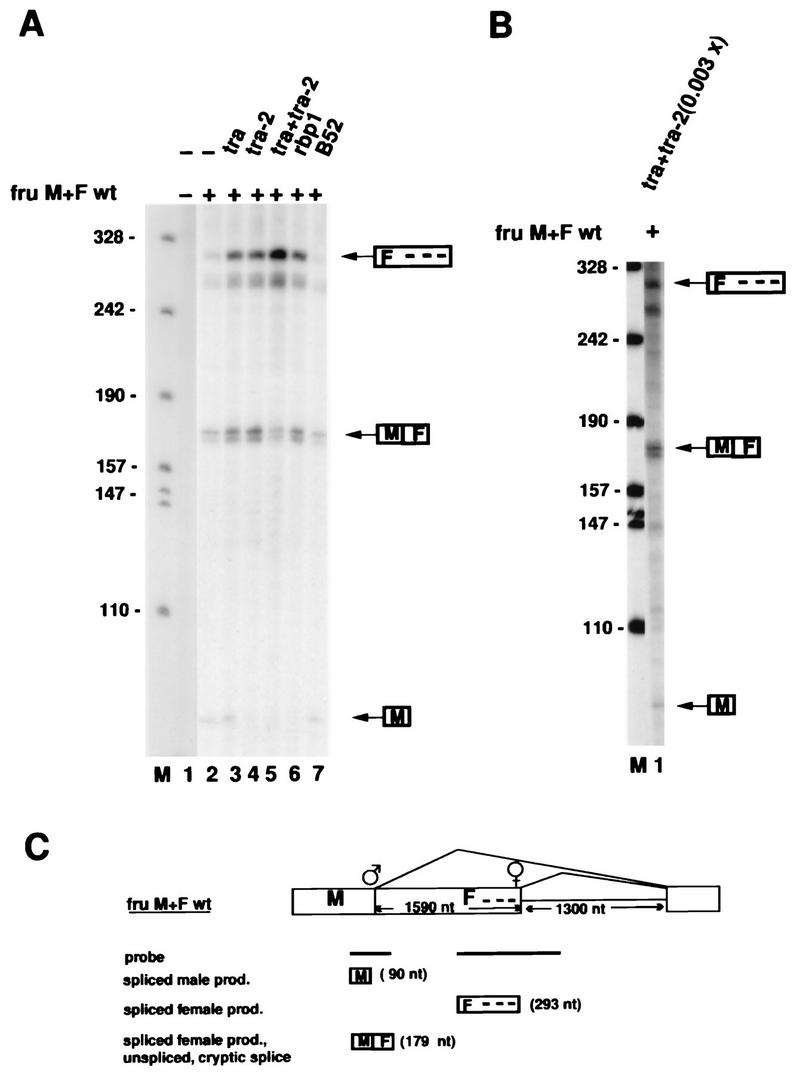
FIG. 2.
Regulation of fru sex-specific splicing in transfected Schneider cells. (A) The fru minigene construct fruM+Fwt was transfected together with various combinations of cDNA expression constructs encoding tra, tra-2, rbp1, and B52 as indicated (28, 53) into Schneider cells, and fru splicing was analyzed by RNase protection assays. The presence (+) or absence (−) of construct fruM+Fwt and of the tra, tra-2, rbp1, and B52 expression constructs in the transfection experiments is indicated above each lane. Splicing products are identified to the right of the autoradiograph. Lane M contains pSK+ DNA cut with HpaII and used as a nucleotide length marker. The positions of size markers (in nucleotides) are shown to the left of the autoradiographs in panels A and B. (B) The amount of cotransfected tra and tra-2 expression constructs was titrated down to 0.3% (0.1 ng) of the amount transfected in panel A. (C) Depiction of construct fruM+Fwt. Exons are represented by boxes, and the intron is represented by a thin horizontal line. The positions of the male-specific and female-specific 5′ SSs are indicated, and male-specific (M) and female-specific (F) exon segments are identified. The three dashes upstream of the female-specific 5′ SS represent the tra/tra-2 repeat elements. The antisense RNase protection probes are indicated directly below the map by horizontal bars. RNase protection products (prod.) derived from male-specific and female-specific fru splicing are given at the bottom of the panel, and the expected product lengths are given in parentheses. The distance between the male-specific and female-specific fru 5′ SSs is 1,590 nt. The intron is 1,300 nt long. Note that the RNase protection probes and the protection products are not drawn to scale. Protection product MF can be obtained as a consequence of female splicing or use of cryptic 5′ splice sites or from unspliced pre-mRNA.
FIG. 1.
Patterns of fru and dsx sex-specific splicing. Schematic drawings of fruitless (fru) and doublesex (dsx) splicing patterns are shown. Only part of the fru gene is depicted. Exons (boxes), introns (thin horizontal lines), and splices (thick lines) are indicated. Female-specific splicing patterns are indicated above and male-specific splicing patterns are indicated below each gene. Male- (M) and female (F)-specific exon segments and common exons (C) are indicated. The tra/tra-2 repeat elements are indicated by thick dashes, and distances of the tra/tra-2 repeat elements to the sex-specific splice sites are given. Female-specific (AUGF) and male-specific (AUGM) translational start codons as well as female-specific (AAF) and male-specific (AAM) polyadenylation sites are shown.
The male-specific fru 5′ SS and the female-specific fru 5′ SS are 1,590 nt apart. To visualize usage of the male and female 5′ SSs simultaneously, the probe used contains two fragments synthesized in one piece which hybridize across the female 5′ SS and the male 5′ SS, respectively (Fig. 2C). Usage of the female and male 5′ SSs are expected to generate the specific RNase protection products F and M, respectively (Fig. 1C). In addition, protection product MF will be generated from transcripts spliced in the female pattern, from unspliced RNA, and also in cases in which there is usage of a cryptic 5′ SS located between the female and male 5′ SSs. No endogenous fru transcript was detected in SL2 cells, as shown in Fig. 2A, lane 1. We observed that in the absence of cotransfected tra/tra-2 expression constructs, the fru minigene fruM+Fwt is spliced mainly in the male pattern (Fig. 2A, lane 2) with a molar ratio of male (M) to female (F) protection products (M/F) of 6. The protection products were quantitated by densitometry, and quantitations were corrected for the different numbers of labeled residues in the protection products. No unspliced fruM+Fwt RNA was detected (not shown). A low ratio of protection product F to protection product MF (F/MF = 0.17) in the absence of cotransfected tra/tra-2 (Fig. 2A, lane 2) probably reflects the presence of cryptic splicing products (Fig. 1C), as also indicated by RT-PCR experiments (data not shown; see Materials and Methods).
