Abstract
The lung microbiota, present in healthy individuals, undergoes alterations in different diseases and is closely linked to changes in both systemic and alveolar immunity. These interactions play a crucial role in the onset and progression of numerous diseases. Acute respiratory distress syndrome (ARDS), one of the most severe conditions encountered in intensive care units (ICU), is characterized by high incidence and mortality rates. The pathophysiology of ARDS involves complex mechanisms, including the activation and dysregulation of overlapping pathways related to injury, inflammation, and coagulation, both locally in the lungs and systemically. Notably, alterations in the microbiota may contribute to the pathogenesis of ARDS. Emerging evidence suggests that changes in the lung microbiota are associated with ARDS development, often marked by increased bacterial burden, reduced microbial diversity, and shifts in microbiota composition. In this review, we focus on the regulatory roles of the lung microbiota in ARDS and their therapeutic potential.
Keywords: lung microbiota, ARDS, corona virus disease 2019 (COVID-19), prognosis, metabolic products
1 Introduction
Advances in culture-independent microbiology, particularly high-throughput sequencing, have revealed that the lungs harbor complex and diverse microbial communities in healthy individuals (Charlson et al., 2011; Dickson et al., 2016a; Dickson et al., 2017), overturned the long-held belief that lungs are sterile (Laurenzi et al., 1961; Pecora, 1963).
Dysbiosis refers to the disruption of the balance between beneficial and harmful microbes, with a relative increase or decrease in certain microbial populations. Lung microbiota dysbiosis can lead to immune imbalances, resulting in excessive inflammation and subsequent tissue damage (Yang et al., 2020). In chronic respiratory diseases, the diversity of the lung microbiome is a predictor of mortality in chronic obstructive pulmonary disease (COPD) (Leitao Filho et al., 2019), and bacterial burden is linked to disease progression and mortality in idiopathic pulmonary fibrosis (IPF) (Molyneaux et al., 2014; O’Dwyer et al., 2019). Additionally, alterations in community composition predict exacerbations in bronchiectasis (Rogers et al., 2014).
In contrast to chronic respiratory diseases, research on the lung microbiome in acute respiratory failure remains in its early stages. ARDS is a common and serious lung problem that often leads to long-term ventilator use and death in critically ill patients. It is characterized by protein-rich pulmonary edema, hypoxemia, and alveolar inflammation. These pathological features may be driven by changes in the pulmonary microbiome. Conversely, the dysbiosis of lung microbiota may result from alveolar nutrient availability following edema onset (Ruff et al., 2020).
Despite advances in optimizing mechanical ventilation settings and antimicrobial therapies, adjunctive treatment options for ARDS remain limited. The lung microbiome represents an underexplored factor contributing to clinical variation in critical illness. A comprehensive understanding of its mechanistic role in ARDS could facilitate the development of targeted therapeutic interventions.
2 Lung microbiota in healthy lung
In most healthy individuals, the lung microbiota harbors a low density but exhibits significant diversity, with various interacting microbial communities. The adult lung microbiota in these individuals is primarily dominated by key genera within the phyla Firmicutes and Bacteroidetes, forming a “core microbiota” of the lungs (Man et al., 2017). This core microbiota, consisting of genera such as Streptococcus, Prevotella, Veillonella, Fusobacterium, and Haemophilus, plays a crucial role in maintaining lung homeostasis (Man et al., 2017; Montassier et al., 2023).
Lung immune cells, especially subpopulations of alveolar macrophages and dendritic cells, exert immunoregulatory functions by promoting the generation of regulatory T cells (Soroosh et al., 2013) and secreting anti-inflammatory molecules such as IL-10, transforming growth factor-beta (TGF-β), and prostaglandin E2 (Hussell and Bell, 2014). Resident microorganisms are integral to maintaining pulmonary immune homeostasis through continuous dialog among lung microbiota, immune cells and airway epithelial cells, which are equipped with pattern recognition receptors. However, the composition of the lung microbiota undergoes significant changes in response to pulmonary pathologies, which can disrupt immune homeostasis and influence the progression of these diseases (Lira-Lucio et al., 2020; Sommariva et al., 2020). Emerging evidence suggests that the lung microbiota plays a critical role in shaping the risk and outcomes of pulmonary diseases by modulating both innate and adaptive immune responses (Khatiwada and Subedi, 2020).
Streptococcus may indirectly promote the excessive production of CXCL8 (interleukin-8) through pulmonary microbiota dysbiosis, with elevated CXCL8 levels in sputum correlating with increased severity of COPD (Wang et al., 2023).
Prevotella, a key part of the airway microbiota, helps activate the innate immune system and protects the respiratory tract. Horn et al. demonstrated that Prevotella facilitated the rapid clearance of Streptococcus pneumoniae from the lungs and improved the outcomes of S. pneumoniae infection by activating neutrophils. Effective neutrophil-mediated clearance requires the recognition of toll-like receptor (TLR) 2, the induction of the pro-inflammatory cytokine tumor necrosis factor-alpha (TNF-α), and the regulation of inflammation by the anti-inflammatory cytokine IL-10 (Horn et al., 2022).
Furthermore, a study showed that specific lung bacteria, including Prevotella spp. and Veillonella spp., were associated with an increased number of lymphocytes in bronchoalveolar lavage fluid (BALF), TH17 cell-mediated lung inflammation, and a reduced TLR4 response by alveolar macrophages (Segal et al., 2016). Although the lung microbiome likely contributes significantly to mucosal immune homeostasis, its precise role in establishing and maintaining respiratory health remains unclear (Man et al., 2017).
Both chronic lung disease and acute lung injury/ARDS are associated with airway dysbiosis and enrichment of Pseudomonas. However, in chronic lung disease, Haemophilus, Streptococcus, and Moraxella are primarily enriched, while in acute lung injury/ARDS, the changes in the lung microbiome are mainly characterized by the enrichment of gut-associated bacteria (i.e., Bacteroides spp.) (Yi et al., 2022).
3 Microbiome features in patients with traditional ARDS
This section provides an overview of the current understanding of the lung microbiome in traditional ARDS and COVID-19-associated ARDS (Table 1), and a few comparisons between bacterial taxa in traditional ARDS and COVID-19 ARDS (Table 2).
TABLE 1.
Studies concerning lung microbiota of mechanically ventilated patient in traditional and COVID-19 ARDS.
