Abstract
MicroRNAs (miRNAs) are important regulators of gene expression and their dysregulation is involved in various diseases, including tumors. Among these, colorectal cancer (CRC) is the result of both genetic and epigenetic alterations with miRNAs playing a key pathogenetic role. Although numerous studies have investigated the most frequently dysregulated miRNAs in CRC, there is still no consensus on the specific role of individual miRNAs in the mechanisms leading to tumorigenesis, tumor progression, and the development of chemoresistance. This lack of clarity highlights the need for a deeper understanding of miRNA functions in CRC. Therefore, this review aims to clarify the role of miRNAs in CRC by examining their involvement in major oncogenic pathways, highlighting key miRNAs implicated in the disease, and exploring their potential as diagnostic biomarkers and therapeutic targets. By providing a comprehensive overview, we hope to shed light on the complex and multifaceted roles of miRNAs in CRC, which could pave the way for more effective CRC monitoring and the development of miRNA-guided therapeutic strategies.
Keywords: microRNA, Colorectal cancer, Biomarker, Epigenetics, Therapeutic target, Drug resistance
Abbreviations
- 5-Aza-2′C
5-aza-2′-deoxycytidine
- 5-FU
5-fluorouracil
- ABC
ATP-binding cassette
- ABCC2
ATP Binding Cassette Subfamily C Member 2
- ABCF1
ATP Binding Cassette Subfamily F Member 1
- ACD
Asymmetric cell division
- ACOX1
Acyl-CoA Oxidase 1
- ACSL1
Acyl-CoA Synthetase Long Chain Family Member 1
- ACSL4
Acyl-CoA Synthetase Long Chain Family Member 4
- ADAM-17
ADAM Metallopeptidase Domain 17
- AFTPH
Aftiphilin
- ALAS1
Aminolevulinate synthase 1
- ALDH1A3
Aldehyde Dehydrogenase 1 Family Member A3
- AMPKα2
Protein Kinase AMP-Activated Catalytic Subunit Alpha 2
- antimiRs
anti-microRNA
- AP4
Transcription Factor AP-4
- APAF1
Apoptotic Peptidase Activating Factor 1
- APC
Adenomatous Polyposis Coli
- ASOs
Antisense oligonucleotides
- ATF3
Activating Transcription Factor 3
- ATG14
Autophagy Related 14
- ATG5
Autophagy Related 5
- ATM
Ataxia telangiectasia mutated
- AXIN1
Axin 1
- AXIN2
Axin 2
- BAG4
BAG Cochaperone 4
- BAK1
BCL2 Antagonist/Killer 1
- BCL2
BCL2 Apoptosis Regulator
- BCL2L1
BCL2 Like 1
- BCL2L2
BCL2 Like 2
- BCL9L
BCL9 Like
- BIM
BCL-2-interacting mediator of cell death
- BIRC5
Baculoviral IAP Repeat Containing 5
- BLM
Bleomycin
- BLNK
B Cell Linker
- BRAF
B-Raf Proto-Oncogene, Serine/Threonine Kinase
- BRG1
SWI/SNF Related BAF Chromatin Remodeling Complex Subunit ATPase 4
- BTBD7
BTB Domain Containing 7
- BTG2
BTG Anti-Proliferation Factor 2
- BTG3
BTG Anti-Proliferation Factor 3
- CA19-9
Carbohydrate Antigen 19-9
- CADM2
Cell Adhesion Molecule 2
- CAV1
Caveolin 1
- CBR3-AS1
CBR3 Antisense RNA 1
- CCND1
Cyclin D1
- CCNE1
Cyclin E1
- CCSCs
Colon cancer stem cells
- CD133
Prominin 1
- CDC42
Cell Division Cycle 42
- CDCA3
Cell Division Cycle Associated 3
- CDDP
Cisplatin
- CDH1
Cadherin 1
- CDH2
Cadherin 2
- CDK19
Cyclin Dependent Kinase 19
- CDK8
Cyclin Dependent Kinase 8
- CDKN1C/p57
Cyclin Dependent Kinase Inhibitor 1C
- CDX1
Caudal Type Homeobox 1
- CEA
Carcinoembryonic Antigen
- ceRNA
competing endogenous RNA
- CFL1
Cofilin 1
- CHD9
Chromodomain Helicase DNA Binding Protein 9
- CHEK2
Checkpoint Kinase 2
- CIMP
CpG island methylator phenotype
- circRNA
circular RNA
- CLCA4
Chloride Channel Accessory 4
- CLL:
Chronic lymphocytic leukemia
- c-Met
MET Proto-Oncogene, Receptor Tyrosine Kinase
- COX2
Cytochrome C Oxidase Subunit 2
- CPA6
Carboxypeptidase A6
- CRC
Colorectal Cancer
- CREB1
CAMP Responsive Element Binding Protein 1
- CRNDE
Colorectal Neoplasia Differentially Expressed
- CSC
Cancer stem cell
- CSF2R
Colony Stimulating Factor 2 Receptor Subunit Alpha
- CSF2RB
Colony Stimulating Factor 2 Receptor Subunit Beta
- CTSS
Cathepsin S
- Cx43
Connexin-43
- CXCL12
C-X-C motif chemokine ligand 12
- CXCL8
C-X-C motif chemokine ligand 8
- CXCR4
C-X-C Motif Chemokine Receptor 4
- CXCR7
C-X-C motif chemokine receptor 7
- DAB2IP
DAB2 Interacting Protein
- DACH1
Dachshund family transcription factor 1
- DCLK1
Doublecortin Like Kinase 1
- DGCR8
DiGeorge Syndrome Critical Region 8 RNA-binding protein
- DKK1
Dickkopf WNT Signaling Pathway Inhibitor 1
- DKK3
Dickkopf WNT Signaling Pathway Inhibitor 3
- DLC1
DLC1 Rho GTPase Activating Protein
- DNAJB4
DnaJ Heat Shock Protein Family (Hsp40) Member B4
- DNMT3A
DNA Methyltransferase 3 Alpha
- DOT1L
Disruptor of telomeric silencing 1-like
- Dox
Doxorubicin
- DSB
Double-strand break
- E2F3
E2F Transcription Factor 3
- E2F5
E2F Transcription Factor 5
- EGFR
Epidermal growth factor receptor
- EIF5A2
Eukaryotic Translation Initiation Factor 5A2
- ELK1
ETS Transcription Factor ELK1
- EMT
Epithelial-mesenchymal transitions
- EREG
Epiregulin
- ERG
ETS Transcription Factor ERG
- FAP
Familial Adenomatous Polyposis
- FBXW7
F-Box and WD Repeat Domain Containing 7
- FFPE
Formalin fixed paraffin embedded
- FIP200
RB1 Inducible Coiled-Coil 1
- FMNL2
Formin Like 2
- FN1
Fibronectin 1
- FOLFIRI
Folinic acid, fluorouracil and irinotecan
- FOLFOX
Folinic acid, fluorouracil and oxaliplatin
- FOXF2
Forkhead Box F2
- Foxj2
Forkhead Box J2
- FOXO1
Forkhead Box O1
- FOXO3a
Forkhead Box O3
- FRA1
FOS Like 1, AP-1 Transcription Factor Subunit
- FSCN1
Fascin Actin-Bundling Protein 1
- GAA
Gossypol-acetic acid
- G-MDSCs
Myeloid-derived granulocyte suppressor cells
- GNA13
G Protein Subunit Alpha 13
- GO
Gene Ontology
- GPX4
Glutathione Peroxidase 4
- GRG5
Groucho-related gene 5
- GSH
Glutathione
- GSK3β
Glycogen Synthase Kinase 3 Beta
- hCNT1
Concentrative nucleoside transporter 1
- HDAC
Histone deacetylase
- HDM4
Human homolog of murine double minute 4
- HER2
