Abstract
Nonsyndromic X-linked mental retardation (NSXLMR) is a very heterogeneous condition, and most of the underlying gene defects are still unknown. Recently, we have shown that ∼30% of these genes cluster on the proximal Xp, which prompted us to perform systematic mutation screening in brain-expressed genes from this region. Here, we report on a novel NSXLMR gene, FTSJ1, which harbors mutations in three unrelated families—one with a splicing defect, one with a nonsense mutation, and one with a deletion of one nucleotide. In two families, subsequent expression studies showed complete absence or significant reduction of mutant FTSJ1 transcripts. FTSJ1 protein is a homolog of Escherichia coli RNA methyltransferase FtsJ/RrmJ and may play a role in the regulation of translation. Further studies aim to elucidate the function of human FTSJ1 and its role during brain development.
X-linked mental retardation (XLMR) is a genetically and clinically heterogeneous group of disorders of the brain affecting 1/600 males (Herbst and Miller 1980). Nonsyndromic XLMR (NSXLMR) accounts for approximately two-thirds of all cases, and as many as 100 different genes on the X chromosome may be involved in this condition (Gecz and Mulley 2000). Most of the genes underlying NSXLMR remain unknown. As can be concluded from the distribution of linkage intervals in 125 families with NSXLMR, ∼30% of all relevant mutations cluster on the proximal short arm of the X chromosome (Ropers et al. 2003). PQBP1 (MIM 300463), which encodes the polyglutamine binding protein 1, and ZNF41 (MIM 314995) were the first two XLMR genes that we identified in this region (Kalscheuer et al. 2003; Shoichet et al. 2003).
Here, we report on the identification of a novel NSXLMR gene in this region. FTSJ1 is a human homolog of the Escherichia coli 2′-O-rRNA methyltransferase FtsJ/RrmJ gene (Ogura et al. 1991; Caldas et al. 2000) and is functionally unrelated to all previously identified NSXLMR genes.
The patient panel for mutation screening included 29 families with XLMR with overlapping linkage intervals and 215 small families with XLMR collected by the European MRX Consortium. All samples were obtained after receiving informed consent.
Family MRX44 has been published previously; all affected males have mild to moderate mental retardation (Hamel et al. 1999).
In family P48, the mother of the index patient is a healthy carrier with normal intelligence and adaptive skills. The index patient was born after normal pregnancy without fetal distress. Chromosome analysis indicated a normal karyotype, and fragile X (FRAXA) screening was negative. The patient first walked at age 18 mo and spoke at age 3–4 years. At the age of 29 years, he is now able to read simple sentences but has difficulties extracting the meaning. Behavior problems, including aggressive outbursts, were treated with antipsychotic pharmaceuticals, including carbamazepine. On recent clinical examination, his height was 170 cm, his weight was 80 kg (mild obesity), and his head circumference was 61 cm (+3 SD). He had a flat midface but otherwise normal features, and neurological examinations, including brain magnetic resonance imaging and electroencephalogram, indicated no abnormalities. Clinical records of his affected uncle are not available, but pregnancy and delivery were normal, and the mental retardation was said to be mild to moderate.
Family A3 has six affected males, all of whom exhibit mild to moderate intellectual disabilities. Individuals II:3, II:12, III:1, and III:5 have all attended special schools, sheltered workshops, or work education courses. Although they are all able to function essentially independently in familiar environments, they all require assistance with finances. In addition, patients II:3, III:1, and III:5 have problems with anger and aggression, and patients II:3 and III:1 are affected by anxiety and depression. Patients III:1 and III:5 have both been treated with paroxetine to relieve these symptoms. Patient II:12 recently received a diagnosis of schizophrenia and has been treated with antipsychotic pharmaceuticals.
Patient III:4 exhibits a somewhat milder phenotype. His disorder was first recognized at the age of 2 years, because of his delayed speech. However, he walked at age 11 mo and never had behavior problems. He attended a regular primary school, and, after participating in a work education program, he obtained employment in the catering industry, where he now undertakes cleaning, dishwashing, and simple food preparation tasks. He is completely independent in managing his finances. On recent examination, patient III:4 had a height of 170 cm (15th percentile), a weight of 94.6 kg (95th percentile), and a head circumference of 57.5 cm (75th percentile). He has mild brachydactyly and a high palate.
