Abstract
Leishmania parasites alternate between hosts, facing environmental changes that demand rapid gene expression adaptation. Lacking canonical RNA polymerase II promoters, transcription in these eukaryotes is polycistronic, with gene regulation occurring post-transcriptionally. Although non-coding RNAs (ncRNAs) have been identified in Leishmania transcriptomes, their functions remain unclear. Recognizing RNA structure's importance, we performed a genome-wide alignment of L. braziliensis and related species, identifying conserved RNA structures, 38 of which overlap with known ncRNAs. One such ncRNA, lncRNA45, was functionally characterized. Using a knockout cell line, we demonstrated that lncRNA45 is crucial for parasite fitness. Reintroducing the wild type lncRNA45 restored fitness, while a version with a single nucleotide substitution in the structured region did not. This mutation also altered RNA-protein interactions. These findings suggest that lncRNA45's regulatory role and protein interactions rely on its secondary structure. This study highlights the significance of structured lncRNAs in Leishmania biology and their potential as therapeutic targets. Further research into these ncRNAs could uncover new parasite regulation mechanisms and inspire novel treatment strategies.
Keywords: Non-coding RNAs, Leishmania braziliensis, RNA structure, Gene expression regulation, CRISPR- Cas9
1. Introduction
Leishmania is a protozoan parasite that causes a group of diseases collectively known as leishmaniasis [1]. Approximately 20 species are pathogenic to humans, causing a range of clinical syndromes from skin lesions to potentially fatal visceral leishmaniasis [1]. Leishmania (Viannia) braziliensis is among the most clinically significant pathogenic species in South America, causing mucocutaneous leishmaniasis—a severe form of the disease that can lead to extensive destruction of mucosal tissues [2]. Currently, there are no vaccines for humans, and available treatments often have severe side effects as well as decreasing efficacy due to parasite resistance [2]. To complete its life cycle, the heteroxenous Leishmania must adapt to significant environmental changes [3]. During a blood meal on an infected mammalian host, female Phlebotomine sandflies ingest phagocytes containing intracellular amastigotes (AMA). Within the insect gut, these amastigotes differentiate into procyclic promastigotes (PRO). After replication, PRO migrate to the anterior gut and further differentiate into intermediate forms until they reach the infective stage—metacyclic promastigotes (META). During the subsequent blood meal, META are regurgitated into the dermis of a vertebrate host, where they are phagocytized by macrophages and differentiate back into AMA [3]. Therefore, rapid and precise regulation of gene expression is essential for efficient transmission and, consequently, parasite survival.
Unlike other eukaryotes, trypanosomatids such as Leishmania exhibit a unique genetic organization, lacking canonical RNA polymerase II (RNA Pol II) promoters for individual genes [4]. Instead, transcription start sites (TSS) drive tandem arrays of genes, resulting in polycistronic transcription in which functionally unrelated genes within the same polycistronic transcriptional units (PTUs) are transcribed together as a single polycistronic mRNA [4]. Each PTU may contain over 200 genes, all of which undergo trans-splicing during RNA processing. In this process, a 39-nucleotide capped exon (spliced leader RNA, SL-RNA) is added to the 5′-end of each mRNA, and the preceding mRNA is polyadenylated at its 3′-end [5]. Mature mRNAs are then exported to the cytoplasm for translation into proteins. Despite this distinctive genetic organization, the mechanisms governing gene expression regulation are not fully understood. Post-transcriptional regulation is crucial involving mRNA processing, stability, nuclear-cytoplasmic transport, and translation [4,6]. RNA-binding proteins (RBPs) are pivotal in recognizing specific sequences or structural motifs in mRNA untranslated regions (UTRs), influencing their stability, localization, or translation [7].
Non-coding RNAs (ncRNAs) were shown to act as gene expression regulators in trypanosomatids [8,9]. They are transcribed but not translated into proteins and are generally classified as either short (<200 bp, sncRNAs) or long (>200 bp, lncRNAs) [10]. This classification, however, is currently under revision [11]. Despite the large number of ncRNAs identified in the transcriptomes of trypanosomatids [8,9,[12], [13], [14]], their biological functions and biogenesis remain incompletely understood. In L. braziliensis, 11,372 putative ncRNAs (3602 of them being differentially expressed across lifecycle stages) have been identified through RNA-seq and comparative transcriptomics suggesting they could be enrolled in gene expression regulation in this parasite [13].
Many ncRNAs, such as tRNAs, rRNAs and miRNAs, are strongly structured and function through structure. LncRNAs appear not too strongly structured but may contain structural regions from which they function [15]. In addition, some lncRNAs were shown to function through structural elements rather than conserved nucleotide sequences, highlighting the importance of secondary structures [16]. Based on the importance of secondary structure for ncRNAs' role in living systems, using a computational approach we searched for conserved secondary structures of ncRNAs in trypanosomatid genomes [17], particularly in L. braziliensis MHOM/BR75/M2903, and identified ncRNAs with conserved secondary structures differentially expressed between morphologies, suggesting their biological relevance. Based on differential expression and genomic localization, we selected a lncRNA with a conserved secondary structure for further investigation into its potential regulatory function. We found that this lncRNA is involved in parasite growth in culture, likely through interactions with proteins implicated in gene expression regulation and cell-cell signaling pathways. Moreover, we found that a single base substitution that alters its secondary structure impairs its function in L. braziliensis. Overall, elucidating the role of ncRNAs—and particularly their structural dynamics—provides important insights into the regulatory mechanisms essential for the survival and transmission of Leishmania parasites.
2. Results
2.1. Assembly of trypanosomatid genomes for comparative analysis
In this study, we selected eleven trypanosomatids genomes to search for RNA conserved secondary structures. We observed that, for most genomes, the percentage of complete Euglenozoa orthologues (from a total of 130) determined by the BUSCO methodology was above 90 %. This confirms the quality of genome assembly and annotation, except for the genomes of Bodo saltans and Trypanosoma cruzi (Table 1). Although these genomes exhibit lower conservation levels, Bodo saltans and Trypanosoma cruzi were included in the analysis to ensure representation of diverse evolutionary lineages; their percentages of complete orthologue remained higher than those of fragmented or missing sequences. The percentage of complete orthologues (C (%), Table 1) exceeded the percentage of fragmented (F (%), Table 1) or missing sequences (M (%), Table 1). We aligned these eleven genomes against the Leishmania braziliensis MHOM/BR75/M2903 genome using Lastz [18]. The resulting pairwise alignments were merged into a L. braziliensis-based multiple alignment using MultiZ [19] and the UCSC tool chain (see Methods for details). Details of genome quality, including percentages of complete, fragmented, and missing orthologues, are presented in Table 1.
Table 1.
BUSCO analysis of trypanosomatids’ genomes. The universal orthologues used for this analysis were obtained from the BUSCO euglenozoan Odb10 dataset.
| Speciesa | BUSCO (euglenozoa – 130)b |
|||
|---|---|---|---|---|
| C (%) | CD (%) | F (%) | M (%) | |
| Leishmania major Friedlin | 98.5 | 0 | 0 | 1.5 |
| Leishmania donovani BPK282A1 | 97.7 | 0 | 0 | 2.3 |
| Leishmania infantum JPCM5 | 97.7 | 0 | 0 | 2.3 |
| Leishmania amazonensis MHOM/BR/1973/M2269 | 96.9 | 0 | 0.8 | 2.3 |
| Leishmania panamensis MHOM/COL/81/L13 | 96.9 | 0 | 1.5 | 1.6 |
| Crithidia fasciculata CF-C1 | 96.2 | 0 | 2.3 | 1.5 |
| Leishmania braziliensis MHOM/BR/75/2903 | 93.8 | 0 | 4.6 | 1.6 |
| Trypanosoma brucei TREU927 | 93.1 | 7.7 | 3.1 | 3.8 |
| Bodo saltans LakeKonstanz | 72.3 | 0.8 | 9.2 | 18.5 |
| Trypanosoma cruzi CL Brener Non-Esmeraldo-like | 66.9 | 0 | 3.8 | 29.3 |
| Trypanosoma cruzi CL Brener Esmeraldo-like | 54.6 | 0 | 3.1 | 42.3 |
Species of Trypanosomatids (genome files from TriTrypDB v44).
BUSCO analysis results: (Complete - C (%)), percentage of 130 Euglenozoa universal single copy orthologues identified as full sequence orthologues in the listed genomes. (Complete Duplicated - CD (%)) full sequence found in more than one position in the genome. (Fragmented – F (%)) only partial sequence was found or; (Missing – M(%)) sequence not found.
2.2. Prediction of conserved structures
To predict conserved RNA structures in the L. braziliensis genome, we used MultiZ/roast to generate a multiple alignment of 10 Leishmania species genomes, covering 99.3 % of the L. braziliensis genome. From the multiple alignment we constructed 12,969 alignment windows that overlap with the lncRNA annotation. On these windows, we predicted conserved RNA structures using RNAz [20]. RNAz yielded 631 ncRNA candidates with a support vector machine (SVM) score ≥0, the threshold for classifying a window as having a conserved RNA structure.
Infernal (version 1.1.2) [21] in conjunction with Rfam (version 14.1) [22] predicted 177 known structured RNAs in the L. braziliensis genome. A comparison between the 177 known structures and the RNAz predictions indicated that 226 of the 12,969 alignment windows overlapped with 64 of the known structures by at least 40 bases. We analyzed the 12,969 alignment windows in several ways and determined that the average pairwise sequence identity and the number of organisms/sequences in each alignment window were the most efficient filters (Supplementary Fig. 1 and Supplementary Fig. 2). We filtered the alignment windows by requiring an average pairwise sequence identity ≥70 % and at least 6 organisms in the alignment (if 7 or more valid sequences were present, RNAzWindow automatically selected the 6 best sequences based on optimal pairwise sequence identity). After this filter, we are left with 6035 alignment windows, of which 206 overlap known RNA structures.
To estimate the false discovery rate (FDR) for the RNAz predictions, we shuffled the 6035 alignment windows 100 times each while preserving di-nucleotide composition. The SVM scores from these shuffled windows provided a background distribution from which p-values for the RNAz predictions were derived (Supplementary Fig. 3). Based on the p-values of the 206 windows overlapping known ncRNAs, we selected an SVM score cut-off of 0.5, which corresponds to a p-value ≤0.004 (Supplementary Figs. 3 and 4). To avoid GC bias for individual candidates, we used the 100 shuffled windows for each candidate as its own background, requiring that all shuffled windows exhibit SVM scores lower than that of the candidate. This ensures that the per-candidate FDR is <0.01. Applying the filtering criteria (background p-value ≤0.004 and per candidate FDR <0.01) yielded a final set of 142 candidates, of which 104 are novel and 38 overlap known ncRNAs. In addition, we recovered 19 out of 54 expressed, highly conserved ncRNAs (38 out of 206 windows, see Fig. 1).
Fig. 1.
Filtering of multiple alignments (99.3 % genome coverage) yielded 12,969 windows, predicting 142 structured RNAs (104 novel, 38 overlapping 177 known RNAs from Infernal/Rfam). Bottom panels show genome coverage, expressed ncRNA coverage, and structure predictions.