Erb-B2 Receptor Tyrosine Kinase 2
- HK II
Hexokinase 2
- HMECs
Human microvascular endothelial cells
- HMGA2
High Mobility Group AT-Hook 2
- hnRNPA1
Heterogeneous Nuclear Ribonucleoprotein A1
- HOTAIR
HOX Transcript Antisense RNA
- HOXB1
Homeobox B1
- HOXB3
Homeobox B3
- HOXB9
Homeobox B9
- HOXD10
Homeobox D10
- hRFI
Human Ring-Finger homologous to Inhibitor of apoptosis protein type
- HSPB2
Heat Shock Protein Family B (Small) Member 2
- IGF1R
Insulin Like Growth Factor 1 Receptor
- IKK-α:
Inhibitor of Nuclear Factor Kappa-B Kinase Subunit Alpha
- IL-17A
Interleukin 17A
- IL-21
Interleukin 21
- IL-6
Interleukin 6
- ING4
Inhibitor Of Growth Family Member 4
- IREB2
Iron Responsive Element Binding Protein 2
- IRS1
Insulin Receptor Substrate 1
- ITGA2
Integrin Subunit Alpha 2
- JAK
Janus Kinas
- JNK2
C-Jun N-Terminal Kinase 2
- KDM4B
Lysine Demethylase 4B
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- KIF14
Kinesin Family Member 14
- KLF4
KLF Transcription Factor 4
- KLF5
KLF Transcription Factor 5
- KLK10
Kallikrein Related Peptidase 10
- KRAS
Kirsten rat sarcoma
- KSR1
Kinase Suppressor of Ras 1
- LASP1
LIM And SH3 Protein 1
- LATS2
Large Tumour Suppressor Kinase 2
- LEF
Lymphoid enhancer factor
- LGR5
Leucine Rich Repeat Containing G Protein-Coupled Receptor 5
- LIN28A
Lin-28 homolog A
- LIN28B
Lin-28 homolog B
- LM
Liver metastasis
- LNAs
Locked nucleic acids
- LncRNA
Long non-coding RNA
- LRP6
LDL Receptor Related Protein 6
- LRPPRC
Leucine-rich pentatricopeptide repeat-containing protein
- MACC1
MET Transcriptional Regulator MACC1
- MAP4K4
Mitogen-Activated Protein Kinase Kinase Kinase Kinase 4
- MAPK
Mitogen-Activated Protein Kinase 1
- MAPK1
Mitogen-Activated Protein Kinase 1
- MAPK7
Mitogen-Activated Protein Kinase 7
- MDE
Exosomes derived from M2 macrophages
- MDM2
E3 Ubiquitin-Protein Ligase Mdm2
- MDR
Multidrug resistance
- MDSCs
Myeloid-derived suppressor cells
- MEKK
Mitogen-Activated Protein Kinase Kinase Kinase 1
- MET
MET Proto-Oncogene, Receptor Tyrosine Kinase
- MFN2
Mitofusin 2
- MHC
Major histocompatibility complex
- MIA3
MIA SH3 Domain ER Export Factor 3
- MICA
MHC Class I Polypeptide-Related Sequence A
- miRNA
microRNA
- MK5
MAPK Activated Protein Kinase 5
- MMP11
Matrix Metallopeptidase 11
- MMP2
Matrix Metallopeptidase 2
- MMP9
Matrix Metallopeptidase 9
- MREs
microRNA response elements
- mRNA
Messenger ribonucleic acid
- MRP-2
Multidrug resistance-associated protein-2
- MSI
Microsatellite instability
- MSI-H:
Microsatellite instability high
- MSS
Microsatellite status
- MST3
Mammalian STE20-Like Protein Kinase 3
- mTOR
mammalian target of rapamycin mTOR
- MUC1
Mucin 1
- MVs
Microvesicles
- MYO6
Myosin VI
- NAMPT
Nicotinamide Phosphoribosyltransferase
- ncRNA
non-coding RNA
- NEAT1
Nuclear Paraspeckle Assembly Transcript 1
- NEDD9
Neural Precursor Cell Expressed, Developmentally Down-Regulated 9
- NF2
Neurofibromin 2
- NF-κB1
Nuclear Factor Kappa B Subunit 1
- NM23-H1
NME/NM23 Nucleoside Diphosphate Kinase 1
- NOTCH3
Notch Receptor 3
- NPEPL1
Aminopeptidase Like 1
- NRP1
Neuropilin 1
- NT5E
5'-Nucleotidase Ecto
- OAZ2
Ornithine decarboxylase 2
- OCLN
Occludin
- OCT4
Octamer-binding transcription factor 4
- P130
RB Transcriptional Corepressor Like 2
- PAK4
P21 (RAC1) Activated Kinase 4
- PBX3
PBX Homeobox 3
- PDCD4
Programmed Cell Death 4
- PDE4D
Phosphodiesterase 4D
- PDH
Pyruvate Dehydrogenase
- PDK1
Pyruvate Dehydrogenase Kinase 1
- PDK4
Pyruvate Dehydrogenase Kinase 4
- PD-L1
Programmed Death Ligand 1
- PFN2
Profilin 2
- PGE2
Prostaglandin E2
- PI3K
Phosphatidylinositol 3-kinase
- PIAS3
Protein Inhibitor Of Activated STAT 3
- PLCD1
Phospholipase C Delta 1
- PPAR
Peroxisome Proliferator Activated Receptor Alpha
- PPP2R5E
Protein Phosphatase 2 Regulatory Subunit B'Epsilon
- PRRX1
Paired Related Homeobox 1
- PTBP1
Polypyrimidine Tract Binding Protein 1
- PTEN
Phosphatase And Tensin Homolog
- PTK6
Protein Tyrosine Kinase 6
- PTP4A
Protein Tyrosine Phosphatase 4A
- PUMA
p53 upregulated modulator of apoptosis
- RAC1
Rac Family Small GTPase 1
- RANBP1
RAN binding protein 1
- Ran-GTPase
nuclear RAS-related protein-guanosine-5'-triphosphate-ase
- RAP1B
RAP1B, Member Of RAS Oncogene Family
- RASA1
RAS P21 Protein Activator 1
- RBL2
RB transcriptional co-repressor like 2
- RCN2
Reticulocalbin 2
- RECK
Reversion Inducing Cysteine Rich Protein With Kazal Motifs
- RFFL
Ring Finger and FYVE Like Domain Containing E3 Ubiquitin Protein Ligase
- RISC
RNA-induced silencing complex
- RMST
Rhabdomyosarcoma 2 Associated Transcript
- RND3
Rho Family GTPase 3
- RNF6
Ring Finger Protein 6
- ROS
Reactive Oxygen Species
- RPL11
Ribosomal Protein L11
- RPS15A
Ribosomal Protein S15a
- RUNX3
RUNX Family Transcription Factor 3
- SATB2
SATB Homeobox 2
- SCD
Stearoyl-CoA Desaturase
- SCD
Symmetrical cell division
- SEMA6D
Semaphorin 6D
- SFRP4
Secreted frizzled-related protein 4
- shRNAs
short hairpin RNAs
- SIP1
SMAD Interacting Protein 1
- siRNA
small interfering RNA
- SIRT1
Sirtuin 1
- SIRT4
Sirtuin 4
- SM
Small molecule
- SMAD3
SMAD Family Member 3
- SMAD4
SMAD Family Member 4
- SMAD7
SMAD Family Member 7
- SMIR
Small inhibitors of miRNA
- SNAIL
Snail Family Transcriptional Repressor 1
- SOCS1
Suppressor of cytokine signaling 1
- SOCS3
Suppressor Of Cytokine Signaling 3
- SOX2
SRY-Box Transcription Factor 2
- SOX4
SRY-Box Transcription Factor 4
- SOX5
SRY-Box Transcription Factor 5
- SPINT1
Serine Peptidase Inhibitor, Kunitz Type 1
- SPOP
Speckle Type BTB/POZ Protein
- SRC
SRC Proto-Oncogene, Non-Receptor Tyrosine Kinase
- SRCIN1
SRC kinase signaling inhibitor 1
- SSH2
Slingshot Protein Phosphatase 2
- ST6GALNAC2
ST6 N-Acetylgalactosaminide Alpha-2,6-Sialyltransferase 2
- STAT
Signal Transducer and Activator Of Transcription
