Abstract
Purpose
Sulfotransferase 1A1 is a member of sulfotransferase family that plays an important role in the biotransformation of numerous carcinogenic and mutagenic compounds through sulfation. The present study has investigated the association between SULT1A1 polymorphism and primary brain tumor incidence.
Methods
SULT1A1 genotypes were successfully detected using the PCR-RFLP assay in 60 primary brain tumor patients and 156 hospital-based healthy control individuals with no history of cancer or precancerous disorder.
Results
There was a significant difference in genotypes distribution (GG vs. GA + AA) between brain tumor patients (GG genotype frequency = 48.3%) and control population (GG genotype frequency = 65.4%; OR = 2.019, 95% CI = 1.103–3.695; P = 0.022). In order to determine the association between SULT1A1 polymorphism and specific types of brain tumors, the patients were classified according to the type of brain tumors they suffer from: glial and non-glial. Results of the statistical analyses of each group of patients in comparison with the control individuals showed a significant difference only between SULT1A1 polymorphism and non-glial brain tumors (OR = 2.615; 95% CI = 1.192–5.739; P = 0.014) but glial tumors (OR = 1.535; 95% CI = 0.688–3.425; P = 0.293). When non-glial tumors were classified as meningiomal and others (pituitary adenoma, craniopharyngioma, acoustic neuroma and hemangioblastoma), statistical analysis showed that this significance is only due to the meningiomal tumors (OR = 3.238; CI = 1.205–8.704; P = 0.015). We also estimated a reduced risk of brain tumor in non-smokers (OR = 1.700; CI = 0.800–3.615) in comparison to smokers (OR = 2.773; CI = 0.993–7.749), but this was not statistically significant.
Conclusion
Our findings have suggested that there was a significant association between brain tumor and SULT1A1*2 allele (A allele that is also known as His allele) and this allele is an important risk factor in the development of meningiomal brain tumors.
Keywords: Sulfotranferase, Epidemiology, Cancer, Polymorphism, Sulfation
Introduction
Considerable circumstantial evidence for a genetic basis of cancer has built up over many years of study of carcinogenesis. Cancer is a complex genetic disease arising from mutations occurring in a number of genes. Biotransformation enzymes, such as P450 families, N-acetyltransferases, Glutathione S-Transferases (GSTs) and sulfotransferases, play an important role in the metabolism of xenobiotics that may be either bioactivated or bioinactivated. The changes in the enzyme activities may affect their abilities of converting procarcinogens to the ultimate form carcinogens, hence influencing the risk of cancers (Wormhoudt et al. 1999).
Sulphation is catalyzed by sulfotransferases (SULT) enzymes encoded by member of the SULT gene superfamily (Weinshilboum et al. 1997). There are at least seven human SULT isoforms (Nagata and Yamazoe 2000) that are differentially expressed in human tissues such as lung, liver, brain, platelet, colon as well as other organs (Hart et al. 1979; Jones et al. 1992; Richard et al. 2001). These enzymes are involved in phase II metabolism of numerous endogenous and exogenous compounds including many drugs, neurotransmitters, thyroid and steroid hormones and procarcinogenic agents (Glatt et al. 2000). Some SULTs are genetically polymorphic, and substantial differences have been observed in the activation of promutagens between allozymes. Therefore, it is important to study whether the SULT genotype will affect health risk in the general population.
SULT1A1 gene mapped to chromosome 16p12.1-p11.2 encodes the SULT1A1 enzyme that is involved in bioactivation of certain procarcinogens, including heterocyclic amines (HCA) and polycyclic aromatic hydrocarbons (PAH), resulting in the production of highly reactive electrophiles that are capable of binding DNA (Glatt 1997; Lewis et al. 1998; McManus et al. 1989). SULT1A1 gene is polymorphic and its alleles encode four different allozymes known as SULT1A1*1, SULT1A1*2, SULT1A1*3 and SULT1A1*4 with varying thermal stability and enzyme activity (Raftogianis et al. 1997). Raftogianis et al. (1997) reported G → A transition at the nucleotide 638 in exon 7 that causes an Arg → His substitution at amino acid 213 of SULT1A1, so called SULT1A1*2. In that in vitro study, genotype–phenotype correlation analysis showed that mutant SULT1A1*2 allozyme (encoded by mutant His allele) was consistently associated with lower thermal stability and enzyme activity than wild-type SULT1A1*1 allozyme (encoded by wildtype Arg allele). Therefore, this enzyme has attracted a great deal of attention of cancer researchers to investigate the association between this mutant allele and certain cancers (Wang et al. 2002; Wu et al. 2003; Zheng et al. 2003; Peng et al. 2003; Han et al. 2004). To our knowledge, no study was undertaken to investigate the relationship between SULT1A1 polymorphism and primary brain tumors to date. Therefore, we aimed to investigate this relationship in a group of Turkish brain tumor patients and healthy controls.