Patient III:6 has the most significant intellectual disability of any member of the family. His IQ has been measured at <55, and he has attended special school since the age of 7 years. Like his affected relatives, he has problems with anger and aggression; for this, he is being treated with amitriptyline. He walked at age 18 mo and first spoke in sentences at ∼10 years of age. On recent examination, he had no significant dysmorphic features. He is 167 cm tall (50th percentile), weighs 57 kg (75th percentile), and has a head circumference of 55 cm (50th percentile).
There are no significant intellectual problems in the obligate female carriers in family A3. For mutation detection, PCR products corresponding to the complete known human FTSJ1 coding sequence (GenBank accession number BC023584) were amplified from genomic DNA using intronic primers (table A [online only]) (GenBank accession number AF196972), and pooled samples were analyzed with the WAVE system (Transgenomic). (Conditions are described in table B [online only]).
Table A.
Primers for Mutation Detection and PCR Amplification Conditions
|
Primer |
|||||
| Exon | Forward | Reverse | MgCl2Concentration(mM) | AnnealingTemperature(°C) | FragmentLength(bp) |
| 2 | gtggtagcccattcatctgg | agtcagcccacctaccacag | 2 | 62 | 301 |
| 3 | gttggagaagtgggtgcag | gcccacatcagcctagtttc | 3 | 63 | 205 |
| 4 | ggagcgaaactaggctgatg | tgcatagaccccaggtaagg | 3 | 63 | 195 |
| 5 | atctgagggcagcagtgg | agggagaggcagggagtaac | 3 | 61 | 247 |
| 6 | aagatgcacagagccagatg | gcaatgttcagagcctgtgg | 3 | 60 | 204 |
| 7 | agtatatgcaggcccagctc | ggtgagaaggcaaagacagc | 2 | 62 | 244 |
| 8 | tggtggagtagagggaggtc | tcatagccctgacagacagc | 4 | 62 | 310 |
| 9 | gctgtctttgccttctcacc | ttgcataaggtgtggcagag | 2 | 59 | 368 |
| 10 | aggcatcctgaccttgtcc | gtcccacccacctactgttg | 3 | 61 | 181 |
| 11 | accatctccctacccctctg | tgggtcagaaaaaggcacac | 3 | 60 | 291 |
| 12 | cctttcctgcctcccaatag | ttgctccctgctctatctcc | 3 | 62 | 393 |
Table B.
Conditions for DHPLC Analysis
|
% Buffer B |
||||
| Exon | Loading | Initial | Final | Temperatures(°C) |
| 2 | 54 | 57 | 67 | 62.0–62.8 |
| 3 | 49 | 52 | 62 | 65.0–65.5 |
| 4 | 48–50 | 51–53 | 61–63 | 61.8–64.8 |
| 5 | 51–52 | 54–55 | 64–65 | 61.0–63.0 |
| 6 | 48–50 | 51–53 | 61–63 | 61.8–64.2 |
| 7 | 51–52 | 54–55 | 64–65 | 63.1–63.9 |
| 8 | 53–54 | 56–57 | 66–67 | 63.5–64.8 |
| 9 | 54–56 | 57–59 | 67–69 | 62.8–64.4 |
| 10 | 48–49 | 51–52 | 61–62 | 61.2–62.7 |
| 11 | 53–54 | 56–57 | 66–67 | 62.0–64.8 |
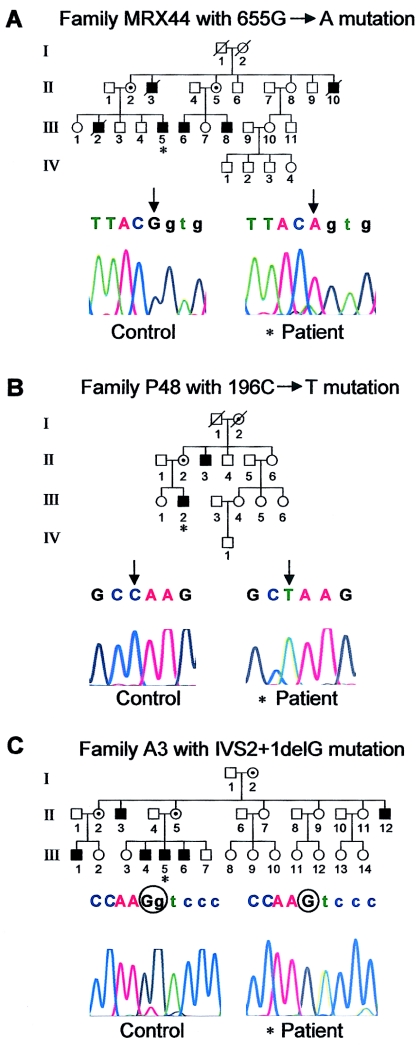
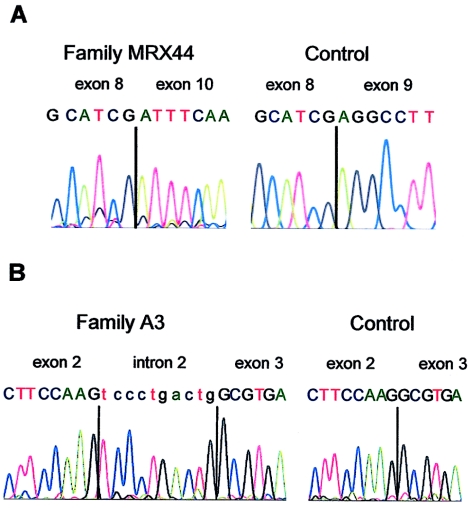