- TAMs
Tumor-associated macrophages
- TBPL1
TATA-Box Binding Protein Like 1
- TCF
β-catenin-T cell factor
- TCF4
Transcription Factor 4
- TCGA
The Cancer Genome Atlas
- TEAD4
TEA Domain Transcription Factor 4
- TGFB2
Transforming Growth Factor Beta 2
- TGFBR2
Transforming growth factor receptor β
- TGFβ
Transforming Growth Factor Beta
- THBS1
Thrombospondin 1
- THBS2
Thrombospondin 2
- TIAM1
TIAM Rac1 Associated GEF 1
- TICs
Tumor-initiating cancer stem cells
- TNFAIP3
TNF Alpha Induced Protein 3
- TNF-α
Tumor Necrotic Factor Alpha
- TP53INP1
Tumor Protein P53 Inducible Nuclear Protein 1
- TPM1
Tropomyosin 1
- TSA
Trichostatin A
- TSP-1
Thrombospondin 1
- TYMS
Thymidylate Synthetase
- VAPA
VAMP Associated Protein A
- VCR
Vincristine
- VEGFA
Vascular Endothelial Growth Factor A
- VIM
Vimentin
- VLDLR
Very Low Density Lipoprotein Receptor
- VOPP1
Vesicular pro-survival protein 1
- WDR43
WD Repeat Domain 43
- WIF1
WNT Inhibitory Factor 1
- XIAP
X-Linked Inhibitor of Apoptosis
- XIST
X Inactive Specific Transcript
- XPO5
exportin5
- YAP1
Yes-associated protein 1
- YES
YES1 Proto-Oncogene, Src Family Tyrosine Kinas
- ZBTB2
Zinc finger and BTB domain containing 2
- ZEB1
Zinc Finger E-Box Binding Homeobox 1
- ZEB2
Zinc Finger E-Box Binding Homeobox 2
- ZNF281
Zinc Finger Protein 281
- ZNRF3
Zinc And Ring Finger 3
1. Introduction
Colorectal cancer (CRC) is the fourth most common cancer and the second leading cause of cancer-related deaths globally. The incidence of CRC varies significantly between geographical areas [1]. Age is one of the main risk factors for CRC development; however, CRC incidence rates have decreased by up to 50 % in older age groups in the US as a result of screening programs [2]. According to Vogelstein's model, truncating mutations affecting the adenomatous polyposis (APC) gene play a crucial role in the regulation of cell adhesion and proliferation due to the alteration of the Wnt/β-catenin axis actively involved in the formation of adenomatous polyps. Following APC mutation, the progression of CRC involves a series of additional genetic changes, notably mutations in KRAS and TP53 involved in cell growth and differentiation and the loss of cell cycle control and increased mutation rates, respectively [3,4]. Serrated polyp pathway is an alternative pathway that leads to CRC development, characterized by the presence of serrated lesions that can give rise to colorectal malignancies. The most frequent initiating event in this pathway is BRAF mutation which promotes cell proliferation and survival. This first molecular trigger leads to extensive methylation of CpG islands, leading to the silencing of critical tumor suppressor genes and further tumor-promoting events. Hypermethylation often affects the promoter region of genes coding for mismatch repair proteins, resulting in a deficiency of the mismatch repair enzyme. These tumors are defined as CIMP+ (CpG island methylator phenotype). Besides CIMP, other typical CRC phenotypes are chromosomal instability (CIN), involving several numerical chromosome aberrations, and microsatellite instability (MSI) CRC [5]. Understanding the molecular mechanisms underlying the development of CRC is essential as it provides the basis for current screening strategies or to predict the prognosis of patients. In this context, several tests, both non-invasive and invasive, are used for CRC screening, however, the diagnosis of CRC is only obtained by histopathological examination [6]. In daily clinical practice, several biomarkers have been proposed for the early detection of CRC and to monitor the disease. Among these, the most relevant include CEA and CA19-9, which have a good predictive value for the monitoring of the disease but have a low specificity and sensitivity for CRC diagnosis [7]. Other studied biomarkers are different antibodies, circulating mutations, specific aberrant RNA transcripts and epigenetic biomarkers, including microRNAs (miRNAs). Concerning CRC therapy, the most curative intervention still relies on surgery for the treatment of localized CRC. Chemotherapy, using 5-fluorouracil (5-FU), capecitabine, irinotecan, oxaliplatin and folic acid (FOLFOX/FOLFIRI regimens) is mainly used in the adjuvant setting after surgery, or as a neoadjuvant treatment to shrink the tumor mass before surgical treatment, especially in rectal cancer and some colon tumors. In addition, these drugs can also be administered in combination with radiotherapy or with immunotherapy in microsatellite instability (MSI) CRC [8,9]. In addition to standard chemotherapy, the detection of specific mutations in the RAS (KRAS) and BRAF genes is important to consider patients eligible for targeted therapy [10]. In the case of wild-type KRAS and BRAF genes, patients can benefit from the FOLFOX/FOLFIRI protocol combined with the anti-EGFR selective inhibitor, named cetuximab [11]. Finally, immunotherapy based on immune checkpoint inhibitors has proven effective for the treatment of metastatic CRC with high microsatellite instability (MSI-H) [12]. Despite the multiple therapeutic options currently available, drug resistance mechanisms may lead to therapeutic failure, affecting the prognosis of patients [13]. In this heterogeneous molecular context, mounting scientific evidence is demonstrating the tumorigenic role of non-coding RNA (ncRNA) in CRC, with miRNAs playing a key role in tumor invasion, metastasis, and chemoresistance. Despite numerous studies, the specific roles of individual miRNAs remain to be fully elucidated, making them a focal point for future CRC research aimed at improving personalized medicine strategies and predicting the development of drug resistance [14,15].