Materials and methods
Study population
The present study, based on the case and control study, was carried out on 60 (26 males and 34 females) brain tumor patients who visited the neurosurgery department at the Tepecik SSK Hospital in İzmir, Turkey between 2002 and 2005. All the cases were newly diagnosed and histologically confirmed, previously untreated (radiotherapy and chemotherapy) primary brain tumor patients. Classification of tumor types was based on the WHO guidelines, 2003. A total of 156 hospital-based individuals (70 males and 86 females) with no history of cancer and precancerous disorder were used as a control group. This study was carried out following ethical approval and under ethical procedures of the Cumhuriyet University. In addition, all cases and controls were informed about the purpose of this study and they gave their informed consent.
Genotyping of SULT1A1
Approximately 2 ml of peripheral blood sample was collected in ethylenediaminetetraasetic acid (EDTA) containing tubes from patients and controls. Total genomic DNA was isolated from white blood cells that were treated with 20 μl proteinase K (10 mg ml−1) and 50 μl SDS (10%) in 500 μl STE buffer (0.1 M NaCl, 0.05 M Tris and 0.01 M EDTA, pH 8) for 2 h at 55°C. After incubation, DNA isolation was proceeded by two steps of phenol-chloroform (25 phenol: 24 chloroform:1 isoamyl alcohol), followed by precipitation with cold absolute ethanol. Precipitated DNA was re-suspended in distilled water and quantified at a wavelength of 260 nm by a spectrophotometer.
PCR was carried out on a 25 μl volume containing 0.2 mM each of the deoxynucleotide triphosphates (dATP, dCTP, dGTP and dTTP), 0.2 mM of each primer (forward: 5′GTT GGC TCT GCA GCG TTT CTA GGA 3′ and reverse: CCC AAA CCC CCT GCT GGC CAG CAC CC 3), approximately 100 ng template DNA, 1 × reaction buffer (75 mM Tris–HCI, pH 8.8 at 25°C, 20 mM (NH4)2SO4, 0.01% Tween20, MBI Fermentas), 1.5 mM MgCl2 and 2 units Taq polymerase (MBI Fermentas). The temperature profile for the 30-cycle amplification reaction using a thermal cycler (Techne, UK) was as follows: initial denaturation at 94°C for 5 min, denaturation at 94°C for 1 min, annealing at 60°C for 1 min and extension at 72°C for 1 min and final extension at 72°C for 5 min.
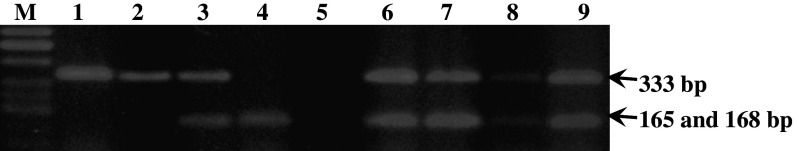
Following the PCR, 10 μl of 333 bp PCR product was digested with 10 units of HaeII restriction enzyme according to the manufacturer’s instructions (MBI, Fermentas). The DNA fragments were separated on 3% agarose gel stained with ethidium bromide. The wildtype homozygous genotype (GG) yielded two bands (165 and 168 bp), the heterozygous genotype (GA) yielded three bands (333, 168 and 165 bp) and finally the mutant genotype (AA) yielded one band (333 bp; Fig. 1).
Fig. 1.
