Abstract
During asymmetric cell division, the membrane-associated Numb protein localizes to a crescent in the mitotic progenitor and is segregated predominantly to one of the two daughter cells. We have identified a putative serine/threonine kinase, Numb-associated kinase (Nak), which interacts physically with the phosphotyrosine binding (PTB) domain of Numb. The PTB domains of Shc and insulin receptor substrate bind to an NPXY motif which is not present in the region of Nak that interacts with Numb PTB domain. We found that the Numb PTB domain but not the Shc PTB domain interacts with Nak through a peptide of 11 amino acids, implicating a novel and specific protein-protein interaction. Overexpression of Nak in the sensory organs causes both daughters of a normally asymmetric cell division to adopt the same cell fate, a transformation similar to the loss of numb function phenotype and opposite the cell fate transformation caused by overexpression of Numb. The frequency of cell fate transformation is sensitive to the numb gene dosage, as expected from the physical interaction between Nak and Numb. These findings indicate that Nak may play a role in cell fate determination during asymmetric cell divisions.
A multicellular organism originates from a single cell via mitosis, leading to the generation of different cell types. The asymmetric cell division which generates two daughter cells of different fates is one of the ways to produce diversity of cell types. Drosophila sensory organ development is well suited for the study of asymmetric cell division (10, 32, 41). These sensory organs are generated through a few rounds of asymmetric cell divisions from sensory organ precursor (SOP) cells. For example, in the external sensory organ lineage (see Fig. 8H and I), a single SOP cell divides to give rise to two different daughter cells, A and B cells. The A cell divides to produce a hair cell and a socket cell, whereas the B cell generates a neuron and a sheath cell. All three divisions are asymmetric, and each generates two daughter cells of different fates.
FIG. 8.
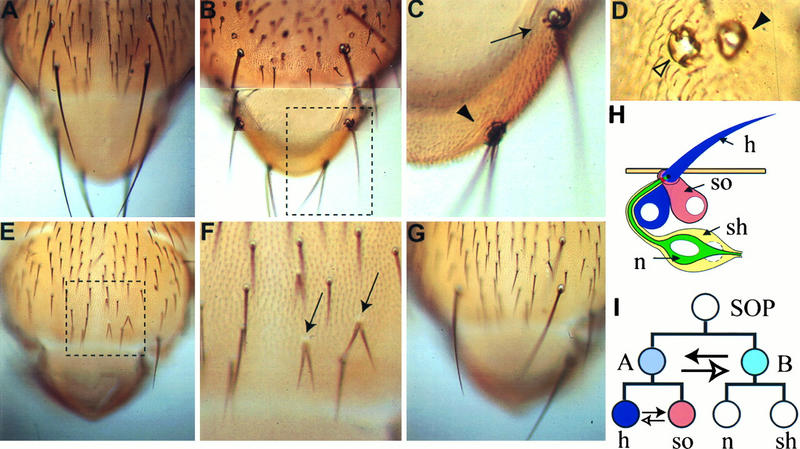
Cell fate transformation in the external sensory organs on the nota. (A) From a wild-type fly; (B) from a Nak overexpression fly; (E) from a Numb overexpression fly; (G) from a fly with both Nak and Numb overexpression; (C and F) from the inlets of panels B and E, respectively. In panel C, the arrowhead indicates a 2-hair–2-socket bristle and the arrow indicates a 1-hair–3-socket bristle. In panel D, the filled arrowhead indicates a 2-socket phenotype and the empty arrowhead indicates a 4-socket phenotype due to overexpression of Nak. In panel F, the 2-hair–no-socket phenotype caused by Numb overexpression is marked with two arrows. (H) Schematic drawing of an adult external sensory organ; (I) external sensory organ lineage, with the solid arrow indicating the direction of cell fate transformation due to overexpression of Nak and the lower, empty arrow indicating the direction caused by overexpression of Numb. n, neuron; sh, sheath cell; h, hair cell, so, socket cell.
Asymmetric cell division may arise from unequal distribution of an intracellular determinant, leading to its preferential segregation into one of the two daughter cells (for a review, see reference 29). The membrane-associated Numb protein is such a determinant for the asymmetric cell divisions of the sensory organ lineage (42). The Numb protein localizes to a cortical crescent in SOP during prophase and segregates preferentally to one of the two daughter cells in telophase (35). Genetic analysis reveals a role of numb in the asymmetric cell divisions (42, 49). Loss of numb function can be induced at different developmental stages and causes both daughter cells of a normally asymmetric cell division to adopt the same fate; the SOP divides to give rise to two A cells (B-cell-to-A-cell transformation; see Fig. 8I for external sensory organ lineage). The A cell divides to produce two hair cells (socket-to-hair cell transformation), and the B cell divides to generate two sheath cells (neuron-to-sheath cell transformation). By contrast, overexpression of Numb results in a transformation opposite that observed in loss-of-function mutants in each of these three divisions, suggesting that the Numb protein level influences cell fate specification. Numb is also localized during the mitosis of neuroblasts in the central nervous system (35, 42), and it is required for the asymmetric division of the MP2 precursors in the central nervous system (47).
Besides intracellular determinants, extracellular cues, such as signaling molecules or cell-cell interactions, can also affect cell fate specification and lead to asymmetric cell division (31). Cell-cell interaction mediated by the membrane Notch receptor represents one mechanism for external cues to affect asymmetric cell division. Notch may be activated by Delta or other ligands to regulate downstream targets, such as Suppresser of Hairless [Su(H)] (2, 36) and Tramtrack (19), which are required for cell fate specification in the sensory organ lineage. Loss or gain of Notch function during the formation of the sensory organs and MP2 neurons causes cell fate transformations, which are opposite those caused by the loss or gain of numb function, respectively (19, 25, 46). Epistasis analysis indicates that Notch acts downstream of numb (19, 46). Direct protein-protein interaction has been observed between Numb and Notch as well as mammalian homologs of Numb and Notch (19, 53). Moreover, Numb inhibits Delta-dependent Notch signaling in cultured Drosophila melanogaster S2 cells, as indicated by the failure of Su(H) to translocate into the nucleus, and ectopically expressed Numb inhibits Notch function during wing development (17). Thus, both the cell-intrinsic factor Numb and the cell-extrinsic influence mediated by Notch are important for proper cell fate specification during asymmetric division, and Numb functions at least in part by antagonizing Notch activity.
The Numb protein contains a phosphotyrosine binding (PTB) domain (also called a phosphotyrosine-interacting domain) (7). The PTB domains of a mouse homolog of Numb (mNumb) (53) and a rat homolog of the mouse Numblike protein (rNbl) (54) show 71.5 and 75.2% amino acid identity with the PTB domain of Drosophila Numb (dNumb). Like dNumb, mNumb is asymmetrically localized during divisions of neural progenitors, and overexpression of either mNumb or rNbl causes cell fate transformation in the Drosophila sensory organ lineage, indicating that the functions of the Numb proteins are conserved through evolution. The PTB domain of dNumb is not necessary for its asymmetric localization but is required for its function (17). Overexpression of PTB domain-deleted dNumb does not cause cell fate transformation in vivo, nor does it inhibit Su(H) nuclear translocation mediated by Notch signaling in cultured S2 cells, even though it still forms a crescent during mitosis.
The PTB domain is also found in signaling molecules such as Shc (5, 34) and insulin receptor substrate (IRS) families (27). As adapter proteins, Shc binds to activated receptors such as epidermal growth factor, nerve growth factor, and antigen receptors, and IRS binds to activated insulin receptor, so as to transduce signals to downstream effector proteins (reviewed in reference 37). When activated, those receptors autophosphorylate at the tyrosine residues, leading to the binding of phosphotyrosines with the Src homology 2 (SH2) or PTB domains of the adapter proteins. Despite the lack of strong sequence similarity between the Shc and IRS PTB domains, interaction of both PTB domains with the NPXpY motifs (in which tyrosine residue is phosphorylated) has been shown (22, 27, 33). However, recognition of the Shc PTB domain and the IRS-1 PTB domain depends on the sequence context of the NPXpY motif (28, 52), suggesting different binding specificity of different PTB domains. Structural analysis further indicates that the topology of the two PTB domains resembles that of the pleckstrin homology (PH) domain (15, 37, 55), with a β-sandwich formed by two nearly antiparallel β-sheets and capped on one end by a C-terminal α-helix. In the complex of the PTB domain and the phosphopeptide-bearing NPXpY motif, the peptide forms a β-strand antiparallel to one of the two β-sheets of the PTB domain and the phosphotyrosine fits into a largely hydrophobic pocket. The Shc PTB domain and the IRS PTB domain exhibit similar mechanisms of ligand binding but use different residues for phosphotyrosine recognition. Although the PH domain is similar in topology to the PTB domains, the mechanism of ligand recognition by PH domain is different from that of PTB domain; the surface of the PH domain of phospholipase Cδ recognized by its ligand, inositol 1,4,5-triphosphate (16), is different from the surface of the PTB domain recognized by the NPXpY motif.
