Abstract
The neuronal PAS domain protein 3 (NPAS3) gene encoding a brain-enriched transcription factor was recently found to be disrupted in a family suffering from schizophrenia. Mice harboring compound disruptions in the NPAS3 and related NPAS1 genes manifest behavioral and neuroanatomical abnormalities reminiscent of schizophrenia. Herein we demonstrate that Npas3-/- mice are deficient in expression of hippocampal FGF receptor subtype 1 mRNA, most notably in the dentate gyrus. In vivo BrdUrd-labeling shows that basal neural precursor cell proliferation in the dentate gyrus of Npas3-/- mice is reduced by 84% relative to wild-type littermates. We propose that a deficiency in adult neurogenesis may cause the behavioral and neuroanatomical abnormalities seen in Npas3-/- mice, and we speculate that impaired neurogenesis may be involved in the pathophysiology of schizophrenia.
Keywords: fibroblast growth factor, hippocampus, schizophrenia, Sprouty4, electroconvulsive seizure
Neuronal PAS domain protein 3 (NPAS3) is a brain-enriched basic helix-loop-helix (bHLH) PAS domain transcription factor that was recently found to be disrupted by genetic translocation in a family with schizophrenia (1). The translocation was present in the affected mother and daughter, absent in the unaffected father, and predicted to generate a truncated NPAS3 polypeptide retaining DNA binding activity but lacking both PAS domains and all functional components C-terminal to the PAS domains.
The human and mouse NPAS3 transcription factors are 90% identical and likely serve related functions in the central nervous system (2). The genomes of mice and humans also encode the highly related NPAS1 transcription factor (3). Both NPAS1 and NPAS3 appear to be orthologous to the Trachealess protein of Drosophila melanogaster. NPAS1 and NPAS3 share, respectively, 45% and 42% amino acid sequence identity with Trachealess, heterodimerize with orthologous partner transcription factors [aryl hydrocarbon receptor nuclear translocator (ARNT) in mammals = Tango in D. melanogaster], and are indistinguishable when tested by a variety of biochemical assays.
We recently demonstrated that mice having either the double-null (Npas1-/- Npas3-/-) or NPAS3-null over NPAS1-heterozygous (Npas1+/- Npas3-/-) genotype exhibit behavioral and neuroanatomical abnormalities related to human schizophrenia. These deficits include impaired social recognition, increased open-field locomotor activity, stereotypic darting behavior, reduced prepulse inhibition, and decreased brain levels of reelin protein (4). The mechanism by which loss of the paralogous NPAS1/3 transcription factors elicits these abnormalities is unknown. We now show that the NPAS3 transcription factor regulates FGF signaling in the dentate gyrus of mammalian brain through controlling expression of the FGF receptor (FGFR) subtype 1 (FGFR1), and in turn also controls neurogenesis emanating from this region. Although aberrations in hippocampal structure and function have been well characterized in schizophrenic patients, their etiology is unknown. We speculate that attenuated or aberrant adult hippocampal neurogenesis may contribute to the pathophysiology of schizophrenia.
Methods
Approval for the animal experiments described herein was obtained by the University of Texas Southwestern Medical Center Institutional Animal Care and Use Committee.
Mice. Npas1+/- Npas3+/- females (F1) mated to Npas1+/- Npas3+/- males (F1) produced 129/SvEv C57BL/6J mixed strain litters. Healthy pups were ear-tagged and tail-snipped when weaned (21 days). Pups that displayed growth deficiencies were group-housed and received special care (nesting squares, isolation foam under the cage, and 11% fat water-soaked food) until viable. Eventually all animals were group-housed until time of testing. Genotypes for the Npas1 and Npas3 loci were established by PCR analysis, as described in ref. 4.
Microarray Expression Analysis. SHEP neuroblastoma cells were cotransfected with ecdysone-inducible expression vectors (pIND) encoding ORFs for NPAS1/3 and ARNT, along with an expression vector (VgRXR) encoding the human retinoid X receptor (RXR) and a modified D. melanogaster ecdysone (VgEcR) receptor. Stable integration of NPAS1/3, ARNT, and VgRXR expression vectors was achieved by selecting cells for neomycin, hygromycin, and zeocin resistance. NPAS1/3 and ARNT expression was induced by treating cells with ponasterone (Invitrogen) for 16 h before RNA was harvested for microarray. RNA was prepared by using RNA STAT-60 (Tel-Test, Friends-wood, TX). Briefly, cells or tissues were homogenized in RNA STAT-60 and RNA was precipitated by 2-propanol and washed with 75% ethanol. RNA pellet was air-dried and dissolved in water. Gene expression analysis was performed at the University of Texas Southwestern Medical Center (Dallas) Microarray Core Facility on Affymetrix GeneChips.
Real-Time Quantitative PCR. For quantification of relative RNA levels by TaqMan real-time PCR technology (Applied Biosystems), 1 μg of DNase-treated total RNA was reverse-transcribed by using random hexamers (Roche, Indianapolis) and Superscript reverse transcriptase (Invitrogen). The cDNA equivalent to 25 ng of total RNA was PCR-amplified in a PRISM 7900HT detection system (Applied Biosystems). Forward and reverse primers were as follows: GAPDH forward, 5′-caaggtcatccatgacaactttg-3′; GAPDH reverse, 5′-ggccatccacagtcttctgg-3′; FGFR1 forward, 5′-tgccaagacggtgaagttca-3′; FGFR1 reverse, 5′-caattcggtggtcaggctta-3′; FGFR2 forward, 5′-ggattgctggcatgctgta-3′; FGFR2 reverse, 5′-gtcttcgaccaactgcttgaa-3′; FGFR3 forward, 5′-ccgatcgtggcaactatacc-3′; FGFR3 reverse, 5′-gtgtatgtctgccggatgct-3′; FGFR4 forward, 5′-tctcgccagcctgtcacta-3′; and FGFR4 reverse, 5′-gaactgtcgggccaaagg-3′. Relative RNA levels were calculated by 2-CT (CT is the cycle number at which the signal reaches the threshold of detection) and normalized to corresponding GAPDH RNA levels. Each CT value used for these calculations was the mean of triplicates of the same reaction. Relative RNA levels were expressed as percentage of the value of first time point for each tissue.