2. microRNAs biogenesis, function and role in cancer
miRNAs are a class of ncRNAs short in size (19–25 nucleotides) that play important roles in regulating the expression of homologous target-gene transcripts through a mechanism known as RNA interference (RNAi) [16]. The biogenesis of miRNAs is a multi-step process that begins in the nucleus, where a long primary transcript (pri-miRNA) is produced and then processed into a precursor miRNA (pre-miRNA) by a multi-protein complex consisting of the DiGeorge syndrome critical region 8 RNA-binding protein (DGCR8) and the ribonuclease III enzyme, Drosha [17]. Subsequently, at the cytoplasmic level, the pre-miRNA is cleaved by the RNase III Dicer endonuclease to form a miRNA duplex, of which one strand will be loaded into the RNA-induced silencing complex (RISC) [18]. Usually, miRNAs interact with the 3′ untranslated region (3′ UTR) of the targeted mRNA to induce mRNA degradation or translational repression. However, miRNAs interacting with other mRNA regions (5′ UTR, coding sequence, and gene promoters) have also been reported [19]. Since a single miRNA can target hundreds of mRNAs and a single target mRNA can be silenced by several miRNAs, the understanding of this epigenetic regulatory network is very intricate and requires high-throughput platforms. miRNAs regulate several biological processes, including oncogenic or tumor suppressor pathways. Therefore, miRNAs aberrant expression can contribute to the development of several pathological conditions including cancer [[20], [21], [22], [23], [24], [25], [26]]. The link between miRNA alteration and cancer development was first demonstrated by Croce and colleagues in 2002. Specifically, Croce's study showed a deletion of the miR-15a/16-1 cluster in chronic lymphocytic leukemia (CLL) associated with tumor progression, thus suggesting the tumor suppressor role of these miRNAs [27]. After this pivotal study, several researchers investigated the role of miRNAs in cancer pathogenesis.
2.1. microRNAs as targets for therapeutic application
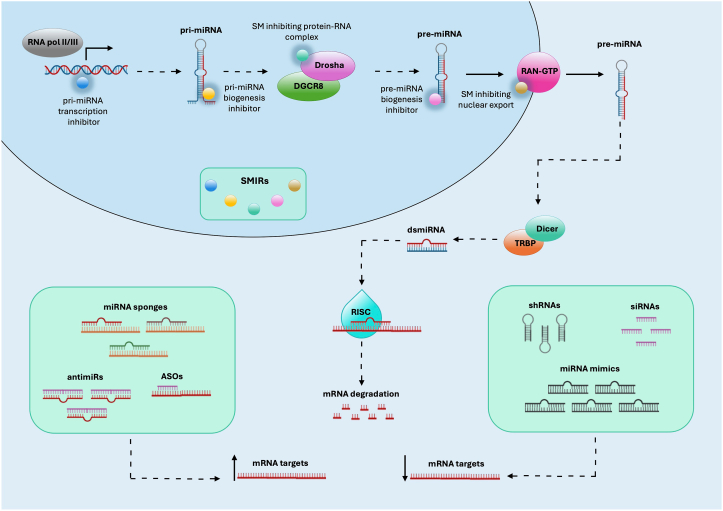
Recently, miRNAs have emerged as attractive targets for therapeutic application. Different miRNA-targeting strategies have been implemented mainly using small molecules (SM) and small inhibitors of miRNAs (SMIRs) (Fig. 1). Among the SM, molecules inhibiting pri-miRNAs, pre-miRNAs, or protein-RNA complexes were developed [28]. An example of SM in Enoxacin that belongs to the family of synthetic antibacterial compounds with a fluoroquinolone skeleton, which enhances RNAi induced by shRNA or siRNA duplexes [29]. Among SM, a subclass is defined as SMIRs since they are able to inhibit miRNA activity. In contrast to oligonucleotide-based therapies targeting mRNAs or miRNAs, SMIRs represent an innovative and promising therapeutic strategy due to their better cellular uptake capacity, greater stability, and the possibility of being administered orally [28]. Examples of SMIRs are azobenzene-2 [30], Targaprimir-96 [31], Benzimidazole [32], Targapremir-210 [33] or AC1MMYR2 [34]. Moreover, bifunctional chimeric molecules obtained by the fusion of a recognition module with a proteolysis-targeted RNA degradation module (ProTaC) have been developed to hinder miRNA biogenesis. For instance, bleomycin (BLM), a natural compound known for its RNA-cleaving properties, was fused with specific ligands to target pri-miR-96, thus allowing specific RNA cleavage and degradation [35]. In addition, several RNA-based therapies have been developed, including antisense oligonucleotides (ASOs), anti-microRNA (antimiRs), small interfering RNAs (siRNAs), short hairpin RNAs (shRNAs), miRNA mimics, miRNA sponges, therapeutic circular RNAs (circRNAs) and CRISPR/Cas9-based gene editing [36] (Fig. 1).
Fig. 1.
Several strategies are available to modulate the levels of miRNAs in the cell. Small miRNA inhibitors (SMIRs) can inhibit the activity of miRNAs by hindering steps in their biogenesis. Antisense oligonucleotides (ASOs), anti-microRNA ASOs (antimiRs) or miRNA sponges are molecules that inhibit miRNA function while small interfering RNAs (siRNAs), short hairpin RNAs (shRNAs) or mimic miRNAs are molecules that mimic miRNA activity.
miRNA mimic molecules are synthetic double-stranded RNA oligonucleotides used for cell transfection. At the cellular level, miRNA mimics are transformed into the single-stranded form by regulating the expression of target genes through a miRNA-like function [37].
Inhibitors of miRNAs, or anti-miRs, bind selected miRNAs by blocking their function [38]. A major problem associated with the use of miRNA inhibitors is their limited half-life. Indeed, naked nucleic acids are susceptible to degradation by nucleases. Another issue is related to targeted delivery, as these nucleic acids must be transported into the cytoplasm or into the nucleus to perform their function [39,40]. Based on these challenges, several chemical modifications of nucleic acids have been developed to resist nuclease degradation, reduce immunogenicity and improve miRNA-target interactions [38,41].
Among miRNA inhibitors, antagomirs are synthetic cholesterol-conjugated RNAs with a 2′-O-methyl bond and a phosphorothioate modification. However, antagomirs require high dosages to effectively block miRNAs [42]. Furthermore, most miRNA-based therapeutic agents employ other modifications, such as locked nucleic acids (LNAs) [43,44].
miRNA sponges are RNAs designed to carry multiple tandem binding sites complementary to a heptamer in the seed sequence of the miRNA of interest. Consequently, a single sponge type can block an entire miRNA seed family. However, for the same reason, miRNA sponges could lead to off-target effects; they also have a relatively low binding affinity and the concentration required to effectively block miRNA function is higher than LNA or antagomir [45].