The involvement of Numb protein in asymmetric cell division raises a number of questions. How does Numb protein become asymmetrically localized during mitosis? How might the physical interaction between Numb and Notch lead to the inhibition of Notch signaling? What is the function of the PTB domain of Numb? Does Numb have other functions in addition to the inhibition of Notch signaling? Given the possibility that the localization or the function of Numb may involve proteins that bind to Numb, we have searched for Numb-interacting proteins by using the yeast two-hybrid system (12) as a first step to answer those questions. We have isolated a novel gene product, Numb-associated kinase (Nak), which showed strong interaction with Numb in biochemical assays. Deletion analysis indicated that the interaction involves the PTB domain of Numb. Whereas the PTB domains of Shc and IRS bind to the NPXpY sequence motif, the interaction of the Numb PTB domain with Nak does not require this motif or a tyrosine. The specificity of Nak interaction with Numb was examined by testing its binding with the PTB domain of Shc. The possible activity of Nak in asymmetric cell division was explored in transgenic flies overexpressing Nak.
MATERIALS AND METHODS
Yeast two-hybrid screening and assays.
The GAL4 activating domain (GAD) library used in the yeast two-hybrid screen was a gift from S. Elledge (Baylor College of Medicine). It was made from third-instar larval cDNA. The methodology in the yeast two-hybrid screening is as described in reference 3. Briefly, the library cDNA was cotransformed with pBHA-Numb (encoding the LexA-Numb fusion protein) into L40 strain (30), and 2 million colonies were selected for His+. The His+ colonies were filter lifted for blue color assay, using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) as the substrate for LacZ activity. After a secondary screening against LexA-Lamin (3) instead of LexA-Numb, five positive clones were obtained; two of the positive clones encode Nak, as shown in Fig. 1A.
FIG. 1.

(A) Schematic representation of the open reading frame of nak, which encodes 1,490 aa. The hatched box indicates the serine/threonine kinase catalytic domain. Also indicated are the starting points of the two positive clones, 13-1 and 7-2, from the two-hybrid screen. (B) Comparison of the kinase catalytic domains of Nak, two proteins from yeast (GenBank accession no. p38080 and p40494), and one from C. elegans (z46242). Only those residues that are identical in these four proteins are boxed. Some of these residues are characteristic of the serine/threonine kinase family and are marked with black dots. The subdomains of the catalytic domain are marked by I to XI. (C) Complete predicted amino acid sequence of Nak. The 11-aa peptide near the C terminus that binds the Numb PTB domain is highlighted.
The LacZ (β-galactosidase) activities given in Fig. 2 to 5 are the averages and standard deviations of six independent samples. β-Galactosidase activity was tested for and activity as described in reference 4, with the following modifications. Four-tenths milliliter of log-phase yeast culture grown in standard yeast medium with 2% galactose, 2% glycerol, and 2% ethanol as carbon sources was mixed with 0.4 ml of Z buffer. After addition of 50 μl of chloroform and 50 μl of 0.1% sodium dodecyl sulfate, the yeast cultures were vortexed for 30 s and preincubated at 30°C for 5 min before the introduction of 160 μl of the substrate, o-nitrophenyl-β-d-galactopyranoside (4 mg/ml). The tubes were further incubated at 30°C until the yellow color developed.
FIG. 2.
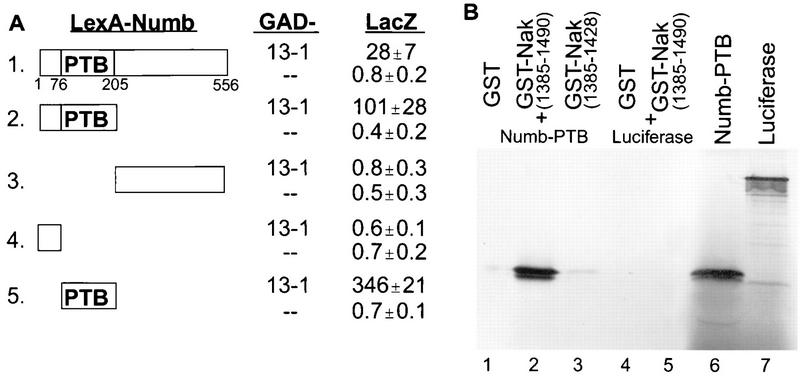
Nak binding to the PTB domain of Numb. (A) Mapping the interaction domain of Numb, using the yeast two-hybrid system. Various Numb fragments were fused with LexA and tested with GAD alone (−−) or the 13-1 clone encoding a fusion of GAD and aa 1088 to 1490 of Nak (13-1). For the assay and calculation of β-galactosidase (LacZ) activity, see Materials and Methods. The means and standard deviations calculated from six independent samples are given. (B) In vitro binding of the Numb PTB domain to the Nak C terminus. GST-Nak(1385–1490) (lane 2), but not GST alone (lane 1) or GST-Nak(1385–1428) (lane 3), binds to 35S-labeled Numb-PTB domain. Neither GST nor GST-Nak(1385-1490) interacts with luciferase (lanes 4 and 5). The loading controls for Numb-PTB and luciferase are shown in lanes 6 and 7.
FIG. 5.
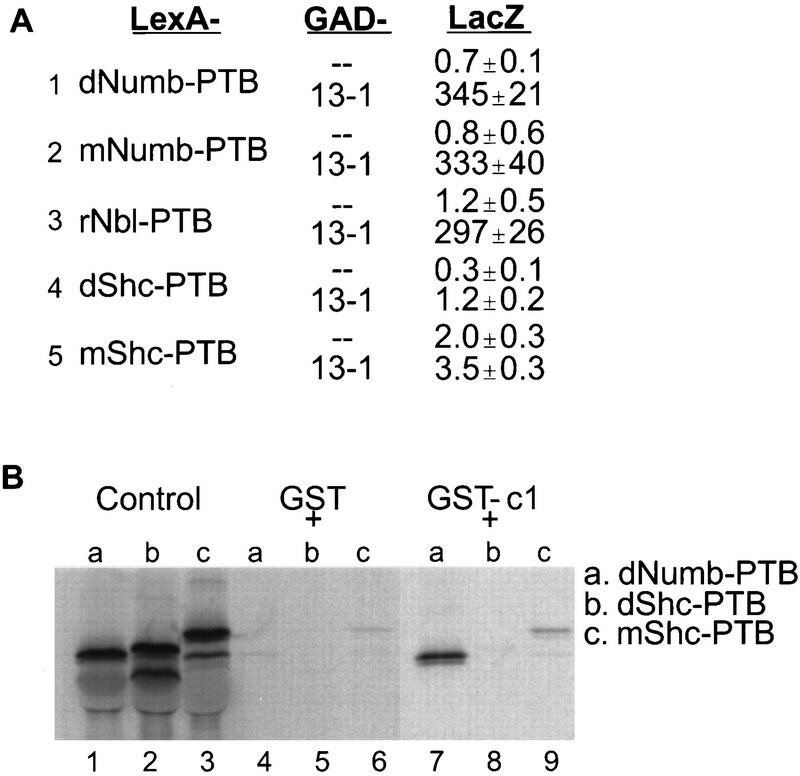
Test for the specificity of interaction between Nak and PTB domains. (A) The PTB domains of mNumb, rNbl, dShc, and mShc were cotransformed with GAD or GAD-Nak (aa 1088 to 1490, the 13-1 clone) into the yeast two-hybrid reporter strain for LacZ activity assay. (B) The PTB domains of dShc and mShc showed no interaction with the c1 fragment (amino acids 1422 to 1490) of Nak in the in vitro binding assay. The control is as for Fig. 2B.
The pBHA vector was modified from pBTM116 (4) with a hemagglutinin tag (YPYDVPDYA) inserted at the beginning of the multiple cloning sites and was used to clone the various fragments of Numb and Shc. The hemagglutinin tag was used to examine the expression of the fusion proteins in yeast (data not shown).
pGAD-GH (23) was used to clone the C-terminal fragments of Nak shown in Fig. 4B. The various deletions of Nak shown in Fig. 4A were done in the original pACT (14) vector used for constructing the library.
FIG. 4.
