Abstract
Linkage of body mass index (BMI) to a broad region of chromosome 7q22-35 has been reported in multiple studies. We previously published a multipoint LOD score of 4.9 at D7S1804 for BMI from the National Heart, Lung, and Blood Institute Family Heart Study. Leptin (LEP), the human homolog of the mouse obesity (ob) gene, is positioned near the linkage peak and is the most prominent candidate gene in this region. Interest in LEP as a susceptibility gene for human obesity has led to numerous linkage and association studies, but the results of these studies are still controversial. In the present study, we employed family-based tests of association with both a quantitative measure of BMI adjusted for age and sex and a dichotomously defined obesity trait. We genotyped 29 single-nucleotide polymorphisms (SNPs) spanning 240 kb around the LEP gene in the 82 extended pedigrees with the strongest evidence for linkage. When the programs TRANSMIT and FBAT were used, a number of SNPs showed association in men but not women, for both the quantitative and qualitative trait definitions (P<.05). Five SNPs (H1328084, H1328083, H1328082, H1328081, and H1328080) positioned 2 kb beyond the previously defined promoter region showed strong association in single-marker and multiple-marker haplotype analysis. This five-marker haplotype (frequency 49% in this sample) is overtransmitted to obese offspring (P=.00005). All five of these SNPs are predicted to modify transcription-factor binding sites. This may indicate new functional variants in an extended promoter region of LEP.
Introduction
In Western societies, the prevalence of obesity has been increasing steadily in recent years. Obesity (MIM 601665) is strongly associated with increased risk for diabetes, lipid disorders, hypertension, and coronary heart disease (CHD). A genetic component in the etiology of BMI has been clearly demonstrated by epidemiological and genetic linkage studies (Borecki et al. 1998; Bouchard et al. 1998). Although many candidate genes have been proposed and numerous genetic studies performed, the resolution of the genetic factors underlying the susceptibility to obesity is far from complete (Chagnon et al. 2003).
Recently, we reported a genomewide linkage scan for BMI in the National Heart, Lung, and Blood Institute (NHLBI) Family Heart Study (FHS) (Feitosa et al. 2002). A LOD score of 4.9 was found at D7S1804 (7q32.3) for age- and sex-adjusted measures of BMI. Leptin (LEP) is positioned close to the center of this linkage peak and is the most prominent candidate gene in the region because of its central role in an adiposity-sensing pathway and its physiological effects on energy balance. The linkage finding at 7q22-36 has been consistently replicated by numerous studies of independent samples and supported by strong evidence of linkage in a meta-analysis of five published studies (Allison and Heo 1998; Li et al. 2003; Platte et al. 2003). The candidate region is large, extending over the 55 Mb between D7S618 and D7S3070. The positive linkage findings appear to be dispersed across three subregions. The first region is flanked by markers D7S1804 and D7S3070 at 7q32.3-q36 and is 10–15 cM downstream of LEP. The second region is near D7S2459/D7S523 at 7q22-31.1 and is 20 cM upstream of LEP. The third region is at 7q31.3, at the LEP gene. It is not clear whether these studies point to one or more than one gene contributing to obesity risk.
Since the identification of the LEP gene at 7q31.3 as the human homolog to the mouse obesity (ob) gene (Zhang et al. 1994), sequence variation in LEP has been studied extensively. Although mutations in the LEP gene are known to cause rare obesity syndromes (Montague et al. 1997a; Strobel et al. 1998) and have been responsible for obesity in several animal models (Zhang et al. 1994; Moon and Friedman 1997; Friedman and Halaas 1998), variation within the coding region of LEP is exceedingly rare in the general population (Considine et al. 1995; Maffei et al. 1996; Carlsson et al. 1997). The interest in LEP as a susceptibility gene for human obesity has also led to the identification of several common polymorphisms in the 5′ regulatory region—G2548A, A19G, and C1887T—that show association with lower LEP levels or obesity in several studies (Hager et al. 1998; Mammes et al. 1998, 2000; Li et al. 1999; Le Stunff et al. 2000). Nevertheless, the results from these studies are still controversial, and the relationship between LEP sequence variation and human body weight remains uncertain (Shigemoto et al. 1997; Lucantoni et al. 2000). In the present study, we describe the results of family-based association analysis of 29 SNPs spanning 240 kb across the LEP region, including the previously published SNPs and recently detected variants. Using linkage disequilibrium (LD) mapping and haplotype analysis, we sought to establish the position of the functionally significant variant(s).
Subjects and Methods
Subjects and Phenotype
The FHS is a multicenter, population-based study of factors influencing risk for CHD. A detailed description of the FHS has been published elsewhere (Family Heart Study Web site). We selected a primary genotyping panel with 548 DNAs derived from the 82 white families with the highest pedigree-specific LOD scores from our previous linkage study (Feitosa et al. 2002). The multipoint LOD score for these 82 families was 17.09 at 136.95 cM (Genetic Map Index Web site, Center for Medical Genetics).
BMI was calculated as weight (in kg) divided by the square of height (in meters). Regression models were used to adjust for sex, age, and center, as reported elsewhere (Feitosa et al. 2002). We also defined a dichotomous phenotype (denoted as “OB”) based on the adjusted BMI residual (BMI-R). A residual cutoff of ⩾0.6 was used to define affected individuals. It is notable that the residual of 0.6 corresponds approximately to a raw BMI score of 30, which is an accepted clinical designation for obesity. The dichotomous phenotype (OB) was coded as affected for those with a BMI-R ⩾0.6, as unaffected for those with a BMI-R ⩽0 (BMI ⩽∼27.5), and as ambiguous or unknown if 0 < BMI-R < 0.6. Of the 82 pedigrees selected on the basis of family-specific LOD scores, 69 contained at least one individual who was classified as affected. The 69 pedigrees contained 186 persons with a BMI-R >0.6 (185 DNAs available), 196 persons with a BMI-R <0 (195 DNAs available), and 88 persons with a BMI-R between 0 and 0.6 (84 DNAs available). Most persons in the ambiguous group are founders without DNA. Nonparametric linkage (NPL) analysis was performed in the 48 families with at least two individuals classified as OB, and an NPL score of 5.16 at 136.95 cM was observed.