We found a single-nucleotide substitution (655G→A) at the last nucleotide of exon 9 in family MRX44 (fig 1A). Subsequent amplification of patient cDNA resulted in a fragment that was smaller than expected; direct sequencing of this specific product revealed that exon 9 is absent in the patient (fig. 2A). The absence of exon 9 introduces no frameshift in the FTSJ1 ORF, but the resulting FTSJ1 protein lacks 28 amino acids, which alters its structure and probably also its function. Subsequently, we searched for FTSJ1 mutations in 215 individuals from unrelated families with putative XLMR (families for which no linkage data was available). This analysis resulted in the detection of a single-nucleotide substitution (196C→T) in exon 4 of family P48 (fig. 1B). The C→T exchange is a nonsense mutation that results in a predicted truncated protein of 65 aa.
Figure 1.
A, Pedigree for family MRX44 with a splice-site mutation in exon 9, shown together with sequence chromatograms from a control and from one affected individual from this family, marked with an asterisk (*) in the pedigree. The affected nucleotide, which results in skipping of exon 9 in male patients, is indicated by a black arrow. Exonic nucleotides are shown in uppercase letters, and intronic nucleotides are shown in lowercase letters. B, Pedigree for family P48 with a nonsense mutation in exon 4 is shown together with the control and patient sequence chromatograms. A black arrow indicates the affected nucleotide. C, Pedigree for family A3 with a single-base-pair deletion affecting splicing of exon 2, together with sequence chromatograms from a control and the patient, who is marked with an asterisk in the pedigree. Affected nucleotides are circled.
Figure 2.
A, Sequence chromatograms from RT-PCR products, indicating exon 9 splicing of FTSJ1 in an affected male from family MRX44 and in a male control. Primers used for amplifications were located in exon 7 and exon 10. Because of the mutation in exon 9, the mutant FTSJ1 transcript lacks exon 9 completely. Vertical black lines indicate the exon boundaries. B, Sequence chromatograms from RT-PCR products, showing exon 2 and exon 3 splicing of FTSJ1 in a male control (A) and in an affected male of family A3. Primers used for amplifications were located in exon 2 and exon 5. Because of a 1-bp deletion in the exon 2/intron 2 splice site of FTSJ1 in the patient, an extra 10 nucleotides of intron 2 are present in the mutant FTSJ1 transcripts. Vertical black lines indicate the boundary between exon 2 and exon 3 in panel A and of intron 2 and exon 3 in panel B. Exonic nucleotides are shown in uppercase letters, and intronic nucleotides are in lowercase.