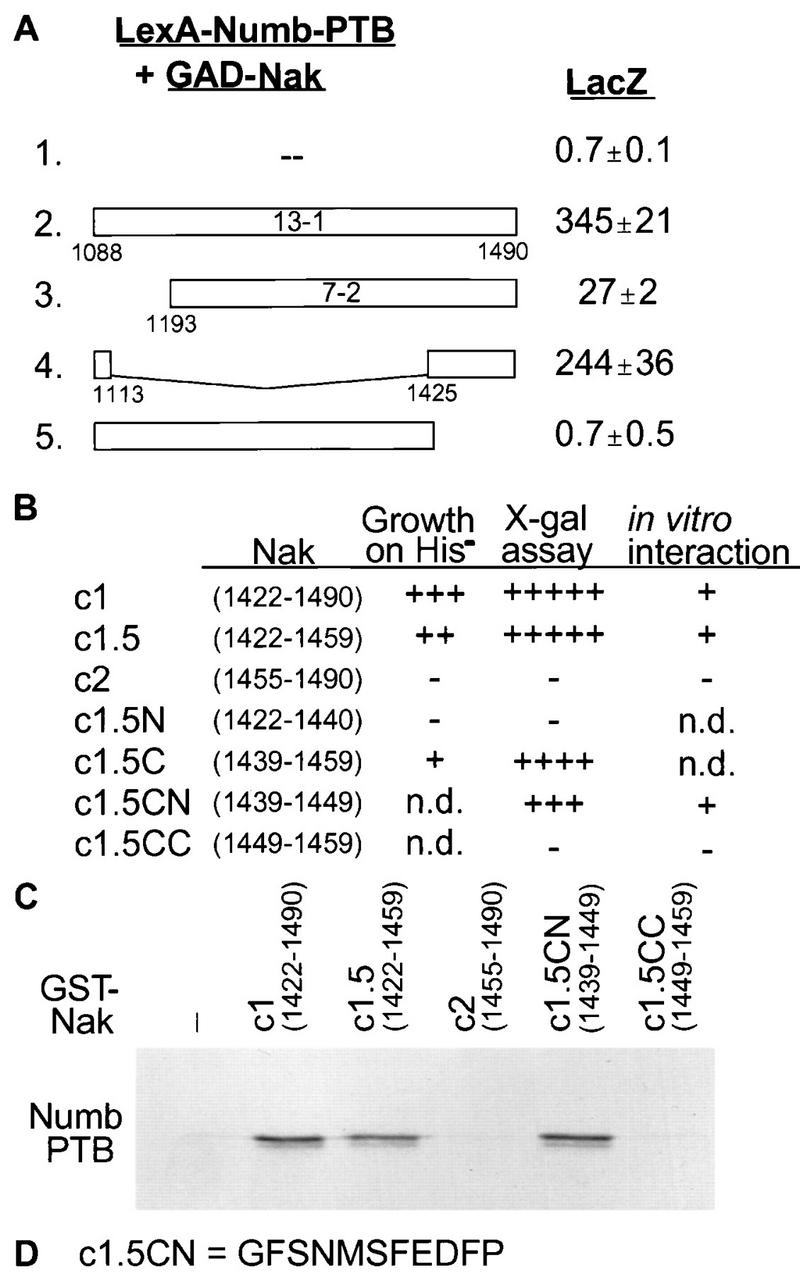
Mapping the interaction region of Nak. (A) Various deletions of the 13-1 constructs affect the interaction with LexA-Numb-PTB to different extent, as indicated by the LacZ activities. (B) Various Nak C-terminal regions were tested for their interaction with LexA-Numb-PTB as indicated by the growth of yeast colonies on a His− plate and by the appearance of blue color in filter lift X-Gal assays. Growth after two days on His− plates was scored, and the number of +’s indicates relative size. −, no growth. For filter lift X-Gal assay, +++++ represents colonies turning blue in 15 min and very dark blue at 300 min, ++++ represents colonies turning blue in 30 min and dark blue at 300 min, +++ represents colonies turning blue in 60 min and medium blue at 300 min, and − represents colonies with no blue color in 300 min. (C) In vitro binding assay to test the interaction between the Nak C-terminal fragments and Numb-PTB. The results of this assay are summarized in the rightmost column of panel B. (D) Amino acid sequence for the smallest region of Nak that interacts with Numb-PTB.
The DNA constructs for fusion proteins of LexA and PTB domains of mNumb and rNbl were provided by W. Zhong, and the DNA fragments for the PTB domains of Drosophila Shc (dShc) and mouse Shc (mShc) were obtained by PCR from a Drosophila and a mouse cDNA library (a gift from Y.-W. Chen, University of California, San Francisco), respectively. The junctions for the PTB domains were as given in reference 55.
Molecular cloning.
The full-length cDNA encoding Nak was obtained by screening both Drosophila λ-ZAP larval and pupal cDNA libraries (gifts from C. S. Thummel, University of Utah).
In vitro binding assays.
Various Nak C-terminal fragments were cloned into in-frame pGEX vectors (Pharmacia Co.) for expression in Escherichia coli DH5α as glutathione S-transferase (GST) fusion proteins. The expression of fusion proteins was induced for 2 to 3 h with 100 μM isopropylthiogalactopyranoside added to log-phase bacteria. Following sonication of the bacteria in phosphate-buffered saline, 0.1% Triton X-100 was added before centrifugation to remove the insoluble pellet. The supernatant were kept in 10% glycerol at −80°C.
The DNA fragments containing coding sequences for PTB domains of Numb and Shc were cloned into pNAC vector (a gift from J. P. O’Connor, University of Penn.), and the 35S-labeled proteins were expressed in the TnT coupled lysate system (Promega Co.). For in vitro binding assay, 2 to 5 μg of GST fusion proteins was incubated at 4°C for 30 min with 5 to 10 μl of lysate containing 35S-labeled proteins. After addition of 20 μl of GST-agarose beads (Sigma Co.), the tubes were incubated at room temperature for 2 min. These beads were washed four times with phosphate-buffered saline–100 mM NaCl–0.1% Triton X-100 and then mixed with protein loading buffer to elude the bound proteins.
To estimate the dissociation constant for binding of the GST-c1 protein fusion to the Numb PTB domain, we used several concentrations of GST-c1 protein to precipitate the in vitro-translated Numb PTB. The concentration of Gst-c1 required to precipitate 50% of labeled Numb PTB is about 1 μM.
Immunoprecipitation.
Embryos were collected overnight from a cross of UAS (upstream activation sequence)-myc-nak males and scabrous-GAL4 females (described below). Embryo extract was prepared according to reference 13. Anti-c-Myc antibody-agarose conjugate (Santa Cruz Biotechnology Co.) was used for coimmunoprecipitation, and the precipitates were run on sodium dodecyl sulfate-polyacrylamide gels for Western blotting with either anti-c-Myc or anti-Numb antibodies.
Fly genetics.
The full-length DNA fragments for nak and numb genes were the NotI-XbaI fragment from pBS-13-1F and KpnI fragment from hs-numb#2 (42), respectively. Each DNA fragment was subcloned into the respective restriction sites of the pUAST vector. The pUAST-nak and pUAST-numb DNAs were injected into w− flies to create transgenic flies (43). These transgenic flies were crossed to Sca-GAL4 (29) or 109-68 flies (17); these GAL4 enhancer trap lines probably have GAL4 coding sequence inserted at the scabrous locus. The larval progenies from these crosses were kept at either 25 or 30°C (normally stronger phenotype is obtained at 30 than at 25°C), and their overexpression phenotypes in the sensory organs were examined. About one-third (10 of 33) of the UAS-nak and most (15 of 18) of the UAS-numb independent transformants gave similar phenotypes, though the strength of the phenotypes varied with the transformant lines. Transformants 15 of UAS-nak and 14 of UAS-numb gave strong and consistent phenotype and were used for the quantitative analysis in this report.
pUAS-myc-nak was created by insertion of nak open reading frame sequence into pBS-βG vector (from I. Clark) which includes 5′ leader sequence of the Xenopus globin gene and a myc tag sequence. The myc-nak fusion sequence was subcloned into pUAST vector for embryo injection.
Phenotype examination.
For examining the embryonic neuron/sheath cell transformation, homozygous UAS-nak and UAS-numb transgenic flies were crossed to homozygous Sca-GAL4 flies. Embryos were collected over a period of 4 h at room temperature and moved to 30°C for another 9.5 h. The fixation and staining procedures were as described in reference 16. Monoclonal antibody 22C10 (56) and rabbit anti-Prospero (50) were used at 1:250 and 1:1,000 dilutions to stain neurons and sheath cells, respectively. The images were analyzed with a Zeiss microscope and a Bio-Rad MRC-600 confocal microscope.
Adult nota were dissected from flies in 80% isopropanol and mounted in Hoyer’s medium (1).
RESULTS
Identification of the nak gene.
