Abstract
Hepatocytes are capable of repeated inducible NO synthase (iNOS) expression, which occurs under inflammatory and stress conditions. This iNOS expression regulates a number of cellular functions as well as cell viability. To better understand the posttranslational mechanisms that regulate the fate of iNOS in these cells, we characterized the iNOS distributed within peroxisomes. The selective permeabilization of membranes (plasma vs. peroxisomal) confirmed that there are cytosolic and peroxisomal pools of iNOS in cytokine-stimulated hepatocytes and that the iNOS protein associates with peroxisome. Detergent solubilization of the membrane fraction released iNOS to the soluble fraction. iNOS localized to membrane fraction is predominantly monomeric, but dimerization is partially reconstituted rapidly upon incubation with tetrahydrobiopterin. The reconstituted iNOS exhibits a lower specific activity than iNOS isolated from the soluble pool. Depletion of intracellular tetrahydrobiopterin with an inhibitor of de novo pterin synthesis resulted in a predominance of monomeric iNOS without a greater relative distribution of iNOS to the peroxisomal pool. Thus, iNOS exists in a least two pools in hepatocytes: a soluble pool composed of both active dimer and monomer and a peroxisomal pool of monomeric iNOS. iNOS might localize to peroxisomes in long-lived cells such as hepatocytes as a protective mechanism to remove incompetent enzyme.
Keywords: subcellular localization, inflammatory and stress conditions, hepatocytes, peroxisomes
Nitric oxide (NO) has critical signaling functions within cells but can also lead to cell dysfunction or toxicity. One mechanism for both regulating and localizing NO production is through distribution of NO synthase (NOS) to structures within the cells. For example, the association of endothelial NOS (eNOS) with caveolin localizes eNOS within structures in the plasma membrane known as caveolae and is a mechanism for regulating eNOS activity (1). The physical interaction of neuronal NOS (nNOS) with other proteins containing PDZ domains localizes this isoform to synaptic junctions in brain and motor endplates in skeletal muscle (2). Comparatively little is known about the posttranslational modifications that regulate inducible NOS (iNOS) distribution and function. In murine macrophages, iNOS has been shown to be both phosphorylated and ubiquitinated (3) and also associates with Rac1 and Rac2 as a mechanism to increase enzyme activity (4). We have previously shown that iNOS localizes to both the cytosol and peroxisomes in hepatocytes in vitro (5) and in vivo (5, 6); however, the structure and function of this cellular pool of iNOS is unknown.
Proteins that localize to peroxisomes function in hydrogen peroxide breakdown, lipid metabolism, and amine synthesis (7). Peroxisomes are single-membrane organelles of 0.2–1.0 μm in size that do not require protein unfolding or disruption of multimeric complexes before protein import (7). These organelles are particularly abundant in hepatocytes, constituting ≈1% of total cell volume. Furthermore, peroxisome numbers markedly increase in various stress conditions (8). Studies were undertaken to determine the structure and functional status of iNOS associated with peroxisomes. Here, we present evidence that only monomeric iNOS is associated with peroxisomes. Peroxisomal iNOS can be reactivated in vitro, but it has a lower specific activity than iNOS in the soluble pool. Thus, uncoupled or deficient iNOS may be targeted to the peroxisome.
