Abstract
Background
Loss of heterozygosity (LOH) in the ING family members has been shown in head and neck squamous cell carcinoma (HNSCC) except for ING2. Like all the other members of ING family, ING2, which is located at chromosome 4q35.1, is a promising tumor suppressor gene (TSG). In this study, we performed LOH analysis of ING2 in HNSCC and compared it with clinicopathological variables.
Materials and methods
We performed LOH analysis in DNAs from 80 paired of normal and HNSCC tissues, using a specifically designed microsatellite marker on chromosome 4q35.1, which detects allelic loss of ING2. TP53 mutation analysis and its relationship with ING2 chromosomal deletion were also performed in available 68 of the samples. The correlation between LOH status and clinicopathological characteristics was evaluated by using statistical methods. The overall survival (OS) and disease free survival (DFS) were also determined.
Results
LOH was detected in 54.6% (30/55) of the informative samples. Statistical significance was obtained between LOH and tumor (T) stage (P = 0.02), application of radiotherapy and chemotherapy. Positive node status (N) appeared to be the only independent prognostic factor for both OS (P = 0.031) and DFS (P = 0.044).
Conclusions
Our study showed allelic loss of 4q35.1 in HNSCC. The high percentage of LOH suggests ING2 as a candidate TSG in HNSCC. High LOH frequency was statistically associated with advanced T stage, suggesting that ING2 LOH might occur in late stages during HNSCC progression.
Keywords: ING2, ING family, Chromosome 4q35.1, Head and neck cancer, LOH, Tumor suppressor gene
Introduction
Tumor suppressor genes (TSG) are defined as genetic elements whose loss or mutational inactivation allows cells to acquire neoplastic growth (Hinds and Weinberg 1994). One of the critical steps in identifying TSG is loss of heterozygosity (LOH) analysis. By this method, well-known TSG such as TP53 and RB were localized and further cloned (Lee et al. 1987; Baker et al. 1989). Previously, we identified several members of the ING family as candidate TSG in oral as well as head and neck cancers (Gunduz et al. 2000, 2002, 2005, 2008a; Cengiz et al. 2007).
Head and neck squamous cell carcinoma (HNSCC) is a heterogeneous disease with complex molecular abnormalities, associated with genetic alterations in either TSG or oncogenes (Perez-Ordoñez et al. 2006). LOH of 9p21 is the most common of all genetic changes occurring in the early progression of these tumors (van der Riet et al. 1994). Moreover, our studies exhibited LOH in several other regions in HNSCC (Gunduz et al. 2000, 2002, 2005, 2008a; Shinno et al. 2005; Beder et al. 2006; Cengiz et al. 2007).
Several studies compared the relationship of allelic loss at various chromosomal locations and inactivation of TSG with clinicopathological data (Mao et al. 1996; Roz et al. 1996; Lydiatt et al. 1998; Cabelguenne et al. 2000; Tannapfel and Weber 2001; Le and Giaccia 2003). These studies focused on finding a reliable and easily applicable marker, which can diagnose cancer early during carcinogenic stage. In such study, LOH at 9p21 and 3p14 was identified in 19 (51%) of 37 patients with oral premalignant lesions. Of these 19 patients, 7 (37%) developed HNSCC during follow-up (Mao et al. 1996). High rate of LOH at chromosome 3p was reported as an early event in oral carcinogenesis (Roz et al. 1996). Other studies focused on molecular analysis, which could give information on the prognosis and behavior of tumor such as metastatic capacity, recurrence, response to therapy, and survival of the patient (Lydiatt et al. 1998; Cabelguenne et al. 2000).
The ING family proteins are tumor suppressors containing a plant homeodomain (PHD) finger, a motif common to many chromatin-regulatory proteins. Five members of the ING family have been identified in humans, ING1 to ING5. They function in cooperation with p53 to induce cell growth arrest and apoptosis (Garkavtsev et al. 1998; Nagashima et al. 2001; Gunduz et al. 2008a). On the other hand, ING proteins were also suggested to function in a p53-independent manner (Cheung et al. 2000; Nagashima et al. 2001; Wang and Li 2006; Coles et al. 2007).
Interestingly, recent array-based comparative genomic hybridization (array-CGH) analyses have identified homozygous deletions at 4q35 loci in oral squamous cell carcinoma (OSCC) cell lines and primary tumors (Nakaya et al. 2007; Nakamura et al. 2008), suggesting the presence of TSG residing at this location. Indeed, a member of the ING family, ING2, cloned by Shimada et al. (1998), is located on chromosome 4q35 (Nagashima et al. 2001). Like other members of the ING family, ING2 is known to function in cooperation with p53 (Wang et al. 2006). It has important functions in senescence (Pedeux et al. 2005), cellular response to DNA damage (Nagashima et al. 2001), and DNA repair (Wang et al. 2006). In addition, ING2 was shown to interact with members of the transforming growth factor (TGF)-β signaling pathways enhancing transcription of target genes and cell cycle arrest (Sarker et al. 2008). In another recent study, decreased expression of ING2 mRNA and protein was observed in hepatocellular carcinoma (HCC) (Zhang et al. 2008). However, there is only one study regarding LOH at 4q35, reporting >20% of deletion in sporadic basal cell carcinomas (BCC) (Sironi et al. 2004). Our previous studies showed significant LOH frequency in various regions including other ING family members in HNSCC (Gunduz et al. 2000, 2002, 2005; Cengiz et al. 2007). Therefore, we sought to perform LOH analysis at chromosome 4q35.1 in HNSCC, and to examine its relationship with TP53 mutation status, clinicopathological characteristics and survival analysis.
Materials and methods
Patients and samples
Paired normal and tumor samples were obtained from 80 patients at the Department of Otolaryngology, Okayama University Hospital. Patients included 66 men and 14 women with a mean age of 65.4 years. The histopathological diagnosis of all cases was squamous cell carcinoma. Bioethics committee of the institution approved the study and informed consents were obtained from the patients. Clinicopathological characteristics of the patients are shown in Table 1.
Table 1.
Clinicopathological characteristics of patients
| Criteria | Number (%) |
|---|---|
| Tumor site | |
| Oral cavity | 37 (46.2) |
| Oropharynx | 10 (12.5) |
| Larynx | 16 (20.0) |
| Hypopharynx | 11 (13.8) |
| Maxilla | 6 (7.5) |
| Age (mean ± SD years) | 65.4 ± 8.3 |
| Gender | |
| Male | 66 (82.5) |
| Female | 14 (17.5) |
| Tumor statusa | |
| T1 | 8 (10.1) |
| T2 | 22 (27.8) |
| T3 | 22 (27.8) |
| T4 | 27 (34.3) |
| Node statusa | |
| N0 | 37 (46.8) |
| N1 | 12 (15.2) |
| N2≤ | 30 (38.0) |
| TNM stagea | |
| I | 6 (7.6) |
| II | 13 (16.5) |
| III | 17 (21.5) |
| IV | 43 (54.4) |
| Recurrence | |
| None | 30 (40.5) |
| Locoregional and/or distant | 44 (59.5) |
| Differentiationa | |
| Well | 24 (31.6) |
| Moderate | 37 (48.7) |
| Poor | 15 (19.7) |
aThe cases with unknown situation for T stage (1), Node status (1), TNM status (1), recurrence (6) and differentiation status (4) were not included
DNA extraction
Genomic DNAs were isolated from frozen tissues by SDS/proteinase K treatment, phenol–chloroform extraction and ethanol precipitation as previously described (Gunduz et al. 2000, 2002).
