Abstract
Bin1 is a Myc-interacting protein with features of a tumor suppressor. The high level of Bin1 expression in skeletal muscle prompted us to investigate its role in muscle differentiation. Significant levels of Bin1 were observed in undifferentiated C2C12 myoblasts, a murine in vitro model system. Induction of differentiation by growth factor withdrawal led to an upregulation of Bin1 mRNA and to the generation of higher-molecular-weight forms of Bin1 protein by alternate splicing. While Bin1 in undifferentiated cells was localized exclusively in the nucleus, differentiation-associated isoforms of Bin1 were found in the cytoplasm as well. To examine the function of Bin1 during differentiation, we generated stable cell lines that express exogenous human Bin1 cDNA in the sense or antisense orientation. Cells overexpressing Bin1 grew more slowly than control cells and differentiated more rapidly when deprived of growth factors. In contrast, C2C12 cells expressing antisense Bin1 showed an impaired ability to undergo differentiation. Taken together, the results indicated that Bin1 expression, structure, and localization are tightly regulated during muscle differentiation and suggested that Bin1 plays a functional role in the differentiation process.
The processes of proliferation, differentiation, and tumorigenesis are intricately related. In normal tissues, immature cells proliferate until environmental signals and intrinsic genetic programs trigger irreversible withdrawal from the cell cycle and terminal differentiation (29, 33). Tumor cells, in contrast, are unable to withdraw from the cell cycle and lack many of the characteristics of differentiated cells (11). This relationship is clinically important, because the degree of dedifferentiation of a tumor cell typically correlates with a poorer prognosis (31). Moreover, interventions that promote differentiation retard tumor growth or even induce tumor regression (7, 9). Thus, proliferation and differentiation are mutually exclusive fates of a cell, and unraveling the mechanisms that control them has clear implications for cancer therapy.
In recent years, many aspects of the genetic programs controlling proliferation and differentiation have been elucidated. In general, these cellular responses are regulated by the opposing actions of two groups of genes, one which promotes cell growth (proto-oncogenes) and the other which opposes it (tumor suppressors) (26). During normal cellular proliferation, growth-promoting genes that control cell cycle entry, DNA synthesis, and cell division are activated by growth factors and by extracellular matrix proteins (4, 32). Inappropriate activation of these genes due to mutation or dysregulation can induce abnormal proliferation and thereby contribute to tumorigenesis (24, 25). During differentiation, many growth-promoting genes (e.g., Myc and cyclin D1) are repressed (36, 43) while many growth-inhibitory genes (e.g., those encoding the retinoblastoma protein and the cyclin-dependent kinase inhibitor p21WAF1) are activated (21, 22). Significantly, differentiation can be inhibited either by forced expression of growth-promoting genes or by inactivation of growth inhibitors (27, 37, 39, 41). Thus, whether a cell grows or differentiates is determined, in large part, by the balance between proto-oncogenes and tumor suppressors.
Bin1 is a novel gene whose features suggest that it may influence this balance (34). Originally identified as a protein that interacts with the N terminus of the Myc oncoprotein, Bin1 is structurally similar to RVS167, a negative regulator of the cell cycle in the yeast Saccharomyces cerevisiae (5). Consistent with the notion that it might play a role in regulating cell growth, Bin1 was found to suppress the cell transforming activity of Myc as well as that of the adenovirus E1A and mutant p53 proteins (19, 34). In addition, Bin1 expression is reduced in carcinoma cells derived from malignancies of the breast and other tissues, and introduction of Bin1 into tumor cell lines lacking endogenous expression reduces their proliferative capacity. Finally, the human Bin1 gene maps to chromosome 2q14 (28), a locus within the mid-2q region that is deleted in >40% of metastatic prostate carcinomas (13). Together, these observations lend strong support to the hypothesis that Bin1 is a tumor suppressor.
Interestingly, analysis of the tissue distribution of Bin1 indicated that the highest levels of expression were in skeletal muscle and brain, tissues which are abundant in postmitotic, terminally differentiated cells (34). Since Bin1 has features of a tumor suppressor, we hypothesized that it might contribute to the regulation of differentiation in these tissues. To investigate this hypothesis, we analyzed Bin1 in an in vitro murine model for muscle differentiation, C2C12 myoblasts (6). In this report, we demonstrate that Bin1 plays a critical role in C2C12 differentiation. After induction of differentiation, Bin1 message and protein levels are dramatically increased and there is a change in the structure of the Bin1 protein due to alternative RNA splicing. This splicing results in a larger form of the protein that localizes in the cytoplasm as well as the nucleus, suggesting a Myc-independent role(s) for Bin1 in differentiated cells. Increased expression appears to be crucial for differentiation, because overexpression of Bin1 promotes myotube formation and upregulation of myosin heavy chain while interference with Bin1 expression significantly impairs these processes.
MATERIALS AND METHODS
Cell culture.
C2C12 cells (kindly provided by David Goldhamer) were carried in growth medium (GM; Dulbecco’s modified Eagle medium supplemented with 15% fetal bovine serum and penicillin-streptomycin). Cells were grown to approximately 70% confluence and then passaged or induced to differentiate. Differentiation was induced by removing the GM, washing the cells with phosphate-buffered saline (PBS), and then culturing the cells in differentiation medium (DM; Dulbecco’s modified Eagle medium supplemented with 5% horse serum and penicillin-streptomycin) for 5 days (or as indicated).
Cells were transfected by using a calcium phosphate precipitation protocol that has been described previously (12). Briefly, 2 × 105 cells seeded in 10-cm-diameter dishes were transfected overnight (18 h) with 15 μg of the appropriate plasmid and 10 μg of pBS+ (Stratagene). The next day, the cells were washed and refed; after an additional 24 h, they were trypsinized and passaged at a 1:25 ratio into new dishes. The following day, G418 was added to 0.8 mg/ml for selection of stable transfectants. The medium was changed every 2 to 3 days, and after 7 to 8 days, individual colonies were ring cloned and expanded into cell lines.
Northern analysis.
Total cytoplasmic RNA was isolated from C2C12 cells as described in reference 35. For Northern analysis, 15 μg of RNA was fractionated on an agarose gel and transferred onto a nylon membrane (Duralon-UV; Stratagene). After UV cross-linking, membranes were prehybridized in Church buffer (35) for 4 h at 65°C and then hybridized overnight with a 32P-labeled human Bin1 cDNA probe (generated by random priming) or with an exon 10-specific oligonucleotide probe, 5′-GGAGAATTCGTTGTCACTGTTCTTCTTTCTG (47), labeled by using T4 polynucleotide kinase (Boehringer Mannheim Biochemicals). Membranes were washed twice in 0.1% sodium dodecyl sulfate (SDS)–0.2% saline sodium citrate for 10 min at 50°C and then exposed to film.
Antibodies and blocking proteins.
The anti-Bin1 monoclonal antibodies (MAbs) 99D and 99F, generated by immunization of mice with a glutathione S-transferase (GST) fusion polypeptide containing amino acids 189 to 398 of human Bin1 (GST-99Pst), are described in detail elsewhere (46). For some immunoprecipitation and Western blotting experiments, 99D was blocked by incubation with a molar excess of a GST fusion polypeptide containing a fragment of murine Bin1 (GST-ATG99) with high affinity for this antibody. Anti-immunoglobulin D (IgD) MAbs (AMS 9.1), used as a negative control for immunoprecipitation and flow cytometry, were a gift from J. Erikson (Wistar Institute). A polyclonal rabbit antiserum to mouse c-Myc (06-213) was obtained from Upstate Biotechnology Inc. MAbs specific for myosin heavy chain (MF20), developed by D. A. Fischman (3), were obtained from the Developmental Studies Hybridoma Bank (Iowa City, Iowa). Fluorescein isothiocyanate (FITC)-coupled goat anti-mouse IgG antiserum, used as a secondary antibody for flow cytometry and immunofluorescence, and horseradish peroxidase (HRP)-coupled goat anti-mouse and anti-rabbit IgGs, used for Western blotting, were obtained from Boehringer Mannheim Biochemicals. For flow cytometry, Western blotting, and immunofluorescence, hybridoma supernatants were diluted 1:20 and secondary antibodies were diluted 1:1,000 (FITC conjugates) or 1:15,000 (HRP conjugates). All antibodies were diluted in PBS plus 0.1% Tween 20 (PBST).
Immunoprecipitation.
