Abstract
We have studied the interaction between recombination signal sequences (RSSs) and protein products of the truncated forms of recombination-activating genes (RAG) by gel mobility shift, DNase I footprinting, and methylation interference assays. Methylation interference with dimethyl sulfate demonstrated that binding was blocked by methylation in the nonamer at the second-position G residue in the bottom strand and at the sixth- and seventh-position A residues in the top strand. DNase I footprinting experiments demonstrated that RAG1 alone, or even a RAG1 homeodomain peptide, gave footprint patterns very similar to those obtained with the RAG1-RAG2 complex. In the heptamer, partial methylation interference was observed at the sixth-position A residue in the bottom strand. In DNase I footprinting, the heptamer region was weakly protected in the bottom strand by RAG1. The effects of RSS mutations on RAG binding were evaluated by DNA footprinting. Comparison of the RAG-RSS footprint data with the published Hin model confirmed the notion that sequence-specific RSS-RAG interaction takes place primarily between the Hin domain of the RAG1 protein and adjacent major and minor grooves of the nonamer DNA.
V(D)J joining is a site-specific recombination process that plays a crucial role in the activation and diversification of antigen receptor genes (44). Joining occurs between two pairs of recombination signal sequences (RSSs): heptamer (CACAGTG) and nonamer (ACAAAAACC) (22, 32). Furthermore, the spacer separating the heptamer and the nonamer is either 12 or 23 bp in length, and recombination takes place between two RSSs in which one contains a 12-bp spacer (12-RSS), and the other contains a 23-bp spacer (23-RSS) (8, 33, 34); this is the so-called 12/23 rule. It has been shown that just two pairs of the heptamer and the nonamer are sufficient for V(D)J type recombination if the 12/23 rule is satisfied (3).
V(D)J type recombination consists of two major processes: site-specific cleavage and ligation of cleaved ends. The former process includes specific recognition of the RSS by DNA-binding components of the recombinase, synaptic complex formation between the two RSSs satisfying the 12/23 rule, and site-specific cleavage of RSSs adjacent to the heptamer (29, 37). The latter process is known to be mediated by DNA repair mechanisms, including DNA-dependent protein kinase, (4, 17, 19), the Ku protein complex (43, 47), XRCC4 (21), and DNA ligases (13, 28). During the process of recombination, nucleotide deletion and addition occur at coding ends. Terminal deoxynucleotidyl transferase is responsible for the insertion of non-germ line nucleotides (11, 18).
Two recombination-activating genes, rag-1 and rag-2, were isolated by their abilities to activate V(D)J type recombination in a fibroblast cell line (26, 36). It was not clear for many years what roles RAG proteins played in the process of V(D)J recombination. The recent demonstration of in vitro RSS cleavage (45) provided a more convincing argument that the RAG proteins were indeed major components of the V(D)J recombinase, rather than simply activators for it. Cleavage occurred in vitro by two successive steps, nicking and hairpin formation (24) following the 12/23 rule (9, 46). A nick is first introduced at the 5′ end of the RSS on the top strand, and the bottom strand is then broken, resulting in a hairpin structure at the coding end and a blunt end at the signal end (24, 29, 45). Several studies have provided information on the roles of various mutations in RAG1 and RAG2 proteins (6, 25, 30, 31, 39).
Detection of specific interactions between RSS and RAG proteins was difficult, probably because the complex dissociates after the cleavage reaction. Two groups reported that the Hin domain in the RAG1 protein interacted with the nonamer of RSS, using a one-hybrid binding assay in vivo (7) and surface plasmon resonance in vitro (42). More recently, Hiom and Gellert (15) detected the RSS-RAG complex with the gel mobility shift assay in the presence of Ca2+ or Mg2+ and a cross-linking chemical, glutaraldehyde (15). Despite similarities between RAG and bacterial Hin systems (7, 42), the differences are sufficient that a real understanding of the RAG protein contacts on RSS DNA cannot be obtained without direct footprinting experiments at the nucleotide level.
Here we report that specific RSS-RAG1 or RSS–RAG1-RAG2 complexes are stable in the absence of protein-DNA cross-linking and that these complexes can be used to evaluate the effect of mutations on RAG complex binding by gel shift assays and DNA footprinting. Heptamer mutants were also useful in identifying the complex, because they prevented the cleavage of the RSS but still allowed binding with RAG proteins. We found that (i) RAG1 can interact with RSS in the absence of RAG2, (ii) the 102-amino-acid (aa) peptide containing the Hin homeodomain of RAG1 gives the same nonamer footprint pattern as those obtained with the RAG1-RAG2 complex, and (iii) the precise contacts have been established by methylation interference and DNase I footprinting in the nonamer region. Comparisons of the footprint data with the Hin model (7, 10, 42) indicate that RAG1 is the major player in strong DNA binding and that the mode of interaction is consistent with homeodomain binding to the nonamer sequence. Presumed non-sequence-dependent interactions with the phosphate backbone may allow for DNA bending or stressing that results in enhanced DNase I cleavage sites.
MATERIALS AND METHODS
Preparation of RAG proteins.
Truncated RAG1 and RAG2 fused with maltose-binding protein were prepared basically as described by van Gent et al. (45). Recombinant murine rag genes were expressed in Spodoptera frugiperda Sf.9 cells (41) by using baculovirus vectors (27). RAG proteins were purified with Ni-nitrilotriacetic acid (Qiagen) and amylose resin columns (New England Biolabs). A Hin homeodomain fusion protein of RAG1 was produced in Escherichia coli from the plasmid vector pMAL-C2 (New England Biolabs) bearing genes coding for maltose-binding protein and 102 aa (aa 376 to 477) of the RAG1 Hin homeodomain. Purities of fusion proteins were examined in Coomassie brilliant blue-stained gels. No detectable contaminant bands were seen when 1 μg of RAG proteins or 10 μg of Hin homeodomain protein was separated.
Gel mobility shift assay.
Gel migration retardation assays were performed as described by Singh et al. (40). End-labeled DNA (0.04 pmol, 2 × 104 to 4 × 104 cpm) was mixed with 0.2 μg of RAG1 and RAG2 in 10 μl of binding buffer containing 25 mM morpholinepropanesulfonic acid (MOPS)-KOH (pH 7.0), 5 mM Tris-HCl (pH 8.0), 2.4 mM dithiothreitol, 90 mM potassium acetate, 30 mM KCl, 1 μM nonspecific oligonucleotide (25-mer; ACTGGAGTTAGTTGAAGCATTAGGT), 10 mM MgCl2 (or 1 mM CaCl2), 0.1 mg of bovine serum albumin per ml, and 2% glycerol. The mixture was incubated at 37°C for 10 min and loaded with glycerol dye mix (25% glycerol, 1 mM EDTA, 0.01% xylene cyanol, 0.01% bromophenol blue) on a 4% polyacrylamide gel (acrylamide bisacrylamide, 19:1) containing 89 mM Tris-borate (pH 8.3).
RSS cleavage reaction assay.
