Abstract
We have typed 275 men from five populations in Algeria, Tunisia, and Egypt with a set of 119 binary markers and 15 microsatellites from the Y chromosome, and we have analyzed the results together with published data from Moroccan populations. North African Y-chromosomal diversity is geographically structured and fits the pattern expected under an isolation-by-distance model. Autocorrelation analyses reveal an east-west cline of genetic variation that extends into the Middle East and is compatible with a hypothesis of demic expansion. This expansion must have involved relatively small numbers of Y chromosomes to account for the reduction in gene diversity towards the West that accompanied the frequency increase of Y haplogroup E3b2, but gene flow must have been maintained to explain the observed pattern of isolation-by-distance. Since the estimates of the times to the most recent common ancestor (TMRCAs) of the most common haplogroups are quite recent, we suggest that the North African pattern of Y-chromosomal variation is largely of Neolithic origin. Thus, we propose that the Neolithic transition in this part of the world was accompanied by demic diffusion of Afro-Asiatic–speaking pastoralists from the Middle East.
Many studies of African genetic diversity have concentrated on sub-Saharan and northeastern Africa, the most likely source region and corridor to the rest of the world (Tishkoff and Williams 2002). North Africa, however, may have followed a distinct evolutionary direction and requires further investigation. Genetic studies of this area, performed using classical markers, have revealed an agreement between genetic and geographic distances (Cavalli-Sforza et al. 1994) and a predominantly east-west structure to the genetic variation (Bosch et al. 1997). A compilation of 185 mtDNAs sampled across North Africa showed (1) that about half of the lineages belonged to the L haplogroups otherwise observed mainly in sub-Saharan Africa and (2) that most of the rest fell into haplogroup U6 (Salas et al. 2002), which perhaps originated in the Near East and spread into North Africa ∼30 thousand years (KY) ago (KYA) (Maca-Meyer et al. 2003). Y-chromosomal studies are potentially highly informative about the origin of male-specific lineages, because of the detailed haplotypes that can be obtained and their high geographical specificity (Jobling and Tyler-Smith 2003), but previous studies have been restricted to limited regions of North Africa (Bosch et al. 1999, 2001; Flores et al. 2001; Manni et al. 2002; Luis et al. 2004). Together, these genetic analyses highlighted the similarity between northeastern Africa and the Middle East and the clear genetic differentiation between northwestern Africa and both sub-Saharan Africa and Europe, including Iberia. The Sahara and Mediterranean, despite the narrow width of the Strait of Gibraltar, seem to have acted as effective long-term barriers to Y-chromosomal gene flow.
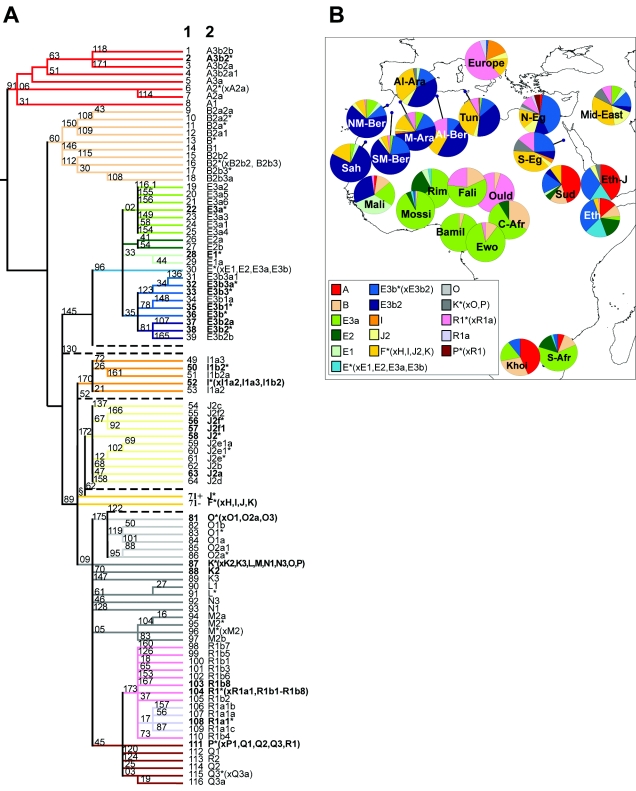
To provide a more complete description of the North African pattern of Y-chromosomal variation, we have analyzed five additional populations: Algerian Arabs, Algerian Berbers, Tunisians, and North and South Egyptians (table 1). Binary polymorphisms (Underhill et al. 2000), including 12f2 (Casanova et al. 1985), were typed in the hierarchical fashion described elsewhere (Rosser et al. 2000; Paracchini et al. 2002), allowing the allelic states at 119 markers defining 117 haplogroups to be measured or inferred from the Y phylogeny (fig. 1A). In the North African sample, 30 binary markers were found to be polymorphic, identifying 23 different haplogroups (fig. 1A) (table A1 [online only]). Phylogenetically related haplogroups were classified into clusters, the frequencies of which are shown schematically in fig. 1B. With the existing data from Morocco (Bosch et al. 2001), the combined set now spans the northern part of the continent. In addition, samples from southern Europe, the Middle East, and sub-Saharan Africa were included in some analyses (Semino et al. 2000; Underhill et al. 2000; Cruciani et al. 2002). Our results reveal four main conclusions about the male-lineage variation in North Africa.
Table 1.
Populations Studied
| Population | Code | Origina | Language | SampleSize | k(SNP)b | k(STR)c |
| Algerian Arabs | Al-Ara | Alger | Arabic | 35 | 10 | 27 |
| Algerian Berbers | Al-Ber | Tizi Ouzu | Berber-Tamazigh | 19 | 5 | 15 |
| North Egyptians | N-Eg | Mansoura | Arabic | 44 | 14 | 37 |
| South Egyptians | S-Eg | Luxor | Arabic | 29 | 9 | 27 |
| Tunisians | Tun | Tunis | Arabic | 148 | 15 | 107 |
Main city in the sampling area.
Number of haplogroups defined by the SNPs.
Number of haplotypes defined by 15 STRs.
Figure 1.
A, Phylogeny of Y-chromosomal haplogroups. The name of each haplogroup is shown at the tip of the lineage (1) according to Underhill et al. (2000) and (2) according to the Y Chromosome Consortium (2002). The polymorphisms screened in this study are shown along the branches. Lineage colors correspond to the haplogroup cluster colors of fig. 1B). Lineages shown by dashed lines were not observed in any of the populations discussed. Haplogroups observed in our samples are shown in boldface. The § symbol denotes 12f2a. B, Frequency distribution of Y haplogroup clusters in African, Middle Eastern, and European samples. Populations include the samples from table 1, as well as Moroccan Arabs (M-Ara), North Central Moroccan Berbers (NM-Ber), Saharawis (Sah), South Moroccan Berbers (SM-Ber) (Bosch et al. 2001); Sudanese (Sud), Ethiopians (Eth), Europeans (Europe), Malians (Mali), Central Africans (C-Afr), South Africans (S-Afr), Khoisan (Khoi), Middle Easterners (Mid-East) (Underhill et al. 2000); Ethiopian Jews (Eth-J), Mossi (Mossi), Rimaibe (Rim), Fali (Fali), Ouldeme (Ould), Bamileke (Bamil), and Ewondo (Ewo) (Cruciani et al. 2002).
First, as shown in fig. 1B, the lineages that are most prevalent in North Africa are distinct from those in the regions to the immediate north and south: Europe and sub-Saharan Africa. This is illustrated by even a cursory examination of the commonest haplogroups: E3b2 is the most common haplogroup in North Africa, forming 42% of the combined sample. In contrast, R1b made up 55% of a mixed European sample (Underhill et al. 2000) and was even higher (77%) in the Iberian sample examined by Bosch et al. (2001), whereas E3a predominates in many sub-Saharan areas, being present at 64% in a pooled sample (Underhill et al. 2000; Cruciani et al. 2002). Such a finding is not surprising, in the light of the earlier genetic studies, but has an important implication: despite haplogroups shared at low frequency, suggesting limited gene flow, North African populations have a genetic history largely distinct from both Europe and sub-Saharan Africa over the timescales needed for the Y-chromosomal differentiation to develop.