Microsatellite analysis
A microsatellite marker named ING2MS forward (5′-TGG ATG TAG GCT CTG GAT AG) and reverse (5′-CTC TCT CCT GTG GTT GAA AG) with an expected polymerase chain reaction (PCR) product size of 225 bp was used. The mapping information and sequences were obtained from the recent genome information (http://www.ncbi.nlm.nih.gov/genome/guide/human). The heterozygosity and number of the tandem nucleotide repeats for the design of ING2MS were acquired from the information site (http://www.gramene.org/db/searches/ssrtool). The primer was designed based on the contiguous genomic sequence (NW_001838921) using GENETYX-MAC 10.1 software (Software Development Co., Ltd, Tokyo, Japan). The sequence of the primer was located at 95 kbp centromere side of ING2 locus. PCR was carried out in 20 μl of reaction mixture with 20 pmol of each primer, 100 ng of genomic DNA, 1× PCR buffer, 200 μM of each deoxynucleotide triphosphate, and 0.5 unit of Taq DNA polymerase (Takara, Kyoto, Japan). Initial denaturation at 94°C for 3 min was followed by 25 cycles of a denaturation step at 94°C for 30 s, an annealing step at 58°C for 30 s, and an extension step at 72°C for 1 min. A final extension step at 72°C for 7 min was also added. After amplification, electrophoresis through an 8% polyacrylamide gel and visualization of DNA bands by silver staining was carried out as described previously (Gunduz et al. 2000, 2002; Cengiz et al. 2007). LOH was scored if one of the heterozygous alleles showed at least 50% reduced intensity in tumor DNA as compared with the corresponding normal DNA as previously described (Gunduz et al. 2000; 2002). This decision was made by direct visualization in the cases with clear deletion as compared to normal allele. If the reduction density of the band was borderline or doubtful, we quantified each band by using computer-based software (Quantity One, Toyobo, Japan). Total LOH frequency was obtained by dividing the number of LOH cases with the total number of informative cases.
Mutation analysis of TP53
Each of the coding regions of exon 4–9 of TP53 gene was amplified by PCR with intron spanning primers designed by using GENETYX-MAC 10.1 software (Software Development Co., Ltd, Tokyo, Japan) as previously described (Gunduz et al. 2005). PCR amplification of genomic DNAs and subsequent direct sequencing were performed as previously described (Gunduz et al. 2005).
Statistical analysis
Pearson’s chi-square test, Fisher’s exact test and Student’s t tests were used to evaluate the correlation between ING2 LOH status and clinicopathological characteristics. Survival curves were calculated according to Kaplan–Meier. For comparison of survival between LOH and retention of ING2, the log-rank test was employed. Overall survival (OS) in months was calculated from the day after surgery to the last follow-up examination or death of the patient. The duration of disease-free survival (DFS) was determined from the day after surgery to the initial recurrence of the cancer. For multivariate analyses, we used the Cox proportional hazards model. All computations were done using the SPSS version 10 for the Windows software system (SPSS, Inc., Chicago, IL) and statistical significance of P < 0.05 was considered.
Results
LOH analysis and mapping of chromosome 4q35.1
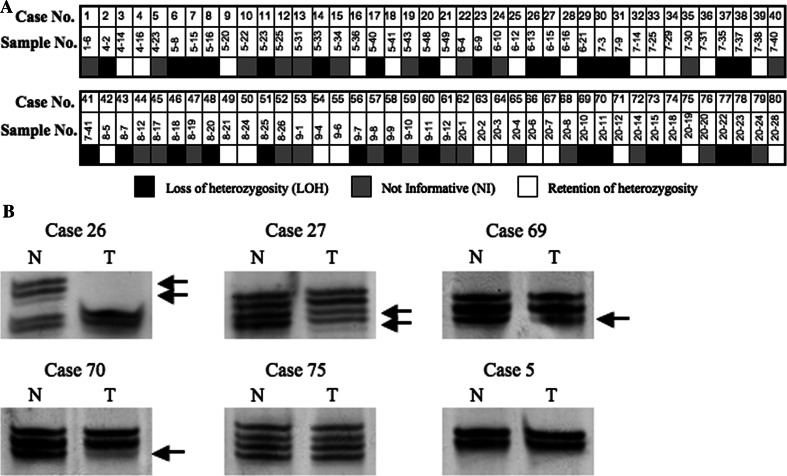
LOH was detected in 30 (54.6%) out of 55 informative cases as summarized in Fig. 1a. Representative examples of LOH are shown in Fig. 1b.
Fig. 1.
LOH analysis on chromosme 4q35.1 in HNSCC. a Schematic representation of LOH distribution. Filled box LOH, open box retention of heterozygosity, shaded box not informative (homozygous). b Representative results of microsatellite analysis. DNAs of tumor (T) and corresponding normal (N) tissues are shown with case numbers on the top. Arrows depict lost alleles in the samples with LOH. Case 75 shows retention of heterozygosity. Case 5 is not informative (homozygous)
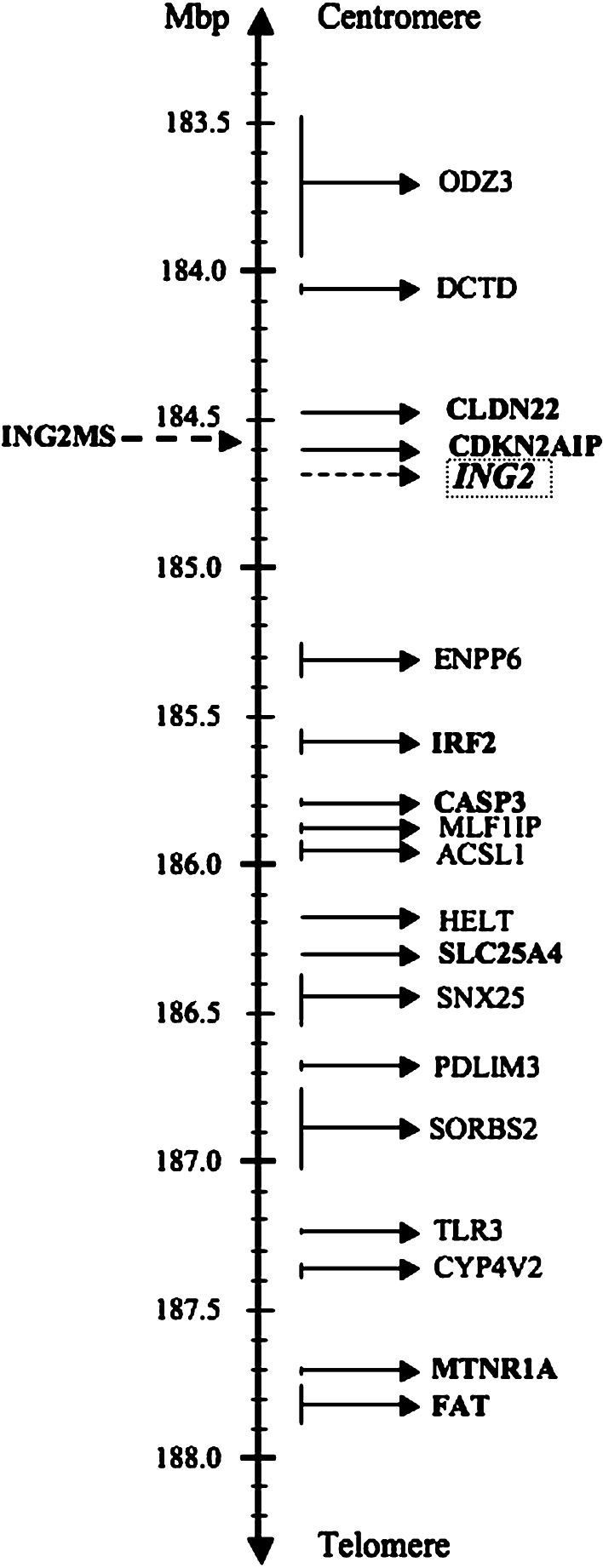
We redefined the mapping of chromosome 4q35.1 by searching the genome database and using contiguous sequences for the locations of our marker and genes (http://www.ncbi.nlm.nih.gov/genome/guide/human/ and http://www.gdb.org/) (Fig. 2).