| Study | Publication, y | Region | Type of patients | Number of patients | Date of sample | Type of samples | Analysis | Main results |
| ARDS-exp | ||||||||
| Schmitt et al. (2020) | 2020 | Germany | Patients with sepsis-induced ARDS following major abdominal surgery | 15 ARDS patients | ARDS onset D0, D5, D10 | BALF | 16S rRNA | Lower α-diversity and increased dominance. A significant reduction in physiological lung flora, particularly anaerobic bacteria such as Prevotella spp. and Veillonella spp. The α-diversity index negatively correlated with the length of ICU stay and the requirement for mechanical ventilation |
| Panzer et al. (2018) | 2018 | United States | Intubated and mechanically ventilated patients with severe blunt trauma | 16 ARDS patients | ICU admission; 48 h after admission; | ETA | 16S rRNA | The development of ARDS was associated with changes in lung community composition at 48 h, characterized by a relative enrichment of Enterobacteriaceae (OTU2119418) and specific taxa, including Prevotella (OTU4319899) and Fusobacterium (OTU4447248). |
| ARDS-p | ||||||||
| Shen et al. (2022) | 2022 | China | Hematological patients with pneumonia-related ARDS | 22 responders; 28 non-responders | Within the first 24 h of admission | BALF | mNGS | Responders exhibited lower α-diversity and had higher ventilator-free days at day 28. Non-responders with co-infection had a significantly lower survival rate compared to other patients |
| Hu et al. (2024) | 2024 | China | Immuno compromised patients with pneumonia-related ARDS |
33“type α”, 12 “type β,” 47 “type γ” | Within 48 h of ICU admission | BALF | mNGS | Patients with type γ exhibited lower microbial α-diversity had higher 30-day mortality and fewer ventilator-free days. |
| Imbert et al. (2024) | 2024 | France | ARDS patients | Including 11 bacterial CAP-related ARDS | Within 24 h of ICU admission | ETA | 16S rRNA | Bacterial α-diversity was significantly lower in the bacterial CAP-related ARDS compared to the COVID-19 ARDS. In bacterial CAP-related ARDS patients, the abundances of Acinetobacter sp. and Actinomyces sp. were higher compared to influenza-related ARDS patients, while the abundances of Prevotella sp. and Pyramidobacter sp. were higher compared to COVID-19-related ARDS patients. |
| Walsh et al. (2017) | 2017 | United States | Patients for burn and inhalation injury | 24 P/F ≤ 300, 24 P/F>300 |
Within 72 h of hospitalization | BALF | 16S rRNA | Enrichment of Prevotella melaninogenica was observed in patients with a PaO2/FiO2 ratio ≤ 300. |
| ARDS | ||||||||
| Zhang et al. (2022) | 2022 | China | Sepsis-Induced ARDS patients | 111 ARDS-p, 45 ARDS-exp | Within 24 h of ARDS diagnosis, D7 | BALF | mNGS | ARDS-p was characterized by decreased microbiome diversity, primarily affecting the normal lung microbes. ARDS-exp was characterized by increased microbiome diversity, particularly in conditionally pathogenic bacteria and intestinal microbes. An increase in the background microbiome Bilophila, a genus of intestinal microbes, is likely a risk factor for mortality in ARDS-exp. |
| Dickson et al. (2016b) | 2016 | American | ARDS patients | 68 ARDS patients; including pneumonia (20, 29.4%), sepsis (18, 26.4%), aspiration (14, 20.6%) |
Following enrollment on D3, D7, D14, D21 | BALF | 16S DNA | The lung microbiota of post-sepsis mice was significantly enriched with numerous bacteria commonly found in the murine gut, including members of the Bacteroidales order, Enterococcus species, and Lachnospiraceae species. A member of the Bacteroidales order (OTU008) was identified as a key microbial driver of the altered lung communities. Alveolar TNF-α was positively correlated with the relative abundance of Proteobacteria. |
| Kyo et al. (2019) | 2019 | Japan | Mechanically ventilated patients | 40 ARDS patients; including 16 non survivors ARDS patients |
within 24 h after intubation | BALF | 16S rRNA, NGS | α-diversity was significantly decreased, and the 16S rRNA gene copy numbers tended to increase. Serum IL-6 levels were significantly elevated and positively correlated with the 16S rRNA gene copy number in non-surviving ARDS patients. The copy numbers and relative abundance of Betaproteobacteria were significantly lower in non-survivors. The copy numbers of Staphylococcus, Streptococcus, and Enterobacteriaceae were significantly correlated with serum IL-6 levels in non-survivors. |
| Dickson et al. (2020) | 2020 | United States | Critically ill patients | 17 ARDS patients; including cardiac arrest (19, 21%), cerebral vascular accident (9, 10%), other (28, 31%) |
24 h of ICU admission | BALF | 16S rRNA | The bacterial DNA burden in BAL specimens was higher in patients with ARDS. ARDS specimens were more commonly characterized by species of the Pasteurellaceae and Enterobacteriaceae families. The Enterobacteriaceae family was significantly more abundant in ARDS specimens. The gut-associated Lachnospiraceae and Enterobacteriaceae families were strongly predictive of fewer ventilator-free days, and the Lachnospiraceae family was significantly predictive of worse clinical outcomes in critically ill patients. |
| Montassier et al. (2023) | 2023 | French | Patients with brain injury requiring invasive mechanical ventilation | 52 ARDS | D0, 4 and 7 after ICU admission | BALF | 16S rRNA | Microbiota signatures associated with ARDS were characterized by an enrichment of potentially pathogenic respiratory microbes, including Pseudomonas and Staphylococcus. Based on the presence of Staphylococcus, Ralstonia, and Enterococcus, samples were classified as either “ARDS” or “no ARDS.” |
| COVID-19 | ||||||||
| Imbert et al. (2024) | 2024 | France | ARDS patients | Including 24 COVID-19 | Within 24 h of ICU admission | ETA | 16S rRNA | In COVID-19-related ARDS patients, the abundances of Prevotella sp. and Streptococcus sp. were higher compared to both influenza-related and bacterial CAP-related ARDS patients, while the abundance of Bacteroides sp. was higher only when compared to influenza-related ARDS patients. |
| Dickson et al. (2016b) | 2020 | China | – | 8 COVID-19 patients | – | BALF | Meta transcriptome Sequencing |
The microbiota in COVID-19 patients was similar to that in CAP patient, characterized by either dominance of pathogens or elevated levels of oral and upper respiratory tract commensal bacteria. |
| Merenstein et al. (2021) | 2021 | United States | – | 83 critically ill patients with COVID-19 | – | ETA | 16S rRNA | Low diversity in the ETA microbiome. Frequent outgrowth of potential respiratory pathogens, particularly Staphylococcus. |
| Zacharias et al. (2022) | 2021 | United States | – | 20 critically ill patients with COVID-19 | A median of 4 days after hospitalization | Lung | 16S rRNA | The lung microbiome exhibited reduced biodiversity. The bacterial community compositions in COVID-19 cases with DAD were significantly different from those in COVID-19 cases with pneumonia. |
| Gaibani et al. (2021) | 2021 | Italy | – | 24 critically ill patients with COVID-19 | – | BALF | 16S rRNA | Low diversity in the lower airway microbiome. The lung microbiome was characterized by enrichment of Pseudomonas alcaligenes, Clostridium hiranonis, Acinetobacter schindleri, Sphingobacterium spp., Acinetobacter spp., and members of the Enterobacteriaceae family. |
| Kullberg et al. (2022) | 2022 | The Netherlands | – | 114 C-ARDS patients | Routine clinical care | BALF | 16S rRNA | Patients with increased lung bacterial were less likely to be extubated and exhibited higher mortality rates. Proinflammatory cytokines, such as TNF-α, were associated with elevated microbial burdens. |
| De Pascale et al. (2024) | 2024 | Italy | – | 70 C-ARDS patients | The first BAL obtained after intubation | BALF | 16S rRNA | COVID-19-related ARDS patients with low Crs/low VR exhibited a lung microbiota dominated by Proteobacteria. Lung microbiota diversity served as a negative predictor for weaning from IMV and survival. Procalcitonin levels were significantly negatively correlated with Firmicutes and positively correlated with Proteobacteria. |
| Sulaiman et al. (2021) | 2021 | United States | 589 C-ARDS patients | – | BALF | 16S rRNA, WGS, RNA meta transcriptome |
A negative association was observed between bacterial burden in the lungs and survival. Enrichment of the lower airway with the oral commensal Mycoplasma salivarium was linked to poor clinical outcomes. |
|
Definition of abbreviations: ARDS-exp, extrapulmonary ARDS; ARDS-p, intrapulmonary ARDS.
TABLE 2.
Comparisons between bacterial taxa in Traditional ARDS and COVID-19 ARDS.
| Traditional ARDS | COVID-19 ARDS | |
| Phylum | Betaproteobacteria ↓ (Kyo et al., 2019) | Proteobacteria ↑ (De Pascale et al., 2024) |
| Class | – | – |
| Order | – | – |
| Family | Pasteurellaceae and Enterobacteriaceae ↑ (Dickson et al., 2020) | – |
| Genus | Bilophila ↑ (Zhang et al., 2022) | Staphylococcus ↑ (Merenstein et al., 2021) |
| Pseudomonas and Staphylococcus ↑ | – | |
| Staphylococcus, Ralstonia, and Enterococcus↑ (Montassier et al., 2023) | – | |
| Species | Prevotella spp. and Veillonella spp. ↓ (Schmitt et al., 2020) | Prevotella sp. and Streptococcus sp. ↑ (Imbert et al., 2024) |
| Prevotella sp. and Pyramidobacter sp. ↑ (Imbert et al., 2024) | Pseudomonas alcaligenes, Clostridium hiranonis, Acinetobacter schindleri, Sphingobacterium spp., Acinetobacter spp., and members of the Enterobacteriaceae ↑ (Gaibani et al., 2021) | |
| Prevotella melaninogenica ↑ (Walsh et al., 2017) | – | |
| OUT | Enterobacteriaceae (OTU2119418), Prevotella (OTU4319899) and Fusobacterium (OTU4447248) ↑ (Panzer et al., 2018) | – |
| Bacteroidales order (OTU008) ↑ (Dickson et al., 2016b) | – |
3.1 Ecological metrics
Several studies have reported that the bacterial DNA burden is higher in patients with ARDS compared to those without ARDS (Dickson et al., 2020; Kyo et al., 2019). Additionally, Kyo et al. found that serum IL-6 levels were significantly elevated in non-survivor ARDS patients and positively correlated with the 16S rRNA copy number (Kyo et al., 2019). Furthermore, Dickson et al. demonstrated that patients with an increased lung bacterial DNA burden had fewer ventilator-free days in critically ill patients (Dickson et al., 2020).
Most studies have shown that alpha diversity is significantly reduced in patients with ARDS (Imbert et al., 2024; Kyo et al., 2019; Schmitt et al., 2020; Zhang et al., 2022). Another study reported that microbial diversity in the lung microbiome of patients with septic ARDS declines progressively over time (Li et al., 2024). However, Zhang et al. found that patients with extrapulmonary infection-induced ARDS exhibited increased microbial diversity (Zhang et al., 2022).