We also found a single-nucleotide deletion in family A3 (fig. 1C). This deletion affects one of the two guanine nucleotides of the exon 2/intron 2 boundary (IVS2+1delG). Direct sequencing of mutant RT-PCR products from the patient showed that, at the boundary between exon 2 and exon 3, the transcript carries an insertion of a 10-bp sequence of intron 2 (fig. 2B), which indicates that the IVS2+1delG deletion affects splicing by cryptic splice-site activation. The incorporation of the intronic sequence leads to a frameshift in the FTSJ1 mRNA, which results in the use of a premature stop codon in exon 3 at position 138, presumably producing a truncated protein of 49 aa.
Sequencing of PCR-amplified FTSJ1 transcripts from available family members showed that all affected males carry the mutation and that all mutations cosegregated with the disease (data not shown). None of these changes was found in 200 control X chromosomes.
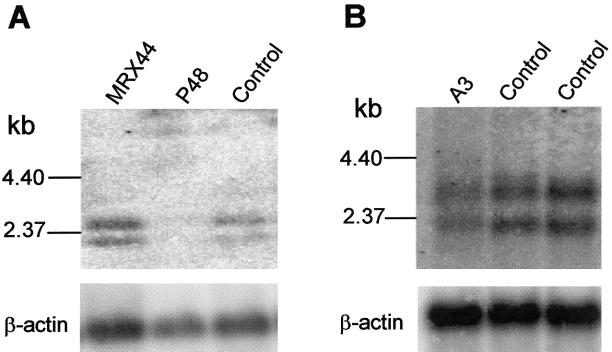
To gain more insight into the pathomechanism underlying NSXLMR in these three families, we performed northern blot hybridizations of poly-A+ cell line RNA from one affected male of each family, using a 744-bp cDNA probe corresponding to FTSJ1 exons 2–10. In the patient from family MRX44 with exon 9 skipping, FTSJ1 RNA level was identical to control RNA, indicating that the mutated transcripts are stable (fig. 3A). In contrast, in mRNA from an affected male from family P48, who carries the C→T substitution in exon 4, FTSJ1 transcripts were almost undetectable (fig. 3A). Likewise, in family A3 with a premature stop codon in exon 3, FTSJ1 transcripts were detected at a significantly reduced level (fig. 3B), suggesting that the phenotype in these families most likely results from functional loss of FTSJ1 protein.
Figure 3.
A, Northern blot analysis of poly-A+ RNA from patient and control lymphoblastoid cell lines. The blot was sequentially hybridized with an FTSJ1 cDNA probe spanning exons 2–10 and a full-length β-actin cDNA probe. B, Northern blot analysis of poly-A+ RNA from a patient lymphoblastoid cell line (A3) and two control cell lines. Hybridization was performed exactly as described in panel A.
When we performed northern blotting with an FTSJ1-specific probe, a transcript of ∼2.4 kb was found in all fetal and adult tissues examined, including brain, lung, liver, and kidney. Remarkably, expression was highest in fetal brain (fig. 4A). In the adult brain, the highest expression was observed in the caudate nucleus and the lowest expression in the corpus callosum (fig. 4B).
Figure 4.
Northern blot hybridization of multiple human tissues, using a probe spanning exons 2–10 of human FTSJ1. A, Fetal tissues, including brain, lung, liver, and kidney. B, Adult brain tissues, including amygdala, caudate nucleus, corpus callosum, hippocampus, whole brain, and thalamus. β-actin served as control for RNA loading.
