Abstract
PURPOSE
NOTCH3 is increasingly implicated for its oncogenic role in many malignancies, including meningiomas. While prior work has linked NOTCH3 expression to higher-grade meningiomas and treatment resistance, the metabolic phenotype of NOTCH3 activation remains unexplored in meningioma.
METHODS
We performed single-cell RNA sequencing on NOTCH3 + human meningioma cell lines. Using the CH157-MN meningioma cell model, we overexpressed NOTCH3 intracellular domain (ICD) and performed untargeted metabolomic, lipidomic, and bulk RNA sequencing analyses as well as functional metabolic assays.
RESULTS
We show that NOTCH3 mediates a metabolic shift towards fatty acid oxidation (FAO), depleting lipid availability and conferring resistance to ferroptosis. Single-cell RNA sequencing revealed a correlation with CD36, a key fatty acid transporter. Furthermore, patient-derived primary meningioma lines stratified by NOTCH3 expression confirmed higher CD36 expression and increased maximal mitochondrial respiration in NOTCH3-high cells in the presence of palmitate, supporting enhanced FAO. NOTCH3 ICD overexpression (OE) exhibited depletion of fatty acid pools, alongside transcriptional upregulation of canonical FAO genes. Functional mitochondrial assays confirmed elevated oxidative respiration in the presence of palmitate compared with controls. Additionally, NOTCH3 OE cells exhibit increased resistance to RSL3-induced ferroptosis, a phenotype that was reversed with CPT1.
CONCLUSION
These data establish a link between NOTCH3 signaling, lipid metabolic reprogramming, and ferroptosis evasion in aggressive meningioma cells. This metabolic shift may contribute to the malignant behavior observed in NOTCH3 + meningiomas, offering new insight into the biochemical vulnerabilities of these tumors.
Keywords: NOTCH3, Meningioma Metabolism, Fatty Acid Oxidation, Ferroptosis, Tumor Microenvironment
Introduction
The NOTCH signaling pathway plays a fundamental role in cell fate determination, differentiation, and tissue homeostasis. Among the four receptors (NOTCH1–4), NOTCH3 expression is more restricted and is predominantly found in vascular smooth muscle, the central nervous system, and thymocytes [1–3]. While deletions of NOTCH1 and NOTCH2 are embryonically lethal, NOTCH3 deletion is not [4, 5]. The NOTCH3 transmembrane receptor undergoes a series of cleavage events, via ADAM10 metalloproteases and γ-secretase complex, yielding an extracellular domain (ECD) and an intracellular domain (ICD) [6, 7]. The active ICD then translocates to the nucleus, where it participates in gene transcription regulation.
NOTCH3 plays an important role in the survival of vascular smooth muscle cells, which explains its expression pattern in this tissue [8]. NOTCH3 has been implicated in vascular pathologies, including cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) as well as multiple cancers, including T-cell lymphoma, breast cancer, pancreatic adenocarcinoma, and others [9–12]. In gliomas, NOTCH3 overexpression has been associated with the promotion of glioma cell proliferation and therapeutic resistance, suggesting a conserved oncogenic role across diverse tumor types [13, 14].
Recently, the NOTCH3 gene has been identified as a driver for tumor growth and resistance to radiation in meningiomas. [15] Choudhury et al. proposed that, due to its conserved expression in meningioma and potential to promote angiogenesis, NOTCH3 is a potential therapeutic target for meningioma. [15] However, the metabolic consequences of NOTCH3 activation in meningiomas remain poorly understood. Tumor metabolism is a key determinant of cancer cell survival and therapeutic response, yet meningiomas remain largely understudied in this context.
Through metabolomic and lipidomic profiling, this study demonstrates that NOTCH3 activation influences lipid metabolism through the depletion of fatty acids, which are used by the tumor cells for FAO. This metabolic profile protects cells from ferroptosis and may be a mechanism to both enhance malignancy and resistance to therapeutic interventions such as sorafenib or cisplatin, portending a poor prognosis in NOTCH3-expressing meningiomas.
Materials And Methods
Cell Culture
Meningioma cell lines and NOTCH3 overexpression
The CH157-MN cell lines was obtained as a gift from Professor David Raleigh at UCSF and were cultured in DMEM (11960069, Life Technologies) supplemented with 10% FBS, 1× GlutaMAX (35050–061, Thermo Fisher Scientific) and 1× penicillin/streptomycin (15140122, Life Technologies). All cells were cultured at 37°C and at 5% CO2 and 21% O2. Cell lines were confirmed to be mycoplasma-free at regular intervals.
The generation of NOTCH3 ICD expressing meningioma cell lines was accomplished by creating pLVX-Puro plasmid containing pCMV6-NOTCH3ICD as previously described [15]. Lentiviral particles were produced by transfecting HEK293T cells with standard packaging vectors using the TransIT-Lenti Transfection Reagent (6605, Mirus). CH157-MN cells were stably transduced with lentiviral particles to generate NOTCH3 ICD OE (CH157-MNNOTCH3 ICD) or empty pLVX vector (CH157-MNEV) cells. Successfully transduced cells were isolated using Puromycin selection, and NOTCH3 overexpression was confirmed using RT-qPCR.
Patient samples
Human PDX samples were obtained under protocol #STU00095863 approved by the Northwestern Institutional Review Board (IRB). Meningioma tissue samples, PDX1 (WHO Grade 2) and PDX2 (WHO Grade 2), were obtained within one hour of surgical resection from 2 different patients and enzymatically dissociated in RPMI 1640 medium (Roswell Park Memorial Institute; Corning) containing 20 μL/mL collagenase (Roche) and 1 μL/mL DNase (Roche), supplemented with 2% fetal bovine serum (FBS). Single-cell suspensions were washed, seeded in culture plates, and expanded at 37°C in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with GlutaMAX™ (Gibco) and 10% FBS. NOTCH3 expression was validated using RT-qPCR.
Real Time – Quantitative PCR
RNA extraction was performed using either the Qiagen RNEasy kit (Qiagen, Hilden, Germany) or the Omega E.Z.N.A. RNA Isolation Kit (R6834–02, Omega Biol-tek, Norcross, GA). The cDNA was generated from RNA samples with an iScript kit (BioRad, Hercules, CA, USA). Reactions were set up using standard amounts of cDNA, SybrGreen (Biorad), and forward and reverse primers (IDT, Newark, NJ, USA). The readout was performed on a CFX96 qPCR machine (Biorad). All primers were generated from Primer-BLAST using the fixed settings. Reactions were performed in triplicate. The following primers were used: NOTCH3 (human) (forward 5’- GCCAAGCGGCTAAAGGTAGA-3’; reverse 5’- GGATGTCAGCAGCAACAAGA-3’), CD36 (human) (forward 5’-CAGGTCAACCTATTGGTCAAGCC-3’; reverse 5’- GCCTTCTCATCACCAATGGTCC-3’), and b-Actin for baseline expression level (human) (forward 5’- CACCATTGGCAATGAGCGGTTC-3’; reverse 5’- AGGTCTTTGCGGATGTCCACGT-3’). Fold changes in gene expression relative to untreated control were calculated by the ΔΔCt method using mouse actin as an endogenous control for mRNA expression.
