Abstract
Inverted repeats (IRs) and trinucleotide repeats (TNRs) that have the potential to form secondary structures in vivo are known to cause genome rearrangements. Expansions of TNRs in humans are associated with several neurological disorders. Both IRs and TNRs stimulate spontaneous unequal sister-chromatid exchange (SCE) in yeast. Secondary structure-associated SCE events occur via double-strand break repair. Here we show that the rate of spontaneous IR-stimulated unequal SCE events in yeast is significantly reduced in strains with mutations in the mismatch repair genes MSH2 or MSH3, but unaffected by a mutation in the nucleotide excision-repair gene RAD1. Non-IR-associated unequal SCE events are increased in both MMR- and rad1-mutant cells; however, SCE events for both IR- and non-IR-containing substrates occur at a higher level in the exo1 background. Our results suggest that spontaneous SCE occurs by a template switching mechanism. Like IRs, TNRs have been shown to generate double-strand breaks (DSBs) in yeast. TNR expansions in mice are MSH2-dependent. Since IR-mediated SCE events are reduced in msh2 cells, we propose that TNR expansion mutations arise when DSBs are repaired using the sister or the homolog as a template.
INTRODUCTION
Unusual DNA sequences or structures promote genetic instability. Inverted repeats (IRs) and trinculeotide repeats (TNRs) are examples of such sequence arrangements. IRs are frequently observed at the breakpoints of gross chromosomal rearrangements, such as deletions, translocations, inversions and large inverted duplications (1–4). In addition, IRs stimulate homologous recombination (5–7). Genomic regions containing TNRs, like IRs, are unstable; the repeated sequences undergo frequent changes in the number of repeats, resulting in either expansions or contractions of the repeat tract (8). While the expansion of TNR tracts is associated with several human genetic diseases (8,9), the mechanisms of TNR expansion are not clearly understood.
Both IRs and several TNRs have been shown to form secondary structures by complementary base pairing within single-stranded DNA (2,10). It is generally believed that formation of stem–loop (hairpin or cruciform) structures by intra-strand base pairing in single-stranded DNA is responsible for the genetic instability induced by IRs and TNRs. Such structures can form during normal DNA metabolism. For example, during DNA replication, single-stranded DNA that may be present on the lagging strand may facilitate hairpin formation. Hairpin formation may cause DNA polymerase slippage, resulting in DNA sequence alterations within and/or near the repeated sequences. The stem–loop structures are also substrates for structure-specific nucleases in vivo, as proposed for the SbcCD complex of Escherichia coli (11). Consistently, it has been shown that large IRs and long CAG-repeat tracts induce double-strand breaks (DSBs) in yeast (5,7,12,13).
The formation of secondary structures by TNRs and IRs also compromises DNA replication in vitro, and in vivo studies indicate that disease-associated TNRs attenuate the progression of the DNA replication fork (14–17). Stalling of replication at secondary structures may facilitate transient dissociation and reassociation of the replication fork. During reassociation, the nascent strand, instead of pairing with the parental template, may pair with the sister-chromatid or the homolog, resulting in a homologous recombination event. Alternatively, the secondary structure can be cleaved by structure-specific nucleases; the resulting lesion is repaired when replication restarts via recombination using the sister or the homolog as a template. Consistently, it has been shown that both IRs and TNRs stimulate spontaneous mitotic unequal sister-chromatid exchange (SCE) in yeast (18). IR-stimulated SCE events are dependent on genes that are required for DSB repair, suggesting that these SCE events occur via DSB repair. It has been proposed that DSBs are generated by cleavage of secondary structures that are formed in single-stranded DNA during DNA replication (18).
In the previous study (18), DSB repair involving sister chromatids can lead to both equal and unequal SCE (19). During DSB repair, the broken ends must be processed to generate single-stranded DNA containing a free 3′ end. The single-stranded tails then invade the intact homologous chromatid and prime DNA synthesis, using the invaded DNA as a template. Any non-homologous DNA present at the 3′ end must be removed for proper annealing and subsequent replication initiation. Proteins that remove non-homologous tails at the 3′ end include the mismatch repair (MMR) proteins Msh2 and Msh3, and the nucleotide excision-repair (NER) proteins Rad1 and Rad10 (20).
It is believed that Msh2/Msh3 binding to duplex DNA with 3′ non-homologous tails stabilizes the intermediate, and allows the Rad1/Rad10 endonuclease to cleave the 3′ tails. No other proteins in the MMR pathway or the NER pathway are necessary for removal of the non-homologous tails (20). During IR-associated SCE, if the break occurs within the IRs, then the unequal SCE events will be dependent on the Msh2/Msh3 and Rad1/Rad10 endonuclease activities to remove the non-homologous DNA from the 3′ tails. In this report we show that IR-stimulated SCE events are MSH2-dependent, but RAD1-independent. Our results suggest that most IR-associated genome rearrangements occur through a common step that is MSH2-dependent.