In Situ Hybridization. In situ hybridization was performed at the University of Texas Southwestern Medical Center Molecular Pathology Core Facility. RNA probes for FGFR1 (5′-ttcagtggctgaagcacatcgaggtgaacgggagtaagatcgggccagacaacttgccgtatgtccagatcctgaagactgctggagttaataccaccgacaaggaaatggaggtgcttcatctacggaatgtctcctttgaggatgcgggggagtatacgtgcttggcgggtaactctatcggactctcccatcactctgcatggttgaccgttctggaagccctggaagagagaccagctgtgatgacctcaccgctctacctggagatcattatctactgcaccggggccttcctgatctcctgcatgttgggctctgtcatcatctataagatgaagagcggcaccaagaagagcgacttcca-3′), FGFR2 (5′-atatgcatcgaaaggcaacctccgggaatacctccgagcccggaggccacctggcatggagtactcctatgacattaaccgtgtccccgaggagcagatgaccttcaaggacttggtgtcctgcacctaccagctggctagaggcatggagtacttggcttcccaaaaatgtatccatcgagatttggctgccagaaacgtgttggtaacagaaaacaatgtgatgaagatagcagactttggcctggccagggatatcaacaacatagactactataaaaagaccacaaatgggcgacttccagtcaagtggatggctcctgaagccctttttgatagagtttacactcatcagagcgatgtctggtccttcggggtgttaatgtgggagatctttactttagggggctcaccctacccagggattcccgtggaggaactttttaagctgctcaaagagggacacaggatggacaagcccaccaactgcaccaatgaac-3′), and FGFR3 (5′-caccgacaaggagctagaggttctgtccttgcacaatgtcacctttgaggacgcgggggagtacacctgcctggcgggcaattctattgggttttcccatcactctgcgtggctggtggtgctgccagctgaggaggagctgatggaaactgatgaggctggcagcgtgtacgcaggcgtcctcagctacggggtggtcttcttcctcttcatcctggtggtggcagctgtgatactctgccgcctgcgcagtcccccaaagaagggcttgggctcgcccaccgtgcacaaggtctctcgcttcccgcttaagcgacaggtgtccttggaatctaactcctctatgaactccaacacaccccttgtccggattgcccggctgtcctcaggagaaggtcctgttctggccaatgtt-3′) were generated through incorporation of PCR-amplified gene-specific fragments into pGEM-T plasmid vectors (Promega) with subsequent in vitro transcription by using an Ambion MAXIscript kit and 35S-labeled thio-UTP. Each labeling reaction used 1 μg of linearized template, 50 μCi of [α-35S]thio-UTP (1 Ci = 37 GBq) (Amersham Pharmacia) and was transcribed by using T7 RNA polymerase (Ambion). In situ hybridization was performed on brain sections from wild-type, Npas3-/-, and Npas1-/- male mice. Three-month-old male mice were anesthetized with methoxyflurane and transcardially perfused with heparinized saline followed by 4% paraformaldehyde. Brains were dissected and immersed in 4% paraformaldehyde overnight at 4°C. Tissue was placed in 70% ethanol, dehydrated through graded ethanol solutions, cleared in xylene, and infused with paraffin. Coronal and parasagittal sections were cut at 4-μm intervals and mounted on Vectabond-treated slides (Vector Laboratories). Sections were probed with antisense transcripts of FGFR1, FGFR2, or FGFR3. Paraffin was removed from the sections with xylene and subsequent graded ethanol dehydration. Postfixation in 4% paraformaldehyde was followed by Pronase digestion (20 μg/ml Pronase for 7.5 min) and acetylation (0.1 M triethanolamine·HCl, pH 7.5/0.25% acetic anhydride for 5 min). Hybridization was conducted for 12 h at 55°C in hybridization solution (50% formamide/0.3% dextran sulfate/1× Denhardt's solution/0.5 mg/ml tRNA/7.5 × 106 cpm/ml riboprobe). After hybridization, slides were washed in 5× SSC (1× SSC = 0.15 M sodium chloride/0.015 M sodium citrate, pH 7) at 55°C for 40 min, followed by high-stringency wash in 50% formamide/2× SSC supplemented by 10 mM DTT at 65°C for 30 min. K.5 nuclear emulsion (Ilford) was applied to slides before exposure at 4°C for 21-28 days, depending on individual probe signal detected on 14-day timer slides. Sections were counterstained with hematoxylin. Sense controls showed no signal (data not shown). Results shown are representative of results from five sections from each of three mice of each genotype.