Extensive sequence comparisons of human FTSJ1 protein (GenBank accession numbers CAA06749 and AAH23584) (also called “TRM7” and “JM23”; see below) with homologous proteins in different species showed that it belongs to a large phylogenetically conserved family of RNA methyltransferases (Bugl et al. 2000; Feder et al. 2003). The counterpart of FTSJ1 in E. coli is FtsJ/RrmJ, an RNA-binding heat shock protein structurally related to S-adenosyl-L-methionine–dependent methyltransferases. The substrate of FtsJ/RrmJ is the 23S ribosomal RNA of ribosomal particles (Bugl et al. 2000). Furthermore, three yeast proteins exhibit striking sequence similarities to E. coli FtsJ/RrmJ—namely, Spb1 (suppressor of poly-A+-binding protein Pab1p), Mrm2 (mitochondrial rRNA methyltransferase 2), and Trm7 (tRNA methyltransferase 7). These proteins are present in different cell compartments—the nucleolus, the mitochondria, and the cytoplasm, respectively—and have different substrates. Spb1 protein is required for 25S rRNA synthesis (Pintard et al. 2000). Mrm2 is a mitochondrial protein that is involved in 21S rRNA methylation (Pintard et al. 2002a). Trm7 catalyzes the formation of 2′-O-methylribose at two positions in the anticodon loops of various tRNAs (Pintard et al. 2002b). All yeast strains with a deletion of any one of these genes show growth reduction due to reduced translation efficiency (Pintard et al. 2000, 2002a, 2002b). As in yeast, the human genome contains three FtsJ/RrmJ homologs. According to Feder et al. (2003), the human FTSJ1 protein is a member of the Trm7 subfamily, which suggests that it may have a role in the posttranscriptional modification of tRNA. Experimental evidence on the function of human FTSJ1 is still lacking, but, on the basis of sequence homology, there is good reason to believe that this protein also plays a role in translation.
In affected males from family MRX44 (Hamel et al. 1999), the mutation leads to skipping of exon 9 and results in a predicted protein that lacks the C-terminal part of the S-AdoMet–binding domain. The deletion probably affects the conserved protein structure, which consists, in the homologous E. coli Ftsj/Rrmj protein, of alternating α helices and β sheets (Bugl et al. 2000).
Despite the ubiquitous expression of FTSJ1, all mutations identified in this study result in a relatively mild phenotype that is essentially brain specific. It is possible that the activity of FTSJ1 is most critical during brain development, which is supported by relative high expression of FTSJ1 in fetal brain. It is also plausible that brain structures are more sensitive to defects in the translation machinery than are other organs. Alternatively, another protein may partially compensate for the loss of FTSJ1, thereby preventing FTSJ1 mutations from causing more severe disorders. In E. coli, overexpression of two small GTPases was found to rescue a null mutation in the FtsJ/RrmJ gene, which restored the impaired ribosome assembly process and/or stability of 70S ribosomes (Tan et al. 2002). Similarly, in yeast, functional redundancy of the methyltransferase activity of Spb1 and a small nucleolar RNA-dependent mechanism has been reported for the 2′-O-methylation of a conserved nucleotide in the peptidyl-transferase center of the ribosome (Bonnerot et al. 2003).
In summary, we have shown that mutations in the methyltransferase FTSJ1 cause NSXLMR. Homologous methyltransferases modify untranslated RNAs and thereby play critical roles in protein translation. The association of FTSJ1 with mental retardation highlights the importance of this process, specifically in brain development and cognitive processes. Current studies aim to further characterize the function of FTSJ1.