In the final selection, we focused on candidates located between two CDSs on the same strand, which is true for a good portion of the known ncRNAs (∼55 % within 10,000 and ∼38 % within 5000 bases). We found that 74 of our 104 novel candidates are located between two CDSs (≤5000). The ncRNAs differentially expressed in L. braziliensis morphologies, were chosen to be functionally analyzed. This choice was based on the possibility that these ncRNA play a role in parasite differentiation being essential for life cycle progression and parasite survival, providing novel insights into the mechanisms orchestrating parasite-host interactions.
2.3. Predicted point mutations that can affect the structures of target ncRNAs
Considering its coverage, size, genomic location, and differential expression, the long non-coding RNA LbrM2906_26_lncRNA45 (hereafter lncRNA45) was selected as a candidate for functional characterization. This lncRNA contains a conserved structured region, referred to as Locus 709.
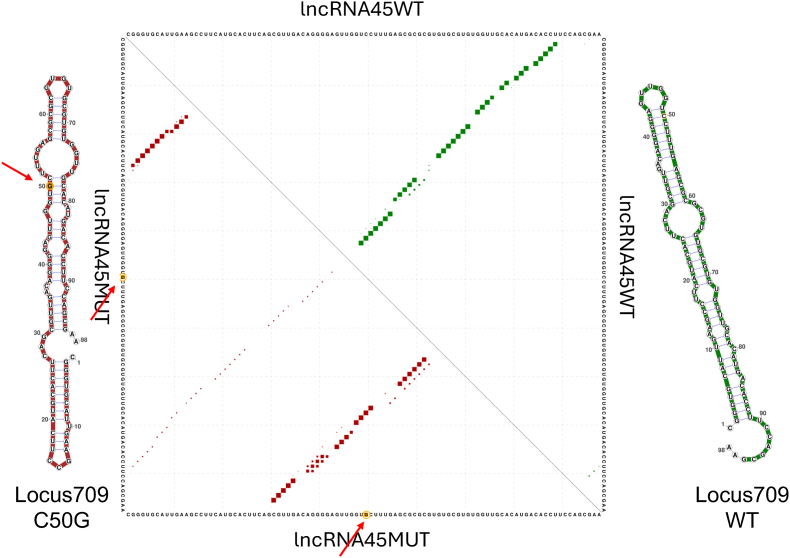
To assess potential mutations affecting RNA secondary structure, stability, or interactions with other macromolecules, in silico predictions were performed using the RNAsnp tool. Among several potential mutations, the cytosine-to-guanine substitution at position 50 (C50G) within Locus 709 was predicted to significantly disrupt the secondary structure of lncRNA45 (p-value = 0.004). Interestingly, although many other single nucleotide polymorphisms (SNPs) resulted in structures markedly different from the wild-type —such as the guanine-to-adenine substitution at position 76 (G76A) (Fig. 2)—it is possible that the functional conformation corresponds to the G76A variant. If so, the C50G mutation might destabilize this structure. However, confirming this hypothesis would require further investigation beyond the scope of the present study.
Fig. 2.
RNAsnp analysis of a C-to-G mutation at position 50: comparison between base-pair probabilities of lncRNA45MUT (dark red) and lncRNA45WT (green); MFE structure of lncRNA45MUT (left – dark red) and lncRNA45 (WT) MFE structure (right – green), and G-to-A mutation at position 76 (bottom right). Red arrows and orange circles indicate the C50G substitution.
2.4. lncRNA45 is processed and is predominantly localized to the cytoplasm of L. braziliensis
lncRNA45 was identified in the L. braziliensis M2903 transcriptome and predicted to be a non-coding RNA (ncRNA) by two predictors—ptRNApred (classified as RNaseP family) and RNAcon (classified as Intron-gp-1) [13]. The predicted size of lncRNA45 is 408 nt, it is located in an intergenic region on chromosome 26 (coordinates: 296,265–296,672) on the minus strand, the same strand sense of the PTU it belongs to (Supplementary Fig. 5A) [13]. LncRNA45 is situated between two coding sequences (CDSs): an upstream gene encoding a putative NMD3 family protein (LbrM2903_260014600.1), and a downstream gene coding for a hypothetical protein (LbrM2903_260014500.1) [23] (Supplementary Fig. 5B). The RNA coverage profile strongly suggests that lncRNA45 is not part of the untranslated regions (UTRs) of either coding sequence (CDS) (Supplementary Fig. 5B). In addition, the RNA-seq coverage profile indicates that lncRNA45 is preferentially expressed in amastigotes (Supplementary Fig. 5A and 5B), showing a 1.53-fold higher abundance in amastigotes compared to other morphological forms. However, RT-qPCR analysis did not confirm the RNA-seq results, showing instead that lncRNA45 is more abundant in promastigotes (PRO) than in other morphological forms (Supplementary Fig. 5C).
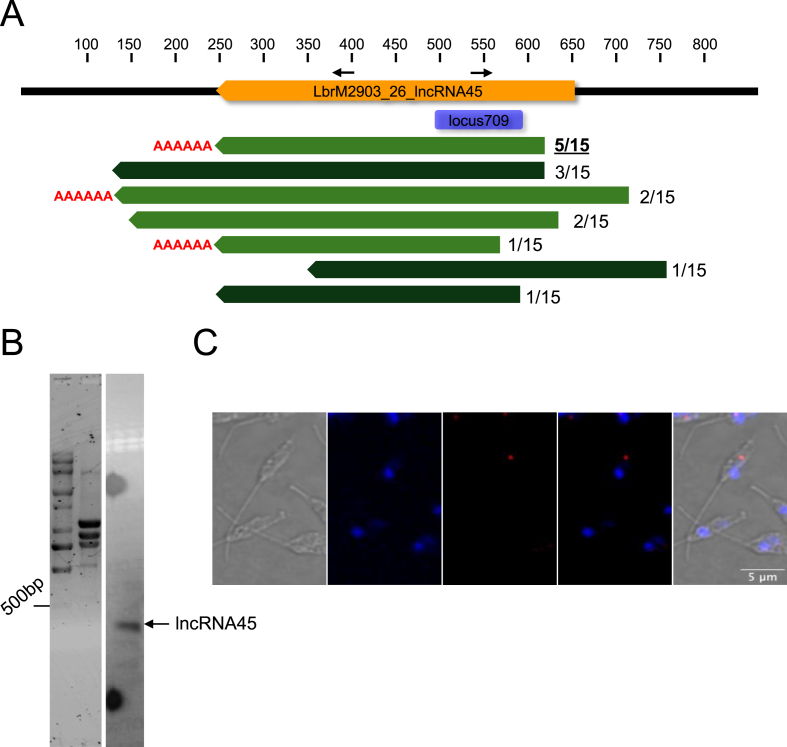
To confirm the size of lncRNA45 and to investigate its processing following polycistronic transcription, we employed an RNA circularization-based protocol [24] (Supplementary Fig. 5D). Total RNA from L. braziliensis M2903 was decapped (TAP+) or not (TAP−) using tobacco acidic phosphatase (TAP), circularized, and reverse transcribed using target-specific primers. Primers directed towards the 5′ and 3’ ends were used to amplify a product containing the transcript ends and its modifications (poly(A) tail and spliced leader, if present). A distinct and specific band was observed in both TAP+ and TAP− reactions (Supplementary Fig. 5E). These bands were cloned into pGEM-T and sequenced. Fifteen clones (ten from TAP+ and five from TAP− reactions) were collected, amplified with M13-F and M13-R primers, and sequenced. Nine clones presenting different fragment sizes after amplification were sequenced and mapped to the locus of lncRNA45 on L. braziliensis M2903 chromosome 26.
While in silico predictions suggested that lncRNA45 is 407 nucleotides long, the circularization assay revealed seven distinct transcript profiles ranging from 325 to 585 bp (Fig. 3A). Among these, four cDNA clones lacked a poly(A) tail in the 3′ UTR, whereas three clones possessed a poly(A) tail consisting of 14–17 adenines (Fig. 3A). No spliced leader sequence was observed in the 5’ UTR of lncRNA45, suggesting that its capping may occur via a mechanism distinct from spliced leader-mediated capping. Interestingly, three different transcript sizes lacking polyadenylation were identified in the TAP- reactions, whereas three out of four clones sequenced from the TAP + reactions were polyadenylated. The most abundant transcript in TAP + samples (5 out of 10 clones) was 376 nucleotides long and had a 13-nucleotide poly(A) tail (indicated by the black arrow in Fig. 3A), corresponding to the region of highest coverage observed for lncRNA45 (Fig. 3A). Notably, except for lncRNA45_TAP+_Clone1, all other identified lncRNA45 isoforms contain the conserved structured Locus 709 identified in comparative analysis.
Fig. 3.
LncRNA45 is a 407-nt, structured and polyadenylated long noncoding RNA, observed in the cytoplasm of Leishmania braziliensis. (A) RNA circularization assay confirms polyadenylation and transcript processing. Total RNA was treated with (TAP+, light green) or without (TAP−, dark green) tobacco acid pyrophosphatase, circularized, reverse-transcribed, PCR-amplified, and sequenced. Among 15 sequenced clones, 7 distinct transcript sizes were identified. Poly(A) tails were found in 8 out of 15 clones (3 transcript profiles), all derived from TAP + RNA. The most frequently observed transcript profile is highlighted (bold and underlined). Positioning of the structured locus 709 is depicted in purple. Orange bar indicates the computationally predicted ncRNA45. Slim black arrows above the orange bar indicates positioning of primers used in the circularization assay. (B) Northern blot analysis detected a single transcript <500 nt corresponding to lncRNA45 in total RNA from promastigotes. White lines observed in the upper portion of the blot are photographic artifacts. Sybr Green staining shows RNA marker and rRNA bands as loading control. (C) RNA FISH reveals lncRNA45 (red), is in the cytoplasm of L. braziliensis. Parasites' nuclei were stained with DAPI (blue). Images acquired at 100 × magnification using a Zeiss multiphoton microscope.
The existence of lncRNA45 as an independent transcript in L. braziliensis M2903 total RNA extract was also confirmed by northern blotting using sequence-specific radiolabeled probes (Fig. 3B). Northern blotting revealed a specific, unique band smaller than 500 bp for lncRNA45, confirming the presence of this ncRNA in L. braziliensis and consistent with its expected size of 408 nucleotides.
The localization of lncRNA45 was determined using RNA fluorescence in situ hybridization (FISH) with probes designated to bind along its entire length. FISH results revealed that lncRNA45 was predominantly observed in the cytoplasm of L. braziliensis (Fig. 3C and Supplementary Fig. 6). In some cells, multiple fluorescence signals were observed; however, due to experimental limitations, we could not determine whether each signal corresponded to a single lncRNA45 molecule or to multiple copies.