Untargeted Metabolomic Profiling
Metabolite extraction
CH157-MNNOTCH3 ICD and the CH157-MNEVcells were scraped and washed twice with PBS before pellets were flash-frozen and stored at − 80°C until metabolite extraction. Pellets were resuspended in 80% methanol/20% H2O and then lysed by 3× cycles of heat shock (liquid nitrogen freezing followed by 42°C water bath). Samples were then spun at 14,000 rpm for 15 min. The supernatants were collected and analyzed as described below.
Method for sample reconstitution after extraction
The extracted supernatants were dried using SpeedVac. 60% acetonitrile was added to the tube for reconstitution, followed by overtaxing for 30 sec. The sample solution was then centrifuged for 30 min at 20,000g, 4°C. Samples were analyzed by High-Performance Liquid Chromatography and High-Resolution Mass Spectrometry and Tandem Mass Spectrometry (HPLC-MS/MS) as previously described [16].
Bulk RNA Sequencing
RNA was extracted using the RNeasy Plus Mini Kit (Qiagen, catalog no. 74134) according to the manufacturer’s protocol. RNA sample quality control, library preparation (polyA selection, nonstranded), sequencing of 20 M paired-end reads, and analysis were performed by Novogene.
Human Single Cell Sequencing
Single-cell RNA-seq data from 22 meningioma samples was extracted from publicly available repositories [17, 18] and processed according to prior descriptions in [18]. Briefly, the Seurat R Package using the scRNA-seq Seurat10x genomic workflow was used for all analyses unless noted otherwise [19]. Since data came from two different repositories, batch correction was performed using the standard Harmony algorithm [20]. Tumor cells were identified using CONICSmat [21] to determine copy-number loss of chromosome 22q using repetitions = 100 and postProb = 0.75. Cells with postProb < 0.15 were considered tumor and > 08.5 as normal. Clusters with more than 80% normal cells were considered non-meningioma clusters. NOTCH3 + meningioma cells were classified based on NOTCH3 expression > 0. Seurat’s FindMarkers was used to find differentially expressed genes between NOTCH3 + meningioma cells and NOTCH3− meningioma cells. Data was displayed using a volcano plot.
Targeted Fatty Acid Analyses
Cellular fatty acids were measured as previously described [22]. Briefly, cells were seeded at an initial density of 90,000 cells per well in a six-well plate in 2 ml of DMEM medium. A parallel plate of cells was scanned with an Incucyte Live-Cell Analysis System (Sartorius) and analyzed for confluence to normalize extraction buffer volumes based on cell number. An empty well was also extracted for a process control. After incubating cells for 24 h, lipids were extracted using an extraction buffer consisting of chloroform:methanol (containing 25 mg/L of butylated hydroxytoluene (Millipore Sigma, B1378)):0.88% KCl (w/v) at a final ratio of 8:4:3. The final extraction buffer also contained 0.75 μg/ml of norvaline and 0.7 μg/ml of cis-10-heptadecenoic acid as internal standards. For extraction, the medium was aspirated from cells, and cells were rapidly washed in ice-cold saline three times. The saline was aspirated, and methanol:0.88% KCl (w/v) (4:3 v/v) was added. Cells were scraped on ice, and the extract was transferred to 1.5 ml Eppendorf tubes (Dot Scientific, RN1700-GMT) before adding chloroform (Supelco, 1.02444). The resulting extracts were vortexed for 10 min and centrifuged at maximum speed (17000 × g) for 10 min. Lipids (organic fraction) were transferred to glass vials (Supelco, 29651-U) and dried under nitrogen gas for further analysis.
Fatty acids were analyzed as pentafluorobenzyl-fatty acid (PFB-FA) derivatives. Fatty acids were saponified from dried lipid pellets by adding 800 μl of 90% methanol/0.3 M KOH, vortexing, and incubating at 80°C for 60 min. Each sample was then neutralized with 80 μl of formic acid (Supelco, FX0440). Fatty acids were extracted twice with 800 μl of hexane and dried under nitrogen gas. To derivatize, fatty acid pellets were incubated with 100 μl of 10% pentafluorobenzyl bromide (Sigma Aldrich, 90257) in acetonitrile and 100 μl of 10% N,N-diisopropylethylamine (Sigma Aldrich, D125806) in acetonitrile at room temperature for 30 min. PFB-FA derivatives were dried under nitrogen gas and resuspended in 50 μl of hexane for GC-MS analysis.
GC-MS was conducted with a TRACE TR-FAME column (ThermoFisher, 260M154P) installed in a Thermo Scientific TRACE 1600 gas chromatograph coupled to a Thermo ISQ 7610 mass spectrometer. Helium was used as the carrier gas at a constant flow of 1.8 ml/min. One microliter of sample was injected at 250°C at a 4:1 split. After injection, the GC oven was held at 100°C for 0.5 min, increased to 200°C at 40°C/min, held at 200°C for 1 min, increased to 250°C at 5°C/min, and held at 250°C for 11 min. The MS system operated under negative chemical ionization mode with methane gas at a flow rate of 1.25 ml/min, and the MS transfer line and ion source were held at 255°C and 200°C, respectively. The detector was used in scanning mode with an ion range of 150–500 m/z. Total ion counts were determined by integrating appropriate ion fragments for each PFB-FA using Skyline software [23, 24].
Ferroptosis Sensitivity and Cell Viability
CH157-MNNOTCH3 ICD and CH157-MNEV cells were plated at a density of 5 × 103 cells/well in a flat-bottom, black 96-well plate treated with the designated amounts of RSL3 for 24 hours as indicated within the figures. Cells were analyzed using the Cell Counting Kit-8 (CCK-8) (Sigma), and absorbance was measured at 450 nm using the Biotek Cytation 5 (Agilent). Cell viability was then normalized to untreated cells at each time point. Etomoxir, a CPT1a inhibitor, was utilized to inhibit mitochondrial function and FAO [25–27]; For assays with etomoxir, wells were pretreated with 5 μM of etomoxir for 4 hours before treatment with the indicated concentrations of RSL3 for 24 hours. Cells were again analyzed using CCK-8 as described above.