DNA end labeled with [γ-32P]ATP (0.1 pmol) was incubated with 0.2 μg of RAG1 and RAG2 proteins at 37°C for 1 h in 10 μl of 25 mM MOPS-KOH (pH 7.0)–30 mM potassium glutamate–2.4 mM dithiothreitol–1 mM MnCl2 or MgCl2–5 mM Tris-HCl (pH 8.0)–30 mM KCl–2% glycerol. After the reaction, 10 μl of formamide dye mix (96% formamide, 20 mM EDTA, 0.01% xylene cyanol, 0.01% bromophenol blue) was added. The sample was heated at 95°C for 2 min and separated in a 12.5% denaturating polyacrylamide gel containing 89 mM Tris-borate (pH 8.3), 2 mM EDTA, and 7 M urea (45).
Preparation of DNA for footprinting and the methylation interference assay.
Various RSSs were chemically synthesized and subcloned into the plasmid vector pKI13, a derivative of pBluescript II SK+ (Stratagene), using SalI and HindIII sites. Synthetic wild-type sequences (top strand) are as follows: 12-RSS, 5′-TCACAGTGCTCCAGGGCTGAACAAAAACCGTCGA-3′; and 23-RSS, 5′-TC ACAGTGGTAGTACTCCACTGTCTGGGTGTACAAAAACCGTCGA-3′ (heptamer and nonamer are underlined). For DNase I footprinting, plasmid DNA was cleaved with either BssHII (for the top strand) or SfaNI (for the bottom strand) and labeled with α-32P-labeled deoxynucleoside triphosphates (dNTPs) by Klenow polymerase. To obtain one-end-labeled RSS fragments, plasmid DNA was then cleaved with either EcoRI (for the top strand) or FspI (for the bottom strand). For the methylation interference assay, plasmid was cleaved with either EcoO109I (for the top strand) or NotI (for the bottom strand) and labeled with 32P. The second cleavage was at either the SfaNI site (for the top strand) or the BssHII site (for the bottom strand). DNA fragments were separated by electrophoresis in a 6% polyacrylamide gel, eluted from gel slices with an elution buffer containing 0.2 M NaCl, 1 mM EDTA, and 20 mM Tris-HCl (pH 7.5), and purified by reversed-phase column chromatography (Elutip-d; Schleicher & Schuell).
Methylation interference assay.
End-labeled DNA (1 pmol, 106 cpm) was treated with 1 μl of dimethyl sulfate (DMS) at 25°C for 2 min in 200 μl of 50 mM sodium cacodylate–1 mM EDTA. Methylation was terminated by adding 50 μl of stop solution containing 1.5 M sodium acetate, 1.0 M β-mercaptoethanol, and 100 μg of yeast tRNA per ml. The DNA was precipitated with ethanol twice and concentrated to 0.1 pmol/μl in 10 mM Tris-HCl (pH 8.0)–0.5 mM EDTA–50 mM KCl. The gel shift experiment was scaled up 10-fold for the methylation interference assay. After electrophoresis, protein-bound DNA and unbound DNA were separately isolated from the polyacrylamide gel. DNA was transferred from the gel slices to a DEAE membrane (NA45; Schleicher & Schuell) by electrophoresis and eluted with 1.0 M NaCl–1 mM EDTA–20 mM Tris-HCl (pH 8.0) at 65°C for 30 min. DNA eluted from the membrane was precipitated and rinsed with ethanol and then treated with 50 μl of 10% piperidine at 90°C for 30 min. DNA was precipitated with 500 μl of n-butanol, solubilized with 50 μl of 1% sodium dodecyl sulfate, and precipitated again with 500 μl of n-butanol. DNA was lyophilized and loaded on an 8% polyacrylamide gel containing 7 M urea. After electrophoresis in 89 mM Tris-borate (pH 8.3)–2 mM EDTA, the gel was dried and subjected to autoradiography by a BAS-2000 bioimage analyzer (Fujifilm).
DNase I footprinting.
Double-stranded DNA was labeled by filling in one 3′ end with 32P-labeled NTPs, using Klenow polymerase. DNA (0.04 pmol, 2 × 104 to 4 × 104 cpm) was mixed with RAG1 (1.7 μg), RAG1-RAG2 (2.6 μg), or Hin homeodomain of RAG1 (40 μg) at 37°C for 10 min in 100 μl of the binding buffer described above. Then 5 U of DNase I (Stratagene) was added, and the incubation was continued for another 2 min at 37°C. DNA was extracted once with phenol-chloroform-isoamyl alcohol (25:24:1), precipitated with ethanol, and washed with 70% ethanol. The sample was suspended in 5 μl of formamide dye mix and separated by electrophoresis in a 6% polyacrylamide sequencing gel containing 7 M urea.
RESULTS
Detection of RSS-RAG complex by gel shift assay.
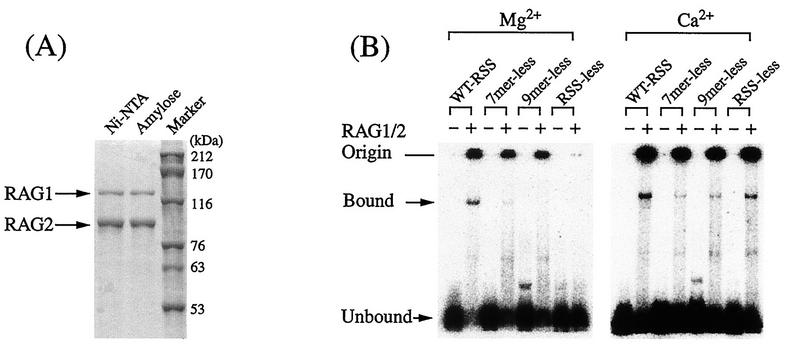
To study the RSS-RAG interaction by methylation interference, we examined binding conditions with the gel mobility shift assay (40). Truncated RAG1 and RAG2 proteins (45) were coexpressed in Sf.9 cells (41) by using baculovirus vectors (27). Purities of RAG proteins were examined in Coomassie brilliant blue-stained gels (Fig. 1A). We first tested the RSS-RAG interaction by gel shift assay in the presence of either Ca2+ or Mg2+ (Fig. 1B). Although protein-bound RSS was found in the presence of Ca2+, binding was seen even with RSS-less DNA. Since nonspecific interaction was found for Ca2+, Mg2+ was used throughout our study. It was also found that glutaraldehyde was not essential in the gel shift assay and did not increase the amount of RSS-RAG complex.
FIG. 1.
Purification and RSS-binding activities of RAG proteins. (A) RAG proteins were expressed with baculovirus vectors and purified by passage through Ni-nitrilotriacetic acid (Ni-NTA) and amylose resin columns. Purities of truncated proteins were examined in Coomassie brilliant blue-stained gels. (B) RSS-binding activities were examined by the gel mobility shift assay in the presence of either Ca2+ or Mg2+. To test the specificity of binding, heptamerless, nonamerless, and RSS-less DNAs were used as probes. WT-RSS, wild-type RSS.
