Abstract
Purpose and Experimental Design: MCM4 is a member of Minichromosome maintenance protein family. MCM2–7 proteins play an essential role in eukaryotic DNA replication and have been identified as components of DNA replication licensing factors. So far, no research on MCM4 has been reported in esophageal cancer. In this study, we detected via RT-PCR the expression status of MCM4 in esophageal cancer from southern China and therefore disclose the relationship between MCM4 and esophageal cancer. Results: 65% (39/60) cases showed increased expression of MCM4 in the carcinomas when compared with normal esophageal epithelia in which no or low MCM4 expression was detected in most cases. Twenty of sixty cases (33%) showed increased expression of MCM4 in the adjacent epithelia. Furthermore, MCM4 expression in esophageal carcinomas was significantly higher than the one in the adjacent epithelia (chi square value is 12.037, P<0.001). Significant difference for the expression status of MCM4 was found between the patients with histopathological stage T3 and stage T1 (chi square value=4.038, P<0.05). Conclusions: The increased expression of MCM4 might be associated with pathological staging of esophageal cancer. The alterations of MCM4 are possibly related to the earlier event of esophageal carcinogenesis. MCM4 is probably a valuable molecular marker involved in the development and/or genesis of esophageal cancer.
Keywords: Esophageal cancer, MCM4, Molecular marker
Introduction
Esophageal carcinoma (EC) is one of the most common malignant tumors worldwide (Russo et al. 1996). It is the forth most prevalent malignancy in China (Tang et al. 2000). Esophageal squamous cell carcinoma (ESCC) is the predominant histologic subtype of esophageal cancer and has higher incidence than adenocarcinomas in Chinese people. The genetic changes in esophageal cancer have been reported as point mutations of p53 and p16, amplification and/or overexpression of int-2, hst-1, cyclin D1 and EGFR, as well as loss of heterozygosity of genes in many chromosomal regions (Montesano et al. 1996; Huang et al. 2002; Liu et al. 2004; Li et al. 2004). However, the molecular events in detail that cause the malignant transformation of esophageal epithelia are still unclear.
MCM4 is mapped to a region on 8q11.2 head-to-head next to the PRKDC/DNA-PK, a DNA-activated protein kinase involved in the repair of DNA double-strand breaks. MCM4 is one of the highly conserved mini-chromosome maintenance proteins that are essential for initiating eukaryotic genome replication. The MCM proteins are the key components of the pre-replication complex and may be involved in the formation of replication forks and in the recruitment of other DNA replication related proteins (Labib et al. 2001).
MCM2 might be used as a prognostic marker in breast cancer (Gonzalez et al. 2003). Aberrant expression of MCM2 and 5 in dysplastic squamous esophageal epithelia and Barrett’s mucosa implied that MCM2 and 5 are possibly diagnostic markers that cause the disruption of cell cycle control (Alison et al. 2002; Going et al. 2002; Sirieix et al. 2003). Immunohistochemical studies showed that MCM3 and 4 are ubiquitously expressed in the cancer cells of human uterine cervix. Further, the expression level of MCM3 and 4 were higher in cancer cells than in the normal proliferating cells of the uterine cervix and dysplastic cells, suggesting that they might be useful markers to distinguish these cells (Ishimi et al. 2003). So far, no research on MCM4 has been reported in esophageal cancer. In order to reveal whether MCM4 is involved in the genesis and/or progression of esophageal cancer, we explored the expression status of MCM4 in ESCCs from southern China via RT-PCR. We also observed the relationship between the changes of MCM4 and clinical/pathological data such as stage, grade, lymph node metastasis, age and sex.
Materials and methods
Tumor samples
For this study, fresh tissues including ESCCs and matched adjacent (less than 2 cm away from the carcinomas) and distant normal epithelia (more than 5 cm away from the carcinomas) from a total of 60 patients were procured from surgical resection specimens collected by the Department of Pathology in Cancer Center, Sun Yat-Sen University, Guangzhou, China. Primary matched esophageal tissues from the same patients were separately excised by experienced pathologists and immediately placed in RNA protection reagents at 4°C overnight, and were thereafter put in −86°C refrigerator or liquid nitrogen. All the patients received no treatment before surgery, and signed separate informed consent forms for sampling, RNA isolation and storage. The experimental procedures were reviewed and accepted by the ethics committees of the Cancer Center of the Sun Yat-Sen University. Data were recorded concerning the clinical/pathological parameters of tumors, including histopathological stage, grade, lymph node metastasis, and clinical data such as age and sex (Table 1).