Seahorse Extracellular Flux Analysis and Oxygen Consumption Rate (OCR)
The OCR was measured in a XF96 extracellular flux analyzer (Agilent Bioscience). The patient-derived meningioma cells, CH157-MNNOTCH3 ICD and CH157-MNEV, were plated at a density of 2 × 104 cells in 150 μL per well in a 96-well XF96 plate and allowed to adhere overnight. The cartridge from the Seahorse XFp FluxPak (Agilent; 103022–100) was hydrated overnight with XF Calibrant solution in a non-CO2 incubator. The plate was washed once with Agilent Seahorse XF Base Medium (Agilent; 103334-100) containing 2 mM glutamine (Agilent 103579–100) and 25 mM glucose prior to the addition of 150 μL Agilent Base Medium with supplements. The plate was incubated for 40 minutes in a non-CO2 incubator prior to analysis. The Seahorse assay was then run with standard injections of the Seahorse XF Cell Mito Stress Test Kit (Agilent; 103015–100). Basal and maximal respiration rates were corrected by subtracting rotenone/antimycin A correction factor via previously reported methods [28, 29]
Results
NOTCH3 + Human Meningioma Cells Upregulate CD36 Expression
To understand the transcriptional profile of NOTCH3-expressing meningiomas, 14,080 NOTCH3 + human meningioma cells from publicly available [17, 18] were analyzed. The scRNA-seq data revealed a correlation between NOTCH3 + meningioma cells and clusters-of-differentiation 36 (CD36) expression, indicating that CD36 is enriched in NOTCH3 + cells (Fig. 1A). The lipid metabolism enrichment observed in NOTCH3-expressing patient-derived meningioma cells led us to investigate the direct role of NOTCH3 overexpression using a controlled in vitro model.
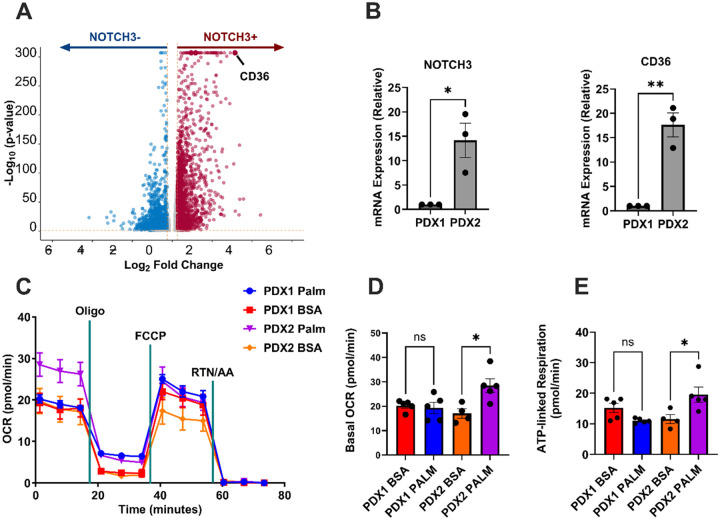
Figure 1. NOTCH3 expression controls lipid uptake and oxidation in meningioma cells.
A. Volcano plot of 14,080 NOTCH3+ human meningioma cells from 22 meningiomas showing upregulation of CD36. P-values were calculated using DESeq2. B. Relative mRNA expression of NOTCH3 and CD36 in 2 patient-derived meningioma lines PDX1 and PDX2 (PDX1, n = 3; PDX2, n = 3). Experiments were performed in triplicate; significance was calculated using an unpaired Student’s t-test. C. Seahorse flux tracing of PDX1 and PDX2 under BSA or palmitate incubation conditions. D. Basal oxygen consumption rate (OCR) in PDX1 and PDX2 cells treated with palmitate-BSA or BSA control (PDX1, n = 3 in each condition; PDX2, n = 3 in each condition). E. ATP-linked respiration in PDX1 and PDX2 cells treated with palmitate-BSA or BSA control (PDX1, n = 3 in each condition; PDX2, n = 3 in each condition). In C-En=2 independent experiments were performed. In D-E, a one-way ANOVA was performed with a Tukey’s post-hoc to calculate significance. P<0.05*; P<0.01**, P<0.001***.
NOTCH3 + Primary Meningioma Cells Exhibit Increased Lipid Metabolism
To further investigate the consequences of NOTCH3 expression on lipid metabolism in human meningioma cells, two patient-derived meningioma lines were profiled for NOTCH3 expression via RT-qPCR. PDX1 exhibited low and PDX2 exhibited high NOTCH3 expression (Fig. 1B). The expression of CD36 was higher in PDX2 than PDX1 (Fig. 1B), consistent with the scRNA-seq data analysis. Since CD36 is associated with the uptake of saturated fatty acids for FAO,25 we hypothesized that the NOTCH3high PDX2 would be more proficient at FAO than NOTCH3low PDX1. To test this, Seahorse extracellular flux analysis was performed on both PDX lines with the inclusion of a FA palmitate-BSA conjugate, or a BSA alone control (Fig. 1C–D). In NOTCH3lo PDX1, there was no difference in basal OCR with the addition of palmitate; whereas PDX2 (NOTCH3high) responded with a significantly higher basal OCR with palmitate (17.0 ± 1.9 pmol O2/min in BSA incubated vs. 28.5 ± 3.2 in the palmitate-treated group; p < 0.05) (Fig. 1C). Furthermore, ATP-linked respiration was also increased in the NOTCH3high PDX2, whereas no change occurred in the NOTCH3low PDX1 (Fig. 1D). This data suggests that NOTCH3 controls lipid metabolism in meningioma cells and prompted the use of an overexpression line to verify that the increase in FAO is directly related to NOTCH3 activity.
