Abstract
The hypomyelinating leukodystrophies X-linked Pelizaeus-Merzbacher disease (PMD) and Pelizaeus-Merzbacher–like disease (PMLD) are characterized by nystagmus, progressive spasticity, and ataxia. In a consanguineous family with PMLD, we performed a genomewide linkage scan using the GeneChip Mapping EA 10K Array (Affymetrix) and detected a single gene locus on chromosome 1q41-q42. This region harbors the GJA12 gene, which encodes gap junction protein α12 (or connexin 46.6). Gap junction proteins assemble into intercellular channels through which signaling ions and small molecules are exchanged. GJA12 is highly expressed in oligodendrocytes, and, therefore, it serves as an excellent candidate for hypomyelination in PMLD. In three of six families with PMLD, we detected five different GJA12 mutations, including missense, nonsense, and frameshift mutations. We thereby confirm previous assumptions that PMLD is genetically heterogeneous. Although the murine Gja12 ortholog is not expressed in sciatic nerve, we did detect GJA12 transcripts in human sciatic and sural nerve tissue by reverse-transcriptase polymerase chain reaction. These results are in accordance with the electrophysiological finding of reduced motor and sensory nerve conduction velocities in patients with PMLD, which argues for a demyelinating neuropathy. In this study, we demonstrate that GJA12 plays a key role in central myelination and is involved in peripheral myelination in humans.
Introduction
The hypomyelinating X-linked leukodystrophy Pelizaeus-Merzbacher disease (PMD [MIM #312080]) (fig. 1) is characterized by nystagmus, impaired motor development, ataxia, choreoathetotic movements, dysarthria, and progressive spasticity. The disease is caused by mutations in the PLP1 gene, which encodes proteolipid protein 1. PLP1 is the major component of myelin in the CNS and is also expressed in myelin of the peripheral nervous system (for review, see Koeppen and Robitaille [2002] and Hudson [2003]). Patients with the PMD phenotype but without mutations or duplications of the PLP1 gene are considered to have Pelizaeus-Merzbacher–like disease (PMLD [MIM 311601]). No gene locus has been described for this disease entity.
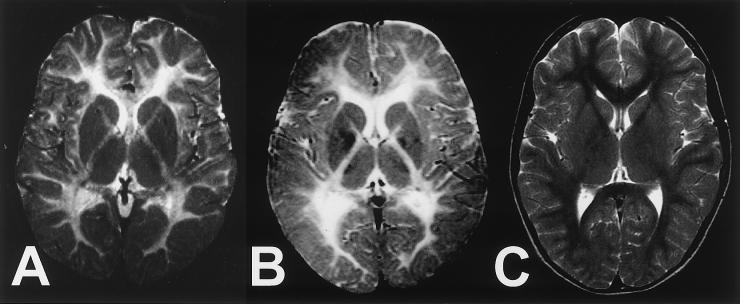
Figure 1.
Axial T2-weighted magnetic resonance images of the brain at the level of the basal ganglia. A, Patient with PMLD at 6 years of age (patient III:3; fig. 3). B, Patient with PMD at 7 years of age. The patients in panels A and B show nearly identical hypomyelination patterns of central white matter, as indicated by diffusely enhanced signal intensity. C, Low signal of normal myelination in an unaffected child.
Gap junction proteins are members of a large family of homologous connexins and comprise four transmembrane, two extracellular, and three cytoplasmic domains (fig. 2) (Willecke et al. 2002; Nagy and Rash 2003; Nagy et al. 2003). They have been identified in a broad range of mammalian tissues, and most tissues express more than one species of connexin protein. The formation of homo- or heteromeric hemichannels (or connexons) by six connexins permits a high level of diversity in channel composition. Two connexons span the plasma membrane of closely apposed cells and align to intercellular channels, thus allowing for the exchange of small molecules, including second messengers and ions. Gap junctional intercellular communication fulfils multiple functions to meet the specific needs of tissues. Several neurological diseases, such as oculodentodigital dysplasia (Paznekas et al. 2003) and heterogeneous sensorineural deafness (Rouan et al. 2001), are associated with mutations of genes that encode single members of the gap junction protein family.
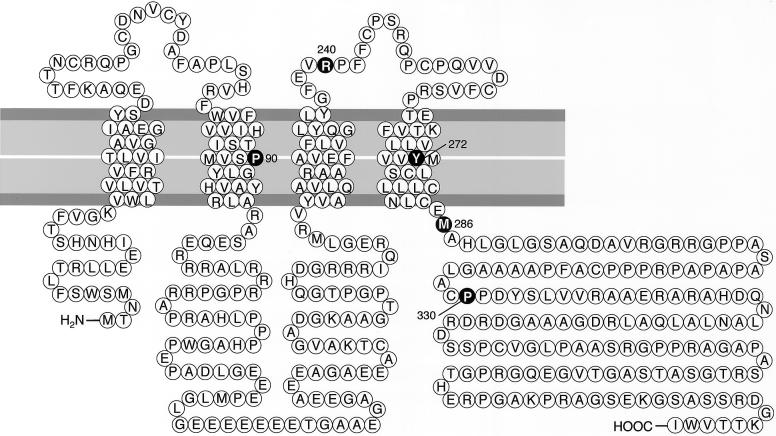
Figure 2.
Domains of the GJA12 gene product. Blackened circles depict the locations of mutations. The missense mutations lie within the second (P90S) and fourth (Y272D) transmembrane and third cytoplasmic (M286T) domains.