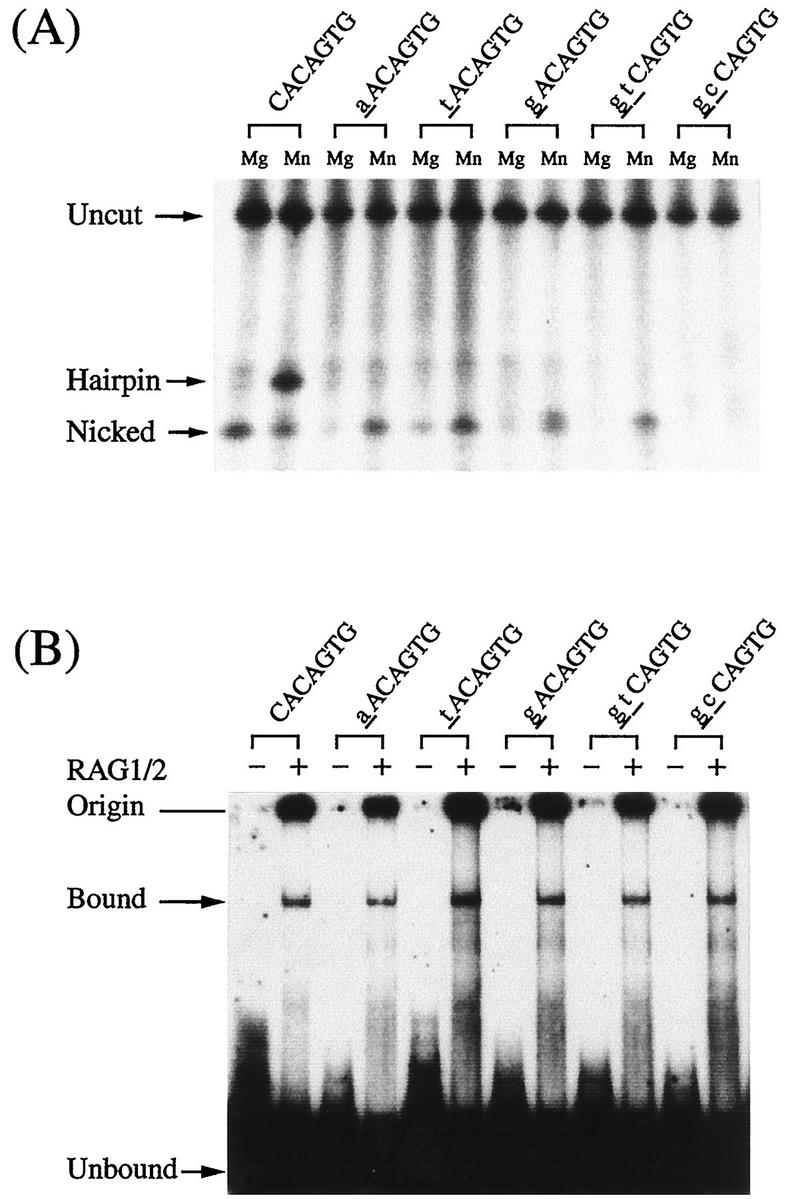
In this study, we analyzed heptamer mutants in parallel with the wild-type RSS, because nicking or cleavage may cause dissociation of the complex and it also eliminates the footprint pattern beyond the cleavage site. We examined five different heptamer mutants for nicking and hairpin formation with RAG proteins. Mutations at the first and second positions in the heptamer greatly reduced hairpin formation (Fig. 2A). In contrast, binding with RAG proteins was not affected by the heptamer mutations (Fig. 2B). In an attempt to prevent the cleavage reaction, we tested a synthetic 12-RSS that contained a thioester bond at the cleavage site. Although the complex was detected in the gel shift assay, nicking and cleavage also took place normally (data not shown). Therefore, introduction of the thioester was not helpful in inhibiting cleavage to accumulate the RSS-RAG complex.
FIG. 2.
Cleavage and binding activities of mutant heptamers. To obtain stable RSS-RAG complexes, heptamer mutated RSSs were tested for their abilities to block the cleavage reaction but still allow binding with RAG proteins. Heptamer mutants containing base substitutions at the first and second positions (noted by lowercase underlined letters) were examined for their abilities to block RAG cleavage (A) and to bind with RAG proteins (B). To detect the nicked or hairpin structures generated at the cleavage site, reaction products were separated in an polyacrylamide gel under denaturing conditions.
Methylation interference assay with DMS.
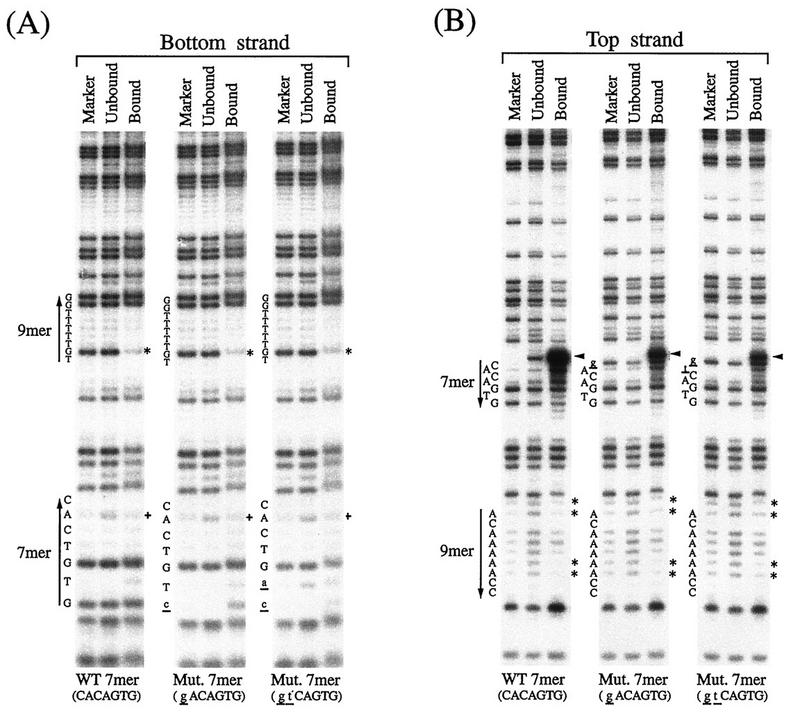
The RSS-RAG interaction was studied at the nucleotide level by the methylation interference assay (38). End-labeled RSS DNA was partially methylated with DMS and mixed with truncated RAG1 and RAG2 proteins. The mixture was electrophoresed in a polyacrylamide gel to separate the RAG-RSS complex from the unbound probe DNA. DNA was eluted from the gel, cleaved with piperidine, and loaded on a DNA sequencing gel. Three DNA samples are compared in Fig. 3: a G-reacted marker, unbound DNA, and RAG-bound DNA. Both the top and the bottom strands of the 12-RSS DNA were analyzed. In this interference assay, G and A residues actually involved in the RSS-RAG interaction are detected as fainter bands in the bound lanes of the sequencing gel than in the unbound lanes because methylated residues at those sites prevent the interaction with protein.
FIG. 3.
Methylation interference in the RSS-RAG interaction. Methylation interference assays demonstrate interference at the second position (G) on the bottom strand (asterisks in panel A) and at positions −1 (A), 1 (A), 6 (A), and 7 (A) on the top strand (asterisks in panel B). Partial interference was seen at positions −2 (G), 3 (A), and 5 (A) on the top strand. In the heptamer, partial interference was seen at the sixth position on the bottom strand (+ in panel A). Nicking occurs only on the top strand (arrowhead) and is reduced in the mutants. The wild-type (WT) 12-RSS (CACAGTG) and two mutant (Mut.) heptamers (gACAGTG and gtCAGTG) were studied in parallel. The heptamer (residues 1 to 7) and the nonamer (residues 1 to 9) are indicated by arrows. Protein-bound and unbound DNAs eluted from the gel were chemically degraded with piperidine and separated in an 8% DNA sequencing gel with the G-reacted 12-RSS as a control.