C2C12 cells were metabolically labeled by incubation for 4 h in methionine- and cysteine-free medium (Life Technologies, Gaithersburg, Md.) containing 100 μCi of [35S]methionine–[35S]cysteine (EXPRESS label; NEN) per ml and then lysed in 1 ml of Nonidet P-40 (NP-40) buffer (50 mM Tris, pH 8.0; 150 mM NaCl; 1% NP-40) containing aprotinin, antipain, leupeptin (2 μg/ml each), and phenylmethylsulfonyl fluoride (100 μg/ml). Lysates were spun in a microcentrifuge (Eppendorf) for 15 min at maximum speed to remove insoluble matter, and protein (0.5 mg per sample) was precleared by incubation for 1 h at 4°C with 40 μl of protein G-Sepharose beads. Proteins were immunoprecipitated by incubating lysates for 2 h with 20 μl of protein G-Sepharose beads that had been precoated with 100 μl of hybridoma supernatant (anti-IgD, 99D or 99F plus blocking proteins, added where indicated in the figures). Immunoprecipitates were washed four times in NP-40 buffer, resuspended in 2× SDS-polyacrylamide gel electrophoresis (PAGE) gel loading buffer, boiled for 5 min, and fractionated on a 10% polyacrylamide gel. Labeled proteins were visualized by fluorography.
Western analysis.
Cells were lysed in NP-40 buffer, and the lysate was centrifuged to remove insoluble material. Protein (50 μg per sample) was then resuspended in 2× SDS-PAGE gel loading buffer, boiled for 5 min, and fractionated on a 10% polyacrylamide gel. Proteins were transferred onto nitrocellulose membranes (Hybond-ECL; Amersham), which were subsequently blocked overnight in PBST containing 5% nonfat dried milk. To detect Bin1 and myosin heavy chain, membranes were incubated for at least 1 h in primary antibody (99D or MF20) and 1 h in secondary antibody (HRP-conjugated goat anti-mouse IgG). To detect c-Myc, membranes were incubated similarly except that anti-c-Myc antibody and HRP-conjugated goat anti-rabbit IgG were used. Membranes were then incubated for 5 min in a chemiluminescent HRP substrate (Pierce) and exposed to film.
Flow cytometry.
Proliferating C2C12 cells (106 per sample) were trypsinized and washed with PBS. Cells were fixed in PBS containing 0.25% paraformaldehyde for 1 h at 4°C and permeabilized in PBST for 15 min at 37°C. Cells were then stained with primary antibody (99D) for 1 h at 4°C and with secondary antibody (FITC-conjugated goat anti-mouse IgG) for 1 h at 4°C. After being stained, cells were washed three times in PBST and analyzed on a Becton Dickinson FACScan using CellQuest software.
Immunofluorescence.
For immunofluorescence analysis, cells were grown on glass coverslips in GM or DM, as indicated in the figures. At the end of the culture period, cells were fixed for 10 min with ice-cold PBS containing 1% paraformaldehyde and then permeabilized for 10 min with ice-cold PBS containing 0.2% Triton X-100. After being washed with PBS, cells were stained for 1 h (at room temperature) with primary antibody (99D or MF20) and for 1 h with secondary antibody (FITC-conjugated goat anti-mouse IgG). Coverslips were washed three times in PBS after each staining step and then mounted on slides with VectaStain mounting medium. Slides were examined and photographed by using a Leitz microscope.
RT-PCR.
A murine Bin1 cDNA has been described previously (40). DNA sequences from this cDNA were used to generated the following primers for analysis of the endogenous Bin1 message in C2C12 cells: mNTsen1 (5′-CAGTGCGTCCAGAATTTC) and mNTanti1 (5′-AACACCTTCTGGGCTTTG), mMIDsen1 (5′-AAGCCCAGAAGGTGTTCGAG) and 5′ATG99 (5′-TGGCTGAGATGGGGACTT), and mCTsen1 (5′-CTGAGATCAGAGTGAACCATG) and mCTanti1 (5′-CACCCGCTCTGTAAAATTC). To detect exogenous Bin1 in transfected cells, the human Bin1-specific primers hX7.1 (5′-GCCAAAATTGCCAAGGCCGAG) and hAntiNLS2 (5′-GTTGTCACTGTTCTTCTTTCTGC) were used. Reverse transcription-(RT)-PCR was performed as follows. Two micrograms of total cytoplasmic RNA was mixed with 50 pmol of the appropriate primer, heated to 70°C, and cooled rapidly on wet ice. RNA and primers were added to a mixture of Moloney murine leukemia virus reverse transcriptase (Life Technologies) and reaction buffer provided by the manufacturer and incubated at 42°C for 1 h to allow first-strand synthesis. From this reaction mixture, 3 μl was removed and added to a solution containing fresh primers, PCR buffer, and Taq polymerase. Samples were subjected to 30 cycles of denaturation (30 s at 94°C), annealing (45 s at 55°C) and polymerization (60 s at 72°C). For each reaction, 10 μl of the product was removed, mixed with sample buffer, and separated on an agarose gel containing ethidium bromide. For further analysis, bands were subcloned into the vector pCR+ (Invitrogen). The DNA sequences of subcloned fragments were determined and analyzed with MacVector and AssemblyLIGN software.
RESULTS
Expression of Bin1 in C2C12 myoblasts.
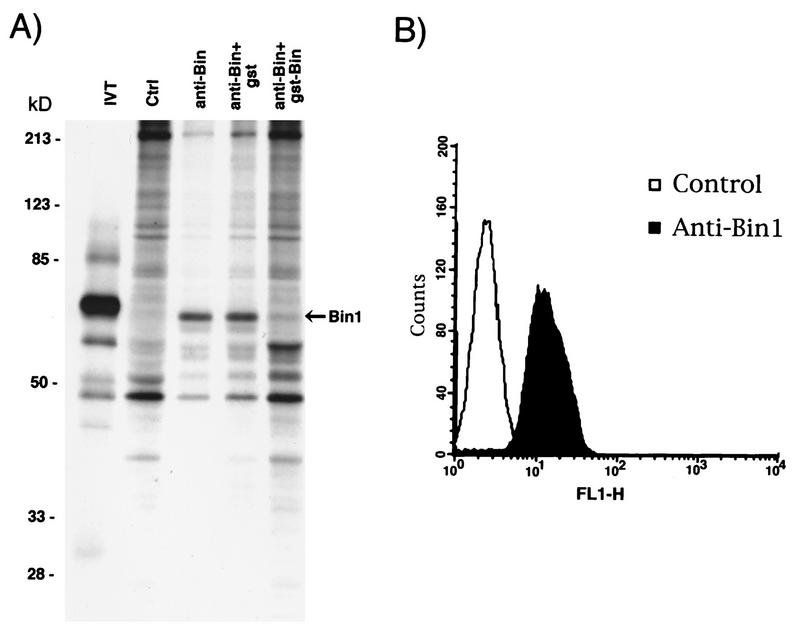
Previous work had indicated that Bin1 mRNA levels in murine skeletal muscle were higher than those in most other tissues (34), suggesting that Bin1 might have a role in this tissue. To begin to address this issue, we examined Bin1 expression in C2C12 cells, a nontransformed myoblast cell line derived from murine skeletal muscle (6). In serum-rich medium, C2C12 cells proliferate rapidly, but when cultured at high density in growth factor-deficient medium, the cells stop dividing, align with one another, express muscle-specific genes, and fuse into multinucleate myotubes (2, 6). Bin1 was immunoprecipitated from extracts of metabolically labeled, proliferating C2C12 cells with 99D, a MAb raised against a human Bin1-GST fusion protein (46). Samples of lysate were also immunoprecipitated with a control antibody (anti-IgD) or with 99D that had been preincubated with a molar excess of nonspecific or specific blocking proteins. Immunoprecipitates were subjected to SDS-PAGE and fluorography (Fig. 1A). 99D specifically recognized a polypeptide of ∼65 kDa, similar in size to that generated by in vitro translation of a full-length Bin1 cDNA (34). The ∼65 kDa protein from C2C12 cells was not recognized by isotype-matched control antibodies or by 99D that was preincubated with the GST-Bin1 fusion protein (incubation with unfused GST had no effect). We concluded that 99D recognized murine Bin1 in C2C12 cells.
FIG. 1.
Expression of Bin1 in C2C12 cells. (A) Immunoprecipitation. C2C12 cells were metabolically labeled with [35S]methionine-[35S]cysteine and lysed in NP-40 lysis buffer. Lysates were precleared and then subjected to immunoprecipitation with anti-IgD antibodies (control [Ctrl]), with 99D (anti-Bin), or with 99D that had been preincubated with GST (anti-Bin+gst) or a protein consisting of GST fused to a murine Bin1 polypeptide (GST-Bin [anti-Bin+gst-Bin]). Immunoprecipitates, along with a 35S-labeled human Bin1 polypeptide generated by in vitro translation (IVT), were analyzed by SDS-PAGE and visualized by fluorography. The positions of molecular mass markers (in kilodaltons) are shown on the left. (B) Fluorescence-activated cell sorter analysis. C2C12 cells were trypsinized to generate a cell suspension and then stained with anti-IgD (control) or 99D (anti-Bin1) antibodies followed by FITC-coupled goat anti-mouse IgG. Cells were then washed and analyzed by flow cytometry. FL1-H, fluorescence channel 1 (FITC).
To determine whether Bin1 was expressed throughout the C2C12 population, cells stained with 99D were examined by flow cytometry. A suspension of proliferating cells was generated by trypsinization, then fixed, permeabilized, and stained with 99D or control antibodies followed by fluorescein-conjugated secondary antibodies. Flow cytometric analysis of the stained cell suspension demonstrated that essentially all cells in the population fluoresced above background (Fig. 1B). We concluded that proliferating C2C12 cells universally expressed Bin1 protein.