Genotyping
Genomic DNA was prepared from whole blood by using the Puregene system (Gentra Systems) and was purified using QIAEX II kits (Qiagen). All of the samples were quantified using the PicoGreen DNA quantification method (Ahn et al. 1996) and were diluted to 2.5 ng/μl before making 96-well PCR source plates.
Five SNPs were selected from Assays-on-Demand SNP Genotyping Products, which are validated against genomic DNAs from four different populations and each of which has a corresponding allele frequency of at least 10% in whites. Four SNPs—G2548A, C1887T, G1387A, and A19G—were selected on the basis of previous publications of association studies involving LEP and BMI (Mammes et al. 1998; Li et al. 1999; Le Stunff et al. 2000). An additional 20 SNPs retrieved from the human SNP database in the Celera Discovery System (Celera Discovery System Web site) were also genotyped. Twenty of these 29 SNPs are in the 22-kb LEP region. An additional five SNPs are from the intergenic 5′ region, two are in the P100 gene 5′ upstream from LEP, and two are from the 3′ region distal to LEP in an unknown gene, FLJ10377.
SNPs were genotyped using the TaqMan technology (Holland et al. 1991; Livak et al. 1995) implemented on an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). PCR was performed using TaqMan Universal Master Mix (Applied Biosystems), 5 ng DNA, 900 nM of each primer, and 200 nM of each probe in a 5-μl reaction.
Bioinformatic Analysis
Transcription-factor binding sites (TFBS) were queried for the 50-bp sequence surrounding each 5′ candidate SNP by comparing the alternative sequence of the two SNP alleles with the TRANSFAC database by use of BLAST (Wingender et al. 2000).
Statistical Analysis of Association
The genotypes were checked for Mendelian inheritance errors and Hardy-Weinberg equilibrium in an integrated database. MERLIN (Abecasis et al. 2002) was used for detection of apparent double recombinants in the data and to calculate NPL scores based on OB. Version 2.5 of the program TRANSMIT (Clayton 1999; Clayton and Jones 1999) was used to perform the transmission/disequilibrium test (TDT) of association with OB on single SNPs and multiple-marker haplotypes. P values for all TDT analyses performed with TRANSMIT were empirically derived from 100,000 bootstrap replicates. To protect against misleading results due to rare alleles or haplotypes, the command-line switches (i.e., flags) “-agg3” and “-c3” were used to aggregate all alleles or haplotypes with frequencies <0.03 before haplotype construction. To test for association in the presence of linkage, the switches “-1” and “-nomf” were used to select one trio from each pedigree, to minimize the dependency on family structure and to maximize the association signal relative to the linkage signal.
The software package FBAT v1.4 (Horvath et al. 2001) was used to perform the family-based association tests for the quantitative trait BMI-R in the 69 pedigrees that included at least one member who was classified as affected. The null hypothesis of linkage but no association was tested using the -e flag, which computes the test statistic through use of the empirical variance, because the sample contains multiple nuclear families in some pedigrees and multiple affected individuals in these nuclear families, and the markers are in an area of known linkage. Multiallelic tests were performed using an additive genetic model.
Since the markers are tightly linked, results for individual SNPs may be highly correlated; thus, a simple Bonferroni correction is unduly conservative, and we report the empirical P values as described above. We used the Benjamini-corrected false-discovery rate (FDR) to control for multiple hypothesis testing (Benjamini and Hochberg 1995). The FDR is the expected proportion of true null hypotheses rejected out of the total number of null hypotheses rejected. Multiple comparison procedures controlling the FDR are more powerful than the commonly used multiple comparison procedures based on the familywise error rate. After adjustment for multiple testing and with an FDR level of 0.05, the cutoff for significant association is P=.01 for analyses of “all individuals,” P=.033 for analyses of “all males,” P=.026 for analyses of “one trio per pedigree with affected male offspring” via a TDT, and P=.031 in FBAT analysis of the families with affected male offspring.
LD block structure was examined by the program Haploview (Haploview Web site, Whitehead Institute). The D′ for all pairs of SNPs was calculated and the haplotype blocks estimated using the confidence-interval method (Gabriel et al. 2002). SNPs with low rare-allele frequencies may inflate estimates of D′, and the use of confidence-bound estimates for D′ reduces this bias. The default settings were used in these analyses, which invoke a one-sided upper 95% confidence bound of D′>0.98 and a lower bound of >0.7 to define SNP pairs in strong LD. A block is identified when at least 95% of SNP pairs in a region meet these criteria for strong LD. Haplotypes were reconstructed and their frequencies estimated using an accelerated expectation-maximization (EM) algorithm similar to the partition/ligation method (Qin et al. 2002) implemented in Haploview.
Results
Characteristics of the study sample are presented in table 1. The unadjusted mean BMI in men and women was significantly different, as assessed by a t test (P=.04). This difference in mean BMI occurred in both the affected (P=.0008) and unaffected (P<.0001) groups, where women had a higher and lower mean BMI, respectively.
Table 1.
Characteristics of Study Subjects Genotyped in the 69 Pedigrees
|
Observation in |
||
| Characteristic | Male Subjects(N=225) | Female Subjects(N=245) |
| Mean ± SD BMI: | ||
| Total sample | 29.2 ± 5.1 | 30.5 ± 7.6 |
| Affected | 34.8 ± 4.5 (n=76) | 37.4 ± 5.3 (n=110) |
| Unaffected | 25.2 ± 2.0 (n=97) | 23.5 ± 2.6 (n = 99) |
| Ambiguous | 28.6 ± 1.2 (n=52) | 28.6 ± 2.2 (n = 36) |
| Mean ± SD age (in years) | 51.5 ± 14.3 | 52.6 ± 13.1 |
| % of sample from study center: | ||
| NC | 17.3 | 12.5 |
| MN | 15.0 | 15.8 |
| MA | 19.5 | 23.1 |
| UT | 48.2 | 48.6 |
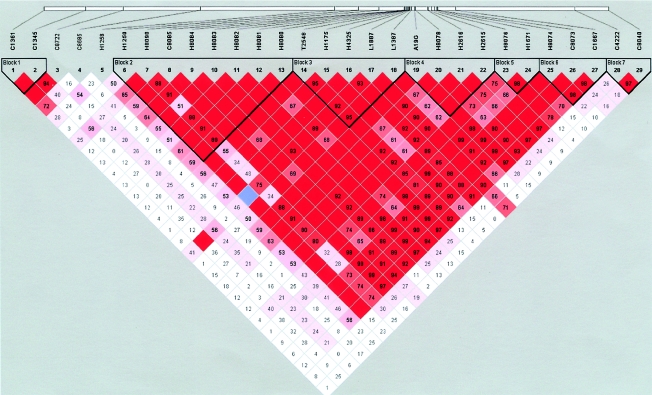
Table 2 shows the map locations and marker characteristics for the 29 SNPs in and around LEP that were genotyped. Although 4 SNPs—G2548A, A19G, C1887T, and G1387A—have been previously reported, the remaining 25 have not been studied for LEP association. The markers are presented according to their physical location, with D7S3061 centromeric. SNPs tested in LEP are intronic except for A19G, which is in the first exon, 5′ UTR region of the LEP gene. The rare-allele frequency for those SNPs is between 16% and 50% except for C1887T, which has a rare-allele frequency of 9%. Strong LD extends from the 5′ region to the first intron of LEP but does not extend to SNPs in the genes distal and proximal to LEP (fig. 1).