Table 1.
Basic clinical/pathological data and messenger RNA expression status of MCM4 in 60 esophageal squamous cell carcinomas from southern China carcinoma
| Case no. | Age /sex | Stage | Grade | LNM | Elevated expression of MCM4 | |
|---|---|---|---|---|---|---|
| Adjacent epithelia | Carcinoma | |||||
| 1 | 69/M | T3 | G3 | + | yes | |
| 2 | 69/M | T3 | G1/G2 | − | × | × |
| 3 | 61/M | T1 | G1 | − | × | × |
| 4 | 45/M | − | G2 | × | yes | |
| 5 | 64/F | T2 | G1 | + | × | yes |
| 6 | 60/M | T3 | G1 | − | × | × |
| 7 | 58/M | − | − | − | × | × |
| 8 | 57/M | T3 | G3/G4 | + | yes | yes* |
| 9 | 53/M | T4 | G1 | + | no | yes |
| 10 | 45/F | T2 | G2/G3 | − | yes | yes* |
| 11 | 42/M | T3 | G2 | − | yes | |
| 12 | 63/M | T2 | G2 | − | × | yes |
| 13 | 52/M | T3 | G2 | + | × | × |
| 14 | 67/F | − | G2/G3 | yes | yes | |
| 15 | 56/M | T1 | G2 | − | × | × |
| 16 | 74/M | T3 | G2/G3 | + | yes | yes* |
| 17 | 68/F | T2 | G1 | − | × | × |
| 18 | 42/M | T2 | − | + | No | |
| 19 | 63/M | T3 | − | − | no | yes |
| 20 | 64/M | T3 | G1 | + | no | No |
| 21 | 70/F | T3 | G2 | + | × | × |
| 22 | 63/M | T3 | G1/G2 | − | × | × |
| 23 | 56/M | T3 | + | × | yes | |
| 24 | 55/M | T3 | G1 | − | yes | yes |
| 25 | 46/M | T3 | G1 | − | yes | yes* |
| 26 | 53/M | T3 | G3 | + | no | No |
| 27 | 65/M | T3 | G2/G3 | − | yes | yes |
| 28 | 60/F | T2 | G2 | − | yes | yes |
| 29 | 50/M | T3 | G2 | − | no | No |
| 30 | 56/M | T2 | G1 | + | no | No |
| 31 | 63/M | T2 | G1/G2 | − | no | no |
| 32 | 72/M | T2 | G2/G3 | − | no | no |
| 33 | 72/M | T3 | G2/G3 | − | yes | yes |
| 34 | 39/F | T2 | G1/G2 | − | yes | |
| 35 | 70/M | T4 | G2/G3 | + | no | no |
| 36 | 54/M | T2 | G1 | − | no | no |
| 37 | 50/M | T3 | G2 | + | no | no |
| 38 | 78/M | T2 | G1/G2 | + | no | no |
| 39 | 45/M | T3 | − | yes | yes | |
| 40 | 49/F | T3 | G1 | − | yes | yes* |
| 41 | 53/F | T3 | − | − | yes | |
| 42 | 59/M | T3 | G1 | − | yes | yes |
| 43 | 45/F | T3 | G2/G3 | + | yes | |
| 44 | 48/M | T3 | − | + | yes | yes* |
| 45 | 60/M | T2 | G3 | − | no | yes |
| 46 | 65/F | T3 | − | − | yes | yes * |
| 47 | 37/M | T3 | − | + | yes | yes |
| 48 | 63/M | T3 | G3 | + | yes | |
| 49 | 50/M | T3 | G2 | − | no | yes |
| 50 | 44/M | T3 | G2/G3 | + | yes | yes |
| 51 | 70/F | T3 | G2 | + | no | yes |
| 52 | 59/M | − | − | − | yes | yes* |
| 53 | 48/M | T2 | G2 | + | yes | |
| 54 | 55/M | T2 | G3 | − | yes | yes |
| 55 | 56/M | Tis | − | − | × | × |
| 56 | 62/M | T3 | G2 | + | no | yes |
| 57 | 67/F | T2 | G1 | − | yes | yes* |
| 58 | 53/M | T3 | G2 | − | × | yes |
| 59 | 55/M | T3 | G2/G3 | + | yes | yes* |
| 60 | 50/M | T3 | G2 | + | no | yes |
Note: LNM stand for lymph node metastasis; − stand for negative; + stand for positive; × stand for no expression detected; *MCM4 expressed at a higher level in the carcinomas than in their matched adjacent epithelia