Shen and colleagues observed that responders to early corticosteroid therapy in hematological patients with pneumonia-associated ARDS had lower alpha diversity compared to non-responders. They also found a negative correlation between alpha diversity and levels of inflammatory markers (serum IL-6, IL-8, TNF-α) and CRP in the responders (Shen et al., 2022). In contrast, the microbiota signatures of immunocompromised patients with no obvious inflammatory symptoms and more severe oxygenation failure were characterized by lower alpha diversity and distinct microbial compositions compared to other immunocompromised patients. Hu et al. reported that gut-associated bacteria were more abundant in patients with type α, which is characterized by more active inflammation (Hu et al., 2024).
Interestingly, Walsh et al. found significant differences not in the species dominating the overall community but in the less abundant taxa in patients with burn and inhalation injuries (Walsh et al., 2017). While these taxa did not differ in microbial diversity between patient groups, they exhibited differences in functional diversity, which ultimately had a greater impact on patient outcomes (Dickson et al., 2016a).
Several studies have reported that lower alpha diversity in the lung microbiota is associated with fewer ventilator-free days (De Pascale et al., 2024; Hu et al., 2024; Schmitt et al., 2020). However, Shen and colleagues detected that the responders of corticosteroids had lower alpha diversities and higher ventilator free days in hematological patients with pneumonia-associated ARDS (Shen et al., 2022). One possible explanation is that corticosteroid treatment may alter the lung microbiota composition, though this aspect was not further explored in their study.
3.2 Differential flora
In ARDS patients, the abundance of Prevotella spp. was found to increase in both direct and indirect lung injury-induced ARDS (Imbert et al., 2024; Panzer et al., 2018; Walsh et al., 2017). However, Schmitt et al. reported a significant reduction in physiological lung flora and mostly anaerobic bacteria, such as Prevotella spp. or Veillonella spp., in patients with sepsis-induced ARDS (Schmitt et al., 2020).
In patients with bacterial community-acquired pneumonia (CAP)-related ARDS, the levels of Acinetobacter spp. and Actinomyces spp. were higher compared to those with influenza-related ARDS. When compared to COVID-19-related ARDS patients, only the abundances of Prevotella spp. and Pyramidobacter spp. were found to be elevated (Imbert et al., 2024).
Additionally, several studies have identified gut-associated Bacteroides genus (OTU009) (Dickson et al., 2016b), Pasteurellaceae (Dickson et al., 2020), Enterobacteriaceae (Dickson et al., 2020; Panzer et al., 2018), and Fusobacterium (OTU4319899) (Panzer et al., 2018) were common and abundant in ARDS patients. Furthermore, Dickson et al. found that the relative abundance of Proteobacteria was positively correlated with alveolar TNF-α levels, while the typically abundant Bacteroidetes phylum showed a negative correlation with alveolar TNF-α concentrations. Furthermore, the gut-associated Bacteroides genus (OTU009) was associated with serum TNF-α levels (Dickson et al., 2016b). Four years later, the same team identified the gut-associated Lachnospiraceae and Enterobacteriaceae families as the taxa most strongly predictive of fewer ventilator-free days (Dickson et al., 2020).
Panzer et al. explored the relationship between microbiota composition at 48 h and inflammation, finding that this relationship was driven by the presence or absence of specific taxa. They suggested that the loss or overgrowth of certain bacteria during the first 48 h in the ICU might contribute to the inflammation observed at this time. Their study revealed that lung bacterial community variation at 0 h was significantly linked to endothelial injury (measured by soluble intercellular adhesion molecule-1), epithelial injury [vascular endothelial growth factor (VEGF)], and inflammation (IL-8) in critically ill blunt trauma patients. At 48 h, community variation was also associated with elevated IL-6 and IL-8 levels. Interestingly, the composition of the microbiota at 0 h was significantly correlated with 48-hour levels of VEGF, receptor for advanced glycation end-products (RAGE), angiopoietin-2 (ANG-2), pentraxin 3 (PENT3), and IL-8 (Panzer et al., 2018).
Montassier et al. identified microbiota signatures associated with ARDS, characterized by an enrichment of potentially pathogenic respiratory microbes, including Pseudomonas and Staphylococcus, through data harmonization and the pooling of individual patient data. Furthermore, patients with the presence of Staphylococcus, Ralstonia and Enterococcus, had a lower probability of successful extubation (Montassier et al., 2023). Zhang et al. observed that an increase in Escherichia coli and Staphylococcus aureus in the lungs may serve as risk factors for mortality in ARDS induced by intrapulmonary infections. While no specific pathogens were linked to prognosis, they demonstrated that the increased presence of Bilophila, a genus of intestinal bacteria, could be a potential risk factor for death in ARDS caused by extrapulmonary infections. In contrast, Hydrobacter might play a protective role in ARDS induced by intrapulmonary infections (Zhang et al., 2022). However, it remains unclear whether Hydrobacter is part of the normal lung microbiota or whether it functions independently or synergistically with other microbiomes.
In addition, Kyo et al. discovered that the ratio of the relative abundance of Betaproteobacterial operational taxonomic units (OTUs) to the maximum relative abundance of three other OTUs (Staphylococcus, Streptococcus, and Enterobacteriaceae) was significantly associated with hospital mortality in ARDS patients. They found that the Betaproteobacteria class was strongly negatively correlated with serum IL-6 levels in the non-survivor. Conversely, Staphylococcus and Streptococcus at the genus level and Enterobacteriaceae at the family level were significantly positively correlated with serum IL-6 levels in the non-survivors (Kyo et al., 2019).
4 Microbiome features in severe COVID-19 ARDS patients
4.1 Ecological metrics
Dysbiosis of the lung microbiome has been observed in COVID-19 patients (Merenstein et al., 2021; Shen et al., 2020). Compared to healthy lung samples, lung microbiota from mechanically ventilated COVID-19 patients exhibited lower diversity (Gaibani et al., 2021; Merenstein et al., 2021; Zacharias et al., 2022). Pascale et al. further reported that patients with reduced lung microbiota diversity had a longer duration of invasive mechanical ventilation (IMV) and higher mortality (De Pascale et al., 2024).
Interestingly, Kullberg et al. found a significant association between bacterial burden and beta diversity in COVID-19-related ARDS among mechanically ventilated patients, suggesting that bacterial overgrowth may drive compositional changes in critically ill lungs. They also discovered that increased bacterial DNA burden correlated with elevated alveolar concentrations of proinflammatory cytokines (TNF-α, IL-6, IL-1). Furthermore, they reported that the overall lung microbiota composition, rather than individual bacterial genera, was linked to successful extubation in COVID-19-related ARDS (Kullberg et al., 2022). Several studies have also indicated that higher bacterial burden is negatively associated with successful extubation (Kullberg et al., 2022) and survival (Sulaiman et al., 2021) in mechanically ventilated patients with COVID-19-related ARDS. These findings are consistent with those observed in traditional ARDS.
4.2 Differential flora
Merenstein et al. found that lung microbiome specimens from COVID-19 patients, sampled via endotracheal aspirates (ETA), showed frequent outgrowth of potential respiratory pathogens, particularly Staphylococcus (Merenstein et al., 2021). Additionally, Gaibani et al. reported that the lung microbiota of critically ill COVID-19 patients was enriched with Pseudomonas alcaligenes, Clostridium hiranonis, Acinetobacter schindleri, Sphingobacterium spp., Acinetobacter spp., and Enterobacteriaceae. In contrast, COVID-19-negative patients displayed a higher abundance of lung commensal bacteria, such as Haemophilus influenzae, Veillonella dispar, Granulicatella spp., Porphyromonas spp., and Streptococcus spp. (Gaibani et al., 2021). Compared to influenza-related and bacterial CAP-related ARDS patients, Prevotella and Streptococcus were more abundant in COVID-19-related ARDS patients, while Bacteroides was only more abundant compared to influenza-related ARDS patients (Imbert et al., 2024). However, Shen et al. reported that the microbiota in COVID-19 patients was similar to that in CAP patients, characterized by either the dominance of pathogens or elevated levels of oral and upper respiratory tract commensal bacteria (Shen et al., 2020).
Moreover, Pascale et al. observed that Firmicutes dominated the lung microbiota of patients with high respiratory system compliance/predicted body weight (Crs) phenotype, while in COVID-19-related ARDS patients with low Crs/low ventilatory ratio (VR), Proteobacteria predominated. They also found a positive correlation between serum procalcitonin and Proteobacteria, and a negative correlation with Firmicutes (De Pascale et al., 2024). Additionally, Sulaiman et al. discovered that the oral commensal Mycoplasma salivarum was enriched in patients who died or required mechanical ventilation for more than 28 days. In contrast, Prevotella oris was more abundant in patients with mechanical ventilation duration ≤ 28 days (Sulaiman et al., 2021).
4.3 Microbiome features in acute lung injury animal model
This section reviews current understanding of the lung microbiome in ALI/ARDS animal model (Table 3).
TABLE 3.