MATERIALS AND METHODS
Yeast strains and plasmids
Yeast strains (Table 1) were derived from AS13 (18) and YB163 (21). The construction of DNY380 and DNY393 are described previously (18). In DNY strains, the sister-chromatid recombination substrate was inserted within the ARG4 locus. YB163 strain has been described previously (21). The sister-chromatid recombination substrate in the YB163 strain was inserted at the TRP1 locus. All genetic manipulations were carried out following standard procedures. The msh2-Tn10LUK7-7-mutant allele was introduced into the chromosome using the plasmid pII-2::Tn10LUK7-7 (22). This plasmid was digested with SpeI before introducing into yeast to release the 9.6 kb fragment, which was separated by gel electrophoresis. The msh3, msh6 and mlh1 alleles were introduced into the chromosome, using pEAI88, pEAI108 and pEAI105 plasmids, respectively. pEAI88 was digested with EcoRI, pEAI108 with EcoRI and SphI, and pEAI105 with Asp718 and SphI, respectively, before transformation. The pEAI plasmids were kindly provided by Eric Alani (Cornell University, Ithaca, NY). The rad1 allele was introduced using plasmid pDG18, where the RAD1 coding sequence is replaced by the URA3 gene. The exo1-disruption (exo1::URA3) plasmid p245 was kindly provided by Kirill Lovachev (Georgia Institute of Technology, Atlanta, GA). The construction of p245 is described previously (23). p245 was digested with HindIII and Asp718 before transformation.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| AS13 | MATaleu2-Bst ura3 ade6 | (18) |
| DNY380 | AS13 lys2 arg4 his3Δ arg4::his3-SCScontrol | (18) |
| DNY383 | DNY380 rad1 | This study |
| DNY413 | DNY380 msh2 | This study |
| DNY420 | DNY380 msh6 | This study |
| DNY423 | DNY380 msh3 | This study |
| DNY429 | DNY380 pms1 | This study |
| DNY431 | DNY380 mlh1 | This study |
| DNY440 | DNY380 exo1 | This study |
| DNY393 | AS13 lys2 arg4 his3Δ arg4::his3-SCSpal140 | (18) |
| DNY407 | DNY393 rad1 | This study |
| DNY415 | DNY393 msh2 | This study |
| DNY418 | DNY393 msh3 | This study |
| DNY421 | DNY393 msh6 | This study |
| DNY430 | DNY393 pms1 | This study |
| DNY432 | DNY393 mlh1 | This study |
| DNY405 | DNY393 rad50 | This study |
| DNY406 | DNY393 mre11 | This study |
| DNY441 | DNY393 exo1 | This study |
| YB163 | MATa-inc ura3-52 his3-Δ200 ade2-101 lys2-801 trp1-Δ1 trp1::[his3-Δ3′::Hocs, his3-Δ5′] | (21) |
| YB221 | YB163 rad1::KanMX | This study |
| YB364 | YB163 msh2 | This study |
Genetic analysis of unequal SCE
Spontaneous SCE rates were determined by the method of median, as described in Nag et al. (18), except that the colonies were counted after 7 days of incubation at 30°C (Table 2). For each strain more than three independent rate calculations were performed, and the significance was determined by Student's t-test. HO-induced DSB-initiated SCE events were measured using a galactose-inducible HO gene. pGHOT-GAL3, containing the HO gene under GAL control (21), was introduced into wild-type, msh2 and rad1 mutants by selecting for Trp+ transformants. After growth in minimal media lacking tryptophan (CSM-TRP), cells were diluted in the ratio of 1:10 in YPLactate and incubated for a minimum of 12 h. At a density of 107 cells/ml, glucose or galactose was added to a final concentration of 2%, to either repress or induce the expression of HO endonuclease, respectively. HO endonuclease digests the 117 bp HO-cut site present at the BglII site in his3-Δ3′ (21). After 2 h, cells were plated directly on YPD medium for viability and on SC-HIS to measure recombination. Colonies appearing on YPD medium were replica plated on SC-TRP to measure the number of Trp+ colonies containing the pGHOT-GAL3 plasmid.
Table 2.
Rates of unequal sister-chromatid exchange in repair-deficient mutants
| his3 Substrate | Genotype | Rate of recom bination (× 106)a | Fold difference in rate, relative to wild-type |
|---|---|---|---|
| his3-SCScontrol | Wild-type | 0.72 ± 0.06 (5) | 1.0 |
| his3-SCScontrol | rad1 | 4.27 ± 0.84 (4) | 5.93 ↑ (P < 0.0001) |
| his3-SCScontrol | msh2 | 1.87 ± 0.33 (8) | 2.59 ↑ (P < 0.0001) |
| his3-SCScontrol | msh3 | 1.27 ± 0.28 (5) | 1.76 ↑ (P = 0.003) |
| his3-SCScontrol | msh6 | 0.71 ± 0.08 (4) | 1.0 |
| his3-SCScontrol | pms1 | 0.99 ± 0.17 (5) | 1.37 ↑ (P = 0.005) |
| his3-SCScontrol | mlh1 | 0.99 ± 0.11 (6) | 1.37 ↑ (P = 0.001) |
| his3-SCScontrol | exo1 | 2.80 ± 0.67 (6) | 3.88 ↑ (P < 0.0001) |
| his3-SCSpal140 | Wild-type | 6.66 ± 0.44 (4) | 1.0 |
| his3-SCSpal140 | rad1 | 9.13 ± 1.4 (4) | 1.37 ↑ (P = 0.015) |
| his3-SCSpal140 | msh2 | 4.18 ± 0.46 (8) | 0.62 ↓ (P < 0.0001) |
| his3-SCSpal140 | msh3 | 3.35 ± 0.25 (7) | 0.50 ↓ (P < 0.0001) |
| his3-SCSpal140 | msh6 | 6.63 ± 0.52 (4) | 1.0 |
| his3-SCSpal140 | pms1 | 6.61 ± 0.85 (4) | 1.0 |
| his3-SCSpal140 | mlh1 | 9.13 ± 2.04 (5) | 1.37 ↑ (P = 0.048) |
| his3-SCSpal140 | exo1 | 26.9 ± 5.93 (5) | 4.03 ↑ (P < 0.0001) |
aThe number in the parenthesis indicate the number of independent rate measurements for each strain.