Methods
Hepatocyte Isolation and Culture. Primary rat hepatocytes were isolated by collagenase perfusion (9). After overnight culture, fresh medium was added with or without test reagents as indicated (5). Cells were treated with cytokine mixture [CM = 100 units/ml IFN-γ (HyCult Biotechnology, Canton, MA), 200 units/ml IL-1 (Leinco Technologies, St. Louis), 500 units/ml TNFα; R&D Systems)] to induce iNOS. Primary culture monolayers were washed in cold PBS, and cells were harvested in lysis buffer (LB; 250 mM sucrose/10 mM Tris·HCl, pH 7.4/5 μg/ml pepstatin A/1 μg/ml chymostatin/5 μg/ml aprotinin/100 μM PMSF; Sigma). The cell lysates were disrupted by three cycles of freezing and thawing, followed by centrifugation at 12,000 × g for 15 min at 4°C to obtain a cleared lysate (CL) fraction. The CL fraction was further centrifuged at 100,000 × g for 30 min at 4°C to obtain the soluble (Sn) and membrane (Mn) fractions. The membrane fraction was resuspended in LB to a final protein concentration of 5 mg/ml. For detergent solubilization experiments, the membrane fraction (M1) was centrifuged as before and the pellet was resuspended in the solubilization buffer {SB; LB + 10 mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS)} to release proteins associated with membranes, and subsequently incubated for 30 min on ice. The samples were then centrifuged again at 100,000 × g to create the second soluble and membrane fractions (S2 and M2). In cultures where tetrahydrobiopterin (BH4) was minimized, cells were stimulated for iNOS in the presence and absence of 2,4-diamino-6-hydroxypyrimidine (DAHP; Sigma) (5 mM). Total biopterins in cell lysates were determined as described in ref. 10. The samples were processed for protein determination [bicinchoninic acid (BCA); Pierce], NOS activity, and immunoblots.
Immunoblots. Proteins were separated on SDS/7% or 10% PAGE in denaturing or nondenaturing conditions and transferred to nitrocellulose membranes. The membranes were hybridized with antibodies for iNOS (1:1,000; BD Transduction Laboratories), peroxisomal membrane protein 70 (PMP70) (1:2,000; Alexis, San Diego), and β-actin (1:10,000; Chemicon) in Tris-buffered saline, pH 7.6, plus 0.01% Tween-20 (TBST) and 1% nonfat milk for 1 h. The immunoblots were developed with WestPico chemiluminescence solution (Pierce) and exposed to x-ray film (Kodak).
Enzyme Assays. iNOS enzyme activity was assayed in 20 mM Tris·HCl, pH 7.4/4 μM flavin adenine dinucleotide/4 μM BH4/3 mM DTT/2 mM l-arginine/2 mM NADPH. Samples were incubated at 37°C for 1 hr and subsequently quenched by preventing NADPH reoxidation. Accumulated NO–2 was measured by the Griess reaction (11).
Immunofluorescence Microscopy. Primary rat hepatocytes were seeded onto collagen-crosslinked 12-mm coverslips and treated with reagents as described in ref. 5. Selective permeabilization of the plasma vs. peroxisomal membranes was accomplished according to Dammai and Subramani (12). After permeabilization, cells were processed for immunofluorescent labeling as described (5) and viewed by using a Fluoview 1000 scanning confocal microscope (Olympus, Melville, NY).
Results
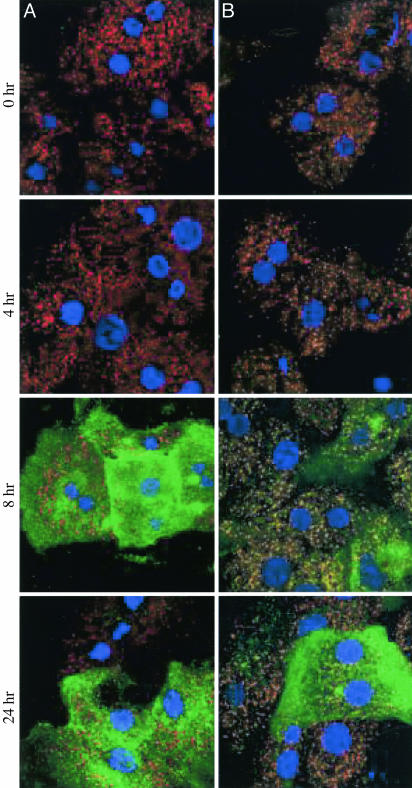
We have previously shown that iNOS partially localizes to peroxisomes in hepatocytes in vivo (5, 6) and in vitro (5). This localization was shown to be most prominent at 8 h after iNOS induction with cytokines. To further characterize how iNOS associates with peroxisomes, selective permeabilization of cytokine-treated cultured hepatocytes was performed. PMP70 was used as a peroxisomal surface marker because it labels only the periphery of the peroxisome. Cytokine-stimulated hepatocytes permeabilized with digitonin (a detergent that permeabilizes only the plasma membrane) were immunopositive for cytosolic iNOS, which did not colocalize with PMP70 (Fig. 1) (13). When cytokine-stimulated hepatocytes were permeabilized with Triton X-100 (a detergent that affects both plasma and peroxisomal membranes), we found that iNOS-positive and PMP70-positive punctate structures colocalize, indicating that iNOS was embedded within the peroxisomal membrane or, more likely, the peroxisome matrix.