For efficient conduction, axons are surrounded by multilamellar myelin membranes that are synthesized by oligodendrocytes and Schwann cells (for review, see Garbern et al. [1997]). Gap junction protein β1 (GJB1, or connexin 32) is crucial for peripheral myelination, and GJB1 mutations lead to X-linked demyelinating peripheral neuropathy Charcot-Marie-Tooth type 1 (CMTX1 [MIM #302800]) (Bergoffen et al. 1993). Similar to PLP1, GJB1 is expressed in both the peripheral and the central nervous system. In consequence, it is not surprising that patients with CMTX1 and specific GJB1 mutations have both peripheral neuropathy and a mild or transient brain disorder (Paulson et al. 2002; Hanemann et al. 2003; Takashima et al. 2003). Vice versa, a few patients with PMD who have specific PLP1 mutations have demyelinating peripheral neuropathy in addition to their hypomyelinating leukodystrophy (Garbern et al. 1997). Here, we report that mutations of the gap junction protein α12 gene (GJA12, or connexin 46.6) are associated with one type of PMLD and that this type is also accompanied by a mild peripheral neuropathy.
Patients and Methods
Families with PMLD and Collection of DNA
We collected blood samples from patients and family members after obtaining written informed consent, and we isolated DNA from peripheral blood lymphocytes according to standard procedures. The study was approved by the Ethics Review Board at the Charité in Berlin and collaborating institutions, according to the Declaration of Helsinki. We studied a total of nine patients and 20 relatives from six unrelated families. The diagnosis of PMLD was made on the basis of clinical criteria and magnetic-resonance-imaging findings (Cailloux et al. 2000; Schiffmann and Boespflug-Tanguy 2001; Koeppen and Robitaille 2002; Hudson 2003; Plecko et al. 2003). The sensory- and motor-nerve conduction velocities (SCVs and MCVs, respectively) were investigated in patients (table 1) and in heterozygous family members II:2, II:3, and III:2 of family 1 (fig. 3). In all patients with PMLD, PMD was ruled out by mutational analysis of the PLP1 coding region, the flanking intron sequences, and its promoters. Duplications of PLP1 were ruled out by Southern blot or real-time PCR. In addition, we found different haplotypes at the PLP1 gene locus in the two affected brothers (III:1 and III:3; fig. 3) of index family 1 (data not shown). This finding excludes a causative role of PLP1 in this family.
Table 1.
Mutations of GJA12 (or CX46.6) and Clinical Data of Patients with PMLD
|
MCV and SCV, in m/s (Age-Corrected Minimal Normal Value; Age)c |
|||||||||||
| Median Nerve |
Tibial Nerve |
||||||||||
| Family(Individual) | Mutation (Amino Acid Substitution; Class of Mutation) | Age at Onset of Nystagmus(wk) | Age at Unaided Sittinga(mo) | Age at Unaided Walkingb(years) | Age Wheelchair Bound(years) | Type of Seizures (Age at Onset, in Years) | MCV | SCV | MCV | SCV | Peroneal Nerve MCV |
| 1 (III:1) | c.857T→C (M286T; missense) | 7 | 15 | Not achieved | 5.5 | Complex partial (8) | 59 (51; 21 years) | 53 (40; 21 years) | 38 (43; 21 years) | 34 (41; 21 years) | 41 (49; 21 years) |
| 1 (III:3) | 7 | 6 | Not achieved | 5.5 | Simple partial (12) | 39 (51; 14 years) | 45 (40; 14 years) | 36 (43; 14 years) | ND | 37 (48; 14 years) | |
| 1 (III:4) | 12 | 12 | 5 | 6.5 | Complex partial (7) | 67 (51; 18 years) | 64 (40; 18 years) | ND | 40 (44; 14 years)d | 48 (49; 18 years) | |
| 2 | c.268C→T (P90S; missense) c.989delC (frameshift) | Congenital | 8 | Not achieved | 6 | Complex partial (7) | ND | ND | 42 (43; 10 years) | 68 (44; 10 years) | ND |
| 3 | c.718C→T (R240X; nonsense) c.814T→G (Y272D; missense) | Congenital | Not achieved | Not achieved | 1 | No seizures | ND | ND | 37 (30; 8 mo) | ND | ND |
Normal age is 7–8 mo.
Normal age is 13–15 mo.
ND = not determined.
SCV is for the sural nerve.
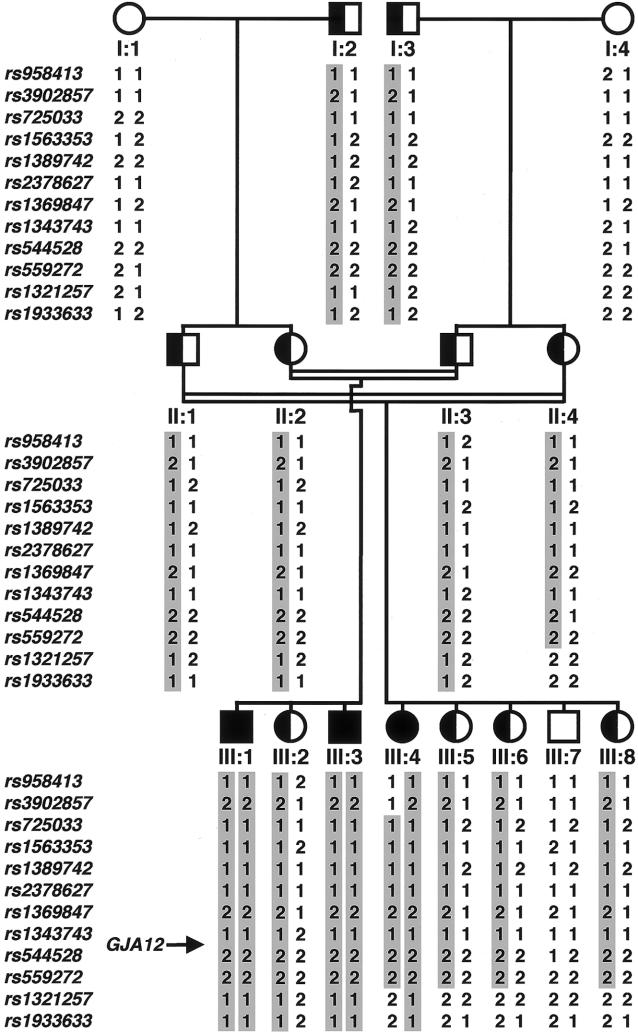
Figure 3.