To identify genes encoding proteins which interact physically with the Numb protein, we used the yeast two-hybrid system to screen a library of third-instar larval cDNA fused to the coding sequences of GAD (see Materials and Methods). From 2 million colonies, we isolated two independent clones (13-1 and 7-2 [Fig. 1A]) that correspond to a novel gene, nak, with an open reading frame of 1,490 amino acids (aa) (Fig. 1C). Nak protein includes a putative serine/threonine kinase domain at the N terminus (Fig. 1A), which has about 40% amino acid identity with the kinase domains encoded by a gene from Caenorhabditis elegans and two genes from Saccharomyces cerevisiae (Fig. 1B). The functions of these genes are not known. Nak and the three related putative kinases contain the characteristic residues that are conserved in the subdomains of known kinases with the exception of domain I: instead of the GXGXXG nucleotide binding motif present in domain I of most kinases, Nak and related kinases contain a GGFA motif. The GGFA motif is also present in the polo kinase family, plo1+ of Schizosaccharomyces pombe, CDC5 of S. cerevisiae, polo of D. melanogaster, and polo-like kinase of mammals (39), suggesting that these proteins may represent a new class of kinases.
To determine the expression pattern of nak, we performed Northern analysis, which showed a 5.5-kb mRNA expressed in all stages (embryo, larvae, pupae, and adult) of development, a 4.5-kb mRNA expressed at least in 0- to 12-h embryos and a 3-kb mRNA expressed in 0- to 3-h, but not 3- to 12-h, embryos. All three messages hybridized to a probe containing coding sequence of the kinase domain, but only the 5.5-kb mRNA hybridized to a probe corresponding to the Nak C terminus (aa 1088 to 1490). A ubiquitous expression pattern in whole mount embryos was observed (data not shown).
Binding of Nak to the PTB domain of Numb.
The Nak-interacting domain of Numb was mapped by using the yeast two-hybrid system. Either the N-terminal part (Fig. 2A, line 2) or the PTB domain (Fig. 2A, line 5) of Numb interacted with the 13-1 clone, which includes the Nak C-terminal region (aa 1088 to 1490). Fragments on either end of the Numb PTB domain did not show any interaction with 13-1 in the two-hybrid assay (Fig. 2A, lines 3 and 4). Thus, the PTB domain of Numb is necessary and sufficient for the interaction with the C-terminal fragment of Nak.
The interaction between the Numb PTB domain and Nak C terminus was also evident in an in vitro binding assay, namely, the coprecipitation of the bacterial fusion protein GST-Nak (aa 1385 to 1490), which includes the Numb-interacting region of Nak (see below), and in vitro-translated PTB domain of Numb (Fig. 2B, lane 6) by glutathione-agarose beads (Fig. 2B, lane 2). This coprecipitation was not observed with either GST or GST-Nak (aa 1385 to 1428), which lacks the Numb-interacting region of Nak (see below). In another control, both GST and GST-Nak (aa 1385 to 1490) failed to coprecipitate with in vitro-translated luciferase protein (Fig. 2B, lanes 4 and 5). Therefore, the PTB domain of Numb interacts specifically with the Nak C terminus.
Novel interactions of Nak with the PTB domain of Numb.
An NPXpY motif containing a phosphotyrosine is recognized by the PTB domains of Shc and IRS-1 (22, 27, 33) and possibly also the Numb PTB domain (51). In contrast, the Numb-interacting region of Nak (aa 1088 to 1490) does not include any NPXY motif, suggesting that the binding of the PTB domain of Numb with Nak is mechanistically different from interactions of PTB domains with the NPXY motif. To test this possibility, we examined the effects of point mutations in the Numb PTB domain which correspond to point mutations in the Shc PTB domain known to disrupt the interaction between Shc and tyrosine-phosphorylated p145 (55). The S148A mutation of the Numb PTB domain corresponds to a mutation in the Shc PTB domain which reduces the interaction between Shc and tyrosine-phosphorylated p145 by 60%, but it did not affect interaction between the Numb PTB domain and the Nak C terminus (aa 1385 to 1490) (Fig. 3A, lane 8). Another point mutation of the Numb PTB domain, R171N, corresponds to a mutation in the Shc PTB domain which eliminates the interaction between Shc and tyrosine-phosphorylated p145 completely. Like S148A, the R171N mutation did not abolish the interaction between Numb PTB domain and Nak C terminus (Fig. 3A, lane 5). Moreover, in the two-hybrid assay, these two mutants were indistinguishable from wild-type Numb in their interaction with Nak C-terminal fragment encoded by the 13-1 clone (Fig. 3B, lines 2, 4, and 6). Taken together, these observations indicate that Nak interacts with the Numb PTB domain in a manner different from interactions involving the Shc PTB domain and NPXY motif.
FIG. 3.
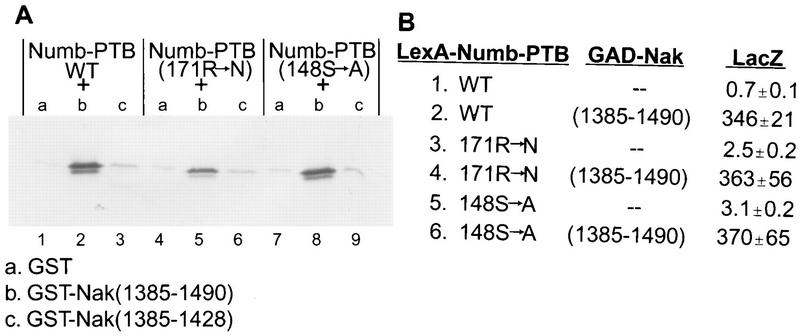
Numb PTB domain mutations predicted to disrupt the interaction with the NPXpY motif do not affect the interaction with Nak. (A) The mutation R171N (substitution of arginine with asparagine at position 171) slightly reduces the interaction between Numb-PTB and GST-Nak(1385–1490) (the middle three lanes). The mutation S148A (substitution of serine with alanine at position 148) does not affect the interaction at all (the last three lanes). The first three lanes are wild-type controls as shown in Fig. 2B. (B) Two-hybrid assay for the interactions between Numb-PTB (wild type and two mutants) and Nak(1385–1490). Both mutants, R171N and S148A, yielded LacZ activities similar to that of the wild-type control.
Identification of the Numb-binding sequence in Nak.
The C-terminal fragment of Nak is sufficient for interaction with Numb, as indicated by the isolation of the 13-1 clone (aa 1088 to 1490 of Nak) and 7-2 clone (aa 1193 to 1490 of Nak) from the two-hybrid screen. Both C-terminal fragments showed interaction with the PTB domain of Numb (Fig. 4A, lines 2 and 3) and the entire Numb protein (Fig. 2A and data not shown). A large internal deletion of the 13-1 clone that leaves the C-terminal 66 residues joined with its N-terminal 26 residues (aa 1088 to 1113 and 1425 to 1490) did not abolish the interaction with the Numb PTB domain (Fig. 4A, line 4). On the other hand, deletion of the most C-terminal 66 aa (Fig. 4A, line 5) eliminated the interaction completely. These data indicate that the C-terminal 66 aa of Nak are responsible for most of the interaction with Numb PTB domain.
To further define the Numb-binding region of Nak, we tested a series of small fragments from the Nak C terminus for their interaction with the Numb PTB domain in the two-hybrid system and in in vitro binding assays (Fig. 4B). A peptide of 11 aa (aa 1439 to 1449 of Nak, c1.5CN) is sufficient for the interaction with the PTB domain of Numb. The Numb PTB domain also recognized three other Nak constructs that contain these 11 aa (c1, c1.5, c1.5C, and c1.5CN) but not those Nak fragments without this sequence (c2, c1.5N, and c1.5CC) (Fig. 4B and C). The sequence of the 11 aa of c1.5CN, as shown in Fig. 4D, does not contain any tyrosine residues, though it is conceivable that the two serine residues in this peptide could be phosphorylated.
Specificity of Nak interaction with the PTB domain of Numb.
To examine the ability of Nak to interact with PTB domains of different proteins, we looked for its interaction with mammalian homologs of Numb as well as the PTB domains of Shc from mouse and from fly proteins. The PTB domains of fly and mammalian Numb proteins show about 70 to 75% amino acid identity (54). PTB domains of both mNumb and rNbl interacted with 13-1 clone (aa 1088 to 1490 of Nak) as strongly as the dNumb PTB domain (Fig. 5A, lines 1 to 3), suggesting that the interaction between Numb and Nak may also be conserved in mammals. In contrast, the PTB domains of dShc and mShc did not show any interaction with this Nak fragment in the two-hybrid assay (Fig. 5A, lines 4 and 5), nor did we observe any binding of the PTB domains of dShc and mShc to the c1 fragment of Nak in the in vitro binding assays (Fig. 5B, lanes 8 and 9). These results indicate that Nak interacts with PTB domain of Numb specifically. The dissociation constant for GST-c1 binding to the dNumb PTB domain was estimated to be ∼1 μM (see Materials and Methods).
Nak interacts with Numb in vivo.