2.5. Knockout of lncRNA45 results in reduced parasite density in culture
To investigate the biological function of lncRNA45, L. braziliensis M2903 cell lines either knocked out (ΔlncRNA45) for lncRNA45 or overexpressing (OE_lncRNA45) this transcript, were generated. Using CRISPR/Cas9, null mutants lacking lncRNA45 were successfully generated, and the complete loss of lncRNA45 was confirmed by PCR (Supplementary Fig. 7A and Supplementary Fig. 7E – PCR A). L. braziliensis M2903 overexpressing lncRNA45 (OE_lncRNA45) was obtained by transfecting the parasites with the pSSU-lncRNA45-Sat plasmid, followed by selection with nourseothricin (Supplementary Fig. 7B and Supplementary Fig. 7F – PCR B). As a mock control, parasites were transfected with the pSSU-GFP-Sat plasmid (OE_mock) (Supplementary Fig. 7C and Supplementary Fig. 7F – PCR C).
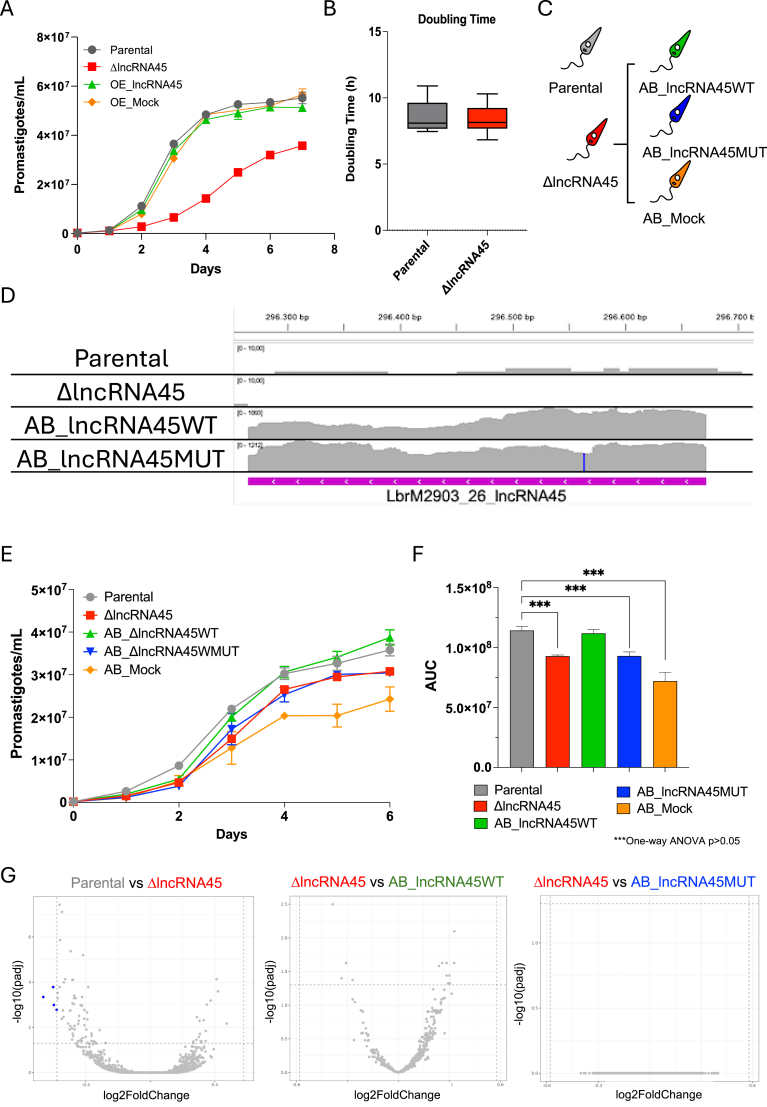
To investigate whether parasite fitness was impaired, these transfectants (ΔlncRNA45, OE_lncRNA45, and OE_mock) along with the parental cell line, were subjected to assays mimicking key steps of the Leishmania life cycle. Promastigote growth was assessed by adjusting the culture to 2 × 105 promastigotes/mL and counting cells for at least 7 days or until reaching the stationary phase. Area under the curve analysis showed significantly reduced growth for ΔlncRNA45, with a cell density of 3.6 × 107 promastigotes/mL on the 7th day of culture, compared to 5.5 × 107 promastigotes/mL in the parental cell line (Fig. 4A). No significant differences were observed for OE_lncRNA45 or OE_mock, compared to the parental cell line (One-way ANOVA and Tukey's multiple-comparison test, p < 0.05).
Fig. 4.
Impact of lncRNA45 knockout on L. braziliensis fitness. (A) Growth curves of L. braziliensis M2903 OE_lncRNA45 (green), OE_Mock (orange), and parental cells (gray). ΔlncRNA45 cells exhibited reduced growth compared to the parental line. The assay was performed with three biological replicates, and similar results were obtained in at least three independent experiments. Due to the low variability among replicates, a representative experiment is shown. Area Under the Curve (AUC) analysis was performed using one-way ANOVA followed by Tukey's test (p < 0.05). (B) Doubling time of ΔlncRNA45 vs. parental cells (non-parametric t-test, p < 0.05). (C) ΔlncRNA45 was complemented with lncRNA45WT (green) sequence, the sequence carrying the C50G mutation (lncRNA45MUT - blue), and the GFP-coding gene (AB-Mock - orange) using pSSU-SAT plasmid. (D) IGV visualization of transcriptomic data from Parental, ΔlncRNA45, AB_lncRNA45WT, and AB_lncRNA45MUT cell lines. Integrated Genomics Viewer (IGV) tracks show normalized RNA-seq read coverage (gray pileups) across the lncRNA45 genomic region. The ΔlncRNA45 track displays a complete absence of reads, confirming successful knockout of lncRNA45. In contrast, both AB_lncRNA45WT and AB_lncRNA45MUT show restored expression of the transcript. The blue vertical line over RNA-reads map of AB_lncRNA45MUT represents the position of the substituted nucleotide. The pink bar corresponds to the annotation of lncRNA45. (E) Growth curves of L. braziliensis ΔlncRNA45 and add-back cell lines over a 7-day period. The ΔlncRNA45 line consistently exhibited reduced growth compared to the parental strain. (F) Quantification of growth by area under the curve (AUC) analysis. Statistical significance was assessed using one-way ANOVA followed by Tukey's post hoc test (p < 0.05). Data represent the mean of three biological replicates. (G) Transcriptomic analysis of lncRNA45 knockout and add-back cell lines. Volcano plots of differential expression (DESeq2, p < 0.05, FC > 1.5x) analysis between Parental vs. knockout (ΔlncRNA45), knockout vs. add-back complemented with the WT sequence of lncRNA45 (AB_lncRNA45WT), and knockout vs. add-back complemented with the lncRNA45 sequence containing a C50G substitution at Locus 709 (AB_lncRNA45C50G). Blue dots represent genes with significant differential expression, while gray dots indicate non-significant differences.
To determine if the reduced cell density observed after 7 days for the ΔlncRNA45 cell line was due to reduced replication capacity, doubling time was measured by adjusting the culture to 1 × 106 promastigotes/mL and counting after 24 h for 4 days. No significant differences in doubling time were observed for ΔlncRNA45 compared to the parental cell line (Fig. 4B) (non-parametric t-test, p < 0.05).
When inside the vector, procyclic promastigotes migrate to the anterior portion of the phlebotomine gut and face an environment with reduced nutrient availability. The ability to withstand nutritional stress was evaluated for ΔlncRNA45 and OE_lncRNA45 by incubating these cells in PBS for 4 h, followed by recovery in M199 for 24 h. The percentage of viable cells after stress was determined relative to non-stressed cells and compared to the parental line. No significant differences were observed in the recovery capacity from nutritional stress for any of the transfectant lines compared to the parental cell line (Supplementary Fig. 8A) (One-way ANOVA and Tukey's multiple-comparison test, p < 0.05).
To infect the mammalian host, procyclic promastigotes need to differentiate into infective metacyclic promastigotes. Since procyclic and metacyclic promastigotes differ in size and density, using a Ficoll gradient, we determined the percentage of metacyclic promastigotes in relation to procyclic forms for ΔlncRNA45, OE_lncRNA45, as well as the parental and OE_mock cell lines. The percentages of metacyclic promastigotes in culture (% of metacyclogenesis) observed for the parental, ΔlncRNA45, OE_lncRNA45, and OE_mock cell lines were 5.7 % ± 1.7 (mean ± SEM), 6.3 % ± 1.5, 5.3 % ± 1.9, and 5.3 % ± 0.6, respectively, with no significant differences observed between the cell lines (Supplementary Fig. 8B) (One-way ANOVA and Tukey's multiple-comparison test, p < 0.05).
The capacity to infect THP-1-derived macrophages and the replication of intracellular amastigotes were also assessed for ΔlncRNA45, OE_lncRNA45, and OE_mock in comparison to the parental cell line but no significant differences were observed in the percentage of infected macrophages (Supplementary Fig. 8C) or in the number of amastigotes per macrophage (Supplementary Fig. 8D) were observed up to 96 h post-infection for any transfectant compared to the parental cell line (One-way ANOVA and Tukey's multiple-comparison test, p < 0.05).
Once inside the macrophages, Leishmania parasites are exposed to reactive oxygen species, which they must survive to replicate and establish infection. The ability to recover from oxidative stress induced by hydrogen peroxide (H2O2) was evaluated for ΔlncRNA45, OE_lncRNA45, and OE_mock in comparison to the parental cell line. The percentage of recovery was determined in comparison to non-stressed parasites, and no significant differences were observed for the transfectant lines compared to the parental cell line (Supplementary Fig. 8E).
2.6. Complementation of ΔlncRNA45 with lncRNA45WT restores promastigote growth
To investigate whether the biological function of lncRNA45 is related to the conserved secondary structure at Locus709 the wild-type (lncRNA45WT) and mutant (lncRNA45MUT, carrying the C50G substitution at Locus 709) sequences of lncRNA45 were cloned into the pSSU-Sat plasmid, the same vector used to overexpress this lncRNA in OE_lncRNA45 transfectants. After cloning, the plasmids were sequenced to ensure C50G was the only SNP present in the sequence and transfected into the L. braziliensis M2903 ΔlncRNA45 cell line. The plasmid carrying the GFP gene instead of the lncRNA45 sequence (pSSU-Mock) was also transfected into ΔlncRNA45 as a control. Thus, three clonal lines were generated over the L. braziliensis M2903 ΔlncRNA45 background (Fig. 4C).
The absence of endogenous lncRNA45 was confirmed by PCR in all the transfectants (Supplementary Fig. 7G - PCR A). The presence of the plasmid containing lncRNA45WT or lncRNA45MUT was confirmed in the add-back cell lines AB_lncRNA45WT and AB_lncRNA45MUT, respectively (Supplementary Fig. 7G - PCR B). The presence of the plasmid pSSU-GFP was confirmed in the AB_Mock cell line (Supplementary Fig. 7G - PCR C). Amplification of a 0.32 kb fragment of hsp70 CDS was used as control for DNA presence and integrity (Supplementary Fig. 7G - PCR D). RNA-seq data analysis also confirmed the absence of reads mapping to lncRNA45 in the ΔlncRNA45 cell line, proving that this lncRNA was indeed deleted (Fig. 4D). The RNA-seq data further confirmed that expression of lncRNA45 was re-established in the add-back and that C50G is the only nucleotide substitution in the lncRNA45MUT sequence (Fig. 4D – blue line).