Studies concerning lung microbiota of ALI/ARDS in animal model and Therapeutic options.
| Study | Publication, y | Inducers of ALI/ARDS model | Therapeutic options; dose; routes of administration | Animals; sex; age | Date of sample | Type of samples | Analysis | Main results |
|
Tian et al. (2022)
|
2022 | LPS; i.p. | – | Sprague-Dawley rats; Male; 8-weeks old | LPS12 h; LPS48 h | Lung | 16S rRNA |
Brevibacterium was found to be correlated with the cytokines TNF–α, IL-10, and IL-6, as well as the hematological percentage of neutrophils. The Wnt, Notch, and chronic myeloid leukemia signaling pathways were associated with IL-1β. The Mitogen-activated protein kinase (MAPK) signaling pathway (specifically yeast) showed a correlation with IL-10. Ascorbate and aldarate metabolism pathways, along with basal transcription factors, were linked to platelet-related indicators. |
| Poroyko et al. (2015) | 2015 | LPS; i.p. | – | C57BL/6J mice; – | BAL 72 h after treatment | BALF | 16S rRNA | The loss of Firmicutes, represented by the Alicyclobacillaceae family, and the proliferation of Proteobacteria, represented by the Brucellaceae and Xanthomonadaceae families. |
| Mukherjee et al. (2022) | 2022 | Plasmodium berghei; i.p. | – | C57BL/6J; DBA/2 mice; Male | – | Lung | 16S rRNA | The parasite in the lung activates the immune system, causing T cells to produce the cytokine IL-10, which disrupts microbial control and promotes MA-ARDS. |
|
Jin et al. (2021)
|
2022 | LPS; – | Vitamin D3; 150 μg/kg; gavages | C57BL/6 mice; Male; 8-10 weeks old | After LPS for 6 h | BALF | 16S rDNA, non-targeted metabolomics |
The abundance of Rodentibacter was positively correlated with the gene expression of IL-1β, IL-6, and TNF-α. Short-term vitamin D3 treatment effectively reduced the abundance of Rodentibacter. The abundance of Rodentibacter was positively correlated with the gene expression of IL-1β, IL-6, and TNF-α. Short-term vitamin D3 treatment effectively reduced the abundance of Rodentibacter. Short-term vitamin D3 supplementation prevents LPS-induced ALI by inhibiting the production of pro-inflammatory cytokines, including IL-1β, IL-6, and TNF-α. Short-term vitamin D3 significantly reduces the phosphorylation of signal transducer and activator of transcription and suppressor of cytokine signaling 3, while upregulating the phosphorylation of the inhibitor of NF-κB. |
| Lv et al. (2023) | 2023 | LPS; i.p. | HUC-MSCs; 0.5 mL PBS containing 1 × 106 HUC-MSCs; i.p. |
C57BL/6 mice; Male; 6-8 weeks old | D3 after HUC-MSC | BALF | 16S rDNA | HUC-MSCs alleviated pulmonary edema and injury in both the lung and ileum, and reduced mononuclear cell and neutrophil counts, protein concentrations in BALF, and inflammatory cytokine levels in the serum, lung, and ileum of ALI mice. Compared to the LPS group, five metabolites were upregulated, and 11 metabolites were downregulated in the LPS + MSC group. |
| Mohammed et al. (2020) | 2020 | SEB; intranasally; i.p. | Δ9-tetrahydro cannabinol; 20 mg/kg; i.p. |
C3H/HeJ mice; Female; 8–10 weeks old | D3 after SEB administration | Lung | 16S rRNA | THC significantly increased the abundance of the beneficial bacterial species Ruminococcus gnavus, while reducing the abundance of the pathogenic microbiota Akkermansia muciniphila. THC treatment led to an increase in SCFAs, with propionic acid being identified as a key factor in inhibiting the inflammatory response. THC upregulated several genes, including lysozyme 1 and 2, β-defensin-2, claudin, zonula occludens-1, occludin-1, Mucin2, and Muc5b, while downregulating β-defensin-1 |
| Sultan et al. (2021) | 2021 | SEB; intranasally | AEA; 40 mg/k; i.p. | C57BL/6 mice; Female; 6–8 weeks old | 48 h after SEB | Lung | 16S rRNA | AEA treatment attenuates SEB-mediated ARDS by suppressing inflammation and preventing dysbiosis through the induction of AMPs, tight junction proteins, and SCFAs. |
| Alghetaa et al. (2021) | 2023 | SEB; intranasally; i.p. | Resveratrol; 100 mg/kg; oral-gavage | C3H/HeJ mice; Female; 6-weeks old | 48 h after 2th SEB | Lung | 16S rRNA | RES-mediated attenuation of ARDS occurs, at least in part, through the induction of beneficial bacteria, such as L. reuteri. |
| Sapra et al. (2024) | 2024 | LPS; intranasally/ CLP | Lactobacillus rhamnosus; 109CFU/day; orally/SCFA (Butyrate); 200 mM; i.p. | BALB/c and C57BL/6 J mice; Male; 8–10 weeks | 24 h after LPS or CLP; | - | - | Pretreatment with LR significantly alleviated pulmonary edema by modulating neutrophil, accompanied by a marked suppression of inflammatory cytokine production in the BALF, lung tissue, and serum. Short-chain fatty acids produced by LR, especially butyrate, targets the phagocytic and neutrophils extracellular traps releasing potential of neutrophils. |
4.4 Ecological metrics
Poroyko et al. observed a fivefold increase in bacterial load in a mouse model 72 h after intratracheal administration of sterile bacterial wall lipopolysaccharide (LPS) (Poroyko et al., 2015). In addition, Tian et al. found a significant reduction in the volatility of α-diversity 48 hours after LPS was injected intraperitoneally in rats (Tian et al., 2022).
In contrast, Dickson et al. reported that the lungs of post-sepsis mice exhibited increased bacterial diversity 3 days after exposure in a mouse model of lung injury induced by cecal ligation and puncture (CLP) (Dickson et al., 2016b).
4.5 Differential flora
In addition, Dickson et al. found that the lung microbiota of post-sepsis mice were significantly enriched with numerous bacteria commonly found in the murine gut, including members of the Bacteroidales order, Enterococcus species, and Lachnospiraceae (sp.). Notably, a member of the Bacteroidales order (OTU008) was identified as a key driver of community alterations (Dickson et al., 2016b).
Poroyko et al. observed that the primary microbial response to LPS-induced ALI was characterized by a loss of Firmicutes—represented by the family Alicyclobacillaceae—and a proliferation of Proteobacteria, particularly from the Brucellaceae and Xanthomonadaceae families (Poroyko et al., 2015).
Previous studies have shown that Stenotrophomonas directly contributes to the inflammatory process and impairs respiratory function (Di Bonaventura et al., 2010). Similarly, Ochrobactrum has been implicated in lung inflammation (Dawkins et al., 2006; Naik et al., 2013). Additionally, Poroyko et al. detected bacterial substrates suitable for Ochrobactrum anthropi and Stenotrophomonas maltophilia in BALF, identifying 35 lung metabolites that could serve as substrates. Notably, they found eight metabolites shared by both pathogens, including four carbohydrates, two nucleotide/nucleoside compounds, and two organic acids (citrate and lactate). Furthermore, six metabolites—putrescine, desmosterol, tetracosanoic acid, uric acid, 3-hydroxycholestane, and lactic acid—were significantly elevated in samples from injured lungs (Poroyko et al., 2015).
Additionally, Mukherjee et al. found that the adherence of Plasmodium-infected red blood cells to the vascular endothelium induces persistent immune activation, leading to the production of the anti-inflammatory cytokine IL-10 by T cells in the lung. Elevated IL-10 levels promote bacterial expansion in the lung, thereby contributing to the development of malaria-associated acute respiratory distress syndrome (MA-ARDS) (Mukherjee et al., 2022).
Tian et al. found that Brevibacterium was positively correlated with serum cytokines TNF-α, IL-10, and IL-6 levels. Additionally, they observed that the functionality of the lung microbiota appeared to be more stable and less variable compared to its composition, with only three differential pathways identified in terms of functional abundance. Changes in the abundance of the ABC transporter pathway may reflect the adaptation of lung bacteria to inflammatory conditions.
The upregulation of bacterial chemotaxis functions in the lung microbiota may result from bacterial adaptation to the edema environment. The reduction in the pentose phosphate pathway could indicate a microbiota with lower antioxidant capacities and diminished DNA repair abilities following lung injury. Regarding lung microbiota functionality, the abundances of four signaling pathways—Wnt (ko04310), Notch (ko04330), chronic myeloid leukemia (ko05220), and MAPK-yeast (ko04011)—were strongly negatively correlated with serum IL-1β and IL-10 levels, suggesting that these pathways may play a role in the interaction between the lung microbiota and host immunity (Tian et al., 2022).