Table 2.
Markers and Map Locations
| MarkerNumber | Marker | Gene | SNP | Frequencya | CeleraLocation(bp) | NCBILocation(bp) | IntermarkerIntervalb(bp) | SNPType | dbSNP rsNumber |
| D7S3061 | NA | 118093192 | 122844090 | … | … | … | |||
| 1 | C1331361 | P100 | T/C | .348 | 122524910 | 127275987 | 4,431,897 | Exon | rs322825 |
| 2 | C1331345 | P100 | G/A | .190 | 122543069 | 127294146 | 18,159 | Intron | rs6953698 |
| 3 | C618722 | A/G | .456 | 122559253 | 127310271 | 16,125 | Intergenic | rs322785 | |
| 4 | C618685 | G/C | .272 | 122592487 | 127343162 | 32,891 | Intergenic | rs53125 | |
| 5 | H1331258 | A/G | .351 | 122650302 | 127400977 | 57,815 | Intergenic | rs11772985 | |
| 6 | H1331250 | G/A | .160 | 122660615 | 127411290 | 10,313 | Intergenic | rs6947095 | |
| 7 | H1328090 | C/A | .389 | 122669402 | 127420076 | 8,786 | Intergenic | rs791601 | |
| 8 | C1328085 | LEP | T/G | .340 | 122680314 | 127430988 | 10,912 | 5′ flank/TFBS | rs10249476 |
| 9 | H1328084 | LEP | G/A | .411 | 122680501 | 127431175 | 187 | 5′ flank/TFBS | rs1349419 |
| 10 | H1328083 | LEP | G/A | .495 | 122681032 | 127431706 | 531 | 5′ flank/TFBS | NA |
| 11 | H1328082 | LEP | C/A | .345 | 122681386 | 127432060 | 354 | 5′ flank/TFBS | rs12535708 |
| 12 | H1328081 | LEP | C/T | .404 | 122681555 | 127432229 | 169 | 5′ flank/TFBS | rs11770725 |
| 13 | H1328080 | LEP | C/A | .341 | 122681623 | 127432297 | 68 | 5′ flank | rs12535747 |
| 14 | G2548A | LEP | G/A | .495 | 122682071 | 127432745 | 448 | Promoter | rs7799039 |
| 15 | H6501175 | LEP | A/G | .446 | 122682636 | 127433310 | 565 | Promoter | rs6467166 |
| 16 | H2944325 | LEP | G/A | .493 | 122682656 | 127433330 | 20 | Promoter | rs12536535 |
| 17 | C1887T | LEP | C/T | .092 | 122682732 | 127433406 | 76 | Promoter | NA |
| 18 | G1387A | LEP | A/G | .446 | 122683231 | 127433905 | 499 | Promoter | NA |
| 19 | A19G | LEP | G/A | .342 | 122684637 | 127435311 | 1,406 | Exon1/5′ UTR | rs2167270 |
| 20 | H1328078 | LEP | T/C | .407 | 122685139 | 127435813 | 502 | Intron 1 | rs2278815 |
| 21 | H1432616 | LEP | G/T | .423 | 122690353 | 127441030 | 5,217 | Intron 1 | NA |
| 22 | H1432615 | LEP | G/A | .426 | 122690424 | 127441101 | 71 | Intron 1 | NA |
| 23 | H1328076 | LEP | A/T | .476 | 122691974 | 127442651 | 1,550 | Intron 1 | rs10244329 |
| 24 | H3001671 | LEP | A/G | .477 | 122693347 | 127444024 | 1,373 | Intron 1 | rs11763517 |
| 25 | H1328074 | LEP | G/A | .356 | 122694372 | 127445049 | 1,025 | Intron 1 | rs11760956 |
| 26 | C1328073 | LEP | G/A | .363 | 122694725 | 127445402 | 353 | Intron 1 | rs10954173 |
| 27 | C3001667 | LEP | A/G | .424 | 122702403 | 127453091 | 7,689 | Intron 2 | rs2060715 |
| 28 | C1574222 | FLJ10377 | G/A | .353 | 122753843 | 127504687 | 51,596 | 3′ UTR | rs12850 |
| 29 | C1328040 | FLJ10377 | C/G | .306 | 122768526 | 127519371 | 14,684 | Intron | rs10279576 |
| D7S1804 | NA | 127031883 | 131246812 | 3,727,441 | … | … |
Allele frequency for the second allele is shown.
Marker intervals are calculated on the basis of NCBI locations (National Center for Biotechnology Information Web site).
Figure 1.
LD block structure around LEP. Haplotype block structure, as depicted by Haploview, is shown. The five-color scheme (white to red) represents the increasing strength of LD. Values for D′ (×100) are shown, but those boxes with D′=1 are shaded in bright red and are empty. Cells with D′<1 are shades of pink or red. Blue represents D′=1 but with a low confidence estimate for D′.
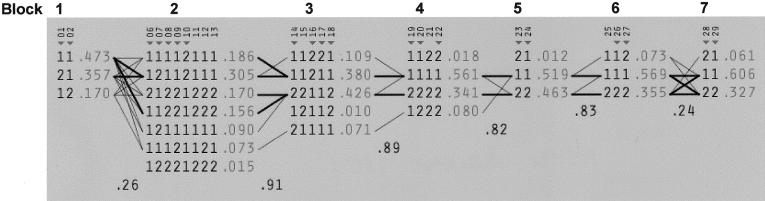
Seven LD blocks are identified across the 240-kb region (see fig. 1). We explored the haplotype diversity in this region through use of Haploview. Figure 2 depicts the values for D′ between the multi-SNP blocks defined in figure 1. Haploview identified tag SNPs for parsimonious haplotypes for each block (Johnson et al. 2001; Patil et al. 2001).