NOTCH3 Overexpression Alters Lipidomic Profile
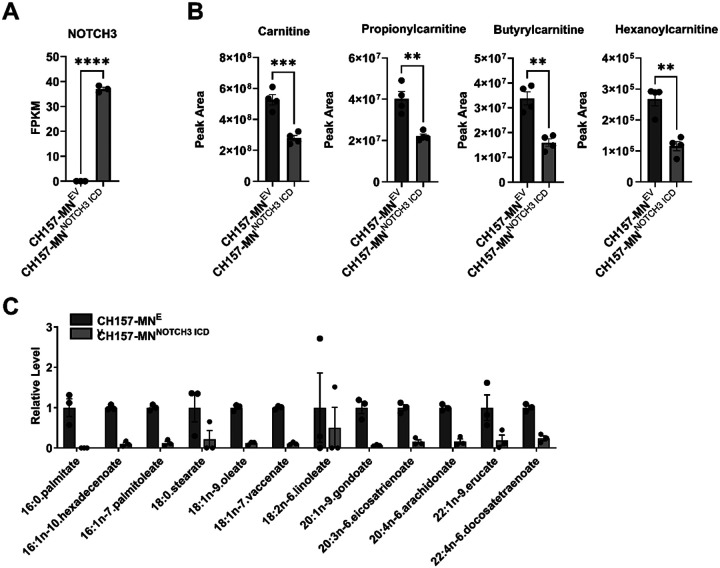
To isolate and directly study the role of NOTCH3 and its effect on metabolism in meningiomas, the CH157-MNNOTCH3 ICD and CH157-MNEV cells were used for further in vitro studies (Fig. 2A). Metabolomic profiling was performed on CH157-MNNOTCH3 ICD and CH157-MNEV cells to determine the metabolite differences between the two cell lines (Fig. 2B). Compared to EV control, CH157-MNNOTCH3 ICD exhibits decreases in several fatty acyl carnitine species required for fatty acid transport into the mitochondria for FAO (Fig. 2C). This was further validated with targeted fatty acid profiling that confirmed decreased saturated and unsaturated fatty acid levels in CH157-MNNOTCH3 ICD compared with the EV control (Fig. 2C).
Figure 2. NOTCH3 overexpression alters fatty acid composition in meningioma cells.
A. Bulk RNA sequencing FPKM values of NOTCH3 in CH157-MNEV and CH157-MNNOTCH3 ICD (CH157-MNEV, n = 3; CH157-MNNOTCH3 ICD, n = 3). B. Untargeted metabolomic analysis showing an increase in acyl carnitine species carnitine, propionylcarnitine, butyrylcarnitine, and hexanoylcarnitine in CH157-MNNOTCH3 ICD compared with CH157-MNEV (CH157-MNEV, n = 4; CH157-MNNOTCH3 ICD, n = 4). Significance was calculated using unpaired Student’s t-tests. P<0.05*; P<0.01**, P<0.001***; P<0.0001****. C. Histogram of targeted lipidomic analysis of CH157-MNEV and CH157-MNNOTCH3 ICD (CH157-MNEV, n = 3; CH157-MNNOTCH3 ICD, n = 3).
NOTCH3 Controls Transcriptional Expression of Lipid Metabolic Genes
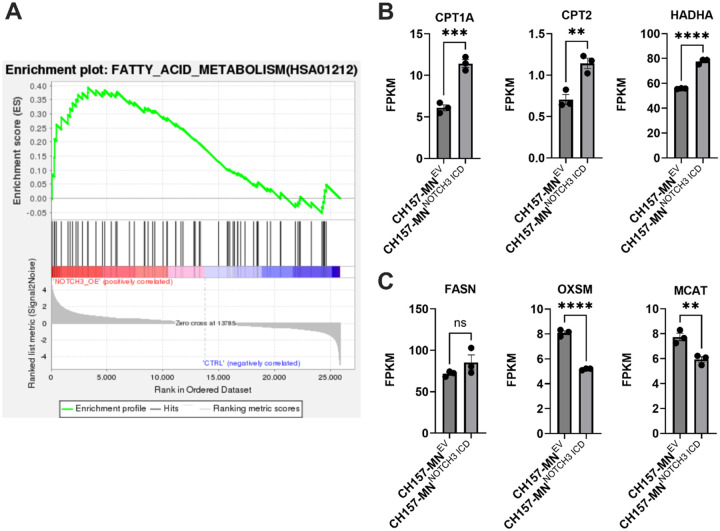
RNA sequencing of CH157-MNNOTCH3 ICD and CH157-MNEV cell lines showed increased enrichment of fatty acid metabolism genes in CH157-MNNOTCH3 ICD cells (Fig. 3A–B). Specifically, key FAO genes such as CPT1A and CPT2, which import long-chain fatty acids into mitochondria for oxidation, and HADHA, which encodes for a mitochondrial enzyme that is essential for β-oxidation, were upregulated in CH157-MNNOTCH3 ICD cells (Fig. 3C). In contrast, genes essential to fatty acid biosynthesis/anabolism, including FASN (fatty acid synthase), OXSM (3-oxoacyl-ACP synthase), and MCAT (malonyl-CoA acyltransferase), were downregulated or unchanged (Fig. 3D). These data suggest that NOTCH3 promotes a shift from fatty acid synthesis to FAO in meningioma.
Figure 3. NOTCH3 overexpression enhances transcription of fatty acid oxidation genes in meningioma cells.
A. Gene set enrichment analysis (GSEA) of bulk RNA sequencing showing increased activation of fatty acid metabolism pathways in CH157-MNNOTCH3 ICD compared with CH157-MNEV (CH157-MNEV, n = 3; CH157-MNNOTCH3 ICD, n = 3). B. Bulk RNA sequencing FPKM values of CH157-MNEV and CH157-MNNOTCH3 ICD showing upregulation of FAO genes CPT1A, CPT2, and HADHA (CH157-MNEV, n = 3; CH157-MNNOTCH3 ICD, n = 3). C. Bulk RNA sequencing FPKM values of CH157-MNEV and CH157-MNNOTCH3 ICD showing no difference or decrease in fatty acid synthesis genes FASN, OXSM, and MCAT (CH157-MNEV, n = 3; CH157-MNNOTCH3 ICD, n = 3). In B-C, significance was calculated using unpaired Student’s t-tests. P<0.05*; P<0.01**, P<0.001***; P<0.0001****.
NOTCH3 Overexpression Increases Maximal Oxidative Respiration in the Presence of Fatty Acids
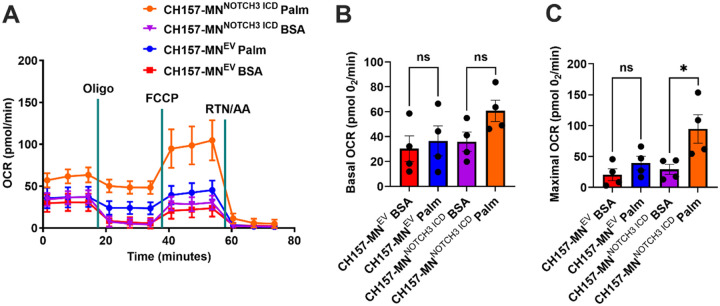
To understand the functional metabolic impact of NOTCH3 expression in meningioma cells, the OCR of CH157-MNNOTCH3 ICD versus CH157-MNEV was compared using the Seahorse extracellular flux assay in the presence of palmitate and BSA palmitate control (Fig. 4A). As in the PDX experiments, the presence of palmitate trended towards an increase in basal OCR only in NOTCH3 overexpressing lines (p = 0.06, Fig. 4B). Unlike the PDX lines, FCCP (FCCP is a mitochondrial uncoupler that increases maximal respiration by collapsing the proton gradient, forcing cells to operate at their maximum respiratory capacity) increased the maximal respiration after the injection (29.3 ± 8.1 pmol O2/min in BSA incubated vs. 94.8 ± 23.0 in the palmitate-treated group; p < 0.05) (Fig. 4C). This data supports the hypothesis that NOTCH3 upregulates FAO in meningioma.