RESULTS
IR-stimulated SCE events are RAD1-independent but MSH2-dependent
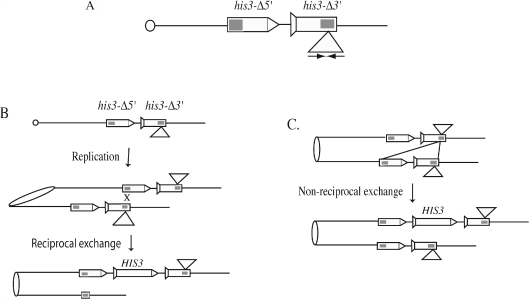
We monitor unequal SCE between two tandem his3 fragments; one fragment lacks the 5′ end (his3-Δ5′) and the other the 3′ end (his3-Δ3′), and there is shared 508 bp homology should between the two deletion constructs (Figure 1). An unequal SCE will generate a wild-type HIS3 gene, thereby allowing the cell to grow on minimal medium lacking histidine. To determine the effect of IRs on SCE, a 140 bp palindromic sequence was inserted within the region of homology in the his3-Δ3′ construct (18).
Figure 1.
Unequal SCE assay. (A) The his3 unequal SCE substrate. The his3-Δ3′ construct is marked with a tail, and the his3-Δ5′ construct is marked with an arrowhead. The shaded region indicates the region of homology between the two truncated his3 fragments. A wild-type HIS3 gene can form by unequal exchange (B) or by a non-reciprocal gene conversion event (C).
IR-stimulated SCE events occur by DSB repair. If DSBs occur within the secondary structures that are formed involving various extents of the IR, then the 3′ end, after DSB formation and exonucleolytic processing, will contain non-homologous tails that must be removed, if a wild-type gene is to be generated by unequal SCE. Since Rad1/Rad10 endonuclease, aided by the Msh2/Msh3 complex, removes the non-homologous ends, we expect that IR-stimulated SCE events that generate His+ cells will be Msh2- and Rad1-dependent. To determine the roles of Msh2 and Rad1 in unequal SCE, we introduced msh2- or rad1-mutations into our strains containing either a control SCE substrate (his3-SCScontrol) that has an insertion of non-repeated sequences containing the HO-cut site within the region of homology in the his3-Δ3′ construct, or the IR-containing substrate (his3-SCSpal140).
The rate of SCE for his3-SCScontrol in rad1 cells was 6-fold greater than the wild-type rate (P < 0.0001), while the rate for his3-SCSpal140 in the rad1 background was increased by only 1.4-fold (P = 0.015) (Table 2). In the msh2 background, the SCE rate for his3-SCScontrol was increased 2.6-fold relative to wild-type cells (P < 0.0001); however, the rate for his3-SCSpal140 was nearly 2-fold reduced (P < 0.0001). Our results suggest that there forms several recombinogenic lesion during the course of normal DNA metabolism that are repaired by Rad1. To generate IR-stimulated unequal SCE events, either there is a Rad1-redundant function that eliminates the non-homologous tails from the free 3′ end, or the generation and subsequent processing of DSBs at IRs occur by an as-yet unidentified mechanism.
Only Msh2/Msh3 of the MMR system is required for IR-stimulated SCE
Since IR-stimulated SCE events are MSH2-dependent and RAD1-independent, it is possible that DSBs at IRs are generated with the help of MMR proteins. Most of our knowledge on MMR comes from extensive studies in E.coli, which employs three dedicated proteins: MutS, MutL and MutH, in addition to a helicase, single-strand binding protein, four exonucleases, polymerase and ligase (24). MMR is initiated by the binding of MutS to the mismatch. MutL couples the mismatch recognition by MutS and downstream processing steps, including the strand-specific cleavage by MutH. Once the nick is made on the newly synthesized strand, single-strand exonucleases generates a gap, then replicative DNA polymerases fills the gap.