Second, just two haplogroups predominate within North Africa, together making up almost two-thirds of the male lineages: E3b2 and J* (42% and 20%, respectively). E3b2 is rare outside North Africa (Cruciani et al. 2004; Semino et al. 2004 and references therein), and is otherwise known only from Mali, Niger, and Sudan to the immediate south, and the Near East and Southern Europe at very low frequencies. Haplogroup J reaches its highest frequencies in the Middle East (Semino et al. 2004 and references therein), whereas the J-276 lineage (equivalent to J* here) is most frequent in Palestinian Arabs and Bedouins. Lineages can rise to high frequency because of biological selection, social selection, and/or neutral drift. There is a suggestion that weak negative selection due to partial deletion of genes needed for spermatogenesis could act on both E3b2 and J (Repping et al. 2003), but this would tend to decrease their frequency, and there is no evidence for positive selection. It therefore seems likely that their increase was due to drift despite any negative selection, implying that male effective population size has been small. Indeed, gene diversity values increase along a latitudinal axis from west to east (fig. 2), and much of this variation is accounted for by haplogroup E3b2, which decreases in frequency in a corresponding fashion from ∼76% in the Saharawis in Morocco to ∼10% in Egypt (fig. 2). The same haplogroup has increased in frequency in many different populations within North Africa, so there must have been gene flow between them.
Figure 2.
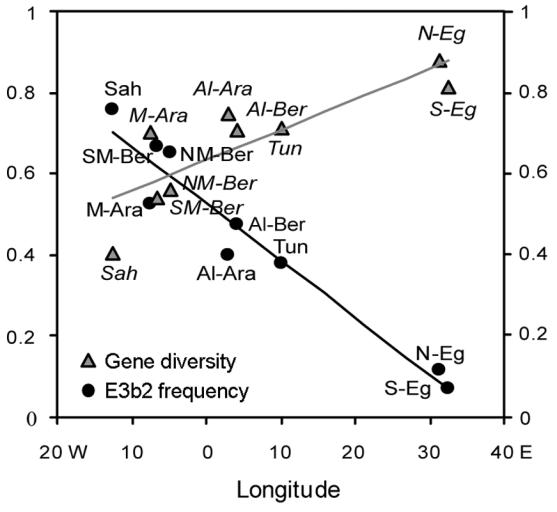
Population gene diversity and haplogroup E3b2 frequency in North Africa as a function of longitude. The linear regressions of gene (i.e., haplogroup) diversity (grey line) and E3b2 frequency (black line) onto longitude are shown. The population codes are reported in table 1 and figure 1 (in italics for gene diversity and in roman type for E3b2 frequency).
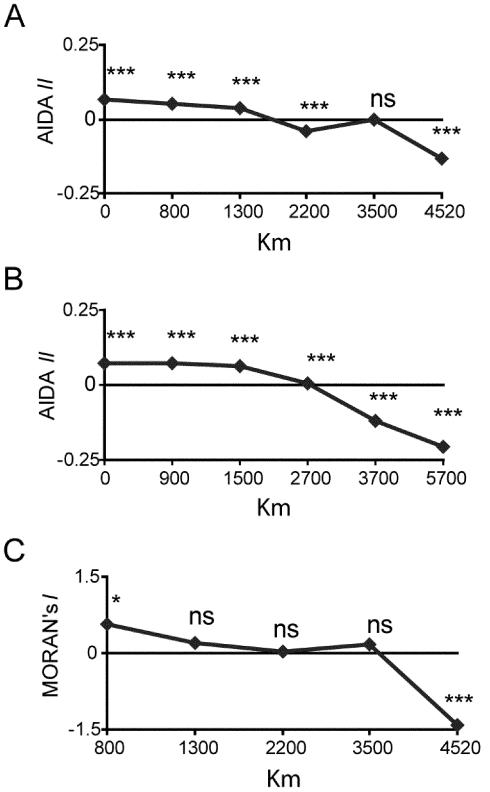
Third, there is strong geographical structure to the Y-chromosomal variation within the region. There is a high and significant correlation observed between genetic and geographical distances (r=0.55, P<.0005). Multidimensional scaling (MDS) analysis of genetic distances (Slatkin 1995) based on pairwise ΦST estimates (calculated using the program Arlequin) between 17 of the samples in fig. 1B showed a close correspondence with their relative geographical locations (fig. 3). Indeed, the positions of the samples in the MDS plot describe a latitudinal axis, from North Africa and the Middle East in the upper part to Central and southern Africa in the lower part. Furthermore, the pattern of genetic affinities among the North African samples parallels the west-east orientation quite precisely, from Morocco on the left-hand side to Egypt and the Middle East on the right. Spatial autocorrelation analysis (by AIDA; Bertorelle and Barbujani 1995) shows a clinal pattern of variation, more marked when Middle Eastern samples are included (fig. 4A and 4B). Haplogroup E3b2 itself shows a significant correlogram in a SAAP analysis (Sokal and Oden 1978) (fig. 4C). Furthermore, diversity within this haplogroup, measured using 15 Y-STRs (Thomas et al. 1999; Ayub et al. 2000), declines substantially towards the west (table A2 [online only]). These findings, together with the gene diversity pattern described above, are consistent with the hypothesis of a demic expansion from the Middle East.
Figure 3.
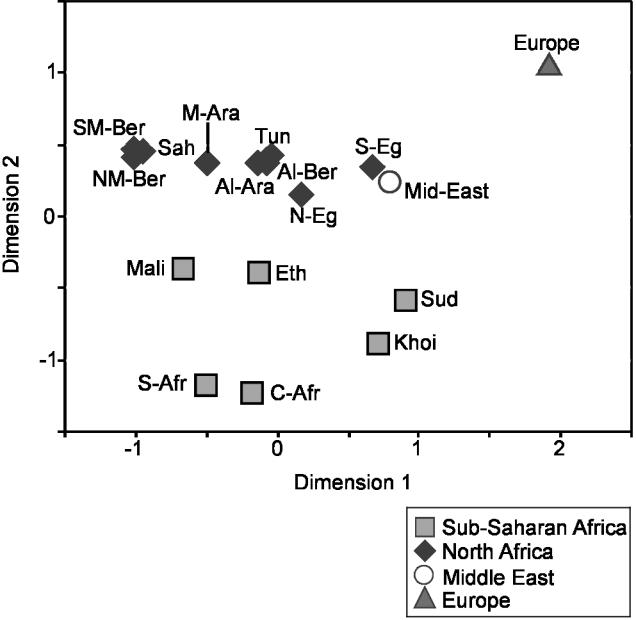
MDS analyses based on binary marker genetic distances among the North African, sub-Saharan African, European, and Middle Eastern populations from fig. 1B, tested using 116 binary markers (stress value 0.15). The population codes are reported in table 1 and figure 1.
Figure 4.
Spatial autocorrelation analyses. A, Correlogram of the AIDA II indices calculated for North Africa. B, Correlogram of the AIDA II indices for North Africa and the Middle East (Syrian, Turkish, and Lebanese samples of Semino et al. [2000]). C, Correlogram of the Moran’s I index for the haplogroup E3b2* (overall correlogram significance P=.001). ***, P<.001; ns, not significant.
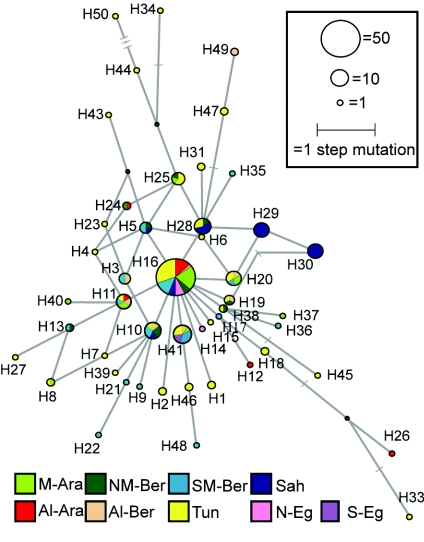
Fourth, the time depth associated with the most common Y-chromosomal haplogroups in North Africa is shallow. Y-STR data (15 loci) were obtained for 256 Y chromosomes and revealed 201 different haplotypes (table A3 [online only]). Of these, only 16 were observed in more than one individual, but two were particularly frequent: one was present in 24 chromosomes from the Algerian Arab, Tunisian, and northern Egyptian populations, belonging, with one exception, to haplogroup E3b2*(xE3b2a); the second haplotype (observed in nine Tunisians) was associated with haplogroup J*. STR variability was used to estimate the TMRCA of North African chromosomes from individual haplogroups using the program BATWING (Wilson and Balding 1998), using either 15 loci (table A4 [online only]) or, to incorporate the Moroccan data (Bosch et al. 2001), 8 loci (table 2). The TMRCA of haplogroup E3b2 was estimated to be ∼4.2 KY (95% CI 2.8–6.0 KY), using the mutation rate measured in father-son pairs (Kayser et al. 2000) and assuming 30 years per generation, or 6.9 (5.9–8.2) KY using the deduced “effective” mutation rate calibrated by historical events (Zhivotovsky et al. 2004) (table 2). The times for haplogroup J, the second-most-common haplogroup observed in North Africa (6.8 KY, 95% CI 4.4–11.1 KY; or 7.9 KY, 95% CI 6.6–9.1 KY) were also quite recent (table 2), supporting the idea of a recent demographic event. A network (Bandelt et al. 1999) of the E3b2*(xE3b2a) chromosomes, calculated using the program NETWORK, based on eight loci, showed a widespread high-frequency central haplotype (32%) and a starlike structure (fig. A1 [online only]). The Moroccan samples display low variability, and their chromosomes often occupy more-peripheral positions in the network. These findings together support our second conclusion, that genetic drift must have shaped the North African Y-chromosomal landscape.