Quantification of Basal in Vivo BrdUrd-Labeled Proliferating Neural Precursor Cells. Basal levels of neural precursor cell proliferation in dentate gyrus granular layer (GL)/subgranular zone (SGZ) were quantified in brains of 3-month-old adult wild-type littermate (n = 8), Npas3-/- (n = 8), and Npas1-/- (n = 11) male mice through immunohistochemical detection of incorporation of the thymidine analog BrdUrd (Sigma-Aldrich) in dividing cells in the brain. Mice were housed individually in standard cages with ad libitum access to food and water and were injected i.p. once daily with BrdUrd (50 μg/g of body weight) for 12 days, according to established methods (5). On day 13, mice were anesthetized and transcardially perfused with 4% paraformaldehyde in PBS at pH 7.4. Brains were dissected, immersed in 4% paraformaldehyde overnight at 4°C, and then sliced coronally on a Vibratome into 40-μm-thick free-floating sections that were subsequently processed for immunohistochemical staining with mouse monoclonal anti-BrdUrd (1:100, Roche). Omission of primary antibodies served as negative controls (data not shown). Unmasking of BrdUrd antigen was achieved through incubating tissue sections for 2 h in 50% formamide/2× SSC at 65°C, followed by 5 min wash in 2× SSC and subsequent incubation for 30 min in 2 M HCl at 37°C. Diaminobenzidine was used as a chromogen. Images were analyzed with a Nikon Eclipse 90i motorized research microscope with Plan Apo lenses coupled with Metamorph Image Acquisition software (Nikon). Sectioning, immunohistochemistry, and counting of immunopositive cells were executed by separate investigators in a double-blinded manner. Immunopositive cells in the dentate gyrus GL and adjacent SGZ, defined as a two-cell-body-wide zone of the hilus along the base of the GL, were counted in both hemispheres of every third section (≈10-15 sections per animal) progressing posteriorly from the point where the suprapyramidal and infrapyramidal blades are joined at the crest region and the dentate gyrus is oriented horizontally beneath the corpus callosum.
Morphometric Analysis of Hippocampal Size. Paraformaldehyde-fixed wild-type (n = 5), Npas3-/- (n = 5), and Npas1-/- male mouse brains (n = 5) were sectioned in the coronal plane and hematoxylin/eosin stained. Histological sections were obtained at 50-μm intervals. Measurements of the hippocampus, dentate granular cell layer, and forebrain were taken at the coronal level in which CA1 approaches the midline and the upper blade of the dentate gyrus runs parallel to the surface of the brain.
Intracerebroventricular (i.c.v.) Infusion of Basic FGF (bFGF). Per established methods (6), male mice were anesthetized and implanted with osmotic minipumps (Alzet 1007D; Cupertino, CA) with cannulas placed in the left lateral ventricle 3 mm deep to the pial surface, -0.3 mm anteroposterior relative to bregma, and 1.3 mm lateral to midline. Commercially available recombinant FGF2 (Sigma-Aldrich) was dissolved in artificial cerebrospinal fluid (aCSF) (128 mM NaCl/2.5 mM KCl/0.95 mM CaCl2/1.9 mM MgCl2) at a concentration of 10 μg/ml. Mice were treated for 3 days with FGF2 in aCSF or with aCSF alone and administered 50 mg/kg BrdUrd i.p. daily for the same 3 consecutive days. Minipumps were surgically removed on day 4 and mice were transcardially perfused on day 6 for immunohistochemistry. Immunopositive cells were quantified in 10-15 sections from each mouse, progressing posteriorly from the point where the suprapyramidal and infrapyramidal dentate gyrus blades are joined at the crest region and the dentate gyrus is oriented horizontally beneath the corpus callosum. BrdUrd-positive cells were quantified as described above but only in the hemisphere contralateral to the site of cannula placement to avoid false signal due to surgical artifact.
Electroconvulsive Seizure (ECS). Three-month-old adult wild-type littermate (n = 3 sham-ECS, n = 3 ECS), Npas3-/- (n = 3 sham-ECS, n = 6 ECS), and Npas1-/- (n = 3 sham-ECS, n = 3 ECS) male mice were housed under standard conditions with free access to lab chow and water. Animals were given repeated seizures (once daily for 5 days) (9-15 mA, 0.1 s) by means of earclip electrodes. Wild-type and Npas1-/- mice required from 13 to 15 mA to elicit generalized tonic-clonic seizures. Npas3-/- mice were found to have a lower seizure threshold, requiring from 9 to 12 mA to elicit generalized tonic-clonic seizures. All mice experienced either sham-ECS or an ECS-induced generalized tonic-clonic seizure of 5- to 10-s duration during the same time period (between 10:00 and 11:00 a.m.) daily for 5 days. Mice were injected i.p. once daily between 2:00 and 3:00 p.m. with BrdUrd (50 μg/g of body weight) for 7 days, starting on the first day of ECS delivery and concluding 2 days after the fifth day of ECS delivery. On day 7, mice were killed by means of transcardial perfusion with 4% paraformaldehyde (pH 7.4), and brains were processed for immunohistochemical staining of incorporated BrdUrd as described above.
Results
We used DNA microarrays to interrogate mRNA expressed from neuroblastoma cells programmed to inducibly express NPAS3 and its obligate heterodimeric partner ARNT. Such studies revealed an NPAS3/ARNT-dependent inhibition of Sprouty4 mRNA expression that was confirmed by real-time quantitative PCR (data not shown). This finding was paralleled by comparably increased level of Sprouty4 mRNA in Npas3-/- mouse brain, relative to wild type, as assayed by both real-time quantitative PCR and analysis of raw microarray expression data (data not shown).