In eukaryotes, there are multiple MutS (MSH) and MutL (MLH) homologs. In Saccharomyces cerevisiae three MSH genes (MSH2, MSH3 and MSH6) and four MLH genes (MLH1-3 and PMS1) are required for nuclear DNA MMR (24). The mismatch recognition is accomplished by Msh2-Msh3 and Msh2-Msh6 heterodimers; the two complexes bind to distinct but overlapping spectra of mismatches. The Msh2/Msh6 complex exhibits strong selectivity for base-base and single insertion/deletion mismatches, and the Msh2/Msh3 complex preferentially recognizes small loops up to 12 nt in size.
Msh2/Msh3 may bind to the hairpin loop, and then generate a nick in the secondary structure with the help of an endonuclease that may or may not be part of MMR. To determine the role of MMR genes in IR-mediated SCE, we introduced mutations in MSH3, MSH6, MLH1 and PMS1 genes in our strains containing the SCE substrate (his3-SCScontrol or his3-SCSpal140) and then measured SCE rates in each strain.
Relative to SCE rates in wild-type cells, the SCE rate for the control substrate in msh3 was increased 1.8-fold (P = 0.003), whereas the rate for the IR-containing substrate in msh3 cells was reduced 2-fold (P < 0.0001) (Table 2). In msh6, however, the rates for both substrates remain nearly the same as the corresponding rates in the wild-type strain. In the pms1 and mlh1 backgrounds, the SCE rates for the his3-SCScontrol were significantly increased (P < 0.05), but not to the level observed in the msh2 background. The rate for his3-SCSpal140 remained unaltered in both mlh1 and pms1 backgrounds (Table 2). These results suggest that only the Msh2/Msh3 complex is required for IR-stimulated SCE events.
The exo1 mutation increases the rate of spontaneous SCE for both the control and the IR-containing substrate
Another component of the MMR pathway that is also required for DSB repair and recombination is the 5′ - 3′ exonuclease Exo1 [reviewed in ref. (25)]. Exo1 physically interacts with Msh and Mlh complexes and is believed to play a structural role in MMR (25–27). In addition, Exo1 is known to process stalled replication forks (28). Since Exo1 is involved in processes that modulate SCE and, since the rate of spontaneous SCE is significantly reduced in the msh2 background, we investigated whether the exo1 mutation would alter the rate of spontaneous SCE for the IR-containing substrate. At the same time, we determined the rate of spontaneous SCE for the control substrate in the exo1 background.
The rates of SCE for both the control substrate and IR-containing substrate were increased by 4-fold in the exo1 background compared to wild-type cells (Table 2). In our SCE assay, DSBs generated at the IR can also be repaired by single-strand annealing that requires excessive degradation by an exonuclease such as Exo1. It is likely that in the absence of Exo1, the DSBs that are normally repaired by single-strand annealing events are channeled into the SCE-repair pathway, resulting in an increase in the rate of SCE. Alternatively, the increase in SCE events may reflect the failure to restart the stalled replication fork in exo1 cells.
Unequal SCE events due to HO-induced DSBs are MSH2-independent
Since IR-stimulated SCE events are reduced in msh2 cells, we wished to determine whether non-IR-mediated DSB-induced SCE events are also MSH2-dependent. We tested whether HO-induced SCE events are MSH2-dependent. If Msh2 is required for repair but not for the generation of DSBs, then HO-induced DSB events would be MSH2-dependent. Alternatively, if Msh2 is required before DSB formation, then HO-generated SCE events will be MSH2-independent. Since the AS13 strain is Gal−, we measured frequencies of DSB-initiated SCE in YB163 and in congenic rad1 and msh2 strains, containing the plasmid (pGHOT-GAL3) with galactose-inducible HO. In the YB163 strain, the sister-chromatid recombination substrate, which is identical to the control substrate used in experiments described above and contains the HO-cut site within the his3-Δ3′ fragment, was inserted within the TRP1 locus. The rate of spontaneous SCE for the control substrate in both YB strains and DNY strains are similar (18,21).
In the wild-type background, unequal SCE frequency increased 12-fold after HO induction (Table 3). In rad1 and msh2 cells, the SCE frequencies were increased 12- and 14-fold, respectively, after HO induction, suggesting that DSB-initiated SCE that occurs after HO induction requires neither Msh2 nor Rad1; Msh2 is required for the IR-stimulated events at or before the DSB formation stage.
Table 3.
HO endonuclease-stimulated sister-chromatid exchange frequencies in wild-type, rad1 and msh2 mutants
| Genotype (strain)a | %Viability after HO inductionb | His+ recombinants/Trp+ CFU × 105 | Ratioe | % Trp+ CFU/total CFU | ||
|---|---|---|---|---|---|---|
| Before HO inductionc | After HO inductiond | Before HO induction | After HO induction | |||
| Wild type (YB163) | 78 ± 15 | 6.5 ± 3 | 79 ± 16 | 12 | 95 ± 0.7 | 83 ± 1.4 |
| msh2 | 41 ± 7 | 6.4 ± 5.4 | 91 ± 50 | 14 | 87 ± 6 | 71 ± 5 |
| rad1 | 27 ± 4 | 6.4 ± 2 | 76 ± 13 | 12 | 97 ± 2 | 81 ± 8 |
| Wild type (YB163) | 78 ± 15 | 6.5 ± 3 | 79 ± 16 | 12 | 95 ± 0.7 | 83 ± 1.4 |
aFor complete genotype, see Table 1.
bTrp+ CFU after HO induction/Trp+ CFU before HO induction × 100.
cHis+ recombinants before HO induction/Trp+ CFU before HO induction.
dHis+ recombinants after HO induction/Trp+ CFU after HO induction.
eHis+ frequency after HO induction/His+ frequency before HO induction.