Table 2.
TMRCA Estimates and 95% CIs of Y-Chromosomal Lineages in North Africa[Note]
|
TMRCA [in KY] (95% CI) |
||||
| SampledLineages | Mutation | No. of Chromosomes | 30 Years/Generation)a | 25 Years/Generation)b |
| All | 390 | 13.95 (8.08–24.67) | 25.87 (20.47–30.99 | |
| E | M145 | 237 | 9.70 (5.98–14.33) | 19.66 (15.31–21.12) |
| E3a | M2 | 8 | 2.96 (1.83–4.25) | 5.06 (3.01–7.61) |
| E1 | M33 | 3 | 3.38 (2.11–5.25) | 5.6 (2.79–9.38) |
| E3b1 | M78 | 38 | 4.48 (3.01–6.16) | 8.10 (5.42–10.71) |
| E3b | M35 | 226 | 8.26 (5.18–12.37) | 14.33 (9.32–19.19) |
| E3b2 | M81 | 165 | 4.15 (2.84–5.97) | 6.90 (5.91–8.19) |
| F | M89 | 152 | 9.77 (6.47–14..84) | 19.05 (15.30–22.61) |
| J | 12f2 | 93 | 6.78 (4.38–11.10) | 7.86 (6.63–9.13) |
| J2 | M172 | 14 | 3.67 (2.50–5.25) | 4.98 (4.10–6.06) |
| R | M173 | 27 | 4.44 (3.04–6.62) | 10.24 (8.54–12.94) |
| I | M170 | 2 | 2.83 (1.68–4.32) | 5.29 (3.19–9.16) |
| K | M9 | 38 | 7.10 (4.38–11.1) | 15.09 (11.56–18.16) |
Note.— TMRCA estimates based on 8 STRs (DYS19, DYS388, DYS389b, DYS389I, DYS390, DYS391, DYS392, and DYS393) in nine North African populations (those from the present study and the Moroccan samples of Bosch et al. 2001). In these calculations, we incorporated the information from binary markers.
The two parameters describing the population growth (alpha and beta) have been set as alpha prior uniform (0.03, 0.05) and beta prior uniform (0.10, 0.20), the microsatellite mutation rates used were from Weale et al. (2001) or gamma (2,1000) for the loci for which published estimates of mutation rates were not available.
Which historical or prehistorical demographic processes could explain the characteristics of the variation of Y-chromosomal lineages in North Africa? The current physical barriers, the Mediterranean Sea to the north and Sahara Desert to the south, could have provided genetic barriers leading to the separate evolutionary paths of the regions, although for the Sahara, episodes of more favorable climatic conditions could have relaxed this barrier at times, particularly during some intervals between ∼10 KYA and ∼5 KYA (Muzzolini 1993). There is no evident reason why it should have acted as a strong genetic barrier at such times, so, if there was substantial gene flow, the genetic differentiation between North and sub-Saharan Africa may postdate this period. A clinal pattern of haplogroup variation like the one we observe can be expected from an east-to-west population expansion, and the finding of lower E3b2 STR variation in the west than in central North Africa (table A2 [online only]), accompanied by a substantial increase in frequency of this haplogroup, is most readily explained by expansion into virtually uninhabited terrain by populations experiencing increasing drift (Barbujani et al. 1994).
The current distributions of the haplogroups can suggest geographical origins, and their TMRCAs provide some constraints on the times of their spread. The M35 lineage (see the phylogeny in fig. 1A for marker locations) is thought to have arisen in East Africa, on the basis of its high frequency and diversity there (Cruciani et al. 2004; Semino et al. 2004), and to have given rise to M81 in North Africa. The TMRCAs for E3b (8.3 KY, 95% CI 5.2–12.4 KY; or 14.4 KY, 95% CI 9.3–19.3 KY; table 2) and E3b2 (2.8–8.2 KY) should thus bracket the spread of E3b2 in North Africa. These times contrast sharply with estimates of 53 ± 21 KYA for the M35 lineage and 32 ± 11 KYA for the M81 lineage, by use of a constant-sized population model, or 30 ± 6 and 19 ± 4 KYA, respectively, by use of an expanding population model (Bosch et al. 2001). They are, however, more in accordance with times of 26.5 KYA (without a useful CI) for the M215 mutation (intermediate between M35 and M96 in the phylogeny; see fig. 1A) and 5.6 KYA for M81 (Cruciani et al. 2004) or of 29.2 ± 4.1 KYA for M35 and 8.6 ± 2.3 KYA for M81 (Semino et al. 2004). An origin for haplogroup J in the Middle East has been proposed (Semino et al. 2004 and references therein); the TMRCA of the J-M267 branch, found in both the Middle East and North Africa (and including our J* chromosomes), was estimated at 24.1 ± 9.4 KY and must predate its spread. This is consistent with our 95% TMRCA estimate of 4.4–11.1 KY for the North African chromosomes. Thus, although Moroccan Y lineages were interpreted as having a predominantly Upper Paleolithic origin from East Africa (Bosch et al. 2001), according to our TMRCA estimates, no populations within the North African samples analyzed here have a substantial Paleolithic contribution.
Early Neolithic sites are documented in the eastern part of North Africa and later ones in the west, which would be compatible with an east-to-west movement at this time, and this is also the case for the Arab expansion. Historical records of the Arab conquest, however, suggest that its demographic impact must have been limited (McEvedy 1980). In addition, genetic evidence shows that E3b2 is rare in the Middle East (Semino et al. 2004), making the Arabs an unlikely source for this frequent North African lineage. Parallel analyses between North Africa and Southern Europe have revealed strikingly similar patterns of Y chromosome variation which would support a scenario in which the Neolithic expansion, originating in the Middle East branched into two flows separated by the geographical barrier of the Mediterranean Sea. Indeed, as in North Africa, Y-chromosome variability in Southern Europe is clinal, gene diversity decreases from east to west, and genetic distances between North Africa and Southern Europe increase in a regular fashion from the Middle East toward the west (results not shown). Under the hypothesis of a Neolithic demic expansion from the Middle East, the likely origin of E3b in East Africa could indicate either a local contribution to the North African Neolithic transition (Barker 2003) or an earlier migration into the Fertile Crescent, preceding the expansion back into Africa.
In conclusion, we propose that the Y-chromosomal genetic structure observed in North Africa is mainly the result of an expansion of early food-producing societies. Moreover, following Arioti and Oxby (1997), we speculate that the economy of those societies relied initially more on herding than on agriculture, because pastoral economies probably supported lower numbers of individuals, thus favoring genetic drift, and showed more mobility than agriculturalists, thus allowing gene flow. Some authors believe that languages families are unlikely to be >10 KY old and that their diffusion was associated with the diffusion of agriculture (Diamond and Bellwood 2003). Since most of the languages spoken in North Africa and in nearby parts of Asia belong to the Afro-Asiatic family (Ruhlen 1991), this expansion could have involved people speaking a proto–Afro-Asiatic language. These people could have carried, among others, the E3b and J lineages, after which the M81 mutation arose within North Africa and expanded along with the Neolithic population into an environment containing few humans.
Acknowledgments
We thank the donors of the DNA samples, for making this work possible, and Guido Barbujani, Jaume Bertranpetit, Giovanni Destro-Bisol, Matthieu Honegger, Matt E. Hurles, Mark A. Jobling, Vincenzo Mandarino, Alicia Sanchez-Mazas, and Francesco Vecchi, for useful discussions and comments on the manuscript. B.A. was supported by a Ph.D. fellowship from the Università Cattolica del Sacro Cuore di Roma, E.S.P. was supported by Swiss National Science Foundation grant 39-59375.99, V.L.P. was supported by grant PRIN-MIUR 2002 N.2002063871 from the Progetti Rilevanza Nazionale–Ministero dell'Istruzione, dell'Università e della Ricerca, A.N. was supported by grant PRIN-MIUR 2002, and C.T.-S. was supported by The Wellcome Trust.
Appendix: Supplementary Material
Figure A1.