In the bottom strand of 12-RSS, strong interference was seen at the second position G in the nonamer sequence (Fig. 3A). Interestingly, two other G residues, at positions 8 and 9 in the nonamer, were not affected. Since the bottom strand was not nicked, basically the same interference pattern was obtained with both wild-type RSS and heptamer mutants. In the top strand, methylation of A residues in the nonamer caused strong interference at positions −1, 1, 6, and 7 (Fig. 3B).
In the heptamer region of the top strand, the band adjacent to the first position represents the nicking product generated by RAG proteins. This band is present in both RAG-bound and unbound DNAs of the wild-type RSS, suggesting that some fraction of the RSS-RAG complex dissociated after the nicking reaction. In the bottom strand, partial interference of the sixth-position A in the heptamer occurred (Fig. 3A).
DNase I footprinting.
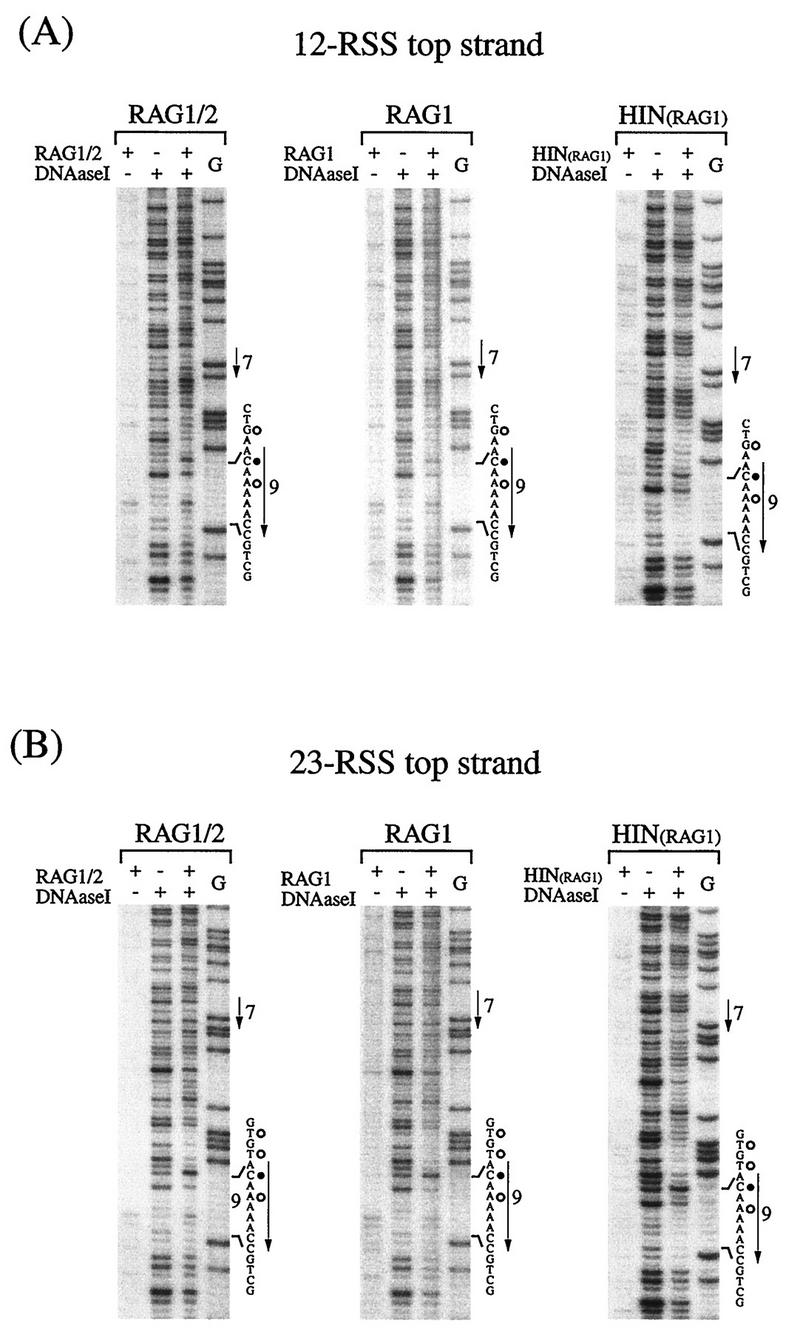
The RSS-RAG interaction was also studied by DNase I footprinting (5). Double-stranded RSS DNA was labeled with 32P on either the top or bottom strand and subjected to footprinting either with RAG1 alone or with the RAG1-RAG2 complex (Fig. 4). In the gel mobility shift assay, the RSS-RAG1 complex was not detected in the absence of RAG2 protein. Therefore, the methylation interference assay could not be performed for the RSS-RAG1 interaction. Since DNase I footprinting does not require the separation of protein-bound DNA, it was useful for the analysis of the RAG1-RSS interaction and subsequently employed for the analysis of RSS mutations.
FIG. 4.
DNase I footprint analysis of the RSS-RAG complex. End-labeled 12-RSS (A) and 23-RSS (B) were analyzed by footprinting. DNA was bound either with the RAG1-RAG2 complex or with RAG1 alone, partially digested with DNase I, and electrophoresed in a 6% polyacrylamide DNA sequencing gel. RSS binding was also examined with a 102-aa peptide (residues 376 to 477) covering the Hin homeodomain (residues 384 to 446) of RAG1 protein. Protein-bound DNA was not separated from unbound DNA prior to or after DNase I digestion. G-reacted DNA was used as a position marker, and a control RSS-RAG mixture without DNase I treatment was used to detect background nuclease activity associated with the RAG protein fraction. The heptamer (residues 1 to 7) and the nonamer (residues 1 to 9) are indicated by arrows. Residues in the nonamer region that became hypersensitive (•) or protected (○) after the addition of RAG proteins are marked. The DNase I cleavage site is assigned to the nucleotide containing the 5′-phosphate from the cleavage reaction, and as is standard for DNase I footprinting, the position marker (G) bands are one residue shorter than the corresponding DNase I products.
It was found that protection patterns of wild-type RSSs with RAG1 alone were essentially the same in the nonamer region as those obtained with the RAG1-RAG2 complex in both strands (Fig. 4). Furthermore, the 102-aa peptide (aa 376 to 477) containing the Hin homeodomain of RAG1 gave very similar footprint patterns (Fig. 4). These footprinting results confirmed the notion that the Hin homeodomain within RAG1 interacts with the nonamer region as well as the adjacent spacer region in a manner similar to that of the RAG1-RAG2 complex. In the nonamer, both inhibition and enhancement of DNase I cleavage were observed. For example, the second position in the top strand became hypersensitive in both 12-RSS and 23-RSS, while the third position in the bottom strand was protected (Fig. 4 and see Fig. 6). The appearance of new bands in the middle of the nonamer in the top strand is probably due to background nuclease activity, because these bands are also seen in the RAG control sample without DNase I. In this report, the DNase I cleavage site is assigned to the nucleotide containing the 5′-phosphate from the cleavage reaction, and as is standard for DNase I footprinting, the position marker (G) bands are one residue shorter than the corresponding DNase I products (23).
FIG. 6.