Figure 2.
Haplotype structure and diversity. Haplotype blocks and their frequencies were estimated using an accelerated EM algorithm implemented in Haploview. SNP numbers corresponding to figure 1 are listed above each column of alleles, and “▾” denotes the tag SNPs that designate a parsimonious haplotype for each block. Recombination rates from one block to the next are defined by a multiallelic value of D′. Haplotypes in adjacent blocks are connected by a thick line if they occur together with a frequency >10% and by a thin line if they occur together with a frequency >1%. For each SNP, “1” represents the common allele, and “2” represents the rare allele.
In each analysis, the TDT was performed for all individuals as well as for male and female family members separately. The SNPs that remain significant after Benjamini correction are indicated with an asterisk (*). In the TDT conducted on the entire sample (the “All” column in table 3), 5 of the 20 LEP SNPs showed significant association with OB (P<.05) after Benjamini correction, but none of these markers remained significant when a “one trio only” TDT analysis was used. When sex-specific analyses were conducted, the association was seen almost exclusively among men. Overall, among all men, 16 SNPs in the “all individual” analysis (P<.013) and 13 in the “one trio” analysis (P<.038) showed significant association with OB. Although the level of significance was slightly lower in the FBAT analysis compared with the TRANSMIT results, the same pattern of association was also observed when the quantitative trait BMI-R was used. Seventeen of 20 SNPs in LEP showed significant association (P<.031 with Benjamini correction). In both analyses, SNPs with strong association are seen in men and are clustered between markers H1331250 and H1328076. The 117 male offspring (64 affected, 36 unaffected, and 17 ambiguous) are very similar to the sample of all 225 male subjects described in table 1, except that they are slightly heavier (mean BMI 31.1 vs. 29.2 in the total sample) and younger (mean age 45.8 years vs. 51.5 years in the total sample).
Table 3.
TDT of Association to OB as a Qualitative Trait in TRANSMIT, and BMI-R as a Quantitative Trait in FBAT
| TRANSMIT P Valueb |
|||||||||
| All Individuals |
One Trio per Pedigree |
BMI-R (-e) FBAT P Value |
|||||||
| SNP IDa | Allc(n=470) | Femaled(n=423) | Malee(n=315) | Allf(n=198) | Femaleg(n=165) | Maleh(n=114) | Allc(n=470) | Femaled(n=423) | Malee(n=315) |
| C1361 | .261 | .692 | .220 | .018 | .261 | .138 | .173 | .649 | .093 |
| C1345 | .974 | .806 | .821 | .976 | .704 | .889 | .558 | .614 | .634 |
| C8722 | .346 | .775 | .255 | .245 | .682 | .016* | .577 | .718 | .100 |
| C8685 | .027 | .026 | .420 | .493 | .092 | .703 | .147 | .104 | .550 |
| H1258 | .400 | .948 | .093 | .463 | .318 | .391 | .426 | .424 | .730 |
| H1250 | .023 | .625 | .0003* | .288 | .887 | .039 | .238 | .626 | .022* |
| H8090 | .136 | .259 | .345 | .050 | .183 | .532 | .258 | .648 | .138 |
| C8085 | .005* | .254 | .002* | .082 | .650 | .0002* | .095 | .893 | .009* |
| H8084 | .017 | .265 | .008* | .179 | .958 | .005* | .077 | .736 | .007* |
| H8083 | .075 | .947 | .001* | .618 | .836 | .007* | .231 | .340 | .003* |
| H8082 | .001* | .173 | .0004* | .107 | .803 | .011* | .059 | .967 | .005* |
| H8081 | .022 | .388 | .005* | .173 | .789 | .009* | .147 | .917 | .010* |
| H8080 | .004* | .240 | .001* | .201 | .671 | .034 | .113 | .708 | .008* |
| 2548A | .114 | .714 | .00007* | .714 | .311 | .00002* | .246 | .325 | .003* |
| H1175 | .036 | .665 | .0008* | .074 | .663 | .0001* | .339 | .317 | .008* |
| H4325 | .132 | .990 | .007* | .302 | .838 | .048 | .551 | .109 | .007* |
| 1887T | .344 | .121 | .370 | .862 | .417 | .0002* | .327 | .160 | .361 |
| 1387A | .064 | .975 | .0002* | .782 | .679 | .0003* | .741 | .152 | .028* |
| A19G | .002* | .113 | .004* | .126 | .769 | .017* | .049 | .631 | .010* |
| H8078 | .006* | .307 | .001* | .133 | .796 | .00002* | .073 | .888 | .003* |
| H2616 | .057 | .560 | .011* | .028 | .774 | .134 | .299 | .586 | .014* |
| H2615 | .035 | .442 | .006* | .009 | .385 | .004* | .138 | .955 | .007* |
| H8076 | .175 | .879 | .005* | .150 | .207 | .015* | .751 | .091 | .014* |
| H1671 | .327 | .819 | .031 | .363 | .576 | .052 | .486 | .241 | .013* |
| H8074 | .055 | .418 | .031 | .240 | .423 | .061 | .232 | .777 | .034 |
| C8073 | .072 | .350 | .065 | .475 | .855 | .384 | .218 | .975 | .041 |
| C1667 | .936 | .304 | .113 | .599 | .081 | .107 | .682 | .069 | .067 |
| C4222 | .072 | .021 | .613 | .028 | .249 | .556 | .248 | .071 | .970 |
| C8040 | .104 | .020 | .717 | .057 | .498 | .988 | .158 | .067 | .852 |
Celera SNP IDs are abbreviated to the final four digits.
P values are generated by simulation of 100,000 bootstrap sampling. P values <.05 are in boldface italic, and P values significant after FDR adjustment are indicated by an asterisk (*).
470 individuals in 69 pedigrees.
423 individuals in 62 pedigrees with female affected offspring.
315 individuals in 44 pedigrees with male affected offspring.
198 individuals in 66 trios with affected offspring.