Network of haplogroup E3b2*(xE3b2a) microsatellite haplotypes in North Africa. Circles represent haplotypes; their areas are proportional to their frequencies in the total North African sample, and their colors indicate the population of origin. Population codes are reported in table 1 and fig. 1. Lines across the network branches represent haplotypes that were not observed.
Table A1.
SNP Haplogroup Frequencies in the Five North African Population Samples and Other Published Samples
|
Population Samplea |
|||||||||||||||||
| Present Study | Bosch et al. (2001) | Underhill et al. (2000) | |||||||||||||||
| Haplogroupb | Al-Ara (n=35) | Al-Ber (n=19) | N-Eg (n=44) | S-Eg (n=29) | Tun (n=148) | M-Ara (n=44) | NM-Ber (n=63) | Sah (n=29) | SM-Ber (n=39) | Sud (n=40) | Eth (n=88) | Europe (n=60) | Mali (n=44) | C-Afr (n=36) | S-Afr (n=54) | Khoi (n=39) | Mid-East (n=23) |
| A3b2b | 6 | ||||||||||||||||
| A3b2* | 1 | 17 | 5 | ||||||||||||||
| A3b2a | 1 | ||||||||||||||||
| A3b1 | 3 | 11 | |||||||||||||||
| A3a | 1 | ||||||||||||||||
| A2*(xA2a) | 5 | ||||||||||||||||
| A2a | 1 | ||||||||||||||||
| A1 | 1 | ||||||||||||||||
| B2a2a | 3 | ||||||||||||||||
| B2a2* | 3 | ||||||||||||||||
| B2a* | 1 | 1 | 1 | ||||||||||||||
| B2a1 | 5 | 4 | 1 | 7 | |||||||||||||
| B | 1 | 1 | |||||||||||||||
| B1 | 1 | ||||||||||||||||
| B2b2 | 1 | ||||||||||||||||
| B2*(xB2b2,B2b3) | 7 | 11 | |||||||||||||||
| B2b3* | 1 | ||||||||||||||||
| B2b3a | 1 | ||||||||||||||||
| E3a2 | 1 | ||||||||||||||||
| E3a5 | 1 | ||||||||||||||||
| E3a6 | 1 | ||||||||||||||||
| E3a* | 2 | 3 | 6 | 1 | 1 | 3 | 7 | 20 | 28 | 7 | 2 | ||||||
| E3a3 | 1 | ||||||||||||||||
| E3a1 | 3 | ||||||||||||||||
| E3a4 | 2 | ||||||||||||||||
| E2a | 2 | 15 | |||||||||||||||
| E2b | 3 | 8 | |||||||||||||||
| E1* | 1 | 1 | 2 | 1 | 13 | 1 | |||||||||||
| E1a | 1 | 2 | |||||||||||||||
| E*(xE1,E2,E3a,E3b) | 16 | 1 | |||||||||||||||
| E3b3a* | 2 | 4 | 2 | 2 | 2 | 1 | 1 | ||||||||||
| E3b3* | 2 | ||||||||||||||||
| E3b1* | 4 | 12 | 5 | 8 | 5 | 1 | 5 | 7 | 20 | 2 | |||||||
| E3b* | 1 | 2 | 5 | 1 | 5 | 3 | 6 | 1 | 4 | ||||||||
| E3b2a | 1 | 1 | |||||||||||||||
| E3b2* | 13 | 9 | 5 | 2 | 56 | 2 | 12 | ||||||||||
| E3b2b | 23c | 41c | 22c | 26c | 1 | ||||||||||||
| I1a3 | 1 | ||||||||||||||||
| I1b2* | 1 | 1 | |||||||||||||||
| I*(xI1a2,I1a3,I1b2) | 1 | 8 | |||||||||||||||
| J2c | 1 | ||||||||||||||||
| J2f* | 1 | 2 | |||||||||||||||
| J2f1 | 1 | ||||||||||||||||
| J2* | 1 | 3 | 1 | 4 | 2 | 2 | |||||||||||
| J2a | 1 | 1d | 2d | 1d | |||||||||||||
| J2e1a | 1 | ||||||||||||||||
| F*(xH,I,J,K) | 4 | 2 | 3 | 5 | 7 | 2 | 8e | ||||||||||
| J* | 8 | 3 | 4 | 6 | 48 | 6 | 3 | 5 | 2 | 4e | 4e | 9e | |||||
| O*(xO1,O2a,O3) | 1 | ||||||||||||||||
| K*(xK2,K3,L,M,N1,N3,O,P) | 1 | 1 | |||||||||||||||
| K2 | 1 | 3 | 1 | ||||||||||||||
| L1 | 2 | ||||||||||||||||
| R1b7 | 3 | ||||||||||||||||
| R1b5 | 1 | ||||||||||||||||
| R1b8 | 1 | ||||||||||||||||
| R1*(xR1a1,R1b1-R1b8) | 3 | 4 | 4 | 9 | 3 | 1 | 1 | 1 | 29 | 1 | 2 | ||||||
| R1a1* | 1 | 3 | |||||||||||||||
| P*(xP1,Q1,Q2,Q3,R1) | 2 | 1 | |||||||||||||||
Nomenclature reported in figure 1.
These are E3b2*(xE3b2a) haplogroups that were not further resolved (marker M165 not tested).
These are J2*(xJ2b-J2f) haplogroups that were not further resolved (marker M47 not tested).
These are F*(xH,I,J,K) haplogroups that were not further resolved (marker 12f2 not tested).
Table A2.
Gene Diversity in the Five North African Samples and Other Published Samples, on the Basis of Y-Chromosomal SNPs and STRs
|
Population Samplea |
|||||||||||||||||
| Present Study |
Bosch et al. (2001) |
Underhill et al. (2000) |
|||||||||||||||
| Variable | Al-Ara | Al-Ber | N-Eg | S-Eg | Tun | M-Ara | NM-Ber | Sah | SM-Ber | Sud | Eth | Europe | Mali | C-Afr | S-Afr | Khoi | Mid-East |
| Sample size (SNPs) | 35 | 19 | 44 | 29 | 148 | 44 | 63 | 29 | 39 | 40 | 88 | 60 | 44 | 36 | 54 | 39 | 23 |
| Gene diversity (SNPs) ± b | .748 ± .049 | .708 ± .074 | .880 ± .029 | .813 ± .053 | .714 ± .024 | .700 ± .067 | .561 ± .071 | .406 ± .101 | .543 ± .090 | .776 ± .053 | .876 ± .017 | .730 ± .052 | .821 ± .035 | .660 ± .077 | .697 ± .059 | .802 ± .029 | .866 ± .054 |
| Sample size (8 STRs) | 32 | 17 | 44 | 28 | 139 | 43 | 19 | 29 | 39 | ||||||||
| Gene diversity (8 STRs) ± SDc | .953 ± .031 | .994 ± .019 | .989 ± .008 | .995 ± .011 | .976 ± .007 | .919 ± .035 | .977 ± .027 | .897 ± .030 | .960 ± .020 | ||||||||
| Sample size (15 STRs) | 32 | 17 | 43 | 28 | 138 | ||||||||||||
| Gene diversity (15 STRs ± SD)d | .979 ± .020 | .994 ± .019 | .991 ± .008 | 1.000 ± .009 | .984 ± .006 | ||||||||||||
| Number of STR-typed E3b2 chromosomes | 14 | 9 | 5 | 2 | 55e | 22f | 10g | 22 | 26 | ||||||||
| E3b2 STR varianceh | .21 | .26 | .02 | - | .30 | .10 | .11 | .11 | .18 | ||||||||
| E3b2 STR variability ± SDi | 3.32 ± 2.00 | 4.17 ± 1.53 | .40 ± .52 | .00 | 4.82 ± 1.95 | 1.74 ± 1.14 | 1.78 ± 1.11 | 1.72 ± 1.11 | 2.83 ± 1.19 | ||||||||
| Number of STR-typed J* chromosomes | 5 | 2 | 4 | 6 | 44 | 6 | 3 | 5 | 2 | ||||||||
| J* STR varianceh | .14 | .25 | .62 | .71 | .37 | .06 | .83 | .11 | .31 | ||||||||
| J* STR variability ± SDi | 2.20 ± .84 | 4.00 ± .00 | 10.00 ± 2.10 | 11.33 ± 3.36 | 5.90 ± 2.43 | 1.47 ± .98 | 13.33 ± 2.52 | 1.80 ± 1.51 | 5.00 ± .00 | ||||||||
Based on SNP-haplogroup frequencies.
Based on eight-STR-haplotype frequencies.
Based on 15-STR-haplotypes frequencies.
Fifty-five out of 56 E3b2 chromosomes were fully STR-typed.