Fig. 2.
A physical map of the ING2MS location. The location of the marker and genes are based upon the latest mapping information derived from the National Center for Biotechnology Information (NCBI) and the Genome Database (GDB) homepages (http://www.ncbi.nlm.nih.gov/genome/guide/human/, http://www.gdb.org/)
Mutation analysis of TP53 gene
Mutation analysis of TP53 gene was performed in genomic DNAs from 68 patients with HNSCC by PCR-direct sequencing. TP53 mutations were identified in 26 (38.2%) out of 68 cases. The mutations observed were 15 missense, 4 nonsense, 1 both nonsense and missense mutation and 6 frameshift mutations (Table 2).
Table 2.
TP53 mutation status
| Cases | Codon change | Aminoacid change | Exon | Cases | Codon change | Aminoacid change | Exon |
|---|---|---|---|---|---|---|---|
| 1 | (−) | 41 | (−) | ||||
| 2 | 306.CGA/TGA | Arg/Early stop | 8 | 42 | 155.ACC/GCC, 213.CGA/TGA | Thr/Ala, Arg/Early stop | 5, 6 |
| 3 | (−) | 43 | (−) | ||||
| 4 | 168.-AG-Ins | Early stop | 5 | 44 | 191.CCT/CGT | Pro/Arg | 6 |
| 5 | (−) | 45 | (−) | ||||
| 6 | 193.CAT/CTT | His/Leu | 6 | 46 | (−) | ||
| 7 | (−) | 47 | (−) | ||||
| 8 | (−) | 48 | 175.CGC/CAC | Arg/His | 5 | ||
| 9 | (−) | 49 | (−) | ||||
| 10 | (−) | 50 | (−) | ||||
| 11 | (−) | 51 | (−) | ||||
| 12 | ND | 52 | 237.ATG/ATA | Met/Ile | 7 | ||
| 13 | (−) | 53 | (−) | ||||
| 14 | Del | Frameshift | 7 | 54 | 220.TAT/TGT | Tyr/Cys | 7 |
| 15 | (−) | 55 | 192.CAG/TAG | Gln/Early stop | 6 | ||
| 16 | (−) | 56 | (−) | ||||
| 17 | 193.CAT/CTT | His/Leu | 6 | 57 | ND | ||
| 18 | (−) | 58 | 151.CCC/CGC | Pro/Arg | 5 | ||
| 19 | (−) | 59 | (−) | ||||
| 20 | 502. 2 bp Ins | Frameshift | 5 | 60 | 244.GGC/GTC | Gly/Val | 7 |
| 21 | (−) | 61 | (−) | ||||
| 22 | 278.CCT/CTT | Pro/Leu | 8 | 62 | (−) | ||
| 23 | (−) | 63 | 193.CAT/AAT | His/Asn | 6 | ||
| 24 | 278.CCT/TCT | Pro/Ser | 8 | 64 | ND | ||
| 25 | (−) | 65 | ND | ||||
| 26 | 220.TAT/TGT | Tyr/Cys | 6 | 66 | ND | ||
| 27 | 245.GGC/AGC | Gly/Ser | 7 | 67 | Del/Ins | Frameshift | 7 |
| 28 | 135.-A-Ins | Early stop | 5 | 68 | 193. 1 bp Del | Frameshift | 6 |
| 29 | 179.CAT/TAT | His/Tyr | 5 | 69 | ND | ||
| 30 | (−) | 70 | ND | ||||
| 31 | 72. 2 bp Del | Frameshift | 4 | 71 | (−) | ||
| 32 | (−) | 72 | (−) | ||||
| 33 | (−) | 73 | (−) | ||||
| 34 | (−) | 74 | ND | ||||
| 35 | (−) | 75 | ND | ||||
| 36 | 173.GTG/ATG | Val/Met | 5 | 76 | ND | ||
| 37 | (−) | 77 | ND | ||||
| 38 | (−) | 78 | (−) | ||||
| 39 | 207 1 bp Del | Frameshift | 7 | 79 | ND | ||
| 40 | (−) | 80 | (−) |
ND not done, Del deletion, Ins insertion
Relationship between ING2 LOH and TP53 mutation status
Table 3 summarizes the ING2 LOH status and TP53 mutation. Out of 26 cases with TP53 mutation, 13 cases (50%) had ING2 LOH. Further, 7/26 (26.9%) cases with mutation showed retention of heterozygosity and 6/26 (23%) were not informative cases. Although high percentages of ING2 LOH and TP53 mutations were observed, no statistical significance was observed when they were compared.
Table 3.
ING2 LOH and TP53 mutation status
| Case | Localization | ING2 LOH | TP53 Mut | Case | Localization | ING2 LOH | TP53 Mut |
|---|---|---|---|---|---|---|---|
| 1 | Oropharynx | LOH | − | 41 | Oral cavity | Retention | − |
| 2 | Oropharynx | LOH | + | 42 | Oral cavity | NI | + |
| 3 | Oral cavity | LOH | − | 43 | Oropharynx | LOH | − |
| 4 | Oropharynx | Retention | + | 44 | Hypopharynx | LOH | + |
| 5 | Larynx | NI | − | 45 | Oropharynx | NI | − |
| 6 | Maxilla | LOH | + | 46 | Hypopharynx | Retention | − |
| 7 | Oral cavity | NI | − | 47 | Oral cavity | Retention | − |
| 8 | Larynx | LOH | − | 48 | Oral cavity | NI | + |
| 9 | Oral cavity | NI | − | 49 | Larynx | Retention | − |
| 10 | Larynx | Retention | − | 50 | Oral cavity | Retention | − |
| 11 | Oral cavity | LOH | − | 51 | Oropharynx | LOH | − |
| 12 | Oral cavity | Retention | ND | 52 | Oral cavity | LOH | + |
| 13 | Maxilla | NI | − | 53 | Oral cavity | Retention | − |
| 14 | Maxilla | LOH | + | 54 | Larynx | NI | + |
| 15 | Oral cavity | NI | − | 55 | Larynx | LOH | + |
| 16 | Maxilla | LOH | − | 56 | Oral cavity | LOH | − |
| 17 | Oral cavity | NI | + | 57 | Hypopharynx | Retention | ND |
| 18 | Oral cavity | Retention | − | 58 | Larynx | NI | + |
| 19 | Maxilla | LOH | − | 59 | Oropharynx | LOH | − |
| 20 | Oral cavity | LOH | + | 60 | Hypopharynx | LOH | + |
| 21 | Oral cavity | Retention | − | 61 | Oral cavity | NI | − |
| 22 | Larynx | LOH | + | 62 | Oral cavity | NI | − |
| 23 | Oral cavity | LOH | − | 63 | Larynx | Retention | + |
| 24 | Oropharynx | Retention | + | 64 | Oral cavity | Retention | ND |
| 25 | Oral cavity | NI | − | 65 | Oral cavity | NI | ND |
| 26 | Oral cavity | Retention | + | 66 | Larynx | NI | ND |
| 27 | Hypopharynx | LOH | + | 67 | Oral cavity | Retention | + |
| 28 | Larynx | LOH | + | 68 | Oral cavity | LOH | + |
| 29 | Larynx | Retention | + | 69 | Oral cavity | Retention | ND |
| 30 | Oral cavity | NI | − | 70 | Hypopharynx | LOH | ND |
| 31 | Hypopharynx | Retention | + | 71 | Oropharynx | NI | − |
| 32 | Hypopharynx | LOH | − | 72 | Oral cavity | NI | − |
| 33 | Hypopharynx | NI | − | 73 | Oral cavity | Retention | − |
| 34 | Hypopharynx | NI | − | 74 | Larynx | LOH | ND |
| 35 | Larynx | NI | − | 75 | Larynx | Retention | ND |
| 36 | Maxilla | LOH | + | 76 | Larynx | LOH | ND |
| 37 | Oral cavity | Retention | − | 77 | Oral cavity | LOH | ND |
| 38 | Oral cavity | Retention | − | 78 | Oral cavity | LOH | − |
| 39 | Hypopharynx | NI | + | 79 | Oral cavity | NI | ND |
| 40 | Oral cavity | Retention | − | 80 | Oropharynx | NI | − |
LOH loss of heterozygosity, NI not informative, ND not done, Mut mutation
Relationship between ING2 LOH status and clinicopathological characteristics
Table 4 summarizes the LOH status and clinicopathological characteristics. Among the clinical predictors, statistical significance was obtained in T stage (P = 0.02), radiation therapy (P = 0.02) and chemotherapy (P = 0.03). High LOH was detected in tumors in the late TNM stage, where 23/29 (79.3%) cases with LOH were at advanced TNM stage. However, no statistical significance was obtained.