SULT1A1 genotypes determined by PCR-RFLP (HaeII) analysis: homozygous A/A (lanes 1 and 2), heterozygous G/A (lanes 3, 6–9) and homozygous G/G (lane 4). M is molecular weight marker (50 bp ladder) and lane 5 is a negative control (PCR reaction performed in the absence of template)
Statistical analysis
SPSS (Statistical Package for Social Sciences) Release 10.0.1 software was used to perform the statistical analyses. Hardy–Weinberg equilibrium, genotype frequencies and allele frequencies were tested by Pearson X 2 test. The statistical significance of the differences in SULT1A1 genotypes among brain tumor patients and healthy controls was performed by X 2 test. Probability values <0.05 were regarded to be statistically significant. The association between SULT1A1 polymorphism and risk of brain tumors was estimated by odd ratios (ORs) with a 95% confidence interval (CI), using a multivariate logistic regression analysis that included several confounding variables (age, sex and smoking status, with ever smokers and never smokers separately).
Results
The demographic characteristics of the cases and controls are summarized in Table 1. The mean ages of the cases and controls were 48.90 ± 16.17 and 41.43 ± 13.39, respectively. The percentages of males and females in the case and control groups were close to each other. However, the percentage of cigarette smoking in case subjects (38.33%) was higher than in controls (30.13%).
Table 1.
Frequency distribution of select characteristics by case–control status
| Variables | Cases | Controls |
|---|---|---|
| Age | ||
| Range | 1–83 | 16–82 |
| Means ± SD | 48.90 ± 16.17 | 41.43 ± 13.39 |
| Males | 52.46 ± 14.52 | 43.76 ± 14.69 |
| Females | 46.18 ± 17.04 | 39.53 ± 11.99 |
| Sex | ||
| Males | 26 (43.33%) | 70 (44.9%) |
| Females | 34 (56.67%) | 86 (55.1%) |
| Smoking status | ||
| Smokers | 23 (38.33%) | 47 (30.13%) |
| Males | 18 (78.26%) | 29 (61.70%) |
| Females | 5 (21.74%) | 18 (38.30%) |
| Non-smokers | 37 (61.67%) | 109 (69.87%) |
| Males | 8 (21.62%) | 41 (37.61%) |
| Females | 29 (78.38%) | 68 (62.39%) |
| Histological type | ||
| Glial | 29 (48.33%) | – |
| Non-glial | 31 (51.67%) | – |
| Meningioma | 19 | – |
| Pituitary adenoma | 6 | – |
| Craniopharyngioma | 4 | – |
| Acoustic neuroma | 1 | – |
| Hemangioblastoma | 1 | – |
In the cases, the frequencies of the homozygous wild-type (GG), and the heterozygous (GA) and the homozygous mutant (AA) genotypes were 48.33, 43.33 and 8.33%, respectively; in the controls, the frequencies were 65.39, 32.05 and 2.56, respectively. The distribution of SULT1A1 genotypes was not significantly different in the cases and the controls, although there was a slightly higher deviation from the Hardy–Weinberg equilibrium for the cases. However, there was a significant deviation in genotype distribution between cases and controls (X 2 = 7.107; P = 0.029). In addition, there was also a significant difference in the frequencies of the alleles between cases and controls in question (X 2 = 6.628; P = 0.01). The frequencies of allele A in the control and patient groups were 18.59 and 30.00%, respectively.
In order to estimate the risk of brain tumor in individuals carrying A allele, we have combined the GA and AA genotypes. Although the frequencies of GA + AA (51.67%) and GG (48.33%) groups of genotypes were close to each other in the cases, a significantly higher proportion of GG (65.38) genotype was observed in the control with an OR = 2.019 (95% CI = 1.103–3.695; P = 0.022; Table 2). We have also evaluated the association between the SULT1A1 polymorphism in healthy controls and the specific type of brain tumors, glial (48.33%) and non-glial tumors (51.67%). As shown in Table 2, a statistically significant difference was only found between groups of SULT1A1 genotypes (GG vs. GA/AA) of healthy controls and non-glial brain tumor patients (p = 0.014; OR = 2.615; CI = 1.192–5.739), but not patients with glial tumors (P = 0.293; OR = 1.535; CI = 0.688–3.425). When non-glial tumors were classified as meningiomal and others (pituitary adenoma, craniopharyngioma, acoustic neuroma and hemangioblastoma), statistical analysis showed that this significance is only due to the meningiomal tumors (OR = 3.238; CI = 1.205–8.704; P = 0.015).
Table 2.