Twenty-two out of 23 E3b2 chromosomes were fully STR-typed.
Ten out of 41 E3b2 chromosomes were fully STR-typed.
Average of variances in repeat numbers over eight STR loci.
Estimated as the average of pairwise sums (over eight STR loci) of squared single-step differences between alleles.
Table A3.
Frequencies of Y-Chromosome Haplotypes (15 STRs) in Tunisians, Algerians, and Egyptians
|
STR Locus |
Population |
||||||||||||||||||||
| 19 | 388 | 389b | 389I | 390 | 391 | 392 | 393 | 426 | 434 | 435 | 436 | 437 | 438 | 439 | Associated Haplogroup | Al-Ara | Al-Ber | Tun | N-Eg | S-Eg | Total |
| 11 | 12 | 17 | 10 | 24 | 10 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | E3b1* | 1 | 1 | ||||
| 11 | 12 | 18 | 9 | 24 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 12 | 11 | E3b1* | 1 | 1 | ||||
| 13 | 10 | 16 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 13 | 10 | 16 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 14 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 14 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 14 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 16 | 9 | 23 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 16 | 9 | 23 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 2 | 2 | ||||
| 13 | 12 | 16 | 10 | 23 | 10 | 11 | 13 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | P*(xP1,Q1,Q2,Q3,R1) | 1 | 1 | ||||
| 13 | 12 | 16 | 10 | 24 | 9 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | F*(xH,I,J,K,) | 1 | 1 | ||||
| 13 | 12 | 16 | 10 | 24 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 16 | 10 | 25 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 16 | 11 | 23 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2*and E3b1* | 1 | 1 and 1 | 3 | |||
| 13 | 12 | 16 | 11 | 23 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 13 | E3b* | 1 | 1 | ||||
| 13 | 12 | 16 | 11 | 23 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 16 | 11 | 23 | 11 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 16 | 11 | 24 | 9 | 10 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 16 | 11 | 24 | 9 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 16 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2*and E3b* | 5 | 13 and 2 | 4 | 24 | ||
| 13 | 12 | 16 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b2* | 1 | 1 | 1 | 3 | ||
| 13 | 12 | 16 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 11 | 10 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 16 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 12 | 12 | 8 | 10 | 11 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 16 | 11 | 24 | 9 | 11 | 13 | 11 | 10 | 11 | 14 | 8 | 10 | 11 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 16 | 11 | 24 | 9 | 11 | 15 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 2 | 2 | ||||
| 13 | 12 | 16 | 11 | 24 | 9 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 16 | 11 | 24 | 9 | 12 | 13 | 11 | 8 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 16 | 11 | 24 | 10 | 11 | 14 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 16 | 11 | 24 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b2* | 1 | 1 | 1 | 3 | ||
| 13 | 12 | 16 | 11 | 25 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2*and E3b2a | 1 | 1 | 2 | 4 | ||
| 13 | 12 | 17 | 9 | 22 | 10 | 11 | 13 | 11 | 10 | 11 | 12 | 8 | 10 | 10 | E3b1* | 1 | 1 | ||||
| 13 | 12 | 17 | 9 | 23 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b1* | 1 | 1 | ||||
| 13 | 12 | 17 | 9 | 23 | 10 | 11 | 14 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b1* | 1 | 1 | ||||
| 13 | 12 | 17 | 9 | 24 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b1* | 1 | 1 | ||||
| 13 | 12 | 17 | 9 | 24 | 10 | 11 | 15 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 17 | 10 | 22 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | E3b3a* | 2 | 2 | ||||
| 13 | 12 | 17 | 10 | 23 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | E3b1* | 1 | 1 | ||||
| 13 | 12 | 17 | 10 | 23 | 10 | 11 | 13 | 11 | 9 | 11 | 14 | 8 | 10 | 13 | E3b* | 1 | 1 | ||||
| 13 | 12 | 17 | 10 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 2 | 2 | ||||
| 13 | 12 | 17 | 10 | 24 | 9 | 11 | 13 | 11 | 8 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 17 | 10 | 24 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 17 | 10 | 24 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 10 | E3b1* | 1 | 1 | ||||
| 13 | 12 | 17 | 10 | 24 | 11 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 13 | E3b3a* | 1 | 1 | ||||
| 13 | 12 | 17 | 10 | 25 | 9 | 11 | 14 | 11 | 9 | 11 | 14 | 8 | 10 | 11 | E3b* | 1 | 1 | ||||
| 13 | 12 | 17 | 10 | 25 | 10 | 11 | 13 | 11 | 9 | 12 | 12 | 8 | 10 | 11 | E3b3a* | 1 | 1 | ||||
| 13 | 12 | 17 | 11 | 23 | 10 | 11 | 15 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 17 | 11 | 24 | 9 | 11 | 13 | 10 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 17 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 17 | 11 | 24 | 10 | 11 | 13 | 11 | 11 | 10 | 12 | 8 | 11 | 11 | E3b1* | 1 | 1 | 2 | |||
| 13 | 12 | 17 | 11 | 24 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | E3b1* | 1 | 1 | ||||
| 13 | 12 | 17 | 11 | 24 | 10 | 13 | 14 | 12 | 9 | 11 | 12 | 9 | 10 | 12 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 13 | 12 | 17 | 11 | 24 | 11 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 17 | 12 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 17 | 12 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 17 | 12 | 24 | 9 | 12 | 15 | 11 | 9 | 12 | 12 | 8 | 10 | 11 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 18 | 9 | 23 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b1* | 1 | 1 | ||||
| 13 | 12 | 18 | 10 | 24 | 10 | 11 | 12 | 10 | 9 | 11 | 12 | 8 | 11 | 10 | E3b2* | 1 | 1 | ||||
| 13 | 12 | 18 | 10 | 24 | 10 | 11 | 14 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | E3b3a* | 1 | 1 | ||||
| 13 | 12 | 18 | 10 | 24 | 11 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 13 | E3b1* | 1 | 1 | ||||
| 13 | 12 | 18 | 11 | 23 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b3a* | 1 | 1 | ||||
| 13 | 12 | 18 | 11 | 24 | 10 | 10 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 13 | E3b3a* | 1 | 1 | ||||
| 13 | 12 | 18 | 11 | 24 | 10 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b* | 1 | 1 | ||||
| 13 | 12 | 19 | 10 | 24 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 13 | E3b3a* | 1 | 1 | ||||
| 13 | 12 | 19 | 10 | 24 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | E3b3a* | 1 | 1 | ||||
| 13 | 13 | 16 | 9 | 24 | 10 | 11 | 13 | 10 | 9 | 11 | 12 | 8 | 10 | 12 | E3b1* | 1 | 1 | ||||
| 13 | 13 | 16 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 13 | 14 | 16 | 11 | 24 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | F*(xH,I,J,K) | 1 | 1 | ||||
| 13 | 17 | 16 | 10 | 23 | 11 | 12 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 13 | 17 | 17 | 10 | 23 | 12 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 12 | 15 | 11 | 23 | 10 | 10 | 14 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | O*(xO1,O2a,O3) | 1 | 1 | ||||