A 7-day growth curve was generated, as described previously. As observed before, reduced growth was seen in ΔlncRNA45 compared to the parental cell line (Fig. 4A and 4E–F). The add-back of the lncRNA45WT sequence restored the growth of ΔlncRNA45 to the levels of the parental cell line (Fig. 4E–F). Supporting the significance of Locus 709 and requirement of lncRNA45, neither the lncRNA45MUT nor mock add-back control (AB_Mock) could restore ΔlncRNA45 growth to the levels of the parental line (Fig. 4E–F).
2.7. lncRNA45 does not regulate other mRNA levels
To investigate if lncRNA45 regulates the abundance of other transcripts in L. braziliensis, RNA-seq was conducted for the parental, ΔlncRNA45, AB_lncRNA45WT, and AB_lncRNA45MUT cell lines. Only two genes were found to be differentially expressed in the knockout (ΔlncRNA45) compared to the parental cell line (DESeq2 p < 0.05, FC > 1.5) (Fig. 4G). No significant differences in gene expression were observed between the knockout and add-back cell lines, suggesting that lncRNA45 does not regulate global mRNA levels in L. braziliensis.
2.8. lncRNA45-protein interaction is modified in the presence of C50G substitution in locus 709
The fact that lncRNA45 is differentially expressed between morphologies and has a conserved structured locus together with the fact that its deletion from L. braziliensis genome impacts promastigote fitness, strengthened the hypothesis that this lncRNA might be involved in gene expression regulation in this parasite. To investigate which pathways lncRNA45 might regulate, an in vitro pulldown assay was conducted to identify the proteins interacting with this lncRNA.
The pulldown was performed using both the wild-type sequence of lncRNA45 (lncRNA45WT) and a sequence containing the C50G substitution (lncRNA45MUT) aiming to determine if the RNA-protein interaction is dependent on the secondary structure formed by Locus 709. The lncRNA45WT and lncRNA45MUT sequences were fused to a 4xS1m aptamer and sequenced to ensure that C50G was the only divergence between the sequences. Since S1m has an affinity for streptavidin-coated beads (SA-beads), lncRNA45 was immobilized and exposed to log-phase L. braziliensis M2903 promastigote protein lysate. As a control, an S1m sequence without the lncRNA45 fusion was used.
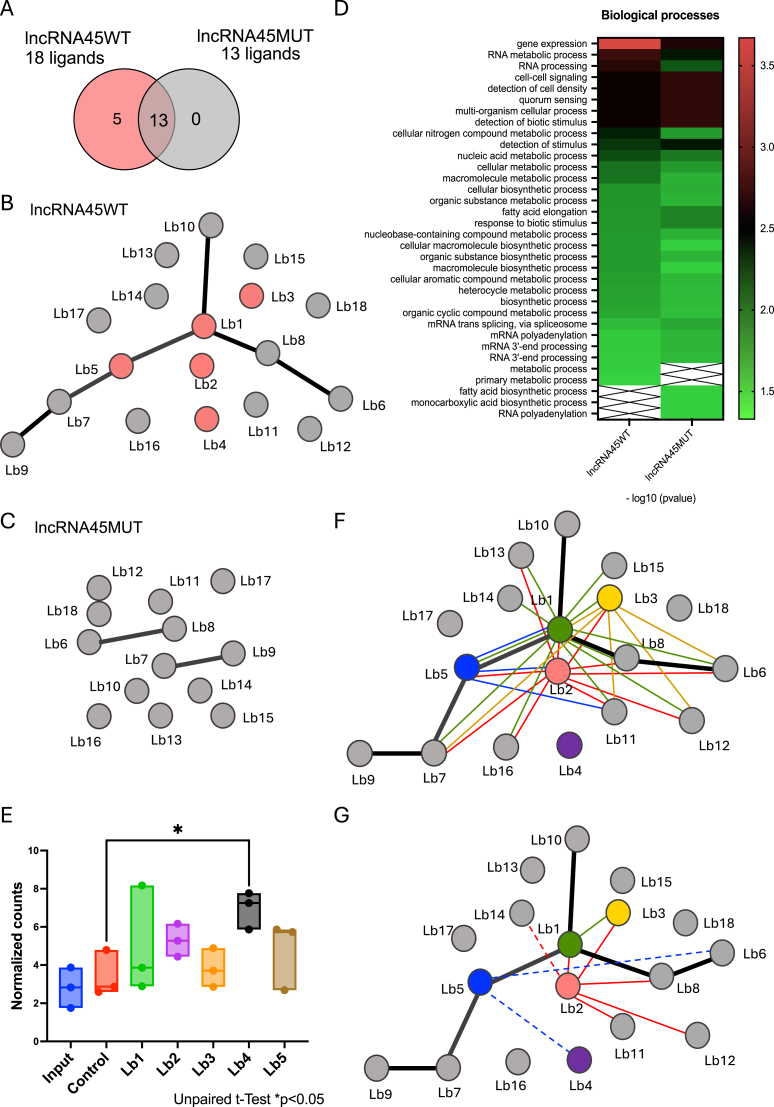
Proteins recovered after filter selection and significantly enriched in all three replicates of lncRNA45WT and lncRNA45MUT compared to the empty control filter were considered binding partners of these sequences. A total of 18 and 13 proteins were identified as binders for lncRNA45WT and lncRNA45MUT, respectively (Table 2, Fig. 5A). Interestingly, the 13 proteins recovered from the lncRNA45MUT pulldown were common to the lncRNA45WT pulldown (Table 2). The other five proteins were lost in the presence of the C50G substitution in the Locus 709 sequence of lncRNA45 (lncRNA45MUT) (Table 2, Fig. 5A).
Table 2.
lncRNA45-protein interactions observed in S1m in vitro pull-down.
| lncRNA45 | ||||||
|---|---|---|---|---|---|---|
| Code | Descriptiona | Accession Numberb | GeneID M2904c | GeneID M2903c | Sized | pValuee |
| Lb1 | 40S ribosomal protein S9, putative | A4H519 | LbrM.07.0750 | LBRM2903_070014500 | 22 kDa | 95 % (0.0029) |
| Lb2 | XRN 5′-3′ exonuclease N-terminus, putative | A4H8G4 | LbrM.16.0400 | LBRM2903_160009500 | 108 kDa | 95 % (0.011) |
| Lb3 | Lupus La protein homolog, putative | A4HBQ1 | LbrM.21.0600 | LBRM2903_210010600 | 37 kDa | 95 % (0.029) |
| Lb4 | hypothetical protein, conserved | A4HGJ6 | LbrM.28.1600 | LBRM2903_280022100 | 80 kDa | 95 % (0.014) |
| Lb5 | Nop53 (60S ribosomal biogenesis), putative | A4HMS1 | LbrM.34.2420 | LBRM2903_340031800 | 38 kDa | 95 % (0.0080) |
| Lb6 | RNA-binding protein, putative | A4HI65 | LbrM.30.1230 | LBRM2903_300017800 | 67 kDa | 95 % (0.014) |
| Lb7 | tRNA (Uracil-5-)-methyltransferase, putative | E9AIL9 | LbrM.20.5870 | LBRM2903_200073900 | 66 kDa | 95 % (0.047) |
| Lb8 | DNA-directed RNA polymerase II subunit 3, putative | A4HH22 | LbrM.29.0150 | LBRM2903_290006800 | 37 kDa | 95 % (0.037) |
| Lb9 | dihydrouridine synthase (Dus), putative | A4H8X8 | LbrM.17.0330 | LBRM2903_170009100 | 66 kDa | 95 % (0.037) |
| Lb10 | Mitochondrial small ribosomal subunit Rsm22, putative | A4HD89 | LbrM.24.0330 | LBRM2903_240008300 | 110 kDa | 95 % (0.031) |
| Lb11 | ISWI complex protein | A4HEU2 | LbrM.26.0880 | LBRM2903_260013600 | 80 kDa | 95 % (0.026) |
| Lb12 | kinetoplast-associated protein, putative | A4HQA1 | LbrM.35.6130 | LBRM2903_350072700 | 14 kDa | 95 % (0.041) |
| Lb13 | acetyl-CoA carboxylase | A4HJT6 | LbrM.31.3340 | LBRM2903_310043300 | 241 kDa | 95 % (0.0053) |
| Lb14 | N-terminal region of Chorein, a TM vesicle-mediated sorter, putative | A4H8K0 | LbrM.16.0770 | LBRM2903_160014400 | 604 kDa | 95 % (0.049) |
| Lb15 | Symplekin tight junction protein C terminal, putative | A4HJL9 | LbrM.31.2640 | LBRM2903_310035900 | 165 kDa | 95 % (0.0028) |
| Lb16 | replication factor C, subunit 3, putative | A4HMZ2 | LbrM.34.3160 | LBRM2903_340040300 | 40 kDa | 95 % (0.011) |
| Lb17 | hypothetical protein, conserved | A4HHV4 | LbrM.30.0100 | LBRM2903_300006000 | 24 kDa | 95 % (0.042) |
| Lb18 | hypothetical protein, conserved | A4HPR3 | LbrM.35.4190 | LBRM2903_350052100 | 51 kDa | 95 % (0.0021) |
Bold labelling refers to proteins specifically binding to lncRNA45WT.
Protein description based on TritrypDB annotation.
Uniprot accession number.
Gene identification in TritrypDB.
Protein size in kilodalton (kDa).
pValue and % of confidence obtained in the statistical analysis. One-Way ANOVA was used to compare S1m results of the negative control, the lncRNA45WT and lncRNA45MUT pull-downs. Unpaired T-test was used to compare S1m results of the negative control with the lncRNA45 pull-down. The differences were considered significant when p < 0.05.
Fig. 5.
In vitro pulldown of lncRNA45WT and lncRNA45MUT sequences. (A) Comparison of proteins binding to lncRNA45WT (18 proteins) and lncRNA45MUT (13 proteins), highlighting five lost interactions due to the C50G substitution at locus 709. Thirteen proteins bind to both variants. (B, C) Protein-protein interaction networks for lncRNA45WT (B) and lncRNA45MUT (C) determined using String. The interactive networks can be accessed for lncRNA45WT (https://www.ndexbio.org/#/network/0175a6c3-274c-11ef-9621-005056ae23aa?accesskey=e4fe291b38d4e92ee1b1585d11e0ca5bf624fdf6e669a3155c711dde39d39089) and for lncRNA45MUT (https://www.ndexbio.org/#/network/d3df8921-274a-11ef-9621-005056ae23aa?accesskey=e505803df77aa75dbf287c55f80c2226fb71bfc82ac437ab93cd47d4975ec1db). (D) Heat map of gene ontology (GO) enrichment analysis of binding proteins. Terms are classified by significance (p < 0.05). (E) RIP analysis confirms lncRNA45-protein interaction, with significant enrichment in Lb4 eluate (p < 0.05). (F) Network analysis of predicted and RIP-identified lncRNA45 binders. Circles represent proteins identified in the S1m assay (Lb1-Lb18), with colored circles indicating RIP-validated interactions. (G) Comparison of proteins in RIP eluates with and without RNase treatment. Dotted lines indicate indirect RNA-dependent interactions absent in negative control comparisons.