Fig. 1.
Scanning laser confocal immunofluorescent detection of the cytosolic iNOS in hepatocyte cultures. Primary rat hepatocytes stimulated with cytokines were permeabilized with digitonin (25 μg/ml) (A) or Triton X-100 (0.1%) (B). Immunofluorescence was performed with antibodies against the PMP70 and iNOS proteins. Green, iNOS; red, PMP70; yellow, colocalized; blue, nucleus.
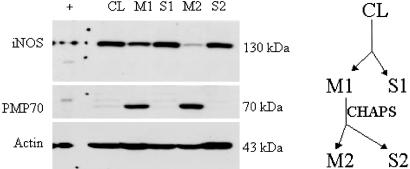
To characterize the structure and activity of peroxisomal iNOS, soluble and membrane fractions were isolated from cytokine-stimulated hepatocytes. Our results here and our previous analysis show all of the membrane-associated pool with iNOS to be within peroxisomes, so we equate membrane iNOS with peroxisomal enzyme. Shown in Fig. 1 is the distribution of iNOS between cytosolic and peroxisomal pools over time after cytokine treatment. As expected, iNOS protein was absent before stimulation, becomes easily detected by 4 h, and remains elevated beyond 24 h. At all time points, iNOS could be recovered from both soluble and membrane fractions (data not shown). Treatment of the membrane fraction with lysis buffer plus 10 mM 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate for 30 min resulted in release of iNOS into the soluble pool (Fig. 2). This iNOS was then easily recovered in the second S2 fraction and thus readily released from peroxisomes by membrane solubilization. This release of iNOS is in contrast to PMP70, a protein known to be an integral membrane protein in peroxisomes (14), showing that iNOS associates more loosely than this integral protein.
Fig. 2.
iNOS immunoblot of cytokine-stimulated hepatocytes. Membrane fraction (M1) solubilized with 10 mM of 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS) releases iNOS from the M1 to the soluble fraction (S2). There is no iNOS remaining in the membrane fraction (M2) of the solubilized sample. +, iNOS positive control; CL, cleared lysate, supernatant of 12,000 × g; Sn, supernatant of 100,000 × g; Mn, pellet of 100,000 × g.
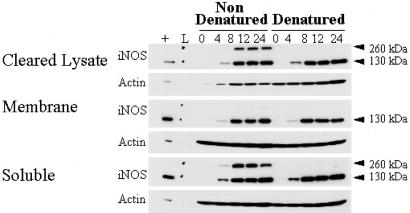
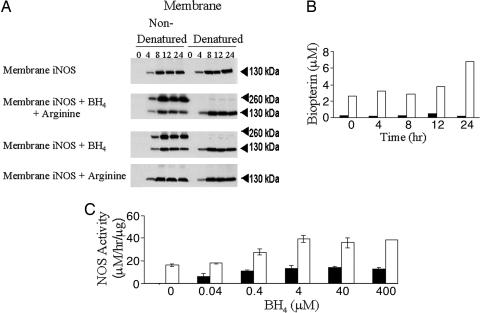
iNOS may exist in cells either as inactive monomer or as active homodimers. To define the structural status of peroxisomal iNOS, we carried out low-temperature, partially denaturing, 7% SDS/PAGE. This procedure preserves the dimeric structure of iNOS while separating the proteins by molecular mass. Fig. 3 shows that under nondenaturing conditions there was a similar distribution of dimeric (260-kDa) to monomeric enzyme in the total cell lysates over a time course of 4–24 h after cytokine stimulation. As expected, iNOS dimer was nearly absent when the samples were denatured by boiling. The dimeric iNOS band was preserved in the soluble fraction but absent in the membrane fraction, where only monomer could be detected. Thus, iNOS localized to peroxisomes is monomeric. The relative amounts of iNOS appearing as monomer appeared to be highest at the earliest time point, suggesting that dimerization may become more efficient as iNOS concentrations increase or essential cofactors are coproduced.