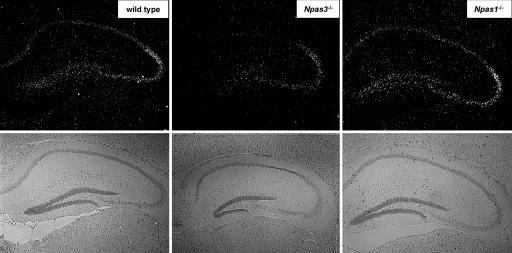
Extensive genetic and molecular biological studies have shown that the Sprouty proteins of D. melanogaster and mammals inhibit receptor tyrosine kinase signaling downstream of FGF (7-13). Our observations of a potential relationship between NPAS3 and Sprouty4 expression led us to investigate whether NPAS3 or NPAS1 might be connected to FGF signaling in mammals. Knowing that Trachealess, the D. melanogaster ortholog of NPAS3 and NPAS1, controls branching morphogenesis by means of direct regulation of the D. melanogaster gene encoding FGFR (14), we hypothesized that NPAS3 might regulate an analogous pathway in mammals. Real-time quantitative PCR analysis of the mRNAs specified by the four mammalian FGFR genes gave evidence of abundant expression of subtypes 1, 2, and 3 in adult mouse forebrain. Subsequent in situ hybridization to localize FGFR subtypes 1, 2, and 3 in adult mouse brain tissue revealed that the expression of FGFR subtype 1 (FGFR1) mRNA is selectively enriched in the hippocampus and dentate gyrus relative to FGFR subtypes 2 and 3 (data not shown). FGFR1 mRNA expression in the hippocampus of Npas1-/- animals was indistinguishable from wild type. By contrast, FGFR1 mRNA expression in Npas3-/- animals was significantly attenuated relative to wild type, especially in the dentate gyrus and CA1 regions of the hippocampus (Fig. 1).
Fig. 1.
Dark-field illumination in situ hybridization pattern for FGF receptor subtype 1 (FGFR1) mRNA showing attenuation in the hippocampus of Npas3-/- mice relative to wild-type and Npas1-/- mice. Bright-field illumination shown directly below the in situ hybridization panels illustrates the neuroanatomy of each section
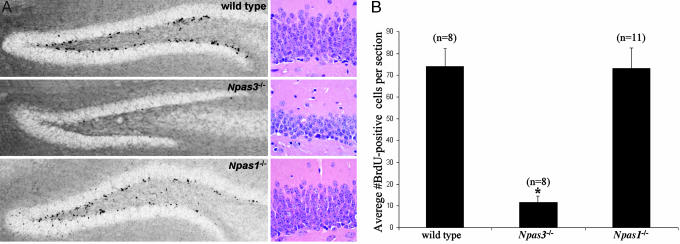
FGFR1 and its ligand, bFGF (also known as FGF-2), are known to play a pivotal, stimulatory role in adult neurogenesis (15-21). Neurogenesis arises in the mammalian brain predominantly from neural precursor cells residing in the hippocampal dentate gyrus SGZ and the subventricular zone (SVZ) of the lateral ventricle. These regions provide cells throughout adult life to the dentate gyrus GL and the olfactory bulb, respectively (22). We used previously established methods (5) to quantify neural precursor cell proliferation in brain tissue of 3-month-old adult mice through immunohistochemical detection of incorporation of the thymidine analog BrdUrd. Relative to wild-type littermates and Npas1-/- animals, Npas3-/- mice showed a profound deficit in neural precursor cell proliferation in the hippocampal dentate gyrus, with levels reduced ≈84% (P < 0.001, Student's t test) relative to wild-type and Npas1-/- mice (Fig. 2 and Table 1). Average number of BrdUrd-positive cells per section in the hippocampal dentate gyrus was 74 ± 8 in wild-type mice, 73 ± 9 in Npas1-/- mice, and 12 ± 3 in Npas3-/- mice.
Fig. 2.
Basal neural precursor cell proliferation in the hippocampal dentate gyrus SGZ and GL showing statistically significant decreases in Npas3-/- (n = 8) adult mice relative to wild-type littermate (n = 8) and Npas1-/- (n = 11) animals (*, P < 0.0001, Student's t test). (A) Mice received daily i.p. injection of BrdUrd (50 μg/g of body weight) for 12 days and representative immunohistochemical staining of incorporated BrdUrd on day 13 is shown in hippocampal dentate gyrus of wild-type, Npas3-/-, and Npas1-/- adult mouse brain. Hematoxylin/eosin staining showed markedly reduced thickness of the dentate gyrus granular layer in Npas3-/- adult mice relative to wild-type and Npas1-/- adult mice. (B) Average numbers of BrdUrd-positive cells per section within the dentate gyrus were 74 ± 8 in wild-type mice, 12 ± 3 in Npas3-/- mice, and 73 ± 9 in Npas1-/- mice
Table 1. Statistical comparison of hippocampal neuroanatomy between Npas3–/– and wild-type mice.
| Measurement | Npas3–/– (n = 5) | Wild type (n = 5) |
|---|---|---|
| Dentate thickness (SEM), μm | 73.9 (2.6)* | 105.6 (2.5) |
| CA1 thickness (SEM), μm | 67.7 (2.2) | 69.2 (3.5) |
| CA3 thickness (SEM), μm | 79.1 (4.5) | 78.1 (5.8) |
| HP width (SEM), mm | 2.79 (0.07) | 3.07 (0.026) |
| FP width (SEM), mm | 4.82 (0.04) | 5.18 (0.06) |
| Width ratio | 0.58 | 0.59 |
| HP height (SEM), mm | 1.06 (0.03) | 1.16 (0.01) |
| FB height (SEM), mm | 1.80 (0.07) | 1.99 (0.02) |
| Height ratio | 0.59 | 0.58 |
The statistically significant difference in measured thickness was not related to differences in overall neuroanatomy between genotypes, because there were no significant differences in the ratio of hippocampus to forebrain size between genotypes. Within the hippocampus, the difference in granule cell layer thickness in Npas3–/– mice (*, P < 0.001, Student's t test) as specific for the dentate gyrus because there were no significant differences in pyramidal cell layer thickness in CA1 and CA3 regions between wild-type and Npas3–/– mice. HP, hippocampus; FB, forebrain.