To identify in vivo interaction between Nak and Numb, we created transgenic flies expressing Myc-tagged Nak protein (see Materials and Methods). Embryo extracts were prepared from the strains with or without Myc-Nak. Using anti-c-Myc antibody, Myc-Nak was detected as doublet of 200- and 180-kDa proteins (Fig. 6, lane 2), similar to the size of in vitro-expressed Nak proteins (data not shown), but not in the control lane 1. Immunoprecipitation was performed with the anti-c-Myc antibody. The two extracts contained similar amounts of Numb protein (Fig. 6, lanes 3 and 4). Numb protein was coprecipitated with Myc-Nak, detected with anti-Numb antibody (Fig. 6, lane 6), but not from embryo extract prepared from the strain without expressing Myc-Nak (Fig. 6, lane 5), indicating an in vivo interaction between Numb and Nak.
FIG. 6.
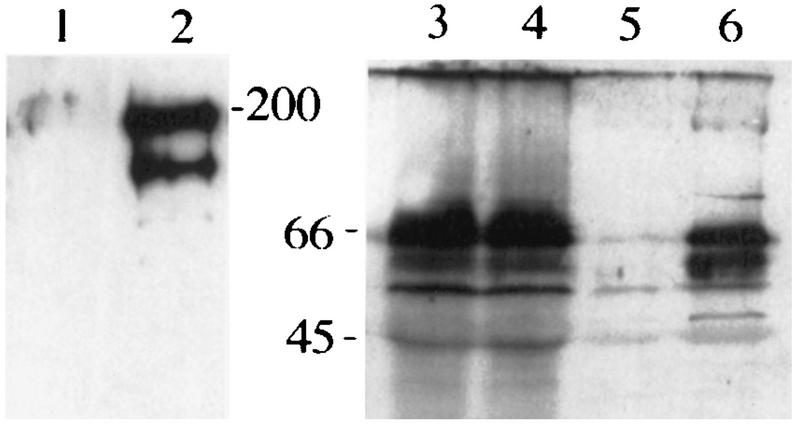
Western blot showing the coimmunoprecipitation of Numb and Nak in embryo extract. Lanes 1, 3, and 5 are embryo extracts prepared from scabrous-GAL4 strains; lanes 2, 4, and 6 are extracts from the cross of scabrous-GAL4 and UAS-myc-nak strains. After immunoprecipitation with anti-c-Myc antibody, lanes 1 and 2 were probed with anti-c-Myc antibody and lanes 5 and 6 were probed with anti-Numb antibody. Control lanes 3 and 4 are embryo extracts before immunoprecipitation and probed with anti-Numb antibody.
The cell fate transformation due to Nak overexpression is opposite to that caused by Numb overexpression.
By doing in situ hybridization to third-instar larval salivary gland polytene chromosomes, we mapped the nak gene to the cytological location 37B4-7 on the left arm of the second chromosome. The smallest available chromosomal deficiency, Df(2L)TW3 (36F7-37A1;37B2-37B7), deletes a number of genes located in the 37A and 37B region, including nak. We found that the homozygous Df(2L)TW3 embryos are quite disorganized, thus precluding us from assessing whether loss of nak function would lead to cell fate change in sensory organs. Another confounding problem to the analysis of zygotic mutant embryos is the presence of a significant maternal contribution of the nak gene activity in early embryos. Thus, we were unable to infer the loss of function phenotype by analyzing homozygous Df(2L)TW3 embryos. We therefore turned to gain-of-function experiments.
We use the GAL4-UAS system (8) to express the Nak protein and to test its function in vivo. The UAS-nak flies were crossed to scabrous-GAL4 enhancer trap lines (17, 29), which leads to the expression of Nak protein in the neural precursor cells. Overexpression of Nak during sensory organ development resulted in several phenotypes suggestive of cell fate transformations within the sensory organ lineage. In the wild-type embryos, five chordotonal neurons and five sheath cells are aligned in the lateral 5 region (lch5 [Fig. 7A, D, G, and J]) of each abdominal hemisegment. In addition, there are one chordotonal organ and three other sensory organs (Fig. 7J) in the same region. When Nak is overexpressed, we observed cell fate transformation from neuron to sheath cell in the lineage of chordotonal organs (Fig. 7B, E, and F), as shown by the increased number of sheath cells and the reduced number of neurons. This is in contrast to the overexpression of Numb protein, which increases the number of neurons and reduces the number of sheath cells in the same lineage (Fig. 7C, F, and I).
FIG. 7.
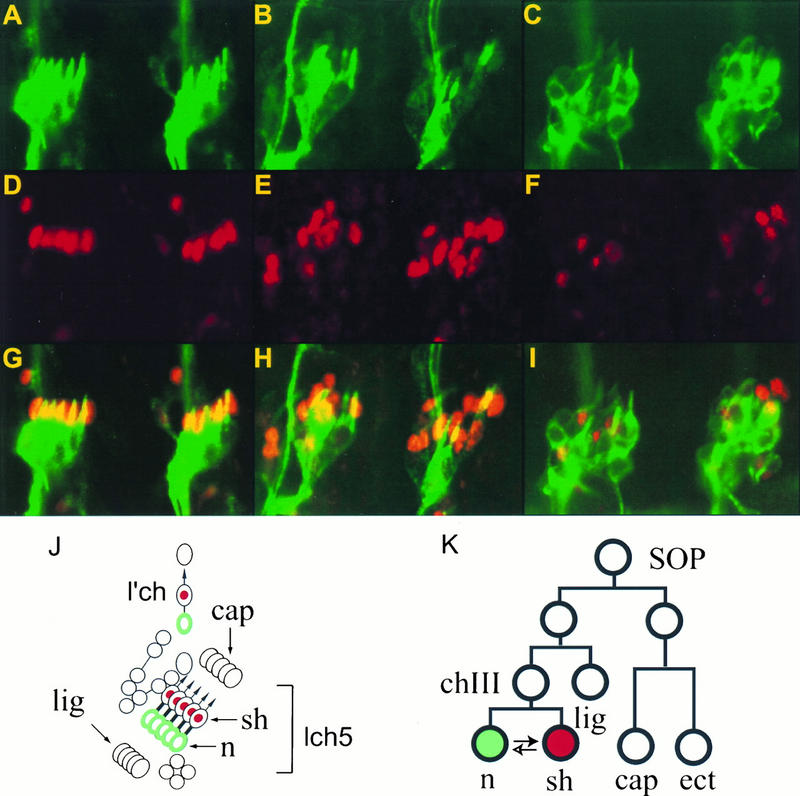
Overexpression of Nak or Numb causes opposite cell fate transformation between neurons and sheath cells in the embryonic lateral chordotonal organs. (A, D, and E) From a wild-type embryo; (B, E, and H) from an embryo with Nak overexpression; (C, F, and I) from an embryo with Numb overexpression. (A to C) Staining of neurons with antibody 22C10; (D to F) staining of the sheath cells with anti-Prospero antibody; (G to I) superimposition of the neuron and the sheath cell staining patterns; (J) Schematic drawing of the lch5 chordotonal organs and their neighboring sensory organs; (K) lineage of lch5 (9), with the solid arrow indicating the direction of cell fate transformation due to overexpression of Nak and the lower, empty arrow indicating the direction of transformation due to overexpression of Numb. n, neuron; sh, sheath cell; cap, cap cell; ect, ectodermal cell; lig, ligament cell; chIII, tertiary chordotonal precursor cell.
Similar to the embryonic phenotypes, Nak overexpression in the adult sensory organs caused cell fate transformations in the bristles in various parts of the flies, including the head, notum, and wing margin. Instead of the normal appearance of a hair and a socket (with a neuron and a sheath cell underneath), the mutant bristles of the transgenic flies sometimes contain two hairs and two sockets (Fig. 8B and C, arrowhead), indicating that the SOP cell divides symmetrically to two A cells (i.e., a transformation of B to A cell [Fig. 8I]), instead of one A and one B cell. In some cases, the sensory bristle contained two sockets but no hair (Fig. 8D, solid arrowhead), indicating that the SOP divided asymmetrically to produce an A and a B cell, but the A cell divided symmetrically to give rise to two sockets but no hair (i.e., a transformation of hair to socket [Fig. 8I]). Other phenotypes are characterized by four sockets (Fig. 8D, empty arrowhead) or one hair and three sockets (Fig. 8C, arrow), suggesting that the SOP cell divided symmetrically and at least one of the two A cells that it generated also divided symmetrically. Besides examination of the external sensory structures, we carried out immunocytochemical studies using neuron-specific antibody and found that neurons were missing under these mutant bristles (data not shown). The absence of neurons could arise either from a transformation of B cell to A cell during SOP division or from neuron-to-sheath cell transformation during the B-cell division. All of the cell fate transformations due to overexpression of Nak are opposite those caused by overexpression of Numb, which resulted in twin hair (socket-to-hair cell transformation [Fig. 8E and F]) and balding (no hair and no socket; A-to-B cell transformation [Fig. 8E]). Thus, during sensory organ development, overexpression of Nak and overexpression of Numb lead to opposite cell fate transformation in every cell division of the sensory organ lineage examined (Fig. 8K and I).