Protein interaction network analysis was performed for the interactors of lncRNA45WT (Fig. 5B) and lncRNA45MUT (Fig. 5C). A network consisting of six proteins was identified when the binding partners of lncRNA45WT were analyzed (Fig. 5B). However, this network was disrupted when the interactors of lncRNA45MUT were imputed (Fig. 5C). Notably, two of the five proteins that exclusively bound to the native lncRNA45 sequence (LbrM.07.0750 and LbrM.34.2420) are components of this network (Fig. 5A).
To identify the pathways lncRNA45 might be involved in, the list of binding partners for lncRNA45WT and lncRNA45MUT sequences was submitted to gene ontology (GO) enrichment analysis (Fig. 5D). Twelve and seventeen terms were retrieved for lncRNA45WT and lncRNA45MUT, respectively. Eleven of these terms were shared between both protein lists. RNA processing (GO:0006396) was exclusively enriched in lncRNA45WT, while six terms were exclusively enriched in lncRNA45MUT (Fig. 5D).
Since genome annotation can significantly impact GO analysis (56), we identified the orthologs of the L. braziliensis genes retrieved in the pulldown analysis within the L. major Friedlin genome, which has a more complete annotation to date, to compare the outcomes of this analysis (Supplementary Fig. 9). In this case, 13 and 20 terms were retrieved for lncRNA45WT and lncRNA45MUT, respectively, with 12 terms shared between both protein lists. Consistently, RNA processing (GO:0006396) was exclusively enriched in lncRNA45WT.
RNA/Protein immunoprecipitation (RIP:IP) was conducted to confirm in vivo relevance of lncRNA45-protein interactions identified in the S1m in vitro pulldown assay. The five proteins that interact only with lncRNA45WT and not with lncRNA45MUT were tagged with HA at the N-terminus (Supplementary Fig. 10) and immunoprecipitated using anti-HA magnetic beads. The L. braziliensis M2903 parental cell line (untagged) was used as the negative control.
RNA that immunoprecipitated with the tagged candidate interacting proteins was extracted, rRNA-cleared and sequenced to confirm interaction with lncRNA45 and identify other potential transcripts in the complex. Surprisingly, lncRNA45 was enriched only in the eluate of the hypothetical protein (Lb4 - LBRM2903_280022100). A higher number of counts of lncRNA45 transcript was also observed for XRN 5′-3′ exonuclease N-terminus (Lb2 - LBRM2903_160009500) in comparison to the negative control, though this difference was not statistically significant (Fig. 5E).
In the same assay, protein-protein interactions were investigated by mass spectrometry to evaluate potential networks involving lncRNA45. Interestingly, four out of the five analyzed proteins were found to interact with at least one of the 18 proteins identified as lncRNA45WT-binding in the S1m pulldown assay (Fig. 5F), confirming some of the interactions predicted by STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) (57).
To determine if these protein-protein interactions are RNA-dependent, a fraction of the eluate material was treated with RNAse and compared with the untreated eluate. Our findings show that the interaction of Lb1 (40S ribosomal protein) with Lb3 (Lupus La protein homolog) is lost after RNase treatment (Fig. 5G). The same is true for the interaction of Lb2 with Lb1, Lb3, Lb8, Lb11, and Lb12 (Fig. 5G). Three other protein-protein interactions (Lb5-Lb4, Lb5-Lb6, Lb2-Lb14) that were lost in RNase-treated compared to untreated samples were not significantly different from the negative control and may thus be artifacts (Fig. 5G). Notably, none of these proteins were detected in the negative controls, confirming their specific interaction with identified targets. This supports the significance of lncRNA45 in regulatory protein complex interactions and functional cohesion.
3. Discussion
In this study, we used innovative in silico methodologies to classify putative ncRNAs of Leishmania based on the evolutionary conservation of their structure. Using reverse genetics, we identified a novel regulatory ncRNA, lncRNA45, with a conserved secondary structure. Our findings demonstrate that this lncRNA impact cellular fitness and that its structure is essential for its function.
Our applied computational approach integrated two strategies for genome-wide screenings of structured RNAs: simultaneous use of sequence and structural alignment along with evaluation of evolutionary conservation of secondary structure. To identify RNA families with shared structural features, we applied clustering methods based on conserved RNA structures. This structure-based RNA clustering conferred a rational basis for selecting ncRNAs for further investigation using reverse genetics. Among these, we analyzed the functional impact of a SNP that modifies the secondary structure of lncRNA45.
It is well established that gene expression modulation occurs throughout the parasite lifecycle and differences in RNA abundance between clinical isolates or under distinct environmental conditions have been demonstrated for Leishmania and other trypanosomatids [4,7,25]. How these parasites modulate gene expression in a context of polycistronic transcription and lack of canonical promoters, or the factors and mechanisms used to regulate gene expression are still not completely understood [4]. Only recently have studies began to suggest the existence of ncRNAs in trypanosomatids. Interestingly, computational and functional assays have demonstrated that among trypanosomatids only Trypanosoma brucei and Leishmania Viannia species possess a functional RNAi machinery [26]. But so far functional exploration and discovery of putative regulatory ncRNAs in T. brucei and Leishmania spp. is still incipient [12,27]. The recent identification of ncRNAs in trypanosomatids [8,9,[12], [13], [14]] introduced a new layer to gene expression regulation, complementing the more extensively characterized RNA-binding proteins [7,28]. Until recently, the biological and regulatory functions of ncRNAs in trypanosomatids remained unclear. However, a study published in 2022 [9], demonstrated that overexpression of the lncRNA Grumpy promotes T. brucei differentiation into stumpy forms, enhancing survival rates in infected mice.
While lncRNAs typically show limited sequence conservation across different organisms, specific structures regions within these transcripts—some even syntenically conserved—have been linked to both transcript stability and biological function [29]. To identify such conserved structured elements within trypanosomatid genomes, we performed a computational analysis at uncovering evolutionary conservation of RNA secondary structures. From this analysis, we selected one candidate, lncRNA45, for functional characterization. Here, we report, for the first time, both the in silico and experimental characterization of a structured lncRNA in L. braziliensis.
LncRNA45 was identified in a comprehensive transcriptomic study aimed at deciphering gene expression regulation mechanisms in L. braziliensis [13]. This lncRNA was selected from among several candidates with a conserved secondary structure, because the RNA-seq analysis previously conducted indicated that it was differentially expressed throughout the lifecycle and is preferentially expressed in the intracellular, proliferative form that infects mammalian macrophages [13]. LncRNA45 contains a structured motif (Locus 709), conserved in 6 Leishmania species (L. braziliensis, L. panamensis, L. major Friedlin, L. donovani, L. infantum and L. amazonensis) suggestive that this lncRNA structure may have a conserved essential function throughout Leishmania. In contrast, the full lncRNA45 sequence is conserved only in four Leishmania species, all belonging to the Viannia subgenus.
The presence of lncRNA45 in L. braziliensis was confirmed by multiple experimental approaches including, RNA-seq, RT-qPCR, RNA FISH, circularization assay and northern blotting, confirming that this transcript is consistently expressed and not an artifact of the RNA-seq data [13]. Characterization of lncRNA45 shows it does not undergo the same processing routes as mRNAs in L. braziliensis, a hallmark feature of lncRNAs in diverse organisms [30], and one that appears to be shared by ncRNAs in Leishmania as well [8]. LncRNA45 has a short (17 nt) poly (A) tail but lacks a 5′ spliced-leader sequence, a characteristic of mature mRNAs in Leishmania. Treatment with a decapping enzyme (TAP) suggests that the 5′end of lncRNA45 is protected and that the transcript is not monophosphorylated. Further studies must be conducted to elucidate whether lncRNA45 undergoes alternative capping and to characterize it biogenesis, maturation and stabilization mechanisms, factors that are likely critical for its protection from degradation and its functional activity in the parasite [31].
Loss-of-function analysis showed that lncRNA45 plays a direct role in parasite growth in culture, consistent with its increased expression during this life stage. Interestingly, the reduced cell density observed in the knockout line, compared to the parental strain was not due to an increased doubling time. This suggests that the growth defect may stem from an increased death rate or disruption of a signaling pathway, potentially affecting the parasite's quorum sensing capacity. The hypothesis that these mutants might be more susceptible to nutritional stress was ruled out, as no differences in their ability to recover from nutritional stress were observed.
To determine whether this biological role is structure-dependent, we used in silico predictions to design a single nucleotide substitution in Locus 709 (C50G) which was expected to disrupt its secondary structure. The inability of the modified lncRNA45 (lncRNA45MUT) to rescue the growth phenotype of the knockout background—unlike the wild-type sequence (lncRNA45WT)—confirmed that the structural integrity of Locus 709 is essential for lncRNA45 function. This observation is consistent with findings from other structured lncRNAs, such as lnc-31 and Braveheart, where point mutations compromise critical molecular interactions, including proteins or RNA scaffolding, thereby impairing their function [16,32].
The functional mechanism in which a lncRNA is involved is closely related to its subcellular localization [10]. Cytoplasmic lncRNAs are typically involved in regulating mRNA stability, RNA processing, translation rates or protein function, whereas nuclear lncRNAs act by modulating transcriptional activity [33]. RNA FISH revealed that lncRNA45 was mostly observed in the cytoplasm, without a preferential subcellular distribution, being spotted in different regions of the cytoplasm of L. braziliensis. Additionally, RNA-seq analysis indicated that lncRNA45 does not modulate the expression of specific mRNAs in this parasite. Based on these findings, we hypothesize that lncRNA45 might influence RNA processing, or translation efficiency; directly interacting with proteins to modulate their activity or stability. Similar mechanisms were described for lnc-31 which positively regulate Rock1 protein levels, without affecting its mRNA levels, suggesting a potential positive role of lnc-31 in promoting translation of Rock1 mRNA [32].
In vitro pulldown assays using both lncRNA45WT and lncRNA45MUT sequences as bait confirmed the importance of the secondary structure formed by Locus 709 for lncRNA45 function. The presence of the C50G substitution disrupted the binding of five proteins to this lncRNA, underscoring the structural dependency of these interactions. The identity of the proteins pulled down with lncRNA45 suggests this transcript may be involved in regulating RNA processing or cell-cell signaling pathways, particularly quorum sensing, which corroborates the reduced parasite density phenotype observed in loss-of-function assays.
Although lncRNA-protein interactions were not statistically confirmed for all five proteins in RIP assays, our RNA-seq data revealed higher lncRNA45 read counts in the eluates compared to the negative control for XRN 5′-3′ exonuclease (Lb2) and for two biological replicates of nucleolar protein Nop53 (Lb5). For the hypothetical protein (Lb4), lncRNA45 was significantly enriched in the eluate. It is important to note that lncRNA45 is not an abundant transcript in L. braziliensis M2903, as observed in the parental cell line RNA-seq data. This low abundance may have affected the sensitivity of the statistical analysis, as this limitation was not observed for other more abundant lncRNAs investigated in the same system. Interestingly, the interaction between Lb4 and Lb5 proteins was shown to be RNA-dependent as was the interaction of Lb2 with six additional proteins (Lb1, Lb3, Lb8, Lb11, Lb12 and Lb14). These findings raise the possibility that lncRNA45 may play a scaffolding role, stabilizing the protein complex via RNA-mediated interactions.