4.6 Prospect of therapy
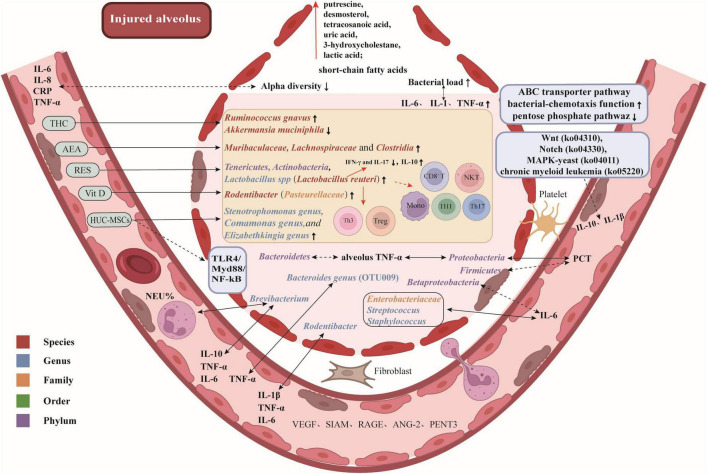
Here are some potential therapeutic targets and promising drugs for attenuating ALI/ARDS by modulating the lung microbiota, as illustrated in Figure 1.
FIGURE 1.
Potential therapeutic targets and promising drugs for attenuating ALI/ARDS by modulating lung microbiota. Gut-associated bacteria, bacteria that are enriched in the gut, such as Bacteroides spp.; Responders to early corticosteroid therapy, patients who showed a significant improvement in their PaO2/FiO2 ratio after receiving corticosteroid treatment. Arterial blood gas measurements were obtained upon ICU admission, prior to the initial administration of corticosteroids, and repeated 24 hours later, following (or prior to) corticosteroid treatment. The difference in PaO2/FiO2 between these two time points, referred to as ΔPaO2/FiO2, was calculated. Patients with a ΔPaO2/FiO2 ≥ 150 mmHg were categorized as responders, while those with a ΔPaO2/FiO2 < 150 mmHg were classified as non-responders.
4.7 The target of potential treatment: microbial community
4.7.1 Anandamide
Endocannabinoids, which are host-derived lipid hormones, possess potent anti-inflammatory properties (Ahmed et al., 2021; Jackson et al., 2014; Osafo et al., 2021; Pandey et al., 2009; Sido et al., 2016). Some studies have suggested that endocannabinoids can modulate the diversity of the gut microbiota (Finn et al., 2021; Minichino et al., 2021). Sultan et al. demonstrated that staphylococcal enterotoxin B (SEB) induces alterations in both the lung and gut microbiota, particularly leading to an overgrowth of pathogenic bacteria such as Pseudomonas. However, AEA treatment reversed these changes by promoting the induction of antimicrobial peptides (AMPs) and tight junction proteins. Additionally, AEA treatment enhanced the abundance of beneficial bacteria in the lungs of mice, including Muribaculaceae, Lachnospiraceae, and Clostridia, which are known to produce butyrate (Sultan et al., 2021).
4.8 The target of drug intervention: microbial community and immune system
4.8.1 Δ9-tetrahydrocannabinol
Δ9-Tetrahydrocannabinol (THC), a cannabinoid found in Cannabis sativa L., is well known for its anti-inflammatory properties (Nagarkatti et al., 2009). Studies have shown that THC and cannabidiol can influence gut microbiota in experimental models of autoimmune diseases (Al-Ghezi et al., 2019; Cluny et al., 2015). Mohammed et al. found that THC administration protected mice from SEB-induced mortality and attenuated ARDS by suppressing lung inflammation and modulating microbial dysbiosis and their metabolites in the lungs. Specifically, they observed that THC increased the abundance of the beneficial bacterial species Ruminococcus gnavus in Firmicutes phylum, while decreasing the abundance of Akkermansia muciniphila (Mohammed et al., 2020). R. gnavus is a human gut symbiont known to enhance the expression of glycoproteins (Graziani et al., 2016), which are critical for maintaining lung tissue integrity, preventing vascular leaks, and defending against bacterial pneumonia (Johansson et al., 2011; Roy et al., 2014). In contrast, A. muciniphila, commonly found in the intestines of humans and animals, can elevate the expression of pro-inflammatory cytokines (Ganesh et al., 2013) and has a specialized ability to degrade mucin (Derrien et al., 2017; Ganesh et al., 2013).
Additionally, THC was found to significantly increase the levels of short-chain fatty acids (SCFAs) such as propionic acid, butyric acid, and acetic acid in the colon. Notably, propionic acid inhibited the production of pro-inflammatory cytokines while promoting the release of anti-inflammatory cytokines. Furthermore, investigations have shown that R. gnavus produces propionate and propanol as end products of metabolism (Crost et al., 2013). These findings suggest that THC may promote the abundance of beneficial bacterial species like R. gnavus, enhance the production of SCFAs such as propionic acid, and suppress pro-inflammatory responses, thereby attenuating acute lung injury.
4.8.2 Human umbilical cord mesenchymal cells
A previous study has demonstrated that HUC-MSCs promote the expression of PD-L1 on macrophages and attenuate ALI in mice (Tu et al., 2022). Lv et al. observed that HUC-MSCs can improve pulmonary edema, lung injury, and endothelial barrier function in LPS-induced ALI mice, while also reducing the expression of inflammatory cytokines in both serum and lung tissue. Additionally, HUC-MSC treatment enhanced the integrity of the endothelial barrier in the lungs. More importantly, they found that HUC-MSCs ameliorated ALI by reducing the abundance of pathogenic bacteria, including Stenotrophomonas, Comamonas, and Elizabethkingia genera, in the BALF of mice. Furthermore, Haemophilus may play a critical role in the improvement of ALI by HUC-MSCs. Moreover, Lactobacillus, Bacteroides, and an unidentified Rikenellaceae genus were identified as potential biomarkers for evaluating the therapeutic efficacy of HUC-MSCs. The study also revealed that HUC-MSC treatment of ALI mice significantly attenuated the expression of the TLR4/Myd88/NF-κB signaling pathway in lung tissue. Metabolomic analysis further indicated that MSC treatment upregulated 5 metabolites and downregulated 11 metabolites, primarily related to purine metabolism and taste transduction signaling pathways. Notably, metabolites involved in drug metabolism, tyrosine metabolism, autophagy, and endocytosis were significantly associated with the improvement of ALI by HUC-MSC treatment (Lv et al., 2023).
4.8.3 Vitamin D3
Jin et al. observed an increased relative abundance of Rodentibacter in the BALF of LPS-treated mice. Short-term vitamin D3 treatment was found to effectively reduce Rodentibacter abundance and attenuate ALI. Furthermore, they noted a positive correlation between the abundance of Rodentibacter and serum levels of pro-inflammatory cytokines, including IL-1β, IL-6, and TNF-α (Jin et al., 2021).
Rodentibacter, primarily found in rodents, belongs to the Pasteurellaceae family (Adhikary et al., 2017; Benga et al., 2018), and has been reported as an opportunistic pathogen, or even a primary pathogen, under certain conditions (Benga et al., 2015).
4.9 The target of potential treatment: microbial community and immune system
4.9.1 Resveratrol
Resveratrol (RES), a stilbenoid, is well known for its potent anti-inflammatory and antioxidant properties (Chen et al., 2020; Salehi et al., 2018). Studies have shown that RES can significantly alter gut microbiota composition (Alrafas et al., 2019; Li F. et al., 2020), and alleviate SEB-induced ARDS in mice (Alghetaa et al., 2018; Rieder et al., 2012). Alghetaa et al. reported that SEB exposure promotes the growth of pathogenic bacteria, such as those from the Proteobacteria phylum and Propionibacterium acnes species, in lung microbiota, contributing to ARDS pathogenesis. Their findings indicated that RES treatment significantly enhanced lung microbiota diversity in SEB-exposed mice, increasing the abundance of beneficial bacteria, including the Tenericutes and Actinobacteria phyla, as well as the Lactobacillus genus, particularly Lactobacillus reuteri. Colonic microbiota transplantation (CMT) experiments further confirmed that RES-induced enrichment of beneficial bacteria, particularly L. reuteri, plays a crucial role in mitigating ARDS. Specifically, L. reuteri reduced the number of lung-infiltrating mononuclear cells, cytotoxic CD8+ T cells, NKT cells, Th1 cells, and Th17 cells, while increasing the proportions of regulatory T cells (Tregs) and Th3 cells. Furthermore, L. reuteri inhibited the SEB-induced production of pro-inflammatory cytokines such as IFN-γ and IL-17, while promoting the production of the anti-inflammatory cytokine IL-10 (Alghetaa et al., 2021).
L. reuteri is a well-studied probiotic bacterium found in various human organs and tissues. It has been shown to produce antimicrobial molecules, which inhibit the colonization of pathogenic microbes (Mu et al., 2018). Studies also suggest that L. reuteri can suppress the production of pro-inflammatory cytokines while enhancing the development and function of regulatory T cells (Tregs) (Li L. et al., 2020). Additionally, L. reuteri has been linked to increased butyric acid concentrations in the gut, contributing to improved gut health and immune regulation (Li L. et al., 2020; Liu et al., 2017).