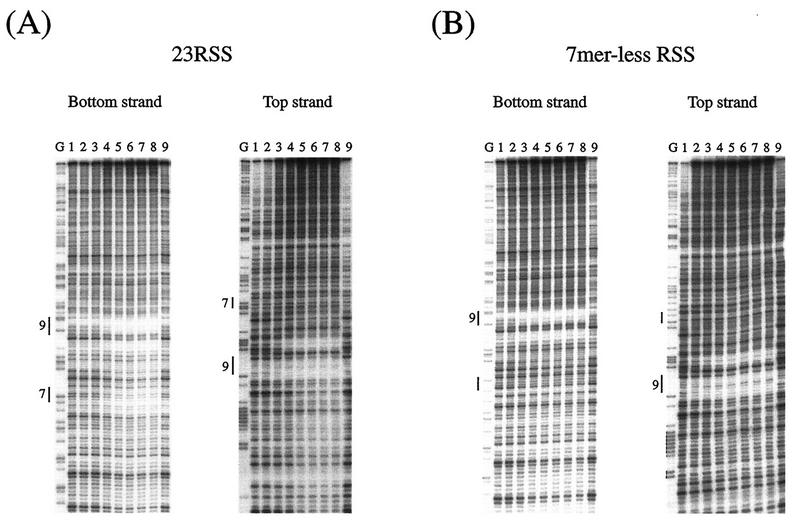
DNase I footprinting of 23-RSS with increasing amounts of RAG1 protein. (A) Titration of RAG1 binding to 23-RSS. Lanes 1 to 9 contained 0.013, 0.027, 0.053, 0.11, 0.21, 0.43, 0.85, 1.7, and 0 μg of RAG1 protein in 100-μl reaction mixtures. (B) The heptamer-less RSS was analyzed in parallel. The heptamer sequence of 23-RSS was totally changed from CACAGTG to GCTGACA. Lanes G, G-reacted RSS position marker; 9 and 7, conserved nonamer and heptamer.
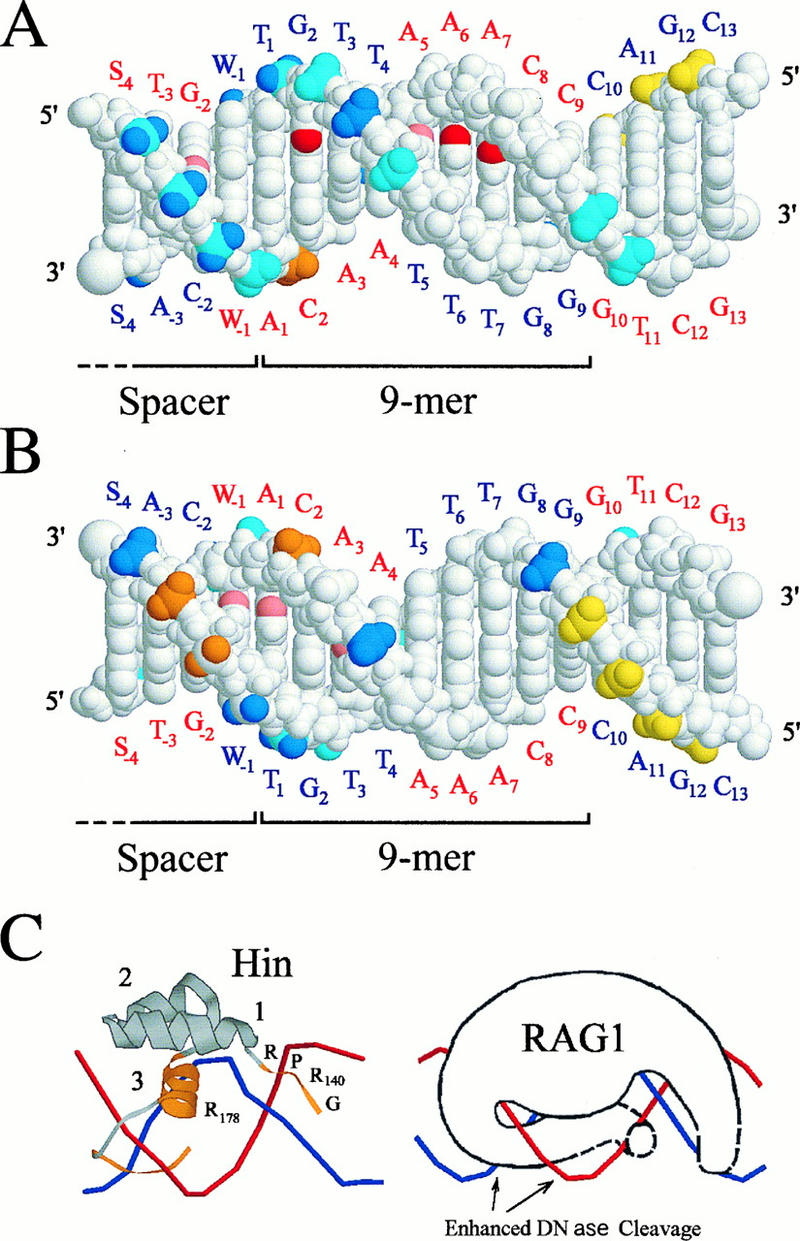
Changes were also found in the spacer region adjacent to the nonamer. For example, in the top strand, residues at positions −2 of 12-RSS and −1 and −3 of 23-RSS were blocked. We also examined 18-RSS, whose spacer sequence was different, and found similar changes (data not shown). Since sequences are not conserved in the spacer, these changes are probably due to the secondary conformational change caused by the specific RAG1 interaction with the nonamer. Alternatively, it is possible that a part of the spacer region is covered by the RAG1 protein which is primarily binding to the nonamer sequence. Therefore, some of the interaction seen in the spacer region may represent nonspecific binding. Protected and hypersensitive residues are summarized in the B-form DNA for the nonamer and a portion of the spacer region (Fig. 5).
FIG. 5.
RAG1-DNA contacts in the nonamer region. (A) Side of the DNA with major groove contacts and the majority of DNase I protected sites; (B) opposite side. The top-strand labels are in red, and those for the bottom strand are in blue. Critical base contacts revealed by methylation interference and protection at N-7 of G2 and N-3 of A6 and A7 are shown in red (A), while partial effects at positions −2 (G), −1 (A), 1 (A), 3 (A), and 5 (A) are in pink (A and B). DNase I-protected (strong in blue and weak in cyan) and -hypersensitive (strong in orange and weak in yellow) phosphate residues are indicated 5′ of the corresponding base. The structure shown is for the 12-RSS, with the data for both 12- and 23-RSSs superimposed (W = A or T; S = G or C). Phosphates with different DNase I footprints for these RSSs are multicolored. Footprint effects (S, strong; W, weak; E, enhanced; N, no effect) for these positions are as follows: top strand −3 to −1, WSW for 12RSS and SWS for 23RSS; bottom strand −2 to +1, NSW for 12RSS and ENS for 23RSS. Only weak DNase I footprinting interactions were observed outside the pictured region. (C) The Hin complex DNA (10) is shown at the left for comparison with the hypothetical RAG1-RSS interaction on the right. The regions of Hin involved in DNA binding are colored orange, and the alpha helices are numbered. Strands are colored the same as in panel A.