Haplotypes in family 1 with PMLD. Haplotype analysis indicated that the cosegregating segment of the PMLD locus is flanked proximally by marker rs3902857 and distally by marker rs1321257 on chromosome 1q41-q42.
Genome Scan and Haplotype Analysis
For the whole-genome scan, we used the GeneChip Mapping EA 10K Array (Affymetrix) according to the guidelines of the manufacturer. This early-access version of the Mapping 10K comprised a total of 10,043 SNPs. The mean intermarker distance was 250 kb, equivalent to 0.34 cM. Individuals I:1–I:4, II:1–II:4, and III:1–III:8 of family 1 were genotyped (fig. 3). The following 12 informative SNP markers were used for haplotype reconstruction and analysis: rs958413, rs3902857, rs725033, rs1563353, rs1389742, rs2378627, rs1369847, rs1343743, rs544528, rs559272, rs1321257, and rs1933633 (dbSNP Home Page). PedCheck was used for detection of Mendelian errors (O’Connell and Weeks 1998). Non-Mendelian errors were identified by MERLIN (Abecasis et al. 2002). Parametric linkage and haplotype analysis were performed by a modified version of GENEHUNTER (Strauch et al. 2000), through use of a sliding window with sets of 50 SNPs. All physical positions are derived from National Center for Biotechnology Information build 34, July 2003.
Sequence Analysis
We designed a set of three primer pairs to amplify the single exon of GJA12 (GenBank accession number NM_020435) and its flanking sequences from genomic DNA (GenBank accession number NT_004559): 1F (5′ TTT AAG GCG GTA AGC TCC AC 3′) and 1R (5′ CAG CAT GGG CTC CTC CTC 3′), 2F (5′ CTG CGA CAA CGT CTG CTA TG 3′) and 2R (5′ GCC ATC TCA CAG AGG TTG AG 3′), and 3F (5′ CCG ACC GGG CAA CAC GAT G 3′) and 3R (5′ GAG TCT GCC TGA GGC CAC CG 3′). For GC-rich fragments, 1 M betaine was used as an additive. For amplification of the coding region of GJB1 (GenBank accession number NM_000166) and its flanking sequences from genomic DNA (GenBank accession number NT_011669), we used the following oligonucleotides: 4F (5′ TGA CCA TCC TTC CTT TCC TG 3′), 4R (5′ ACA TGA AGA CGG CCT CAA AC 3′), 5F (5′ GGC TCA CCA GCA ACA CAT AG 3′), and 5R (5′ AGT AGC CAG GGA AGG AAG GT 5′).
PCR products were directly sequenced using an ABI PRISM 3730 DNA Analyzer and BigDye Terminator Cycle Sequencing Kit version 1.1, according to the protocol of the manufacturer (Applied Biosystems).
We verified the intrafamilial segregation of all mutations by RFLP analysis (fig. 4). We used primer-induced restriction analysis for all mutations without natural restriction sites (the size [in bp] of the fragments yielded after restriction with each endonuclease is given; primer mismatches are underlined): c.268C→T: primers 2F and 1R, HpyCh4IV, mutant = 9 + 33 + 38 + 179 bp, wild type = 9 + 33 + 217 bp; c.718C→T: primers 3F and 3R, MaeIII, mutant = 716 + 137 bp, wild type = 853 bp; c.814T→G: primers 3F and 3R, RsaI, mutant = 9 + 57 + 58 + 729 bp, wild type = 9 + 57 + 58 + 111 + 618 bp; c.857T→C: primers 6F (5′ GTA CCT GCT GTA CGG CTT CG 3′) and 6R (5′ GCT GCC CAA GCC CAG GTC GGC C 3′), EagI, mutant = 166 + 21 bp, wild type = 187 bp; and c.989delC: primers 3F and 7R (5′ CGC ACC ACC AGG CTG TAG TCG GCC G 3′), SfiI, mutant = 108 + 326 bp, wild type = 25 + 83 + 326 bp. Primer sequences of TGFB2, MARK1, CAPN2, DEGS, ENAH, and IMAGE3451454 are available from the authors on request.