| 14 | 12 | 16 | 9 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 14 | 12 | 16 | 10 | 23 | 9 | 13 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | K2 | 1 | 1 | ||||
| 14 | 12 | 16 | 10 | 24 | 10 | 13 | 12 | 12 | 9 | 11 | 12 | 9 | 12 | 12 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 14 | 12 | 16 | 10 | 24 | 11 | 12 | 13 | 12 | 9 | 11 | 12 | 9 | 13 | 12 | R1b8 | 1 | 1 | ||||
| 14 | 12 | 16 | 10 | 25 | 10 | 13 | 12 | 12 | 9 | 11 | 12 | 8 | 12 | 12 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 14 | 12 | 16 | 11 | 23 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 14 | 12 | 16 | 11 | 23 | 10 | 14 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | F*(xH,I,J,K) | 1 | 1 | ||||
| 14 | 12 | 16 | 11 | 23 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 11 | F*(xH,I,J,K) | 1 | 1 | ||||
| 14 | 12 | 16 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 2 | 3 | |||
| 14 | 12 | 16 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b2* | 1 | 1 | ||||
| 14 | 12 | 16 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 12 | 12 | E3b2* | 1 | 1 | ||||
| 14 | 12 | 16 | 11 | 24 | 9 | 11 | 13 | 11 | 11 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 14 | 12 | 16 | 11 | 24 | 9 | 11 | 15 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 14 | 12 | 17 | 9 | 23 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b1* | 1 | 1 | ||||
| 14 | 12 | 17 | 9 | 23 | 11 | 11 | 14 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | F*(xH,I,J,K) | 1 | 1 | ||||
| 14 | 12 | 17 | 9 | 24 | 11 | 13 | 12 | 12 | 9 | 11 | 12 | 9 | 12 | 12 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 14 | 12 | 17 | 10 | 23 | 11 | 13 | 13 | 12 | 9 | 11 | 12 | 8 | 10 | 11 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 14 | 12 | 17 | 10 | 24 | 9 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 14 | 12 | 17 | 10 | 24 | 10 | 12 | 12 | 12 | 8 | 11 | 12 | 8 | 12 | 12 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 14 | 12 | 17 | 10 | 24 | 10 | 13 | 12 | 12 | 9 | 11 | 12 | 9 | 12 | 11 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 14 | 12 | 17 | 10 | 25 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 13 | E3b1* | 1 | 1 | ||||
| 14 | 12 | 17 | 10 | 25 | 11 | 13 | 13 | 12 | 9 | 11 | 12 | 10 | 12 | 12 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 14 | 12 | 17 | 11 | 23 | 10 | 10 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | K2 | 1 | 1 | ||||
| 14 | 12 | 17 | 11 | 23 | 10 | 14 | 12 | 12 | 9 | 11 | 12 | 9 | 10 | 13 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 14 | 12 | 17 | 11 | 24 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b1* | 1 | 1 | ||||
| 14 | 12 | 18 | 9 | 24 | 11 | 13 | 12 | 11 | 9 | 11 | 12 | 9 | 12 | 13 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 14 | 12 | 18 | 9 | 25 | 10 | 11 | 14 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b3a* | 1 | 1 | ||||
| 14 | 12 | 18 | 10 | 23 | 10 | 11 | 13 | 11 | 8 | 11 | 12 | 8 | 10 | 12 | E3b1* | 1 | 1 | ||||
| 14 | 12 | 18 | 10 | 24 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 14 | E3b1* | 1 | 1 | ||||
| 14 | 12 | 18 | 11 | 24 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | E3b1* | 1 | 1 | 2 | |||
| 14 | 12 | 19 | 10 | 23 | 10 | 10 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | K2 | 1 | 1 | ||||
| 14 | 12 | 19 | 10 | 22 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | F*(xH,I,J,K) | 1 | 1 | ||||
| 14 | 13 | 16 | 10 | 23 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | J2* | 1 | 1 | ||||
| 14 | 13 | 17 | 9 | 24 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 9 | 10 | 12 | E3b1* | 1 | 1 | ||||
| 14 | 13 | 17 | 10 | 25 | 9 | 12 | 12 | 12 | 9 | 11 | 12 | 9 | 9 | 10 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 14 | 14 | 16 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 14 | 14 | 16 | 11 | 24 | 10 | 11 | 14 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b* | 1 | 1 | ||||
| 14 | 14 | 16 | 11 | 24 | 10 | 11 | 12 | 11 | 10 | 12 | 14 | 9 | 10 | 12 | J* | 1 | 1 | ||||
| 14 | 14 | 17 | 11 | 23 | 11 | 11 | 12 | 11 | 10 | 11 | 12 | 10 | 9 | 11 | J2* | 1 | 1 | ||||
| 14 | 14 | 17 | 10 | 22 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 15 | 16 | 10 | 23 | 10 | 11 | 12 | 11 | 9 | 11 | 12 | 9 | 10 | 11 | J2a | 1 | 1 | ||||
| 14 | 15 | 16 | 10 | 23 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 9 | 13 | J2* | 2 | 2 | ||||
| 14 | 15 | 16 | 11 | 23 | 10 | 11 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | J* | 1 | 1 | ||||
| 14 | 15 | 18 | 10 | 25 | 9 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | F*(xH,I,J,K) | 1 | 1 | ||||
| 14 | 16 | 16 | 10 | 23 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | J* | 1 | 1 | ||||
| 14 | 16 | 16 | 10 | 23 | 10 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 16 | 17 | 10 | 23 | 10 | 10 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 16 | 17 | 10 | 23 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 16 | 17 | 10 | 23 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 16 | 17 | 10 | 23 | 11 | 12 | 12 | 11 | 9 | 12 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 16 | 17 | 10 | 23 | 12 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 14 | J* | 1 | 1 | ||||
| 14 | 16 | 17 | 11 | 23 | 13 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 17 | 15 | 10 | 23 | 12 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 17 | 15 | 10 | 23 | 10 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 17 | 16 | 10 | 23 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 2 | 2 | ||||
| 14 | 17 | 16 | 10 | 23 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 11 | J* | 1 | 1 | ||||
| 14 | 17 | 16 | 10 | 23 | 12 | 11 | 10 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 17 | 16 | 10 | 24 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 9 | 11 | 11 | J* | 1 | 1 | ||||
| 14 | 17 | 16 | 11 | 22 | 10 | 10 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 11 | J* | 1 | 1 | ||||
| 14 | 17 | 16 | 11 | 23 | 12 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 17 | 17 | 9 | 23 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 17 | 17 | 10 | 23 | 9 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 17 | 17 | 10 | 23 | 10 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | J* | 1 | 1 | 2 | |||
| 14 | 17 | 17 | 10 | 23 | 10 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | 2 | |||
| 14 | 17 | 17 | 10 | 23 | 10 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 11 | J* | 1 | 1 | ||||
| 14 | 17 | 17 | 10 | 23 | 11 | 11 | 12 | 11 | 10 | 12 | 14 | 9 | 10 | 12 | J* | 1 | 1 | ||||
| 14 | 17 | 17 | 10 | 23 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 13 | J* | 1 | 1 | ||||
| 14 | 17 | 17 | 10 | 23 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 12 | J* | 1 | 1 | ||||
| 14 | 17 | 17 | 10 | 23 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 9 | 9 | ||||
| 14 | 17 | 17 | 10 | 23 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 12 | 13 | J* | 1 | 1 | ||||
| 14 | 17 | 17 | 10 | 23 | 12 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | J* | 1 | 1 | ||||
| 14 | 17 | 17 | 10 | 23 | 12 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 3 | 4 | |||