An in-depth literature review of the five proteins shown to interact with lncRNA45 in a structure-dependent manner revealed that at least one of them, the Lupus La homolog, has previously been identified as an interactor of two other long non-coding RNA: XIST in mouse and Grumpy in T. brucei. Notably, both Lupus La homolog and the XRN 5′-3′ exonuclease have been implicated in regulating T. brucei growth in loss-of-functions studies demonstrating their impact on parasite growth [34], mirroring the phenotype observed in L. braziliensis lncRNA45 knockout parasites.
Interestingly, three of these proteins (40S ribosomal protein S9, Nop53 and Lupus La) have also been associated with ribosome biogenesis and translation initiation [[35], [36], [37]], supporting the hypothesis that lncRNA45 may be involved in post-transcriptional regulation. RIP-seq MS data not only confirmed these interactions but also expanded lncRNA45-protein network to include 18 putative partners. Importantly, interactions between the Lupus La homolog interaction a both the 40S ribosomal protein and Nop53 were disrupted upon RNAse-treatment of RIP samples, suggesting that lncRNA45 and/or other RNAs are required to mediate or stabilize these protein-protein interactions.
To our knowledge, this is the first demonstration of a secondary structure-dependent lncRNA activity in trypanosomatids, and the first functional characterization of a regulatory ncRNA in Leishmania. This study encourages the search for other conserved, structured regulatory lncRNAs that could serve as potential therapeutic targets or vaccine candidates.
In conclusion, our study underscores the relevance of long non-coding RNAs (lncRNAs) in the regulation of gene expression in trypanosomatids. Through the characterization of lncRNA45 in Leishmania braziliensis, we demonstrated that the conserved secondary structure in this lncRNA is essential for both its interaction with proteins and biological function. Loss-of-function analyses revealed that lncRNA45 plays a crucial role in promastigote growth, highlighting the importance of its secondary structure for its regulatory role.
The protein interaction network identified via pulldown assays and RIP-seq implicates lncRNA45 in RNA processing and cell signaling pathways, notably quorum sensing. Moreover, the interaction of lncRNA45 with proteins as Lupus La homolog, 40S ribosomal protein S9, and Nop53, previously linked to ribosome biogenesis and translation initiation, further supports its involvement in post-transcriptional regulation.
Altogether, our findings provide compelling evidence for the functional relevance of regulatory ncRNAs in Leishmania and demonstrate, for the first time, a lncRNA whose activity is dependent on the secondary structure in trypanosomatids. This study contributes to a deeper understanding of the complexity of RNA-based regulation in these parasites and reinforces the potential of structured lncRNAs as novel molecular targets in the fight against leishmaniasis.
4. Materials and methods
4.1. Quality analysis of trypanosomatids genome
Genomic assembly completeness was assessed using BUSCO v4.0.0 (Benchmarking Universal Single-Copy Orthologs) [38]. BUSCO identifies candidate regions via local alignment against amino acid consensus sequences [39], extracts gene models using block profiles [40], and evaluates them against profile Hidden Markov Models (HMMs) to quantify genome completeness.
BUSCO tool suite (v.4.0.0) metrics were used in genome mode with the euglenozoan_odb10 dataset (creation date: 2019-11-21; 31 species; 130 universal orthologs) to assess the quality and completeness of the assemblies of Trypanosomatids genomes. The analysis was performed in genome mode, and Leishmania tarentolae was selected as the AUGUSTUS parameter, as it is the only Trypanosomatid species with an available training annotation file.
4.2. Genome alignment and search for conserved regions
Lastz (v1.04.03) [18] was used to align ten trypanosomatid genomes to Leishmania braziliensis MHOM/BR/75/M2903, covering 99 % of its genome. After filtering for positive scores, ≥3 sequences per alignment, and a minimum alignment size of 20, coverage was reduced to 92 %. Alignment windows of 40–120 nt (step size: 40, ≥3 sequences) further filtered coverage to 77 %. Overlapping lncRNA annotations yielded 12,969 windows, analyzed by RNAz on both strands. False-discovery rates were estimated by shuffling windows 100 × with SISSIz [41] while preserving di-nucleotide content.
4.3. Predictions of mutations that affect the RNA structure
RNAsnp tool with parameters Model1 p.value 0.01, Model2 p.value 0.01, winsize 200, winsize extension 200 [42,43] were used to predict point mutations that can affect the stability of RNA secondary structures.
4.4. Cell lines culture
L. braziliensis M2903 expressing Cas9 and T7 RNA polymerase from pTB007 plasmid, and tdTomato in the SSU locus was used [44,45]. Promastigotes were cultured at 25 °C in M199 medium supplemented with 2.2 g/L NaHCO3, 0.1 mM adenine hemi sulfate, 0.005 % haemin, 40 mM HEPES pH 7.4, 100 μg/mL penicillin/streptomycin (Pen/Strep), 1 μM Biopterin and 10 % heat-inactivated FBS 10 %. The selection drugs were added at 32 μg/mL hygromycin B (Gibco™), 50 μg/mL Nourseothricin (Sigma-Aldrich®), and 20 μg/mL puromycin dihydrochloride (Gibco™).
Axenic amastigotes were obtained by incubating metacyclic promastigotes in 100 % FBS at 33 °C with 5 % CO2. THP-1 cells for infectivity assays were maintained in RPMI + 10 % FBS at 37 °C with 5 % CO2, passaged every 3 days or when cell density was close to 106 cell/mL.
4.5. DNA extraction
L. braziliensis total DNA extraction for cloning and sequencing purposes was done using DNeasy Blood & Tissue Kits (Qiagen) according to the manufacturer's specifications. For colony screening the protocol described by Rotureau, Gego and Carme [46] was employed.
4.6. RNA extraction
Total RNA was extracted from L. braziliensis cultures using the Directzol kit Quick-RNA Miniprep (Zymo Research) according to the manufacturer's instructions. After extraction, RNA samples were treated with TURBO™ DNAse to remove any DNA remains.
4.7. Real-time quantitative PCR (RT-qPCR)
Differential expression was assessed by RT-qPCR using primers specific for lncRNA45 (lncRNA45_RT-F and lncRNA45_RT-R). The 7SL RNA gene served as a normalizer, amplified with primers 7SL_RT-F and 7SL_RT-R (Supplementary File 1). RT-qPCR was performed using SYBR Green/ROX qPCR MasterMix, on a StepOnePlus™ Real-Time PCR System (95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 60 s, and 72 °C for 20 s). Ct values were normalized to 7SL RNA, and differential expression was calculated relative to the procyclic promastigote stage using the 2ΔΔCt method [47].
4.8. RNA circularization
The RNA circularization assay was performed as described by Hang, Deng, Liu, Mo and Cao [24], with modifications. Total RNA (5 μg) from L. braziliensis M2903 was either treated (TAP+) or not (TAP−) with Tobacco Acid Pyrophosphatase (FirstChoice® RLM-RACE - Invitrogen™). T4 RNA ligase (NEB), ATP, and RNasin® (Promega) were added for circularization. The resulting circular RNA (cRNA) was precipitated and reverse transcribed using lncRNA45_cRT primer and TransScript® II First-Strand cDNA Synthesis SuperMix kit (TransGen Biotech). PCR amplification was conducted using Platinum™ Taq DNA Polymerase High Fidelity (Invitrogen™) and the primers lncRNA45_cF1 and lncRNA45_cR1 followed by a nested PCR using primers lncRNA45_cF2 and lncRNA45_cR2 primers (Supplementary File 1). PCR products were cloned into pGEM®-T Easy Vector (Promega) for transformation in E. coli DH5-α. Positive clones were confirmed by PCR with M13 primers and analyzed by Sanger sequencing. Sequences were mapped to the L. braziliensis M2903 genome using Geneious Software.
4.9. Northern blotting
Total RNA was extracted from log-phase parasites using the mirVana™ Total RNA Isolation Kit (ThermoFisher Scientific) and treated with Turbo DNase. 4 μg of total RNA was mixed with glyoxal mix, denatured at 50 °C for 40 min and electrophoresed on a 2 % agarose gel stained with SYBR Green II. The gel was imaged on an iBright imager (Invitrogen™) after running at 150 V for 90 min. RNA was transferred to a Hybond®-N+ membrane by capillarity in SSC buffer overnight, UV cross-linked (20,000 μJ/cm2, 1 min), and dried at 80 °C for 1 h. For radioactive probing, a PCR-generated amplicon from the ncRNA target cloned into pJET II was labeled using the DIG Northern Starter Kit (Sigma). Hybridization occurred at 42 °C for 18 h in a buffer containing tetrasodium pyrophosphate, SSC, Denhardt's solution, SDS, and heparin, followed by washes with 0.1 % SDS in 2× SSC. The membrane was exposed to photographic film at −80 °C for seven days, developed, and imaged with a conventional camera.
4.10. RNA FISH
Approximately 1 × 107 cells were centrifuged at 2000×g for 10 min, washed once with PBS and resuspended in a solution of 4 % formaldehyde in PBS. Cells were spread in a 0.01 % poly-lysine-coated slide (Sigma-Aldrich®) for 30 min and washed once with PBS. Hybridization of Type I probes, labeled with Alexa Fluor 546 fluorochrome (Thermo Fisher™) was conducted using ViewRNA™ ISH Cell Assay Kit (Invitrogen™) according to the manufacturer instructions and the protocol described by Kramer [48]. Cells were imaged in a LSM 780/LSM 7 MP Carl Zeiss multiphoton microscope with 0.75 magnification.
4.11. Generation of transfectant line
Transfections. L. braziliensis M2903 Cas9/T7 promastigotes were transfected using Amaxa™ Nucleofactor™ (Lonza) X-001 program in Tb-BSF 1× buffer. After 16 h in M199 medium, cells were cloned on M199-agar plates with biopterin (1 μM) and selection drug. Genomic modifications were confirmed by PCR.
Knockout (ΔlncRNA45). A lncRNA45 knockout (ΔlncRNA45) line was generated using CRISPR/Cas9, following the method from Ref. [44]. sgRNAs were designed using EuPaGDT [49], targeting sequences near the 5′ and 3′ ends of lncRNA45. Homology arms (30 nt) flanking the Cas9 cleavage site were included in the donor DNA. Transfected clones were screened by PCR using primers Conf-F and Conf-R. Homozygous knockouts showed only the donor DNA band, while heterozygous clones displayed both lncRNA45 and donor DNA bands.
Overexpression (OE-lncRNA45). For overexpression, lncRNA45 was amplified from L. braziliensis M2903 genomic DNA using OE_lncRNA45_NotI-F and OE_lncRNA45_BamHI-R primers. The fragment was cloned into pSSU-Sat to generate pSSU-OElncRNA45-Sat. Transfected clones were screened by PCR using lncRNA45-F and Sat-R primers. A control plasmid (pSSU-GFP-Sat) was verified using GFP-F and Sat-R primers.