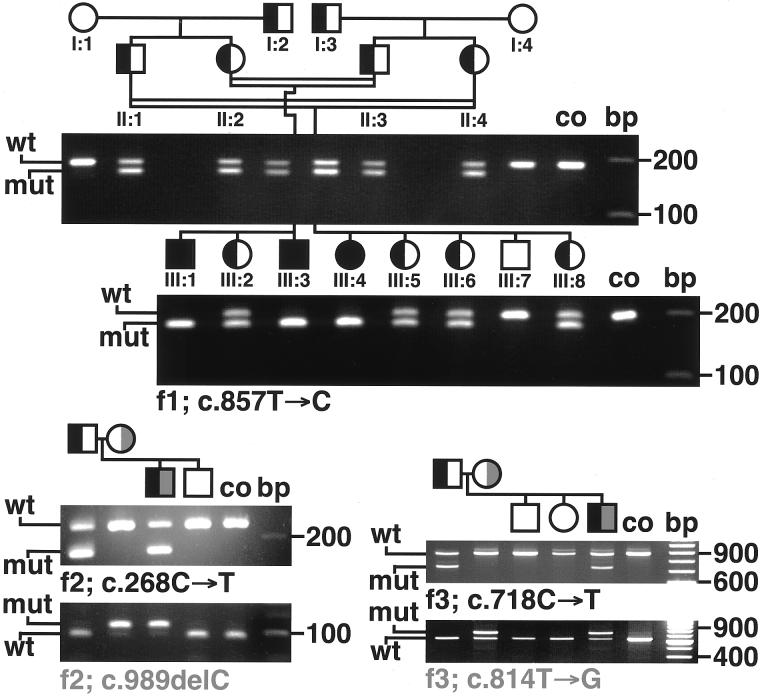
Figure 4.
Segregation of GJA12 mutations (RFLP analysis). In family 2 (f2), the affected boy was compound heterozygous, carrying the paternal c.268C→T missense mutation and the maternal c.989delC frameshift deletion. In family 3 (f3), the affected boy carried the paternal c.718C→T nonsense mutation and the maternal c.814T→G missense mutation. Family numbers correspond to those in table 1. co = control; f1–f3 = families 1–3; mut = mutated GJA12; wt = wild-type GJA12.
RT-PCR
Total RNA was extracted from fresh frozen sural and sciatic nerve specimens from adult healthy individuals by the TRIzol reagent (Invitrogen). RNA from adult brain, spinal cord, and skeletal muscle was purchased from CLONTECH. Integrity of RNA was verified on an agarose gel by visualization of the 5S, 18S, and 28S rRNA bands. Two micrograms of total RNA were subjected to reverse transcription by use of the RevertAid First Strand cDNA Synthesis Kit (Fermentas). Equal loading of template was verified by simultaneous amplification of ACTB as an internal standard. GJA12 was amplified by primers 2F and 2R; for GJB1, we used primers 8F (5′ ACT CCC CCT GCA CAG ACA T 3′) and 8R (5′ TCT CAT CAC CCC ACA CAC TC 3′). To exclude the provenience of the PCR bands from genomic DNA, we performed control-PCR runs with the above mentioned primers on RT-reaction products in which reverse transcriptase had been omitted. None of the reactions yielded a band. For a relative estimate of the amounts of cDNA, we interrupted amplification of GJA12 and GJB1 after 28, 34, and 40 cycles.
Protein Alignment
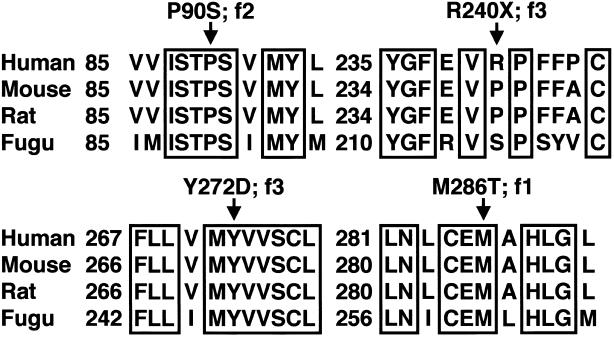
We performed multiple alignments (fig. 5) (Gja12_fugu [GenBank accession number CAAB01002259.1], GJA12_human [GenBank accession number NP_065168], Gja12_mouse [UniProt TrEMBL accession number Q9EPM1], and Gja12_rat [UniProt TrEMBL accession number Q80XF7]) with the CLUSTAL W program, version 1.74 (Thompson et al. 1994).
Figure 5.
Alignment of selected regions of human GJA12 with orthologs of other species, including the Japanese pufferfish Fugu rubripes. Arrows indicate positions of the missense and nonsense mutations in patients with PMLD. Family numbers correspond to those in table 1. f1–f3 = families 1–3.
Results
Phenotypic Features
All patients with PMLD with GJA12 mutations (families 1, 2, and 3) showed the characteristic clinical symptoms of the hypomyelinating leukodystrophies PMD and PMLD (fig. 1), such as nystagmus, impaired motor development, ataxia, choreoathetotic movements, dysarthria, and progressive spasticity (Nezu et al. 1996; Lazzarini et al. 1997; Cailloux et al. 2000; Schiffmann and Boespflug-Tanguy 2001; Koeppen and Robitaille 2002; Hudson 2003; Plecko et al. 2003). Nystagmus and poor head and trunk control were the presenting symptoms in early infancy (table 1). By the age of 8–15 mo, an impaired motor development became apparent when developmental milestones like unaided sitting and/or walking were delayed or could not be achieved. Only one patient was able to walk a few steps at the age of 5 years. All patients experienced facial weakness. In addition, the patients in families 1 and 2 had focal epileptic seizures prior to adolescence (table 1). In addition to the involvement of the CNS, PMLD associated with GJA12 mutations seems to be accompanied by a mild peripheral neuropathy. MCVs and SCVs of the lower limb nerves were reduced or slightly below the age-corrected normal values in most patients (table 1). Nerve conduction velocities of the median nerve, available only in members of family 1, were reduced only in patient III:3. Heterozygous individuals had no neurologic symptoms. In contrast to the cranial magnetic resonance imaging (cMRI) of all patients, the cMRIs of two heterozygous healthy members of family 1 (II:2 and II:3; fig. 3) did not reveal any abnormality (data not shown).