RNA extraction and first strand cDNA synthesis
Total RNAs were prepared using Trizol reagent (Gibco) according to the manufacturer’s instructions. 50 μg of total RNAs from cell line EC9706, tumor tissues, adjacent and distant normal epithelia were treated with DNase I (Takara) following the manufacturer’s instruction. 5 μg of each RNA was reverse transcribed with 200 units MMLV (Promega) in the presence of 50 μM oligo (dT) primer and 25 units rRNasin Ribonuclease inhibitor in 20 μl reverse transcription reaction buffer (200 mM Tris-HCl, pH8.4, 500 mM KCl, 2.5 mM MgCl2, 10 mM DTT, 0.5 mM dNTP), at 42°C for 50 min, followed by heating to 70°C for 15 min to inactivate reverse transcriptase.
RT-PCR analysis
RT-PCR was performed in a 20 μl reaction containing 1 μl of first-stranded cDNA, 1X PCR buffer, 1.5 mM MgCl2, 200 μM dNTPs, 0.5 μM primers of MCM4 and Beta-actin, 1.5 U Taq DNA polymerase (Takara). PCR was started by heating at 94°C for 5 min, followed by 35 cycles of 94°C for 40 s (MCM4)/30 s (Beta-actin), 59°C (MCM4)/55°C (Beta-actin) for 40 s (MCM4)/30 s (Beta-actin), and 72°C for 1 m (MCM4)/40 s (Beta-actin), with a final extension at 72°C for 5 min. The RT-PCR products were resolved by 1.2% agarose (containing 0.1 mg/ml ethidium bromide) gel electrophoresis. The lengths of the PCR products of MCM4 and Beta-actin were 697 bp and 315 bp, respectively. The primer sequences of MCM4 were as follows: upstream primer 5′CCG AAT CAA CAT GGA AAC CT 3′; downstream primer 5’AGT CCA CCT GGC GAG TAG C 3′. The upstream and downstream primer sequences of Beta-actin were 5′ CAT CTC TTG CTC GAA GTC CA 3′ and 5′ ATC ATG TTT GAG ACC TTC AAC A 3′, respectively. Using beta-actin as an internal control, we know the expression status of MCM4 in ESCCs.
Sequencing of MCM4 RT-PCR products
In order to exclude the possibility of the PCR products as non-specific amplification, firstly, RT-PCR of MCM4 was done with the high-fidelity DNA polymerase pfu. RT-PCR was done in a 50 μl reaction mixture containing 2 μl of first-stranded cDNA, 1X pfu buffer, 200 μM dNTPs, 0.5 μM primers of MCM4, 2 U pfu DNA polymerase (Promega). Other PCR reaction conditions were same as above-mentioned. The RT-PCR products were electrophoresized on 1% agarose gel. The target gene products were reclaimed with Agarose Gel DNA Purification Kit (Takara) according to the manufacturer’s protocol. Sequencing of the PCR products was done in Sangon Incorporation.
Statistical analysis
Chi-square tests were used in statistical analysis to evaluate the relationship between alterations of MCM4 and clinical/pathological parameters (tumor grade or stage or lymph node metastasis or age or sex). All P values were two-sided and were considered statistically significant if P<0.05.