Table 4.
Relationship between ING2 LOH status and clinicopathological characteristics
| Predictors | ING2 LOH status | TP53 mutation status | ||||
|---|---|---|---|---|---|---|
| Retention (%) | LOH (%) | P value | Wild (%) | Mutant (%) | P value | |
| n = 25 (45.4) | n = 30 (54.6) | n = 42 (61.8) | n = 26 (38.2) | |||
| Gender | ||||||
| Male | 22 (88.0) | 24 (80.0) | 0.49b | 34 (81.0) | 22 (84.6) | 0.76b |
| Female | 3 (12.0) | 6 (20.0) | 8 (19.0) | 4 (15.4) | ||
| Age | ||||||
| Mean ± SD (years) | 66.0 ± 8.3 | 64.8 ± 8.6 | 0.62d | 64.4 ± 9.4 | 65.4 ± 7.1 | 0.63d |
| Smokinge | ||||||
| Yes | 18 (75.0) | 21 (72.4) | 0.83c | 28 (68.3) | 20 (80.0) | 0.30c |
| No | 6 (25.0) | 8 (27.6) | 13 (31.7) | 5 (20.0) | ||
| Alcohol consumptione | ||||||
| Yes | 14 (63.6) | 14 (51.9) | 0.41c | 21 (53.8) | 14 (60.9) | 0.59c |
| No | 8 (36.4) | 13 (48.1) | 18 (46.2) | 9 (39.1) | ||
| TNM stagea | ||||||
| Early stage (I-II) | 8 (32.0) | 6 (20.7) | 0.34c | 11 (26.2) | 6 (23.1) | 0.77c |
| Late stage (III-IV) | 17 (68.0) | 23 (79.3) | 31 (73.8) | 20 (76.9) | ||
| Tumor stagea | ||||||
| Early T (T1-T2) | 15 (60.0) | 8 (27.6) | 0.02 c | 16 (38.1) | 10 (38.5) | 0.97c |
| Late T (T3-T4) | 10 (40.0) | 21 (72.4) | 26 (61.9) | 16 (61.5) | ||
| Nodal stagea | ||||||
| N(0) | 12 (48.0) | 17 (58.6) | 0.43c | 20 (47.6) | 12 (46.2) | 0.90c |
| N(+) | 13 (52.0) | 12 (41.4) | 22 (52.4) | 14 (53.8) | ||
| TP53 statuse | ||||||
| Wild | 13 (65.0) | 13 (50.0) | 0.31c | – | – | |
| Mutated | 7 (35.0) | 13 (50.0) | – | – | ||
| Differentiatione | ||||||
| Well | 7 (29.2) | 7 (24.1) | 0.68c | 13 (31.0) | 8 (34.8) | 0.75c |
| Moderate-poor | 17 (70.8) | 22 (75.9) | 29 (69.0) | 15 (65.2) | ||
| Radiation therapye | ||||||
| Performed | 7 (30.4) | 19 (63.3) | 0.02 c | 19 (46.3) | 13 (54.2) | 0.54c |
| Not performed | 16 (69.6) | 11 (36.7) | 22 (53.7) | 11 (45.8) | ||
| Chemotheraphye | ||||||
| Performed | 4 (16.7) | 13 (44.8) | 0.03 c | 13 (31.7) | 9 (36.0) | 0.72c |
| Not performed | 20 (83.3) | 16 (55.2) | 28 (68.3) | 16 (64.0) | ||
| Previous cancer historye | ||||||
| Exist | 1 (04.8) | 7 (25.0) | 0.12b | 5 (13.2) | 6 (26.1) | 0.20c |
| Not exist | 20 (95.2) | 21 (75.0) | 33 (86.8) | 17 (73.9) | ||
| Location | ||||||
| Oral cavity | 15 (60.0) | 9 (30.0) | 23 (54.8) | 8 (30.8) | ||
| Oropharynx | 2 (08.0) | 5 (16.67) | 7 (16.7) | 3 (11.5) | ||
| Larynx | 5 (20.0) | 6 (20.0) | 5 (11.9) | 7 (27.0) | ||
| Maxilla | 0 (00.0) | 5 (16.67) | 3 (07.1) | 3 (11.5) | ||
| Hypopharynx | 3 (12.0) | 5 (16.67) | 4 (09.5) | 5 (19.2) | ||
aAccording to the International Union Against Cancer 1997 TNM classification system
bFisher’s exact test
cPearson’s chi-square test
dStudent’s t test
eThe cases with unknown situation for smoking (2), alcohol consumption (6), TP53 mutation status (9 including not done cases), differentiation (2; 3 for TP53), TNM (1), radiation therapy (2; 3 for TP53), chemotherapy (2) and previous cancer history (6; 7 for TP53) were not included for evaluation
ING2 LOH status and survival analysis
All patients were enrolled in a follow-up program with follow-up time of 1–72 months. Valid follow-up data were available for 74/80 (92.5%) cases. Disease relapses (locoregional recurrence and/or distant metastases) occurred in 44 patients (59.5%) and death in 40 patients (54.1%). The median OS was 30 months (1–72 months) and the median DFS was 11 months (1–72 months). The mean duration of DFS and OS were 25.9 and 36.5 months, respectively. Five-year DFS and OS were 30.6 and 40.8% respectively.