Ultimately, other possible therapeutic applications of miRNA targeting can be focused on the inhibition of oncogenic miRNAs by using miRNA antagonists [46] or restoring miRNA expression using a tumor suppressor mimic miRNA to recover its loss of function [47]. The efficacy of miRNA- and siRNA-based therapies has been tested in several clinical trials. Examples are TargomiR (miR-16 mimic-based therapy) used for the treatment of mesothelioma, Cobomarsen (anti-miR-155) in T-cell leukemia/lymphoma, and Miravirsen (anti-miR-122) in individuals with hepatitis C infection. Nusinersen (Spinraza), is a fully MOE-modified 18-mer ASO that redirects the splicing of SMN2 gene, approved for the treatment of spinal muscular atrophy [48,49]. An example of siRNA-based drug is Onpattro (Patisiran) containing 2′-O-methyl modified and unmodified ribonucleosides, with 2′-deoxythymidine dinucleotide overhangs at the 3′ ends, which is encapsulated in lipid-based nanoparticle approved by the FDA for the treatment of amyloidosis, marking a significant milestone in the history of RNAi technology and establishing a new therapeutic class [40,50]. Givosiran (Givlaari™) is another siRNA-based drug that targets aminolevulinate synthase 1 (ALAS1) and is covalently linked to a ligand that drives it into hepatocytes. Downregulation of ALAS1 mRNA by Givosiran prevents the accumulation of neurotoxic δ-aminolevulinic acid and porphobilinogen both responsible for acute porphyria attacks [51].
The identification and targeting of the mainly dysregulated miRNAs in CRC that play a crucial role in cancer progression and the development of chemoresistance is of major importance. Therefore, miRNA- and siRNA-based therapies may represent an innovative therapeutic approach with the potential to improve treatment outcomes and to overcome drug resistance.
3. Role of microRNAs in colorectal cancer
As previously mentioned, the alteration of miRNA expression levels may depend on several factors. Tumorigenesis is often accompanied by chromosomal aberrations such as deletions, amplifications or translocations. In many cases, miRNA alterations result from a variation in the copy number of specific genomic loci. Other factors that can impact miRNA expression are epigenetic modifications. DNA methylation plays a key role in regulating the expression of suppressor miRNAs in cancer cells. The hypermethylation of the promoters of let-7, miR-34, miR-342, miR-345, miR-9, miR-129, and miR-137 was associated with a reduced expression of these miRNAs and consequently with CRC development [52]. The hypermethylation of CpG islands in miRNA coding promoter regions results in their transcriptional silencing as demonstrated in CRC models for the miR-143, miR-145, and miR-133b. In particular, histone acetylation and DNA methylation were investigated in early and late-stage CRC cells (SW1116 and DLD1, respectively) by treatment with 5-aza-2′-deoxycytidine (5-Aza-2′C) and the histone deacetylase (HDAC) inhibitor trichostatin A (TSA). The epigenetic modulation of miRNA expression induced by these treatments demonstrated that miRNA expression is sensitive to DNA demethylation in both early- and late-stage CRC cells, whereas histone acetylation has a moderate influence on miRNA expression only in early-stage CRC [53].
The altered expression of miRNAs may be related to the impaired activity of transcription factors that regulate the transcription of pri-miRNAs [54]. Another mechanism that may affect the expression of miRNAs is mediated by competing endogenous RNA (ceRNAs). Salmena and colleagues in 2011 first formulated the ceRNA hypothesis, according to which there is a cross-talk between both coding and non-coding RNAs via microRNA response elements (MREs) whose alteration could influence disease onset [55]. This category of ncRNAs includes circRNAs and lncRNAs. CircRNAs are a class of closed-loop RNA molecules that play an important regulatory role in modulating miRNA functions through sponge adsorption. Besides sponging, the circRNA-miRNA interaction mechanisms also include storage and transport of miRNAs and the interference with their expression. LncRNAs can also sponge miRNAs and compete with them for interaction with mRNA [56,57]. For instance, HOX transcript antisense intergenic RNA (HOTAIR) negatively regulates the expression of miR-203a-3p, miR-545 and miR-218, leading to the up-regulation of their targets, such as β-catenin, groucho-related gene 5 (GRG5), epidermal growth factor receptor (EGFR), and vesicular pro-survival protein 1 (VOPP1), involved in the proliferation of CRC cells [[58], [59], [60]]. In addition to the endogenous “sponge” effect, lncRNA-miRNA interaction can accelerate miRNA degradation [61]. In the context of CRC, miRNAs can also play a role in physiological and pathological processes by influencing cancer stem cell biology and angiogenesis, epithelial-mesenchymal transitions (EMT), and drug resistance [62,63]. The expression of miRNAs can be altered at different tumor stages, including tumor initiation, progression and metastasis. Specific tumor histotypes have a distinct signature of altered miRNAs compared to matched normal tissue and other tumors. When altered, miRNAs promote tumor progression by affecting the mechanisms of cell growth, cell motility, alteration in hormonal stress response, proliferation, evasion of tumor suppression, apoptosis, metastasis, angiogenesis and drug resistance [64].
A comprehensive assessment of differentially expressed miRNAs and target genes between CRC samples and healthy controls may facilitate the identification of miRNAs functionally related to this tumor. Specific CRC miRNA expression profiles have been identified through several differential analysis studies. Such studies are generally based on miRNA profiling performed on different sources of samples (cells, fresh tissue, formalin-fixed paraffin-embedded tissue, body fluids), analyzed by using RT-qPCR panels, microarray or RNA sequencing platforms. After wet analyses, the data obtained are processed by using both statistical and bioinformatics approaches [65]. A RT-qPCR study analyzing isolated colonic crypts from 24 CRC patients identified 13 differentially expressed miRNAs in tumor glandular cells and surrounding stromal cells. Specifically, miR-130a-3p, miR-143-3p, miR-206, miR-31-5p, miR-27a-3p and miR-27b-3p were found to be upregulated in gland cells isolated from CRC compared to non-tumor samples, while miR-21-5p, miR-195-5p, miR-19a-3p, miR-34b-3p, miR-186-5p, miR-191-5p and let-7a-5p were downregulated [66]. Another microarray and RT-qPCR study comparing miRNA expression levels in 12 CRC tissue samples and 9 adjacent normal tissues found that miR-31 was significantly upregulated in CRC. Notably, this miRNA plays a significant role in activating the RAS signaling pathway by inhibiting the translation of RASA1, thereby increasing the growth of CRC cells and promoting tumorigenesis [67]. Likewise, miR-31, together with miR-18a and miR-21-5p, was identified among the most upregulated miRNAs associated with APC gene alterations by another differential expression study conducted on 40 CRC tumor samples and in their paired normal counterpart. In this study, it was found that miR-31 expression levels correlate with the expression of the tumor biomarker CA19-9 [68]. Using miRNA-Seq, Shaath H et al., performed a miRNA expression profiling on 15 CRC tissues compared to the corresponding normal adjacent mucosa. miR-133a-3p, miR-363-3p, miR-145-5p, and miR-195-3p, were found to be the most downregulated miRNAs while miR-135b-5p, miR-552-5p, miR-224-5p, miR-183-5p and miR-552-3p were found among those miRNAs upregulated in CRC [69]. Almeida MI et al., evaluated the expression levels of miR-28-5p and miR-28-3p in 108 CRC and 49 normal colorectal samples, of which 47 were paired, finding that both miRNAs were downregulated in CRC compared to normal tissues. They also conducted analyses on HCT116, RKO and SW480 cells, demonstrating how miR-28-5p restoration altered the expression of CCND1 and HOXB3 and reduced the proliferation, migration and invasion of CRC cells, while miR-28-3p modulated the expression of NM23-H1 and increased the migration and invasion of CRC cells in vitro [70]. Ling H et al. performed a study based on miRNA microarray profiling in primary CRC tissues of patients with (N = 4) and without (N = 8) metastases. In particular, the expression of miR-224 increased with a positive correlation with tumor burden and microsatellite stability status. SMAD4, a target of miR-224, shows a negative correlation with miR-224 expression in clinical samples. Thus, miR-224 might, in part, promote CRC metastasis through the regulation of SMAD4 [71]. Another differential expression analysis based on microarray showed that miR-139 is downregulated in 34 CRC tissues compared with corresponding noncancer tissues. Restoration of miR-139 did not inhibit CRC cell growth but suppressed CRC cell metastasis and invasion in vitro and in vivo by inhibiting the IGF-IR/MEK/ERK axis and down-regulating the matrix metalloproteinase 2 (MMP-2) [72].