| 14 | 17 | 17 | 10 | 24 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 17 | 17 | 10 | 25 | 11 | 15 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 17 | 17 | 11 | 23 | 10 | 10 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 17 | 17 | 11 | 23 | 10 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 17 | 17 | 11 | 23 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | J* | 1 | 1 | ||||
| 14 | 17 | 18 | 9 | 23 | 11 | 10 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 17 | 18 | 10 | 23 | 10 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J2* | 1 | 1 | ||||
| 14 | 17 | 18 | 10 | 23 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 17 | 18 | 10 | 23 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 11 | 11 | J* | 1 | 1 | ||||
| 14 | 18 | 17 | 11 | 23 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 14 | 19 | 17 | 10 | 23 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 15 | 11 | 16 | 9 | 22 | 10 | 11 | 13 | 12 | 9 | 11 | 11 | 9 | 10 | 11 | A3b2* | 1 | 1 | ||||
| 15 | 12 | 16 | 10 | 22 | 10 | 10 | 13 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | K2 | 1 | 1 | ||||
| 15 | 12 | 16 | 10 | 24 | 10 | 13 | 13 | 12 | 9 | 11 | 12 | 8 | 9 | 12 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 15 | 12 | 16 | 10 | 24 | 11 | 14 | 13 | 12 | 9 | 11 | 14 | 8 | 11 | 11 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 15 | 12 | 16 | 11 | 22 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 12 | F*(xH,I,J,K) | 1 | 1 | ||||
| 15 | 12 | 16 | 11 | 22 | 11 | 11 | 14 | 11 | 8 | 11 | 12 | 10 | 10 | 12 | F*(xH,I,J,K) | 1 | 1 | ||||
| 15 | 12 | 16 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 15 | 12 | 16 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b2* | 1 | 1 | ||||
| 15 | 12 | 16 | 11 | 24 | 10 | 12 | 14 | 12 | 9 | 11 | 12 | 8 | 10 | 12 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 15 | 12 | 16 | 11 | 24 | 11 | 13 | 13 | 12 | 9 | 11 | 12 | 8 | 9 | 14 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 15 | 12 | 17 | 9 | 24 | 10 | 11 | 14 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | F*(xH,I,J,K) | 1 | 1 | ||||
| 15 | 12 | 17 | 10 | 22 | 11 | 11 | 13 | 11 | 8 | 11 | 12 | 8 | 10 | 12 | E1* | 1 | 1 | ||||
| 15 | 12 | 17 | 10 | 23 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 12 | F*(xH,I,J,K) | 1 | 1 | ||||
| 15 | 12 | 17 | 10 | 23 | 11 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b1* | 1 | 1 | ||||
| 15 | 12 | 17 | 10 | 24 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | E3b1* | 1 | 1 | ||||
| 15 | 12 | 17 | 10 | 25 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | E3b1* | 1 | 1 | ||||
| 15 | 12 | 17 | 10 | 25 | 11 | 12 | 15 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b1* | 1 | 1 | ||||
| 15 | 12 | 17 | 11 | 23 | 10 | 10 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 12 | P*(xP1,Q1,Q2,Q3,R1) | 1 | 1 | ||||
| 15 | 12 | 17 | 11 | 23 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | J* | 1 | 1 | ||||
| 15 | 12 | 17 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 15 | 12 | 18 | 9 | 21 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 11 | 10 | E3a* | 1 | 1 | ||||
| 15 | 12 | 18 | 9 | 23 | 10 | 10 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 11 | F*(xH,I,J,K) | 1 | 1 | ||||
| 15 | 12 | 18 | 10 | 21 | 10 | 11 | 14 | 11 | 9 | 11 | 12 | 8 | 10 | 13 | E3a* | 1 | 1 | ||||
| 15 | 12 | 18 | 10 | 24 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | E3b1* | 1 | 1 | ||||
| 15 | 12 | 18 | 10 | 26 | 10 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | E3b3* | 1 | 1 | ||||
| 15 | 13 | 18 | 10 | 25 | 11 | 11 | 13 | 12 | 9 | 11 | 12 | 8 | 11 | 10 | R1a1* | 1 | 1 | ||||
| 15 | 14 | 18 | 10 | 24 | 10 | 11 | 16 | 11 | 10 | 11 | 12 | 8 | 10 | 11 | F*(xH,I,J,K) | 1 | 1 | ||||
| 15 | 15 | 16 | 9 | 24 | 9 | 11 | 12 | 11 | 9 | 11 | 12 | 9 | 9 | 11 | J2* | 2 | 2 | ||||
| 15 | 15 | 16 | 10 | 23 | 10 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 13 | J2f* | 1 | 1 | ||||
| 15 | 15 | 16 | 10 | 23 | 10 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 9 | 12 | J2* | 1 | 1 | ||||
| 15 | 15 | 16 | 10 | 24 | 10 | 13 | 13 | 11 | 9 | 11 | 12 | 8 | 9 | 12 | F*(xH,I,J,K) | 1 | 1 | ||||
| 15 | 15 | 17 | 10 | 22 | 11 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 12 | J* | 1 | 1 | ||||
| 15 | 16 | 17 | 10 | 23 | 10 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 11 | F*(xH,I,J,K) | 1 | 1 | ||||
| 15 | 16 | 17 | 10 | 24 | 10 | 10 | 13 | 11 | 9 | 11 | 12 | 10 | 10 | 12 | I*(xI1a2,I1a3,I1b2) | 1 | 1 | ||||
| 15 | 15 | 17 | 11 | 22 | 10 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 9 | 11 | J2f1 | 1 | 1 | ||||
| 16 | 12 | 16 | 11 | 23 | 11 | 13 | 14 | 12 | 9 | 11 | 12 | 8 | 11 | 12 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 16 | 12 | 16 | 11 | 23 | 11 | 13 | 14 | 12 | 9 | 11 | 12 | 8 | 12 | 12 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 16 | 12 | 16 | 11 | 24 | 10 | 12 | 14 | 12 | 9 | 11 | 12 | 8 | 11 | 12 | P*(xP1,Q1,Q2,Q3,R1) | 1 | 1 | ||||
| 16 | 12 | 16 | 11 | 24 | 10 | 12 | 14 | 12 | 9 | 11 | 12 | 8 | 11 | 13 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 16 | 12 | 16 | 11 | 24 | 10 | 12 | 15 | 12 | 9 | 11 | 12 | 8 | 10 | 10 | R1*(xR1a1,R1b1-b8) | 1 | 1 | ||||
| 16 | 12 | 17 | 9 | 23 | 10 | 12 | 13 | 11 | 8 | 11 | 12 | 10 | 10 | 13 | F*(xH,I,J,K) | 1 | 1 | ||||
| 16 | 12 | 17 | 10 | 23 | 10 | 11 | 14 | 11 | 9 | 11 | 12 | 10 | 11 | 11 | F*(xH,I,J,K) | 1 | 1 | ||||
| 16 | 12 | 17 | 11 | 24 | 9 | 11 | 13 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
| 16 | 12 | 19 | 10 | 21 | 11 | 12 | 13 | 11 | 9 | 11 | 12 | 8 | 11 | 12 | E3b* | 1 | 1 | ||||
| 16 | 12 | 19 | 10 | 25 | 11 | 11 | 13 | 12 | 9 | 11 | 12 | 8 | 11 | 11 | K2 | 1 | 1 | ||||
| 17 | 14 | 18 | 10 | 22 | 10 | 11 | 13 | 11 | 8 | 11 | 12 | 11 | 9 | 14 | E1* | 1 | 1 | ||||
| 17 | 14 | 18 | 10 | 24 | 10 | 11 | 12 | 11 | 10 | 11 | 12 | 8 | 10 | 11 | J* | 1 | 1 | ||||
| 18 | 12 | 17 | 10 | 24 | 9 | 11 | 12 | 11 | 9 | 11 | 12 | 8 | 10 | 10 | E3b2* | 1 | 1 | ||||
Table A4.
TMRCA Estimates (in KYA) and 95% CIs of Y-Chromosomal Lineages in North Africa[Note]
|
TMRCA [in KY] (95% CI) |
||||
| Lineage | Mutation | No. of Chromosomes | 30 Years/Generationa | 25 Years/Generationb |
| All | 256 | 13.25 (9.56–18.50) | 35.51 (25.12–54.40) | |
| E | M145 | 135 | 9.63 (7.39–12.68) | 21.76 (16.51–27.77) |
| E3a | M2 | 2 | 2.97 (1.49–5.02) | 6.18 (3.15–8.99) |
| E1 | M33 | 2 | 4.10 (2.37–6.74) | 8.12 (5.32–14.46) |
| E3b1 | M78 | 27 | 4.70 (3.43–6.37) | 10.05 (8.48–11.90) |
| E3b | M35 | 131 | 8.70 (6.53–11.51 | 19.54 (15.06–25.71 |
| E3b2 | M81 | 85 | 3.87 (2.94–5.10 | 7.61 (6.62–8.68 |
| F | M89 | 122 | 9.75 (7.42–12.64) | 22.87 (19.87–26.33) |
| J | 12f2 | 72 | 6.78 (4.85–9.52) | 15.43 (11.91–19.34) |
| J2 | M172 | 11 | 3.70 (2.72–4.86) | 7.64 (6.42–9.96) |
| R | M173 | 22 | 5.48 (4.01–7.17) | 12.03 (9.39–14.82) |
| I | M170 | 1 | … | … |
| K | M9 | 32 | 7.94 (5.88–10.51) | 17.80 (13.63–23.05) |
Note.— Estimates are based on the 15 STRs in the five North African samples of the present study.
The two parameters describing the population growth (alpha and beta) have been set as in footnote a of table 2, by use of the same microsatellite mutation rates as in that footnote.