Antagonistic actions of the nak transgene and the endogenous numb gene.
We then investigated the genetic interaction between nak and numb by examining the effect of numb copy number on Nak overexpression phenotype. The phenotype caused by overexpression of Nak is enhanced by reducing the copy number of numb gene from two to one (Table 1; compare rows 1 and 2). Whereas eight macrochaetes are present at the dorsocentral and scutellar positions on the notum of a wild-type fly, overexpression of Nak caused 31.7% of these macrochaetes to exhibit phenotypes indicative of cell fate transformation when the transgenic flies contained two copies of the wild-type numb gene. The fraction of mutant macrochaetes with indication of cell fate transformation was increased to 67.1% in transgenic flies with overexpression of Nak but only one copy of wild-type numb gene (numb1/+); the 2/4-socket phenotype (see Table 1, footnotes b to d, for definitions of phenotypes) was increased from 15.0 to 30.4%, and the 1-hair–3-socket and 2-hair–2-socket phenotypes were increased from 16.7 to 36.7%. In the absence of the nak transgene, the numb1/+ flies have no bristle phenotype. Thus, the effect of nak transgene in the sensory organ development could be enhanced by reducing the gene dosage of numb.
TABLE 1.
Phenotypes of external sensory organs on the nota with different dosages of nak and numba
| Genotype (n) | Temp (°C) | % of bristles with indicated phenotype
|
|||
|---|---|---|---|---|---|
| Wild type | 2/4-socketb (without hair) | 1-hair–3-socketc/ 2-hair–2-socket | Othersd | ||
| Sca-GAL4, UAS-nak (30) | 30 | 62.5 | 15.0 | 16.7 | 5.8 |
| Sca-GAL4, UAS-nak; numb1/+ (30) | 30 | 25.0 | 30.4 | 36.7 | 7.9 |
| 109-68, UAS-numb (33) | 25 | 6.4 | 93.6 | ||
| 109-68, UAS-numb; UAS-nak (42) | 25 | 44.3 | 55.7 | ||
| 109-68, UAS-numb/TW3 (32) | 25 | 0.5 | 99.5 | ||
The flies were maintained at the indicated temperatures, and the bristle phenotypes were scored under the dissecting microscope.
Includes two-socket and four-socket mutant bristles; the exact number of bristles is sometimes difficult to determine due to fusion of the sockets.
One of the two hairs in the 2-hair–2-socket phenotype is sometimes very small due to partial transformation; therefore, we included both phenotypes in one category.
This category includes balding (no hair and no socket) and one hair without socket. The last three rows represent the balding phenotype only.
Not only did a reduction of numb gene enhance the cell fate transformation phenotype caused by Nak overexpression, overexpression of Numb as well as Nak suppressed most of the bristle phenotypes due to overexpression of either Numb or Nak alone, including the 2-socket (the phenotype of Nak overexpression), the 2-hair (the phenotype of Numb overexpression) (Fig. 8G), and the 2-hair–2-socket or 1-hair–3-socket phenotypes due to overexpression of Nak. The balding phenotype of macrochaetes caused by overexpression of Numb was also significantly reduced; 44.3% of the bristles (or 3.6 bristles per fly on average) were normal in flies overexpressing Nak and Numb, compared to only 6.4% (or 0.5 bristle per fly) in flies overexpressing Numb alone (Fig. 8E and G; Table 1, rows 3 and 4). Similarly, most of the balding phenotype of microchaetes was also suppressed in flies expressing Numb as well as Nak (Fig. 8E and G). Thus, the phenotype of the opposite cell fate transformation due to the action of Nak is sensitive to both increase and decrease of the Numb protein level.
To further test the genetic interaction between numb and nak, we used the Df(2L)TW3 deficiency, which deletes nak and several other genes, and tested if overexpression phenotype of Numb is sensitive to the gene dosage of nak. Indeed, heterozygous Df(2L)TW3 embryos that overexpressed Numb showed enhanced Numb overexpression phenotype (Table 1; compare row 3 with row 5). This result suggests that the Numb overexpression phenotype is sensitive to the gene dosage of a certain gene(s) in the region deleted in Df(2L)TW3. nak is likely to be the gene that is responsible for this genetic interaction.
DISCUSSION
In this study, we have used the yeast two-hybrid system to isolate a novel Numb-interacting protein which includes a putative serine/threonine kinase domain at the N terminus and a Numb-binding region at the C terminus. Interaction between Numb and Nak requires the PTB domain of Numb and an 11-aa peptide of Nak; this interaction is specific for the Numb PTB domain and may represent a novel interaction independent of tyrosine phosphorylation. Overexpression of Nak in transgenic flies affects asymmetric cell division in a manner that is antagonistic to the actions of Numb. The characteristics of Nak binding to the PTB domain of Numb and the possible functions of Nak in vivo are discussed below.
Nak binding to the PTB domain of dNumb, mNumb, and rNbl but not the PTB domain of dShc and mShc.
Several observations suggest a novel interaction of Nak with the PTB domain of Numb. First, unlike interactions involving the PTB domains of proteins in the Shc family or the IRS family (22, 27, 33), Nak interaction with the PTB domain of Numb does not involve an NPXpY motif (Fig. 4D). In fact, there are no tyrosine residues in the 11-aa peptide from Nak that is sufficient for the binding to the Numb PTB domain. Second, Nak binding was not affected by mutations of the Numb PTB domain that corresponds to those mutations of the Shc PTB domain known to disrupt Shc interaction with the tyrosine-phosphorylated p145 (55) (Fig. 3). Moreover, the Nak interaction is specific for the PTB domain of Numb; no interaction with the Shc PTB domains can be detected (Fig. 5).
The requirement of a short peptide of 11 aa from Nak for binding to the Numb PTB domain is suggestive of peptide-surface association, similar to the interactions between the SH2 domains and tyrosine-phosphorylated peptides, the PDZ domain and a peptide containing the Ser/Thr-X-Val motif, and the PTB domains of Shc or IRS families and a peptide containing the NPXpY motif (for a review, see reference 24). In another screen for Numb-interacting proteins, we found that the intracellular domain of the Drosophila epidermal cell surface receptor encoded by stranded at second (sas) (44) binds Numb PTB domain. The 37 aa of the intracellular domain of Sas contains the NPXY motif, and point mutations of either the Asn or Tyr residues abolished the interaction between Sas and Numb in the two-hybrid assay (11a). This result suggests that the Numb PTB domain is capable of binding to the NPXY motif. Also, a mammalian homolog of Numb was shown to bind tyrosine-phosphorylated proteins (51), although the motif which mediates the specific interaction was not defined. The versatility of the PTB domain binding specificity was further shown by the binding of the dNumb PTB domain to GPpY motifs (38) and the Shc PTB domain to NPLH motifs (11). In addition, the PTB domains of two neuronal proteins, X11 and FE65, are capable of binding to the YENPTY motif of the cytoplasmic domain of the amyloid precursor protein (6). This binding is not tyrosine phosphorylation dependent even though this sequence contains an NPXY motif. It remains to be determined whether the binding surfaces of the PTB domain to various motifs are distinct from each other and if the PTB domain can utilize multiple binding regions for its interaction with different proteins simultaneously or these binding events are mutually exclusive.
Possible functions of Nak in asymmetric cell division.
Whereas the specific physical interaction between Nak and Numb suggests that Nak may be part of the Numb pathway in specifying daughter cell fate during asymmetric division, it will be necessary to test this possibility by examining both the loss-of-function phenotype and the gain-of-function phenotype of the nak gene. No loss-of-function mutations of the nak gene are currently available. It is worth noting, however, that a number of genes known to be involved in the Numb pathway for asymmetric division exhibit overexpression phenotypes which correspond to cell fate transformations opposite those caused by loss of gene function. Hence, overexpression of the protein products of Delta, Notch, tramtrack, Suppresser of Hairless, or enhancer of split causes transformation of the B cell to the A cell, the hair cell to the socket cell, and the neuron to the sheath cell (18, 26, 40, 45, 48), opposite their respective loss-of-function phenotypes. Phenotypes due to overexpression of these genes are similar to the numb null mutant phenotype, whereas overexpression of Numb causes the opposite cell fate transformation (42). The overexpression phenotypes of nak are very similar to those of Notch, tramtrack, and other downstream genes of numb and are therefore highly suggestive of the involvement of nak in asymmetric divisions.