4.9.2 Lactobacillus rhamnosus
Pretreatment with LR notably improved lung vascular permeability (edema) by modulating neutrophils, while also significantly decreasing the expression of inflammatory cytokines in the BALF, lungs, and serum in both pulmonary and extrapulmonary mouse models. From a mechanistic perspective, LR, through its short-chain fatty acids (with butyrate being the most potent and effective in improving the pathophysiology of both pulmonary and extrapulmonary ARDS), influences neutrophil phagocytosis and their capacity to release extracellular traps. Moreover, butyrate demonstrates enhanced potential in alleviating the pathophysiology of ARDS by reducing neutrophil infiltration into the lungs (Sapra et al., 2024).
5 Conclusion and prospects
The lung microbiome undergoes significant alterations in ARDS compared to healthy individuals, with these changes closely linked to disease severity. While the literature partially agrees on the specific microbial changes, certain patterns are consistently observed. ARDS is characterized by an increased bacterial burden and reduced microbial diversity. Patients with lower alpha diversity and higher bacterial loads in their lung microbiota tend to have fewer ventilator-free days and higher mortality rates. Physiological lung flora, such as Firmicutes, is notably diminished, while respiratory pathogens, including Staphylococcus, Streptococcus, Enterobacteriaceae, Pseudomonas, and Proteobacteria, proliferate in ARDS patients. Furthermore, the lung microbiome is closely associated with immune and inflammatory responses in ARDS. However, the impact of different initial infection sites on lung microbiome diversity and composition remains unclear, and study heterogeneity may significantly influence findings. We summarized that these studies involved populations with different causes of ARDS, varying in sample sizes, countries and regions, all of which may contribute to the inconsistency of the findings. Given that lower airway microbiota composition can vary depending on the underlying cause of ARDS, future research and experimental models should account for the etiology when investigating the role of the lung microbiome in ARDS pathophysiology.
Funding Statement
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by 1.3.5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (ZYGD23012).
Abbreviations
ALI, acute lung injury; AEA, anandamide; AMPs, antimicrobial peptides; ANG-2, angiopoietin-2; ARDS, acute respiratory distress syndrome; ARDS-exp, extrapulmonary ARDS; ARDS-p, intrapulmonary ARDS; BALF, bronchoalveolar lavage fluid; CLP, cecal ligation and puncture; CMT, colonic microbiota transplantation; COPD, chronic obstructive pulmonary disease; COVID-19, corona virus disease 2019; Crs, respiratory system compliance; ETA, endotracheal aspirates; HUC-MSCs, human umbilical cord mesenchymal cells; ICU, intensive care unit; IMV, invasive mechanical ventilation; i.p., intraperitoneally; LPS, lipopolysaccharide; MA-ARDS, malaria-associated acute respiratory distress syndrome; MV, mechanical ventilation; OTUs, operational taxonomic units; PENT3, pentraxin 3; RAGE, receptor for advanced glycation end-products; RES, resveratrol; SCFAs, short-chain fatty acids; SEB, staphylococcal enterotoxin-b; THC, Δ9-tetrahydrocannabinol; TNF-α, tumor necrosis factor-α; TLR, toll-like receptor; VEGF, vascular endothelial growth factor; VR, ventilatory ratio.
Author contributions
YT: Writing – original draft. BL: Writing – original draft. AM: Writing – original draft. BW: Writing – review & editing. HX: Writing – original draft. YZ: Writing – review & editing. JY: Writing – review & editing. YK: Writing – review & editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Adhikary S., Nicklas W., Bisgaard M., Boot R., Kuhnert P., Waberschek T., et al. (2017). Rodentibacter gen. nov. including Rodentibacter pneumotropicus comb. nov., Rodentibacter heylii sp. nov., Rodentibacter myodis sp. nov., Rodentibacter ratti sp. nov., Rodentibacter heidelbergensis sp. nov., Rodentibacter trehalosifermentans sp. nov., Rodentibacter rarus sp. nov., Rodentibacter mrazii and two genomospecies. Int. J. Syst. Evol. Microbiol. 67 1793–1806. 10.1099/ijsem.0.001866 [DOI] [PubMed] [Google Scholar]
- Ahmed I., Rehman S. U., Shahmohamadnejad S., Zia M. A., Ahmad M., Saeed M. M., et al. (2021). Therapeutic attributes of endocannabinoid system against neuro-inflammatory autoimmune disorders. Molecules 26:3389. 10.3390/molecules26113389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alghetaa H., Mohammed A., Sultan M., Busbee P., Murphy A., Chatterjee S., et al. (2018). Resveratrol protects mice against SEB-induced acute lung injury and mortality by miR-193a modulation that targets TGF-β signalling. J. Cell Mol. Med. 22 2644–2655. 10.1111/jcmm.13542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alghetaa H., Mohammed A., Zhou J., Singh N., Nagarkatti M., Nagarkatti P. (2021). Resveratrol-mediated attenuation of superantigen-driven acute respiratory distress syndrome is mediated by microbiota in the lungs and gut. Pharmacol. Res. 167:105548. 10.1016/j.phrs.2021.105548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Ghezi Z. Z., Busbee P. B., Alghetaa H., Nagarkatti P. S., Nagarkatti M. (2019). Combination of cannabinoids, delta-9-tetrahydrocannabinol (THC) and cannabidiol (CBD), mitigates experimental autoimmune encephalomyelitis (EAE) by altering the gut microbiome. Brain Behav. Immun. 82 25–35. 10.1016/j.bbi.2019.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alrafas H. R., Busbee P. B., Nagarkatti M., Nagarkatti P. S. (2019). Resveratrol modulates the gut microbiota to prevent murine colitis development through induction of Tregs and suppression of Th17 cells. J. Leukoc. Biol. 106 467–480. 10.1002/JLB.3A1218-476RR [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benga L., Benten W. P., Engelhardt E., Gougoula C., Sager M. (2015). Spontaneous bacterial and fungal infections in genetically engineered mice: Is Escherichia coli an emerging pathogen in laboratory mouse? Berl. Munch Tierarztl. Wochenschr. 128 278–284. [PubMed] [Google Scholar]
- Benga L., Sager M., Christensen H. (2018). From the [Pasteurella] pneumotropica complex to Rodentibacter spp.: An update on [Pasteurella] pneumotropica. Vet. Microbiol. 217 121–134. 10.1016/j.vetmic.2018.03.011 [DOI] [PubMed] [Google Scholar]
- Charlson E. S., Bittinger K., Haas A. R., Fitzgerald A. S., Frank I., Yadav A., et al. (2011). Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med. 184 957–963. 10.1164/rccm.201104-0655OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Fu Q., Song X., Muhammad A., Jia R., Zou Y., et al. (2020). Preparation of resveratrol dry suspension and its immunomodulatory and anti-inflammatory activity in mice. Pharm. Biol. 58 8–15. 10.1080/13880209.2019.1699123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluny N. L., Keenan C. M., Reimer R. A., Le Foll B., Sharkey K. A. (2015). Prevention of diet-induced obesity effects on body weight and gut microbiota in mice treated chronically with Δ9-Tetrahydrocannabinol. PLoS One 10:e0144270. 10.1371/journal.pone.0144270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crost E. H., Tailford L. E., Le Gall G., Fons M., Henrissat B., Juge N. (2013). Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One 8:e76341. 10.1371/journal.pone.0076341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins P., Robertson A., Robertson W., Moore V., Reynolds J., Langman G., et al. (2006). An outbreak of extrinsic alveolitis at a car engine plant. Occup. Med. (Lond) 56 559–565. 10.1093/occmed/kql110 [DOI] [PubMed] [Google Scholar]
- De Pascale G., Posteraro B., De Maio F., Pafundi P. C., Tanzarella E. S., Cutuli S. L., et al. (2024). Lung microbiota composition, respiratory mechanics, and outcomes in COVID-19-related ARDS. Microbiol. Spectr. 12:e0357423. 10.1128/spectrum.03574-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien M., Belzer C., de Vos W. M. (2017). Akkermansia muciniphila and its role in regulating host functions. Microb. Pathog. 106 171–181. 10.1016/j.micpath.2016.02.005 [DOI] [PubMed] [Google Scholar]