The phenotype of the patients with PMLD without GJA12 mutations was not clearly different from that of patients with GJA12 mutations. However, two of four patients without GJA12 mutations did not present with nystagmus in infancy.
PMLD Locus on Chromosome 1q41-q42
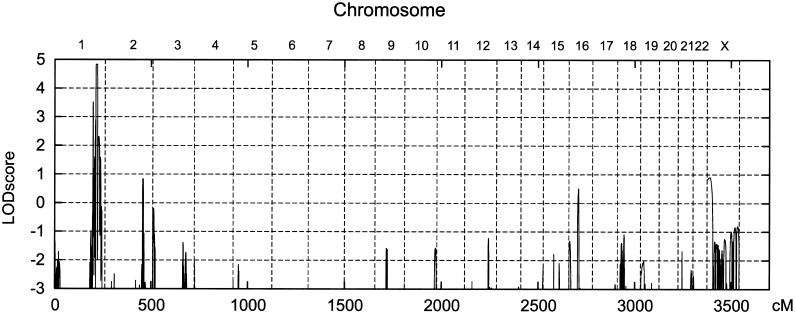
Genomewide linkage scanning revealed linkage of PMLD to SNPs on chromosome 1q41-q42. Multipoint linkage analysis yielded a maximum LOD score of 4.83 for SNP markers rs1563353–rs544528. No additional peak with a LOD score value >1 was found in the genome (fig. A1 of the appendix [online only]). Haplotype analysis disclosed recombination events in individual III:4 of family 1 that confined the candidate locus to a region distal to SNP rs3902857 (physical map position 215.5 Mb) and proximal to SNP rs1321257 (physical map position 227.3 Mb) (fig. 3). Consistent with parental consanguinity, the affected siblings III:1 and III:3 and the affected cousin III:4 were autozygous for all SNPs within the cosegregating segment. The expression of Gja12 in murine oligodendrocytes (Menichella et al. 2003; Odermatt et al. 2003) led to the singling out of GJA12 (physical map position 225.3 Mb) as one of the most promising candidate genes. Mutations in the coding region and its flanking intronic sequences of TGFB2, MARK1, CAPN2, DEGS, ENAH, and IMAGE3451454 were ruled out by sequence analysis.
PMLD Gene GJA12
GJA12 is composed of a single exon. We analyzed DNA samples from six families with PMLD. In one Turkish consanguineous (family 1; fig. 3) and two German nonconsanguineous families, we identified five different GJA12 mutations (fig. 2 and table 1). We could not find GJA12 mutations in three other affected families. As expected, patients from the consanguineous family displayed a homozygous mutation, and patients from the nonconsanguineous families were compound heterozygous for the GJA12 mutations. A homozygous c.857T→C transition predicts a substitution of threonine for methionine in family 1 (M286T). A heterozygous c.268C→T transition (paternal allele) predicts a substitution of serine for proline (P90S), and a heterozygous c.989delC 1-bp deletion (maternal allele) leads to a frameshift and a nonsense peptide of 141 amino acids after amino acid 329 (cysteine) in family 2. A heterozygous c.718C→T transition (paternal allele) represents a nonsense mutation (R240X), and a heterozygous c.814T→G transversion (maternal allele) leads to the replacement of tyrosine by aspartic acid (Y272D) in family 3.
Several findings supported the hypothesis that mutations in GJA12 are the primary cause for a subgroup of PMLD and are consistent with an autosomal recessive mode of inheritance: (i) no missense mutations were detected in 220 alleles of 110 unaffected individuals, which rules out common polymorphisms; (ii) GJA12 mutations segregated with the disease phenotype in all families (fig. 4); and (iii) all residues affected by the three missense mutations are highly conserved (fig. 5).
A silent c.594C→T GJA12 polymorphism was found in families 3 and 6 and in 3 of 20 healthy individuals. Furthermore, since Gja12 and Gjb1 reveal a high level of functional redundancy in murine oligodendrocytes (Menichella et al. 2003; Odermatt et al. 2003), we analyzed not only GJA12 but also GJB1. We did not find mutations in the coding region of GJB1 or in its flanking intronic sequences in any of the six families with PMLD.
Expression of GJA12 in the Central and Peripheral Nervous System
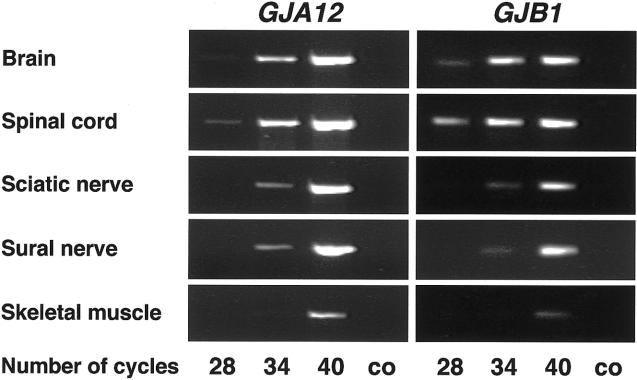
To further investigate the reduced MCVs in two patients with PMLD (table 1), we examined the GJA12 expression in comparison with that of GJB1 by RT-PCR analysis in human sciatic and sural nerve specimens. GJA12 cDNA could be amplified from sciatic and sural nerves of healthy adults (fig. 6). Intensities of PCR products were compared after various numbers of PCR cycles, to estimate relative amounts of GJA12 and GJB1 transcripts. Although GJA12 and GJB1 bands both appeared after 28 cycles in brain and spinal cord, they were not detectable in sciatic and sural nerve before 34 cycles or in skeletal muscle before 40 cycles (fig. 6). Thus, the highest GJA12 and GJB1 transcript numbers were found in brain and spinal cord, and the lowest were found in skeletal muscle.