Bin1 is upregulated during C2C12 differentiation.
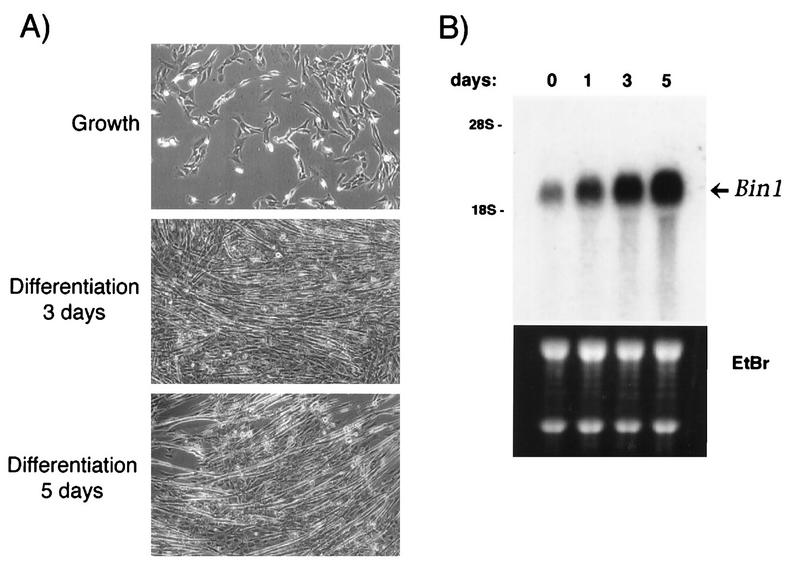
We next investigated whether Bin1 expression was affected by differentiation. C2C12 cells grown to near confluence and then shifted to DM undergo a pronounced change in morphology; cells elongate, align with one another, and fuse into myotubes (Fig. 2A). In our cultures, morphological differentiation (alignment and fusion) typically began 2 to 3 days following addition of DM, biochemical differentiation (expression of myosin heavy chain; see below) was detectable by days 3 to 4, and myotube generation was maximal (50 to 70% fusion) by day 5 to 6. To assess Bin1 expression during this period, RNA was isolated from cells at various times and subjected to Northern analysis. As shown in Fig. 2B, the level of Bin1 message in C2C12 cells increased dramatically during differentiation (days 1, 3, and 5). Expression began to increase as early as day 2 and reached its highest level at 5 days, when cell differentiation was maximal.
FIG. 2.
Bin1 message is upregulated during differentiation. (A) Differentiation of C2C12 cells. Cells were photographed after being cultured in medium containing 15% FCS (Growth) or after 3 or 5 days of culture in medium containing 5% horse serum (Differentiation). (B) Expression of Bin1 mRNA during differentiation. Total cytoplasmic RNA from proliferating (day 0) or differentiating (days 1, 3, and 5) C2C12 cells was separated by agarose gel electrophoresis and blotted onto a nylon membrane. The membrane was probed with a 32P-labeled human Bin1 cDNA and then exposed to film (top panel). RNA integrity and quantity were confirmed by ethidium bromide (EtBr) staining of the gel before transfer (bottom panel).
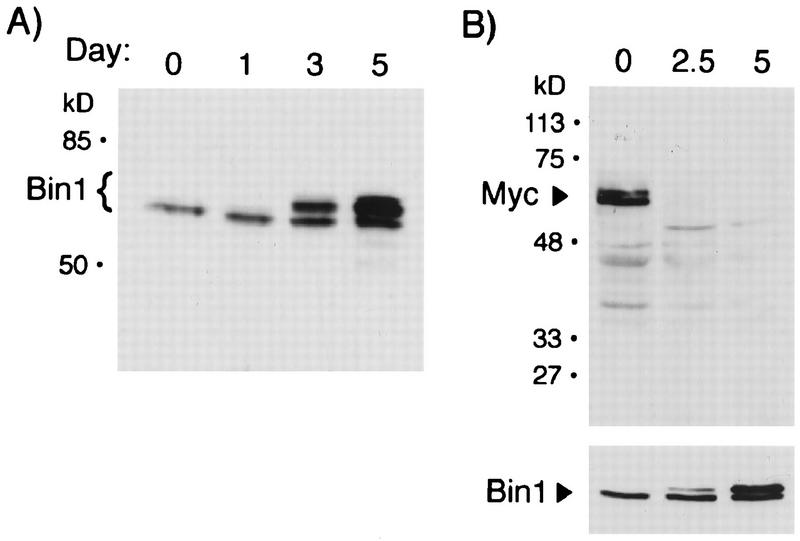
To confirm that the upregulation of Bin1 message was associated with an increase in Bin1 protein, lysates from proliferating or differentiating C2C12 cells were analyzed by Western blotting with 99D (Fig. 3A). Proliferating cells contained a ∼65-kDa polypeptide similar to that observed after immunoprecipitation. Following induction of differentiation, the level of this protein increased a few fold. In addition, differentiated cells contained higher-molecular-weight proteins (68 to 70 kDa) that were recognized by 99D. These proteins appeared to be Bin1 related, since they were also observed in immunoprecipitates from differentiated cells (see below) and they were not detected when blots were probed with an isotype-matched control antibody or with 99D that had been preincubated with a GST-Bin1 blocking protein (data not shown).
FIG. 3.
Induction of novel isoforms of Bin1 and downregulation of Myc during differentiation. (A) Western analysis of Bin1. NP-40 lysates from proliferating (day 0) or differentiating (days 1, 3, and 5) C2C12 cells were separated by SDS-PAGE and then transferred onto a nitrocellulose membrane. The membrane was probed with the anti-Bin1 antibody 99D followed by HRP-conjugated goat anti-mouse IgG. Proteins were detected by chemiluminescence. (B) Western analysis of Myc. Lysates from proliferating (day 0) or differentiating (days 2.5 and 5) cells were analyzed as above, except that a rabbit anti-Myc antibody and HRP-conjugated anti-rabbit IgG were used. The bottom panel shows Bin1 induction on a parallel blot of the same lysates. The positions of molecular mass markers (in kilodaltons) are shown to the left of both panels A and B.
Since we initially identified Bin1 through its ability to interact with Myc (34), we examined Myc expression in a second population of C2C12 cells that were growing or that had been subjected to serum withdrawal for 2.5 or 5 days. As observed in other cell systems, Myc was rapidly downregulated after induction of differentiation, such that it was undetectable at 2.5 days after serum withdrawal (Fig. 3B). In contrast, it was at this time that one could begin to detect the altered forms of Bin1 that were induced by serum withdrawal. Thus, the increased expression and apparent alteration of Bin1 occurred in cells lacking Myc. We concluded that during C2C12 differentiation, Myc levels decreased whereas Bin1 mRNA and protein levels increased and novel Bin1 species were generated.
Bin1 mRNA is subject to alternate splicing.
Although the larger polypeptides that appeared during C2C12 differentiation were immunologically related to Bin1, their structural relationship to Bin1 was not clear. If they represented alternate forms of Bin1, rather than related proteins, the larger and smaller species would be expected to have similar peptide maps. To examine this, 99D immunoprecipitates were fractionated by SDS-PAGE and the larger and smaller species were isolated from gels and subjected to V8 protease mapping. We observed that the different species had virtually identical peptide maps (data not shown), suggesting that they represented different isoforms of Bin1.
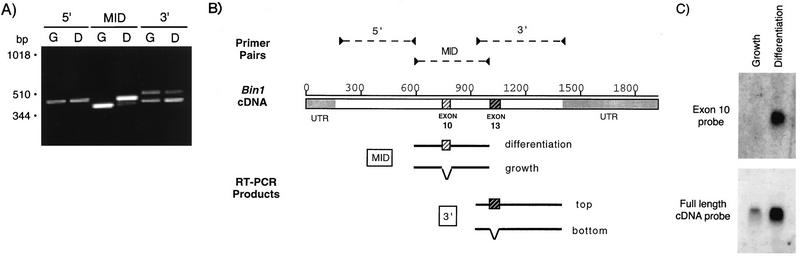
Since one explanation for the different sizes of Bin1 was alternate RNA splicing, we compared Bin1 mRNA structure in proliferating and differentiated cells by RT-PCR. Segments representing the 5′ end, middle, and 3′ end of the Bin1 RNA were amplified with separate sets of primers. The results are shown in Fig. 4A. RT-PCR with the 5′-end primers, corresponding to the N-terminal region of the polypeptide, generated a single band of ∼450 bp from RNA from both proliferating and differentiated cells. In contrast, RT-PCR with the midsection primers yielded fragments of ∼400 bp from proliferating cells and of 445 bp from differentiated cells. Finally, RT-PCR with the 3′-end primers, corresponding to the C-terminal region of the polypeptide, yielded products of 425 and 515 bp that were present at similar levels in both proliferating and differentiated cells.