DISCUSSION
Several outcomes are possible, when a replication fork has stalled at a secondary structure generated by an IR. First, replication stalling may facilitate slippage of the replication fork, resulting in a deletion, or else it may cause template switching, resulting in a homologous recombination event. The template switching or slippage of the replication fork may occur between sequences with microhomologies (e.g. 2–8 bp), when a large sequence homology is unavailable (2). Second, the replisome may disassemble at the pause site, allowing the newly synthesized strand to become the loading site for recombination proteins. Finally, a DSB may occur at the pause site due to cleavage of the secondary structure by a structure-specific nuclease. Several recent studies suggest that most IR-associated rearrangements are due to cleavage of secondary structures and subsequent repair of these DNA lesions (4,5,7,18). A DSB may also occur due to fork collapse. Consistently, it has been shown that the slowly replicating regions generate a high level of DSBs in certain yeast mutants (29).
In mammalian cells, genomic regions known as fragile sites often break when DNA replication is perturbed. Several chromosomal translocations have breakpoints within the fragile sites. In yeast, regions containing large IRs behave similarly to fragile sites in mammalian cells, causing an elevated level of chromosomal translocations via homologous recombination between repeated sequences (4). The results described here demonstrate that IR-stimulated unequal SCE is significantly reduced in msh2 and msh3 cells. None of the other MMR proteins, tested in our assay, is required for IR-associated SCE events. IR-stimulated ectopic recombination is also 2-to 4-fold reduced in the msh2 and msh3 backgrounds, but remains unaltered in msh6 cells (30). These results suggest that all IR-induced DSB-associated chromosomal rearrangements go through a common Msh2/Msh3-dependent step and that IR-associated translocation events are likely to be inefficient in the msh2 background.
Haber and colleagues studied the effect of the presence of non-homologous DNA ends on DSB repair and they showed that the repair efficiency is reduced, but not eliminated, in rad1 and msh2 backgrounds (31), suggesting that there is a MSH2- and RAD1-independent process that can remove non-homologous tails from the broken end. Our results are different from previously published observations in two ways: first, IR-stimulated events were reduced only in the msh2 background (Table 2), but not in rad1 cells. Second, HO-mediated DSBs also generate non-homologous ends that must be removed to generate His+ cells by unequal SCE. These events are both RAD1 and MSH2-independent (Table 3). These results suggest that a Rad1/Msh2-independent process removes HO-generated non-homologous ends, and that DSBs at IRs are processed or generated by a Msh2-dependent pathway.
The genetic requirements for spontaneous unequal SCE for his3-SCScontrol suggest that spontaneous SCE events with the his3 substrate do not occur via DSB repair (18,21). However, spontaneous SCE for his3-SCScontrol was significantly increased in all MMR-mutants tested, except msh6. It is possible that one mechanism of spontaneous SCE is template switching (Figure 2A). Although the his3 sequences in our system may not contain any strong replication-pause site that would generate DSBs, they may contain weak pause sites sufficient for template switching. Alternatively, spontaneous SCE events occur by single-strand gap repair. Generation of His+ cells by template switching or single-strand gap repair would require the formation of a large loop (Figure 2A), which must be preserved until the next round of replication. Such loops are normally repaired by NER and MMR proteins (32). The rate of spontaneous SCE was increased in rad1 and MMR-deficient backgrounds, suggesting the formation of a loop as an intermediate during unequal SCE. An increased rate of unequal SCE in msh2/msh3 cells, which is involved in loop repair, and in other MMR-deficient cells including exo1 (Table 2) supports this conclusion. Interestingly, SCE rates for both the control substrate and the IR-containing substrate were increased by 4-fold in the exo1 background (Table 2), suggesting that exo1 cells generate a high level of SCE-initiation events with both substrates. Exo1 is known to process stalled replication forks (28). It is possible that unprocessed fork leads to a higher level of template switching for the control substrate, and produces a higher level of DSBs at the IR-generated secondary structure.