The severe attenuation of neural precursor cell proliferation in Npas3-/- mice correlated with reduced thickness of the granular layer of the dentate gyrus (Figs. 1 and 2 and Table 1). In wild-type mice, the upper blade of the dentate gyrus averaged 105.6 ± 2.5 μm, whereas its thickness in Npas3-/- mice was only 73.9 ± 2.7 μm(P < 0.001, Student's t test). The size of individual granule cells was indistinguishable in Npas3-/- and wild-type mice, and thickness of the granule cell layer did not differ between Npas1-/- and wild-type mice. Furthermore, there was no significant difference between Npas3-/- and wild-type animals in thickness of the CA1 or CA3 hippocampal pyramidal cell layers.
Apparently, genetic deficiency of NPAS3 conferred a specific and profound deficit in proliferation of adult hippocampal neural precursor cells as well as thickness of the dentate gyrus granule cell layer, the site to which these proliferating cells migrate. The finding that deficient adult neural precursor cell proliferation in Npas3-/- mice is associated with selectively reduced thickness of the dentate gyrus is consistent with the conclusion that NPAS3 mediates production of new neurons critical to growth or maintenance of this structure throughout development or adulthood.
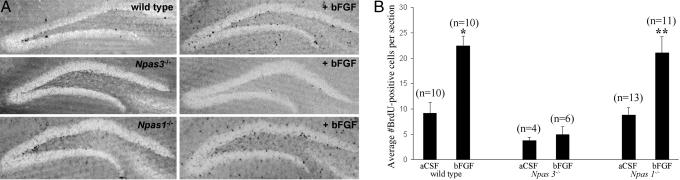
We next compared the ability of i.c.v. infusion of bFGF to stimulate adult hippocampal neural precursor cell proliferation in the hemisphere contralateral to the site of infusion in 3-month-old adult wild-type, Npas3-/-, and Npas1-/- mice. As expected from previous studies in rats (6, 18, 20, 23), bFGF significantly increased adult neural precursor cell proliferation in wild-type animals. Average number of BrdUrd-positive cells per section in the dentate gyrus in wild-type mice increased from 9 ± 2 for vehicle control up to 22 ± 2 after bFGF administration (P < 0.001, Student's t test). In Npas1-/- mice, the average number of BrdUrd-positive cells per section similarly increased from 9 ± 2 for vehicle control up to 23 ± 3 after bFGF administration (P < 0.001, Student's t test). In Npas3-/- mice, however, i.c.v. infusion of bFGF failed to stimulate neural precursor cell proliferation. The average number of BrdUrd-positive cells per section in Npas3-/- hippocampal dentate gyrus was 4 ± 1 for vehicle control and 5 ± 2 after bFGF administration (Fig. 3). The measured basal level of BrdUrd-positive cells in the FGF paradigm was substantially less than the basal level in mice evaluated in the paradigm pertaining to Fig. 1. This outcome is likely because of the shorter duration of BrdUrd injection in the FGF paradigm (3 vs. 12 days) as well as the fact that in the FGF paradigm only BrdUrd-positive cells in the hemisphere contralateral to the site of injection were counted, as opposed to the first paradigm, in which both hemispheres were analyzed. In addition, the FGF paradigm used anesthesia, which might affect neural precursor cell proliferation.
Fig. 3.
Proliferation of neural precursor cells was increased in a statistically significant manner in the hippocampal dentate gyrus GL and SGZ of adult wild-type and Npas1-/- mice after lateral ventricle i.c.v infusion of bFGF dissolved in aCSF. Neural precursor cell proliferation in Npas3-/- mice was not affected by i.c.v. infusion of bFGF. (A) Representative immunohistochemical staining of BrdUrd is shown in hippocampal dentate gyrus of wild-type, Npas3-/-, and Npas1-/- adult mice after i.c.v. infusion of either aCSF vehicle control or bFGF. BrdUrd-positive cells were quantified within the dentate gyrus GL and SGZ in the hemisphere contralateral to the site of cannula placement to avoid false signal due to surgical artifact. (B) Average numbers of BrdUrd-positive cells per section significantly increased from 9 ± 2 in wild-type mice infused with aCSF alone (n = 10) to 22 ± 2 in wild-type mice infused with bFGF dissolved in aCSF (n = 10) (*, P < 0.0001, Student's t test), and from 9 ± 2 in Npas1-/- mice infused with aCSF alone (n = 13) to 21 ± 3 in Npas1-/- mice infused with bFGF dissolved in aCSF (n = 11) (**, P < 0.0001, Student's t test). Average numbers of BrdUrd-positive cells per section were 4 ± 1 in Npas3-/- mice infused with aCSF alone (n = 4) and 5 ± 2 in Npas3-/- mice infused with bFGF dissolved in aCSF (n = 6)
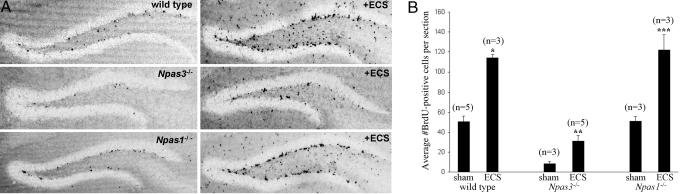
Enhanced proliferation of Npas3-/- neural precursor cells was, however, achieved by ECS, a well characterized model for stimulation of neurogenesis in rats (24-27). ECS potently stimulated neural precursor cell proliferation in the hippocampal dentate gyrus in animals of all three genotypes. The average number of BrdUrd-positive cells per section in the hippocampal dentate gyrus increased from 50 ± 6 in sham-ECS up to 121 ± 6 in ECS-treated wild-type mice (P < 0.001, Student's t test), from 9 ± 2 in sham-ECS up to 31 ± 6 in ECS-treated Npas3-/- mice (P < 0.001, Student's t test), and from 51 ± 5 in sham-ECS up to 122 ± 15 in ECS-treated Npas1-/- mice (P < 0.001, Student's t test) (Fig. 4). The measured basal level of BrdUrd-positive cells in the ECS paradigm was less than the basal level in mice in the paradigm pertaining to Fig. 1, probably because of the shorter duration of BrdUrd injection in the ECS paradigm (7 vs. 12 days).