Figure 6.
PCR products of GJA12 and GJB1 cDNA after various numbers of PCR cycles in human brain, spinal cord, sciatic and sural nerve, and skeletal muscle. GJA12 and GJB1 are more highly expressed in brain and spinal cord than in peripheral nerve tissue. Untranscribed RNA was used as a control (co).
Discussion
In this study, we demonstrate that mutations of the GJA12 gene are associated with one form of autosomal recessive PMLD. In three of six affected families with a similar phenotype, we could not find GJA12 mutations, a finding that complies with previous reports that PMLD is a group of genetically heterogeneous diseases (Lazzarini et al. 1997; Schiffmann and Boespflug-Tanguy 2001). In addition to the characteristic clinical symptoms, patients with PMLD with GJA12 mutations had peripheral neuropathy and seizures, both of which occur only sporadically in PMD (Garbern et al. 1997; Cailloux et al. 2000; Koeppen and Robitaille 2002).
Some patients showed reduced nerve conduction velocities, which indicate the presence of a mild peripheral demyelinating motor neuropathy, predominantly of the lower limbs. This complies with our finding that GJA12 is expressed not only in the CNS but also in sural and sciatic nerve tissue. However, in the murine sciatic nerve, Gja12 was not found to be expressed, as demonstrated by the absence of fluorescence from the EGFP reporter gene, which replaced the Gja12 coding DNA (Odermatt et al. 2003), as well as by northern blot analysis of the sciatic nerve in wild-type mice (Teubner et al. 2001). Amplification of small amounts of GJA12 and GJB1 from human skeletal muscle RNA might be explained by amplification from muscle tissue itself or—more likely—from small amounts of nerve fibers within skeletal muscle. Furthermore, seizures were a frequent finding in our subgroup of patients with PMLD. This observation supports the hypothesis of Samoilova et al. (2003) that gap junctional communication plays an important role in the incidence of seizures. The finding that GJA12 mutations are associated with one subgroup of PMLD provokes questions concerning those cellular mechanisms that lead to the phenotype of hypomyelinating leukodystrophy. Our data and the studies of other groups suggest mechanisms other than merely a loss of GJA12 function.
Gja12-knockout mice are completely Gja12-deficient but clinically normal (Odermatt et al. 2003). In contrast, our patients are affected by missense mutations on one or both alleles and therefore should synthesize mutant GJA12 protein. It has been shown that mutant gap junction proteins may become toxic. For instance, the GJB1 mutant S85C forms functional cell-cell channels, and its hemichannels show an increased opening (Abrams et al. 2002). The GJA12 missense mutation P90S of family 2 (table 1) is located close to the CMTX1-associated S85C missense mutation of the paralogous GJB1. If, in fact, GJA12 mutant proteins became toxic in PMLD, a gain of toxic function might depend on mutant gene dosage, since all heterozygous members of families 1–3 were healthy.
In coexpression studies, it has been shown that mutant gap junction proteins can inhibit functions of other wild-type connexins. Most cell types express more than one gap junction protein species and form gap junctions by single or different protein species (Rouan et al. 2001). Similar to Gja12 and Gjb1 in murine oligodendrocytes (Menichella et al. 2003), GJB2 (or connexin 26) and GJA1 (or connexin 43) colocalize in human hyperkeratotic skin (Rouan et al. 2001). In coexpression studies in Xenopus oocytes, GJB2 mutants partially blocked the function of wild-type GJA1. Similar negative effects have been shown by Gja1 mutants on wild-type Gjb1 (Lagree et al. 2003). The use of site-directed mutagenesis of GJA12 and GJB1 for cotransfection in cellular systems will help to clarify the pathogenicity of each connexin mutant.
Both Gjb1 and Gja12 are functionally redundant in murine oligodendrocytes (Menichella et al. 2003; Odermatt et al. 2003). Although Gja12 or Gjb1 null mutants show no clinical abnormalities, mice lacking both Gja12 and Gjb1 develop severe oligodendrocyte death and present with tremor and tonic seizures (Scherer et al. 1998; Menichella et al. 2003; Odermatt et al. 2003). For this reason, we carefully ruled out mutations in the coding region of GJB1 in all families with PMLD. Under the hypothesis that GJB1 can compensate for the loss of GJA12 not only in mice (Menichella et al. 2003; Odermatt et al. 2003) but also in humans, mechanisms other than loss of GJA12 function should be proposed for our patients with PMLD.
GJB1 mutations lead to demyelinating peripheral neuropathy CMTX1 (Bergoffen et al. 1993; Lin et al. 1999; Hahn et al. 2000). Patients with CMTX1 and specific GJB1 mutations also present with CNS symptoms (Paulson et al. 2002; Hanemann et al. 2003; Takashima et al. 2003) similar to those seen in PMLD. This is in contrast to patients with deletions of the entire GJB1 coding sequence who are not affected by brain disorders (Kleopa et al. 2002; Hanemann et al. 2003). These data suggest that other gap junction proteins may compensate for a loss of central GJB1 function and that distinct GJB1 mutants may have negative effects on oligodendrocytes in humans. We assume that the pathomechanisms in GJA12-related PMLD are similar.
GJA12 seems to be more important for oligodendrocyte homeostasis than GJB1. In fact, the central symptoms in CMTX1 (Paulson et al. 2002; Hanemann et al. 2003; Takashima et al. 2003) are transient or milder than in PMLD, and, in contrast to Gja12-knockout animals, which show subtle ultrastructural myelin abnormalities in the optic nerve system (Odermatt et al. 2003), Gjb1-knockout mice do not display pathological changes of myelination in optic nerves (Scherer et al. 1998).