FIG. 4.
Differentiation-associated isoforms are generated by alternate splicing. (A) Detection of splicing by RT-PCR. Total cytoplasmic RNA from growing (G) and from differentiated (D) C2C12 cells was reverse transcribed, and the resulting cDNA was amplified by PCR with primers designed to hybridize to the 5′ end, middle (MID), or 3′ end of the murine Bin1 mRNA (see Materials and Methods). PCR products were separated by agarose gel electrophoresis, stained with ethidium bromide, and photographed. The positions of molecular size markers are shown on the left. (B) Summary of RT-PCR results. The PCR products shown in panel A were sequenced, and the sequences from growing and differentiated cells were compared. 5′ fragments from each population were identical to one another (data not shown). Midregion (MID) fragments from differentiated cells contained a 45-bp sequence (homologous to human exon 10) that was absent in fragments from proliferating cells. Each cell population contained two 3′ fragments; these differed from one another in the presence or absence of a 60-bp sequence homologous to human exon 13. UTR, untranslated region. (C) Detection of alternate splicing by Northern blotting. RNA from growing and from differentiated cells was separated by electrophoresis, transferred onto a nylon membrane, and probed with a 32P-labeled oligonucleotide fragment derived from exon 10 of human Bin1 (top panel) or with a full-length human Bin1 cDNA probe (bottom panel). Membranes were exposed to film for 1 week. No exon 10-positive RNA was detected on film exposed for up to ∼3 weeks.
DNA sequence analysis of the 5′ and 3′ RT-PCR products indicated no change in the structures of these regions in proliferating and differentiated cells. The two 3′ products (detected in RNA from both sources) differed in the presence or absence of a 90-bp segment encoding part of the Myc-binding domain (MBD) of Bin1 (19, 34). Significantly, this 90-bp fragment corresponded exactly to an exon conserved in the human gene, exon 13 (47). This result strongly suggested that a murine exon corresponding to human exon 13 was subject to alternate splicing in both proliferating and differentiated C2C12 cells.
A similar analysis of the RT-PCR products amplified with the midsection primers showed that the 400- and 445-bp products found in proliferating and differentiated cells, respectively, were identical except for the presence of an additional 45-bp segment in the latter. This segment is absent from a murine Bin1 cDNA isolated from an embryo library (40) but is present in a human cDNA isolated from a skeletal muscle library (34). As had been observed with the 3′ segment, the 45-bp segment spliced into the midsection was found to correspond to a discrete exon in the human gene, exon 10. Thus, a murine exon corresponding to human exon 10 is alternately spliced into Bin1 mRNA, and this event is regulated during C2C12 differentiation. The splice forms of Bin1 identified in this analysis are summarized in Fig. 4B.
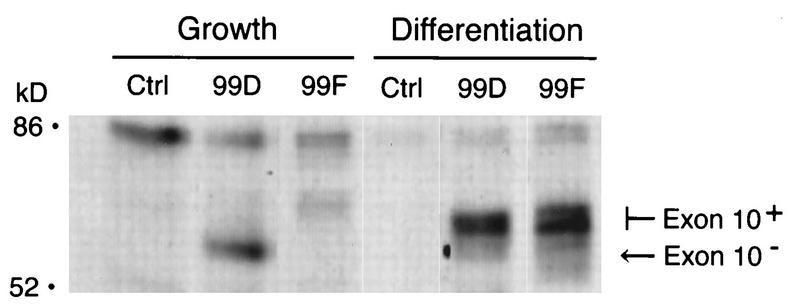
Two additional experiments were performed to verify that exon 10 was expressed only in differentiated C2C12 cells. First, total cytoplasmic RNA from proliferating and differentiated cells was subjected to Northern analysis with an oligonucleotide probe specific for exon 10 sequences. While a full-length cDNA probe recognized Bin1 mRNA from either population, the exon 10-specific probe detected message only in differentiated cells (Fig. 4C). Second, to confirm this difference at the protein level, we used a Bin1 MAb, 99F, that had been determined to recognize an exon 10-encoded epitope (46). 99F was found to bind in vitro-translated Bin1 polypeptides that included exon 10 sequences but not those that lacked such sequences. Moreover, 99F failed to detect Bin1 protein present in a variety of tumor cell lines, suggesting that the exon 10 epitope was masked or absent in these cells. We employed 99F as a probe to examine the exon 10-containing Bin1 species identified in differentiated C2C12 cells. As shown in Fig. 5, immunoprecipitation of extracts from 35S-labeled C2C12 cells indicated that 99D recognized Bin1 proteins from both proliferating and differentiated cells. In contrast, 99F failed to detect Bin1 in proliferating cells but recognized the larger Bin1 species in differentiated cells. Both the smaller and larger species detected in differentiated cells were heterogeneous. The reason for this was unclear but might reflect differences in phosphorylation states since Bin1 has been shown to be a phosphoprotein (46). We concluded that exon 10 sequences were spliced into Bin1 message during differentiation and that the higher-molecular-weight species of Bin1 protein observed in differentiated cells were due to the expression of exon 10-encoded residues.
FIG. 5.
Differentiation-associated Bin1 proteins can be detected with an exon 10-specific antibody. Growing and differentiated C2C12 cells were metabolically labeled with [35S]methionine-[35S]cysteine, lysed in NP-40 buffer, and subjected to immunoprecipitation with anti-IgD antibodies (Ctrl), 99D, or the exon 10-specific antibody 99F. Immunoprecipitates were separated by SDS-PAGE and visualized by fluorography. The proliferation-associated (Exon 10−) and differentiation-associated (Exon 10+) forms of Bin1 are indicated; note that 99F recognizes only the Exon 10+ form. The positions of molecular mass markers are shown on the left.
Changes in Bin1 structure correlate with changes in cellular localization.
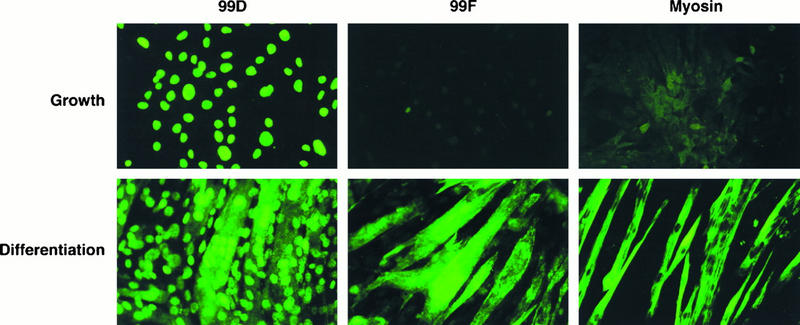
To begin to assess the significance of alternate splicing of exon 10 in differentiated cells, we used 99D and 99F to compare the localization of Bin1 in C2C12 cells before and after differentiation (Fig. 6). Consistent with the results described above, in proliferating cells (top panels), Bin1 was detected by 99D but not by 99F. In these cells, as had been observed in other human and rodent cells (34, 46), Bin1 was localized exclusively in the nucleus. In contrast, in differentiated myotubes (bottom panels), Bin1 was detected by 99D as well as 99F, and the pattern of staining with each of these MAbs was distinct. 99D staining was observed in both the nucleus and cytoplasm, while 99F staining appeared predominantly in the cytoplasm, in a fibrous pattern along the length of the myotube. These staining patterns were specific for Bin1, because they were not observed with isotype-matched control antibodies and because they were blocked by preincubation with specific blocking proteins (data not shown). In addition, staining with an antibody specific for myosin heavy chain confirmed that extensive differentiation had taken place in these cultures. Taken together, these results indicated that the low-molecular-weight form of Bin1 in proliferating C2C12 cells was confined to the nucleus whereas the high-molecular-weight, differentiation-associated Bin1 species were found predominantly in the cytoplasm.
FIG. 6.
Localization of Bin1 changes during differentiation. C2C12 cells were plated onto glass coverslips and cultured in GM for 1 day or in GM for 5 days. Cells were then stained with the anti-Bin1 antibody 99D, the exon 10-specific antibody 99F, or the antimyosin antibody MF20, in each case followed by FITC-conjugated goat anti-mouse IgG antibodies. Cells were photographed by using a Leitz microscope.
Bin1 is necessary for C2C12 differentiation.