Figure 4. NOTCH3 overexpression increases maximal aerobic respiration in the presence of lipids.
A. Oxygen consumption rate (OCR) of CH157-MNEV and CH157-MNNOTCH3 ICD treated with palmitate-BSA or BSA control (CH157-MNEV, n = 4 in each condition; CH157-MNNOTCH3 ICD, n = 4 in each condition). B. Basal corrected oxygen consumption rate (OCR) in CH157-MNEV and CH157-MNNOTCH3 ICD cells treated with palmitate-BSA or BSA control (CH157-MNEV, n = 4 in each condition; CH157-MNNOTCH3 ICD, n = 4 in each condition). C. Maximal corrected oxygen consumption rate (OCR) in CH157-MNEV and CH157-MNNOTCH3 ICD cells treated with palmitate-BSA or BSA control (CH157-MNEV, n = 4 in each condition; CH157-MNNOTCH3 ICD, n = 4 in each condition). In B-C, a one-way ANOVA was performed with a Tukey’s post-hoc to calculate significance. P<0.05*; P<0.01**, P<0.001***.
NOTCH3 Overexpression Confers Resistance to Ferroptosis
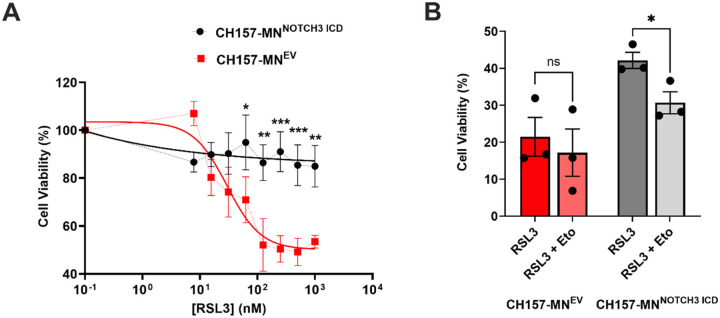
To understand the impact of increased FAO in CH157-MNNOTCH3 ICD cells on ferroptosis, a series of cell death experiments utilizing RSL3, a ferroptosis inducer, was conducted. Utilizing RSL3, which inhibits glutathione peroxidase 4 (GPX4) and induces ferroptosis, the percent viability of CH157-MNNOTCH3 ICD was compared with EV cells. At increasing concentrations of RSL3, CH157-MNNOTCH3 ICD cells exhibit complete resistance to ferroptosis induction when compared with CH157-MNEV (Fig. 5A). Considering the protection from RSL3 observed in the CH157-MNNOTCH3 ICD, and the low levels of intracellular lipids in this line, we hypothesized NOTCH3 protects meningioma cells from ferroptosis via their increased FAO. To test this hypothesis, we treated CH157-MNEV and CH157-MNNOTCH3 ICD cells with 5 μM of etomoxir (a FAO inhibitor) co-incubated with 125 nM RSL3 for 24 hours. While the addition of etomoxir did not potentiate the cell death of CH157-MNEV, there was a significant decrease in viability in the CH157MN-NOTCH3 ICD (42.2 ± 2.2% in RSL3 compared to 30.7 ± 3.0% in RSL3 with Eto; p < 0.05), demonstrating that enhanced FAO is protective against ferroptosis in meningioma cells.
Figure 5. NOTCH3-driven lipid depletion confers resistance to ferroptosis.
A. Cell viability of CH157-MNEV and CH157-MNNOTCH3 ICD treated with increasing concentration of RSL3 for 24 hours (CH157-MNEV, n = 5; CH157-MNNOTCH3 ICD, n = 5). To test for significance in curves, a two-way ANOVA was performed, and Sidak’s multiple comparison post hoc test was used to test for individual significance across rows. B. Cell viability of CH157-MNEV and CH157-MNNOTCH3 ICD treated with or without 5 μM of etomoxir with 32.3 nM RSL3 (CH157-MNEV, n = 3 in each condition; CH157-MNNOTCH3 ICD, n = 3 in each condition). A one-way ANOVA was performed with a Tukey’s post-hoc to calculate significance. P<0.05*; P<0.01**, P<0.001***.
Discussion
In this study, we demonstrate that NOTCH3 promotes a metabolic shift towards FAO, lipid depletion, and resistance to ferroptosis in meningioma cells. In previous studies, NOTCH3 expression has been associated with higher grade, radiation resistance, and recurrence in meningioma [15]. However, the metabolic role of NOTCH3 in meningiomas has not been established. In multiple tumor types, alterations in lipid metabolism have contributed to treatment resistance and malignancy [30–32]. This study is the first to identify an association between NOTCH3 expression, FAO regulation, and ferroptosis resistance in meningioma cells.
Through scRNA-seq, we demonstrate that NOTCH3 + human meningioma cells exhibit enriched expression of CD36. CD36 is a membrane-bound fatty acid transporter that facilitates the uptake of long-chain fatty acids (LCFAs), directing them toward FAO [33, 34]. CD36-driven FAO enhances tumor cell survival in stress-induced environments by providing an alternative energy source, reducing reactive oxygen species (ROS)-mediated cell death, and maintaining cancer stem cell (CSC) populations, contributing to chemotherapy, radiotherapy, and immunotherapy resistance [35–37]. CD36 has been shown to facilitate evasion of therapy-induced metabolic stress in tumor cells [38, 39]. We established NOTCH3 high and low patient-derived primary meningioma lines and validated NOTCH3 correlates with CD36 expression. By comparing OCR between the NOTCH3high PDX2 and NOTCH3low PDX1 primary lines, we show an increase in maximal mitochondrial respiration in the presence of palmitate in NOTCH3high primary cells, suggesting a functional upregulation of lipid metabolism in NOTCH3-expressing meningioma cells.