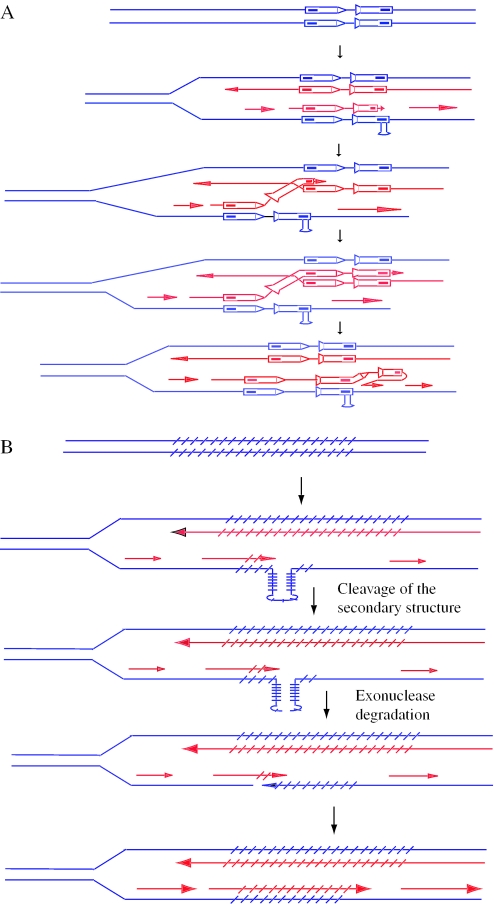
Figure 2.
Consequences of replication fork stalling at secondary structures. (A) Pausing at the replication fork can lead to template switching. Template switching can form a wild-type HIS3 gene. The large loop must be preserved from mismatch repair until the next round of DNA replication, or else be repaired in favor of loop retention, to generate a His+ cell. (B). A DSB formed due to nicking of the secondary structure within the triplet repeat sequence may be repaired by an intra-chromosomal single-strand annealing-like event, leading to contraction of the repeat tract. The hatch marks represent the repeated units.
Although rad1 increases the rate of SCE for his3-SCScontrol, the rate is much higher than that in the MMR mutants, suggesting that Rad1 deficiency in loop repair generates several recombinogenic intermediates that are resolved via sister-chromatid recombination, resulting in a higher level of SCE. Both IR- and HO-induced SCE events are Rad1-independent (Tables 2 and 3), suggesting that Rad1 is required neither for generation nor for processing of DSBs at IRs. The SCE rate for his3-SCSpal140 in the msh2/msh3 background was reduced by 2-fold. It is possible that IR-stimulated SCE events occur by template switching and DSB repair. Only the DSB repair events are MSH2-dependent. In the absence of DSBs at secondary structures, stalled replication forks are processed by channeling into the template switching pathway.
Like IRs, CAG repeats stimulate unequal SCE in yeast (18). While IRs increase SCE by 10- to 12-fold, the TNRs increased SCE by only 2-fold. One explanation for the observed difference between IR- and TNR-simulated SCEs is that DSBs formed within the TNR-generated hairpin structure are repaired by intra-molecular single-strand annealing events (Figure 2B). A minor proportion of DSBs are repaired via SCE. This model is consistent with the observations that most tract-length alterations in yeast are contractions, and that long CAG-repeat tracts also act as fragile sites in mitotic yeast cells (13). It has been proposed that expansions occur when replication-associated DSBs in TNRs are repaired using either the sister-chromatid or the homolog as a template (18).
Studies with human patients have indicated that TNR expansions arise via multiple DNA transactions during both meiosis and mitosis (33). The effect of MMR on TNR instability has been extensively studied in model systems (34–41). Expansion mutations in mice are Msh2-dependent, and only the Msh2/Msh3 complex is required for expansion (37–41). In vitro, Msh2 has been shown to bind hairpin structures containing TNRs (42). One proposed role for Msh2 is that it facilitates expansion by binding, and thus stabilizing the hairpin structure that may form during DNA damage repair (38). Since secondary structure-associated DSB repair events are decreased in the msh2 background [(30), and the data described above], it is possible that Msh2 binds at the secondary structure, generated by IRs and TNRs, that is formed during various DNA transactions, and then recruits an endonuclease to generate a nick in the stem–loop structure. Alternatively, Msh2 may recruit an exonuclease to the DSB site for processing of the broken ends.
In summary, we provide here the genetic evidence that the MMR proteins promote chromosomal rearrangements, and that a MSH2-dependent step exists in all IR-induced DSB-associated chromosomal rearrangements.