Risk estimates for SULTA1 genotypes categorized by gender, smoking status and age
| Variable | Genotype combinations | No. of cases (%) | No. of controls (%) | X 2 | P value | OR (95% CI) | aOR (95% CI) |
|---|---|---|---|---|---|---|---|
| Total | GG | 29 (48.33) | 102 (65.38) | 5.279 | 0.022 | 2.019 (1.103–3.695) | 1.954 (1.048–3.645) |
| GA + AA | 31 (51.67) | 54 (34.62) | |||||
| Gender | |||||||
| Females | GG | 18 (52.94) | 58 (66.67) | 1.972 | 0.160 | 1.778 (0.793–3.987) | 1.487 (0.641–3.449) |
| GA + AA | 16 (47.06) | 29 (33.33) | |||||
| Males | GG | 11 (42.31) | 44 (63.77) | 3.568 | 0.059 | 2.288 (0.877–5.972) | 2.784 (1.060–7.312) |
| GA + AA | 15 (57.69) | 25 (36.23) | |||||
| Smoking status | |||||||
| Smokers | GG | 11 (47.83) | 32 (68.09) | 3.896 | 0.052 | 2.773 (0.993–7.749) | 2.874 (0.984–8.395) |
| GA + AA | 12 (52.17) | 15 (31.91) | |||||
| Non-smokers | GG | 19 (51.35) | 70 (64.22) | 1.922 | 0.166 | 1.700 (0.800–3.615) | 1.569 (0.717–3.434) |
| GA + AA | 18 (48.65) | 39 (35.78) | |||||
| Histological type of tumor | |||||||
| Glial brain tumors | GG | 16 (55.17) | 1.104 | 0.293 | 1.535 (0.688–3.425) | 1.493 (0.641–3.479) | |
| GA + AA | 13 (44.83) | ||||||
| Non-glial brain tumors | GG | 13 (41.94) | 6.056 | 0.014 | 2.615 (1.192–5.739) | 2.571 (1.159–5.703) | |
| GA + AA | 18 (58.06) | ||||||
| Meningioma | GG | 7 (36.84) | 5.874 | 0.015 | 3.238 (1.205–8.704) | 3.186 (1.136–8.935) | |
| GA + AA | 12 (63.16) | ||||||
| Others (pituitary adenoma, craniopharyngioma, acoustic neuroma and hemangioblastoma) | GG | 6 (50.00) | 1.149 | 0.284 | 1.889 (0.581–6.139) | 2.056 (0.617–6.853) | |
| GA + AA | 6 (50.00) | ||||||
aAdjusted for age, gender and smoking status, where appropriate
Risk of brain tumor related to SULT1A1 genotype was further examined with stratification by gender and smoking status. When we examined the data by gender, although there was no significant association between gender and brain tumor risk, results showed that men (OR = 3.568; CI = 0.877–5.972) were more prone to brain tumor than women (OR = 1.778; CI: 0.793–3.987). We also estimated a reduced risk of brain tumor in non-smokers (OR = 1.700; CI = 0.800–3.615) in comparison to smokers (OR = 2.773; CI = 0.993–7.749), but this was not statistically significant (Table 2).
Discussion
The distribution of G (Arg) and A (His) alleles is highly variable among ethnic groups. The variant His allele was studied in Chinese (Han et al. 2004), Taiwanese (Wu et al. 2003), Korean (Kim et al. 2005), Nigerian and Caucasian (Coughtrie et al. 1999) populations and their frequencies were 9.5, 5.5, 0.124, 26.9 and 32.1%, respectively. The Turkish population studied in the present work had a much higher frequency of His allele (18.59%) than those of Chinese, Taiwanese and Korean populations, but lower than those reported for other Caucasian and Nigerian populations.
Although no study has been carried out on the relationship of SULTA1 polymorphism and brain tumors, there are a number of studies on a variety of cancers. The findings of these studies are highly variable and inconsistent in some cases. A positive association was found between the SULT1A1 variant His allele with lung (Wang et al. 2002), breast (Han et al. 2004; Zheng et al. 2001), esophageal (Wu et al. 2003) and gastric (Boccia et al. 2005) cancers. On the other hand, several studies reported no significant association between SULT1A1 polymorphism and breast (Seth et al. 2000), colorectal (Peng et al. 2003; Wong et al. 2002; Bamber et al. 2001), hepatic, lung, oral, gastric, renal and cervical (Peng et al. 2003) cancers. In contrast to most of those previous studies that have shown an increased risk or null findings associated with His allele genotype, Zheng et al. (2003) found that His variant allele was associated with statistically significant reduced risk of bladder cancer.