Fig. 4.
Proliferation of neural precursor cells was increased in a statistically significant manner in the hippocampal dentate gyrus of adult wild-type, Npas3-/-, and Npas1-/- mice after ECS. (A) Representative immunohistochemical staining of BrdUrd is shown in the hippocampal dentate gyrus of wild-type, Npas3-/-, and Npas1-/- mice after ECS. (B) Average numbers of BrdUrd-positive cells per section were increased in a statistically significant manner from 50 ± 6 in sham-ECS (n = 5) up to 121 ± 6 in ECS-treated (n = 3) wild-type mice (*, P < 0.001, Student's t test), from 9 ± 2 in sham-ECS (n = 3) up to 31 ± 6 in ECS-treated (n = 5) Npas3-/- mice (**, P < 0.001, Student's t test), and from 51 ± 5 in sham-ECS (n = 3) up to 122 ± 15 in ECS-treated (n = 3) Npas1-/- mice (***, P < 0.001, Student's t test)
Discussion
We conclude that NPAS3 operates in the adult mouse brain in a manner analogous to its D. melanogaster homolog Trachealess, which controls branching morphogenesis of the larval tracheal system by regulating the FGF signaling pathway (14). We hereby show that adult Npas3-/- mice are significantly deficient in expression of FGFR1 mRNA in the hippocampal dentate gyrus, and that this finding is associated with marked attenuation in adult hippocampal neural precursor cell proliferation. Furthermore, unlike wild-type and Npas1-/- mice, neural precursor cell proliferation in Npas3-/- mice was not stimulated by i.c.v. administration of bFGF. The inability of bFGF to stimulate hippocampal neural precursor cell proliferation in Npas3-/- mice likely relates to attenuated expression of FGFR1 in the dentate gyrus of the adult brain. Hippocampal neural precursor cells in Npas3-/- mice did, however, retain proliferative ability in response to ECS. Although the mechanisms of ECS-stimulated neurogenesis are unknown, genetic microarray data have implicated diverse and distinct signaling pathways, including but not restricted to those mediated by FGF, brain-derived neurotrophic factor, neuropeptide Y, vascular endothelial growth factor, thyroid-stimulating hormone-releasing hormone, stem cell factor, and the arachidonic acid pathway (28, 29). Thus, the observed attenuation in neural precursor cell proliferation in Npas3-/- mice may be specifically related to a diminished endogenous response of cellular proliferation to FGF signaling during adulthood. The majority of newly born cells in the adult dentate gyrus differentiate into neurons (30), and the effect of NPAS3 deficiency on adult neurogenesis could be influenced by events during embryogenesis, development, or adulthood. It will be important in future studies to distinguish the roles of NPAS3 in hippocampal neurogenesis at these different stages.
The recent finding of a genetic translocation within the NPAS3 gene in a family with schizophrenia, coupled with the present report of reduced neurogenesis in Npas3-/- mice, raises the possibility that impaired hippocampal neurogenesis may be relevant to some aspects of hippocampal pathology that are associated with schizophrenia. Proliferating neural precursor cells in the hippocampal dentate gyrus SGZ are confined locally to the granule cell layer, which shows dramatic structural and functional plasticity in adulthood through ongoing neurogenesis (31). Aberrations in hippocampal structure and function such as reduced hippocampal volume, altered hippocampal neuronal activity, and predisposition to greater hippocampal injury after anoxia have been extensively documented in schizophrenic patients (32-41). Although the function of adult hippocampal neurogenesis is poorly understood, correlative observations suggest that it facilitates integration of novel environmental stimuli and adaptive remodeling of the dentate gyrus throughout development and adulthood (42-45). Aberrations in hippocampal neurogenesis could be involved in the pathophysiology of schizophrenia by conferring an inability to appropriately remodel neural connections in hippocampal circuitry in response to environmental stimuli or psychological challenges. Our observations compel further investigation of the role of NPAS3 in neurogenesis. Such efforts may expand our understanding of the pathophysiology of schizophrenia with hopes of identifying new therapeutic opportunities or clarifying mechanisms of older therapies such as electroconvulsive therapy for treatment of this devastating form of mental illness.
Acknowledgments
We thank Fred H. Gage, Huda Y. Zoghbi, and Solomon H. Snyder for critical review of the manuscript; Eric J. Nestler, Carol A. Tamminga, Steven G. Kernie, and Amelia J. Eisch for critical input and encouragement; and E. W. Brunskill and S. S. Potter for providing heterozygous Npas3+/- mice. A.A.P. is supported as the David Nathan Meyerson Fellow by the Morton H. Meyerson Tzedakah Fund. This work was also supported by funds provided to S.L.M. by National Institute of Mental Health Grant R01-4R37MH59388, the McKnight Foundation for Neurosciences, the National Institutes of Health Director's Pioneer Award, and unrestricted funds from an anonymous donor.