165 individuals in 55 trios with affected female offspring.
114 individuals in 38 trios with affected male offspring.
We sought to establish the boundary of the significant association, which extended from 25 kb upstream of LEP to its first intron, by applying the TDT to single markers. We confirmed the previously published association with BMI at markers A19G and G2548A (Hager et al. 1998; Mammes et al. 1998, 2000; Li et al. 1999); G2548A showed a strong association with OB in both the “all individuals” (P=.00007) and the “one trio per pedigree” (P=.00002) analyses, as well as with BMI-R (P=.003) in the FBAT analysis. All of these associations were found solely in men. A19G showed a strong association in the TDT conducted on the entire sample (P=.004) but only a moderate association for the “one trio per pedigree” analysis in men (P=.017).
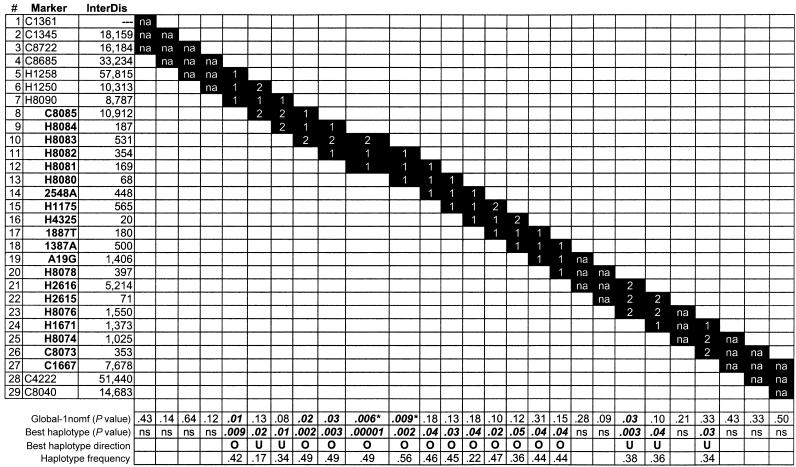
We also analyzed two-, three-, four-, and five-marker haplotypes for the 29 SNPs, and figure 3 shows the results for the three-marker haplotypes tested. The “global” P value represents the overall significance when the observed versus expected transmissions of all of the haplotypes are considered together. Although multiple LEP haplotypes showed association after correction for multiple testing, only two common haplotypes show significance globally: H1328083-H1328082-H1328081, with global P=.006, and H1328082-H1328081-H1328080, with global P=.009. The haplotype with the strongest association is a composition of allele A of H1328083, allele C of H1328082, and allele C of H1328081 (frequency 49%), overtransmitted from parents to their affected offspring (P=.00001).
Figure 3.
Haplotype analyses using one trio in each pedigree for a three-marker sliding window. Below each triad is the P value for that haplotype, the haplotype frequency, the direction of the transmitted haplotype (either overtransmitted [“O”] or undertransmitted [“U”]), and the global P value (median value of 20 runs), derived by evaluation of the transmissions of all haplotypes simultaneously. SNPs within the LEP gene are in boldface and are offset, and the approximate location within the LEP gene is shown below the haplotype frequencies. 1 = common allele; 2 = rare allele; InterDis = marker-to-marker distance (in bp); na = no haplotype resulted in P<.05; ns = not significant; Global-1nomf (P value) = global P value in the “one trio per pedigree” analysis. Celera SNP IDs are abbreviated to the final four digits. P values significant after FDR adjustment are indicated with an asterisk (*).
Results of two-marker haplotype analysis resembled those for single-marker analysis (data not shown), and the four-marker and five-marker haplotype results were similar to the findings for the reported three-marker haplotypes. Among the four-marker haplotypes, two showed significant association globally: H1328084-H1328083-H1328082-H1328081 and H1328083-H1328082-H1328081-H1328080.
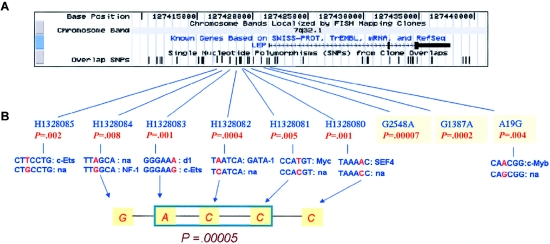
One common five-SNP haplotype, representing alleles G, A, C, C, and C from SNPs H1328084, H1328083, H1328082, H1328081, and H1328080, respectively, was found to be overtransmitted to affected offspring (P=.00005) and spans a region ∼2 kb up from the previously defined promoter region (Gong et al. 1996). This five-marker haplotype can be simplified to three tag SNPs—H1328081 (which is equivalent to H1328084), H1328082 (which is equivalent to C1328085 and H1328080), and H1328083—but it is not clear which combination of three is most informative. Bioinformatic analysis revealed that all five of these SNPs are predicted to modify TFBS (see fig. 4). Although G2548A shows a strong individual SNP association to OB (table 3), it is in strong LD with H1328083 (r2=0.97), and, thus, G2548A may not add significantly to the information provided by that SNP to the common five-SNP haplotype.
Figure 4.
Genomic view of the LEP 5′ region, its polymorphisms, and the best five-marker haplotype. A, Genomic view of LEP mRNA and SNPs surrounding LEP (UCSC Genome Browser Web site). B, Polymorphisms at the 5′ region of LEP, with single-marker P values from TRANSMIT analysis and their respective putative TFBS, with the polymorphic site highlighted in red. No TFBS changes were found for G2548A and G1387A. na = no TFBS site identified for a specific short sequence.
Discussion
To assess the role of LEP in association with BMI, we performed a family-based study for both a dichotomous obesity characterization (OB) and a quantitative BMI residual measurement (BMI-R). A number of recently identified SNPs showed strong association with both of these measures in the 82 pedigrees with the strongest evidence for linkage (P<.01). The association signals came exclusively from men, and a sex effect of this magnitude in LEP has not been previously reported. The boundary of significant association extended from 25 kb upstream of LEP to its first intron. We identified a common risk haplotype with frequency of 49%, which is overtransmitted to obese offspring (P=.00005) and positioned 2 kb up from the previously defined LEP promoter. The five SNPs (H1328084-H1328083-H1328082-H1328081-H1328080) in this high-risk haplotype are predicted to modify TFBS, and, thus, any one of them may be functionally implicated for modifying the transcription of LEP. Because the SNPs are in very high LD and span a very small region (∼1.2 kb), it is difficult to discern which SNP(s) is most likely implicated in LEP transcription. Future functional analyses using various combinations of these variants may help to confirm these findings and provide insight into the function of each variant.