Acknowledgments
We thank Eric Alani and Kirill Lobachev for providing plasmids used in this study. We thank the reviewer for valuable comments on the manuscript. We also thank L. Zeng for assistance in HO-induced DSB analysis, Andrew Reilly for his help in statistical analysis, and Ashley Hart for her comments on the manuscript. The research in MF's laboratory was funded by NIH grant CA70105. Funding to pay the Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ford M., Fried M. Large inverted duplications are associated with gene amplification. Cell. 1986;45:4425–4430. doi: 10.1016/0092-8674(86)90328-4. [DOI] [PubMed] [Google Scholar]
- 2.Leach D.R.F. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- 3.Rattray A.J., Shafer B.K., Neelam B., Strathern J.N. A mechanism of palindrome gene amplification in Saccharomyces cerevisiae. Genes Dev. 2005;19:1390–1399. doi: 10.1101/gad.1315805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemoine F.J., Degtyareva N.P., Lobachev K., Petes T.D. Chromosomal translocations in yeast induced by low levels of DNA polymerase: a model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- 5.Nag D.K., Kurst A. A 140-base-pair long palindromic sequence induces double-strand breaks during meiosis in the yeast Saccharomyces cerevisiae. Genetics. 1997;146:835–847. doi: 10.1093/genetics/146.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farah J.A., Hartsuiker E., Mizuno K., Ohta K., Smith G.R. A 160 bp palindrome is Rad50.Rad32-dependent mitotic recombination hotspot in Schizosaccharomyces pombe. Genetics. 2002;161:461–468. doi: 10.1093/genetics/161.1.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lobachev K.S., Gordenin D.A., Resnick M.A. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 8.Nag D.K. Trinucleotide repeat expansions: timing is everything. Trends Mol. Med. 2003;9:455–457. doi: 10.1016/j.molmed.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 9.Cummings C.J., Zoghbi H.Y. Fourteen and counting: unraveling trinucleotide repeat diseases. Hum. Mol. Genet. 2000;9:909–916. doi: 10.1093/hmg/9.6.909. [DOI] [PubMed] [Google Scholar]
- 10.Sinden R.R., Potaman V.D., Oussatcheva E.A., Pearson C.E., Lyubchenko Y.L., Shlyakhtenko L.S. Triplet repeat DNA structures and human genetic disease: dynamic mutations from dynamic DNA. J. Biosci. 2002;27:53–65. doi: 10.1007/BF02703683. [DOI] [PubMed] [Google Scholar]
- 11.Connelly J.C., Kirkham L.A., Leach D.R.F. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl Acad. Sci. USA. 1998;95:7969–7974. doi: 10.1073/pnas.95.14.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jankowski C., Nasar F., Nag D.K. Meiotic instability of CAG repeat tracts occurs by double-strand break repair in yeast. Proc. Natl Acad. Sci. USA. 2000;97:2134–2139. doi: 10.1073/pnas.040460297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Callahan J.L., Andrews K.J., Zakian V.A., Freudenreich C.H. Mutations in yeast replication proteins that increase CAG/CTG expansions also increase repeat fragility. Mol. Cell. Biol. 2003;23:7849–7860. doi: 10.1128/MCB.23.21.7849-7860.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pelletier R., Krasilnikovam M.M., Samadashwily G.M., Lahue R., Mirkin S.M. Replication and expansion of trinucleotide repeats in yeast. Mol. Cell. Biol. 2003;23:1349–1357. doi: 10.1128/MCB.23.4.1349-1357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang S., Ohshima K., Shimizu M., Amirhaeri S., Wells R.D. Pausing of DNA synthesis in vitro at specific loci in CTG and CGG triplet repeats from human hereditary disease genes. J. Biol. Chem. 1995;270:27014–27021. doi: 10.1074/jbc.270.45.27014. [DOI] [PubMed] [Google Scholar]
- 16.Usdin K., Woodford K.J. CGG repeats associated with DNA instability and chromosome fragility from structures that block DNA synthesis in vitro. Nucleic Acids Res. 1995;23:4202–4209. doi: 10.1093/nar/23.20.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang C.C., Hearst J.E. Pauses at positions of secondary structure during in vitro replication of single-stranded fd bacteriophage DNA by T4 DNA polymerase. Anal. Biochem. 1980;103:127–139. doi: 10.1016/0003-2697(80)90246-8. [DOI] [PubMed] [Google Scholar]
- 18.Nag D.K., Suri M., Stenson E.K. Both CAG repeats and inverted DNA repeats stimulate spontaneous unequal sister-chromatid exchange in Saccharomyces cerevisiae. Nucleic Acids Res. 2004;32:5677–5684. doi: 10.1093/nar/gkh901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gonzalez-Barrera S., Cortes-Ledesma F., Wellinger R.E., Aguilera A. Equal sister chromatid exchange is a major mechanism of double-strand break repair in yeast. Mol. Cell. 2003;11:1661–1671. doi: 10.1016/s1097-2765(03)00183-7. [DOI] [PubMed] [Google Scholar]
- 20.Surtees J.A., Argueso J.L., Alani E. Mismatch repair proteins: key regulators of genetic recombination. Cytogenet. Genome Res. 2004;107:146–159. doi: 10.1159/000080593. [DOI] [PubMed] [Google Scholar]