Acknowledgments
We sincerely thank the families for participation in this study, G. Dellatolas for help with collecting samples, and S. Shoichet for critical reading of the manuscript. This work was supported by German Human Genome Program grant 01KW99087, National Genome Research Network grant 01GR0105, the Australian National Health and Medical Research Council, and 5th European Union Framework grant QLG3-CT-2002-01810.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for FTSJ1 mRNA sequences [accession number BC023584], FTSJ1 protein [accession numbers CAA06749 and AAH23584], and genomic BAC clone [accession number AF196972])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PQBP1 and ZNF41)
References
- Bonnerot C, Pintard L, Lutfalla G (2003) Functional redundancy of Spb1p and a snR52-dependent mechanism for the 2′-O-ribose methylation of a conserved rRNA position in yeast. Mol Cell 12:1309–1315 10.1016/S1097-2765(03)00435-0 [DOI] [PubMed] [Google Scholar]
- Bugl H, Fauman EB, Staker BL, Zheng F, Kushner SR, Saper MA, Bardwell JC, Jakob U (2000) RNA methylation under heat shock control. Mol Cell 6:349–360 10.1016/S1097-2765(00)00035-6 [DOI] [PubMed] [Google Scholar]
- Caldas T, Binet E, Bouloc P, Costa A, Desgres J, Richarme G (2000) The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J Biol Chem 275:16414–16419 10.1074/jbc.M001854200 [DOI] [PubMed] [Google Scholar]
- Feder M, Pas J, Wyrwicz LS, Bujnicki JM (2003) Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2′-O-methyltransferases. Gene 302:129–138 10.1016/S0378-1119(02)01097-1 [DOI] [PubMed] [Google Scholar]
- Gecz J, Mulley J (2000) Genes for cognitive function: development on the X. Genome Res 10:157–163 10.1101/gr.10.2.157 [DOI] [PubMed] [Google Scholar]
- Hamel BC, Smits AP, van den Helm B, Smeets DF, Knoers NV, van Roosmalen T, Thoonen GH, Assman-Hulsmans CF, Ropers HH, Mariman EC, Kremer H (1999) Four families (MRX43, MRX44, MRX45, MRX52) with nonspecific X-linked mental retardation: clinical and psychometric data and results of linkage analysis. Am J Med Genet 85:290–304 [DOI] [PubMed] [Google Scholar]
- Herbst DS, Miller JR (1980) Nonspecific X-linked mental retardation II: the frequency in British Columbia. Am J Med Genet 7:461–469 [DOI] [PubMed] [Google Scholar]
- Kalscheuer VM, Freude K, Musante L, Jensen LR, Yntema HG, Gecz J, Sefiani A, et al (2003) Mutations in the polyglutamine binding protein 1 gene cause X-linked mental retardation. Nat Genet 35:313–315 10.1038/ng1264 [DOI] [PubMed] [Google Scholar]
- Ogura T, Tomoyasu T, Yuki T, Morimura S, Begg KJ, Donachie WD, Mori H, Niki H, Hiraga S (1991) Structure and function of the ftsH gene in Escherichia coli. Res Microbiol 142:279–282 10.1016/0923-2508(91)90041-8 [DOI] [PubMed] [Google Scholar]
- Pintard L, Kressler D, Lapeyre B (2000) Spb1p is a yeast nucleolar protein associated with Nop1p and Nop58p that is able to bind S-adenosyl-L-methionine in vitro. Mol Cell Biol 20:1370–1381 10.1128/MCB.20.4.1370-1381.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L, Bujnicki JM, Lapeyre B, Bonnerot C (2002a) MRM2 encodes a novel yeast mitochondrial 21S rRNA methyltransferase. Embo J 21:1139–1147 10.1093/emboj/21.5.1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintard L, Lecointe F, Bujnicki JM, Bonnerot C, Grosjean H, Lapeyre B (2002b) Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. Embo J 21:1811–1820 10.1093/emboj/21.7.1811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropers HH, Hoeltzenbein M, Kalscheuer V, Yntema H, Hamel B, Fryns JP, Chelly J, Partington M, Gecz J, Moraine C (2003) Nonsyndromic X-linked mental retardation: where are the missing mutations? Trends Genet 19:316–320 10.1016/S0168-9525(03)00113-6 [DOI] [PubMed] [Google Scholar]
- Shoichet SA, Hoffmann K, Menzel C, Trautmann U, Moser B, Hoeltzenbein M, Echenne B, Partington M, Van Bokhoven H, Moraine C, Fryns JP, Chelly J, Rott HD, Ropers HH, Kalscheuer VM (2003) Mutations in the ZNF41 gene are associated with cognitive deficits: identification of a new candidate for X-linked mental retardation. Am J Hum Genet 73:1341–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J, Jakob U, Bardwell JC (2002) Overexpression of two different GTPases rescues a null mutation in a heat-induced rRNA methyltransferase. J Bacteriol 184:2692–2698 10.1128/JB.184.10.2692-2698.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]