Add-back. Wild-type (lncRNA45WT) and mutant (lncRNA45MUT) sequences were synthesized and amplified using lncRNA45_NotI-F and lncRNA45_BamHI-R. The fragments were cloned into pSSU-Sat, generating pSSU-lncRNA45WT-Sat and pSSU-lncRNA45MUT-Sat, and transfected into ΔlncRNA45 promastigotes. Verification was done by PCR using lncRNA45-F and Sat-R, while pSSU-GFP-Sat was used as a mock control.
HA-tagged cell line. Target proteins were HA-tagged using CRISPR/Cas9 as described in Refs. [44,50]. sgRNA and donor DNA primers were designed using LeishGEdit, which also provided primers for confirmation. The complete list of primers used in this study is provided in Supplementary File 1.
4.12. Phenotypic assays
Growth Curve. Parental and transfectant cell lines' growth rates were compared using AUC analysis over a 7-day growth curve. Cultures of 2 × 105 promastigotes/mL in M199 medium at 25 °C were counted daily for 7 days. Biological triplicates were analyzed using GraphPad Prism 8.0.2. Significance was assessed via non-parametric t-test (two groups) or One-Way ANOVA with Tukey's test (multiple groups).
Doubling Time. Doubling time (DT) was measured by adjusting culture density to 1 × 106 promastigotes/mL in M199, incubating at 25 °C, and counting parasites daily for 4 days. DT was calculated at the end.
Nutritional Stress. Recovery from nutritional stress was assessed by incubating 1 × 106 promastigotes in M199 (unstressed) or PBS (stressed) for 4 h. After centrifugation, fresh M199 with MTT was added, and after 24 h, formazan crystals were dissolved with DMSO. Absorbance at 570 nm was measured to calculate cell viability. Experiments were performed in triplicate with five technical replicates. Data were analyzed using One-Way ANOVA and Tukey's test.
Metacyclogenesis. Promastigotes were cultured for 5 days at 25 °C, centrifuged, and resuspended in M199. A Ficoll gradient (20 %, 10 %) was used to isolate metacyclic promastigotes. The percentage of metacyclogenesis was calculated and compared to the parental line using a non-parametric t-test.
THP-1 Infection. THP-1 monocytes were differentiated into macrophages using PMA and infected with metacyclic promastigotes (10:1 ratio). Infection rates and amastigotes per macrophage were measured at 24 h and 48 h using the ImageXpress Micro XLS System. Data were analyzed using One-Way ANOVA and Tukey's test.
Oxidative Stress. Axenic amastigotes were cultured in M199 pH 5.4 at 33 °C for 72 h, then exposed to 300 mM H2O2 (stressed) or no H2O2 (non-stressed) for 24 h. MTT was added, and after 24 h, formazan crystals were dissolved with DMSO. Absorbance at 570 nm was measured to calculate viability. Experiments were performed in triplicate with five technical replicates. Data were analyzed using One-Way ANOVA and Tukey's test.
4.13. S1m-mediated RNA pull-down
LncRNA45 (native/mutated) was amplified, digested (PacI, NcoI), and cloned into pUC57-T7-4xS1m. RNA was transcribed in vitro (MEGAscript™ T7), immobilized on streptavidin-coated magnetic beads (SA-beads), and incubated overnight with L. braziliensis M2903 protein lysate, pre-cleared with SA-beads at 4 °C. After washing, bound proteins were eluted, boiled in Laemmli buffer, and analyzed by SDS-PAGE. Protein digestion and MS analyses were performed at CHU de Québec. Gel bands were excised, processed, and peptides analyzed by nano-LC-MS/MS (Dionex UltiMate 3000-Orbitrap Fusion). MS data were acquired in data-dependent mode (XCalibur 4.3, 120,000 resolution, 350–1800 m/z). MS/MS spectra were searched against the L. braziliensis Uniprot database (8153 entries) using Mascot 2.5.1, with fixed carbamidomethylation and variable modifications (deamidation, oxidation, methylation/dimethylation). Identifications were validated in Scaffold 5.0.1 (FDR <1 %, ≥2 peptides). Statistical significance was assessed using One-Way ANOVA (p < 0.05).
4.14. Protein/RNA immunoprecipitation (RIP)
Immunoprecipitations were conducted as described previously by Walrad et al. [35,36] with modifications. L. braziliensis promastigotes (1 × 109) were lysed in IP-Lysis Buffer, frozen, sonicated, and centrifuged. Supernatant was incubated with 1 mg of Pierce™ Anti-HA Magnetic Beads (Thermo Scientific™) for 2 h at 4 °C. Negative control beads were blocked with HA peptide. Beads were washed, split, and processed for RNA extraction, protein elution (Laemmli buffer, 95 °C), or RNase treatment. Immunoblotting was performed with anti-HA (1:5000) and anti-rabbit IgG (1:10,000) antibodies, detected using Amersham ECL (Cytiva) on a BioRad ChemiDoc.
Mass Spectrometry Analysis of RIP Samples. SDS-PAGE-separated samples were in-gel digested with trypsin, peptides extracted, and loaded onto EvoSep C18 columns for separation (100 SPD gradient, EvoSep One). MS analysis used timsTOF HT (Bruker) with PASEF-DIA acquisition. Data were analyzed with DIA-NN (1 % FDR) against L. braziliensis TriTrypDB, and quantification was performed using Quant-UMS and KINME (q < 0.01, ≥2 peptides). Differential abundance was assessed with Limma in FragPipe-Analyst (adjusted p < 0.05).
RNA-seq. RNA libraries were prepared (Macrogen, Inc., Illumina Ribo-Zero & TruSeq) and sequenced (∼40M 101 bp paired-end reads). Reads were mapped with bowtie2 (TriTrypDB v66) and counted using htseq-count. Differential expression analysis was performed with DESeq2 (p < 0.05, fold-change >1.5).
4.15. GO analysis
The list of accession numbers was converted into Gene ID on the Uniprot website. A search for the list of IDs was conducted on the TriTrypDB website, and the results were sent to gene ontology enrichment of biological processes. A p-value of 0.05 was used as the cutoff for significance. The retrieved terms, fold-change enrichment, number of genes in the term and p-value were used to generate a heat map using GraphPad Prism 10.
4.16. Interactions networks
Interactions networks for the proteins identified in the pull-down assay for both lncRNA45WT (18 proteins) and lncRNA45MUT (13 proteins) were retrieved from the STRING database [46] using all available evidence types. Cytoscape [51] was used to highlight proteins and interactions shared between the networks. An interactive version of the networks was submitted to the NDEx database [52].
CRediT authorship contribution statement
Caroline R. Espada: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Christian Anthon: Writing – review & editing, Writing – original draft, Methodology, Formal analysis, Data curation, Conceptualization. Rubens D.M. Magalhães: Writing – review & editing, Formal analysis, Data curation, Conceptualization. José Carlos Quilles Junior: Writing – review & editing, Methodology, Investigation, Data curation. Natalia M.M. Teles: Writing – review & editing, Investigation. Fabiano S. Pais: Writing – review & editing, Methodology, Formal analysis. Lissur A. Orsine: Writing – review & editing, Methodology, Investigation, Formal analysis, Data curation. Letícia de Almeida: Writing – review & editing, Formal analysis, Data curation. Tânia P.A. Defina: Writing – review & editing, Methodology. Adam Dowle: Writing – review & editing, Methodology, Formal analysis, Data curation. Jan Gorodkin: Writing – review & editing, Supervision, Formal analysis, Conceptualization. Pegine B. Walrad: Supervision, Funding acquisition, Formal analysis, Conceptualization. Angela K. Cruz: Writing – review & editing, Writing – original draft, Supervision, Resources, Project administration, Funding acquisition, Conceptualization.
Funding
This study was funded by FAPESP (grants 2018/14398-0, 2015/13618-8), CNPq (305775/2013-8, 152584/2022-6), and CAPES (Finance Code 001) to A.K.C. Fellowships were awarded by FAPESP to C.R.E. (2020/00087-2 and 2022/10270-4), R.D.M.M. (2019/18607-5) J.C.Q.J. (2020/00088-9, 2021/15182-3), L.A. (2017/19040-3), and L.A.O. (2021/10043-5, 2023/18057-0) and by the Medical Research Council to N.M.M.T. and P.B.W. InTEGRL (MR/V031511/1). Funding agencies had no role in study design, data collection, analysis, or publication.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Dr. Susanne Kramer for assistance with Northern blotting, Viviane Ambrosio for technical support, and Prof. Michael Plevin for input on Leishmania pull-down assays. Proteomic and RNA analyses were conducted at the Bioscience Technology Facility, University of York. We acknowledge the Microscopy Facility (Roberta Rosales and Elizabete Milani - FMRP-USP, Brazil), the Proteomics Platform (CHU de Québec, Canada), and TriTrypDB. We thank ChatGPT (OpenAI) for assistance with grammatical revision and language improvement.