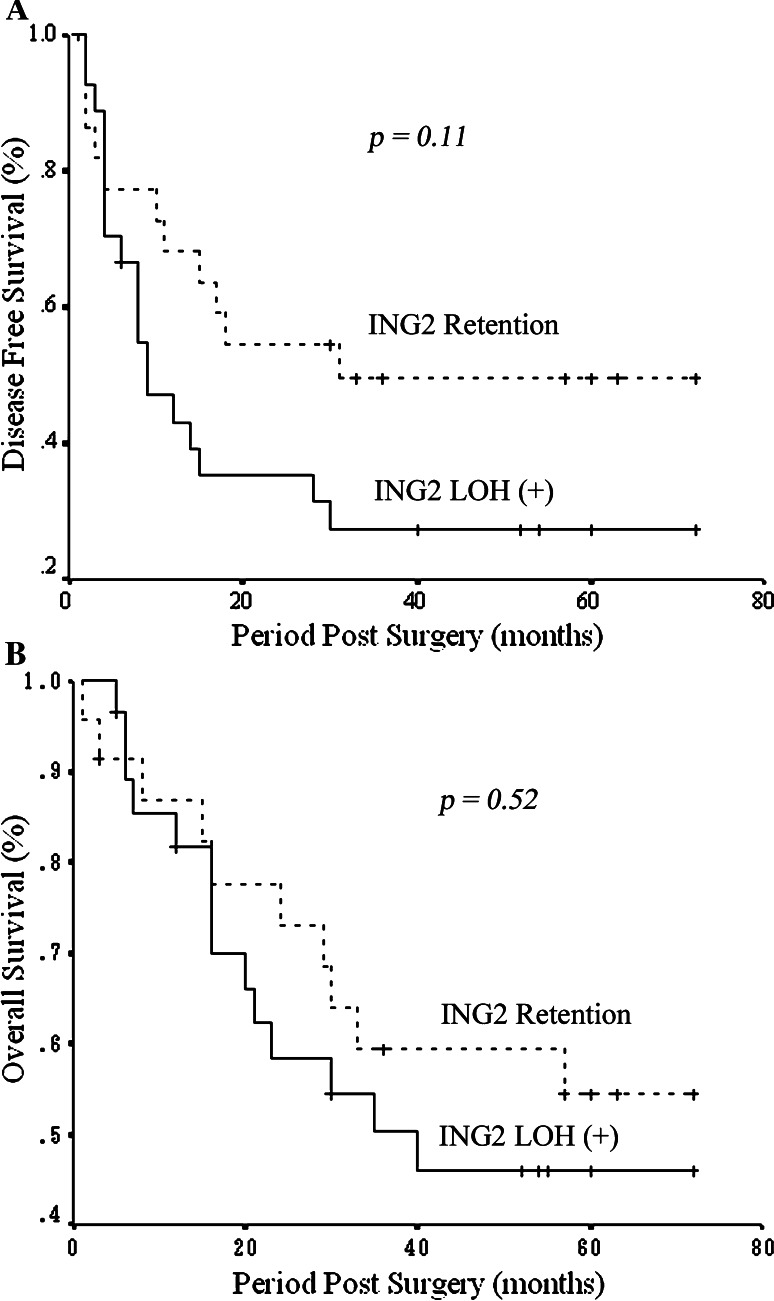
Correlations between ING2 LOH status and the patients’ OS and DFS were analyzed using the univariate Kaplan–Meier method. In a period of 5-year follow up, 54% of patients with retention of ING2 survived. In Fig. 3a, the mean DFS was 27 ± 6 months (95% CI: 15–38 months) in the LOH group and 41 ± 7 months (95% CI: 28–54 months) in the retention group. The log rank test showed that patients with LOH of ING2 had a shorter DFS than the retention group (Fig. 3a). In Fig. 3b, the mean OS was 43 ± 5 months (95% CI: 32–54 months) in the LOH group and 49 ± 6 months (95% CI: 38–61 months) in the retention group (Fig. 3b). Although the number of cases with higher survival rates occurred in the retention group, no significant difference was observed.
Fig. 3.
Overall and disease-free survivals in the groups of HNSCC patients with ING2 LOH and retention of heterozygosity. Kaplan–Meier survival curves for the total number of cases (n = 74) stratified by ING2 LOH. The cases were divided into LOH positive (+) and LOH negative (−) (retention). Statistical significance was defined as P < 0.05. a The mean disease-free survival in the LOH (+) group was 27 ± 6 months (95% CI: 15–38 months) and in the LOH (−) group was 41 ± 7 months (95% CI: 28–54 months) (P = 0.11). b The mean overall survival in the LOH group was 43 ± 5 months (95% CI: 32–54 months) and in the retention group was 49 ± 6 months (95% CI: 38–61 months) (P = 0.52)
A multivariate analysis was used to test the independent value of each parameter predicting OS and DFS (Table 5). Positive node status (N) appeared to be the only independent prognostic factor for both poor DFS and OS (P = 0.044 and P = 0.031, respectively).
Table 5.
Cox proportional hazard model for survival analysis
| Variables | Disease Free Survival | Overall Survival | ||||||
|---|---|---|---|---|---|---|---|---|
| P value | RR | 95.0% CI | P value | RR | 95.0% CI | |||
| Lower | Upper | Lower | Upper | |||||
| Age | 0.392 | 0.786 | 0.453 | 1.364 | 0.911 | 0.963 | 0.502 | 1.850 |
| Smoking | 0.368 | 0.649 | 0.254 | 1.661 | 0.887 | 0.921 | 0.294 | 2.887 |
| T stage | 0.263 | 2.053 | 0.583 | 7.229 | 0.163 | 2.473 | 0.692 | 8.837 |
| N stage | 0.044 | 2.946 | 1.030 | 8.432 | 0.031 | 3.873 | 1.132 | 13.249 |
| TNM stage | 0.084 | 0.525 | 0.253 | 1.092 | 0.914 | 0.954 | 0.404 | 2.253 |
| RT | 0.272 | 0.554 | 0.193 | 1.591 | 0.976 | 0.980 | 0.262 | 3.661 |
| CT | 0.955 | 0.965 | 0.277 | 3.362 | 0.554 | 1.566 | 0.355 | 6.914 |
| Localization | 0.740 | 1.073 | 0.708 | 1.626 | 0.580 | 0.885 | 0.576 | 1.362 |
| TP53 mutation | 0.357 | 0.602 | 0.204 | 1.772 | 0.777 | 1.173 | 0.387 | 3.557 |
| ING2 LOH | 0.102 | 2.493 | 0.833 | 7.458 | 0.691 | 1.275 | 0.385 | 4.226 |
95.0% CI confidence interval, RR risk ratio
Discussion
HNSCC is a worldwide common cancer. Our previous studies showed several ING family members as candidate TSG in head and neck and oral cancers (Gunduz et al. 2000, 2002, 2005, 2008b; Cengiz et al. 2007). We demonstrated that ING1 LOH and tumor-specific mutations took place in HNSCC (Gunduz et al. 2000). We recently showed decreased expression of one of the ING family members, ING3, and its relationship with poor prognosis in HNSCC (Gunduz et al. 2008b). Being structurally homologous to ING1, we speculated ING2 to have a similar tumor suppressor function in HNSCC. Therefore, we evaluated ING2 LOH and correlated it with the clinicopathological characteristics.
Recent array-CGH studies revealed few candidate TSG to be located at 4q35 in OSCC (Nakaya et al. 2007; Nakamura et al. 2008). Presently, only one LOH study at 4q32–35 region reported involvement of ING2 in sporadic BCC carcinogenesis (Sironi et al. 2004). Previous investigations demonstrated that ING2 induced fibroblast senescence in a p53-dependent manner (Pedeux et al. 2005). However, only few studies analyzed ING2 gene in human cancer. For instance, decreased ING2 expression was shown to correlate with poor prognosis in HCC (Zhang et al. 2008). It has also been reported that ING2 regulates cell proliferation by enhancing p53 acetylation in vitro (Nagashima et al. 2001). Another study showed down-regulation of ING2 mRNA expression in lung cancer cell lines, suggesting ING2 as a TSG (Okano et al. 2006).