To test for regulation of fru sex-specific splicing by tra and tra-2, we cotransfected tra and tra-2 expression constructs. Significantly, when both tra and tra-2 expression constructs were cotransfected together with the fru minigene, a shift to purely female-specific 5′ SS usage occurs (Fig. 2A, lane 5), as indicated by an increase in spliced female product F, by the absence of detectable product M, and by a ratio of protection product F to protection product MF (F/MF) of 1.1. Switching from the male-specific 5′ SS to the female-specific 5′ SS by tra/tra-2 was also confirmed by RT-PCR assay (data not shown; see Materials and Methods) and depends on the concentrations of the tra/tra-2 expression constructs (compare Fig. 2B, lane 1, and Fig. 2A, lane 5). In Fig. 2B, lane 1, the ratio of product M to product F is 1.38 and the ratio of product F to product MF is 0.19. Expression of tra alone (Fig. 2A, lane 3) or tra-2 (Fig. 2A, lane 4) induces only a slight shift to female-specific fru splicing, as indicated by a ratio of protection product F to protection product MF (F/MF) of 0.28. Thus, transfection of both tra and tra-2 is required to switch fru splicing precisely and efficiently from the male-specific 5′ SS to the female-specific 5′ SS in cultured Schneider cells. These results in our assay system are consistent with results obtained in Drosophila, where female flies require both tra and tra-2 for female-specific fru splicing (54).
We also cotransfected the Drosophila SR protein RBP1 in our fru splicing assay. Expression of RBP1 in transfected Schneider cells was shown to induce female-specific dsx splicing (28). We found that transfection of RBP1 can also induce some female-specific fru splicing (Fig. 2A, lane 6), suggesting that RBP1 may also play a role in the regulation of fru splicing. A potential RBP1 target site (ATCCCCA) (28) is present 12 nt upstream of the female-specific fru 5′ SS. As a control, transfection of the Drosophila SR protein B52 (16, 52) did not affect fru splicing (Fig. 2A, lane 7).
The tra/tra-2 repeat elements are required for fru splicing regulation by tra/tra-2.
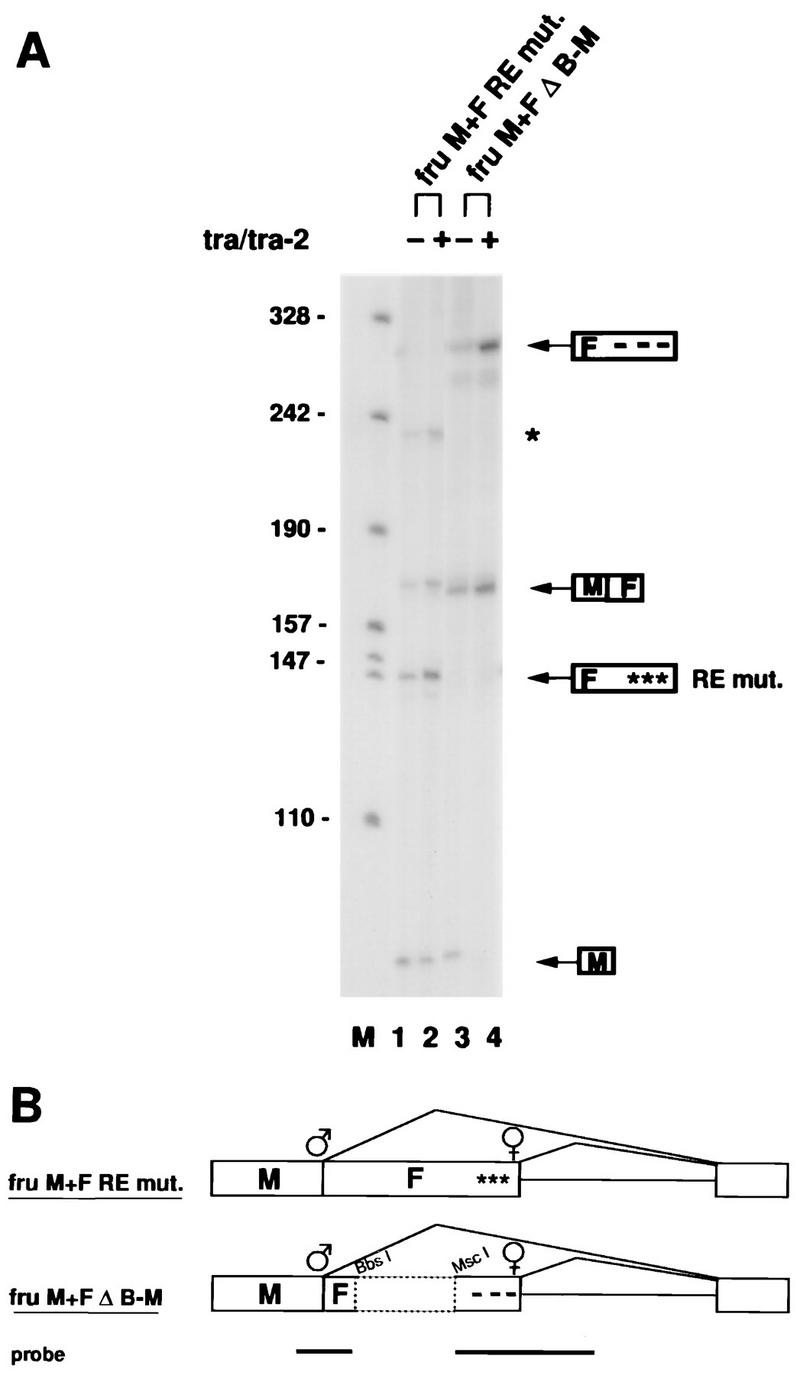
To determine whether the tra/tra-2 repeat elements are essential for regulation of fru splicing by tra and tra-2, we tested a construct, fruM+FREmut, in which the sequence of the conserved part of the tra/tra-2 repeat elements was changed from TCAATCAACA to GGCAGCTTAC. The probe used is the same as in Fig. 2, and since it contains wt tra/tra-2 repeat elements, generates short loop-outs when hybridized to the mutant tra/tra-2 repeat elements in fruM+FREmut, resulting in a shortened female-specific RNase protection product (Fig. 3A, lanes 1 and 2). Inefficient cutting of these short loop-outs by RNase A also generates a larger protection product derived from spliced female product which is marked by an asterisk (Fig. 3A). We observed that in construct fruM+FREmut, switching to female fru splicing by tra and tra-2 was almost completely blocked, as indicated by the presence of significant amounts of male splicing product M in the presence of cotransfected tra and tra-2 (Fig. 3A, lane 2). In contrast, deletion of a 1-kb fragment between the repeat elements and the male-specific 5′ SS, as in construct fruM+FΔB-M (Fig. 3B), did not affect regulation of fru splicing by tra and tra-2 (Fig. 3A, lanes 3 and 4). These findings show that the tra/tra-2 repeat elements are required for regulation of fru splicing by tra and tra-2, suggesting that tra and tra-2 promote female-specific fru splicing by acting through these elements.