In addition to the five SNPs mentioned above, we also confirmed the previously reported (Mammes et al. 1998, 2000; Li et al. 1999; Le Stunff et al. 2000) strong association between G2548A and BMI (P=.00002) in a single-marker TDT, but the association with this marker is not detected in any multimarker haplotype analysis. Since this SNP is neither at a conserved region among human, mouse, and rat, nor at a predicted TFBS (Li et al. 1999), its functional importance is speculative. Nevertheless, we cannot rule out the possibility that more than one functional polymorphism in LEP affects variation in BMI. The finding that G2548A shows strong association in single-marker analysis but does not show association in combination with other SNPs, such as those found in the five-SNP haplotype, may indicate that it is associated with an independent functional polymorphism.
The previously reported A19G SNP, in the 5′ UTR of the first exon of LEP (Hager et al. 1998; Li et al. 1999), shows only a modest association with OB in a stringent TDT using only one trio per pedigree (P=.017 in men) and a similar modest association with BMI-R in FBAT analysis. The finding that only men show LEP association with BMI may explain the low frequency of replication for the association of BMI with LEP. Furthermore, the modest effect occasionally observed for the A19G polymorphism may be attributable to strong LD between this SNP and SNPs in the haplotype (r2=0.984 for A19G and H1328084; r2=0.681 for H1328083), which appear to have a much stronger association with obesity.
The characteristics of promoter region for LEP have been reported elsewhere (Gong et al. 1996). Of note is that all five SNPs in the risk haplotype are in a 2-kb region 5′ to the reported promoter that may represent an enlarged promoter region for LEP. All five SNPs in this haplotype modify predicted TFBS (see fig. 4). For example, the modification of the A allele to a C allele of SNP H1328082 results in the loss of a GATA-1 binding site. Recently, functional variation for complex phenotypes has been detected for changes other than those altering the structure of the protein encoded by the responsible gene (Toma et al. 2002). Much of the genetic component of human phenotypic diversity, including susceptibility to disease, is hypothesized to be the result of cis-acting influences on gene expression. If this hypothesis is correct, it implies that cis-acting regulatory variation may be a common phenomenon. It is also supported by some of the recent successes in the fields of diabetes (Horikawa et al. 2000), inflammatory bowel disease (Rioux et al. 2001), and schizophrenia (Stefansson et al. 2002; Straub et al. 2002), in which no obvious pathogenic coding changes have yet been identified. These observations have re-emphasized the possibility that inherited variation in gene expression may play an important role in susceptibility to complex traits such as obesity (Lander 1996; Peltonen and McKusick 2001). However, our current knowledge of regulatory elements in the human genome is far from comprehensive, and, beyond their coding sequences, most genes are not well annotated.
We genotyped the two previously reported LEP mutations, Met110Val and Arg105Trp (Karvonen et al. 1998; Strobel et al. 1998), and neither of them was present in our FHS samples, confirming that these mutations are exceedingly rare in the general population. Although the high-risk haplotype we identified is very common in FHS families showing linkage to the 7q22-35 region (49%), it is less clear how common this haplotype may be in the general population or in other genetic studies of obesity.
Sex-specific analyses show that the significant association for LEP was seen almost exclusively among men in this sample. This finding lends support to an unexpected sex effect between the LEP gene and BMI. Only two studies have included sex-stratified analyses for LEP and BMI or obesity (Mammes et al. 1998, 2000). These studies found that the G allele of SNP G2548A was more frequent in overweight subjects of both sexes (P<.01). However, only male carriers of this allele had lower LEP concentrations after adjusting for fat mass (P=.05). The T allele of C1887T was associated with a decrease in LEP levels in response to a restrictive diet (P=.005) but was not associated with BMI or LEP concentration. However, because of the small number of T-allele carriers, one cannot rule out the possibility that this decrease represents random variability. Nevertheless, these results are consistent with our finding that the common G allele of G2548A is overtransmitted in the OB offspring. A sex-specific genetic component in BMI has been reported in Norwegian twins (Harris et al. 1995) and Mexican Americans (Comuzzie et al. 1995), as well as in whites in the Framingham Study (Atwood et al. 2003). Sex-specific differences in human LEP mRNA expression have been implicated by previous studies (Montague et al. 1997b). The HERITAGE family study demonstrated that the association between the LEP receptor gene and adiposity was found only in white males (Chagnon et al. 2000). We may speculate that a candidate for the connection between LEP and sex-specific phenotypic differences is testosterone, which is down-regulated by leptin (Tena-Sempere et al. 2001). The finding that the effect of LEP is mainly in men, while linkage to the region is seen in both male and female subgroups in our study, suggests that there may be other important BMI-related genes in the 7q22-35 region that are worthy of additional investigation.
In summary, the strong association of a number of common variants and haplotypes in the 5′ region of LEP with both a quantitative measure of BMI adjusted for age and sex and a dichotomously defined obesity trait in the present study suggest that common LEP sequence variants are associated with obesity in the general population. The common risk haplotype identified from this study may be clinically useful for identifying a subgroup of people who may have a LEP deficiency derived from altered regulatory elements and may predispose this subgroup to obesity. It will be valuable to replicate these findings in other family sets showing linkage to this region and to test for association to BMI in independent studies.
Acknowledgments
NHLBI grant HL68891-03 supported this project. This article is presented on behalf of the investigators of the NHLBI FHS.