In addition to these experimental studies, several bioinformatics investigations based on the integrated analysis of multiple profiling data were performed to establish CRC-associated signatures. For instance, Jevšinek Skok D et al. analyzed the high-throughput molecular profiling data of 295 CRC samples from The Cancer Genome Atlas (TCGA) database. The genes FN1, TGFB2, RND3, ZEB1 and ZEB2 and the miRNAs miR-200a/b/c-3p, miR-141-3p and miR-429 were selected as the most associated with CRC, while a negative correlation was found between the miRNA miR-200b/c-3p and its target gene FN1 and between miR-200a-3p and its target TGFB2 [73]. Falzone L et al., analyzed the miRNA expression levels observed in CRC samples and normal tissues from different miRNA microarray expression datasets obtained from the Gene Expression Omnibus DataSets database. In this analysis, 19 differentially expressed miRNAs were identified. In addition, it was shown that the up-regulated miRNAs miR-183-5p and miR-21-5p and the down-regulated miRNAs miR-195-5p and miR-497-5p play important roles in the regulation of the mismatch repair mechanism as well as in the Wnt, RAS, MAPK, PI3K, TGF-β and p53 signaling pathways involved in the development and progression of CRC [74]. A bioinformatics study further identified 874 targets for tissue-specific miRNAs and 157 for circulating miRNAs most frequently altered in CRC. In particular, this analysis showed that miR-424-5p, miR-96-5p, miR-1290, miR-224, miR-133a and miR-363-3p target genes known to play a role in CRC, including BRAF, KRAS, EGFR, APC. Moreover, miR-133a and miR-96-5p regulate the PI3K-AKT signaling pathway, which is known to be associated with CRC progression [75]. The data obtained on miR-133a were also confirmed in other tumors of the gastrointestinal tract, including oral cancer and gastric cancer [76,77].
In all mammals, several miRNAs are organized in genomic clusters on single polycistronic transcripts containing two or more miRNAs with similar sequences. Usually, a cluster corresponds to a single transcriptional unit; thus, the members of a miRNA cluster, whether up-regulated or down-regulated, are involved in the regulation of functionally associated genes [78]. Some miRNA clusters are typically up-regulated in CRC, including miR-106a/363, miR-106b/93/25, miR-17/92a-1, miR-181a-1/181b-1, miR-181a-2/181b-2, miR-181c/181d, miR-183/96/182, miR-191/425, miR-200c/141, miR-203a/203b, miR-222/221, miR-29b-1/29a, miR-301b/130b and miR-452/224 [79]. Others clusters are often downregulated in CRC including clusters like the miR-100/let-7a-2/miR-125b-1, miR-99a/let-7c, miR-99b/let-7e/miR-125a, miR-1-2/133a-1, miR-1-1/133a-2 and miR-206/133b, miR-192/194-2 and miR-215/194-1, miR-15a/16-1 and miR-15b/16-2, miR-143/145, miR-302b/302c/302a/302d/367, miR-497/195. Regarding the miR-23a/27a/24-2 cluster, there are conflicting data as it is usually reported as upregulated in CRC, whereas other studies demonstrated its downregulation with a consequent tumor suppressor role (Table 1). A thorough understanding of the transcriptional regulation of these clusters occurring in CRC could lead to a multi-target specific therapeutic approach [80]. Furthermore, all the aforementioned miRNAs can have a potential value for CRC diagnosis, prognosis and susceptibility [81]. For instance, the analysis of the expression levels of the circulating miR-17-3p, miR-92a, and miR-29a analyzed in liquid biopsy sample of individuals at risk for CRC has been proposed as a diagnostic strategy for the early detection of this tumor. Similarly, miR-20a, miR-21, miR-106a, miR-181b, and miR-203 were associated with poor survival [82], while increased levels of miR-155, miR-223, miR-31 and miR-26b were correlated with MSI-H status [83].
Table 1.
Summary of the most frequently dysregulated miRNA clusters in CRC.