Utilizing the CH157-MN meningioma model, we demonstrate a shift in lipid metabolism with NOTCH3 ICD overexpression. Untargeted metabolomic profiling of CH157-MNNOTCH3 ICD revealed decreased fatty acyl carnitine species when compared with CH157-MNEV controls. Upon bulk RNA sequencing, CH157-MNNOTCH3 ICD cells exhibit upregulation of canonical FAO and simultaneous downregulation of FAS genes, confirming enhancement of lipid metabolism with NOTCH3 overexpression [40–42]. Complementary targeted fatty acid analyses revealed depletion of intracellular fatty acids in CH157-MNNOTCH3 ICD cells. Functionally, extracellular flux assays demonstrate elevated maximal mitochondrial respiration in the presence of palmitate, confirming increased mitochondrial oxidative capacity in CH157-MNNOTCH3 ICD cells.
Lipid bioavailability is critical to ferroptosis, a non-apoptotic form of cell death caused by toxic iron accumulation and lipid peroxidation [43–45] Through the generation of reactive oxygen species (ROS), ferroptosis acts as a tumor suppressor mechanism [46]. This study demonstrates reduced sensitivity to RSL3, a ferroptosis inducer, in CH157-MNNOTCH3 ICD cells. Co-treatment with etomoxir, an inhibitor of CPT1-dependent mitochondrial fatty acid import, restored RSL3-induced ferroptosis in CH157-MNNOTCH3 ICD cells. In a similar study, investigators report NOTCH3 expression negatively regulates ROS-mediated lipid peroxidation in non-small cell lung carcinoma (NSCLC), increasing tumorigenesis [47]. These results suggest that NOTCH3-driven lipid depletion protects cells from ferroptosis, which may confer a survival advantage in meningioma.
These findings suggest that NOTCH3 is a central mediator of fatty acid metabolism in meningiomas. By upregulating FAO pathways, NOTCH3-expressing meningioma cells may enhance ATP generation while reducing susceptibility to ferroptototic death processes. The combined benefit of improved fatty acid utilization and cell survival may confer an advantage under conditions of metabolic or oxidative stress, particularly in the context of radiation. These data serve as one potential mechanism of why NOTCH3 expression portends a poor prognosis as NOTCH3 + meningiomas are higher grade, more likely to recur, and exhibit resistance to radiation therapy [15]. FAO, with adjuvant CPT1a inhibitors, may restore ferroptosis vulnerability in NOTCH3 expressing meningiomas. This approach could be efficacious when combined with ferroptosis inducers or radiation therapy, both of which rely on oxidative stress as a mechanism of cytotoxicity.
This study has several limitations. All experiments were performed in an in vitro setting using primary and established meningioma cell lines. While this study focuses on mechanistic investigation, further in vivo studies are needed to assess the role of NOTCH3 on meningioma metabolism. Additionally, this study did not assess clinical outcomes patient samples stratified by NOTCH3 expression as this was outside the scope of this investigation. Future studies should incorporate transcriptomic and metabolomic profiling of patient-derived tumors stratified by WHO grade to validate the clinical implication of our findings.
Conclusions
These findings highlight the role of NOTCH3 in mediating FAO in meningiomas. Increased utilization of lipids in NOTCH3 expressing meningiomas poses a metabolic advantage was well as contributes to resistance to ferroptosis, leading increased cell survival in the setting of oxidative stress. These data highlight a potential mechanism for increased malignant potential of NOTCH3 + meningiomas.
Key Points.
NOTCH3 expression in meningioma upregulates FAO.
Enhanced FAO contributes to ferroptosis resistance in NOTCH3-overexpressing (OE) meningioma cells.
Importance of the study.
This study investigates the metabolic reprogramming by NOTCH3 in meningiomas, identifying increased FAO as a key adaptation that contributes to ferroptosis resistance. Given the limited treatment options for aggressive or recurrent meningiomas, uncovering metabolic vulnerabilities could offer new opportunities for therapeutic intervention. Our findings suggest that NOTCH3-expressing meningiomas upregulate FAO for survival, highlighting a previously unrecognized aspect of meningioma metabolism.
ACKNOWLEDGEMENTS
We would like to thank the Research-Intensive Scholarly Emphasis (RISE) program at Northwestern University for financial support of N.S.S and the Metabolomics Core Facility of the Robert H. Lurie Comprehensive Cancer Center supported by NCI CCSG P30 CA060553 of Northwestern University.
Funding
This study was supported by National Institutes of Health grants CA279686, CA262311, CA118816, and P50CA221747.
Funding Statement
This study was supported by National Institutes of Health grants CA279686, CA262311, CA118816, and P50CA221747.
Footnotes
Ethics
Human PDX samples were obtained under protocol #STU00095863 approved by the Northwestern Institutional Review Board (IRB).
Conflict of interest
The authors declare no conflicts of interest
Contributor Information
Nishanth S. Sadagopan, Northwestern University Feinberg School of Medicine
Mateo Gomez, Northwestern University Feinberg School of Medicine.
Shashwat Tripathi, Northwestern University Feinberg School of Medicine.
Leah K. Billingham, Northwestern University Feinberg School of Medicine
Susan L. DeLay, Northwestern University Feinberg School of Medicine
Martha A. Cady, University of California
Harrshavasan T. S. Congivaram, Northwestern University Feinberg School of Medicine
Tzu-yi Chia, Northwestern University Feinberg School of Medicine.
Hanxiao Wan, Northwestern University Feinberg School of Medicine.
Si Wang, Northwestern University Feinberg School of Medicine.
David R. Raleigh, University of California
Faith C. Kaluba, Van Andel Institute
Evan C. Lien, Van Andel Institute
Amy B. Heimberger, Northwestern University Feinberg School of Medicine
Catalina Lee-Chang, Northwestern University Feinberg School of Medicine.
Mark W. Youngblood, Northwestern University Feinberg School of Medicine
Stephen T. Magill, Northwestern University Feinberg School of Medicine
Jason M. Miska, Northwestern University Feinberg School of Medicine
Data availability
Any data is available at the request of the corresponding author, Jason Miska (Jason.miska@northwestern.edu). RNA-sequencing data can be accessed via BioProject accession number PRJNA1124134 (https://www.ncbi.nlm.nih.gov/bioproject/1124134).