FIG. 3.
cis-acting elements required for regulation of fru splicing by tra/tra-2. (A) Constructs carrying mutations within the tra/tra-2 repeat elements (construct fruM+FREmut) or a deletion of sequences between the male-specific fru 5′ SS and the tra/tra-2 repeat sequences (construct fruM+FΔB-M) were transfected in the absence (−) or presence (+) of cotransfected tra/tra-2 as indicated above the RNase protection assay autoradiograph. RNase protection products are indicated to the right of the autoradiograph and are as described in the legend to Fig. 2C. An additional RNase protection product (RE mut.) is derived from female-specific splicing of construct fruM+REmut. An incompletely cleaved protection product derived from female-specific splicing of construct fruF+MRE mut is indicated by an asterisk. The positions of size markers (in nucleotides) are shown to the left of the autoradiograph. Lane M contains pSK+ DNA cut with HpaII. (B) Illustration of constructs fruM+FREmut and fruM+FΔB-M. fruM+FREmut contains mutated tra/tra-2 repeat elements indicated by asterisks. In fruM+FΔB-M, a 1-kb BbsI-MscI fragment indicated by a broken-line box was deleted. The probe used in these experiments is as described in the legend to Fig. 2.
Evidence for activation of the female-specific fru 5′ SS as the mechanism of fru splicing regulation.
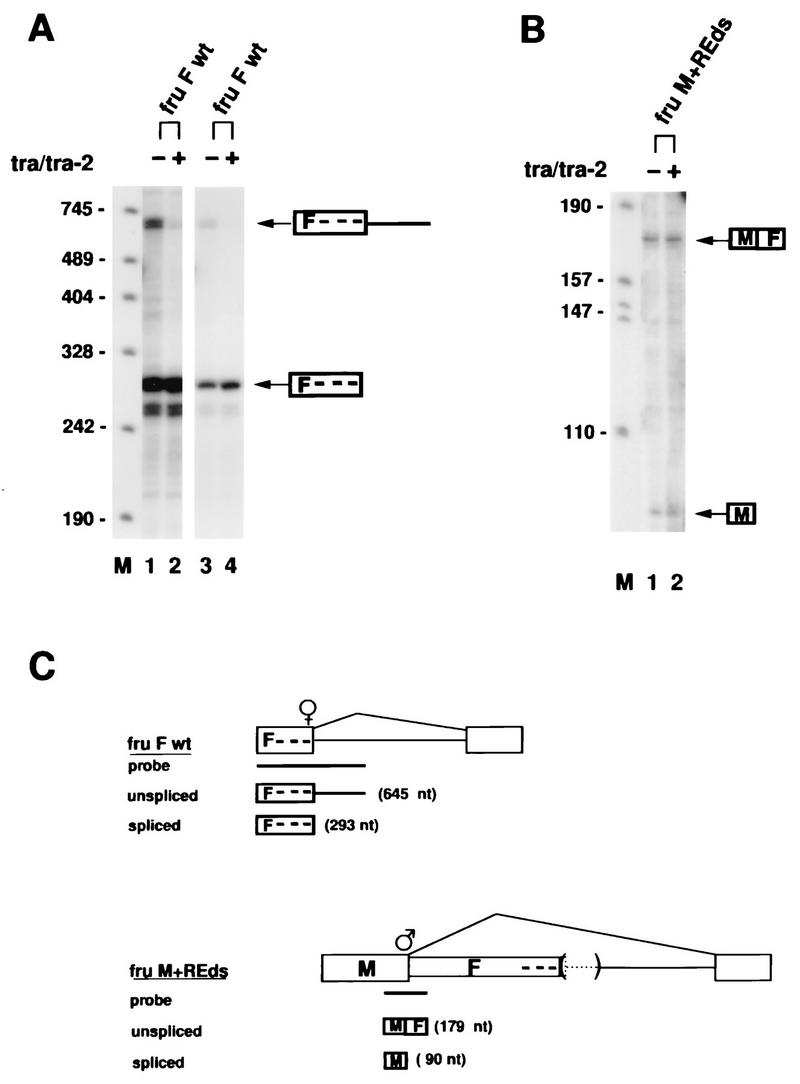
To determine whether tra and tra-2 induce female-specific fru splicing by blocking the male-specific 5′ SS or by activating the female-specific 5′ SS, we tested constructs that contain upstream of the common 3′ SS (i) either the male-specific 5′ SS, including the tra/tra-2 repeats (construct fruM+REds [Fig. 4C]), or (ii) the female-specific 5′ SS, including the tra/tra-2 repeats (construct fruFwt [Fig. 4C]). The tra/tra-2 repeat elements were included at the authentic position relative to the splice sites, 1,352 nt downstream of the male-specific 5′ SS and 38 nt upstream of the female-specific 5′ SS, respectively. We did observe activation of the isolated female-specific 5′ SS by tra/tra-2, as indicated by the disappearance of unspliced RNA upon cotransfection with tra/tra-2 (Fig. 4A, lanes 1 and 2). A shorter exposure of the same gel (Fig. 4A, lanes 3 and 4) also shows an increase in spliced female product by tra/tra-2 (Fig. 4A, lane 4), suggesting that the effect observed in Fig. 4A, lane 2, is not due to instability of unspliced pre-mRNA. While spliced female product is seen to increase in Fig. 4A, lane 4, the background bands below the spliced female product in Fig. 4A, lanes 3 and 4, remain constant, suggesting equal loading of RNA. Usage of the isolated female-specific 5′ SS in the absence of cotransfected tra and tra-2 (Fig. 4A, lane 1) is probably due to the lack of the competing male-specific fru 5′ SS, since the deletion of sequences located between the male-specific fru 5′ SS and the fru repeat region, as in construct fruM+FΔB-M (Fig. 3B), does not affect fru splicing. Similarly, the isolated female-specific dsx 3′ SS is also used in the absence of cotransfected tra/tra-2, as shown previously (53) and as also reproduced in this study (see Fig. 6). In contrast, the male-specific fru 5′ SS was found to be unaffected by cotransfection with tra/tra-2 (Fig. 4B, lanes 1 and 2). Taken together, these results suggest that tra/tra-2 induce female-specific fru splicing by activating the female-specific fru 5′ SS, as also suggested by the proximity of the tra/tra-2 repeat elements to the female-specific 5′ SS. Thus, in fru, the tra/tra-2 repeat region functions as an upstream splicing enhancer.