Fig. 3.
iNOS dimer assembly in cytokine-induced hepatocytes. At each of the four time points (h), dimeric iNOS protein (nondenatured) was visualized by immunoblot at ≈260 kDa. These samples exhibited comparable monomeric (denatured) protein based on an iNOS-positive band migrating at 130 kDa. L, molecular weight ladder; +, iNOS positive control.
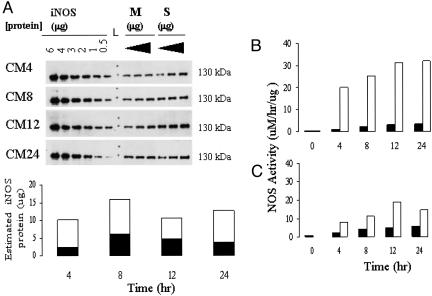
To estimate specific activity, iNOS protein content in the soluble and membrane fraction was assessed. iNOS content was measured by using immunoblots of membrane and soluble fraction at multiple total protein concentrations at each time point and compared with known amount of iNOS purified protein (Fig. 4A). Calculation of iNOS activity (measured with BH4 and arginine present) related to either total protein (Fig. 4B) or the estimated amount of iNOS protein (Fig. 4C) showed markedly lower levels of specific activity in the iNOS isolated from peroxisomes. This low level of NOS activity suggests that the membrane-associated iNOS is not structurally replete.
Fig. 4.
Quantitation of iNOS protein content. (A) Membrane (M) and soluble (S) iNOS protein was measured in comparison with a range of iNOS protein concentrations over the established cytokine time course. CMn, cytokine mixture exposure, in hours. (B and C) Comparable total protein (B) and comparable iNOS protein (C) NOS activity assays. Black bars, M; white bars, L, molecular weight ladder.
We next sought to determine whether the peroxisomal iNOS could form dimers. Both BH4 and arginine have been shown to participate in NOS dimerization (15, 16). To determine the requirements for dimerization of peroxisomal iNOS, the membrane fraction was incubated with BH4 or arginine alone or in combination. As shown in Fig. 5A, BH4 alone was adequate to induce dimerization. Concentrations of BH4 as low as 40 nM were sufficient to induce dimerization of peroxisomal iNOS from the membrane fraction (Fig. 5B). Measurement of total biopterin content revealed the presence of biopterin only in the soluble fraction and not in the membrane fraction (Fig. 5C). Thus, the lack of BH4 may limit iNOS dimerization in the peroxisome. NOS enzyme activity was measured in the membrane and soluble fractions in the absence and presence of BH4. As shown in Fig. 5B, no activity was recovered in the membrane fraction until BH4 was added. Activity was easily recovered in the soluble fraction even in the absence of exogenous BH4, and there was a further increase in activity when BH4 was included in the reaction mixture.
Fig. 5.
iNOS protein expression was monitored by low-temperature, partially denaturing, 7% SDS/PAGE. (A) Membrane protein (100 μg) incubated with BH4 (4 μM) or BH4 and arginine (2 μM) was immunopositive for dimeric iNOS protein. (B) Reconstitution of NOS activity and dimerization in membrane and soluble fractions upon the addition of BH4 (40 nM to 400 μM). (C) Biopterin measurement of the membrane and soluble fractions of cytokine-stimulated hepatocytes. Black bars, membrane; white bars, soluble.