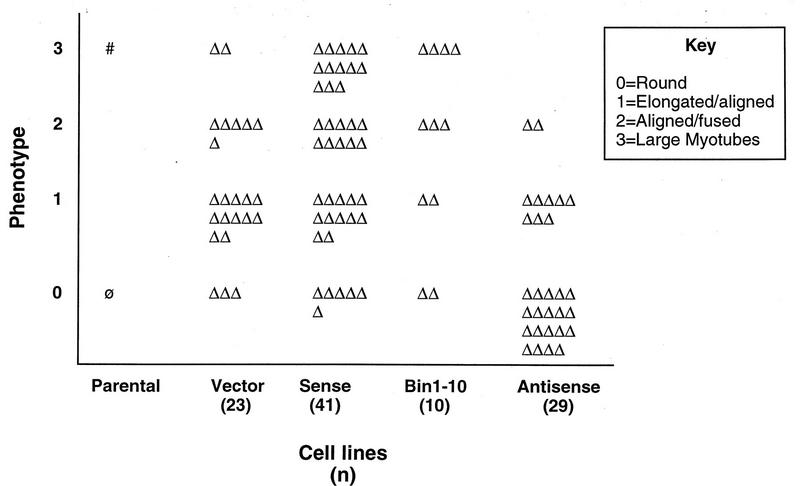
The complex regulation of Bin1 structure and localization during differentiation suggested that it might play a role in the differentiation process. To test this hypothesis, we investigated the effects of overexpressing sense and antisense forms of human Bin1 cDNA in C2C12 cells. Since alternate splicing of exon 10 (but not exon 13) sequences was tightly associated with differentiation, we also examined the effects of overexpressing a Bin1 species lacking exon 10 sequences to distinguish whether exon 10-encoded information, rather than upregulation of Bin1 expression per se, might be important. Cells were transfected with an expression vector encoding a neomycin resistance gene or the same vector containing a full-length human Bin1 cDNA (sense or antisense). Cell lines derived from G418-resistant colonies were screened for expression of exogenous Bin1 by RT-PCR, using primers specific for the human cDNA that was introduced. To rule out any effects of clonal variation, at least 10 cell lines derived from each vector were generated. A summary of the phenotypes exhibited by each set of cell lines is depicted in Fig. 7.
FIG. 7.
Phenotypes of C2C12 cell lines. This figure summarizes the phenotypes of clonal cell lines generated by transfection of the indicated vectors. Cells were incubated for 5 to 6 days in DM and then assessed for the phenotypic characteristics noted in the key. Each triangle represents a single cell line. The total number of cell lines examined (n) is indicated beneath the type of vector transfected. The range phenotypes represent clonal variation in the set of cell lines examined; the trend on the y axis represents a greater or lesser tendency toward a differentiated character following incubation in DM. Φ, phenotype of parental cells in GM; #, phenotype of parental cells in DM.
We observed that sense and antisense lines differentiated better and worse, respectively, than the vector control lines. Only a limited number (10 to 20%) of the cell lines derived from sense cDNA-transfected cells showed elevated expression of Bin1. In addition, cells showing exogenous Bin1 expression grew more slowly than control cell lines, both during and after the selection period (data not shown). These observations argued that Bin1 overexpression might interfere with the growth of C2C12 cells, consistent with results in other cell lines (19, 34). Notably, lines overexpressing Bin1-10 did not show this growth deficit, although they shared with the sense lines a propensity to differentiate more strongly than controls (see below). To further examine the effects of Bin1 overexpression on differentiation, several sense lines were selected for further analysis (from a total of 41 lines generated and phenotypically examined), two of which are reported here (Fig. 8A). Relative to control lines, these cells had significant amounts of exogenous Bin1 mRNA detectable by RT-PCR (top panel). Western analysis of extracts derived from these cells showed two- to fourfold-higher levels of Bin1 protein, as detected with 99D (second panel). Despite the presence of elevated levels of Bin1, however, their morphology in GM was similar to that of control cells (data not shown), with no evidence of premature alignment or fusion. We concluded that Bin1 overexpression impeded C2C12 proliferation to some extent through a mechanism requiring exon 10-derived sequences but that on its own, Bin1 was insufficient to drive differentiation in GM.
FIG. 8.
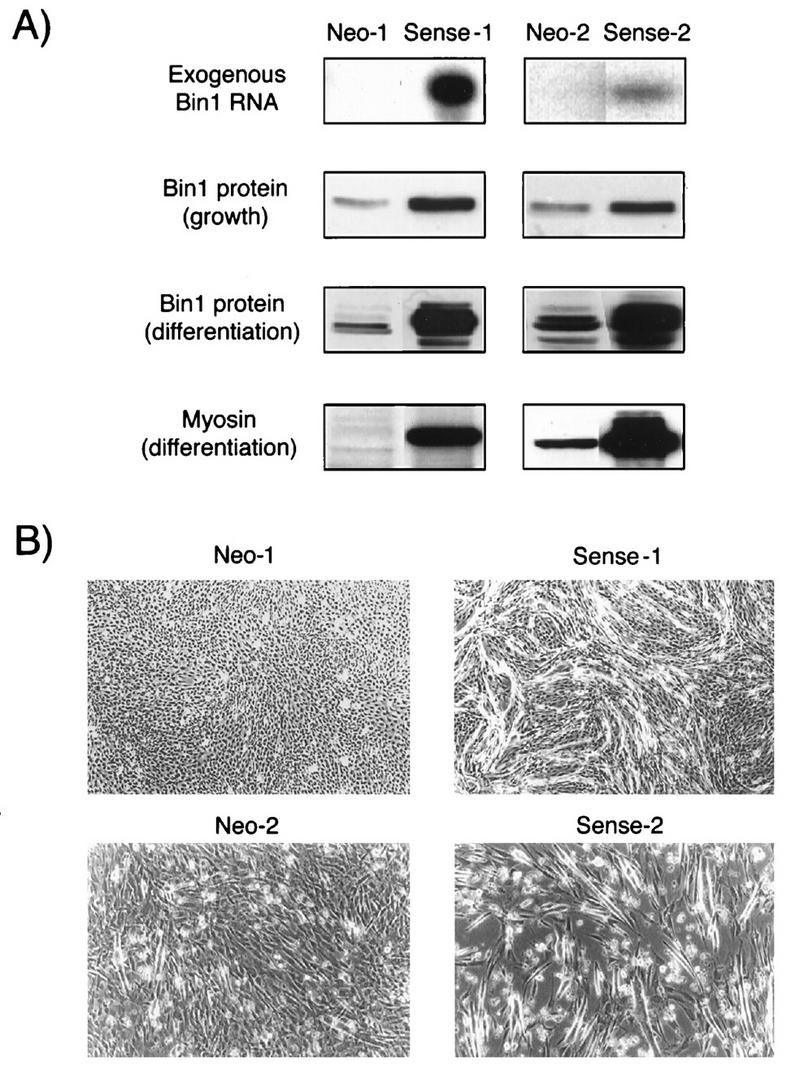
Overexpression of Bin1 accelerates differentiation. (A) Bin1 and myosin expression. (Top panel) Stable cell lines transfected with empty vector (Neo-1 and Neo-2) or with human Bin1 cDNA (Sense-1 and Sense-2) were selected in antibiotic-containing medium and analyzed by RT-PCR with primers specific for human (exogenous) Bin1. (Second panel) Cells cultured in GM were analyzed for Bin1 protein expression by Western blotting with 99D. (Third panel) Cells were cultured in DM for 3 days, and then Bin1 protein levels were assessed by Western blotting. (The high-molecular-weight forms of Bin1 are presumably generated by alternate splicing of the endogenous mRNA.) (Bottom panel) Differentiated cells were analyzed for myosin heavy chain expression by Western blotting with MF20. (B) Morphology of control and Bin1-overexpressing cells after 3 days in DM. Note the extensive cell fusion in Bin1-overexpressing (Sense-1 and Sense-2) cells compared to controls (Neo-1 and Neo-2).
After 3 days in DM, control cells became elongated and aligned but showed limited fusion into myotubes (Fig. 7 and 8B). Consistent with these observations, only modest increases in expression of differentiation-associated isoforms of endogenous Bin1 and myosin heavy chain were observed (Fig. 8A, third and fourth panels). Although control cells showed increased alignment and fusion after longer culturing in DM (see below), they seldom displayed the rate or degree of differentiation observed in parental (nontransfected) cells. This blunted differentiation response in transfected cells might have been due to the high density at which cells were cultured during the drug selection period.
In comparison to control cells, cells overexpressing Bin1 underwent a more rapid and pronounced differentiation in DM (Fig. 7 and 8A). An examination of 12 cell lines overexpressing Bin1 10 showed a similar response trend (data not shown), suggesting that it was the overexpression of Bin1 rather than exon 10-encoded sequences per se that mediated the effect. Notably, cells expressing sense Bin1 or Bin1-10 differentiated even more vigorously than parental C2C12 cells. Within 2 to 3 days of culture in DM, cells exhibited sharp increases in their overall level of Bin1 protein (due to increases in endogenous expression), with significant accumulation of the high-molecular-weight differentiation-associated species (Fig. 8A, third panel). In parallel with this upregulation, there was a dramatic increase in myosin heavy chain levels (Fig. 8A, fourth panel), efficient cell alignment, and extensive cell fusion into myotubes (Fig. 8B). This rapid and efficient differentiation was not vector dependent, because similar phenomena were observed in cells that were transfected with two other Bin1 vectors (data not shown). We concluded that elevation of the levels of Bin1, either containing or lacking exon 10-encoded sequences, was insufficient to induce C2C12 differentiation but accelerated or enhanced the differentiation program once it was initiated.