FIG. 4.
Testing the activation and the blockage model of fru regulation. (A) Construct fruFwt containing the female-specific fru 5′ SS and the tra/tra-2 repeat elements was transfected in the absence (−) or presence (+) of cotransfected tra/tra-2 as indicated above the RNase protection assay autoradiograph. Lanes 3 and 4 show a shorter exposure of lanes 1 and 2, respectively. Lane M contains pSK+ DNA cut with HpaII. The positions of size markers (in nucleotides) are shown to the left of the autoradiographs in panels A and B. (B) Construct fruM+REds containing the male-specific fru 5′ SS and the tra/tra-2 repeat elements was transfected in the absence (−) or presence (+) of cotransfected tra/tra-2 as indicated above the RNase protection assay autoradiograph. RNase protection products derived from spliced and unspliced pre-mRNAs of constructs fruFwt and fruM+REds are indicated to the right of the autoradiograph. (C) Illustration of constructs fruFwt and fruM+REds. fruFwt contains the female-specific fru 5′ SS, and fruM+REds contains the male-specific fru 5′ SS. The tra/tra-2 repeat elements were included at the correct distance upstream of the female-specific 5′ SS and downstream of the male-specific 5′ SS, respectively. The deletion of the female-specific fru 5′ SS in construct in fruM+REds is indicated by parentheses. The probes used in the RNase protection experiments are represented by horizontal bars below each construct. RNase protection products corresponding to spliced and unspliced pre-mRNAs are shown below each construct.
FIG. 6.
The fru and dsx repeat regions are functionally interchangeable. (A) Construct dsxF+dsxRE containing the dsx repeat region downstream of the female-specific dsx 3′ SS and construct dsxF+fruRE containing the fru repeat region downstream of the female-specific dsx 3′ SS were transfected in the absence (−) or presence (+) of cotransfected tra/tra-2 as indicated above the RNase protection assay autoradiograph. RNase protection products derived from spliced and unspliced pre-mRNAs are indicated to the right of the autoradiograph and are as shown in panel B. The slightly lower mobility of the protection product F derived from construct dsxF+fruRE (lanes 3 and 4) compared to that of product F derived from construct dsxF+dsxRE (lanes 1 and 2) is due to a short stretch of polylinker present in construct dsxF+fruRE and in the probe, but not in construct dsxF+dsxRE. Lane M contains pSK+ DNA cut with HpaII. The positions of size markers (in nucleotides) are indicated to the left of the autoradiograph. (B) Illustration of constructs dsxF+dsxRE and dsxF+fruRE. The fru repeat region is indicated by three dashes, and the dsx repeat region is indicated by six dashes. The common (C) and the female (F) dsx exon are indicated (compare diagram with that in Fig. 1). The probe used in the RNase protection experiments is represented by a horizontal bar below the constructs. RNase protection products corresponding to spliced and unspliced pre-mRNAs are shown at the bottom of the panel.
The fru repeat region is sufficient to promote the activation of a 5′ SS.
We next wanted to address the question of whether the tra/tra-2 repeat region in fru is sufficient to promote the activation of a 5′ SS by tra/tra-2. Since we showed that the male-specific fru 5′SS is normally unaffected by tra/tra-2 (Fig. 4), we inserted a 300-bp fragment of fru containing the tra/tra-2 repeats 4 nt upstream of the male-specific fru 5′ SS (construct fruM+REwt [Fig. 5B]). Interestingly, spliced male product was detected upon cotransfection with tra/tra-2 (Fig. 5A, lane 2), suggesting activation of the male-specific fru 5′ SS by tra/tra-2 in this hybrid construct. For a control, when the same fragment carrying mutations within the tra/tra-2 repeat elements was inserted 4 nt upstream of the male-specific fru 5′ SS, as in construct fruM+REmut (Fig. 5B), no spliced male product in the presence of tra/tra-2 was detected. Two separate probes, complementary to the constructs fruM+REwt and fruM+REmut, respectively, were used in these experiments. Thus, the tra/tra-2 repeat elements are essential to promote the activation of a heterologous 5′ SS by tra/tra-2. Lack of usage of the male-specific 5′ SS in the constructs fruM+REwt and fruM+REmut in the absence of cotransfected tra/tra-2 (Fig. 5A, lanes 1 and 3) was found to be due to the deletion of a stretch of sequence upstream of the male-specific 5′ SS in these constructs. The insertion of the fru repeat region in either orientation does not affect the usage of the male-specific fru 5′ SS in the absence of cotransfected tra/tra-2 (data not shown).
FIG. 5.
Activation of a heterologous 5′ SS by tra/tra-2. (A) Construct fruM+REwt containing wt tra/tra-2 repeat elements upstream of the male-specific fru 5′ SS and construct fruM+REmut containing mutant tra/tra-2 repeat elements upstream of the male-specific fru 5′ SS were transfected in the absence (−) or presence (+) of cotransfected tra/tra-2 as indicated above the RNase protection assay autoradiograph. RNase protection products derived from spliced and unspliced pre-mRNAs are indicated to the right of the autoradiograph and are as described in the legend to Fig. 5B. Lane M contains pSK+ DNA cut with HpaII. The positions of size markers (in nucleotides) are shown to the left of the autoradiograph. (B) Illustration of constructs fruM+REwt and fruM+REmut. The wt fru repeat region is indicated by three dashes, and the mutated fru repeat region is indicated by three asterisks. The repeat mutations in construct fruM+REmut are as in construct fruM+FREmut. For each construct, a specific RNase protection probe hybridizing across the splice sites was generated and is indicated by a horizontal bar below the constructs. RNase protection products corresponding to spliced and unspliced pre-mRNAs are shown below the constructs.
The fru repeat region and the dsx repeat region are functionally interchangeable.