The present study analyzed for the first time LOH at chromosome 4q35.1 in HNSCC. We found allelic deletion in more than half of the cases. The proximity of the marker to ING2 highly suggests ING2 as a strong candidate TSG in HNSCC. However, there are other genes closely situated at 4q35.1 region, which may also be considered as candidate TSG (Fig. 2). For instance, CDKN2AIP (also known as CARF) is located at 34kpb from the marker used. It encodes a protein that has been shown to cooperate with CDKN2A in p53 activation (Hasan et al. 2002). CLDN22 was also found close to our marker. Although there are no reports about CLDN22 in cancer, it belongs to the Claudin family of tight-junction forming transmembrane proteins, known to have tumor suppressor roles (Morin 2005; Oliveira and Morgado-Díaz 2007). IRF2 is a well-known oncogene. However, a recent study suggested a possible tumor suppressor function in myeloid cells (Huang et al. 2007). A member of the caspases, CASP3 is also localized in the area. It has an important apoptotic effect, reported in different types of cancers (Winter et al. 2001; O’Donovan et al. 2003), and with therapeutic implications related to its activation by chemotherapeutic agents. (Devarajan et al. 2002; Jin et al. 2007). The SLC25A4 gene, which encodes the protein ANT1, has also been reported to regulate apoptosis in vitro (Zamora et al. 2004; 2006). Finally, the most telomeric located genes, MTNR1A and FAT, have been identified as candidate TSG in OSCC (Nakaya et al. 2007; Nakamura et al. 2008).
TP53 gene encodes a protein that regulates transcription of a number of downstream targets in order to carry out tumor suppressor functions such as cell cycle control and apoptosis (Lane 1992). As in most types of cancer, TP53 alterations have been implicated in HNSCC. TP53 mutations are known to frequently take place in HNSCC (Bradford et al. 1997; Högmo et al. 1999). Furthermore, some studies demonstrated TP53 mutations to have clinical and prognostic significance in HNSCC (Erber et al. 1998; Cabelguenne et al. 2000). In the current study, coexistence of ING2 LOH ratio and TP53 mutation status were high, indicating that TP53 mutation and ING2 LOH simultaneously occur as common genetic alterations in HNSCC. However, the lack of correlation between these two events may also support TP53 mutation be an early event in HNSCC progression (Le and Giaccia 2003), and ING2 allelic loss might preferentially occur in late stages. Recent studies showed other ING members to function in a p53-independent manner (Wang and Li 2006; Coles et al. 2007). Considering that, most cases with TP53 mutations show inactivation of p53, our result indicates a p53-independent ING2 pathway in HNSCC. Nevertheless, characterization of each mutation type could give a better evaluation for its cellular effects and the relationship with ING2 LOH.
We examined the relationship between ING2 LOH status and clinicopathological data. LOH status appeared to be statistically correlated with late stage tumors compared to early stage patients. Several chromosomal loci are sequentially deleted during carcinogenic process. Le and Giaccia (2003) reported a stepwise allelic loss involving 9p21, 3p, 17p13, 13q21, 14q24, 6p, 8p23 and 4q26–28 in order, from hyperplasia to dysplasia, carcinoma in situ and invasive cancer. There is only one report considering LOH at the chromosome region 4q35, in sporadic BCC (Sironi et al. 2004), which did not include the clinicopathological parameters. Our data suggests that allelic loss at chromosome 4q35.1 is likely to be a late event in HNSCC. Regarding to cancer treatment, the ratio of patients that received radiotherapy and chemotherapy was higher in LOH than retention cases, showing statistical significant values (Table 4). Although we do not know the exact reason for these associations, the fact that most cases subjected to chemotherapy and radiation therapy were in advanced T stage could indicate higher aggressiveness in tumors presenting ING2 LOH. Several studies reported allelic loss at different chromosomal locations to associate with therapeutic response of malignant tumors (Iuchi et al. 2002; Jamieson et al. 2003). Due to limited clinical information, we could not assess this possibility by the current analysis. However, such investigations would contribute to a better understanding of the prognosis significance of ING2 in HNSCC.
When analyzing correlations between ING2 LOH and the patients OS and DFS, no relationship was detected in terms of OS. On the other hand, a near significant relationship was shown between ING2 LOH and DFS (P = 0.11). This result suggests the potential prognostic significance of ING2, and supports the tumor suppressor character of the gene. Further analysis, with more samples might clarify this point.
To the best of our knowledge, this is the first report considering ING2 in HNSCC. The focus of the study was to analyze LOH at 4q35.1 using a highly specific microsatellite marker closely located to ING2 locus. Nevertheless, further studies, on the candidate TSG ING2, are required in order to completely elucidate its role in HNSCC.
We can conclude that the high percentage of LOH suggests ING2 as a candidate TSG in HNSCC. Moreover, the association of ING2 LOH and advanced T stage marks the relevance of ING2 in HNSCC carcinogenesis. Finally, the lack of correlation between ING2 LOH and TP53 mutation status suggests that ING2 LOH occurs independently of TP53 status in HNSCC.
Acknowledgments
This work was partially supported by grants-in-aid for scientific researches from the Ministry of Education, Culture, Sports, Science and Technology [19592109 (to HN), 18-06262 (to EG), 17406027 (to NN)], Seed Innovation Research from Japan Science and Technology Agency (to MG), from Sumitomo Trust Haraguchi Memorial Cancer Research Promotion (to MG) and Astrazeneca Research Grant (to MG).
Conflict of interest statement
None of the authors has any potential conflict of interest.