An examination of antisense cDNA-expressing cell lines suggested that Bin1 was a necessary component of the differentiation program (Fig. 7). Unlike sense transfectants, a significant proportion (50 to 60%) of the G418-resistant cell lines transfected with the antisense vector exhibited expression of the exogenous construct by RT-PCR (Fig. 9A, top panel). Moreover, whereas the sense cDNA-expressing cells were observed to grow more slowly than controls, antisense cDNA-expressing cells proliferated somewhat more rapidly, such that more frequent passaging was necessary to avoid confluence. Several antisense cell lines were selected for further analysis (from a total of 29 lines generated and phenotypically examined), two of which are reported here (Fig. 9A). Western blotting revealed a two- to fourfold decrease in basal levels of Bin1 protein in these cell lines relative to controls (second panel). Similar to sense cDNA-expressing cell lines, the morphology of antisense cDNA-expressing cells in GM was indistinguishable from that of control cells, and these cells did not show an increased tendency to undergo alignment or fusion.
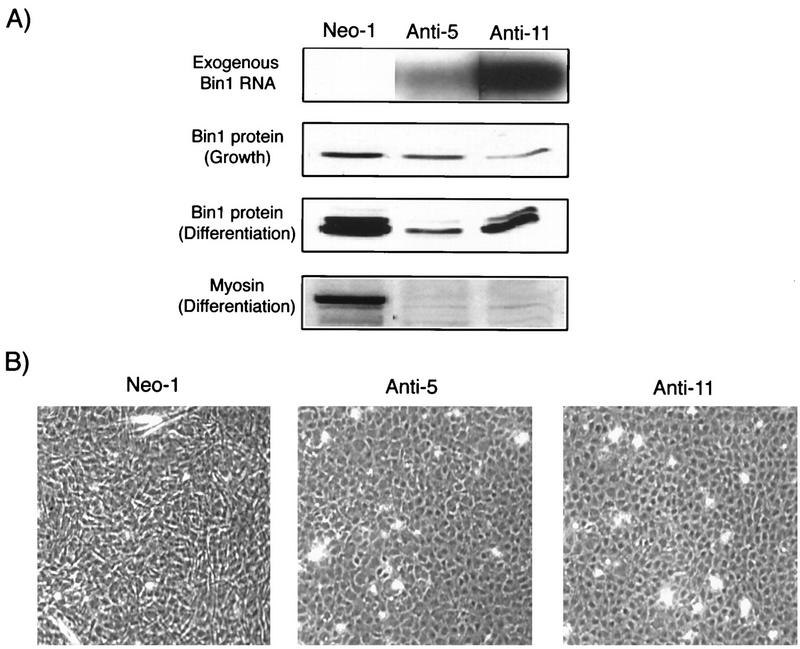
FIG. 9.
Antisense Bin1 expression impairs differentiation. (A) Bin1 and myosin expression. (Top panel) Stable cell lines transfected with empty vector (Neo-1) or with antisense Bin1 (Anti-5 and Anti-11) were selected in antibiotic-containing medium and analyzed by RT-PCR for expression of exogenous Bin1. (Second panel) Bin1 protein expression in cells cultured in GM (detected by Western blotting with 99D). (Third panel) Bin1 protein levels in cells cultured in DM for 6 days. (Bottom panel) Myosin heavy chain expression (detected by Western blotting with MF20) in cells cultured in DM for 6 days. (B) Morphology of control (Neo-1) and antisense Bin1 cDNA-expressing (Anti-5 and Anti-11) cells after 6 days in DM. Antisense cDNA-expressing cells appear round and unfused, while control cells show substantial alignment and fusion at this stage.
The effects of antisense cDNA expression on differentiation were determined by examining the same set of biochemical and morphological features as before, in cells cultured in DM for up to 6 days (a time point at which control cells exhibited maximal morphological differentiation). Compared to control lines, antisense lines showed significantly less upregulation of differentiation-associated Bin1 species (Fig. 9, third panel). In addition, while control cells exhibited increased levels of myosin heavy chain, antisense cDNA-expressing cells showed little upregulation of this marker (Fig. 9, bottom panel). Finally, while control cells showed substantial alignment and some fusion after 6 days in DM, antisense lines showed little if any alignment, instead retaining the rounded morphology that is characteristic of undifferentiated cells. Taken together with the sense results, these data led us to conclude that upregulation of Bin1 is necessary for differentiation of C2C12 cells.
DISCUSSION
Many genes originally identified through their action in cancer cells have since been shown to play a role in regulating normal cellular growth and differentiation (1, 23, 38). Bin1 was originally identified through its interaction with the N terminus of the Myc oncoprotein (34). Bin1 inhibits the oncogenic and transcriptional properties of Myc but also displays the ability to inhibit cell growth by at least two other Myc-independent mechanisms (19, 34). In this study, we demonstrated that the differentiation of C2C12 myoblasts is accompanied by upregulation and alternate splicing of Bin1 mRNA. This splicing results in the generation of differentiation-specific isoforms of the Bin1 protein which are characterized by their higher molecular weights and distinct patterns of cellular localization. By modulating the amount of Bin1 protein in C2C12 cells, we also demonstrated that Bin1 has an integral role in the muscle differentiation program.
Regulation of Bin1 structure and expression during muscle cell differentiation.
Interest in Bin1 in muscle cells was initially stimulated by our observation that murine skeletal muscle expressed higher levels of Bin1 mRNA than most other tissues (34). Consistent with this observation, we found that C2C12 cells contain at least 10-fold-higher levels of Bin1 protein than other cell lines that have been examined. It is noteworthy that these cells express relatively high levels of Bin1 even before myotube differentiation. One possible reason for this comes from studies of the human Bin1 promoter, which have revealed that Bin1 transcription is activated by the myogenic transcription factor myoD (47). Since C2C12 cells are committed to the muscle lineage and already express myoD (1), they may express relatively higher amounts of Bin1 for this reason. Whether Bin1 has a distinct role in the early stages of myogenic commitment in addition to its role in differentiation remains to be determined.
In examining the expression of Bin1 during C2C12 differentiation, we found that Bin1 mRNA levels were dramatically upregulated within 2 days of growth factor withdrawal, at approximately the same time as morphological differentiation was first detectable. Thereafter, Bin1 expression continued to increase as greater numbers of cells aligned and fused into myotubes. In addition to changes in mRNA levels, we observed changes in mRNA splicing during differentiation, with an exon corresponding to human exon 10 (47) being introduced into Bin1 message in differentiated cells. Notably, upregulation and splicing of Bin1 mRNA did not take place when cells were allowed to reach confluence in GM or when growth factors were withdrawn from subconfluent cultures, conditions that do not promote complete morphological or biochemical differentiation (45). Thus, upregulation of Bin1 is intimately linked to activation of a differentiation program.
Several species of Bin1 were found to be generated in C2C12 cells by alternate splicing. Approximately half of Bin1 mRNAs in both proliferating and differentiated cells contained a 3′ sequence corresponding to human exon 13 (47). In differentiated cells, several Bin1 bands were detected by immunoprecipitation and Western blotting, and it is possible that these species differ from one another in expression of exon 13. In proliferating cells, such heterogeneity is not readily apparent; however, longer gels offering higher resolution have revealed closely spaced Bin1 bands that are also consistent with an exon 13 splicing event (45). Interestingly, exon 13 forms part of the MBD of Bin1, which allows it to antagonize Myc-mediated transcriptional activation and cell transformation (34). The fact that exon 13 is subject to alternate splicing suggests that not all Bin1 polypeptides in the cell have Myc-binding capability. Since Bin1 is known to have a Myc-independent as well as a Myc-dependent growth-inhibitory capacity (19, 34), these studies raise the possibility that different functions of Bin1 are mediated by separate species within a cell.
The larger species of Bin1 identified in differentiated cells were shown to result from alternate splicing of a sequence corresponding to human exon 10 (47). While the functional significance of exon 10 splicing remains unclear, its correlation with cytosolic localization suggests that exon 10 sequences may be responsible for targeting of Bin1 to the cytosol. Counterintuitively, exon 10 encodes a highly basic segment which resembles nuclear localization signal motifs (8, 34). In the context of Bin1, however, this motif is neither necessary nor sufficient for nuclear localization, since Bin1 species that lack exon 10 are found in the nucleus of C2C12 cells, as well as other human and rodent cell lines (46), and species that contain exon 10 are present in the cytoplasm of C2C12-derived myotubes. An alternative function for exon 10 may be revealed by an ongoing analysis of a recently identified Bin1-interacting protein whose binding appears to depend on exon 10-encoded sequences (32a).