Author contributions: A.A.P., X.W., T.W.H., S.J.E., C.A.D., J.A.R., D.J.B., and S.L.M. designed research; A.A.P., X.W., T.W.H., S.J.E., Q.D., L.C.W., S.R.-F., C.A.D., J.A.R., and D.J.B. performed research; A.A.P., X.W., T.W.H., S.J.E., S.R.-F., J.A.R., D.J.B., and S.L.M. contributed new reagents/analytic tools; A.A.P., X.W., T.W.H., S.R.-F., C.A.D., J.A.R., D.J.B., and S.L.M. analyzed data; and A.A.P., S.J.E., D.J.B., and S.L.M. wrote the paper.
Abbreviations: NPAS3, neuronal PAS domain protein 3; NPAS1, neuronal PAS domain protein 1; FGFR, FGF receptor; bFGF, basic FGF; ARNT, aryl hydrocarbon receptor nuclear translocator; SGZ, subgranular zone; GL, granular layer; i.c.v., intracerebroventricular; aCSF, artificial cerebrospinal fluid; ECS, electroconvulsive seizure.
References
- 1.Kamnasaran, D., Muir, W. J., Ferguson-Smith, M. A. & Cox, D. W. (2003) J. Med. Genet. 40, 325-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunskill, E. W., Witte, D. P., Shreiner, A. B. & Potter, S. S. (1999) Mech. Dev. 88, 237-241. [DOI] [PubMed] [Google Scholar]
- 3.Zhou, Y.-D., Barnard, M., Tian, H., Li, X., Ring, H. Z., Franke, U., Shelton, J., Richardson, J., Russel, D. W. & McKnight, S. L. (1997) Proc. Natl. Acad. Sci. USA 94, 713-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erbel-Sieler, C., Dudley, C., Zhou, Y., Wu, X., Estill, S. J., Han, T., Diaz-Arrastia, R., Brunskill, E. W., Potter, S. S. & McKnight, S. L. (2004) Proc. Natl. Acad. Sci. USA 101, 13648-13653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kempermann, G., Kuhn, H. G. & Gage, F. H. (1997) Nature 386, 493-495. [DOI] [PubMed] [Google Scholar]
- 6.Jin, K., Sun, Y, Xie, L., Batteur, S., Mao, X. O., Smelick, C., Logvinova, A. & Greenberg. D. A. (2003) Aging Cell 2, 175-183. [DOI] [PubMed] [Google Scholar]
- 7.Sivak, M., Petersen, L. F. & Amaya, E. (2005) Dev. Cell 8, 689-701. [DOI] [PubMed] [Google Scholar]
- 8.Tsang, M. & Dawid, I. B. (2004) Sci. STKE 228, pe17. [DOI] [PubMed] [Google Scholar]
- 9.Hanafusa, H., Torii, S., Yasunaga, T. & Nishida, E. (2002) Nat. Cell. Biol. 11, 850-858. [DOI] [PubMed] [Google Scholar]
- 10.Sasaki, A., Taketomi, T., Wakioka, T., Kato, R. & Yoshimura, A. (2001) J. Biol. Chem. 276, 36804-36808. [DOI] [PubMed] [Google Scholar]
- 11.Reich, A., Sapir, A. & Shilo, B. (1999) Development (Cambridge, U.K.) 126, 4139-4147. [DOI] [PubMed] [Google Scholar]
- 12.Kramer, S., Okabe, M., Hacohen, N., Krasnow, M. A. & Hiromi, Y. (1999) Development (Cambridge, U.K.) 126, 2515-2525. [DOI] [PubMed] [Google Scholar]
- 13.Hacohen, N., Kramer, S., Sutherland, D., Hiromi, Y. & Krasnow, M. A. (1998) Cell 92, 253-263. [DOI] [PubMed] [Google Scholar]
- 14.Ohshiro, T. & Saigo, K. (1997) Development (Cambridge, U.K.) 124, 3975-3986. [DOI] [PubMed] [Google Scholar]
- 15.Gage, F. H., Coates, P. W., Palmer, T. D., Kuhn, H. G., Fisher, L. J., Suhonen, J. O., Peterson, D. A., Suhr, S. T. & Ray, J. (1995) Proc. Natl. Acad. Sci. USA 92, 11879-11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer, T. D., Ray, J. & Gage, F. H. (1995) Mol. Cell. Neurosci. 6, 474-486. [DOI] [PubMed] [Google Scholar]
- 17.Palmer, T. D., Markakis, E. A., Willhoite, A. R., Safar, F. & Gage, F. H. (1999) J. Neurosci. 19, 8487-8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshimura, S., Takagi, Y., Harada, J., Teramoto, T., Thomas, S. S., Waeber, C., Bakowska, J. C., Breakefield, X. O. & Moskowitz, M. A. (2001) Proc. Natl. Acad. Sci. USA 98, 5874-5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng, Y., Black, I. B. & DiCicco-Bloom, E. (2002) Eur. J. Neurosci. 15, 3-12. [DOI] [PubMed] [Google Scholar]