- Di Bonaventura G., Pompilio A., Zappacosta R., Petrucci F., Fiscarelli E., Rossi C., et al. (2010). Role of excessive inflammatory response to Stenotrophomonas maltophilia lung infection in DBA/2 mice and implications for cystic fibrosis. Infect. Immun. 78 2466–2476. 10.1128/IAI.01391-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. P., Erb-Downward J. R., Freeman C. M., McCloskey L., Falkowski N. R., Huffnagle G. B., et al. (2017). Bacterial topography of the healthy human lower respiratory tract. mBio 8:e02287-16. 10.1128/mBio.02287-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. P., Erb-Downward J. R., Martinez F. J., Huffnagle G. B. (2016a). The microbiome and the respiratory tract. Annu. Rev. Physiol. 78 481–504. 10.1146/annurev-physiol-021115-105238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. P., Schultz M. J., van der Poll T., Schouten L. R., Falkowski N. R., Luth J. E., et al. (2020). Lung microbiota predict clinical outcomes in critically Ill patients. Am. J. Respir. Crit. Care Med. 201 555–563. 10.1164/rccm.201907-1487OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. P., Singer B. H., Newstead M. W., Falkowski N. R., Erb-Downward J. R., Standiford T. J., et al. (2016b). Enrichment of the lung microbiome with gut bacteria in sepsis and the acute respiratory distress syndrome. Nat. Microbiol. 1:16113. 10.1038/nmicrobiol.2016.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn D. P., Haroutounian S., Hohmann A. G., Krane E., Soliman N., Rice A. S. C. (2021). Cannabinoids, the endocannabinoid system, and pain: A review of preclinical studies. Pain 162 (Suppl. 1), S5–S25. 10.1097/j.pain.0000000000002268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaibani P., Viciani E., Bartoletti M., Lewis R. E., Tonetti T., Lombardo D., et al. (2021). The lower respiratory tract microbiome of critically ill patients with COVID-19. Sci. Rep. 11:10103. 10.1038/s41598-021-89516-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh B. P., Klopfleisch R., Loh G., Blaut M. (2013). Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLoS One 8:e74963. 10.1371/journal.pone.0074963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziani F., Pujol A., Nicoletti C., Dou S., Maresca M., Giardina T., et al. (2016). Ruminococcus gnavus E1 modulates mucin expression and intestinal glycosylation. J. Appl. Microbiol. 120 1403–1417. 10.1111/jam.13095 [DOI] [PubMed] [Google Scholar]
- Horn K. J., Schopper M. A., Drigot Z. G., Clark S. E. (2022). Airway Prevotella promote TLR2-dependent neutrophil activation and rapid clearance of Streptococcus pneumoniae from the lung. Nat. Commun. 13:3321. 10.1038/s41467-022-31074-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Shen J., An Y., Jiang Y., Zhao H. (2024). Phenotypes and lung microbiota signatures of immunocompromised patients with pneumonia-related acute respiratory distress syndrome. J. Inflamm. Res. 17 1429–1441. 10.2147/JIR.S453123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussell T., Bell T. J. (2014). Alveolar macrophages: Plasticity in a tissue-specific context. Nat. Rev. Immunol. 14 81–93. 10.1038/nri3600 [DOI] [PubMed] [Google Scholar]
- Imbert S., Revers M., Enaud R., Orieux A., Camino A., Massri A., et al. (2024). Lower airway microbiota compositions differ between influenza, COVID-19 and bacteria-related acute respiratory distress syndromes. Crit. Care 28:133. 10.1186/s13054-024-04922-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. R., Hegde V. L., Nagarkatti P. S., Nagarkatti M. (2014). Characterization of endocannabinoid-mediated induction of myeloid-derived suppressor cells involving mast cells and MCP-1. J. Leukoc. Biol. 95 609–619. 10.1189/jlb.0613350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin A., Zhao Y., Yuan Y., Ma S., Chen J., Yang X., et al. (2021). Single treatment of vitamin D3 ameliorates LPS-induced acute lung injury through changing lung Rodentibacter abundance. Mol. Nutr. Food Res. 66:e2100952. 10.1002/mnfr.202100952 [DOI] [PubMed] [Google Scholar]
- Johansson M. E., Ambort D., Pelaseyed T., Schütte A., Gustafsson J. K., Ermund A., et al. (2011). Composition and functional role of the mucus layers in the intestine. Cell Mol. Life Sci. 68 3635–3641. 10.1007/s00018-011-0822-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatiwada S., Subedi A. (2020). Lung microbiome and coronavirus disease 2019 (COVID-19): Possible link and implications. Hum. Microb. J. 17:100073. 10.1016/j.humic.2020.100073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullberg R. F. J., de Brabander J., Boers L. S., Biemond J. J., Nossent E. J., Heunks L. M. A., et al. (2022). Lung microbiota of critically Ill patients with COVID-19 are associated with nonresolving acute respiratory distress syndrome. Am. J. Respir. Crit. Care Med. 206 846–856. 10.1164/rccm.202202-0274OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyo M., Nishioka K., Nakaya T., Kida Y., Tanabe Y., Ohshimo S., et al. (2019). Unique patterns of lower respiratory tract microbiota are associated with inflammation and hospital mortality in acute respiratory distress syndrome. Respir. Res. 20:246. 10.1186/s12931-019-1203-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenzi G. A., Potter R. T., Kass E. H. (1961). Bacteriologic flora of the lower respiratory tract. N. Engl. J. Med. 265 1273–1278. 10.1056/NEJM196112282652601 [DOI] [PubMed] [Google Scholar]
- Leitao Filho F. S., Alotaibi N. M., Ngan D., Tam S., Yang J., Hollander Z., et al. (2019). Sputum microbiome is associated with 1-year mortality after chronic obstructive pulmonary disease hospitalizations. Am. J. Respir. Crit. Care Med. 199 1205–1213. 10.1164/rccm.201806-1135OC [DOI] [PubMed] [Google Scholar]
- Li F., Han Y., Cai X., Gu M., Sun J., Qi C., et al. (2020). Dietary resveratrol attenuated colitis and modulated gut microbiota in dextran sulfate sodium-treated mice. Food Funct. 11 1063–1073. 10.1039/c9fo01519a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Fang Z., Liu X., Hu W., Lu W., Lee Y. K., et al. (2020). Lactobacillus reuteri attenuated allergic inflammation induced by HDM in the mouse and modulated gut microbes. PLoS One 15:e0231865. 10.1371/journal.pone.0231865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N. N., Kang K., Zhou Y., Liu Y. Q., Zhang Q. Q., Luo P. Y., et al. (2024). Throat microbiota drives alterations in pulmonary alveolar microbiota in patients with septic ARDS. Virulence 15:2350775. 10.1080/21505594.2024.2350775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira-Lucio J. A., Falfán-Valencia R., Ramírez-Venegas A., Buendía-Roldán I., Rojas-Serrano J., Mejía M., et al. (2020). Lung microbiome participation in local immune response regulation in respiratory diseases. Microorganisms 8:1059. 10.3390/microorganisms8071059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Hou C., Wang G., Jia H., Yu H., Zeng X. (2017). Lactobacillus reuteri I5007 modulates intestinal host defense peptide expression in the model of IPEC-J2 cells and neonatal piglets. Nutrients 9:559. 10.3390/nu9060559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L., Cui E. H., Wang B., Li L. Q., Hua F., Lu H. D., et al. (2023). Multiomics reveal human umbilical cord mesenchymal stem cells improving acute lung injury via the lung-gut axis. World J. Stem Cells 15 908–930. 10.4252/wjsc.v15.i9.908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man W. H., de Steenhuijsen Piters W. A., Bogaert D. (2017). The microbiota of the respiratory tract: Gatekeeper to respiratory health. Nat. Rev. Microbiol. 15 259–270. 10.1038/nrmicro.2017.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merenstein C., Liang G., Whiteside S. A., Cobián-Güemes A. G., Merlino M. S., Taylor L. J., et al. (2021). Signatures of COVID-19 severity and immune response in the respiratory tract microbiome. mBio 12:e0177721. 10.1128/mBio.01777-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minichino A., Jackson M. A., Francesconi M., Steves C. J., Menni C., Burnet P. W. J., et al. (2021). Endocannabinoid system mediates the association between gut-microbial diversity and anhedonia/amotivation in a general population cohort. Mol. Psychiatry 26 6269–6276. 10.1038/s41380-021-01147-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed A., Alghetaa H. K., Zhou J., Chatterjee S., Nagarkatti P., Nagarkatti M. (2020). Protective effects of Δ9 -tetrahydrocannabinol against enterotoxin-induced acute respiratory distress syndrome are mediated by modulation of microbiota. Br. J. Pharmacol. 177 5078–5095. 10.1111/bph.15226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molyneaux P. L., Cox M. J., Willis-Owen S. A., Mallia P., Russell K. E., Russell A. M., et al. (2014). The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 190 906–913. 10.1164/rccm.201403-0541OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montassier E., Kitsios G. D., Radder J. E., Le Bastard Q., Kelly B. J., Panzer A., et al. (2023). Robust airway microbiome signatures in acute respiratory failure and hospital-acquired pneumonia. Nat. Med. 29 2793–2804. 10.1038/s41591-023-02617-9 [DOI] [PubMed] [Google Scholar]