Figure 6 shows titration experiments of DNase I footprinting of 23-RSS with increasing amounts of RAG1 proteins. Protection was indicated in the bottom strand of 23-RSS not only in the nonamer region but also in the heptamer region when the RAG1 protein was added (Fig. 6A). Similar results were obtained with 12-RSS (data not shown). With the heptamerless mutant (CACAGTG→GCTGACA) of 23-RSS, protection in the mutated heptamer region was not evident, while the nonamer region was clearly protected in both strands (Fig. 6B).
Some nonamer mutations abolish the RSS-RAG interaction.
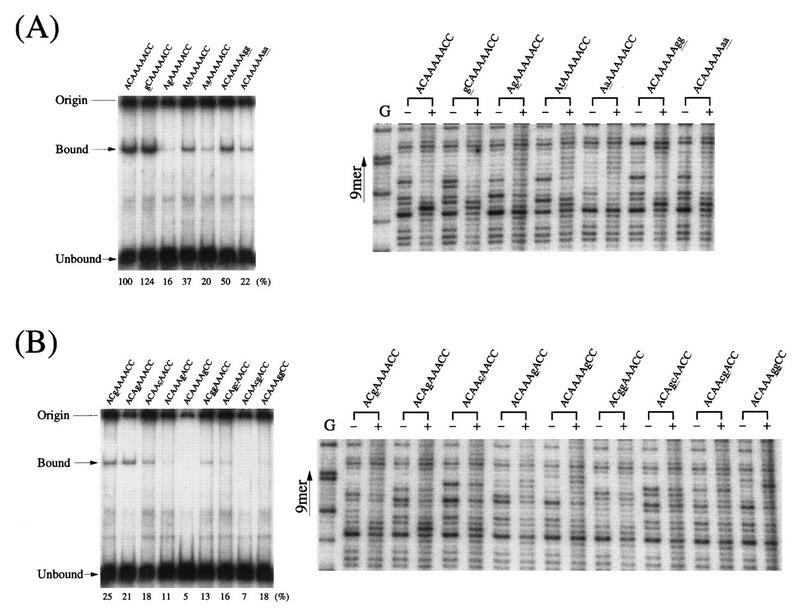
The nonamer signal (ACAAAAACC) is characterized by its A-rich sequence flanked by C residues. We have analyzed various nonamer mutants for their abilities to interact with RAG1 by DNase I footprinting. Figure 7 shows footprint patterns of 12-RSSs containing base substitutions in the A/T core or in the flanking residues. In vivo studies suggested that the flanking C residues in the nonamer are important in V(D)J recombination (2, 14). These flanking residues may play an important role when the recombinase measures lengths of spacers, probably by marking the border of the A stretch in the nonamer. As shown in Fig. 7A, a single substitution, at position 2 from C to G, greatly reduced the interaction in the DNase I footprinting. For the A residues in the nonamer, single- or double-base changes at positions 5, 6, and 7 appeared to be effective in eliminating binding in the gel shift assay (Fig. 7B). This is in a good agreement with previous in vivo observations (2, 14). Generally, double-base substitutions showed stronger effects on the interaction than single substitutions, making their footprint patterns similar to the pattern of unbound DNA.
FIG. 7.
Binding studies of nonamer mutants. Mutants for 12-RSS nonamer were analyzed by both DNase I footprinting and a gel shift assay. (A) Mutants for flanking residues (positions 1, 2, 8, and 9). (B) Mutants for the A/T core (positions 3 to 7). The RSS region within a 257-bp HindIII-PvuII fragment from pUC118 plasmid DNA (24) was obtained by HindIII cleavage, labeling with 32P, and digestion with PvuII. Two DNase I digestion patterns with (+) and without (−) the RAG1 protein are compared for each mutant. Mutations are noted by underlined lowercase letters. The G-reacted 12-RSS (wild type) is shown as a position marker (G). On the left side of the DNase I footprints, gel shifts for the mutants with RAG1-RAG2 are shown. For the gel shift assay, an 83-bp probe DNA fragment (HindIII-EcoRI) was prepared from pUC118 plasmid after labeling at the HindIII site. An EcoRI end was filled in with unlabeled dNTPs by using Klenow DNA polymerase. Relative amounts of shifted bands (mutant/wild type) are shown below the leftmost gels.
DISCUSSION
RAG1 is the major contributor to stable footprints. In this study, the RSS-RAG interaction was analyzed at the nucleotide level by a methylation interference assay and DNase I footprinting. Although it had been established that RAG1 interacts with DNA and requires RAG2 for catalytic activity (7, 42), the precise nature of this interaction was unknown. Since Ca2+ was found to cause nonspecific interaction with DNA and the cross-linker interfered with footprinting, we used Mg2+ instead of Ca2+ and omitted the cross-linker. Heptamer mutants were useful in detecting the complex, because they blocked the cleavage reaction but still allowed the RSS-RAG interaction. Our results indicate that RAG interacts with the nonamer sequence and the adjacent spacer (Fig. 5). Comparison of the RAG1-RAG2 footprint with that of RAG1 alone reveals similar levels of DNase I protection, demonstrating that RAG1 produces the main footprinting pattern. Highly sequence-specific interactions should occur in the nonamer, while non-sequence-specific interactions occur with the DNA sugar-phosphate backbone in the spacer.
The RAG1 footprinting pattern is consistent with a Hin homeodomain-like interaction.
The RAG1-nonamer interaction has been compared with that of the homeodomain of the bacterial Hin recombinase (7, 20, 42), which is involved in DNA inversions associated with Salmonella phase variation (12, 16). This relationship is strengthened by comparison of the data presented here with the crystal structure of the Hin-hix DNA complex (10). In Fig. 5C, the Hin homeodomain consists of the conserved sequence GGRPR, which is located in the minor groove, three alpha helices, two of which support a third alpha helix which acts as a reading head in the DNA major groove, and a final region that extends from the reading head down into the adjacent minor groove (10). In the case of RAG1, the minimal domain required for RSS binding is only 94 aa (residues 384 to 377) and includes a similar homeodomain (42).
In our DMS experiments, the G2 in the bottom strand of the nonamer was protected from methylation (data not shown) and, if methylated, interfered with RAG binding (Fig. 3A). Thus, this single residue is critical for the RAG-nonamer interaction. Because of the preponderance of A-T base pairs in the nonamer sequence, there has been a particular interest in the C-G base pair at position 2. Mutation of C2 (top strand) to each of the other possible residues results in various reduced amounts of recombination in vivo (2, 14). The DNase I footprinting and the gel shift analysis of mutations at this position also revealed the importance of this C-G pair. In the Hin crystal structure, the equivalent G interacts with helix 3 Arg178 (Fig. 5C) (10). However, considerable differences between Hin and RAG1 make any prediction for the amino acid residue involved in the RAG1 interaction at G2 premature.
In the minor groove, methylation at residues A6 and A7 strongly interferes with RAG binding while methylation at A5 partially interferes (Fig. 5A). This is similar to the result of Hin footprinting, where the equivalents of A5 to A7 are protected from methylation and interfere with binding if they are methylated (10, 12). In the Hin-hix interaction, Gly139 and Arg 140 are essential for DNA binding and are intimately associated with the minor groove at positions equivalent to A5 and A6. Methylation or mutation of these nucleotides eliminates Hin binding. In addition to interactions in this minor groove, partial interference at A1 and A3 suggests that RAG1 protein, like Hin, also interacts with the minor grooves on the both sides of the major groove.