Although alternate splicing explains some of the major differences observed in Bin1 species in C2C12 cells, additional complexity of Bin1 structure exists in these and other cells. Work in human cell lines has provided evidence for alternate splicing of another exon in the central region of the Bin1 gene, exon 12A (47), and additional exons are spliced into brain-specific forms of Bin1 (10, 30, 42, 47). While we have not detected any of these exons in mRNA from either human muscle cells or C2C12 cells, they may be relatively rarer and/or spliced at other stages of muscle differentiation or in other cell lineages. Posttranslational modification may also contribute to structural variation, because Bin1 has been found to be phosphorylated in both proliferating and differentiated C2C12 cells (46). In future work, it will be important to analyze the various isoforms of Bin1, since this would provide insights into Bin1 function and into the significance of alternative splicing events in C2C12 and other cell types.
Requirement for Bin1 in muscle cell differentiation.
We found that perturbing Bin1 expression in C2C12 cells altered their growth and their susceptibility to induction of differentiation. Expression of exogenous Bin1 (in the sense orientation) interfered with cell growth and promoted cell differentiation. The effects of Bin1 on growth were inferred from the fact that only a small proportion of G418-resistant Bin1 sense cDNA-transfected cells showed overexpression of the exogenous gene by RT-PCR. One interpretation of this finding was that cells expressing high levels of Bin1 had a growth disadvantage and were diluted out during the selection period by cells that expressed lower levels of the protein. Consistent with this notion, the lines that did survive selection expressed only moderate levels of exogenous Bin1 (two- to fourfold-higher levels of expression relative to controls) and grew more slowly than empty-vector control lines. The ability of Bin1 to inhibit cell growth has been documented previously (34), and as noted above, exon 10-encoded sequences may contribute to this property in certain cell lineages, such as muscle cells.
Notably, exogenous Bin1 expression did not promote differentiation of C2C12 cells in GM but dramatically accelerated and enhanced expression of the differentiation program induced by growth factor withdrawal. This accelerated differentiation was observed both morphologically (in terms of cell alignment and fusion) and biochemically (in terms of increased expression of myosin heavy chain and of endogenous Bin1). The fact that Bin1-expressing cells cultured in DM showed more rapid upregulation of differentiation-associated Bin1 isoforms than control or parental cells suggested that Bin1 may positively regulate its own expression, a possibility which needs further investigation.
The analysis of antisense cDNA-expressing cells also strongly supported a role for Bin1 in differentiation. In these cells, the morphological and biochemical features of differentiation were diminished significantly compared to those of control cells. Although we did not determine precisely where Bin1 acts in the differentiation pathway, the facts that Bin1 upregulation occurs relatively quickly (within 2 days of serum withdrawal) and that antisense Bin1 inhibits the earliest morphological signs of differentiation suggest that it may function rather early. Taken together, the data argued that Bin1 upregulation may be a rate-limiting step in the differentiation program.
Although the exact mechanism(s) by which Bin1 acts is unclear, its ability to promote differentiation may reflect both Myc-dependent and Myc-independent activities. Studies of Bin1 structure and function in cell transformation (19, 34) prompt several testable hypotheses. First, as discussed above, Bin1 can interact with the Myc oncoprotein and can inhibit Myc-mediated transcription and transformation. At very early times, before Myc is effectively removed by downregulation, it is possible that Bin1 directly antagonizes Myc’s growth-promoting effects and thereby relieves cells of one barrier to differentiation. Previous studies on the role of Myc in muscle differentiation indicate that its overexpression can interfere with biochemical differentiation and/or fusion (15, 16, 27, 29). In this light, Bin1 may directly antagonize the growth-promoting activity of Myc at early times after induction of differentiation, thereby relieving cells of one barrier toward this process. At later times, when Myc is absent, Bin1 would have to act by a Myc-independent mechanism(s). Although we did not define the exact point(s) where Bin1 acts, the altered splicing and relocalization that it undergoes at later times suggests some other role, possibly one affecting cell alignment or fusion. The question of whether Bin1 would be dominant to Myc in C2C12 is somewhat moot because enforced Myc expression is compatible with differentiation (though not cell fusion) in this cell system (15).
In addition to its Myc-related functions, Bin1 also can act in a Myc-independent manner. For example, Bin1 can inhibit transformation of primary rat embryo fibroblasts by the adenovirus gene product E1A, in a manner independent of the MBD (34). Since E1A can inhibit differentiation of myoblasts and reactivate the cell cycle in differentiated myotubes (41, 44), it is possible that Bin1 may counteract these effects as well. In this scenario, Bin1 may function in differentiation by directly or indirectly affecting known targets of E1A, such as the retinoblastoma protein, p107, and p300/CBP (17, 18, 20, 41). Similarly, we have observed that Bin1 can inhibit cell transformation by a dominant inhibitory mutant of p53 (19). Although the mechanism of this effect is not clear, the fact that p53 function is required for C2C12 differentiation (39) raises the possibility that Bin1 also exerts its effects on differentiation via p53. Future analysis of stable C2C12 lines that overexpress Bin1 mutants defective in E1A and/or p53 inhibition may shed light on the pathways involved in BIN action.
Our studies provide strong evidence that Bin1 can regulate muscle differentiation. Since Bin1 is expressed ubiquitously (34), it may also contribute to the control of differentiation programs in other cell types. Consistent with this possibility, we have noted that during induction of the monocytic differentiation program in the promyelocytic cell line HL-60 (14), Bin1 expression and splicing patterns are altered in a manner similar to that observed in C2C12 cells (45). Thus, Bin1 may have a general role in cell differentiation. If so, the frequent loss of Bin1 may contribute to malignant development both via the loss of processes required for terminal differentiation and by contributing to Myc deregulation.
ACKNOWLEDGMENTS
We are grateful to David Goldhamer for providing C2C12 cells, helpful discussions, and criticism of the work. We thank Wei Du for providing a human Bin1 cDNA lacking exon 10 sequences. Anti-IgD antibodies were a gift of Jan Erikson. We thank Rudi Grosschedl and Paul Stein for critically reading the manuscript. MAbs specific for myosin heavy chain (MF20), developed by D. A. Fischman (Cornell University, New York, N.Y.), were obtained from the Developmental Studies Hybridoma Bank maintained by the University of Iowa Department of Biological Sciences, Iowa City, IA 52242, under contract NO1-HD-7-3263 from the National Institute of Child Health and Human Development.
This work was supported by grants CN-160 from the American Cancer Society and DAMD17-96-1-6324 from the U.S. Army Breast Cancer Research Program to G.C.P. R.W.-R. is a fellow of the Medical Research Council of Canada. K.J.E. was supported by an NIH training grant. G.C.P. is the recipient of an American Cancer Society Junior Faculty Award and is a Pew Scholar in the Biomedical Sciences.
REFERENCES
- 1.Alemà S, Tatò F. Oncogenes and muscle differentiation: multiple mechanisms of interference. Semin Cancer Biol. 1994;5:147–156. [PubMed] [Google Scholar]
- 2.Andrés V, Walsh K. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J Cell Biol. 1996;132:657–666. doi: 10.1083/jcb.132.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bader D, Masaki T, Fischman D A. Immunochemical analysis of myosin heavy chain during avian myogenesis in vivo and in vitro. J Cell Biol. 1982;95:763–770. doi: 10.1083/jcb.95.3.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bar-Sagi D, Schlessinger J. Activation of Ras and other signaling pathways by receptor tyrosine kinases. Cold Spring Harbor Symp Quant Biol. 1994;59:173–179. doi: 10.1101/sqb.1994.059.01.021. [DOI] [PubMed] [Google Scholar]
- 5.Bauer F, Urdaci M, Aigle M, Crouzet M. Alteration of a yeast SH3 protein leads to conditional viability with defects in cytoskeletal and budding patterns. Mol Cell Biol. 1993;13:5070–5084. doi: 10.1128/mcb.13.8.5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blau H M, Pavlath G K, Hardeman E C, Chiu C P, Silberstein L, Webster S G, Miller S C, Webster C. Plasticity of the differentiated state. Science. 1985;230:758–766. doi: 10.1126/science.2414846. [DOI] [PubMed] [Google Scholar]
- 7.Borden E C, Lotan R, Levens D, Young C W, Waxman S. Differentiation therapy of cancer: laboratory and clinical investigations. Cancer Res. 1993;53:4109–4115. [PubMed] [Google Scholar]
- 8.Boulikas T. Nuclear localization signals. Crit Rev Eukarytic Gene Expr. 1993;3:193–227. [PubMed] [Google Scholar]
- 9.Breitman T R, Chen Z X, Takahashi N. Potential applications of cytodifferentiation therapy in hematologic malignancies. Semin Hematol. 1994;31:18–25. [PubMed] [Google Scholar]
- 10.Butler M H, David C, Ochoa G-C, Freyberg Z, Daniell L, Grabs D, Cremona O, De Camilli P. Amphiphysin II (SH3P9; BIN1), a member of the amphiphysin/RVS family, is concentrated in the cortical cytomatrix of axon initial segments and nodes of Ravier in brain and around T tubules in skeletal muscle. J Cell Biol. 1997;137:1355–1367. doi: 10.1083/jcb.137.6.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carriaga M T, Henson D E. The histologic grading of cancer. Cancer. 1995;75:406–421. doi: 10.1002/1097-0142(19950101)75:1+<406::aid-cncr2820751322>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 12.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cher M L, Bova G S, Moore D H, Small E J, Carroll P R, Pin S S, Epstein J I, Isaacs W B, Jensen R H. Genetic alterations in untreated metastases and androgen-independent prostate cancer detected by comparative genomic hybridization and allelotyping. Cancer Res. 1996;56:3091–3102. [PubMed] [Google Scholar]
- 14.Collins S J, Gallo R C, Gallagher R E. Continuous growth and differentiation of human myeloid leukaemic cells in suspension culture. Nature. 1977;270:347–349. doi: 10.1038/270347a0. [DOI] [PubMed] [Google Scholar]
- 15.Crescenzi M, Crouch D H, Tatò F. Transformation by myc prevents fusion but not biochemical differentiation of C2C12 myoblasts: mechanisms of phenotypic correction in mixed culture with normal cells. J Cell Biol. 1994;125:1137–1145. doi: 10.1083/jcb.125.5.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denis N, Blanc S, Leibovitch M P, Nicolaiew N, Dautry F, Raymondjean M, Kruh J, Kitzis A. c-myc oncogene expression inhibits the initiation of myogenic differentiation. Exp Cell Res. 1987;172:212–217. doi: 10.1016/0014-4827(87)90107-8. [DOI] [PubMed] [Google Scholar]
- 17.Dyson, N. 1994. pRB, p107 and the regulation of the E2F transcription factor. J. Cell Sci. 18(Suppl.):81–87. [DOI] [PubMed]
- 18.Eckner R, Yao T-P, Oldread E, Livingston D M. Interaction and functional collaboration of p300/CBP and bHLH proteins in muscle and B-cell differentiation. Genes Dev. 1996;10:2478–2490. doi: 10.1101/gad.10.19.2478. [DOI] [PubMed] [Google Scholar]
- 19.Elliott, K., D. Sakamuro, W. Du, and G. C. Prendergast. Bin1 inhibits Myc transactivation and cell proliferation by diverse mechanisms.