Our study shows that in fru, the tra/tra-2 repeat region functions as an upstream splicing enhancer required for the activation of the female-specific fru 5′ SS by tra/tra-2. In contrast, tra/tra-2 activate the female-specific dsx 3′ SS by recognizing tra/tra-2 repeat elements present downstream of this 3′ SS (27, 30, 53). To address the question of whether the tra/tra-2 repeat regions in fru and dsx are functionally interchangeable, we inserted the tra/tra-2 repeat region from fru downstream of the female-specific dsx 3′ SS (Fig. 6B). For the activation of the female-specific dsx 3′ SS by tra/tra-2 to occur, the dsx repeat region has to be positioned at a distance of 300 bp downstream of this 3′ SS (65). To ensure positioning of the fru repeat region at this precise location, we placed the fru repeat region downstream of a dsx PCR fragment ending immediately upstream of the dsx repeat region, generating construct dsxF+fruRE (Fig. 6B), which lacks the competing downstream male-specific dsx 3′ SS (compare diagram with that in Fig. 1). For a control, splicing of a construct dsxF+dsxRE (Fig. 6B) containing the dsx repeat region was tested. As shown previously, due to default usage of the isolated female-specific dsx 3′ SS, activation of the female-specific dsx 3′ splice site by tra/tra-2 is indicated by a decrease in unspliced RNA in a construct lacking the male-specific dsx 3′ SS (53) and is also demonstrated here (Fig. 6A, lanes 1 and 2). We found that the fru repeat region promotes the activation of the female-specific dsx 3′ SS by tra/tra-2 almost as efficiently (Fig. 6A, lanes 3 and 4) as the dsx repeat region (Fig. 6A, lanes 1 and 2), demonstrating that the fru and dsx repeat regions can be functionally interchangeable in terms of dsx regulation.
DISCUSSION
Here we report that tra and tra-2 induce female-specific fru splicing by activating the female-specific fru 5′ SS. Our results also suggest that tra and tra-2 regulate fru splicing directly, since we show that fru splicing regulation requires the tra/tra-2 repeat elements recognized by tra and tra-2. Hence, the tra and tra-2 genes are functioning directly upstream of the fru gene. Although our results explain how female-specific fru splicing is achieved, it remains to be determined how male flies ensure exclusively male fru splicing. Several scenarios are conceivable. The female-specific fru 5′ SS could be a weak 5′ SS, causing default usage of the male-specific fru 5′ SS in males when tra is not expressed. This model is analogous to the situation in dsx, where the regulated female-specific dsx 3′ SS is a weak 3′ SS due to a purine-rich polypyrimidine tract (13). Both the sequences of the female (TCG/GTAAGT) and male (TAG/GTAAGC) fru 5′ SSs match the Drosophila 5′ SS consensus sequence MAG/GTRAGT (47) in 7 of 9 nt. However, the precise sequence context of the 9-nt consensus sequence of the 5′ SS has been proposed to affect 5′ SS usage (1). Alternatively, yet unidentified trans-acting factors could either block the female-specific fru 5′ SS or activate the male-specific fru 5′ SS in male flies. Our observation that the deletion of sequences upstream of the male-specific fru 5′ SS inhibits male fru splicing could indicate that a mechanism for activating the male-specific fru 5′ SS exists.
Activation of the proximal female-specific fru 5′ SS by tra and tra-2 represents a previously unknown functional property of tra/tra-2. Although increasing the concentration of the human SR protein ASF/SF2 induces the usage of proximal 5′ SSs (22, 38), the mechanism of action of ASF/SF2 appears to be fundamentally different from what we know about the mechanism that leads to the usage of a proximal 5′ SS in fru. ASF/SF2 has been shown to have a general affinity to 5′ SSs (77) and stabilizes the recognition of 5′ SSs by U1 small nuclear ribonucleoprotein particle (snRNP) (20, 59). Increasing the affinity of U1 snRNP to all 5′ SS in a pre-mRNA molecule has been proposed to lead to the preferential usage of the 5′ SS closest to a 3′ SS (20). Splicing to a distal 5′ SS in the human caldesmon gene is promoted by a purine-rich downstream splicing enhancer, although the regulatory factor(s) involved is not known (31). In contrast, tra/tra-2 specifically select a 5′ SS for activation depending on the presence of the upstream tra/tra-2 repeat elements.
Involvement of tra/tra-2 in both 5′ SS activation in fru and 3′ SS activation in dsx shows that tra and tra-2 are multifunctional splicing regulators. tra-2 (but not tra) is also involved in regulating splicing of the tra-2 pre-mRNA (45) and the exuperantia pre-mRNA (26) in the male germ line, although it is not known whether these tra-2 functions involve tra/tra-2 repeat elements. Other examples of multifunctional splicing factors have been described both in Drosophila and in humans. The Drosophila Sxl protein regulates splicing of the Sxl (9), tra (11, 49, 58), and msl-2 (7, 34, 75) pre-mRNAs and affects msl-2 translation (8, 35). The human U1 snRNP-specific protein A is also involved in pre-mRNA polyadenylation (10, 42). Functional activity of the fru repeat region in promoting dsx 3′ SS activation by tra/tra-2 suggests that the mechanisms of 5′ SS activation and 3′ SS activation are surprisingly similar. 5′ SSs and 3′ SSs are defined by distinct sequence motifs and are specifically recognized early in spliceosome assembly by general splicing factors. While 5′ SSs are recognized by U1 snRNP (76) and by the SR protein ASF/SF2 (77), U2 snRNPs and U2AF bind to the 3′ ends of introns (74). SR proteins have also been shown to engage in protein-protein interactions both with U1 snRNPs and with U2AF (37, 68). Since tra and tra-2 can interact with U2AF and with SR proteins (29, 68), it is conceivable that by binding to tra/tra-2 repeat elements, tra/tra-2 can stabilize the recognition of both a specific 5′ SS and a 3′ SS by general splicing factors. Thus, activation of a 5′ SS versus a 3′ SS by tra/tra-2 could depend solely on the interactions of tra/tra-2 with general splicing factors recognizing 5′ SSs on the one hand and 3′ SSs on the other. Alternatively, additional regulatory factors specific to fru 5′ SS activation and/or dsx 3′ SS activation could be involved. There is evidence that the regulation of dsx splicing involves dsx-specific features. The position of the dsx repeat region 300 nt downstream of the female-specific dsx 3′ SS is conserved between D. melanogaster and Drosophila virilis (12a) and is essential for dsx regulation by tra/tra-2 (65). In contrast, purine-rich downstream splicing enhancers appear to be generally found significantly closer to the upstream intron, at distances ranging from 3 to 117 nt (24, 40, 51, 66), and placing a purine-rich splicing enhancer beyond 293 nt downstream of the 3′ SS inhibits splice site activation (40). Furthermore, activation of the female-specific dsx 3′ SS involves the Drosophila SR protein RBP1 which recognizes evolutionarily conserved RNA target sequences present within the unusual purine-rich polypyrimidine tract of the female-specific dsx 3′ SS and within the dsx repeat region (28). Further experiments are needed to identify possible additional players in fru and dsx splicing regulation and to define the molecular interactions involved.