Results
Elevated expression of MCM4 in ECs
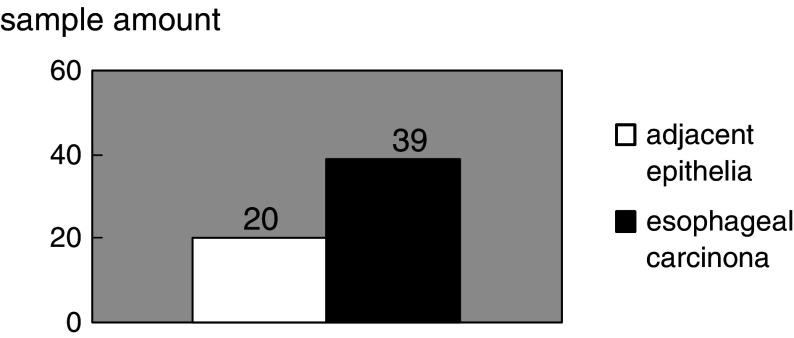
In order to explore the expression status of MCM4, RT-PCR was done in 60 ESCCs from southern China. Figure 1 shows the representative figures of the expression status of MCM4 in ESCCs. The results revealed that, when compared with normal esophageal epithelia in which no or low MCM4 expression was detected, 65% (39/60) and 33% (20/60) cases were observed as increased the expression of MCM4 in the carcinomas and adjacent epithelia, respectively. Moreover, MCM4 expression in ECs was significantly higher than the one in the adjacent epithelia (chi square value is 12.037, P<0.001) (Table 1, Fig 2).
Fig. 1.
Expression status of MCM4 in esophageal squamous cell carcinomas from southern China. M, stands for “molecular weight marker DL2000”; N, stands for “normal esophageal epithelium (more than 5 cm away from the carcinomas)”; P, stands for “adjacent esophageal epithelia (less than 2 cm away from esophageal carcinomas, ECs)”; T, stands for “EC”; “1,24,25,28” stands for “case number”
Fig. 2.
Case number of elevated expression of MCM4 in esophageal carcinoma and its adjacent tissue when compared with normal esophageal epithelia. When compared with the normal epithelia in which no or low expression of MCM4 was detected, 33% (20/60) and 65% (39/60) cases showed increased expression of MCM4 in the adjacent tissue and esophageal cancer, respectively. Furthermore, MCM4 expression in ECs was significantly higher than the one in the adjacent epithelia
Identification of MCM4 via sequencing of RT-PCR products
In order to exclude the possibility of the PCR products as non-specific amplification, firstly, RT-PCR of MCM4 was done with the high-fidelity DNA polymerase pfu. The PCR products were then purified with the DNA Fragment Purification Kit (Takara). We sequenced the PCR products in Sangon Incorporation. No mutation or polymorphism of MCM4 was found in esophageal cancer. However, the PCR products were proven to be MCM4.
Bioinformatic analysis of MCM4
Blast research using the sequence of MCM4 RT-PCR products indicated that except for the sequence of MCM4 itself, no other human genomic or RNA sequence showed significant homology with MCM4.
The relationship between elevated expression of MCM4 and clinical/pathological data of esophageal cancer
Chi square test was used in statistical analysis for the relationship between the expression status of MCM4 and clinical/pathological parameters. There is a tendency of increased expression for MCM4 when we compared the expression status of MCM4 in an earlier stage with the one in a later stage (T3+T4 versus T1+T2, chi square value=3.585, 0.05<P<0.1). Furthermore, significant difference was found when we compared the expression status of MCM4 at T3 with the one at T1 (chi square value=4.038, P<0.05). The chi square values were 1.538, 0.000814, 1.814 and 1.077, respectively, when we compared the expression status of MCM4 with the clinical/pathological parameters such as grade (G3+G4 versus G1+G2), lymph node metastasis (positive versus negative), sex (male versus female) and age (over 60 years old versus younger than 60 years old). All these P values are more than 0.05. Thus, no significant difference was found between the alterations of MCM4 and the clinical/pathological data.