In summary, mutations of GJA12 are associated with one form of autosomal recessive PMLD. Since Gjb1 and Gja12 are functionally redundant in mice, we favor the hypothesis that the missense mutants found in our patients with PMLD display toxic gain of function in oligodendrocytes, as specific GJB1 mutants may do with regard to central functions of GJA12 in CMTX1.
Acknowledgments
The authors thank the patients and their families for participation in the study. Help, provision of tissue samples, stimulating discussions, and critical comments from C. Bassir, C. Becker, W. Brück, S. Cirak, C. Janetzki, B. Lucke, I. Scheer, V. Schneider, J. Senderek, S. Stöckler-Ipsiroglu, H. Witt, and A. Zwirner are gratefully acknowledged. This study has been supported by grants from the “Rahel-Hirsch-Stiftung,” Charité (to B.U.); the German National Genome Research Network (to F.R. and P.N.); and the “Deutsche Forschungsgemeinschaft” (to C.H.); as well as by a Heinrich Heine University Faculty of Medicine research grant (to J.G.).
Appendix
Figure A1.
Genome scan using the GeneChip Mapping EA 10K Array (Affymetrix). Vertical bars separate the chromosomes. The scanning revealed linkage of PMLD to SNPs on chromosome 1q41-q42. Multipoint linkage analysis yielded a maximum LOD score of 4.83 for SNP markers rs1563353–rs544528.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- dbSNP Home Page, http://www.ncbi.nlm.nih.gov/SNP/ (for refSNP ID rs958413, rs3902857, rs725033, rs1563353, rs1389742, rs2378627, rs1369847, rs1343743, rs544528, rs559272, rs1321257, and rs1933633)
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for human GJA12 mRNA [accession number NM_020435], human GJA12 genomic sequence [accession number NT_004559], mouse Gja12 mRNA [accession number NM_080454], Gja12_fugu [accession number CAAB01002259.1], human GJB1 mRNA [accession number NM_000166], human GJB1 genomic sequence [accession number NT_011669])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for CMTX1 [MIM #302800], PMD [MIM #312080], PMLD [MIM 311601])
- UniProt/TrEMBL, http://www.ebi.ac.uk/trembl/ (for Gja12_mouse [accession number Q9EPM1] and Gja12_rat [accession number Q80XF7])
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- Abrams CK, Bennett MV, Verselis VK, Bargiello TA (2002) Voltage opens unopposed gap junction hemichannels formed by a connexin 32 mutant associated with X-linked Charcot-Marie-Tooth disease. Proc Natl Acad Sci USA 99:3980–3984 10.1073/pnas.261713499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergoffen J, Scherer SS, Wang S, Scott MO, Bone LJ, Paul DL, Chen K, Lensch MW, Chance PF, Fischbeck KH (1993) Connexin mutations in X-linked Charcot-Marie-Tooth disease. Science 262:2039–2042 [DOI] [PubMed] [Google Scholar]
- Cailloux F, Gauthier-Barichard F, Mimault C, Isabelle V, Courtois V, Giraud G, Dastugue B, Boespflug-Tanguy O (2000) Genotype-phenotype correlation in inherited brain myelination defects due to proteolipid protein gene mutations. Eur J Hum Genet 8:837–845 10.1038/sj.ejhg.5200537 [DOI] [PubMed] [Google Scholar]
- Garbern JY, Cambi F, Tang XM, Sima AA, Vallat JM, Bosch EP, Lewis R, Shy M, Sohi J, Kraft G, Chen KL, Joshi I, Leonard DG, Johnson W, Raskind W, Dlouhy SR, Pratt V, Hodes ME, Bird T, Kamholz J (1997) Proteolipid protein is necessary in peripheral as well as central myelin. Neuron 19:205–218 10.1016/S0896-6273(00)80360-8 [DOI] [PubMed] [Google Scholar]
- Hahn AF, Ainsworth PJ, Naus CC, Mao J, Bolton CF (2000) Clinical and pathological observations in men lacking the gap junction protein connexin 32. Muscle Nerve Suppl 9:S39–S48 [DOI] [PubMed] [Google Scholar]
- Hanemann CO, Bergmann C, Senderek J, Zerres K, Sperfeld AD (2003) Transient, recurrent, white matter lesions in X-linked Charcot-Marie-Tooth disease with novel connexin 32 mutation. Arch Neurol 60:605–609 10.1001/archneur.60.4.605 [DOI] [PubMed] [Google Scholar]
- Hudson LD (2003) Pelizaeus-Merzbacher disease and spastic paraplegia type 2: two faces of myelin loss from mutations in the same gene. J Child Neurol 18:616–624 [DOI] [PubMed] [Google Scholar]
- Kleopa KA, Yum SW, Scherer SS (2002) Cellular mechanisms of connexin32 mutations associated with CNS manifestations. J Neurosci Res 68:522–534 10.1002/jnr.10255 [DOI] [PubMed] [Google Scholar]
- Koeppen AH, Robitaille Y (2002) Pelizaeus-Merzbacher disease. J Neuropath Exp Neur 61:747–759 [DOI] [PubMed] [Google Scholar]
- Lagree V, Brunschwig K, Lopez P, Gilula NB, Richard G, Falk MM (2003) Specific amino-acid residues in the N-terminus and TM3 implicated in channel function and oligomerization compatibility of connexin43. J Cell Sci 116:3189–3201 10.1242/jcs.00604 [DOI] [PubMed] [Google Scholar]