- 21.Dong Z., Fasullo M. Multiple recombination pathways for sister chromtaid exchange in Saccharomyces cerevisiae: role of RAD1 and the RAD52 epistasis group genes. Nucleic Acids Res. 2003;31:2576–2585. doi: 10.1093/nar/gkg352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reenan R.A.G., Kolodner R.D. Isolation and characterization of two Saccharomyces cerevisiae genes encoding homologs of the bacterial HexA and MutS mismatch repair proteins. Genetics. 1992;132:963–973. doi: 10.1093/genetics/132.4.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tran H.T., Gordenin D., Resnick M.A. The 3′→5′ exonucleases of DNA polymerase δ and ε and the 5′→3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schofield M.J., Hsieh P. DNA mismatch repair: molecular mechanisms and biological function. Ann. Rev. Microbiol. 2003;57:579–608. doi: 10.1146/annurev.micro.57.030502.090847. [DOI] [PubMed] [Google Scholar]
- 25.Tran P.T., Erdeniz N., Symington L.S., Liskay R.M. EXO1-A multi-tasking eukaryotic nuclease. DNA Repair. 2004;3:1549–1559. doi: 10.1016/j.dnarep.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 26.Tishkoff D.X., Boerger A.L., Bertrans P., Filosi N., Gaida G.M., Kane M.F., Kolodner R.D. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an endonuclease that interacts with MSH2. Proc. Natl Acad. Sci. USA. 1997;94:7484–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tran P.T., Simon J.A., Liskay R.M. Interactions of Exo1p with components of MutLalpha in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2001;98:9760–9765. doi: 10.1073/pnas.161175998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cotta-Ramusino C., Fachinetti D., Lucca C., Doksani Y., Lopez M., Sogo J., Foiani M. ExoI processes stalled replication forks and counteracts fork reversal in checkpoint-defective cells. Mol. Cell. 2005;17:153–159. doi: 10.1016/j.molcel.2004.11.032. [DOI] [PubMed] [Google Scholar]
- 29.Cha R.S., Kleckner N. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science. 2002;297:602–606. doi: 10.1126/science.1071398. [DOI] [PubMed] [Google Scholar]
- 30.Lobachev K.S., Stenger J.E., Kozyreva O.G., Jurka J., Gordenin D.A., Resnick M.A. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 2000;19:3822–3830. doi: 10.1093/emboj/19.14.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colaiacovo M.P., Paques F., Haber J.E. Removal of one nonhomologous DNA end during gene conversion by a RAD1- and MSH2-independent pathway. Genetics. 1999;151:1409–1423. doi: 10.1093/genetics/151.4.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirkpatrick D.T., Petes T.D. Repair of DNA loops involves DNA-mismatch and nucleotide-excision repair proteins. Nature. 1997;387:929–931. doi: 10.1038/43225. [DOI] [PubMed] [Google Scholar]
- 33.Yoon S.-R., Dubeau L., de Young M., Wexler N.S., Arnheim N. Huntington disease expansion mutations in humans can occur before meiosis is completed. Proc. Natl Acad. Sci. USA. 2003;100:8834–8838. doi: 10.1073/pnas.1331390100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaworski A., Rosche W.A., Gellibolian R., Kang S., Shimizu M., Bowater R.P., Sinden R.R., Wells R.D. Mismatch repair in Escherichia coli enhances instability of (CTG)n triplet repeats from human hereditary diseases. Proc. Natl Acad. Sci. USA. 1995;92:11019–11023. doi: 10.1073/pnas.92.24.11019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schweitzer J.K., Livingston D.M. Destabilization of CAG trinucleotide repeat tracts by mismatch repair mutations in yeast. Hum. Mol. Genet. 1997;6:349–355. doi: 10.1093/hmg/6.3.349. [DOI] [PubMed] [Google Scholar]
- 36.Miret J.J., Pessoa-Brandao L., Lahue R.S. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1998;95:12438–12443. doi: 10.1073/pnas.95.21.12438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manley K., Shirley T.L., Flaherty L., Messer A. Msh2 deficiency prevents in vivo somatic instability of the CAG repeat in Huntington disease transgenic mice. Nature Genet. 1999;23:471–473. doi: 10.1038/70598. [DOI] [PubMed] [Google Scholar]
- 38.Kovtun I.V., McMurray C.T. Trinucleotide expansion in haploid germ cells by gap repair. Nature Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 39.van den Broek W.J.A.A., Nelen M.R., Wansink D.G., Coerwinkel M.M., te Riele H., Groenen P.J.T.A., Wieringa B. Somatic expansion behavior of the (CTG)n repeat in myotonic dystrophy knock-in mice is differentially affected by Msh3 and Msh6 mismatch-repair proteins. Hum. Mol. Genet. 2002;11:191–198. doi: 10.1093/hmg/11.2.191. [DOI] [PubMed] [Google Scholar]
- 40.Savouret C., Brisson E., Essers J., Kanaar R., Pastink A., te Riele H., Junien C., Gourdon G. CTG repeat instability and size variation timing in DNA repair-deficient mice. EMBO J. 2003;22:2264–2273. doi: 10.1093/emboj/cdg202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gomes-Pereira M., Fortune M.T., Ingram L., McAbney J.P., Monckton D.G. Pms2 is a genetic enhancer of trinucleotide CAG.CTG repeat somatic mosaicism: implications for the mechanism of triplet repeat expansion. Hum. Mol. Genet. 2004;13:1815–1825. doi: 10.1093/hmg/ddh186. [DOI] [PubMed] [Google Scholar]
- 42.Pearson C.E., Ewel A., Acharya S., Fishel R.A., Sinden R.R. Human Msh2 binds to trinucleotide repeat DNA structures associated with neurodegenerative diseases. Hum. Mol. Genet. 1997;6:1117–1123. doi: 10.1093/hmg/6.7.1117. [DOI] [PubMed] [Google Scholar]