The in vivo interaction of Myc-Nak with Numb protein is also indicative of the function of Nak in the asymmetric cell division pathway. In addition to the immunocoprecipitation of Numb and Myc-Nak, we also observed that the ectopically expressed Myc-Nak localize to the cortical membrane where Numb and Notch are distributed (data not shown), suggesting that Nak can localize to the site for participation in the asymmetric cell divisions. Due to the overexpression of Myc-Nak, it is difficult to analyze the segregation of Myc-Nak during cell division. Whether Nak is asymmetrically localized during asymmetric cell divisions awaits the availability of an antibody that is suitable for immunocytochemistry.
The potential involvement of Nak in asymmetric divisions in Drosophila is reminiscent of the involvement of the par-1 gene in asymmetric divisions during early embryonic development of C. elegans. Par-1 also contains a serine/threonine kinase domain and a C-terminal region that binds other proteins; whereas the C terminus of Nak binds Numb, the C terminus of Par-1 binds a nonmuscle myosin (20, 21). A priori, a Numb-binding protein could be involved in asymmetric localization of Numb during asymmetric division or in executing the actions of asymmetrically segregated Numb in specifying daughter cell fate. It appears unlikely that Nak is involved in asymmetric localization of Numb, for the following reasons. First, the Nak overexpression phenotypes could be suppressed by Numb overexpression. This restoration of the proper asymmetric divisions could not have been achieved if overexpression of Nak had abolished asymmetric Numb localization. Second, Nak binds to the PTB domain but not to the rest of the Numb protein. The PTB domain is not necessary for asymmetric localization of Numb in dividing neural precursor cells but is necessary for the ability of Numb to inhibit Notch signaling (17). Third, both mNumb and mouse Numblike (mNbl; homolog of rNbl) contain PTB domains which are 70 to 75% identical to the PTB domain of dNumb at the amino acid level, and when overexpressed in Drosophila, mNumb and mNbl can transform cell fate in the sensory organ lineages. But only mNumb, not mNbl, is asymmetrically localized in transgenic flies (54). It thus appears unlikely that Nak plays a role in asymmetric Numb localization.
The observation of Nak activity thus far is consistent with the possibility that Nak mediates or modulates the action of asymmetrically distributed Numb. For example, Nak may phosphorylate Numb and negatively regulate Numb function. Alternatively, the interaction of Nak and Numb may prevent the binding of Notch to Numb (19, 53), thus relieving the inhibition of Notch from Numb. It is also conceivable that Nak may be recruited to the vicinity of Notch due to physical interactions of Numb with Notch and Nak, so that it could phosphorylate Notch or its downstream effectors, thereby inhibiting Notch signaling. These and other possible scenarios may be tested by future genetic and biochemical studies.
ACKNOWLEDGMENTS
We thank Weimin Zhong for mammalian Numb and Numblike DNA constructs, Yenwen Chen (UCSF) for mammalian cDNA libraries, Steve Elledge (Baylor College of Medicine) for a Drosophila two-hybrid library, Carl S. Thummel (University of Utah) for larval and pupal cDNA libraries, Jue Wang for helping with some of the biochemical assays, and Susan Younger and Alice Turner for chromosomal in situ hybridizations. We also thank Ming Guo for suggestions on the manuscript.
Cheng-ting Chien was supported by the Jane Coffin Childs Memorial Fund for Cancer Research. Shuwen Wang is a research associate and Lily Y. Jan and Yuh Nung Jan are investigators of the Howard Hughes Medical Institute.
REFERENCES
- 1.Ashburner M. Drosophila: a laboratory handbook. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 2.Bailey A M, Posakony J W. Suppressor of Hairless directly activates transcription of Enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 3.Bartel P L, Chien C-T, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. BioTechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- 4.Bartel P L, Chien C-T, Sternglanz R, Fields S. Using the two-hybrid system to detect protein-protein interactions. In: Hartley D A, editor. Cellular interaction in development: a practical approach. Oxford, England: Oxford University Press; 1993. pp. 153–179. [Google Scholar]
- 5.Blaikie P, Immanuel D, Wu J, Li N, Yajnik V, Margolis B. A region in Shc distinct from the SH2 domain can bind tyrosine-phosphorylated growth factor receptors. J Biol Chem. 1994;269:32031–32034. [PubMed] [Google Scholar]
- 6.Borg J P, Ooi J, Levy E, Margolis B. The phosphotyrosine interaction domains of X11 and FE65 bind to distinct sites on the YENPTY motif of amyloid precursor protein. Mol Cell Biol. 1996;16:6229–6241. doi: 10.1128/mcb.16.11.6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bork P, Margolis B. A phosphotyrosine interaction domain. Cell. 1995;80:693–694. doi: 10.1016/0092-8674(95)90347-x. [DOI] [PubMed] [Google Scholar]
- 8.Brand A H, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 9.Brewster R, Bodmer R. Origin and specification of type II sensory neurons in Drosophila. Development. 1995;121:2923–2936. doi: 10.1242/dev.121.9.2923. [DOI] [PubMed] [Google Scholar]
- 10.Campos-Ortega J A. Numb diverts Notch pathway off the Tramtrack. Neuron. 1996;17:1–4. doi: 10.1016/s0896-6273(00)80274-3. [DOI] [PubMed] [Google Scholar]
- 11.Charest A, Wagner J, Jacob S, McGlade C J, Tremblay M L. Phosphotyrosine-independent binding of SHC to the NPLH sequence of murine protein-tyrosine phosphatase-PEST. Evidence for extended phosphotyrosine binding/phosphotyrosine interaction domain recognition specificity. J Biol Chem. 1996;271:8424–8429. doi: 10.1074/jbc.271.14.8424. [DOI] [PubMed] [Google Scholar]
- 11a.Chien, C.-T., M. Rothernberg, L. Y. Jan, and Y. N. Jan. Unpublished data.
- 12.Chien C-T, Bartel P, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cleghon V, Gayko U, Copeland T D, Perkins L A, Perrimon N, Morrison D K. Drosophila terminal structure development is regulated by the compensatory activities of positive and negative phosphotyrosine signaling sites on the Torso RTK. Genes Dev. 1996;10:566–577. doi: 10.1101/gad.10.5.566. [DOI] [PubMed] [Google Scholar]
- 14.Durfee T, Becherer K, Chen P L, Yeh S H, Yang Y, Kilburn A E, Lee W H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 15.Eck M J, Dhe-Paganon S, Trub T, Nolte R T, Shoelson S E. Structure of the IRS-1 PTB domain bound to the juxtamembrane region of the insulin receptor. Cell. 1996;85:695–705. doi: 10.1016/s0092-8674(00)81236-2. [DOI] [PubMed] [Google Scholar]
- 16.Ferguson K M, Lemmon M A, Schlessinger J, Sigler P B. Structure of the high affinity complex of inositol triphosphate with a phospholipase C pleckstrin homology domain. Cell. 1995;83:1037–1046. doi: 10.1016/0092-8674(95)90219-8. [DOI] [PubMed] [Google Scholar]
- 17.Frise E, Knoblich J A, Younger-Shepherd S, Jan L Y, Jan Y N. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc Natl Acad Sci USA. 1996;93:11925–11932. doi: 10.1073/pnas.93.21.11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo M, Bier E, Jan L Y, Jan Y N. tramtrack acts downstream of numb to specify distinct daughter cell fates during asymmetric cell divisions in the Drosophila PNS. Neuron. 1995;14:913–925. doi: 10.1016/0896-6273(95)90330-5. [DOI] [PubMed] [Google Scholar]
- 19.Guo M, Jan L Y, Jan Y N. Control of daughter cell fate during asymmetric division: interaction of Numb and Notch. Neuron. 1996;17:27–41. doi: 10.1016/s0896-6273(00)80278-0. [DOI] [PubMed] [Google Scholar]
- 20.Guo S, Kemphues K J. par-1, a gene required for establishing polarity in C. elegans embryos, encodes a putative ser/thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- 21.Guo S, Kemphues K J. A non-muscle myosin required for embryonic polarity in Caenorhabditis elegans. Nature. 1996;382:455–458. doi: 10.1038/382455a0. [DOI] [PubMed] [Google Scholar]