Discussion
Esophageal cancer is one of the most prevalent malignancy in China. It is also very popular in southern China, for example, Guangdong province. So far, we have not found efficient diagnosis and treatment measures to deal with the disease. Searching for valuable molecular markers or esophageal cancer-related genes will help to disclose the secret of the mechanism underlying the development and/or genesis of esophageal cancer. In order to uncover the mechanism underlying esophageal carcinogenesis, firstly, we detected via RT-PCR the expression status of a great deal of genes, including apoptosis related genes, cell proliferation-related genes, DNA damage and repair-related genes and signal transduction related genes, in ESCCs from southern China. Previous reports showed altered expression pattern of many genes in ESCCs (Zhi et al. 2003; Yue et al. 2003). In our study, MCM4, a gene that is essential for eukaryotic genome replication, is of our particular interest. So far, no reports showed any relationship between the changes of MCM4 and esophageal cancer. Comparing with normal esophageal epithelia in which no or low MCM4 expression was detected, higher frequency of elevated expression of MCM4 was found in 65% (39/60) of ECs. Sequencing of MCM4 was done in Sangon Incorporation. The RT-PCR products were proved to be MCM4. Our sequencing study together with DNase I treatment for the RNA samples excluded the possible influence of contaminated genomic DNA on RT-PCR results. Higher frequency of elevated expression of MCM4 in esophageal squamous cell carcinomas implicates the possibility of MCM4 involved in the genesis and/or progression of esophageal cancer.
Early diagnosis and treatment is an efficient measure to reduce the death rate of esophageal cancer patients. But most patients accepting treatment in hospital have been in a later stage. One of the main reasons leading to this situation is that no efficient molecular markers for earlier stage could be used in the diagnosis and treatment of esophageal cancer. While our intention behind comparing the expression pattern between esophageal cancer tissues and matched distant normal esophageal mucosa (more than 5 cm away from the carcinomas) was to find differentially expressed genes, we also compared the expression status of genes from adjacent mucosa (less than 2 cm away from the carcinomas) with the one from the carcinomas and distant normal tissues, respectively, to search for genes related to earlier stage of esophageal cancer. The results revealed that increased expression of MCM4 in adjacent mucosa was found in 20 of 60 (33%) cases, indicating that changed expression of MCM4 is possibly an earlier event during the development of some ECs. Significant difference for the expression status of MCM4 was found when we compared the ECs (65%) with the adjacent epithelia (33%) (chi square value is 12.037, P<0.001). The results suggested that MCM4 is probably a valuable molecular marker to be involved in the progression of esophageal cancer. It was reported that MCM4 are ubiquitously expressed in cancer cells of the uterine cervix. Further, the positive rate and level of MCM4 expression appeared to be higher in cancer cells than in normal proliferating cells of the uterine cervix and dysplastic cells (Ishimi et al. 2003). Our results were consistent with previous reports on uterine cervix cancer.
The choice of different clinical treatment measures of esophageal cancer depends largely on clinical staging. But patients with esophageal cancer at the same stage experienced varying clinical outcomes despite identical tumor staging by standard diagnostic methods because clinical and histopathological staging failed to reveal the underlying complex biology of cancer. More and more evidence (Hosch et al. 2003; Lambart et al. 2004) showed that molecular staging is possibly a good way to solve this problem. However, we have not yet found meaningful molecular markers used in clinical staging of esophageal cancer. In the near future from now, searching for valuable molecular markers will still be a hotspot project in the study of esophageal cancer. In this study, we observed the relationship between the increased expression of MCM4 and the histopathological stage of esophageal cancer. The results showed a tendency of increased expression for MCM4 when we compared the expression status of MCM4 in an earlier stage with the one in a later stage (T3+T4 versus T1+T2, chi square value=3.585, 0.05<P<0.1). Furthermore, significant difference was found when we compared the expression status of MCM4 at T3 with the one at T1 (chi square value=4.038, P<0.05). Although a bias might exist due to fewer T1 stage sample amounts, we could conclude in the present study that the increased expression of MCM4 might be associated with pathological staging of esophageal cancer. In our future projects, we should enlarge our sample amounts to confirm this conclusion. These results suggested that MCM4 is probably a molecular marker used in distinguishing different stagings of esophageal cancer.
Acknowledgements
This work was supported by the grant from Chinese Scientific and Technological Key Project of Guangdong Province (A3020103) and the grant from Scientific and Technological Project of Guangzhou City (2002J1-C0181).