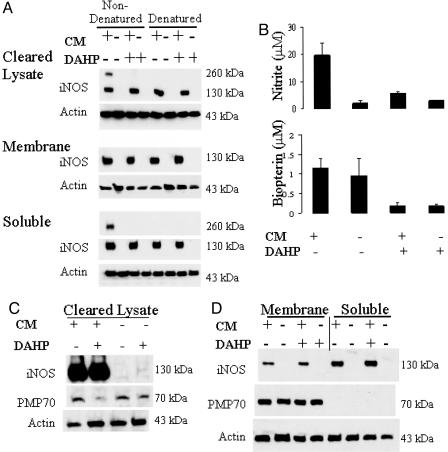
To determine whether levels of monomer iNOS were a determinant of iNOS accumulation in peroxisomes, cells were incubated in the presence of DAHP, a GTP cyclohydrolase I inhibitor, to block BH4 synthesis and dimer formation. DAHP blocked de novo BH4 synthesis leading to only monomer iNOS in all fractions of cytokine-treated hepatocytes (Fig. 6A). As expected, DAHP treatment inhibited nitrite formation and reduced biopterin content (Fig. 6B). Total iNOS content was not influenced by DAHP treatment (Fig. 6C), and the abundance of iNOS distributed to the membrane fraction was unaltered (Fig. 6D). Similar findings were observed when complete monomerization of iNOS was achieved by using a mechanistic inhibitor that suppresses dimerization (data not shown) (17). Thus iNOS monomerization does not force a greater distribution of the enzyme to peroxisomes.
Fig. 6.
Increased iNOS monomer decreases peroxisomes. (A) Diminishing the generation of BH4 with DAHP made cleared lysate and soluble iNOS monomeric. CM, cytokine mixture exposure. (B) These DAHP-treated samples also had diminished biopterin (Lower) and consequently diminished nitrite production (Upper). (C) Cleared lysate fractions have reduced PMP70 levels while maintaining comparable iNOS and actin protein levels. (D) Membrane and soluble fractions. iNOS, PMP70, and actin protein levels are comparable among the treatment groups when comparing similar subcellular fractions. +, CM present; –, noCM.
Discussion
Peroxisomes, like mitochondria, consume molecular oxygen and metabolize fatty acids, cleaving two carbon atoms per cycle to generate acetyl-CoA in a process called β-oxidation. Unlike mitochondrial β-oxidation, peroxisomal β-oxidation is not coupled to ATP synthesis/energy production. The contents of peroxisomes are metabolic and oxidative enzymes that are selectively targeted for import from the cytosol posttranslationally (18). The content and function of the enzymes within the peroxisome vary by cell type. In environments with competing metabolic needs, the immediate physiological needs of the cellular environment dictate the state of the peroxisome population and peroxisomal protein content. Catalase, the predominant peroxisomal enzyme, detoxifies the cellular environment by converting H2O2 into H2O when combined with other organic compounds (phenols, aldehydes, and alcohols). Here we add to the list of peroxisomal enzymes iNOS, which, when produced as a result of stress in hepatocytes, accumulates in peroxisomes in the inactive monomeric form.
Fully functional, dimeric iNOS produces NO in a sustained manner. However, in the absence of adequate substrate, or when in its monomeric form, iNOS can generate 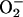 (19–21). Thus, iNOS has the potential to produce damaging oxygen radicals. NO that is produced from dimerized iNOS can combine with
(19–21). Thus, iNOS has the potential to produce damaging oxygen radicals. NO that is produced from dimerized iNOS can combine with 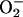 to form the oxidant peroxynitrite (ONOO–), or in the presence of adequate antioxidants can facilitate removal of
to form the oxidant peroxynitrite (ONOO–), or in the presence of adequate antioxidants can facilitate removal of 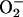 (22). iNOS may function within the heme protein-rich environment of peroxisomes to maximize heme sulfhydryl modification and lipid peroxidation (23). Therefore, there are several possible explanations for the transport of iNOS into peroxisomes. First, uncoupled iNOS might be sequestered by peroxisomes to protect the larger cellular environment from monomeric iNOS-generated superoxide. Second, the peroxisome could represent a pool of enzyme interchangeable with the soluble form. Third, defective iNOS may be disposed through the eventual destruction of the peroxisome.