The results of DNase I footprinting are consistent with a Hin-like interaction. Based on the Hin structure, Arg393 (Hin Arg142) rises out of the minor groove and interacts with the 5′ phosphate of T3 on the nonamer bottom strand, blocking this residue in DNase I footprinting as reported here (Fig. 5A). There are three nucleotides strongly protected from DNase I in the adjacent spacer (positions −1 to −3 on the top strand) which are equivalent to sites sterically blocked by the Hin protein as it extends from the third alpha helix into the minor groove (Fig. 5A and C). These interactions with the spacer region may be important in the bending or deformation of the DNA that results in greater cleavage by DNase I on the opposite side of the DNA helix. The enhanced sites of cleavage (C2 on top strand and residues at −2 and −3 in the spacer on the bottom strand) are consistent with their position on the DNA face away from the main body of the recombinase (Fig. 5B). There are two other sites probably blocked for DNase I cleavage by steric interactions as judged from the Hin model, T1 (Fig. 5A) and position −4 on the bottom strand (Fig. 5B). There are two sites, A4 on the top strand and G8 on the bottom strand (Fig. 5B), that are not candidates for blocking as indicated by the Hin model but would be in appropriate positions for blocking if the protein strands in both minor grooves were extended and folded across the DNA sugar-phosphate backbone (Fig. 5C). Evidence for such minor groove interactions is provided by the methylation interference on the top strand of 12-RSS at positions −1, 1, 3, 5, 6, and 7.
One of the predictions of a Hin-like model for RAG1 is that the conserved GGRPR in the minor groove would not protect G8 and G9 (bottom strand) from major groove methylation. This prediction is borne out by the complete absence of methylation protection or interference for these two nucleotides.
Although most of the strong sites of DNase I inhibition or enhancement are the same for the 12- and 23-RSSs, there is a difference in that the 23-RSS has an additional enhanced DNase I cleavage site at position −2. This cleavage site is adjacent to a residue at position −1 that is protected only in the 12-RSS. One interpretation of this result is that the protein occupies a slightly different position on the 12-RSS, protecting the DNA backbone and eliminating one enhanced site.
Footprinting results are consistent with mutant phenotypes.
Several mutations in the nonamer region were found to affect the local footprinting pattern. Changing C2 to A (top strand) or changing the terminal CC to AA (positions 8 and 9 in the top strand) altered the pattern adjacent to these residues, reduced DNase I-enhanced sites, and diminished the pattern in the spacer region. For the most part, mutations that retain considerable function also show DNase I protection and enhanced cleavage. Mutations with very low activity show neither protection nor enhancement and result in reduced DNA binding in the gel retardation assay.
Although it had been reported that RAG1 interacts with RSS DNA and requires RAG2 for DNA cleavage (7, 42), footprint analysis of the RAG-RSS interaction at the nucleotide level has not been described. Results of the present study demonstrate that RAG1 interacts with specific bases in the nonamer sequence and that the interaction extends into the adjacent spacer (Fig. 5). An unexpected finding was that there was very little evidence of interactions between RAG2 and RSS. Furthermore, protection with RAG1 in the heptamer region was only partial. Stable interaction of the heptamer with RAG proteins may occur only after the synaptic complex is formed between the 12-RSS and the 23-RSS (1). It should be mentioned that in the Mu transposase system, cleavage is mediated by the protein sitting on the counterpart substrate in the synaptic complex (35). Identification of a functional 12/23 complex and its footprint analysis will clarify the issue and shed light on the molecular basis of the 12/23 rule for V(D)J type recombination.
ACKNOWLEDGMENTS
We thank Yoshiharu Matsuura and Masaki Kashiwada (National Institute of Infectious Diseases, Tokyo, Japan) for helpful advice, Taiji Itoh, Masami Kodama, and Satomi Shichijo for expert technical assistance, and Michiko Kimura for excellent secretarial work.
The first three authors contributed equally to this work.
This work was supported by the Special Promotion Research Grant of Monbusho in Japan and by grants from Torey Science Foundation, Nissan Science Foundation, and Mitsubishi Foundation. A.J.O. was a recipient of fellowships from Monbusho and the Japan Society for the Promotion of Science.
REFERENCES
- 1.Agrawal A, Schatz G. RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell. 1997;89:43–53. doi: 10.1016/s0092-8674(00)80181-6. [DOI] [PubMed] [Google Scholar]
- 2.Akamatsu Y, Tsurushita N, Nagawa F, Matsuoka M, Okazaki K, Imai M, Sakano H. Essential residues in V(D)J recombination signals. J Immunol. 1994;153:4520–4529. [PubMed] [Google Scholar]
- 3.Akira S, Okazaki K, Sakano H. Two pairs of recombination signals are sufficient to cause immunoglobulin V-(D)-J joining. Science. 1987;238:1134–1138. doi: 10.1126/science.3120312. [DOI] [PubMed] [Google Scholar]
- 4.Blunt T, Finnie N J, Taccioli G E, Smith G C M, Demengeot J, Gottlieb T M, Mizuta R, Varghese A J, Alt F W, Jeggo P A, Jackson S P. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell. 1995;80:813–823. doi: 10.1016/0092-8674(95)90360-7. [DOI] [PubMed] [Google Scholar]
- 5.Brenowitz M, Senear D F, Shea M A, Ackers G K. Quantitative DNase I footprint titration: a method for studying protein-DNA interactions. Methods Enzymol. 1986;130:132–181. doi: 10.1016/0076-6879(86)30011-9. [DOI] [PubMed] [Google Scholar]
- 6.Cuomo C A, Oettinger M A. Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res. 1994;22:1810–1814. doi: 10.1093/nar/22.10.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Difilippantonio M J, McMahan C J, Eastman Q M, Spanopoulou E, Schatz D G. Rag-1 mediates signal sequence recognition and recruitment of Rag-1 in V(D)J recombination. Cell. 1996;87:253–262. doi: 10.1016/s0092-8674(00)81343-4. [DOI] [PubMed] [Google Scholar]
- 8.Early P, Huang H, Davis M, Calame K, Hood L. An immunoglobulin heavy chain variable region gene is generated from three segments of DNA: VH, D and JH. Cell. 1980;19:981–992. doi: 10.1016/0092-8674(80)90089-6. [DOI] [PubMed] [Google Scholar]
- 9.Eastman Q M, Leu T M J, Schatz D G. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature. 1996;380:85–88. doi: 10.1038/380085a0. [DOI] [PubMed] [Google Scholar]
- 10.Feng J-A, Johnson R C, Dickerson R E. Hin recombinase bound to DNA: the origin of specificity in major and minor groove interactions. Science. 1994;263:348–355. doi: 10.1126/science.8278807. [DOI] [PubMed] [Google Scholar]
- 11.Gilfillan S, Dierich A, Lemeur M, Benoist C, Mathis D. Mice lacking TdT: mature animals with an immature lymphocyte repertoire. Science. 1993;261:1175–1178. doi: 10.1126/science.8356452. [DOI] [PubMed] [Google Scholar]