- 20.Yoshimura, S., Teramoto, T., Whalen, M. J., Irizarry, M. C., Takagi, Y., Qiu, J., Harada, J., Waeber, C., Breakefield, X. O. & Moskowitz, M. A. (2003) J. Clin. Invest. 112, 1202-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohkubo, Y., Uchida, A. O., Shin, D., Partanen, J. & Vaccarino, F. M. (2004) J. Neurosci. 24, 6057-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gage, F. H. (2000) Science 287, 1433-1438. [DOI] [PubMed] [Google Scholar]
- 23.Wagner, J. P., Black, I. B. & DiCicco-Bloom, E. (1999) J. Neurosci. 19, 6006-6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Madsen, T. M., Treschow, A., Bengzon, J., Bolwig, T. G., Lindvall, O. & Tingstrom, A. (2000) Biol. Psychiatry 47, 1043-1049. [DOI] [PubMed] [Google Scholar]
- 25.Madsen, T. M., Yeh, D. D., Valentine, G. W. & Duman, R. S. (2005) Neuropsychopharmacology 30, 27-34. [DOI] [PubMed] [Google Scholar]
- 26.Hellsten, J., Wennstrom, M., Mohapel, P., Ekdahl, C. T., Bengzon, J. & Tingstrom, A. (2002) Eur. J. Neurosci. 16, 283-290. [DOI] [PubMed] [Google Scholar]
- 27.Scott, B. W., Wojtowicz, J. M. & Burnham, W. M. (2000) Exp. Neurol. 165, 231-236. [DOI] [PubMed] [Google Scholar]
- 28.Newton, S. S., Collier, E. F., Hunsberger, J., Adams, D., Terwilliger, R., Selvanayagam, E. & Duman, R. S. (2003) J. Neurosci. 23, 10841-10851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Altar, C. A., Laeng, P., Jurata, L. W., Brockman, J. A., Lemire, A., Bullard, J., Bukhman, Y. V., Young, T. A., Charles, V. & Palfreyman, M. G. (2004) J. Neurosci. 24, 2667-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cameron, H. A., Woolley, C. S., McEwen, B. S. & Gould, E. (1993) Neuroscience 56, 337-344. [DOI] [PubMed] [Google Scholar]
- 31.Taupin, P. & Gage, F. H. (2002) J. Neurosci. Res. 69, 745-749. [DOI] [PubMed] [Google Scholar]
- 32.Lawrie, S. & Abukmeil, S. (1998) Br. J. Psychiatry 172, 110-120. [DOI] [PubMed] [Google Scholar]
- 33.Nelson, M. D., Saykin, A. J., Flashman, L. A. & Riordan, H. J. (1998) Arch. Gen. Psychiatry 55, 433-440. [DOI] [PubMed] [Google Scholar]
- 34.Wright, I. C., Rabe-Hesketh, S., Woodruff, P. W., David, A. S., Murray, R. M. & Bullmore, E. T. (2000) Am. J. Psychiatry 157, 16-25. [DOI] [PubMed] [Google Scholar]
- 35.Eyler Zorilla, L. Y., Jeste, D. V., Paulus, M. P. & Brown, G. G. (2002) Schizophr. Res. 59, 187-198. [DOI] [PubMed] [Google Scholar]
- 36.Heckers, S., Rauch, S. L., Goff, D., Savage, C. R., Schacter, D. L., Fischman, A. J. & Alpert, N. M. (1998) Nat. Neurosci. 4, 318-323. [DOI] [PubMed] [Google Scholar]
- 37.Weiss, A. P., Schacter, D. L., Goff, D. C., Rauch, S. L., Alpert, N. M., Fischman, A. J. & Heckers, S. (2003) Biol. Psychiatry 53, 48-55. [DOI] [PubMed] [Google Scholar]
- 38.Nordahl, T. E., Kusubov, N., Carter, C., Salamat, S., Cummings, A. M., O'Shora-Celaya, L., Eberling, J., Robertson, L., Huesman, R. H., Jagust, W., et al. (1996) Neuropsychopharmacology 6, 541-554. [DOI] [PubMed] [Google Scholar]
- 39.Tamminga, C. A., Thaker, G. K., Buchanan, R., Kirkpatrick, B., Alphs, L. D., Chase, T. N. & Carpenter, W. T. (1992) Arch. Gen. Psychiatry 49, 522-530. [DOI] [PubMed] [Google Scholar]
- 40.McNeil, T. F., Cantor-Graae, E. & Weinberger, D. R. (2000) Am. J. Psychiatry 157, 203-212. [DOI] [PubMed] [Google Scholar]
- 41.Van Erp, T. G., Saleh, P. A., Rosso, I. M., Huttunen, M., Lonnqvist, J., Pirkola, T., Salonen, O., Valanne, L., Poutanen, V. P., Standertskjold-Nordenstam, C.G., et al. (2002) Am. J. Psychiatry 159, 1514-1520. [DOI] [PubMed] [Google Scholar]
- 42.Kempermann, G., Brandon, E. P. & Gage, F. H. (1998) Curr. Biol. 8, 939-942. [DOI] [PubMed] [Google Scholar]
- 43.Nilsson, M., Perfilieva, E., Johansson, U., Orwar, O. & Eriksson, P. (1999) J. Neurobiol. 39, 569-578. [DOI] [PubMed] [Google Scholar]
- 44.Kempermann, G. & Gage, F. H. (1999) Hippocampus 9, 321-332. [DOI] [PubMed] [Google Scholar]
- 45.Brown, J., Cooper-Kuhn, C. M., Kempermann, G., Van Praag, H., Winkler, J., Gage, F. H. & Kuhn, H. G. (2003) Eur. J. Neurosci. 17, 2042-2046. [DOI] [PubMed] [Google Scholar]