ACKNOWLEDGMENT
This work was supported by grants from the National Institutes of Health to B.S.B.
REFERENCES
- 1.Aebi M, Hornig H, Weissman C. 5′ cleavage site in eukaryotic pre-mRNA splicing is determined by the overall 5′ splice region, not by the conserved 5′ GU. Cell. 1987;50:237–246. doi: 10.1016/0092-8674(87)90219-4. [DOI] [PubMed] [Google Scholar]
- 2.Amrein H, Gorman M, Nöthiger R. The sex-determining gene tra-2 of Drosophila encodes a putative RNA binding protein. Cell. 1988;55:1025–1035. doi: 10.1016/0092-8674(88)90247-4. [DOI] [PubMed] [Google Scholar]
- 3.Amrein H, Hedley M L, Maniatis T. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by transformer 2. Cell. 1994;76:735–746. doi: 10.1016/0092-8674(94)90512-6. [DOI] [PubMed] [Google Scholar]
- 4.Ayane M, Preuss U, Köhler G, Nielsen P J. A differentially expressed murine RNA encoding a protein with similarities to two types of nucleic acid binding proteins. Nucleic Acids Res. 1991;19:1273–1278. doi: 10.1093/nar/19.6.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker B S. Sex in flies: the splice of life. Nature. 1989;340:521–524. doi: 10.1038/340521a0. [DOI] [PubMed] [Google Scholar]
- 6.Banfi S, Borsani G, Rossi E, Bernard L, Guffanti A, Rubboli F, Marchitiello A, Giglio S, Coluccia E, Zolla M, Zuffardi O, Ballabio A. Identification and mapping of human cDNAs homologous to Drosophila mutant genes through EST database searching. Nature Genet. 1996;13:167–174. doi: 10.1038/ng0696-167. [DOI] [PubMed] [Google Scholar]
- 7.Bashaw G J, Baker B S. The msl-2 dosage compensation gene of Drosophila encodes a putative DNA-binding protein whose expression is sex specifically regulated by Sex-lethal. Development. 1995;121:3245–3258. doi: 10.1242/dev.121.10.3245. [DOI] [PubMed] [Google Scholar]
- 8.Bashaw G J, Baker B S. The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell. 1997;89:789–798. doi: 10.1016/s0092-8674(00)80262-7. [DOI] [PubMed] [Google Scholar]
- 9.Bell L R, Horabin J I, Schedl P, Cline T W. Positive autoregulation of Sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell. 1991;65:229–239. doi: 10.1016/0092-8674(91)90157-t. [DOI] [PubMed] [Google Scholar]
- 10.Boelens W C, Jansen E J R, van Venrooij W J, Stripecke R, Mattaj I W, Gunderson S I. The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell. 1993;72:881–892. doi: 10.1016/0092-8674(93)90577-d. [DOI] [PubMed] [Google Scholar]
- 11.Boggs R T, Gregor P, Idriss S, Belote J M, McKeown M. Regulation of sexual differentiation in D. melanogaster via alternative splicing of RNA from the transformer gene. Cell. 1987;50:739–747. doi: 10.1016/0092-8674(87)90332-1. [DOI] [PubMed] [Google Scholar]
- 12.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 12a.Burtis, K. C. Personal communication.
- 13.Burtis K C, Baker B S. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56:997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- 14.Caputi M, Casari G, Guenzi S, Tagliabue R, Sidoli A, Melo C A, Baralle F E. A novel bipartite splicing enhancer modulates the differential processing of the human fibronectin EDA exon. Nucleic Acids Res. 1994;22:1018–1022. doi: 10.1093/nar/22.6.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cavaloc Y, Popielarz M, Fuchs J-P, Gattoni R, Stévenin J. Characterization and cloning of the human splicing factor 9G8: a novel 35kDa factor of the serine/arginine protein family. EMBO J. 1994;13:2639–2649. doi: 10.1002/j.1460-2075.1994.tb06554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Champlin D T, Frasch M, Saumweber H, Lis J T. Characterization of a Drosophila protein associated with boundaries of transcriptionally active chromatin. Genes Dev. 1991;5:1611–1621. doi: 10.1101/gad.5.9.1611. [DOI] [PubMed] [Google Scholar]
- 17.Dauwalder B, Amaya-Manzanares F, Mattox W. A human homologue of the Drosophila sex determination factor transformer-2 has conserved splicing regulatory functions. Proc Natl Acad Sci USA. 1996;93:9004–9009. doi: 10.1073/pnas.93.17.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diamond R H, Du K, Lee V M, Mohn K L, Haber B A, Tewari D S, Taub R. Novel delayed-early and highly insulin-induced growth response genes. Identification of HRS, a potential regulator of alternative pre-mRNA splicing. J Biol Chem. 1993;268:15185–15192. [PubMed] [Google Scholar]
- 19.Dominski Z, Kole R. Identification of exon sequences involved in splice site selection. J Biol Chem. 1994;269:23590–23596. [PubMed] [Google Scholar]
- 20.Eperon I C, Ireland D C, Smith R A, Mayeda A, Krainer A R. Pathways for selection of 5′ splice sites by U1 snRNPs and SF2/ASF. EMBO J. 1993;12:3607–3617. doi: 10.1002/j.1460-2075.1993.tb06034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu X-D, Maniatis T. Isolation of a complementary DNA that encodes the mammalian splicing factor SC35. Science. 1992;256:535–538. doi: 10.1126/science.1373910. [DOI] [PubMed] [Google Scholar]
- 22.Ge H, Manley J L. A protein factor, ASF, controls cell-specific alternative splicing of SV40 early pre-mRNA in vitro. Cell. 1990;62:25–34. doi: 10.1016/0092-8674(90)90236-8. [DOI] [PubMed] [Google Scholar]
- 23.Ge H, Zuo P, Manley J L. Primary structure of the human splicing factor ASF reveals similarities with Drosophila regulators. Cell. 1991;66:373–382. doi: 10.1016/0092-8674(91)90626-a. [DOI] [PubMed] [Google Scholar]
- 24.Gontarek R R, Derse D. Interactions among SR proteins, an exonic splicing enhancer, and a lentivirus Rev protein regulate alternative splicing. Mol Cell Biol. 1996;16:2325–2331. doi: 10.1128/mcb.16.5.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goralski T J, Edstrom J E, Baker B S. The sex determination locus transformer-2 of Drosophila encodes a polypeptide with similarity to RNA binding proteins. Cell. 1989;56:1011–1018. doi: 10.1016/0092-8674(89)90634-x. [DOI] [PubMed] [Google Scholar]