The results of this study put forward a strong association between primary brain tumors and His variant allele (Arg/His + His/His) with an OR = 2.019 (95% CI = 1.103–3.695; P = 0.022). This overall significance was only due to the difference between the genotype frequencies of healthy controls and patients with non-glial tumors, but not patients with glial tumors. Subclassification of non-glial brain tumor patients showed that this significance is only due to the meningiomal brain tumors, but not other types (pituitary adenoma, craniopharyngioma, acoustic neuroma and hemangioblastoma). A meta-analysis study also reported a significant relationship between GSTT1 variants and risk of meningioma, though there was not any significant association between any variants of GSTs and glioma (Lai et al. 2005).
These overall findings may be explained in part by the wide substrate specificity of the SULTs and the marked differences in their activity in the different tissues (Jones et al. 1992; Richard et al. 2001). Richard et al. (2001) investigated the expression in SULT1A1 in the developing human brain and found interesting results; SULT1A1 is expressed at low levels in the fetal brain, although choroids plexus is a major site of expression. They suggest that this region of brain is of neuroectodermal origin, as is the rest of the brain, and its primary function is the production of cerebrospinal fluid. It is also the most highly vascularized tissue of the developing brain and therefore, as a potential portal of entry of circulating toxins, it seems reasonable for this tissue to have a high degree of chemical defense. Studies have demonstrated that mutant His allele leads to distinct changes in biochemical properties of the enzyme, including decreased enzymatic activity and low thermal stability (Raftogianis et al. 1999). Pereira et al. (2005) clearly showed that the mutation in SULT1A1 is functional, with subjects carrying the mutant allele presenting lower sulfotransferase activity. Both heterozygous and homozygous individuals for the mutant His allele had significantly decreased sulfotransferase enzymatic activity in cancer patients, although there was no an allelic association with SULT1A1 polymorphism. Nowell et al. (2004) showed that there was a strong association between increased SULT1A1 activity and prostate cancer in Caucasians (OR = 3.04; CI% = 1.81–5.1) as well as African-American patients (OR = 6.7; CI% = 2.1–21). They also suggested that the magnitude of the association might differ by ethnicity and be modified by meat consumption. Recently, Nagar et al. (2006) examined the intracellular stability of the SULT1A1*1 and SULT1A1*2 allozymes in insect as well as mammalian cells. They found that the SULT1A1*2 protein had a shorter half-life than the SULT1A1*1 protein and increased degradation via a proteasome pathway.
The findings of this study also provided an indication of the relationship between genders and smoking status. Although the distribution of His allele genotypes in male and female patients in comparison to those in the control did not differ significantly, men were relatively at a higher risk than women. This finding is not in agreement with those published. Nowell et al. (2000) suggested that platelet SULT activity was higher in women than in men. This contradiction is presumably due to the higher proportion of smoker male patients (78.26%) than smoker males in the control group (61.70%).
In conclusion, overall findings of this study have shown that mutant SULT1A1*2 (His allele) is a significant risk factor for developing meningiomal brain tumors. This allele may be useful as a valuable marker for brain tumors. However, since this is the first study that investigates the relationship between SULT1A1 gene and primary brain tumors, a possible trend towards brain tumor risk and the role of sulfotransferases has emerged, which needs further investigation with more case–control studies.
Acknowledgments
This work was supported financially by the Research Council of Cumhuriyet University, Sivas, Turkey.