- 22.Gustafson T A, He W, Craparo A, Schaub C D, O’Neill T J. Phosphotyrosine-dependent interaction of Shc and IRS-1 with the NPXY motif of the insulin receptor via a novel non-SH2 domain. Mol Cell Biol. 1995;15:2500–2508. doi: 10.1128/mcb.15.5.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hannon G J, Demetrick D, Beach D. Isolation of the Rb-related p130 through its interaction with CDK2 and cyclins. Genes Dev. 1993;7:2378–2391. doi: 10.1101/gad.7.12a.2378. [DOI] [PubMed] [Google Scholar]
- 24.Harrison S C. Peptide-surface association: the case of PDZ and PTB domains. Cell. 1996;86:341–343. doi: 10.1016/s0092-8674(00)80105-1. [DOI] [PubMed] [Google Scholar]
- 25.Hartenstein V, Posakony J W. Development of adult sensilla on the wing and notum of Drosophila melanogaster. Development. 1989;107:389–405. doi: 10.1242/dev.107.2.389. [DOI] [PubMed] [Google Scholar]
- 26.Hartenstein V, Posakony J W. A dual function of the Notch gene in Drosophila sensillum development. Dev Biol. 1990;142:13–30. doi: 10.1016/0012-1606(90)90147-b. [DOI] [PubMed] [Google Scholar]
- 27.He W, Craparo A, Zhu Y, O’Neill T J, Wang L M, Pierce J H, Gustafson T A. Interaction of insulin receptor substrate-2 with the insulin and IGF-1 receptors: evidence for two-distinct phosphotyrosine-dependent interaction domains within IRS-2. J Biol Chem. 1996;271:11641–11645. doi: 10.1074/jbc.271.20.11641. [DOI] [PubMed] [Google Scholar]
- 28.He W, O’Neill T J, Gustafson T A. Distinct modes of interaction of SHC and insulin receptor substrate-1 with insulin receptor NPXY region via non-SH2 domains. J Biol Chem. 1995;270:23258–23262. doi: 10.1074/jbc.270.40.23258. [DOI] [PubMed] [Google Scholar]
- 29.Hinz U, Giebel B, Campos-Ortega J A. The basic-helix-loop-helix domain of Drosophila lethal of scute protein is sufficient for proneural function and activates neurogenic genes. Cell. 1994;76:77–88. doi: 10.1016/0092-8674(94)90174-0. [DOI] [PubMed] [Google Scholar]
- 30.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horvitz H R, Herskowitz I. Mechanisms of asymmetric cell division: two Bs or not two Bs, that is the question. Cell. 1992;68:237–255. doi: 10.1016/0092-8674(92)90468-r. [DOI] [PubMed] [Google Scholar]
- 32.Jan Y N, Jan L Y. Maggot’s hair and bug’s eye: role of cell interactions and intrinsic factors in cell fate specification. Neuron. 1995;14:1–5. doi: 10.1016/0896-6273(95)90235-x. [DOI] [PubMed] [Google Scholar]
- 33.Kavanaugh W M, Turck C W, William L T. PTB domain binding to signaling proteins through a sequence motif containing phosphotyrosine. Science. 1995;268:1177–1179. doi: 10.1126/science.7539155. [DOI] [PubMed] [Google Scholar]
- 34.Kavanaugh W M, William L T. An alternative to SH2 domains for binding tyrosine-phosphorylated proteins. Science. 1994;266:1862–1865. doi: 10.1126/science.7527937. [DOI] [PubMed] [Google Scholar]
- 35.Knoblich J A, Jan L Y, Jan Y N. Asymmetric segregation of Numb and Prospero during cell division. Nature. 1995;377:624–627. doi: 10.1038/377624a0. [DOI] [PubMed] [Google Scholar]
- 36.Lecourtois M, Schweisguth F. The neurogenic Suppressor of Hairless DNA-binding protein mediates the transcriptional activation of the Enhancer of split complex genes triggered by Notch signaling. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- 37.Lemmon M A, Ferguson K M, Schlessinger J. PH domains: diverse sequences with a common fold recruit signaling molecules to the cell surface. Cell. 1996;85:621–624. doi: 10.1016/s0092-8674(00)81022-3. [DOI] [PubMed] [Google Scholar]
- 38.Li S-C, Zhou S, Vincent S J F, Zwahlen C, Wiley S, Cantley L, Kay L E, Forman-Kay J, Pawson T. High-affinity binding of the Drosophila Numb phosphotyrosine-binding domain to peptides containing a Gly-Pro-(p)Tyr motif. Proc Natl Acad Sci USA. 1997;94:7204–7209. doi: 10.1073/pnas.94.14.7204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohkura H, Hagan I M, Glover D M. The conserved Schizosaccharomyces pombe kinase plo1, required to form a biopolar spindle, the actin ring, and septum, can drive septum formation in G1 and G2 cells. Genes Dev. 1995;9:1059–1073. doi: 10.1101/gad.9.9.1059. [DOI] [PubMed] [Google Scholar]
- 40.Parks A, Muskavitch M A. Delta function is required for bristle organ determination and morphogenesis in Drosophila. Dev Biol. 1993;157:484–496. doi: 10.1006/dbio.1993.1151. [DOI] [PubMed] [Google Scholar]
- 41.Posakony J W. Nature versus nurture: asymmetric cell divisions in Drosophila bristle development. Cell. 1994;76:415–418. doi: 10.1016/0092-8674(94)90105-8. [DOI] [PubMed] [Google Scholar]
- 42.Rhyu M, Jan L, Jan Y N. Asymmetric distribution of numb protein during division of the sensory organ precursor confers distinct fates to daughter cells. Cell. 1994;76:477–491. doi: 10.1016/0092-8674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 43.Rubin G M, Spradling A C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 44.Schonbaum C P, Organ E L, Qu S, Cavener D R. The Drosophila melanogaster stranded at second (sas) gene encodes a putative epidermal cell surface receptor required for larval development. Dev Biol. 1992;151:431–445. doi: 10.1016/0012-1606(92)90183-h. [DOI] [PubMed] [Google Scholar]
- 45.Schweisguth F, Posakony J W. Antagonistic activities of Suppressor-of-Hairless and Hairless control alternative cell fates in the Drosophila adult epidermis. Development. 1994;120:1433–1441. doi: 10.1242/dev.120.6.1433. [DOI] [PubMed] [Google Scholar]
- 46.Spana E P, Doe C Q. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 47.Spana E P, Kopczynski C, Goodman C S, Doe C Q. Asymmetric localization of numb autonomously determines sibling neuron identity in the Drosophila CNS. Development. 1995;121:3489–3494. doi: 10.1242/dev.121.11.3489. [DOI] [PubMed] [Google Scholar]
- 48.Tata F, Hartley D A. Inhibition of cell fate in Drosophila by Enhancer of split genes. Mech Dev. 1995;51:305–315. doi: 10.1016/0925-4773(95)00377-0. [DOI] [PubMed] [Google Scholar]
- 49.Uemura T, Shepherd S, Ackerman L, Jan L Y, Jan Y N. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell. 1989;58:349–360. doi: 10.1016/0092-8674(89)90849-0. [DOI] [PubMed] [Google Scholar]
- 50.Vaessin H, Grell E, Wolff E, Bier E, Jan L Y, Jan Y N. prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell. 1991;67:941–953. doi: 10.1016/0092-8674(91)90367-8. [DOI] [PubMed] [Google Scholar]
- 51.Verdi J M, Schmandt R, Bashirullah A, Jacob S, Salvino R, Craig C G, Program A E, Lipshitz H D, McGlade C J. Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr Biol. 1996;6:1134–1145. doi: 10.1016/s0960-9822(02)70680-5. [DOI] [PubMed] [Google Scholar]
- 52.Wolf G, Trub T, Ottinger E, Groninga L, Lynch A, White M F, Miyazaki M, Lee J, Shoelson S E. PTB domains of IRS-1 and Shc have distinct but overlapping binding specificities. J Biol Chem. 1995;270:27407–27410. doi: 10.1074/jbc.270.46.27407. [DOI] [PubMed] [Google Scholar]
- 53.Zhong W, Feder J N, Jiang M-M, Jan L Y, Jan Y N. Asymmetric localization of a mammalian Numb homolog during mouse cortical neurogenesis. Neuron. 1996;17:43–53. doi: 10.1016/s0896-6273(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 54.Zhong W, Jiang M-M, Weinmaster G, Jan L Y, Jan Y N. Differential expression of mammalian Numb homologues and Notch1 suggests distinct roles during mouse cortical neurogenesis. Development. 1997;124:1887–1897. doi: 10.1242/dev.124.10.1887. [DOI] [PubMed] [Google Scholar]
- 55.Zhou M-M, Ravichandran K S, Olejniczak E T, Petros A M, Meadows R P, Sattler M, Harlan J E, Wade W S, Burakoff S J, Fesik S W. Structure and ligand recognition of the phosphotyrosine binding domain of Shc. Nature. 1995;378:584–592. doi: 10.1038/378584a0. [DOI] [PubMed] [Google Scholar]
- 56.Zipursky S L, Venkatesh T R, Teplow D B, Benzer S. Neuronal development in the Drosophila retina: Monoclonal antibodies as molecular probes. Cell. 1984;36:15–26. doi: 10.1016/0092-8674(84)90069-2. [DOI] [PubMed] [Google Scholar]