References
- Baker SJ, Fearon ER, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, van Tuinen P, Ledbetter DH, Barker DF, Nakamura Y, White R, Vogelstein B (1989) Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 244(4901):217–221. doi:10.1126/science.2649981 [DOI] [PubMed] [Google Scholar]
- Beder LB, Gunduz M, Ouchida M, Gunduz E, Sakai A, Fukushima K, Nagatsuka H, Ito S, Honjo N, Nishizaki K, Shimizu K (2006) Identification of a candidate tumor suppressor gene RHOBTB1 located at a novel allelic loss region 10q21 in head and neck cancer. J Cancer Res Clin Oncol 132(1):19–27. doi:10.1007/s00432-005-0033-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford CR, Zhu S, Poore J, Fisher SG, Beals TF, Thoraval D, Hanash SM, Carey TE, Wolf GT (1997) p53 mutation as a prognostic marker in advanced laryngeal carcinoma. Department of Veterans Affairs Laryngeal Cancer Cooperative Study Group. Arch Otolaryngol Head Neck Surg 123(6):605–609 [DOI] [PubMed] [Google Scholar]
- Cabelguenne A, Blons H, de Waziers I, Carnot F, Houllier AM, Soussi T, Brasnu D, Beaune P, Laccourreye O, Laurent-Puig P (2000) p53 alterations predict tumor response to neoadjuvant chemotherapy in head and neck squamous cell carcinoma: a prospective series. J Clin Oncol 18(7):1465–1473 [DOI] [PubMed] [Google Scholar]
- Cengiz B, Gunduz M, Nagatsuka H, Beder L, Gunduz E, Tamamura R, Mahmut N, Fukushima K, Ali MA, Naomoto Y, Shimizu K, Nagai N (2007) Fine deletion mapping of chromosome 2q21-37 shows three preferentially deleted regions in oral cancer. Oral Oncol 43(3):241–247. doi:10.1016/j.oraloncology.2006.03.004 [DOI] [PubMed] [Google Scholar]
- Cheung KJ Jr, Bush JA, Jia W, Li G (2000) Expression of the novel tumour suppressor p33ING1 is independent of p53. Br J Cancer 83(11):1468–1472. doi:10.1054/bjoc.2000.1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles AH, Liang H, Zhu Z, Marfella CG, Kang J, Imbalzano AN, Jones SN (2007) Deletion of p37Ing1 in mice reveals a p53-independent role for Ing1 in the suppression of cell proliferation, apoptosis, and tumorigenesis. Cancer Res 67(5):2054–2061. doi:10.1158/0008-5472.CAN-06-3558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarajan E, Sahin AA, Chen JS, Krishnamurthy RR, Aggarwal N, Brun AM, Sapino A, Zhang F, Sharma D, Yang XH, Tora AD, Mehta K (2002) Down-regulation of caspase 3 in breast cancer: a possible mechanism for chemoresistance. Oncogene 21(57):8843–8851. doi:10.1038/sj.onc.1206044 [DOI] [PubMed] [Google Scholar]
- Erber R, Conradt C, Homann N, Enders C, Finckh M, Dietz A, Weidauer H, Bosch FX (1998) TP53 DNA contact mutations are selectively associated with allelic loss and have a strong clinical impact in head and neck cancer. Oncogene 16(13):1671–1679. doi:10.1038/sj.onc.1201690 [DOI] [PubMed] [Google Scholar]
- Garkavtsev I, Grigorian IA, Ossovskaya VS, Chernov MV, Chumakov PM, Gudkov AV (1998) The candidate tumour suppressor p33ING1 cooperates with p53 in cell growth control. Nature 391(6664):295–298. doi:10.1038/34675 [DOI] [PubMed] [Google Scholar]
- Gunduz M, Ouchida M, Fukushima K, Hanafusa H, Etani T, Nishioka S, Nishizaki K, Shimizu K (2000) Genomic structure of the human ING1 gene and tumor-specific mutations detected in head and neck squamous cell carcinomas. Cancer Res 60(12):3143–3146 [PubMed] [Google Scholar]
- Gunduz M, Ouchida M, Fukushima K, Ito S, Jitsumori Y, Nakashima T, Nagai N, Nishizaki K, Shimizu K (2002) Allelic loss and reduced expression of the ING3, a candidate tumor suppressor gene at 7q31, in human head and neck cancers. Oncogene 21(28):4462–4470. doi:10.1038/sj.onc.1205540 [DOI] [PubMed] [Google Scholar]
- Gunduz M, Nagatsuka H, Demircan K, Gunduz E, Cengiz B, Ouchida M, Tsujigiwa H, Yamachika E, Fukushima K, Beder L, Hirohata S, Ninomiya Y, Nishizaki K, Shimizu K, Nagai N (2005) Frequent deletion and down-regulation of ING4, a candidate tumor suppressor gene at 12p13, in head and neck squamous cell carcinomas. Gene 356:109–117. doi:10.1016/j.gene.2005.02.014 [DOI] [PubMed] [Google Scholar]
- Gunduz M, Gunduz E, Rivera RS, Nagatsuka H (2008a) The inhibitor of growth (ING) gene family: potential role in cancer therapy. Curr Cancer Drug Targets 8(4):275–284. doi:10.2174/156800908784533454 [DOI] [PubMed] [Google Scholar]
- Gunduz M, Beder LB, Gunduz E, Nagatsuka H, Fukushima K, Pehlivan D, Cetin E, Yamanaka N, Nishizaki K, Shimizu K, Nagai N (2008b) Downregulation of ING3 mRNA expression predicts poor prognosis in head and neck cancer. Cancer Sci 99(3):531–538. doi:10.1111/j.1349-7006.2007.00708.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasan MK, Yaguchi T, Sugihara T, Kumar PK, Taira K, Reddel RR, Kaul SC, Wadhwa R (2002) CARF is a novel protein that cooperates with mouse p19ARF (human p14 ARF) in activating p53. J Biol Chem 277(40):37765–37770. doi:10.1074/jbc.M204177200 [DOI] [PubMed] [Google Scholar]
- Hinds PW, Weinberg RA (1994) Tumor suppressor genes. Curr Opin Genet Dev 4(1):135–141. doi:10.1016/0959-437X(94)90102-3 [DOI] [PubMed] [Google Scholar]
- Högmo A, Börresen-Dale AL, Blegen H, Lindholm J, Kuylenstierna R, Auer G, Munck-Wikland E (1999) TP53 mutations do not correlate with locoregional recurrence in stage I tongue carcinomas. Anticancer Res 19(4C):3433–3438 [PubMed] [Google Scholar]
- Huang W, Horvath E, Eklund EA (2007) PU.1, interferon regulatory factor (IRF) 2, and the interferon consensus sequence-binding protein (ICSBP/IRF8) cooperate to activate NF1 transcription in differentiating myeloid cells. J Biol Chem 282(9):6629–6643. doi:10.1074/jbc.M607760200 [DOI] [PubMed] [Google Scholar]
- Iuchi T, Namba H, Iwadate Y, Shishikura T, Kageyama H, Nakamura Y, Ohira M, Yamaura A, Osato K, Sakiyama S, Nakagawara A (2002) Identification of the small interstitial deletion at chromosome band 1p34-p35 and its association with poor outcome in oligodendroglial tumors. Genes Chromosomes Cancer 35(2):170–175. doi:10.1002/gcc.10080 [DOI] [PubMed] [Google Scholar]
- Jamieson TA, Brizel DM, Killian JK, Oka Y, Jang HS, Fu X, Clough RW, Vollmer RT, Anscher MS, Jirtle RL (2003) M6P/IGF2R loss of heterozygosity in head and neck cancer associated with poor patient prognosis. BMC Cancer 3:4–12. doi:10.1186/1471-2407-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin CY, Park C, Cheong J, Choi BT, Lee TH, Lee JD, Lee WH, Kim GY, Ryu CH, Choi YH (2007) Genistein sensitizes TRAIL-resistant human gastric adenocarcinoma AGS cells through activation of caspase-3. Cancer Lett 257(1):56–64. doi:10.1016/j.canlet.2007.06.019 [DOI] [PubMed] [Google Scholar]
- Lane DP (1992) Cancer. p53, guardian of the genome. Nature 358(6381):15–16. doi:10.1038/358015a0 [DOI] [PubMed] [Google Scholar]