Electronic-Database Information
The URLs for data presented herein are as follows:
- Celera Discovery System, http://www.celeradiscoverysystem.com/index.cfm
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/ (for SNPs [rs numbers listed in ])
- Family Heart Study, http://www.biostat.wustl.edu/fhs/
- Genetic Map Index, Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/Map_Markers/maps/IndexMapFrames.html (for the Marshfield genetic map)
- Haploview, Whitehead Institute, http://www.broad.mit.edu/personal/jcbarret/haploview/index.php
- National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for obesity)
- UCSC Genome Browser, http://genome.ucsc.edu/
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- Ahn SJ, Costa J, Emanuel JR (1996) PicoGreen quantitation of DNA: effective evaluation of samples pre- or post-PCR. Nucleic Acids Res 24:2623–2625 10.1093/nar/24.13.2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison DB, Heo M (1998) Meta-analysis of linkage data under worst-case conditions: a demonstration using the human OB region. Genetics 148:859–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood LD, Heard-Costa NL, Cupples LA, Fox CS, Jaquish CE, Wilson PWF, D’Agostino RB (2003) Sex and age dependent effects of chromosomal regions linked to body mass index across 28 years of the Framingham Heart Study. Paper presented at the 53rd Annual Meeting of The American Society of Human Genetics, Los Angeles, California, November 4–8 [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc B 57:289–300 [Google Scholar]
- Borecki IB, Higgins M, Schreiner PJ, Arnett DK, Mayer-Davis E, Hunt SC, Province MA (1998) Evidence for multiple determinants of the body mass index: the National Heart, Lung, and Blood Institute Family Heart Study. Obes Res 6:107–114 [DOI] [PubMed] [Google Scholar]
- Bouchard C, Pérusse L, Rice T, Rao DC (1998) The genetics of human obesity. In: Bray GA, Bouchard C, James WPT (eds) Handbook of obesity. M Dekker, New York, pp 157–190 [Google Scholar]
- Carlsson B, Lindell K, Gabrielsson B, Karlsson C, Bjarnason R, Westphal O, Karlsson U, Sjostrom L, Carlsson LM (1997) Obese (ob) gene defects are rare in human obesity. Obes Res 5:30–35 [DOI] [PubMed] [Google Scholar]
- Chagnon YC, Rankinen T, Snyder EE, Weisnagel SJ, Pérusse L, Bouchard C (2003) The human obesity gene map: the 2002 update. Obes Res 11:313–367 [DOI] [PubMed] [Google Scholar]
- Chagnon YC, Wilmore JH, Borecki IB, Gagnon J, Pérusse L, Chagnon M, Collier GR, Leon AS, Skinner JS, Rao DC, Bouchard C (2000) Leptin receptor gene and adiposity in middle-aged Caucasian males from the HERITAGE family study. J Clin Endocrinol Metab 85:29–34 10.1210/jc.85.1.29 [DOI] [PubMed] [Google Scholar]
- Clayton D (1999) A generalization of the transmission/disequilibrium test for uncertain-haplotype transmission. Am J Hum Genet 65:1170–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton D, Jones H (1999) Transmission/disequilibrium tests for extended marker haplotypes. Am J Hum Genet 65:1161–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comuzzie AG, Blangero J, Mahaney MC, Mitchell BD, Hixson JE, Samollow PB, Stern MP, MacCluer JW (1995) Major gene with sex-specific effects influences fat mass in Mexican Americans. Genet Epidemiol 12:475–488 [DOI] [PubMed] [Google Scholar]
- Considine RV, Considine EL, Williams CJ, Nyce MR, Magosin SA, Bauer TL, Rosato EL, Colberg J, Caro JF (1995) Evidence against either a premature stop codon or the absence of obese gene mRNA in human obesity. J Clin Invest 95:2986–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feitosa MF, Borecki IB, Rich SS, Arnett DK, Sholinsky P, Myers RH, Leppert M, Province MA (2002) Quantitative-trait loci influencing body-mass index reside on chromosomes 7 and 13: the National Heart, Lung, and Blood Institute Family Heart Study. Am J Hum Genet 70:72–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JM, Halaas JL (1998) Leptin and the regulation of body weight in mammals. Nature 395:763–770 10.1038/27376 [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgens J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 10.1126/science.1069424 [DOI] [PubMed] [Google Scholar]
- Gong DW, Bi S, Pratley RE, Weintraub BD (1996) Genomic structure and promoter analysis of the human obese gene. J Biol Chem 271:3971–3974 10.1074/jbc.271.8.3971 [DOI] [PubMed] [Google Scholar]
- Hager J, Clement K, Francke S, Dina C, Raison J, Lahlou N, Rich N, Pelloux V, Basdevant A, Guy-Grand B, North M, Froguel P (1998) A polymorphism in the 5′ untranslated region of the human ob gene is associated with low leptin levels. Int J Obes Relat Metab Disord 22:200–205 10.1038/sj.ijo.0800567 [DOI] [PubMed] [Google Scholar]
- Harris JR, Tambs K, Magnus P (1995) Sex-specific effects for body mass index in the new Norwegian twin panel. Genet Epidemiol 12:251–265 [DOI] [PubMed] [Google Scholar]
- Holland PM, Abramson RD, Watson R, Gelfand DH (1991) Detection of specific polymerase chain reaction products by utilizing the 5′ to 3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA 88:7276–7280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, et al (2000) Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet 26:163–175 10.1038/79876 [DOI] [PubMed] [Google Scholar]
- Horvath S, Xu X, Laird N (2001) The family based association test method: strategies for studying general genotype-phenotype associations. Eur J Hum Genet 9:301–306 10.1038/sj.ejhg.5200625 [DOI] [PubMed] [Google Scholar]
- Johnson GC, Esposito L, Barratt BJ, Smith AN, Heward J, Di Genova G, Ueda H, Cordell HJ, Eaves IA, Dudbridge F, Twells RC, Payne F, Hughes W, Nutland S, Stevens H, Carr P, Tuomilehto-Wolf E, Tuomilehto J, Gough SC, Clayton DG, Todd JA (2001) Haplotype tagging for the identification of common disease genes. Nat Genet 29:233–237 10.1038/ng1001-233 [DOI] [PubMed] [Google Scholar]