(22). iNOS may function within the heme protein-rich environment of peroxisomes to maximize heme sulfhydryl modification and lipid peroxidation (23). Therefore, there are several possible explanations for the transport of iNOS into peroxisomes. First, uncoupled iNOS might be sequestered by peroxisomes to protect the larger cellular environment from monomeric iNOS-generated superoxide. Second, the peroxisome could represent a pool of enzyme interchangeable with the soluble form. Third, defective iNOS may be disposed through the eventual destruction of the peroxisome.
Our findings that the iNOS localized to peroxisomes has less specific activity than soluble iNOS suggests that incompetent or modified enzyme may be targeted to the peroxisome. This finding, combined with our observation that significant levels of soluble monomeric iNOS are available for dimerization, argues against the peroxisome as a reservoir of iNOS monomers. Furthermore, there is no precedence for peroxisomes serving as a reservoir for a soluble enzyme.
Dimerization of heme-containing iNOS monomers can be enhanced by the presence of BH4 by hydrophilic interactions (15, 16, 24, 25). In hepatocytes, BH4 is expressed constitutively at levels sufficient for iNOS activity (10, 26). Consistent with this finding, we show here that soluble iNOS was minimally enhanced by the addition of exogenous BH4. In contrast, peroxisomal iNOS exhibited almost no activity in the absence of BH4. Furthermore, biopterin levels were negligible in the membrane fraction and some dimerization took place on the addition of exogenous BH4 to enzyme isolated from the membrane fraction. These data suggest that defective iNOS could be transferred to the low biopterin environment of the peroxisome to force or maintain inactivation.
It is important to note that monomerization alone is not sufficient to promote iNOS transfer into the peroxisome. We show that there is a stable pool of soluble monomeric iNOS, and complete monomerization did not increase peroxisomal content of iNOS. This sequestration is consistent with our hypothesis that a select pool or subset of iNOS is transferred to the peroxisome. It is also possible that the peroxisomal pool did not further increase by forcing monomerization because the uptake mechanism was saturated.
Peroxisomal membrane and matrix proteins are imported posttranslationally by either a peroxisomal targeting sequence (PTS) or protein–protein associations (e.g., HSP70) (7, 27). Import is conserved evolutionarily, as demonstrated by transfection experiments, wherein proteins containing PTS1 [variations of (Ser/Ala/Cys), (Lys/His/Arg), and (Leu/Met)] (28) from one species are imported into peroxisomes of cells from another species (28). The second PTS (PTS2) is a nine amino acid bipartite [sequence of some variation of (Arg/Lys), (Xaa)5, (His/Gln), and (Leu/Ala)] that may function at varying distances from the amino terminus (29). Import into the peroxisome has also been found to occur in the absence of an established PTS (30). How iNOS is localized to the peroxisome is unclear. Whereas iNOS contains a variation of the known PTSs (1 and 2, PTS1 and PTS2), it is unknown whether these sequences are functional (5). The mechanism for iNOS peroxisomal localization remains to be determined.
Although iNOS localization to vesicles has been shown in macrophages (3), peroxisomal localization in mammalian cells other than hepatocytes has not been reported. Plant NOS has been reported to be localized to the peroxisome, and the subsequent generation of nitrogen radicals has been shown to lead a signal cascade that promotes growth, development, and innate immunity in plants (31, 32). The combination of NO generated by plant-expressed NOS with peroxisomal generated oxygen radicals may easily generate peroxynitrite or S-nitrosoglutathione, both of which may easily diffuse into to cytosol. Within the peroxisome, the combination of nitrogen and oxygen radical suppresses the endogenous antioxidant catalase and promotes overproduction of H2O2 by enhanced fatty acid β-oxidation. Our results, showing that hepatocyte iNOS localized to the peroxisome is not competent to produce NO, point to potential functional differences between mammalian and plant peroxisomal NOS. Hepatocytes are long-lived parenchymal cells and are capable of repeated iNOS expression. As such, hepatocytes may have adopted mechanisms to use peroxisomes in enzyme processing to maximize function and limit toxicity. This view does not exclude a role for iNOS in the regulation of peroxisomal function.