Footnotes
Peer review under the responsibility of Editorial Board of Non-coding RNA Research.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ncrna.2025.05.010.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Burza S., Croft S.L., Boelaert M. Leishmaniasis. Lancet (London, England) 2018;392:951–970. doi: 10.1016/s0140-6736(18)31204-2. [DOI] [PubMed] [Google Scholar]
- 2.Uliana S.R.B., Trinconi C.T., Coelho A.C. Chemotherapy of leishmaniasis: present challenges. Parasitology. 2018;145:464–480. doi: 10.1017/s0031182016002523. [DOI] [PubMed] [Google Scholar]
- 3.Bates P.A. Revising Leishmania's life cycle. Nat. Microbiol. 2018;3:529–530. doi: 10.1038/s41564-018-0154-2. [DOI] [PubMed] [Google Scholar]
- 4.Clayton C. Regulation of gene expression in trypanosomatids: living with polycistronic transcription. Open Biol. 2019;9 doi: 10.1098/rsob.190072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LeBowitz J.H., Smith H.Q., Rusche L., Beverley S.M. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- 6.Haile S., Papadopoulou B. Developmental regulation of gene expression in trypanosomatid parasitic protozoa. Curr. Opin. Microbiol. 2007;10:569–577. doi: 10.1016/j.mib.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 7.de Pablos L.M., Ferreira T.R., Dowle A.A., Forrester S., Parry E., Newling K., Walrad P.B. The mRNA-bound proteome of Leishmania mexicana: novel genetic insight into an ancient parasite. Mol. Cell. Proteomics. 2019;18:1271–1284. doi: 10.1074/mcp.RA118.001307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quilles J.C., Espada C.R., Orsine L.A., Defina T.A., Almeida L., Holetz F., Cruz A.K. A short ncRNA modulates gene expression and affects stress response and parasite differentiation in Leishmania braziliensis. Front. Cell. Infect. Microbiol. 2025;15 doi: 10.3389/fcimb.2025.1513908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guegan F., Rajan K.S., Bento F., Pinto-Neves D., Sequeira M., Gumińska N., Mroczek S., Dziembowski A., Cohen-Chalamish S., Doniger T., Galili B., Estévez A.M., Notredame C., Michaeli S., Figueiredo L.M. A long noncoding RNA promotes parasite differentiation in African trypanosomes. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abn2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- 11.Mattick J.S., Amaral P.P., Carninci P., Carpenter S., Chang H.Y., Chen L.L., Chen R., Dean C., Dinger M.E., Fitzgerald K.A., Gingeras T.R., Guttman M., Hirose T., Huarte M., Johnson R., Kanduri C., Kapranov P., Lawrence J.B., Lee J.T., Mendell J.T., Mercer T.R., Moore K.J., Nakagawa S., Rinn J.L., Spector D.L., Ulitsky I., Wan Y., Wilusz J.E., Wu M. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023;24:430–447. doi: 10.1038/s41580-022-00566-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michaeli S., Doniger T., Gupta S.K., Wurtzel O., Romano M., Visnovezky D., Sorek R., Unger R., Ullu E. RNA-seq analysis of small RNPs in Trypanosoma brucei reveals a rich repertoire of non-coding RNAs. Nucleic Acids Res. 2012;40:1282–1298. doi: 10.1093/nar/gkr786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruy P.C., Monteiro-Teles N.M., Miserani Magalhaes R.D., Freitas-Castro F. Comparative transcriptomics in Leishmania braziliensis: disclosing differential gene expression of coding and putative noncoding RNAs across developmental stages. RNA Biol. 2019;16:639–660. doi: 10.1080/15476286.2019.1574161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freitas Castro F., Ruy P.C., Nogueira Zeviani K., Freitas Santos R., Simoes Toledo J., Kaysel Cruz A. Evidence of putative non-coding RNAs from Leishmania untranslated regions. Mol. Biochem. Parasitol. 2017;214:69–74. doi: 10.1016/j.molbiopara.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Seemann S.E., Mirza A.H., Hansen C., Bang-Berthelsen C.H., Garde C., Christensen-Dalsgaard M., Torarinsson E., Yao Z., Workman C.T., Pociot F., Nielsen H., Tommerup N., Ruzzo W.L., Gorodkin J. The identification and functional annotation of RNA structures conserved in vertebrates. Genome Res. 2017;27:1371–1383. doi: 10.1101/gr.208652.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fazal F.M., Chang H.Y. lncRNA structure: message to the heart. Mol. Cell. 2016;64:1–2. doi: 10.1016/j.molcel.2016.09.030. [DOI] [PubMed] [Google Scholar]
- 17.Gorodkin J., Hofacker I.L. From structure prediction to genomic screens for novel non-coding RNAs. PLoS Comput. Biol. 2011;7 doi: 10.1371/journal.pcbi.1002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris R.S. 2007. Improved Pairwise Alignment of Genomic DNA, PhD thesis, The Pennsylvania State University. [Google Scholar]
- 19.Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., Haussler D., Miller W. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Washietl S., Hofacker I.L., Stadler P.F. Fast and reliable prediction of noncoding RNAs. Proc. Natl. Acad. Sci. U. S. A. 2005;102:2454–2459. doi: 10.1073/pnas.0409169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nawrocki E.P., Eddy S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalvari I., Nawrocki E.P., Ontiveros-Palacios N., Argasinska J., Lamkiewicz K., Marz M., Griffiths-Jones S., Toffano-Nioche C., Gautheret D., Weinberg Z., Rivas E., Eddy S.R., Finn R.D., Bateman A., Petrov A.I. Rfam 14: expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021;49:D192–D200. doi: 10.1093/nar/gkaa1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bühlmann M., Walrad P., Rico E., Ivens A., Capewell P., Naguleswaran A., Roditi I., Matthews K.R. NMD3 regulates both mRNA and rRNA nuclear export in African trypanosomes via an XPOI-linked pathway. Nucleic Acids Res. 2015;43:4491–4504. doi: 10.1093/nar/gkv330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hang R., Deng X., Liu C., Mo B., Cao X. Circular RT-PCR assay using arabidopsis samples. Bio-Protoc. 2015;5 doi: 10.21769/BioProtoc.1533. [DOI] [Google Scholar]
- 25.Inbar E., Hughitt V.K., Dillon L.A., Ghosh K., El-Sayed N.M. The transcriptome of Leishmania major developmental stages in their natural sand fly vector. mBio. 2017;8 doi: 10.1128/mBio.00029-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnes R.L., Shi H., Kolev N.G., Tschudi C., Ullu E. Comparative genomics reveals two novel RNAi factors in Trypanosoma brucei and provides insight into the core machinery. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolev N.G., Franklin J.B., Carmi S., Shi H., Michaeli S., Tschudi C. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Assis L.A., Santos Filho M.V.C., da Cruz Silva J.R., Bezerra M.J.R., de Aquino I., Merlo K.C., Holetz F.B., Probst C.M., Rezende A.M., Papadopoulou B., da Costa Lima T.D.C., de Melo Neto O.P. Identification of novel proteins and mRNAs differentially bound to the Leishmania Poly(A) Binding Proteins reveals a direct association between PABP1, the RNA-binding protein RBP23 and mRNAs encoding ribosomal proteins. PLoS Neglected Trop. Dis. 2021;15 doi: 10.1371/journal.pntd.0009899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang W., Xiong T., Zhao Y., Heng J., Han G., Wang P., Zhao Z., Shi M., Li J., Wang J., Wu Y., Liu F., Xi J.J., Wang Y., Zhang Q.C. Computational prediction and experimental validation identify functionally conserved lncRNAs from zebrafish to human. Nat. Genet. 2024;56:124–135. doi: 10.1038/s41588-023-01620-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Statello L., Guo C.J., Chen L.L., Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021;22:96–118. doi: 10.1038/s41580-020-00315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borden K., Culjkovic-Kraljacic B., Cowling V.H. To cap it all off, again: dynamic capping and recapping of coding and non-coding RNAs to control transcript fate and biological activity. Cell Cycle. 2021;20:1347–1360. doi: 10.1080/15384101.2021.1930929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dimartino D., Colantoni A., Ballarino M., Martone J., Mariani D., Danner J., Bruckmann A., Meister G., Morlando M., Bozzoni I. The long non-coding RNA lnc-31 interacts with Rock1 mRNA and mediates its YB-1-Dependent translation. Cell Rep. 2018;23:733–740. doi: 10.1016/j.celrep.2018.03.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin A., Li C., Xing Z., Hu Q., Liang K., Han L., Wang C., Hawke D.H., Wang S., Zhang Y., Wei Y., Ma G., Park P.K., Zhou J., Zhou Y., Hu Z., Zhou Y., Marks J.R., Liang H., Hung M.C., Lin C., Yang L. The LINK-A lncRNA activates normoxic HIF1alpha signalling in triple-negative breast cancer. Nat. Cell Biol. 2016;18:213–224. doi: 10.1038/ncb3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shan F., Mei S., Zhang J., Zhang X., Xu C., Liao S., Tu X. A telomerase subunit homolog La protein from Trypanosoma brucei plays an essential role in ribosomal biogenesis. FEBS J. 2019;286:3129–3147. doi: 10.1111/febs.14853. [DOI] [PubMed] [Google Scholar]
- 35.Paterou A., Walrad P., Craddy P., Fenn K., Matthews K. Identification and stage-specific association with the translational apparatus of TbZFP3, a CCCH protein that promotes trypanosome life-cycle development. J. Biol. Chem. 2006;281:39002–39013. doi: 10.1074/jbc.M604280200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Walrad P., Paterou A., Acosta-Serrano A., Matthews K.R. Differential trypanosome surface coat regulation by a CCCH protein that co-associates with procyclin mRNA cis-elements. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walrad P.B., Capewell P., Fenn K., Matthews K.R. The post-transcriptional trans-acting regulator, TbZFP3, co-ordinates transmission-stage enriched mRNAs in Trypanosoma brucei. Nucleic Acids Res. 2012;40:2869–2883. doi: 10.1093/nar/gkr1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seppey M., Manni M., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness. Methods Mol. Biol. 2019;1962:227–245. doi: 10.1007/978-1-4939-9173-0_14. [DOI] [PubMed] [Google Scholar]
- 39.Gertz E.M., Yu Y.K., Agarwala R., Schaffer A.A., Altschul S.F. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 2006;4:41. doi: 10.1186/1741-7007-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Keller O., Kollmar M., Stanke M., Waack S. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics. 2011;27:757–763. doi: 10.1093/bioinformatics/btr010. [DOI] [PubMed] [Google Scholar]
- 41.Gesell T., Washietl S. Dinucleotide controlled null models for comparative RNA gene prediction. BMC Bioinf. 2008;9:248. doi: 10.1186/1471-2105-9-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabarinathan R., Tafer H., Seemann S.E., Hofacker I.L., Stadler P.F., Gorodkin J. RNAsnp: efficient detection of local RNA secondary structure changes induced by SNPs. Hum. Mutat. 2013;34:546–556. doi: 10.1002/humu.22273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabarinathan R., Tafer H., Seemann S.E., Hofacker I.L., Stadler P.F., Gorodkin J. The RNAsnp web server: predicting SNP effects on local RNA secondary structure. Nucleic Acids Res. 2013;41:W475–W479. doi: 10.1093/nar/gkt291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beneke T., Madden R., Makin L., Valli J., Sunter J., Gluenz E. A CRISPR Cas9 high-throughput genome editing toolkit for kinetoplastids. R. Soc. Open Sci. 2017;4 doi: 10.1098/rsos.170095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lorenzon L., Quilles J.C., Jr., Campagnaro G.D., Azevedo Orsine L., Almeida L., Veras F., Miserani Magalhaes R.D., Alcoforado Diniz J., Rodrigues Ferreira T., Kaysel Cruz A. Functional study of Leishmania braziliensis protein arginine methyltransferases (PRMTs) reveals that PRMT1 and PRMT5 are required for macrophage infection. ACS Infect. Dis. 2022;8:516–532. doi: 10.1021/acsinfecdis.1c00509. [DOI] [PubMed] [Google Scholar]
- 46.Rotureau B., Gego A., Carme B. Trypanosomatid protozoa: a simplified DNA isolation procedure. Exp. Parasitol. 2005;111:207–209. doi: 10.1016/j.exppara.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 47.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 48.Kramer S. Simultaneous detection of mRNA transcription and decay intermediates by dual colour single mRNA FISH on subcellular resolution. Nucleic Acids Res. 2017;45:e49. doi: 10.1093/nar/gkw1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Peng D., Tarleton R. EuPaGDT: a web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens. Microb. Genom. 2015;1 doi: 10.1099/mgen.0.000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anacleto C., Abdo M.C., Ferreira A.V., Murta S.M., Romanha A.J., Fernandes A.P., Moreira E.S. Structural and functional analysis of an amplification containing a PGPA gene in a glucantime-resistant Leishmania (Viannia) guyanensis cell line. Parasitol. Res. 2003;90:110–118. doi: 10.1007/s00436-002-0798-x. [DOI] [PubMed] [Google Scholar]
- 51.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pratt D., Chen J., Welker D., Rivas R., Pillich R., Rynkov V., Ono K., Miello C., Hicks L., Szalma S., Stojmirovic A., Dobrin R., Braxenthaler M., Kuentzer J., Demchak B., Ideker T. NDEx, the network data exchange. Cell Syst. 2015;1:302–305. doi: 10.1016/j.cels.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.