References
- 1.Anastasi E, Campese AF, Bellavia D, Bulotta A, Balestri A, Pascucci M, et al. Expression of activated Notch3 in transgenic mice enhances generation of T regulatory cells and protects against experimental autoimmune diabetes. Journal Immunol. 2003;171(9):4504–11. doi: 10.4049/jimmunol.171.9.4504. [DOI] [PubMed] [Google Scholar]
- 2.Lardelli M, Dahlstrand J, Lendahl U. The novel Notch homologue mouse Notch 3 lacks specific epidermal growth factor-repeats and is expressed in proliferating neuroepithelium. Mech Dev. 1994;46(2):123–36. doi: 10.1016/0925-4773(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 3.Joutel A, Andreux F, Gaulis S, Domenga V, Cecillon M, Battail N, et al. The ectodomain of the Notch3 receptor accumulates within the cerebrovasculature of CADASIL patients. J Clin Invest. 2000;105(5):597–605. doi: 10.1172/JCI8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamada Y, Kadokawa Y, Okabe M, Ikawa M, Coleman JR, Tsujimoto Y. Mutation in ankyrin repeats of the mouse Notch2 gene induces early embryonic lethality. Development. 1999;126(15):3415–24. doi: 10.1242/dev.126.15.3415. [DOI] [PubMed] [Google Scholar]
- 5.Krebs LT, Xue Y, Norton CR, Sundberg JP, Beatus P, Lendahl U, et al. Characterization of Notch3-deficient mice: normal embryonic development and absence of genetic interactions with a Notch1 mutation. Genesis. 2003;37(3):139–43. doi: 10.1002/gene.10241. [DOI] [PubMed] [Google Scholar]
- 6.Lee SF, Srinivasan B, Sephton CF, Dries DR, Wang B, Yu C, et al. Gamma-secretase-regulated proteolysis of the Notch receptor by mitochondrial intermediate peptidase. The Journal of biological chemistry. 2011;286(31):27447–53. Epub 20110617. doi: 10.1074/jbc.M111.243154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baron M. Combining genetic and biophysical approaches to probe the structure and function relationships of the notch receptor. Mol Membr Biol. 2017;34(1–2):33–49. Epub 20180924. doi: 10.1080/09687688.2018.1503742. [DOI] [PubMed] [Google Scholar]
- 8.Bray SJ. Notch signalling in context. Nat Rev Mol Cell Biol. 2016;17(11):722–35. Epub 20160810. doi: 10.1038/nrm.2016.94. [DOI] [PubMed] [Google Scholar]
- 9.Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383(6602):707–10. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- 10.Bellavia D, Campese AF, Alesse E, Vacca A, Felli MP, Balestri A, et al. Constitutive activation of NF-kappaB and T-cell leukemia/lymphoma in Notch3 transgenic mice. EMBO J. 2000;19(13):3337–48. doi: 10.1093/emboj/19.13.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mezquita B, Mezquita J, Barrot C, Carvajal S, Pau M, Mezquita P, et al. A truncated-Flt1 isoform of breast cancer cells is upregulated by Notch and downregulated by retinoic acid. J Cell Biochem. 2014;115(1):52–61. doi: 10.1002/jcb.24632. [DOI] [PubMed] [Google Scholar]
- 12.Zhou JX, Zhou L, Li QJ, Feng W, Wang PM, Li EF, et al. Association between high levels of Notch3 expression and high invasion and poor overall survival rates in pancreatic ductal adenocarcinoma. Oncol Rep. 2016;36(5):2893–901. Epub 20160909. doi: 10.3892/or.2016.5079. [DOI] [PubMed] [Google Scholar]
- 13.Alqudah MA, Agarwal S, Al-Keilani MS, Sibenaller ZA, Ryken TC, Assem M. NOTCH3 is a prognostic factor that promotes glioma cell proliferation, migration and invasion via activation of CCND1 and EGFR. Plos One. 2013;8(10):e77299. Epub 20131015. doi: 10.1371/journal.pone.0077299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Su LP, Ji M, Liu L, Sang W, Xue J, Wang B, et al. The expression of ASAP3 and NOTCH3 and the clinicopathological characteristics of adult glioma patients. Open Med (Wars). 2022;17(1):1724–41. Epub 20221031. doi: 10.1515/med-2022-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Choudhury A, Cady MA, Lucas CG, Najem H, Phillips JJ, Palikuqi B, et al. Perivascular NOTCH3+ Stem Cells Drive Meningioma Tumorigenesis and Resistance to Radiotherapy. Cancer Discov. 2024;14(10):1823–37. doi: 10.1158/2159-8290.CD-23-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fiehn O. Metabolomics by Gas Chromatography-Mass Spectrometry: Combined Targeted and Untargeted Profiling. Curr Protoc Mol Biol. 2016;114:30 4 1–4 2. Epub 20160401. doi: 10.1002/0471142727.mb3004s114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang AZ, Bowman-Kirigin JA, Desai R, Kang LI, Patel PR, Patel B, et al. Single-cell profiling of human dura and meningioma reveals cellular meningeal landscape and insights into meningioma immune response. Genome Med. 2022;14(1):49. Epub 20220510. doi: 10.1186/s13073-022-01051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choudhury A, Magill ST, Eaton CD, Prager BC, Chen WC, Cady MA, et al. Meningioma DNA methylation groups identify biological drivers and therapeutic vulnerabilities. Nat Genet. 2022;54(5):649–59. Epub 20220509. doi: 10.1038/s41588-022-01061-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–20. Epub 20180402. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nature Methods. 2019;16(12):1289–96. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muller S, Cho A, Liu SJ, Lim DA, Diaz A. CONICS integrates scRNA-seq with DNA sequencing to map gene expression to tumor sub-clones. Bioinformatics. 2018;34(18):3217–9. doi: 10.1093/bioinformatics/bty316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sokol KH, Lee CJ, Rogers TJ, Waldhart A, Ellis AE, Madireddy S, et al. Lipid availability influences ferroptosis sensitivity in cancer cells by regulating polyunsaturated fatty acid trafficking. Cell Chem Biol. 2025;32(3):408–22 e6. Epub 20241022. doi: 10.1016/j.chembiol.2024.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams KJ, Pratt B, Bose N, Dubois LG, St John-Williams L, Perrott KM, et al. Skyline for Small Molecules: A Unifying Software Package for Quantitative Metabolomics. J Proteome Res. 2020;19(4):1447–58. Epub 20200326. doi: 10.1021/acs.jproteome.9b00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quehenberger O, Armando AM, Dennis EA. High sensitivity quantitative lipidomics analysis of fatty acids in biological samples by gas chromatography-mass spectrometry. Biochim Biophys Acta. 2011;1811(11):648–56. Epub 20110720. doi: 10.1016/j.bbalip.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McGirt MJ, Woodworth GF, Coon AL, Frazier JM, Amundson E, Garonzik I, et al. Independent predictors of morbidity after image-guided stereotactic brain biopsy: a risk assessment of 270 cases. Journal of neurosurgery. 2005;102(5):897–901. doi: 10.3171/jns.2005.102.5.0897. [DOI] [PubMed] [Google Scholar]