- Le QT, Giaccia AJ (2003) Therapeutic exploitation of the physiological and molecular genetic alterations in head and neck cancer. Clin Cancer Res 9(12):4287–4295 [PubMed] [Google Scholar]
- Lee WH, Bookstein R, Hong F, Young LJ, Shew JY, Lee EY (1987) Human retinoblastoma susceptibility gene: cloning, identification, and sequence. Science 235(4794):1394–1399. doi:10.1126/science.3823889 [DOI] [PubMed] [Google Scholar]
- Lydiatt WM, Davidson BJ, Schantz SP, Caruana S, Chaganti RS (1998) 9p21 deletion correlates with recurrence in head and neck cancer. Head Neck 20(2):113–118. doi:10.1002/(SICI)1097-0347(199803)20:2<113::AID-HED3>3.0.CO;2-5 [DOI] [PubMed] [Google Scholar]
- Mao L, Lee JS, Fan YH, Ro JY, Batsakis JG, Lippman S, Hittelman W, Hong WK (1996) Frequent microsatellite alterations at chromosomes 9p21 and 3p14 in oral premalignant lesions and their value in cancer risk assessment. Nat Med 2(6):682–685. doi:10.1038/nm0696-682 [DOI] [PubMed] [Google Scholar]
- Morin PJ (2005) Claudin proteins in human cancer: promising new targets for diagnosis and therapy. Cancer Res 65(21):9603–9606. doi:10.1158/0008-5472.CAN-05-2782 [DOI] [PubMed] [Google Scholar]
- Nagashima M, Shiseki M, Miura K, Hagiwara K, Linke SP, Pedeux R, Wang XW, Yokota J, Riabowol K, Harris CC (2001) DNA damage-inducible gene p33ING2 negatively regulates cell proliferation through acetylation of p53. Proc Natl Acad Sci USA 98(17):9671–9676. doi:10.1073/pnas.161151798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura E, Kozaki KI, Tsuda H, Suzuki E, Pimkhaokham A, Yamamoto G, Irie T, Tachikawa T, Amagasa T, Inazawa J, Imoto I (2008) Frequent silencing of a putative tumor suppressor gene melatonin receptor 1 A (MTNR1A) in oral squamous-cell carcinoma. Cancer Sci 99(7):1390–1400. doi:10.1111/j.1349-7006.2008.00838.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakaya K, Yamagata HD, Arita N, Nakashiro KI, Nose M, Miki T, Hamakawa H (2007) Identification of homozygous deletions of tumor suppressor gene FAT in oral cancer using CGH-array. Oncogene 26(36):5300–5308. doi:10.1038/sj.onc.1210330 [DOI] [PubMed] [Google Scholar]
- O’Donovan N, Crown J, Stunell H, Hill AD, McDermott E, O’Higgins N, Duffy MJ (2003) Caspase 3 in breast cancer. Clin Cancer Res 9(2):738–742 [PubMed] [Google Scholar]
- Okano T, Gemma A, Hosoya Y, Hosomi Y, Nara M, Kokubo Y, Yoshimura A, Shibuya M, Nagashima M, Harris CC, Kudoh S (2006) Alterations in novel candidate tumor suppressor genes, ING1 and ING2 in human lung cancer. Oncol Rep 15(3):545–549 [PubMed] [Google Scholar]
- Oliveira SS, Morgado-Díaz JA (2007) Claudins: multifunctional players in epithelial tight junctions and their role in cancer. Cell Mol Life Sci 64(1):17–28. doi:10.1007/s00018-006-6314-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedeux R, Sengupta S, Shen JC, Demidov ON, Saito S, Onogi H, Kumamoto K, Wincovitch S, Garfield SH, McMenamin M, Nagashima M, Grossman SR, Appella E, Harris CC (2005) ING2 regulates the onset of replicative senescence by induction of p300-dependent p53 acetylation. Mol Cell Biol 25(15):6639–6648. doi:10.1128/MCB.25.15.6639-6648.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Ordoñez B, Beauchemin M, Jordan RC (2006) Molecular biology of squamous cell carcinoma of the head and neck. J Clin Pathol 59(5):445–453. doi:10.1136/jcp.2003.007641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roz L, Wu CL, Porter S, Scully C, Speight P, Read A, Sloan P, Thakker N (1996) Allelic imbalance on chromosome 3p in oral dysplastic lesions: an early event in oral carcinogenesis. Cancer Res 56(6):1228–1231 [PubMed] [Google Scholar]
- Sarker KP, Kataoka H, Chan A, Netherton SJ, Pot I, Huynh MA, Feng X, Bonni A, Riabowol K, Bonni S (2008) ING2 as a novel mediator of Transforming Growth Factor-β-dependent responses in epithelial cells. J Biol Chem 283(19):13269–13279. doi:10.1074/jbc.M708834200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada Y, Saito A, Suzuki M, Takahashi E, Horie M (1998) Cloning of a novel gene (ING1L) homologous to ING1, a candidate tumor suppressor. Cytogenet Cell Genet 83(3–4):232–235. doi:10.1159/000015188 [DOI] [PubMed] [Google Scholar]
- Shinno Y, Gunduz E, Gunduz M, Nagatsuka H, Tsujigiwa H, Cengiz B, Lee YJ, Tamamura R, Ouchida M, Fukushima K, Shimizu K, Nagai N (2005) Fine deletional mapping of chromosome 4q22-35 region in oral cancer. Int J Mol Med 16(1):93–98 [PubMed] [Google Scholar]
- Sironi E, Cerri A, Tomasini D, Sirchia SM, Porta G, Rossella F, Grati FR, Simoni G (2004) Loss of heterozygosity on chromosome 4q32-35 in sporadic basal cell carcinomas: evidence for the involvement of p33ING2/ING1L and SAP30 genes. J Cutan Pathol 31(4):318–322. doi:10.1111/j.0303-6987.2004.0187.x [DOI] [PubMed] [Google Scholar]
- Tannapfel A, Weber A (2001) Tumor markers in squamous cell carcinoma of the head and neck: clinical effectiveness and prognostic value. Eur Arch Otorhinolaryngol 258(2):83–88. doi:10.1007/s004050000303 [DOI] [PubMed] [Google Scholar]
- van der Riet P, Nawroz H, Hruban RH, Corio R, Tokino K, Koch W, Sidransky D (1994) Frequent loss of chromosome 9p21-22 early in head and neck cancer progression. Cancer Res 54(5):1156–1158 [PubMed] [Google Scholar]
- Wang Y, Li G (2006) ING3 promotes UV-induced apoptosis via Fas/Caspase-8 pathway in melanoma cells. J Biol Chem 281(17):11887–11893. doi:10.1074/jbc.M511309200 [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang J, Li G (2006) Leucine zipper-like domain is required for tumor suppressor ING2-mediated nucleotide excision repair and apoptosis. FEBS Lett 580(16):3787–3793. doi:10.1016/j.febslet.2006.05.065 [DOI] [PubMed] [Google Scholar]
- Winter RN, Kramer A, Borkowski A, Kyprianou N (2001) Loss of caspase-1 and caspase-3 protein expression in human prostate cancer. Cancer Res 61(3):1227–1232 [PubMed] [Google Scholar]
- Zamora M, Meroño C, Viñas O, Mampel T (2004) Recruitment of NF-kappaB into mitochondria is involved in adenine nucleotide translocase 1 (ANT1)-induced apoptosis. J Biol Chem 279(37):38415–38423. doi:10.1074/jbc.M404928200 [DOI] [PubMed] [Google Scholar]
- Zamora M, Ortega JA, Alaña L, Viñas O, Mampel T (2006) Apoptotic and anti-proliferative effects of all-trans retinoic acid. Adenine nucleotide translocase sensitizes HeLa cells to all-trans retinoic acid. Exp Cell Res 312(10):1813–1819. doi:10.1016/j.yexcr.2006.02.014 [DOI] [PubMed] [Google Scholar]
- Zhang HK, Pan K, Wang H, Weng DS, Song HF, Zhou J, Huang W, Li JJ, Chen MS, Xia JC (2008) Decreased expression of ING2 gene and its clinicopathological significance in hepatocellular carcinoma. Cancer Lett 261(2):183–192. doi:10.1016/j.canlet.2007.11.019 [DOI] [PubMed] [Google Scholar]