References
- Alison MR, Hunt T, Forbes SJ, (2002) Minichromosome maintenance (MCM) proteins may be pre-cancer markers. Gut 50(3):290–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Going JJ, Keith WN, Neilson L, Stoeber K, Stuart RC, Williams GH, (2002) Aberrant expression of minichromosome maintenance proteins 2 and 5, and Ki-67 in dysplastic squamous oesophageal epithelium and Barrett’s mucosa. Gut 50(3):373–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez MA, Pinder SE, Callagy G, Vowler SL, Morris LS, Bird K, Bell JA, Laskey RA, Coleman N, (2003) Minichromosome maintenance protein 2 is a strong independent prognostic marker in breast cancer. J Clin Oncol 21(23):4306–4313 [DOI] [PubMed] [Google Scholar]
- Hosch SB, Stoecklein NH, Izbicki JR, (2003) Molecular markers and staging of early esophageal cancer. Langenbecks Arch Surg 388(2):77–82 [DOI] [PubMed] [Google Scholar]
- Huang XP, Wei F, Liu XY, Xu X, Hu H, Chen BS, Xia SH, Han YS, Han YL, Cai Y, Wu M, Wang MR, (2002) Allelic loss on 13q in esophageal squamous cell carcinomas from northern China. Cancer Lett 185(1):87–94 [DOI] [PubMed] [Google Scholar]
- Ishimi Y, Okayasu I, Kato C, Kwon HJ, Kimura H, Yamada K, Song SY (2003) Enhanced expression of Mcm proteins in cancer cells derived from uterine cervix. Eur J Biochem 270(6):1089–1101 [DOI] [PubMed] [Google Scholar]
- Labib K, Kearsey SE, Diffley JF, (2001) MCM2–7 proteins are essential components of prereplicative complexes that accumulate cooperatively in the nucleus during G1-phase and are required to establish, but not maintain, the S-phase checkpoint. Mol Biol Cell 12(11):3658–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert R, (2004) Diagnosis of esophagogastric tumors. Endoscopy 36(2):110–119 [DOI] [PubMed] [Google Scholar]
- Li XD, Huang XP, Zhao CX, Li QJ, Xu X, Cai Y, Han YL, Rong TH, Wang MR, (2004) Identification of a minimal deletion region on chromosome 5q in Chinese esophageal squamous cell carcinomas. Cancer Lett 215(2):221–228 [DOI] [PubMed] [Google Scholar]
- Liu FX, Huang XP, Zhao CX, Xu X, Han YL, Cai Y, Wu RL, Wu M, Zhan QM, Wang MR (2004) Allelic loss and down-regulation of FHIT gene expression in esophageal squamous cell carcinoma. Article in Chinese Ai Zheng 23(9):992–998 [PubMed] [Google Scholar]
- Montesano R, Hollstein M, Hainaut P, (1996). Genetic alterations in oesophageal cancer and their relevance to etiology and pathogenesis: a review. Int J Cancer (Pred Oncol) 69:225–235 [DOI] [PubMed] [Google Scholar]
- Russo A, Franceschi S (1996) The epidemiology of esophageal cancer. Ann Ist super sanita 32:65–72 [PubMed] [Google Scholar]
- Sirieix PS, O’Donovan M, Brown J, Save V, Coleman N, Fitzgerald RC (2003) Surface expression of minichromosome maintenance proteins provides a novel method for detecting patients at risk for developing adenocarcinoma in Barrett’s esophagus. Clin Cancer Res 9(7):2560–2566 [PubMed] [Google Scholar]
- Tang ZY. Modern Oncology (the second edition), published by FuDan publishing Company in October, 2000. Article in Chinese Page:658–694
- Yue CM, Deng DJ, Bi MX, Guo LP, Lu SH (2003) Expression of ECRG4, a novel esophageal cancer-related gene, downregulated by CpG island hypermethylation in human esophageal squamous cell carcinoma. World J Gastroenterol 9(6):1174–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi H, Zhang J, Hu G. Lu J, Wang X, Zhou C, Wu M, Liu Z (2003) The deregulation of arachidonic acid metabolism-related genes in human esophageal squamous cell carcinoma. Int J Cancer 106(3):327–333 [DOI] [PubMed] [Google Scholar]