| Cluster | Dysregulation | Function/Role | Related Information | miRNA | Target | Reference |
|---|---|---|---|---|---|---|
| miR-143/145 | Downregulated | Tumor suppressors | Frequently downregulated in CRC due to hypermethylation of their CpG islands. | miR-145 | IRS1 | [84] |
| MUC1 | [85] | |||||
| BRAF and CD44 | [86] | |||||
| IGF1R | [87] | |||||
| KLF5 | [86] | |||||
| MDM2 | [88] | |||||
| MYC | [89] | |||||
| NRAS | [90] | |||||
| FSCN1 | [91] | |||||
| CDCA3 | [92] | |||||
| MAPK1 | [93] | |||||
| SIP1 | [94] | |||||
| catenin δ-1 | [95] | |||||
| YES and STAT1 | [96] | |||||
| PAK4 | [97] | |||||
| ERG | [98] | |||||
| SOX2 | [99] | |||||
| miR-143 | BRAF and CD44 | [86] | ||||
| MDM2 | [88] | |||||
| MYO6 | [100] | |||||
| KRAS | [101] | |||||
| MAPK7 | [102] | |||||
| DNMT3A | [103] | |||||
| HKII | [104] | |||||
| MACC1 | [105] | |||||
| BCL2 | [100] | |||||
| miR-1-2/133a-1, miR-1-1/133a-2, miR-206/133b | Downregulated | Tumor suppressors | myo-miRNAs, muscle-specific miRNAs generally down-regulated in CRC cell lines and tissue samples. | miR-133a | AFTPH | [106] |
| FSCN1 | [107] | |||||
| LASP1 | [108] | |||||
| RFFL | [109] | |||||
| miR-133b | PTBP1 | [110] | ||||
| MET | [111] | |||||
| CXCR4 | [112] | |||||
| miR-206 | NOTCH3 | [113] | ||||
| MET | [114] | |||||
| miR-1 | PTBP1 | [110] | ||||
| NOTCH3 | [115] | |||||
| SMAD3 | [116] | |||||
| miR-497/195 | Downregulated | Tumor suppressors | Expression of miR-497-5p and miR-195-5p is down-regulated in CRC. Increased expression of miR-497-5p or miR-195-5p is associated with decreased cell proliferation, migration and EMT. | miR-497 | IGF1-R | [117] |
| FRA1 | [118] | |||||
| IRS1 | [119] | |||||
| KSR1 | [120] | |||||
| miR-195 | CDK8 | [121] | ||||
| YAP1 | [122] | |||||
| BCL2 | [123] | |||||
| BCL2L2 | [124] | |||||
| γ-catenin | [125] | |||||
| CCNE1 | [126] | |||||
| AXIN2 | [127] | |||||
| miR-15a/16–1,miR-15b/16–2 | Downregulated | Tumor suppressors | Although more frequently miR-15/16 in CRC is detected downregulated, there are also studies documenting up-regulation of miR-15/16 expression. Similarly, better survival is generally correlated with high expression of miR-15/16; however, an association of worse survival with high expression of miR-15/16 has also been documented. | miR-15a | AP4 | [128] |
| GPX4 | [129] | |||||
| SIRT4 | [130] | |||||
| KDM4B | [131] | |||||
| miR-15b | DCLK1 | [132] | ||||
| NF-κB1 and IKK-α | [133] | |||||
| ACOX1 | [134] | |||||
| miR-16 | AP4 | [128] | ||||
| BIRC5 | [135] | |||||
| HMGA2 | [136] | |||||
| ALDH1A3 | [137] | |||||
| COX2 | [138] | |||||
| ITGA2 | [139] | |||||
| miR-192/194–2 and miR-215/194–1 | Downregulated | Tumor suppressors | Frequently downregulated in CRC, the reported functions of miR-192/194-2 and miR-215/194-1 clusters indicate their tumour-suppressive roles as cell cycle arrest and inhibition of cell adhesion are often observed after their overexpression. | miR-192 | CAV1 | [140] |
| EIF5A2 | [141] | |||||
| miR-194 | VAPA | [142] | ||||
| KLK10 | [143] | |||||
| MAP4K4 | [144] | |||||
| THBS1 | [145] | |||||
| PDK1, AKT2 and XIAP | [146] | |||||
| SSH2 | [147] | |||||
| SOX5 | [148] | |||||
| SIRT1 | [149] | |||||
| miR-215 | EREG and HOXB9 | [150] | ||||
| Atg14 | [151] | |||||
| CDX1 | [152] | |||||
| miR-183/96/182 | Upregulated | Oncogenes | Frequently downregulated in CRC, miR-183/96/182 cluster promote migration, invasion and metastasis. | miR-183 | ATG5 | [153] |
| FOXO1 | [154] | |||||
| PFN2 | [155] | |||||
| DNAJB4 | [156] | |||||
| RCN2 | [157] | |||||
| miR-96 | TPM1 | [158] | ||||
| CPA6 | [159] | |||||
| AMPKα2 | [160] | |||||
| TP53INP1, FOXO1, FOXO3a | [161] | |||||
| miR-182 | NAMPT | [162] | ||||
| CFL1 | [163] | |||||
| TIAM1 | [164] | |||||
| DAB2IP | [165] | |||||
| SATB2 | [166] | |||||
| ST6GALNAC2 | [167] | |||||
| FoxF2 | [168] | |||||
| TSP-1 | [169] | |||||
| miR-17/92a | Upregulated | Oncogenes | miRNAs of miR-17/92a cluster can act as oncogenes and promote proliferation, angiogenesis and inhibit differentiation and apoptosis. | miR-17 | HSPB2 | [170] |
| MFN2 | [171] | |||||
| BLNK | [172] | |||||
| RUNX3 | [173] | |||||
| PLCD1 | [174] | |||||
| CADM2 | [175] | |||||
| VEGFA | [176] | |||||
| P130 | [177] | |||||
| VIM | [178] | |||||
| SPOP | [179] | |||||
| PTEN | [180] | |||||
| RND3 | [181] | |||||
| hCNT1 | [182] | |||||
| miR-18a | ING4 | [183] | ||||
| BTG3 | [184] | |||||
| PIAS3 | [185] | |||||
| TBPL1 | [186] | |||||
| miR-19a | PTEN | [187] | ||||
| IREB2 | [188] | |||||
| CLCA4 | [189] | |||||
| THBS1 | [190] | |||||
| KRAS | [191] | |||||
| FOXF2 | [192] | |||||
| NPEPL1 | [193] | |||||
| miR-19b | FBXW7 | [194] | ||||
| PPP2R5E | [195] | |||||
| ACSL1, ACSL4, and SCD | [196] | |||||
| miR-20a | ATG5 and FIP200 | [197] | ||||
| CXCL8 | [198] | |||||
| PDCD4 | [199] | |||||
| MICA | [200] | |||||
| FOXJ2 | [201] | |||||
| PTEN | [202] | |||||
| SMAD4 | [203] | |||||
| miR-92a | DKK3 | [204] | ||||
| SOCS3 | [205] | |||||
| NF2 | [206] | |||||
| KLF4 | [207] | |||||
| miR-200c/141 | Upregulated | Biomarkers | miRNA members of the miR-200c/141 cluster are found to be frequently upregulated in CRC at both tissue and circulating levels. | miR-200c | VLDLR | [208] |
| KIF14 | [209] | |||||
| JNK2 | [210] | |||||
| miR-141 | SIP1 | [211] | ||||
| EGFR | [212] | |||||
| ZEB1 and ZEB2 | [213] | |||||
| MAP4K4 | [214] | |||||
| DLC1 | [215] | |||||
| miR-203a/203b | Upregulated | Biomarkers | miR-203a/203b are generally overexpressed in CRC and are associated with poor prognosis. | miR-203a | PTEN | [216] |
| RNF6 | [217] | |||||
| PDE4D | [218] | |||||
| THBS2 | [219] | |||||
| CREB1 | [220] | |||||
| miR-203b | BCL2L1 | [221] | ||||
| miR-222/221 | Upregulated | Circulating biomarkers | Both members of the miR-222/221 cluster are positively correlated with disease recurrence and are frequently detected upregulated in in the circulation of patients with CRC. | miR-222 | SPINT1 | [222] |
| CD4 | [223] | |||||
| ADAM-17 | [224] | |||||
| MIA3 | [225] | |||||
| ATF3 | [226] | |||||
| BRG1 | [227] | |||||
| MST3 | [228] | |||||
| miR-221 | SPINT1 | [222] | ||||
| CD4 | [223] | |||||
| TP53INP1 | [229] | |||||
| CDKN1C/p57 | [230] | |||||
| SOCS3 | [231] | |||||
| RECK | [232] | |||||
| 23a/27a/24–2 | conflicting data | conflicting data | Members of miR-23a/27a/24-2 cluster has been proposed to control the cell cycle, cell proliferation, cell death and cell differentiation. | miR-23a | SEMA6D | [233] |
| APAF1 | [234] | |||||
| miR-27a | c-Met | [235] | ||||
| BTG2 | [236] | |||||
| RMST | [237] | |||||
| miR-24 | NRP1 | [238] | ||||
| miR-29b-1/29a | Upregulated | Biomarkers | miR-29b-1/29a seems to have a biomarker value for risk, recurrence, metastasis and survival outcome of CRC. | miR-29b-1 | SMAD3 | [239] |
| miR-29a | RPS15A | [240] | ||||
| TNFAIP3 | [241] | |||||
| KLF4 | [242] | |||||
| miR-301b/130b | Upregulated | Oncogenes | Members of the miR-301b/130b cluster act as oncogenes by promoting cell growth and migration and may serve as biomarkers for the diagnosis of CRC. | miR-301b | HOXB1 | [243] |
| miR-130b | CHD9 | [244] | ||||
| integrin α5 | [245] |
These and other studies allowed researchers to identify miRNAs potentially involved in the development and progression of tumors, including CRC (Table 1).