Acknowledgments
We thank Debra Williams, Qui She, Angela Green, Yun Sheng, Toshie Yoneyama, Elizabeth Humphris, and Carol Meiers (Department of Surgery, University of Pittsburgh School of Medicine) and Sean Alber, Mark Ross, Jason Devlin, and Stuart Shand (Center for Biologic Imaging, University of Pittsburgh School of Medicine) for their excellent technical help. Dr. Suresh Subramani (University of California, San Diego) was tremendously generous with his time and knowledge. This work was supported by National Institutes of Health General Medicine Grants GM44100 (to T.R.B.) and GM37753 (to R.L.S.) and National Cancer Institute Grant CA76541 (to D.B.S.).
Author contributions: P.A.L., Y.V., R.L.S., and T.R.B. designed research; P.A.L. and D.B.S. performed research; D.B.S. and S.C.W. contributed new reagents/analytic tools; P.A.L. analyzed data; P.A.L. and T.R.B. wrote the paper; and Y.V. provided comments.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NOS, NO synthase; iNOS, inducible NO synthase; BH4, tetrahydrobiopterin; DAHP, 2,4-diamino-6-hydroxypyrimidine; PMP70, peroxisomal membrane protein 70; PTS, peroxisomal targeting sequence; Sn, soluble fractions; Mn, membrane fractions.
References
- 1.Ju, H., Zou, R., Venema, V. J. & Venema, R. C. (1997) J. Biol. Chem. 272, 18522–18525. [DOI] [PubMed] [Google Scholar]
- 2.Brenman, J. E., Chao, D. S., Xia, H., Aldape, K. & Bredt, D. S. (1995) Cell 82, 743–752. [DOI] [PubMed] [Google Scholar]
- 3.Vodovotz, Y., Russell, D., Xie, Q.-W., Bogdan, C. & Nathan, C. (1995) J. Immunol. 154, 2914–2925. [PubMed] [Google Scholar]
- 4.Kuncewicz, T., Balakrishnan, P., Snuggs, M. B. & Kone, B. C. (2001) Am. J. Physiol. 281, F326–F336. [DOI] [PubMed] [Google Scholar]
- 5.Stolz, D. B., Zamora, R., Vodovotz, Y., Loughran, P. A., Billiar, T. R., Kim, Y. M., Simmons, R. L. & Watkins, S. C. (2002) Hepatology 36, 81–93. [DOI] [PubMed] [Google Scholar]
- 6.Collins, J. L., Vodovotz, Y., Hierholzer, C., Villavicencio, R. T., Liu, S., Alber, S., Gallo, D., Stolz, D. B., Watkins, S. C., Godfrey, A., et al. (2003) Shock 19, 117–122. [DOI] [PubMed] [Google Scholar]
- 7.Subramani, S. (1993) Annu. Rev. Cell Biol. 9, 445–478. [DOI] [PubMed] [Google Scholar]
- 8.Purdue, P. E. & Lazarow, P. B. (2001) Annu. Rev. Cell Dev. Biol. 17, 701–752. [DOI] [PubMed] [Google Scholar]
- 9.Seglen, P. O. (1976) Methods Cell Biol. 13, 29–83. [DOI] [PubMed] [Google Scholar]
- 10.Geller, D. A., Di Silvio, M., Billiar, T. R. & Hatakeyama, K. (2000) Biochem. Biophys. Res. Commun. 276, 633–641. [DOI] [PubMed] [Google Scholar]
- 11.Vodovotz, Y., Kwon, N.-S., Popischil, M., Manning, J., Paik, J. & Nathan, C. (1994) J. Immunol. 152, 4110–4118. [PubMed] [Google Scholar]
- 12.Dammai, V. & Subramani, S. (2001) Cell 105, 187–196. [DOI] [PubMed] [Google Scholar]
- 13.Wendland, M. & Subramani, S. (1993) J. Cell Biol. 120, 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kamijo K, Taketani, S., Yokata, S., Osumi, T. & Hashimoto, T. (1990) J. Biol. Chem. 265, 4534–4540. [PubMed] [Google Scholar]