- Karvonen MK, Pesonen U, Heinonen P, Laakso M, Rissanen A, Naukkarinen H, Valve R, Uusitupa MI, Koulu M (1998) Identification of new sequence variants in the leptin gene. J Clin Endocrinol Metab 83:3239–3242 10.1210/jc.83.9.3239 [DOI] [PubMed] [Google Scholar]
- Lander ES (1996) The new genomics: global views of biology. Science 274:536–539 10.1126/science.274.5287.536 [DOI] [PubMed] [Google Scholar]
- Le Stunff C, Le Bihan C, Schork NJ, Bougneres P (2000) A common promoter variant of the leptin gene is associated with changes in the relationship between serum leptin and fat mass in obese girls. Diabetes 49:2196–2200 [DOI] [PubMed] [Google Scholar]
- Li WD, Li D, Wang S, Zhang S, Zhao H, Price RA (2003) Linkage and linkage disequilibrium mapping of genes influencing human obesity in chromosome region 7q22.1-7q35. Diabetes 52:1557–1561 [DOI] [PubMed] [Google Scholar]
- Li WD, Reed RD, Lee JH, Xu W, Kilker RL, Sodam BR, Price RA (1999) Sequence variants in the 5′ flanking region of the leptin gene are associated with obesity in women. Ann Hum Genet 63:227–234 10.1046/j.1469-1809.1999.6330227.x [DOI] [PubMed] [Google Scholar]
- Livak KJ, Flood SAJ, Marmaro J, Giusti W, Deetz K (1995) Ligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl 4:357–362 [DOI] [PubMed] [Google Scholar]
- Lucantoni R, Ponti E, Berselli ME, Savia G, Minocci A, Calo G, de Medici C, Liuzzi A, Di Blasio AM (2000) The A19G polymorphism in the 5′ untranslated region of the human obese gene does not affect leptin levels in severely obese patients. J Clin Endocrinol Metab 85:3589–3591 10.1210/jc.85.10.3589 [DOI] [PubMed] [Google Scholar]
- Maffei M, Stoffel M, Barone M, Moon B, Dammerman M, Ravussin E, Bogardus C, et al (1996) Absence of mutations in the human OB gene in obese/diabetic subjects. Diabetes 45:679–682 [DOI] [PubMed] [Google Scholar]
- Mammes O, Betoulle D, Aubert R, Giraud V, Tuzet S, Petiet A, Colas-Linhart N, Fumeron F (1998) Novel polymorphisms in the 5′ region of the LEP gene: association with leptin levels and response to low-calorie diet in human obesity. Diabetes 47:487–489 [DOI] [PubMed] [Google Scholar]
- Mammes O, Betoulle D, Aubert R, Herbeth B, Siest G, Fumeron F (2000) Association of the G-2548A polymorphism in the 5′ region of the LEP gene with overweight. Ann Hum Genet 64:391–394 [DOI] [PubMed] [Google Scholar]
- Montague CT, Farooqi IS, Whitehead JP, Soos MA, Rau H, Wareham NJ, Sewter CP, Digby JE, Mohammed SN, Hurst JA, Cheetham CH, Earley AR, Barnett AH, Prins JB, O’Rahilly S (1997a) Congenital leptin deficiency is associated with severe early-onset obesity in humans. Nature 387:903–908 10.1038/43185 [DOI] [PubMed] [Google Scholar]
- Montague CT, Prins JB, Sanders L, Digby JE, O’Rahilly S (1997b) Depot- and sex-specific differences in human leptin mRNA expression: implications for the control of regional fat distribution. Diabetes 46:342–347 [DOI] [PubMed] [Google Scholar]
- Moon BC, Friedman JM (1997) The molecular basis of the obese mutation in ob 2J mice. Genomics 42:152–156 10.1006/geno.1997.4701 [DOI] [PubMed] [Google Scholar]
- Patil N, Berno AJ, Hinds DA, Barrett WA, Doshi JM, Hacker CR, Kautzer CR, Lee DH, Marjoribanks C, McDonough DP, Nguyen BT, Norris MC, Sheehan JB, Shen N, Stern D, Stokowski RP, Thomas DJ, Trulson MO, Vyas KR, Frazer KA, Fodor SP, Cox DR (2001) Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294:1719–1723 10.1126/science.1065573 [DOI] [PubMed] [Google Scholar]
- Peltonen L, McKusick VA (2001) Genomics and medicine: dissecting human disease in the postgenomic era. Science 291:1224–1229 10.1126/science.291.5507.1224 [DOI] [PubMed] [Google Scholar]
- Platte P, Papanicolaou GJ, Johnston J, Klein CM, Doheny KF, Pugh EW, Roy-Gagnon M-H, Stunkard AJ, Francomano CA, Wilson AF (2003) A study of linkage and association of body mass index in the Old Order Amish. Am J Med Genet 121C:71–80 [DOI] [PubMed] [Google Scholar]
- Qin Z, Niu T, Liu J (2002) Partition-ligation–expectation-maximization algorithm for haplotype inference with single-nucleotide polymorphisms. Am J Hum Genet 71:1242–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux JD, Daly MJ, Silverberg MS, Lindblad K, Steinhart H, Cohen Z, Delmonte T, et al (2001) Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet 29:223–228 10.1038/ng1001-223 [DOI] [PubMed] [Google Scholar]
- Shigemoto M, Nishi S, Ogawa Y, Isse N, Matsuoka N, Tanaka T, Azuma N, Masuzaki H, Nishimura H, Yoshimasa Y, Hosoda K, Nakao K (1997) Molecular screening of both the promoter and the protein coding regions in the human ob gene in Japanese obese subjects with non-insulin-dependent diabetes mellitus. Eur J Endocrinol 137:511–513 [DOI] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, et al (2002) Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet 71:877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, Cesare AJ, Gibberman A, Wang X, O’Neill FA, Walsh D, Kendler KS (2002) Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am J Hum Genet 71:337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel A, Issad T, Camoin L, Ozata M, Strosberg AD (1998) A leptin missense mutation associated with hypogonadism and morbid obesity. Nat Genet 18:213–215 [DOI] [PubMed] [Google Scholar]
- Tena-Sempere M, Manna PR, Zhang FP, Pinilla L, Gonzalez LC, Dieguez C, Huhtaniemi I, Aguilar E (2001) Molecular mechanisms of leptin action in adult rat testis: potential targets for leptin-induced inhibition of steroidogenesis and pattern of leptin receptor messenger ribonucleic acid expression. J Endocrinol 170:413–423 [DOI] [PubMed] [Google Scholar]
- Toma DP, White KP, Hirsch J, Greenspan RJ (2002) Identification of genes involved in Drosophila melanogaster geotaxis, a complex behavioral trait. Nat Genet 31:349–353 [DOI] [PubMed] [Google Scholar]
- Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F (2000) TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res 28:316–319 10.1093/nar/28.1.316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM (1994) Positional cloning of the mouse obese gene and its human homologue. Nature 372: 425–432 10.1038/372425a0 [DOI] [PubMed] [Google Scholar]