- 12.Glasgow A C, Bruist M F, Simon M I. DNA-binding properties of the Hin recombinase. J Biol Chem. 1989;264:10072–10082. [PubMed] [Google Scholar]
- 13.Grawunder U, Matthias W, Xiantuo W, Kulesza P, Wilson T E, Mann M, Lieber M R. Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature. 1997;388:492–495. doi: 10.1038/41358. [DOI] [PubMed] [Google Scholar]
- 14.Hesse J E, Lieber M R, Mizuuchi K, Gellert M. V(D)J recombination: a functional definition of the joining signals. Genes Dev. 1989;3:1053–1061. doi: 10.1101/gad.3.7.1053. [DOI] [PubMed] [Google Scholar]
- 15.Hiom K, Gellert M. A stable RAG1-RAG2-DNA complex that is active in V(D)J cleavage. Cell. 1997;88:65–72. doi: 10.1016/s0092-8674(00)81859-0. [DOI] [PubMed] [Google Scholar]
- 16.Hughes K T, Gaines P C W, Karlinsey J E, Vinayak R, Simon M I. Sequence-specific interaction of the Salmonella Hin recombinase in both major and minor grooves of DNA. EMBO J. 1992;11:2695–2705. doi: 10.1002/j.1460-2075.1992.tb05335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirchgessner C U, Patil C K, Evans J W, Cuomo C A, Fried L M, Carter T, Oettinger M A, Brown J M. DNA-dependent kinase (p350) as a candidate gene for the murine SCID defect. Science. 1995;267:1178–1183. doi: 10.1126/science.7855601. [DOI] [PubMed] [Google Scholar]
- 18.Komori T, Okada A, Stewart V, Alt F W. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–1175. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]
- 19.Lees-Miller S P, Godbout R, Chan D W, Weinfeld M, Day III R S, Barron G M, Allalunis-Turner J. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science. 1995;267:1183–1185. doi: 10.1126/science.7855602. [DOI] [PubMed] [Google Scholar]
- 20.Lewis S M, Wu G G. The origin of V(D)J recombination. Cell. 1997;88:159–162. doi: 10.1016/s0092-8674(00)81833-4. [DOI] [PubMed] [Google Scholar]
- 21.Li Z, Otevrel T, Gao Y, Cheng H-L, Seed B, Stamato T D, Taccioli G E, Alt F W. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- 22.Max E E, Seidman J G, Leder P. Sequences of five potential recombination sites encoded close to an immunoglobulin kappa constant region gene. Proc Natl Acad Sci USA. 1979;76:3450–3454. doi: 10.1073/pnas.76.7.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maxam A M, Gilbert S. A new method for sequencing DNA. Proc Natl Acad Sci USA. 1977;74:560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McBlane J F, van Gent D C, Ramsden D A, Romeo C, Cuomo C A, Gellert M, Oettinger M A. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 25.McMahan C J, Sadofsky M J, Schatz D G. Definition of a large region of RAG1 that is important for coimmunoprecipitation of RAG2. J Immunol. 1997;158:2202–2210. [PubMed] [Google Scholar]
- 26.Oettinger M A, Schatz D G, Gerka C, Baltimore D. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science. 1990;248:1517–1523. doi: 10.1126/science.2360047. [DOI] [PubMed] [Google Scholar]
- 27.O’Reilly D R, Miller L K, Luckow V A. Baculovirus expression vectors: a laboratory manual. New York, N.Y: Oxford University Press; 1994. [Google Scholar]
- 28.Ramsden D A, Paull T T, Gellert M. Cell-free V(D)J recombination. Nature. 1997;388:488–491. doi: 10.1038/41351. [DOI] [PubMed] [Google Scholar]
- 29.Roth D B, Menetski J P, Nakajima P M, Bosma M J, Gellert M. V(D)J recombination: covalently sealed (hairpin) coding ends in scid mouse thymocytes. Cell. 1992;70:983–991. doi: 10.1016/0092-8674(92)90248-b. [DOI] [PubMed] [Google Scholar]
- 30.Sadofsky M J, Hesse J E, McBlane J F, Gellert M. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 1993;21:5644–5650. doi: 10.1093/nar/21.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sadofsky M J, Hesse J E, Gellert M. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 1994;22:1805–1809. doi: 10.1093/nar/22.10.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sakano H, Huppi K, Heinrich G, Tonegawa S. Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature. 1979;280:288–294. doi: 10.1038/280288a0. [DOI] [PubMed] [Google Scholar]
- 33.Sakano H, Maki R, Kurosawa Y, Roeder W, Tonegawa S. Two types of somatic recombination are necessary for the generation of complete immunoglobulin heavy-chain genes. Nature. 1980;286:676–683. doi: 10.1038/286676a0. [DOI] [PubMed] [Google Scholar]
- 34.Sakano H, Kurosawa Y, Weigert M, Tonegawa S. Identification and nucleotide sequence of a diversity DNA segment (D) of immunoglobulin heavy-chain genes. Nature. 1981;290:562–565. doi: 10.1038/290562a0. [DOI] [PubMed] [Google Scholar]
- 35.Savilahti H, Mizuuchi K. Mu transpositional recombination: donor DNA cleavage and strand transfer in trans by the Mu transposase. Cell. 1996;85:271–280. doi: 10.1016/s0092-8674(00)81103-4. [DOI] [PubMed] [Google Scholar]
- 36.Schatz D G, Oettinger M A, Baltimore D. The V(D)J recombination-activating gene, RAG-1. Cell. 1989;59:1035–1048. doi: 10.1016/0092-8674(89)90760-5. [DOI] [PubMed] [Google Scholar]
- 37.Schlissel M, Constantinescu A, Morrow T, Baxter M, Beug A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- 38.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–716. [PubMed] [Google Scholar]
- 39.Silver D P, Spanopoulou E, Mulligan R C, Baltimore D. Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc Natl Acad Sci USA. 1993;90:6100–6104. doi: 10.1073/pnas.90.13.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh H, Sen R, Baltimore D, Sharp P. A nuclear factor which binds to a conserved sequence motif in transcriptional control elements of immunoglobulin genes. Nature. 1986;319:154–158. doi: 10.1038/319154a0. [DOI] [PubMed] [Google Scholar]
- 41.Smith G E, Ju G, Ericson B L, Moschera J, Lahm H-W, Chizzonite R, Summers M D. Modification and secretion of human interleukin 2 produced in insect cells by a baculovirus expression vector. Proc Natl Acad Sci USA. 1985;82:8404–8408. doi: 10.1073/pnas.82.24.8404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spanopoulou E, Zaitseva F, Wang F-H, Santagata S, Baltimore D, Panayotou G. The homeodomain region of Rag-1 reveals the parallel mechanisms of bacterial and V(D)J recombination. Cell. 1996;87:263–276. doi: 10.1016/s0092-8674(00)81344-6. [DOI] [PubMed] [Google Scholar]
- 43.Taccioli G E, Rathbun G, Oltz E, Stamato T, Jeggo P A, Alt F W. Impairment of V(D)J recombination in double strand break mutants. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 44.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 45.van Gent D C, McBlane J F, Ramsden D A, Sadofsky M J, Hesse J E, Gellert M. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81:925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 46.van Gent D C, Ramsden D A, Gellert M. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell. 1996;85:107–113. doi: 10.1016/s0092-8674(00)81086-7. [DOI] [PubMed] [Google Scholar]
- 47.Zhu C, Bogue M A, Lim D-S, Hasty P, Roth D B. Ku80-deficient mice exhibit severe combined immunodeficiency and defective processing of V(D)J recombination intermediates. Cell. 1996;86:379–390. doi: 10.1016/s0092-8674(00)80111-7. [DOI] [PubMed] [Google Scholar]