- 20.Gu W, Bhatia K, Magrath I T, Dang C V, DallaFavera R. Binding and suppression of the myc transcriptional activation domain by p107. Science. 1994;264:251–254. doi: 10.1126/science.8146655. [DOI] [PubMed] [Google Scholar]
- 21.Gu W, Schneider J W, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72:309–324. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- 22.Halevy O, Novitch B G, Spicer D B, Skapek S X, Rhee J, Hannon G J, Beach D, Lassar A B. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 1995;267:1018–1021. doi: 10.1126/science.7863327. [DOI] [PubMed] [Google Scholar]
- 23.Harper J W, Elledge S J. Cdk inhibitors in development and cancer. Curr Opin Genet Dev. 1996;6:56–64. doi: 10.1016/s0959-437x(96)90011-8. [DOI] [PubMed] [Google Scholar]
- 24.Huebner K, Ohta M, Lubinski J, Berd D, Maguire H C. Detection of specific genetic alterations in cancer cells. Semin Oncol. 1996;23:22–30. [PubMed] [Google Scholar]
- 25.Hunter T. Oncoprotein networks. Cell. 1997;88:333–346. doi: 10.1016/s0092-8674(00)81872-3. [DOI] [PubMed] [Google Scholar]
- 26.Jones C L, Kane M A. Oncogenic signaling. Curr Opin Oncol. 1996;8:54–59. doi: 10.1097/00001622-199601000-00010. [DOI] [PubMed] [Google Scholar]
- 27.La Rocca S A, Crouch D H, Gillespie D A F. c-Myc inhibits myogenic differentiation and myoD expression by a mechanism which can be dissociated from cell transformation. Oncogene. 1994;9:3499–3508. [PubMed] [Google Scholar]
- 28.Negorev D, Reithman H, Wechsler-Reya R, Sakamuro D, Prendergast G C, Simon D. The Bin1 gene localizes to human chromosome 2q1.4 by PCR analysis of somatic cell hybrids and fluorescence in situ hybridization. Genomics. 1996;33:329–331. [PubMed] [Google Scholar]
- 29.Olson E N. Interplay between proliferation and differentiation within the myogenic lineage. Dev Biol. 1992;154:261–272. doi: 10.1016/0012-1606(92)90066-p. [DOI] [PubMed] [Google Scholar]
- 30.Ramjaun A R, Micheva K D, Bouchelet I, McPherson P S. Identification and characterization of a nerve terminal-enriched amphiphysin isoform. J Biol Chem. 1997;272:16700–16706. doi: 10.1074/jbc.272.26.16700. [DOI] [PubMed] [Google Scholar]
- 31.Raza A, Yousuf N, Bokhari S A J, Mehdi A, Masterson M, Lampkin B, Yanik G, Mazewski C, Khan S, Preisler H. Contribution of in vivo proliferation/differentiation studies toward the development of a combined functional and morphologic system of classification of neoplastic diseases. Cancer. 1992;69:1557–1566. doi: 10.1002/1097-0142(19920315)69:6+<1557::aid-cncr2820691309>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 32.Roskelley C D, Srebrow A, Bissell M J. A hierarchy of ECM-mediated signalling regulates tissue-specific gene expression. Curr Opin Cell Biol. 1995;7:736–747. doi: 10.1016/0955-0674(95)80117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a.Rowe, F., R. Buccafusca, and G. C. Prendergast. Unpublished data.
- 33.Sachs L. The control of hematopoiesis and leukemia: from basic biology to the clinic. Proc Natl Acad Sci USA. 1996;93:4742–4749. doi: 10.1073/pnas.93.10.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sakamuro D, Elliott K, Wechsler-Reya R, Prendergast G C. BIN1 is a novel MYC-interacting protein with features of a tumor suppressor. Nature Genet. 1996;14:69–77. doi: 10.1038/ng0996-69. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. I. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Sejersen T, Sumegi J, Ringertz N R. Density-dependent arrest of DNA replication is accompanied by decreased levels of c-myc mRNA in myogenic but not in differentiation-defective myoblasts. J Cell Physiol. 1985;125:465–470. doi: 10.1002/jcp.1041250315. [DOI] [PubMed] [Google Scholar]
- 37.Skapek S X, Rhee J, Spicer D B, Lassar A B. Inhibition of myogenic differentiation in proliferating myoblasts by cyclin D1-dependent kinase. Science. 1995;267:1022–1024. doi: 10.1126/science.7863328. [DOI] [PubMed] [Google Scholar]
- 38.Slack R S, Miller F D. Retinoblastoma gene in mouse neural development. Dev Genet. 1996;18:81–91. doi: 10.1002/(SICI)1520-6408(1996)18:1<81::AID-DVG9>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 39.Soddu S, Blandino G, Scardigli R, Coen S, Marchetti A, Rizzo M G, Bossi G, Cimino L, Crescenzi M, Sacchi A. Interference with p53 protein inhibits hematopoietic and muscle differentiation. J Cell Biol. 1996;134:193–204. doi: 10.1083/jcb.134.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sparks A B, Hoffman N G, McConnell S J, Fowlkes D M, Kay B K. Cloning of ligand targets: systematic isolation of SH3 domain-containing proteins. Nat Biotechnol. 1996;14:741–744. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]
- 41.Tiainen M, Spitkovsky D, Jansen-Dürr P, Sacchi A, Crescenzi M. Expression of E1A in terminally differentiated muscle cells reactivates the cell cycle and suppresses tissue-specific genes by separable mechanisms. Mol Cell Biol. 1996;16:5302–5312. doi: 10.1128/mcb.16.10.5302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsutsui K, Maeda Y, Tsutsui K, Seki S, Tokunaga A. cDNA cloning of a novel amphiphysin isoform and tissue-specific expression of its multiple splice variants. Biochem Biophys Res Commun. 1997;236:178–183. doi: 10.1006/bbrc.1997.6927. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Nadal-Ginard B. Regulation of cyclins and p34cdc2 expression during terminal differentiation of C2C12 myocytes. Biochem Biophys Res Commun. 1995;206:82–88. doi: 10.1006/bbrc.1995.1012. [DOI] [PubMed] [Google Scholar]
- 44.Webster K A, Muscat G E, Kedes L. Adenovirus E1A products suppress myogenic differentiation and inhibit transcription from muscle-specific promoters. Nature. 1988;332:553–557. doi: 10.1038/332553a0. [DOI] [PubMed] [Google Scholar]
- 45.Wechsler-Reya, R. Unpublished data.
- 46.Wechsler-Reya R, Elliott K, Herlyn M, Prendergast G C. The putative tumor suppressor BIN1 is a short-lived nuclear phosphoprotein whose localization is altered in malignant cells. Cancer Res. 1997;57:3258–3263. [PubMed] [Google Scholar]
- 47.Wechsler-Reya, R., D. Sakamuro, J. Zhang, J. DuHadaway, and G. C. Prendergast. 1998. Structural analysis of the human BIN1 gene: evidence of tissue-specific transcriptional regulation and alternate RNA splicing. J. Biol. Chem., in press. [DOI] [PubMed]