- Lazzarini A, Schwarz KO, Jiang S, Stenroos ES, Lehner T, Johnson WG (1997) Pelizaeus-Merzbacher-like disease: exclusion of the proteolipid protein locus and documentation of a new locus on Xq. Neurology 49:824–832 [DOI] [PubMed] [Google Scholar]
- Lin C, Numakura C, Ikegami T, Shizuka M, Shoji M, Nicholson G, Hayasaka K (1999) Deletion and nonsense mutations of the connexin 32 gene associated with Charcot-Marie-Tooth disease. Tohoku J Exp Med 188:239–244 [DOI] [PubMed] [Google Scholar]
- Menichella DM, Goodenough DA, Sirkowski E, Scherer SS, Paul DL (2003) Connexins are critical for normal myelination in the CNS. J Neurosci 23:5963–5973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy JI, Ionescu AV, Lynn BD, Rash JE (2003) Coupling of astrocyte connexins Cx26, Cx30, Cx43 to oligodendrocyte Cx29, Cx32, Cx47: implications from normal and connexin32 knockout mice. Glia 44:205–218 10.1002/glia.10278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy JI, Rash JE (2003) Astrocyte and oligodendrocyte connexins of the glial syncytium in relation to astrocyte anatomical domains and spatial buffering. Cell Commun Adhes 10:401–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezu A, Kimura S, Uehara S, Osaka H, Kobayashi T, Haraguchi M, Inoue K, Kawanishi C (1996) Pelizaeus-Merzbacher-like disease: female case report. Brain Dev 18:114–118 10.1016/0387-7604(95)00078-X [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odermatt B, Wellershaus K, Wallraff A, Seifert G, Degen J, Euwens C, Fuss B, Bussow H, Schilling K, Steinhauser C, Willecke K (2003) Connexin 47 (Cx47)-deficient mice with enhanced green fluorescent protein reporter gene reveal predominant oligodendrocytic expression of Cx47 and display vacuolized myelin in the CNS. J Neurosci 23:4549–4559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson HL, Garbern JY, Hoban TF, Krajewski KM, Lewis RA, Fischbeck KH, Grossman RI, Lenkinski R, Kamholz JA, Shy ME (2002) Transient central nervous system white matter abnormality in X-linked Charcot-Marie-Tooth disease. Ann Neurol 52:429–434 10.1002/ana.10305 [DOI] [PubMed] [Google Scholar]
- Paznekas WA, Boyadjiev SA, Shapiro RE, Daniels O, Wollnik B, Keegan CE, Innis JW, Dinulos MB, Christian C, Hannibal MC, Jabs EW (2003) Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am J Hum Genet 72:408–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plecko B, Stockler-Ipsiroglu S, Gruber S, Mlynarik V, Moser E, Simbrunner J, Ebner F, Bernert G, Harrer G, Gal A, Prayer D (2003) Degree of hypomyelination and magnetic resonance spectroscopy findings in patients with Pelizaeus Merzbacher phenotype. Neuropediatrics 34:127–136 10.1055/s-2003-41276 [DOI] [PubMed] [Google Scholar]
- Rouan F, White TW, Brown N, Taylor AM, Lucke TW, Paul DL, Munro CS, Uitto J, Hodgins MB, Richard G (2001) Trans-dominant inhibition of connexin-43 by mutant connexin-26: implications for dominant connexin disorders affecting epidermal differentiation. J Cell Sci 114:2105–2113 [DOI] [PubMed] [Google Scholar]
- Samoilova M, Li J, Pelletier MR, Wentlandt K, Adamchik Y, Naus CC, Carlen PL (2003) Epileptiform activity in hippocampal slice cultures exposed chronically to bicuculline: increased gap junctional function and expression. J Neurochem 86:687–699 10.1046/j.1471-4159.2003.01893.x [DOI] [PubMed] [Google Scholar]
- Scherer SS, Xu YT, Nelles E, Fischbeck K, Willecke K, Bone LJ (1998) Connexin32-null mice develop demyelinating peripheral neuropathy. Glia 24:8–20 [DOI] [PubMed] [Google Scholar]
- Schiffmann R, Boespflug-Tanguy O (2001) An update on the leukodystrophies. Curr Opin Neurol 14:789–794 10.1097/00019052-200112000-00018 [DOI] [PubMed] [Google Scholar]
- Strauch K, Fimmers R, Kurz T, Deichmann KA, Wienker TF, Baur MP (2000) Parametric and nonparametric multipoint linkage analysis with imprinting and two-locus-trait models: application to mite sensitization. Am J Hum Genet 66:1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima H, Nakagawa M, Umehara F, Hirata K, Suehara M, Mayumi H, Yoshishige K, Matsuyama W, Saito M, Jonosono M, Arimura K, Osame M (2003) Gap junction protein beta 1 (GJB1) mutations and central nervous system symptoms in X-linked Charcot-Marie-Tooth disease. Acta Neurol Scand 107:31–37 10.1034/j.1600-0404.2003.01317.x [DOI] [PubMed] [Google Scholar]
- Teubner B, Odermatt B, Guldenagel M, Sohl G, Degen J, Bukauskas F, Kronengold J, Verselis VK, Jung YT, Kozak CA, Schilling K, Willecke K (2001) Functional expression of the new gap junction gene connexin47 transcribed in mouse brain and spinal cord neurons. J Neurosci 21:1117–1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke K, Eiberger J, Degen J, Eckardt D, Romualdi A, Guldenagel M, Deutsch U, Sohl G (2002) Structural and functional diversity of connexin genes in the mouse and human genome. Biol Chem 383:725–737 [DOI] [PubMed] [Google Scholar]