- 26.Hazelrigg T, Tu C. Sex-specific processing of the Drosophilia exuperantia transcript is regulated in male germ cells by the tra-2 gene. Proc Natl Acad Sci USA. 1994;91:10752–10756. doi: 10.1073/pnas.91.22.10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hedley M L, Maniatis T. Sex-specific splicing and polyadenylation of dsx pre-mRNA requires a sequence that binds specifically to a tra-2 protein in vitro. Cell. 1991;65:579–586. doi: 10.1016/0092-8674(91)90090-l. [DOI] [PubMed] [Google Scholar]
- 28.Heinrichs V, Baker B S. The Drosophila SR protein RBP1 contributes to the regulation of doublesex alternative splicing by recognizing RBP1 RNA target sequences. EMBO J. 1995;14:3987–4000. doi: 10.1002/j.1460-2075.1995.tb00070.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heinrichs V, Baker B S. In vivo analysis of the functional domains of the Drosophila splicing regulator RBP1. Proc Natl Acad Sci USA. 1997;94:115–120. doi: 10.1073/pnas.94.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoshijima K, Inoue K, Higuchi I, Sakamoto H, Shimura Y. Control of doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science. 1991;252:833–836. doi: 10.1126/science.1902987. [DOI] [PubMed] [Google Scholar]
- 31.Humphrey M B, Bryan J, Cooper T A, Berget S M. A 32-nucleotide exon-splicing enhancer regulates usage of competing 5′ splice sites in a differential internal exon. Mol Cell Biol. 1995;15:3979–3988. doi: 10.1128/mcb.15.8.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inoue K, Hoshijima K, Higuchi I, Sakamoto H, Shimura Y. Binding of the Drosophila transformer and transformer-2 proteins to the regulatory elements of doublesex primary transcript for sex-specific RNA processing. Proc Natl Acad Sci USA. 1992;89:8092–8096. doi: 10.1073/pnas.89.17.8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ito H, Fujitani K, Usui K, Shimizu-Nishikawa K, Tanaka S, Yamamoto D. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc Natl Acad Sci USA. 1996;93:9687–9692. doi: 10.1073/pnas.93.18.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelley R L, Solovyeva I, Lyman L M, Richman R, Solovyev V, Kuroda M I. Expression of msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell. 1995;81:867–877. doi: 10.1016/0092-8674(95)90007-1. [DOI] [PubMed] [Google Scholar]
- 35.Kelley R L, Wang J, Bell L, Kuroda M I. Sex-lethal controls dosage compensation in Drosophila by a non-splicing mechanism. Nature. 1997;387:195–199. doi: 10.1038/387195a0. [DOI] [PubMed] [Google Scholar]
- 36.Kim Y-J, Zuo P, Manley J L, Baker B S. The Drosophila RNA binding protein RBP1 is localized to transcriptionally active sites of chromosomes and shows a functional similarity to human splicing factor ASF/SF2. Genes Dev. 1992;6:2569–2579. doi: 10.1101/gad.6.12b.2569. [DOI] [PubMed] [Google Scholar]
- 37.Kohtz J D, Jamison S F, Will C L, Zuo P, Lührmann R, Garcia-Blanco M A, Manley J L. Protein-protein interactions and 5′ splice site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 38.Krainer A R, Conway G C, Kozak D. The essential pre-mRNA splicing factor SF2 influences 5′ splice site selection by activating proximal sites. Cell. 1990;62:35–42. doi: 10.1016/0092-8674(90)90237-9. [DOI] [PubMed] [Google Scholar]
- 39.Krainer A R, Mayeda A, Kozak D, Binns G. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell. 1991;66:383–394. doi: 10.1016/0092-8674(91)90627-b. [DOI] [PubMed] [Google Scholar]
- 40.Lavigueur A, La Branche H, Kornblihtt A R, Chabot B. A splicing enhancer in the human fibronectin alternate ED1 exon interacts with SR proteins and stimulates U2 snRNP binding. Genes Dev. 1993;7:2405–2417. doi: 10.1101/gad.7.12a.2405. [DOI] [PubMed] [Google Scholar]
- 41.Li H, Bingham P M. Arginine/serine-rich domains of the su(wa) and tra RNA processing regulators target proteins to a subnuclear compartment implicated in splicing. Cell. 1991;67:335–342. doi: 10.1016/0092-8674(91)90185-2. [DOI] [PubMed] [Google Scholar]
- 42.Lutz C S, Alwine J C. Direct interaction of the U1 snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev. 1994;8:576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- 43.Lynch K W, Maniatis T. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 1996;10:2089–2101. doi: 10.1101/gad.10.16.2089. [DOI] [PubMed] [Google Scholar]
- 44.Matsuo N, Ogawa S, Imai Y, Takagi T, Tohyama M, Stern D, Wanaka A. Cloning of a novel RNA binding polypeptide (RA301) induced by hypoxia/reoxygenation. J Biol Chem. 1995;270:28216–28222. doi: 10.1074/jbc.270.47.28216. [DOI] [PubMed] [Google Scholar]
- 45.Mattox W, Baker B S. Autoregulation of the splicing of transcripts from the transformer-2 gene of Drosophila. Genes Dev. 1991;5:786–796. doi: 10.1101/gad.5.5.786. [DOI] [PubMed] [Google Scholar]
- 46.Min H, Chan R, Black D L. The generally expressed hnRNP F is involved in a neural-specific pre-mRNA splicing event. Genes Dev. 1995;9:2659–2671. doi: 10.1101/gad.9.21.2659. [DOI] [PubMed] [Google Scholar]
- 47.Mount S M, Burks C, Hertz G, Stormo G D, White O, Fields C. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 1992;20:4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nagoshi R, Baker B S. Regulation of sex-specific RNA splicing at the Drosophila doublesex gene: cis-acting mutations in exon sequences alter sex-specific RNA splicing patterns. Genes Dev. 1990;4:89–97. doi: 10.1101/gad.4.1.89. [DOI] [PubMed] [Google Scholar]
- 49.Nagoshi R N, McKeown M, Burtis K C, Belote J M, Baker B S. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell. 1988;53:229–236. doi: 10.1016/0092-8674(88)90384-4. [DOI] [PubMed] [Google Scholar]