- 26.Raud B, Roy DG, Divakaruni AS, Tarasenko TN, Franke R, Ma EH, et al. Etomoxir Actions on Regulatory and Memory T Cells Are Independent of Cpt1a-Mediated Fatty Acid Oxidation. Cell Metab. 2018;28(3):504–15 e7. Epub 20180628. doi: 10.1016/j.cmet.2018.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Connor RS, Guo LL, Ghassemi S, Snyder NW, Worth AJ, Weng L, et al. The CPT1a inhibitor, etomoxir induces severe oxidative stress at commonly used concentrations. Scientific reports. 2018;8. doi: ARTN 6289 10.1038/s41598-018-24676-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caines JK, Barnes DA, Berry MD. The Use of Seahorse XF Assays to Interrogate Real-Time Energy Metabolism in Cancer Cell Lines. Methods Mol Biol. 2022;2508:225–34. doi: 10.1007/978-1-0716-2376-3_17. [DOI] [PubMed] [Google Scholar]
- 29.Divakaruni AS, Jastroch M. A practical guide for the analysis, standardization and interpretation of oxygen consumption measurements. Nat Metab. 2022;4(8):978–94. Epub 20220815. doi: 10.1038/s42255-022-00619-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin-Perez M, Urdiroz-Urricelqui U, Bigas C, Benitah SA. The role of lipids in cancer progression and metastasis. Cell Metab. 2022;34(11):1675–99. Epub 20221018. doi: 10.1016/j.cmet.2022.09.023. [DOI] [PubMed] [Google Scholar]
- 31.Irshad R, Tabassum S, Husain M. Aberrant Lipid Metabolism in Cancer: Current Status and Emerging Therapeutic Perspectives. Curr Top Med Chem. 2023;23(12):1090–103. doi: 10.2174/1568026623666230522103321. [DOI] [PubMed] [Google Scholar]
- 32.Vishwa R, BharathwajChetty B, Girisa S, Aswani BS, Alqahtani MS, Abbas M, et al. Lipid metabolism and its implications in tumor cell plasticity and drug resistance: what we learned thus far? Cancer Metastasis Rev. 2024;43(1):293–319. Epub 20240305. doi: 10.1007/s10555-024-10170-1. [DOI] [PubMed] [Google Scholar]
- 33.Pepino MY, Kuda O, Samovski D, Abumrad NA. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu Rev Nutr. 2014;34:281–303. Epub 20140516. doi: 10.1146/annurev-nutr-071812-161220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y, Zhang J, Cui W, Silverstein RL. CD36, a signaling receptor and fatty acid transporter that regulates immune cell metabolism and fate. The Journal of experimental medicine. 2022;219(6). Epub 20220419. doi: 10.1084/jem.20211314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang T, Fahrmann JF, Lee H, Li YJ, Tripathi SC, Yue C, et al. JAK/STAT3-Regulated Fatty Acid beta-Oxidation Is Critical for Breast Cancer Stem Cell Self-Renewal and Chemoresistance. Cell Metab. 2018;27(1):136–50 e5. Epub 20171214. doi: 10.1016/j.cmet.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma Y, Temkin SM, Hawkridge AM, Guo C, Wang W, Wang XY, et al. Fatty acid oxidation: An emerging facet of metabolic transformation in cancer. Cancer Lett. 2018;435:92–100. Epub 20180810. doi: 10.1016/j.canlet.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Choi HJ, Jhe YL, Kim J, Lim JY, Lee JE, Shin MK, et al. FoxM1-dependent and fatty acid oxidation-mediated ROS modulation is a cell-intrinsic drug resistance mechanism in cancer stem-like cells. Redox Biol. 2020;36:101589. Epub 20200529. doi: 10.1016/j.redox.2020.101589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Feng WW, Wilkins O, Bang S, Ung M, Li J, An J, et al. CD36-Mediated Metabolic Rewiring of Breast Cancer Cells Promotes Resistance to HER2-Targeted Therapies. Cell Rep. 2019;29(11):3405–20 e5. doi: 10.1016/j.celrep.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruan C, Meng Y, Song H. CD36: an emerging therapeutic target for cancer and its molecular mechanisms. J Cancer Res Clin Oncol. 2022;148(7):1551–8. Epub 20220227. doi: 10.1007/s00432-022-03957-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wedan RJ, Longenecker JZ, Nowinski SM. Mitochondrial fatty acid synthesis is an emergent central regulator of mammalian oxidative metabolism. Cell Metab. 2024;36(1):36–47. Epub 20231220. doi: 10.1016/j.cmet.2023.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao Y, Yang Y, Xiong H, Dong G. The implications of FASN in immune cell biology and related diseases. Cell Death Dis. 2024;15(1):88. Epub 20240125. doi: 10.1038/s41419-024-06463-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin C, Gong S, Liang T, Zhang Z, Thomas J, Deng J, et al. HADHA Regulates Respiratory Complex Assembly and Couples FAO and OXPHOS. Adv Sci (Weinh). 2024;11(47):e2405147. Epub 20241103. doi: 10.1002/advs.202405147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: an iron-dependent form of nonapoptotic cell death. Cell. 2012;149(5):1060–72. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang D, Minikes AM, Jiang X. Ferroptosis at the intersection of lipid metabolism and cellular signaling. Mol Cell. 2022;82(12):2215–27. Epub 20220406. doi: 10.1016/j.molcel.2022.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dixon SJ, Olzmann JA. The cell biology of ferroptosis. Nat Rev Mol Cell Biol. 2024;25(6):424–42. Epub 20240216. doi: 10.1038/s41580-024-00703-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Q, Meng Y, Li D, Yao L, Le J, Liu Y, et al. Ferroptosis in cancer: From molecular mechanisms to therapeutic strategies. Signal Transduct Target Ther. 2024;9(1):55. Epub 20240308. doi: 10.1038/s41392-024-01769-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Z, Xiao J, Liu M, Cui J, Lian B, Sun Y, et al. Notch3 regulates ferroptosis via ROS-induced lipid peroxidation in NSCLC cells. FEBS Open Bio. 2022;12(6):1197–205. Epub 20220318. doi: 10.1002/2211-5463.13393. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Any data is available at the request of the corresponding author, Jason Miska (Jason.miska@northwestern.edu). RNA-sequencing data can be accessed via BioProject accession number PRJNA1124134 (https://www.ncbi.nlm.nih.gov/bioproject/1124134).