- Mu Q., Tavella V. J., Luo X. M. (2018). Role of Lactobacillus reuteri in human health and diseases. Front. Microbiol. 9:757. 10.3389/fmicb.2018.00757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee D., Chora ÂF., Lone J. C., Ramiro R. S., Blankenhaus B., Serre K., et al. (2022). Host lung microbiota promotes malaria-associated acute respiratory distress syndrome. Nat. Commun. 13:3747. 10.1038/s41467-022-31301-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkatti P., Pandey R., Rieder S. A., Hegde V. L., Nagarkatti M. (2009). Cannabinoids as novel anti-inflammatory drugs. Future Med. Chem. 1 1333–1349. 10.4155/fmc.09.93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik C., Kulkarni H., Darabi A., Bhanot N. (2013). Ochrobactrum anthropi: A rare cause of pneumonia. J. Infect. Chemother. 19 162–165. 10.1007/s10156-012-0436-1 [DOI] [PubMed] [Google Scholar]
- O’Dwyer D. N., Ashley S. L., Gurczynski S. J., Xia M., Wilke C., Falkowski N. R., et al. (2019). Lung microbiota contribute to pulmonary inflammation and disease progression in pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 199 1127–1138. 10.1164/rccm.201809-1650OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osafo N., Yeboah O. K., Antwi A. O. (2021). Endocannabinoid system and its modulation of brain, gut, joint and skin inflammation. Mol. Biol. Rep. 48 3665–3680. 10.1007/s11033-021-06366-1 [DOI] [PubMed] [Google Scholar]
- Pandey R., Mousawy K., Nagarkatti M., Nagarkatti P. (2009). Endocannabinoids and immune regulation. Pharmacol. Res. 60 85–92. 10.1016/j.phrs.2009.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzer A. R., Lynch S. V., Langelier C., Christie J. D., McCauley K., Nelson M., et al. (2018). Lung microbiota is related to smoking status and to development of acute respiratory distress syndrome in critically Ill trauma patients. Am. J. Respir. Crit. Care Med. 197 621–631. 10.1164/rccm.201702-0441OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecora D. V. (1963). A comparison of transtracheal aspiration with other methods of determining the bacterial flora of the lower respiratory tract. N. Engl. J. Med. 269 664–666. 10.1056/NEJM196309262691304 [DOI] [PubMed] [Google Scholar]
- Poroyko V., Meng F., Meliton A., Afonyushkin T., Ulanov A., Semenyuk E., et al. (2015). Alterations of lung microbiota in a mouse model of LPS-induced lung injury. Am. J. Physiol. Lung Cell Mol. Physiol. 309 L76–L83. 10.1152/ajplung.00061.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder S. A., Nagarkatti P., Nagarkatti M. (2012). Multiple anti-inflammatory pathways triggered by resveratrol lead to amelioration of staphylococcal enterotoxin B-induced lung injury. Br. J. Pharmacol. 167 1244–1258. 10.1111/j.1476-5381.2012.02063.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers G. B., Zain N. M., Bruce K. D., Burr L. D., Chen A. C., Rivett D. W., et al. (2014). A novel microbiota stratification system predicts future exacerbations in bronchiectasis. Ann. Am. Thorac. Soc. 11 496–503. 10.1513/AnnalsATS.201310-335OC [DOI] [PubMed] [Google Scholar]
- Roy M. G., Livraghi-Butrico A., Fletcher A. A., McElwee M. M., Evans S. E., Boerner R. M., et al. (2014). Muc5b is required for airway defence. Nature 505 412–416. 10.1038/nature12807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff W. E., Greiling T. M., Kriegel M. A. (2020). Host-microbiota interactions in immune-mediated diseases. Nat. Rev. Microbiol. 18 521–538. 10.1038/s41579-020-0367-2 [DOI] [PubMed] [Google Scholar]
- Salehi B., Mishra A. P., Nigam M., Sener B., Kilic M., Sharifi-Rad M., et al. (2018). Resveratrol: A double-edged sword in health benefits. Biomedicines 6:91. 10.3390/biomedicines6030091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapra L., Saini C., Das S., Mishra P. K., Singh A., Mridha A. R., et al. (2024). Lactobacillus rhamnosus (LR) ameliorates pulmonary and extrapulmonary acute respiratory distress syndrome (ARDS) via targeting neutrophils. Clin. Immunol. 258:109872. 10.1016/j.clim.2023.109872 [DOI] [PubMed] [Google Scholar]
- Schmitt F. C. F., Lipinski A., Hofer S., Uhle F., Nusshag C., Hackert T., et al. (2020). Pulmonary microbiome patterns correlate with the course of the disease in patients with sepsis-induced ARDS following major abdominal surgery. J. Hosp. Infect. 10.1016/j.jhin.2020.04.028 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Segal L. N., Clemente J. C., Tsay J. C., Koralov S. B., Keller B. C., Wu B. G., et al. (2016). Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol. 1:16031. 10.1038/nmicrobiol.2016.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Hu Y., Lv J., Zhao H., Wang B., Yang S., et al. (2022). Lung Microbiota Signature and Corticosteroid Responses in Pneumonia-Associated Acute Respiratory Distress Syndrome in Hematological Patients. J. Inflamm. Res. 15 1317–1329. 10.2147/JIR.S353662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Xiao Y., Kang L., Ma W., Shi L., Zhang L., et al. (2020). Genomic diversity of severe acute respiratory Syndrome-Coronavirus 2 in patients with Coronavirus Disease 2019. Clin. Infect. Dis. 71 713–720. 10.1093/cid/ciaa203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sido J. M., Nagarkatti P. S., Nagarkatti M. (2016). Production of endocannabinoids by activated T cells and B cells modulates inflammation associated with delayed-type hypersensitivity. Eur. J. Immunol. 46 1472–1479. 10.1002/eji.201546181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommariva M., Le Noci V., Bianchi F., Camelliti S., Balsari A., Tagliabue E., et al. (2020). The lung microbiota: Role in maintaining pulmonary immune homeostasis and its implications in cancer development and therapy. Cell Mol. Life Sci. 77 2739–2749. 10.1007/s00018-020-03452-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soroosh P., Doherty T. A., Duan W., Mehta A. K., Choi H., Adams Y. F., et al. (2013). Lung-resident tissue macrophages generate Foxp3+ regulatory T cells and promote airway tolerance. J. Exp. Med. 210 775–788. 10.1084/jem.20121849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman I., Chung M., Angel L., Tsay J. J., Wu B. G., Yeung S. T., et al. (2021). Microbial signatures in the lower airways of mechanically ventilated COVID-19 patients associated with poor clinical outcome. Nat. Microbiol. 6 1245–1258. 10.1038/s41564-021-00961-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan M., Wilson K., Abdulla O. A., Busbee P. B., Hall A., Carter T., et al. (2021). Endocannabinoid anandamide attenuates acute respiratory distress syndrome through modulation of microbiome in the gut-lung axis. Cells 10:3305. 10.3390/cells10123305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z., Wu E., You J., Ma G., Jiang S., Liu Y., et al. (2022). Dynamic alterations in the lung microbiota in a rat model of lipopolysaccharide-induced acute lung injury. Sci. Rep. 12:4791. 10.1038/s41598-022-08831-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C., Wang Z., Xiang E., Zhang Q., Zhang Y., Wu P., et al. (2022). Human umbilical cord mesenchymal stem cells promote macrophage PD-L1 expression and attenuate acute lung injury in mice. Curr. Stem Cell Res. Ther. 17 564–575. 10.2174/1574888X17666220127110332 [DOI] [PubMed] [Google Scholar]
- Walsh D. M., McCullough S. D., Yourstone S., Jones S. W., Cairns B. A., Jones C. D., et al. (2017). Alterations in airway microbiota in patients with PaO2/FiO2 ratio = 300 after burn and inhalation injury. PLoS One 12:e0173848. 10.1371/journal.pone.0173848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Huang C., Yang W., Wang C., Wang P., Guo L., et al. (2023). Respiratory microbiota and radiomics features in the stable COPD patients. Respir. Res. 24:131. 10.1186/s12931-023-02434-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Xing Y., Song X., Qian Y. (2020). The impact of lung microbiota dysbiosis on inflammation. Immunology 159 156–166. 10.1111/imm.13139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X., Gao J., Wang Z. (2022). The human lung microbiome-A hidden link between microbes and human health and diseases. Imeta 1:e33. 10.1002/imt2.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias M., Kashofer K., Wurm P., Regitnig P., Schütte M., Neger M., et al. (2022). Host and microbiome features of secondary infections in lethal covid-19. iScience 25:104926. 10.1016/j.isci.2022.104926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Liu B., Zheng W., Chen Y., Wu Z., Lu Y., et al. (2022). Pulmonary microbial composition in sepsis-induced acute respiratory distress syndrome. Front. Mol. Biosci. 9:862570. 10.3389/fmolb.2022.862570 [DOI] [PMC free article] [PubMed] [Google Scholar]