Electronic-Database Information
Accession numbers and URLs for data presented herein are as follows:
- AIDA: Autocorrelation Indices for DNA Analysis, http://web.unife.it/progetti/genetica/Giorgio/giorgio_soft.html
- Arlequin, http://lgb.unige.ch/arlequin/ (for population genetics software package)
- Fluxus Engineering, http://www.fluxus-engineering.com/sharenet.htm (for NETWORK 3.1.1.1 phylogenetic network analysis software)
- Ian Wilson’s download page, http://www.maths.abdn.ac.uk/~ijw/downloads/download.htm (for BATWING program)
- Y Chromosome Consortium, http://ycc.biosci.arizona.edu/nomenclature_system/frontpage.html
References
- Arioti M, Oxby C (1997) From hunter-fisher-gathering to herder-hunter-fisher-gathering in prehistoric times (Saharo-Sudanese region). Nomadic Peoples (New Ser) 1:98–119 [Google Scholar]
- Ayub Q, Mohyuddin A, Qamar R, Mazhar K, Zerjal T, Mehdi SQ, Tyler-Smith C (2000) Identification and characterisation of novel human Y-chromosomal microsatellites from sequence database information. Nucleic Acids Res 28:e8 10.1093/nar/28.2.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Röhl A (1999) Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol 16:37–48 [DOI] [PubMed] [Google Scholar]
- Barbujani G, Pilastro A, De Domenico S, Renfrew C (1994) Genetic variation in North Africa and Eurasia: Neolithic demic diffusion vs. Paleolithic colonisation. Am J Phys Anthropol 95:137–154 [DOI] [PubMed] [Google Scholar]
- Barker G (2003) Transitions to farming and pastoralism in North Africa. In: Bellwood P, Renfrew C (eds) Examining the farming/language dispersal hypothesis. McDonald Institute for Archaeological Research, Cambridge, United Kingdom, pp 151–161 [Google Scholar]
- Bertorelle G, Barbujani G (1995) Analysis of DNA diversity by spatial autocorrelation. Genetics 140:811–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Comas D, Oefner PJ, Underhill PA, Bertranpetit J (2001) High-resolution analysis of human Y-chromosome variation shows a sharp discontinuity and limited gene flow between northwestern Africa and the Iberian Peninsula. Am J Hum Genet 68:1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch E, Calafell F, Perez-Lezaun A, Comas D, Mateu E, Bertranpetit J (1997) Population history of North Africa: evidence from classical genetic markers. Hum Biol 69:295–311 [PubMed] [Google Scholar]
- Bosch E, Calafell F, Santos FR, Perez-Lezaun A, Comas D, Benchemsi N, Tyler-Smith C, Bertranpetit J (1999) Variation in short tandem repeats is deeply structured by genetic background on the human Y chromosome. Am J Hum Genet 65:1623–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova M, Leroy P, Boucekkine C, Weissenbach J, Bishop C, Fellous M, Purrello M, Fiori G, Siniscalco M (1985) A human Y-linked DNA polymorphism and its potential for estimating genetic and evolutionary distance. Science 230:1403–1406 [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A (1994) The history and geography of human genes. Princeton University Press, Princeton, NJ [Google Scholar]
- Cruciani F, La Fratta R, Santolamazza P, Sellitto D, Pascone R, Moral P, Watson E, Guida V, Colomb EB, Zaharova B, Lavinha J, Vona G, Aman R, Calì F, Akar N, Richards M, Torroni A, Novelletto A, Scozzari R (2004) Phylogeographic analysis of haplogroup E3b (E-M215) Y chromosomes reveals multiple migratory events within and out of Africa. Am J Hum Genet 74:1014–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciani F, Santolamazza P, Shen P, Macaulay V, Moral P, Olckers A, Modiano D, Holmes S, Destro-Bisol G, Coia V, Wallace DC, Oefner PJ, Torroni A, Cavalli-Sforza LL, Scozzari R, Underhill PA (2002) A back migration from Asia to sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am J Hum Genet 70:1197–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond J, Bellwood P (2003) Farmers and their languages: the first expansions. Science 300:597–603 10.1126/science.1078208 [DOI] [PubMed] [Google Scholar]
- Flores C, Maca-Meyer N, Pérez JA, Hernández M, Cabrera VM (2001) Y-chromosome differentiation in Northwest Africa. Hum Biol 73:513–524 [DOI] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C (2003) The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet 4:598–612 10.1038/nrg1124 [DOI] [PubMed] [Google Scholar]
- Kayser M, Roewer L, Hedman M, Henke L, Henke J, Brauer S, Kruger C, Krawczak M, Nagy M, Dobosz T, Szibor R, de Knijff P, Stoneking M, Sajantila A (2000) Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am J Hum Genet 66:1580–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis JR, Rowold DJ, Regueiro M, Caeiro B, Cinnioglu C, Roseman C, Underhill PA, Cavalli-Sforza LL, Herrera RJ (2004) The Levant versus the Horn of Africa: evidence for bidirectional corridors of human migrations. Am J Hum Genet 74:532–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maca-Meyer N, González AM, Pestano J, Flores C, Larruga JM, Cabrera VM (2003) Mitochondrial DNA transit between West Asia and North Africa inferred from U6 phylogeography. BMC Genet 4:15 10.1186/1471-2156-4-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manni F, Leonardi P, Barakat A, Rouba H, Heyer E, Klintschar M, McElreavey K, Quintana-Murci L (2002) Y-chromosome analysis in Egypt suggests a genetic regional continuity in Northeastern Africa. Hum Biol 74:645–658 [DOI] [PubMed] [Google Scholar]
- McEvedy C (1980) The Penguin atlas of African history. Penguin Books, New York [Google Scholar]
- Muzzolini A (1993) The emergence of a food-producing economy in the Sahara. In: Shaw T, Sinclair P, Andah B, Okpoko A (eds) The archaeology of Africa: food, metals and towns. Routledge, London, pp 227–239 [Google Scholar]
- Paracchini S, Arredi B, Chalk R, Tyler-Smith C (2002) Hierarchical high-throughput SNP genotyping of the human Y chromosome using MALDI-TOF mass spectrometry. Nucleic Acids Res 30:e27 10.1093/nar/30.6.e27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repping S, Skaletsky H, Brown L, van Daalen SK, Korver CM, Pyntikova T, Kuroda-Kawaguchi T, de Vries JW, Oates RD, Silber S, van der Veen F, Page DC, Rozen S (2003) Polymorphism for a 1.6-Mb deletion of the human Y chromosome persists through balance between recurrent mutation and haploid selection. Nat Genet 35:247–251 10.1038/ng1250 [DOI] [PubMed] [Google Scholar]
- Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, Amorim A, Amos W, et al. (2000) Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet 67:1526–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlen M (1991) A Guide to the World’s Languages. Vol 1. Edward Arnold, London [Google Scholar]
- Salas A, Richards M, De la Fe T, Lareu MV, Sobrino B, Sánchez-Diz P, Macaulay V, Carracedo A (2002) The making of the African mtDNA landscape. Am J Hum Genet 71:1082–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Magri C, Benuzzi G, Lin AA, Al-Zahery N, Battaglia V, Maccioni L, Triantaphyllidis C, Shen P, Oefner PJ, Zhivotovsky LA, King R, Torroni A, Cavalli-Sforza LL, Underhill PA, Santachiara-Benerecetti AS (2004) Origin, diffusion, and differentiation of Y-chromosome haplogroups E and J: inferences on the Neolithization of Europe and later migratory events in the Mediterranean area. Am J Hum Genet 74:1023–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semino O, Passarino G, Oefner PJ, Lin AA, Arbuzova S, Beckman LE, De Benedictis G, Francalacci P, Kouvatsi A, Limborska S, Marcikiae M, Mika A, Mika B, Primorac D, Santachiara-Benerecetti AS, Cavalli-Sforza LL, Underhill PA (2000) The genetic legacy of Paleolithic Homo sapiens sapiens in extant Europeans: a Y chromosome perspective. Science 290:1155–1159 10.1126/science.290.5494.1155 [DOI] [PubMed] [Google Scholar]
- Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokal RR, Oden NL (1978) Spatial autocorrelation in biology. Biol J Linn Soc Lond 10:199–249 [Google Scholar]
- Thomas MG, Bradman N, Flinn HM (1999) High throughput analysis of 10 microsatellite and 11 diallelic polymorphisms on the human Y-chromosome. Hum Genet 105:577–581 10.1007/s004390051148 [DOI] [PubMed] [Google Scholar]
- Tishkoff SA, Williams SM (2002) Genetic analysis of African populations: human evolution and complex disease. Nat Rev Genet 3:611–621 [DOI] [PubMed] [Google Scholar]
- Underhill PA, Shen P, Lin AA, Jin L, Passarino G, Yang WH, Kauffman E, Bonne-Tamir B, Bertranpetit J, Francalacci P, Ibrahim M, Jenkins T, Kidd JR, Mehdi SQ, Seielstad MT, Wells RS, Piazza A, Davis RW, Feldman MW, Cavalli-Sforza LL, Oefner PJ (2000) Y chromosome sequence variation and the history of human populations. Nat Genet 26:358–361 10.1038/81685 [DOI] [PubMed] [Google Scholar]
- Weale ME, Yepiskoposyan L, Jager RF, Hovhannisyan N, Khudoyan A, Burbage-Hall O, Bradman N, Thomas MG (2001) Armenian Y chromosome haplotypes reveal strong regional structure within a single ethno-national group. Hum Genet 109:659–674 10.1007/s00439-001-0627-9 [DOI] [PubMed] [Google Scholar]
- Wilson IJ, Balding DJ (1998) Genealogical inference from microsatellite data. Genetics 150:499–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Y Chromosome Consortium (2002) A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res 12:339–348 10.1101/gr.217602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky LA, Underhill PA, Cinnioglu C, Kayser M, Morar B, Kivisild T, Scozzari R, Cruciani F, Destro-Bisol G, Spedini G, Chambers GK, Herrera RJ, Yong KK, Gresham D, Tournev I, Feldman MW, Kalaydjieva L (2004) The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet 74:50–61 [DOI] [PMC free article] [PubMed] [Google Scholar]