- 50.Peng X, Mount S M. Genetic enhancement of RNA-processing defects by a dominant mutation in B52, the Drosophila gene for an SR protein splicing factor. Mol Cell Biol. 1995;15:6273–6282. doi: 10.1128/mcb.15.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramchatesingh J, Zahler A M, Neugebauer K M, Roth M B, Cooper T A. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15:4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roth M B, Zahler A M, Stolk J A. A conserved family of nuclear phosphoproteins localized to sites of polymerase II transcription. J Cell Biol. 1991;115:587–596. doi: 10.1083/jcb.115.3.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryner L C, Baker B S. Regulation of doublesex pre-mRNA processing occurs by 3′-splice site activation. Genes Dev. 1991;5:2071–2085. doi: 10.1101/gad.5.11.2071. [DOI] [PubMed] [Google Scholar]
- 54.Ryner L C, Goodwin S F, Castrillon D H, Anand A, Villela A, Baker B S, Hall J C, Taylor B J, Wasserman S A. Control of male sexual behaviour and sexual orientation in Drosophila by the fruitless gene. Cell. 1996;87:1079–1089. doi: 10.1016/s0092-8674(00)81802-4. [DOI] [PubMed] [Google Scholar]
- 55.Screaton G R, Cáceres J F, Mayeda A, Bell M V, Plebanski M, Jackson D G, Bell J I, Krainer A R. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Segade F, Hurle B, Claudio E, Ramos S, Lazo P S. Molecular cloning of a mouse homologue for the Drosophila splicing regulator Tra-2. FEBS Lett. 1996;387:152–156. doi: 10.1016/0014-5793(96)00496-6. [DOI] [PubMed] [Google Scholar]
- 57.Siebel C W, Kanaar R, Rio D C. Regulation of tissue-specific P-element pre-mRNA splicing requires the RNA-binding protein PSI. Genes Dev. 1994;8:1713–1725. doi: 10.1101/gad.8.14.1713. [DOI] [PubMed] [Google Scholar]
- 58.Sosnowski B A, Belote J M, McKeown M. Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell. 1989;58:449–459. doi: 10.1016/0092-8674(89)90426-1. [DOI] [PubMed] [Google Scholar]
- 59.Staknis D, Reed R. SR proteins promote the first specific recognition of pre-mRNA and are present with the U1 small nuclear ribonucleoprotein particle in a general splicing enhancer complex. Mol Cell Biol. 1994;14:7670–7682. doi: 10.1128/mcb.14.11.7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun Q, Mayeda A, Hampson R K, Krainer A R, Rottman F. General splicing factor SF2/ASF promotes alternative splicing by binding to an exonic splicing enhancer. Genes Dev. 1993;7:2598–2608. doi: 10.1101/gad.7.12b.2598. [DOI] [PubMed] [Google Scholar]
- 61.Tacke R, Manley J L. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tanaka K, Watakabe A, Shimura Y. Polypurine sequences within a downstream exon function as a splicing enhancer. Mol Cell Biol. 1994;14:1347–1354. doi: 10.1128/mcb.14.2.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taylor B J. Differentiation of a male-specific muscle in Drosophila melanogaster does not require the sex-determining genes doublesex or intersex. Genetics. 1992;132:179–191. doi: 10.1093/genetics/132.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Theissen H, Etzerodt M, Reuter R, Schneider C, Lottspeich F, Argos P, Lührmann R, Philipson L. Cloning of the human cDNA for the U1 RNA-associated 70K protein. EMBO J. 1986;5:3209–3217. doi: 10.1002/j.1460-2075.1986.tb04631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tian M, Maniatis T. A splicing enhancer exhibits both constitutive and regulated activities. Genes Dev. 1994;8:1703–1712. doi: 10.1101/gad.8.14.1703. [DOI] [PubMed] [Google Scholar]
- 66.van Oers C C M, Adema G J, Zandberg H, Moen T, Baas P D. Two different sequence elements within exon 4 are necessary for calcitonin-specific splicing of the human calcitonin/calcitonin gene-related peptide I pre-mRNA. Mol Cell Biol. 1994;14:951–960. doi: 10.1128/mcb.14.2.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vellard M, Sureau A, Soret J, Martinerie C, Perbal B. A potential splicing factor is encoded by the opposite strand of the trans-spliced c-myb exon. Proc Natl Acad Sci USA. 1992;89:2511–2515. doi: 10.1073/pnas.89.7.2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu J Y, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 69.Xu R, Teng J, Cooper T A. The cardiac troponin T alternative exon contains a novel purine-rich positive splicing element. Mol Cell Biol. 1993;13:3660–3674. doi: 10.1128/mcb.13.6.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yeakley J M, Hedjran F, Morfin J-P, Merillat N, Rosenfeld M G, Emeson R B. Control of calcitonin/calcitonin gene-related peptide pre-mRNA processing by constitutive intron and exon elements. Mol Cell Biol. 1993;13:5999–6011. doi: 10.1128/mcb.13.10.5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zahler A M, Neugebauer K M, Lane W S, Roth M B. Distinct functions of SR proteins in alternative pre-mRNA splicing. Science. 1993;260:219–222. doi: 10.1126/science.8385799. [DOI] [PubMed] [Google Scholar]
- 72.Zahler A M, Neugebauer K M, Stolk J A, Roth M B. Human SR proteins and isolation of a cDNA encoding SRp75. Mol Cell Biol. 1993;13:4023–4028. doi: 10.1128/mcb.13.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zahler A M, William L S, Stolk J A, Roth M B. SR proteins: a conserved family of pre-mRNA splicing factors. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 74.Zamore P D, Patton J G, Green M R. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992;355:609–614. doi: 10.1038/355609a0. [DOI] [PubMed] [Google Scholar]
- 75.Zhou S, Yang Y, Scott M J, Pannuti A, Fehr K C, Eisen A, Koonin E V, Fouts D L, Wrightsman R, Manning J E, Lucchesi J C. Male-specific lethal 2, a dosage compensation gene of Drosophila, undergoes sex-specific regulation and encodes a protein with a RING finger and a metallothionein-like cysteine cluster. EMBO J. 1995;14:2884–2895. doi: 10.1002/j.1460-2075.1995.tb07288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhuang Y, Weiner A M. A compensatory base change in U1 snRNA suppresses a 5′ splice site mutation. Cell. 1986;46:827–835. doi: 10.1016/0092-8674(86)90064-4. [DOI] [PubMed] [Google Scholar]
- 77.Zuo P, Manley J L. The human splicing factor ASF/SF2 can specifically recognize pre-mRNA 5′ splice sites. Proc Natl Acad Sci USA. 1994;91:3363–3367. doi: 10.1073/pnas.91.8.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]