References
- Bamber DE, Fryer AA, Strange RC, Elder JB, Deakin M, Rajagopal R, Fawole A, Gilissen RA, Campbell FC, Coughtrie MW (2001) Phenol sulfotransferase SUL1A1*1 genotype is associated with reduced risk of colorectal cancer. Pharmacogenet 11(8):679–685 [DOI] [PubMed] [Google Scholar]
- Boccia S, Persiani R, La Torre G, Rausei S, Arzani D, Gianfagna F, Romano-Spica V, D’Ugo D, Ricciardi G (2005) Sulfotransferase 1A1 polymorphism and gastric cancer risk: a pilot case–control study. Cancer Lett 229(2):235–243 [DOI] [PubMed] [Google Scholar]
- Coughtrie MW, Gilissen RA, Shek B, Strange RC, Fryer AA, Jones PW, Bamber DE (1999) Phenol sulphotransferase SULT1A1 polymorphism: molecular diagnosis and allele frequencies in Caucasian and African populations. Biochem J 337(Pt 1):45–49 [PMC free article] [PubMed] [Google Scholar]
- Glatt H (1997) Sulfation and sulfotransferases 4: bioactivation of mutagens via sulfation. FASEB J 11(5):314–321 [DOI] [PubMed] [Google Scholar]
- Glatt H, Engelke CE, Pabel U, Teubner W, Jones AL, Coughtrie MW, Andrae U, Falany CN, Meinl W (2000) Sulfotransferases: genetics and role in toxicology. Toxicol Lett 112:341–348 [DOI] [PubMed] [Google Scholar]
- Han DF, Zhou X, Hu MB, Wang CH, Xie W, Tan XD, Zheng F, Liu F (2004) Sulfotransferase 1A1 (SULT1A1) polymorphism and breast cancer risk in Chinese women. Toxicol Lett 150(2):167–177 [DOI] [PubMed] [Google Scholar]
- Hart RF, Renskers KJ, Nelson EB, Roth JA (1979) Localization of phenol sulfotransferase in human platelets. Life Sci 24 (2):125–130 [DOI] [PubMed] [Google Scholar]
- Jones AL, Hume R, Bamforth KJ, Coughtrie MW (1992) Estrogen and phenol sulfotransferase activities in human fetal lung. Early Hum Dev 28(1):65–77 [DOI] [PubMed] [Google Scholar]
- Kim KA, Lee SY, Park PW, Ha JM, Park JY (2005) Genetic polymorphisms and linkage disequilibrium of sulfotransferase SULT1A1 and SULT1A2 in a Korean population: comparison of other ethnic groups. Eur J Clin Pharmacol 61(10):743–747 [DOI] [PubMed] [Google Scholar]
- Lai R, Crevier L, Thabane L (2005) Genetic polymorphisms of glutathione S-transferases and the risk of adult brain tumors: a meta-analysis. Cancer Epidemiol Biomarkers Prev 14:1784–1790 [DOI] [PubMed] [Google Scholar]
- Lewis AJ, Walle UK, King RS, Kadlubar FF, Falany CN, Walle T (1998) Bioactivation of cooked mutagen N-hydroxy-2-amino-1-methyl-6-phenylimidazol[4,5-b]pyridine by estrogen sulfotransferase in cultured human mammary epithelial cells. Carsinogenesis 19(11):2049–2053 [DOI] [PubMed] [Google Scholar]
- McManus ME, Felton JS, Knize MG, Burgess WM, Roberts-Thomson S, Pond SM, Stupans I, Veronese ME (1989) Activation of the food-derived mutagen 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine by rabbit and human-liver microsomes and purified forms of cytochrome-p-450. Carcinogenesis 10(2):357–363 [DOI] [PubMed] [Google Scholar]
- Nagar S, Walther S, Blanchard RL (2006) Sulfotransferase (SULT) 1A1 polymorphic variants*1,*2, and*3 are associated with altered enzymatic activity, cellular phenotype, and protein degradation. Mol Pharmacol 69(6):2084–2092 [DOI] [PubMed] [Google Scholar]
- Nagata K, Yamazoe Y (2000) Pharmacogenetics of sulfotransferase. Annu Rev Pharmacol Toxicol 40:159–176 [DOI] [PubMed] [Google Scholar]
- Nowell S, Ambrosome CB, Ozawa S, MacLeod SL, Mrackova G, Williams S, Plaxco J, Kadlubar FF, Lang NP (2000) Relationship of phenol transferase activity (SULT1A1) genotype to sulfotransferase phenotype in platelet cytosol. Pharmacogenet 10(9):789–797 [DOI] [PubMed] [Google Scholar]