- 15.Ghosh, D. K., Wu, C., Pitters, E., Moloney, M., Werner, E. R., Mayer, B. & Stuehr, D. J. (1997) Biochemistry 36, 10609–10619. [DOI] [PubMed] [Google Scholar]
- 16.Crane, B. R., Arvai, A. S., Gachhui, R., Wu, C., Ghosh, D. K., Getzoff, E. D., Stuehr, D. J. & Tainer, J. A. (1997) Science 278, 425–431. [DOI] [PubMed] [Google Scholar]
- 17.Blasko, E., Glaser, C. B., Devlin, J. J., Xia, W., Feldman, R. I., Polokoff, M. A., Phillips, G. B., Whitlow, M., Auld, D. S., McMillan, K., et al. (2002) J. Biol. Chem. 277, 295–302. [DOI] [PubMed] [Google Scholar]
- 18.Fujiki, Y. & Lazarow, P. B. (1985) J. Biol. Chem. 260, 5603–5609. [PubMed] [Google Scholar]
- 19.Xia, Y. & Zweier, J. L. (1997) Proc. Natl. Acad. Sci. USA 94, 6954–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pou, S., Keaton, L., Surichamorn, W. & Rosen, G. M. (1999) J. Biol. Chem. 274, 9573–9580. [DOI] [PubMed] [Google Scholar]
- 21.Stuehr, D., Pou, S. & Rosen, G. M. (2001) J. Biol. Chem. 276, 14533–14536. [DOI] [PubMed] [Google Scholar]
- 22.Thomas, D. D., Espey, M. G., Vitek, M. P., Miranda, K. M. & Wink, D. A. (2002) Proc. Natl. Acad. Sci. USA 99, 12691–12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beckman, J. S., Beckman, T. W., Chen, J., Marshall, P. A. & Freeman, B. A. (1990) Proc. Natl. Acad. Sci. USA 87, 1620–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ghosh, D. K., Crane, B. R., Ghosh, S., Wolan, D., Gachhui, R., Crooks, C., Presta, A., Tainer, J. A., Getzoff, E. D. & Stuehr, D. J. (1999) EMBO J. 18, 6260–6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, H., Raman, C. S., Glaser, C. B., Blasko, E., Young, T. A., Parkinson, J. F., Whitlow, M. & Poulos, T. L. (1999) J. Biol. Chem. 274, 21276–21284. [DOI] [PubMed] [Google Scholar]
- 26.Pastor, C. M., Williams, D., Yoneyama, T., Hatakeyama, K., Singleton, S., Naylor, E. & Billiar, T. R. (1996) J. Biol. Chem. 271, 24534–24538. [DOI] [PubMed] [Google Scholar]
- 27.Walton, P. A., Wendland, M., Subramani, S., Rachubinski, R. A. & Welch, W. J. (1994) J. Cell Biol. 125, 1037–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rachubinski, R. A. & Subramani, S. (1995) Cell 83, 525–528. [DOI] [PubMed] [Google Scholar]
- 29.Tsukamoto, T., Hata, S., Yokota, S., Miura, S., Fujiki, Y., Hijikata, M., Miyazawa, S., Hashimoto, T. & Osumi, T. (1994) J. Biol. Chem. 269, 6001–6010. [PubMed] [Google Scholar]
- 30.McNew, J. A. & Goodman, J. M. (1994) J. Cell Biol. 127, 1245–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barroso, J. B., Corpas, F. J., Carreras, A., Sandalio, L. M., Valderrama, R., Palma, J. M., Lupianez, J. A. & del Rio, L. A. (1999) J. Biol. Chem. 274, 36729–36733. [DOI] [PubMed] [Google Scholar]
- 32.Zeidler, D., Zahringer, U., Gerber, I., Dubery, I., Hartung, T., Bors, W., Hutzler, P. & Durner, J. (2004) Proc. Natl. Acad. Sci. USA 101, 15811–15816. [DOI] [PMC free article] [PubMed] [Google Scholar]