- Nowell S, Ratnasinghe DL, Ambrosone CB, Williams S, Teague-Ross T, Trimble L, Runnels G, Carrol A, Gren B, Stone A, Johnson D, Grene G, Kadlubar FF, Lang NP (2004) Association of SULT1A1 phenotype and genotype with prostate cancer risk in African-Americans and Caucasians. Cancer Epidemiol Biomarkers Prev 13(2):270–276 [DOI] [PubMed] [Google Scholar]
- Peng CT, Chen JC, Yeh KT, Wang YF, Hou MF, Lee TP, Shih MC, Chang JY, Chang JG (2003) The relationship among the polymorphisms of SULT1A1, 1A2 and different types of cancers in Taiwanese. Int J Mol Med 11(1):85–89 [PubMed] [Google Scholar]
- Pereira WO, Paiva AS, Queiroz JW, Toma L, Dietrich CP, Nader HB, Jeronimo SM (2005) Genetic polymorphism in the sulfotransferase SULT1A1 gene in cancer. Cancer Genet Cytogenet 160(1):55–60 [DOI] [PubMed] [Google Scholar]
- Raftogianis RB, Wood TC, Otterness DM, Van Loon JA, Weinshilboum RM (1997) Phenol sulfotransferase pharmacogenetics in human: association of common SULT1A1 alleles with TS PST phenotype. Biochem Biophys Res Commun 239(1):298–304 [DOI] [PubMed] [Google Scholar]
- Raftogianis RB, Wood TC, Weinshilboum RM (1999) Human phenol sulfotransferases SULT1A2 and SULT1A1: genetic polymorphisms, correlations. Biochem Pharmacol 58(4):605–616 [DOI] [PubMed] [Google Scholar]
- Richard K, Hume R, Kaptein E, Stanley EL, Visser TJ, Coughtrie MW (2001) Sulfation of thyroid hormone and dopamine during human development: ontogeny of phenol sulfotransferases and arylsulfatase in liver, lung, and brain. J Clin Endocrinol Metab 86(6):2734–2742 [DOI] [PubMed] [Google Scholar]
- Seth P, Lunetta KL, Bell DW, Gray H, Nasser SM, Rhei E, Kaelin CM, Iglehart DJ, Marks JR, Garber JE, Haber DA, Polyak K (2000) Phenol sulfotransferases: hormonal regulation, polymorphism, and age of onset of breast cancer. Cancer Res 60(24):6859–6863 [PubMed] [Google Scholar]
- Wang YF, Spitz MR, Tsou AM, Zhang K, Makan N, Wu X (2002) Sulfotransferase (SULT) 1A1 polymorphism as a predisposition factor for lung cancer: a case–control analysis. Lung Cancer 35(2):137–142 [DOI] [PubMed] [Google Scholar]
- Weinshilboum RM, Otterness DM, Aksoy IA, Wood TC, Her C, Raftogianis RB (1997) Sulfotransferase molecular biology: cDNAs and genes. FASEB J 11(1):3–14 [PubMed] [Google Scholar]
- Wong CF, Liyou N, Leggett B, Young J, Johnson A, McManus ME (2002) Association of the SULT1A1 R213H polymorphism with colorectal cancer. Clin Exp Pharmacol Physiol 29(9):754–758 [DOI] [PubMed] [Google Scholar]
- Wormhoudt LW, Commandeur JNM, Vermeulen NP (1999) Genetic polymorphisms of human N-acetyltransferase, cytochrome P450, glutathione-S-Transferase, and epoxide hydrolase enzymes: Relevance to xenobiotic metabolism and toxicity. Crit Rev Toxicol 29(1):59–124 [DOI] [PubMed] [Google Scholar]
- Wu MT, Wang YT, Ho CK, Wu DC, Lee YC, Hsu HK, Kao EL, Lee JM (2003) SULT1A1 polymorphism and esophageal cancer in males. Int J Cancer 103(1):101–104 [DOI] [PubMed] [Google Scholar]
- Zheng W, Xie D, Cerhan JR, Sellers TA, Wen W, Folsom AR (2001) Sulfotransferase 1A1 polymorphism, endogenous estrogen exposure, well done meat intake, and breast cancer risk. Cancer Epidemiol Biomarkers Prev 10(2):89–94 [PubMed] [Google Scholar]
- Zheng L, Wang Y, Schabath MB, Grossman HB, Wu X (2003) Sulfotransferase 1A1 (SULT1A1) polymorphism and bladder cancer risk: a case–control study. Cancer lett 202(1):61–69 [DOI] [PubMed] [Google Scholar]