Notably, all these studies report data obtained on tissue or liquid biopsy samples or both, however, it is important to discriminate the reasons behind altered tissue and circulating expression levels of miRNAs. Altered expression of miRNAs in CRC tissues can be determined by intrinsic changes within cancer cells, including genomic alterations (e.g., amplifications, deletions), epigenetic dysregulation (e.g., DNA methylation, histone modifications), disrupted transcription factor activity, and impaired miRNA processing mechanisms. These changes often reflect the biology of the tumor itself, its interaction with the surrounding microenvironment, and the broader pathological transformation of the organ [246,247].
Conversely, changes in the circulating expression of miRNAs may have a more complex and multifactorial origin. They can be actively secreted by tumor cells via exosomes, microvesicles, or protein complexes, or passively released as a consequence of tumor cell death (apoptosis or necrosis). However, not all circulating miRNAs are directly tumor-derived. Some may represent a physiological systemic response to the presence of tumor, involving immune modulation, inflammation, or stress signaling [248]. Therefore, while tissue-derived miRNAs provide insights into the tumor's molecular profile, circulating miRNAs may serve as minimally invasive biomarkers that reflect both tumor biology and the host's systemic response. A comprehensive understanding of these distinct yet interconnected sources of miRNA alterations is essential for the development of robust biomarkers and targeted therapeutic strategies in colorectal cancer.
3.1. microRNAs in colorectal cancer development
In approximately 80 % of cases, the pathogenesis of CRC follows the adenoma-carcinoma sequence. In the vast majority of these cases, the development of CRC starts with an APC mutation responsible for chromosomal instability and the gradual accumulation of molecular and epigenetic changes. The remaining 15–20 % of CRC cases arise via alternative pathways, such as defective mismatch repair systems, CIMP hypermethylation, or BRAF activation. From a molecular point of view, the tumor suppressor genes APC, TP53, PTEN, TGFβ, SMAD4, the oncogenes KRAS, BRAF, HER2, and the tumor-modifying genes COX2, PPAR and CHEK2 play an essential role in the development of CRC [249]. All these genes cause the activation of inflammatory signaling pathways and oncogenic signaling pathways. Besides activating mutations, these signaling pathways are also finely regulated by single miRNAs or by miRNA clusters/groups. If the expression of these miRNAs is altered, the proper functioning of these important signaling pathways may be impaired [250].
Another mechanism promoting CRC development is mediated by chronic inflammation due to inflammatory diseases, including colitis and inflammatory bowel diseases (IBDs). Colitis and chronic inflammation are responsible for immune cell infiltration, oxidative stress and the production of pro-inflammatory cytokines which induce genetic and signal transduction alterations associated with neoplastic transformation [[251], [252], [253]]. In this intricate scenario, different miRNAs regulating interleukin production, oxidative stress and p53 signaling were identified as associated with both IBDs and CR,C suggesting the epigenetic regulation of colitis-mediated carcinogenesis [251].
The miR-143/145 cluster is highly expressed in the colon and is typically reported to be downregulated in CRC and other cancers. Importantly, miR-143/145 cluster is not expressed in colon epithelial cells but in mesenchymal cells such as fibroblasts and smooth muscle cells. Through regulation of multiple targets, these miRNAs exert potent effects on cancer cell growth and tumorigenesis [254]. miR-145 plays a crucial role in cancer biology by directly targeting the pluripotency factors OCT4, SOX2, and KLF4. These factors are integral to the maintenance of stem cell pluripotency, which is also regulated by transcription factors like NANOG, SOX2, OCT4, KLF4, LIN28, and c-MYC. In this context, the loss of miR-145 impairs differentiation and leads to increased levels of OCT4, SOX2, and KLF4 [255]. Furthermore, miR-145 is an inhibitor of the embryonic stem cell program, promoting cell differentiation and inhibiting the proliferation of SW48 cells harboring KRAS mutation [256].
Another mechanism responsible for the development of CRC is the inactivation of the APC gene. APC encodes a large scaffolding protein that is part of the AXIN destruction complex, which is required for phosphorylation and degradation of β-catenin. β-catenin is a key effector of Wnt signaling that interacts with the HMG-box DNA-binding factor TCF4 (TCF/L2) to drive transcription of target genes. If APC loses its function, β-catenin levels increase. Most mutations in APC generate premature stop codons that lead to the production of truncated proteins depleted of β-catenin binding sites. Consequently, β-catenin accumulates and stimulates the Wnt signaling pathway, leading to active transcription of target genes. In this scenario, miRNAs can modulate Wnt signaling through the repression of some of the components of this pathway. A study assessed the relationship between the downregulation of the miR-143/145 cluster and genetic aberrations in APC. In particular, it has been proposed that the downregulation of the miR-143/145 cluster often occurs before the osnet of APC gene aberrations. Thus, it may be considered an important epigenetic event in the early phase of CRC development [257]. The miR-143/145 cluster can also modulate the Ras-MAPK pathway; specifically, miR-145 targets EGFR, RASA1, MEKK, and RREB1, while miR-143 targets KRAS, ERK1/2, and ELK1. Furthermore, the miR-143/145 proximal promoter is negatively regulated by the K-Ras-RREB1 feedback loop. Specifically, RREB1 is activated by the MAPK pathway and negatively represses the miR-143/145 promoter through the interaction with two Ras-responsive elements (RREs) [254,258]. Other recognized targets of miR-145 are insulin receptor substrate 1 [84], Src-related tyrosine kinase YES [96], c-MYC and ERK5 [259], catenin δ-1 [95], PXN [260], FSCN1 [91], MUC1 [85]. Many studies have also identified several targets for miR-143, such as MDM2 [88], HKII [104], DNMT3A [103], MAPK7 [102], KRAS [101], BRAF and CD44 [86]. All these factors, when dysregulated, promote CRC development by increasing cell cycle progression, cell proliferation, cell metabolism, cell survival, immune evasion, and metastasis formation.
The miR-23b/27b/24 cluster has two paralogs in humans, the miR-23b/27b/24-1 cluster, which is encoded within an intron on the C9orf3 gene located on chromosome 9, and miR-23a/27a/24-2 located on chromosome 19. The miR-23b/27b/24 cluster seems to have a role in cell migration by targeting FOXP2 through miR-23b and miR-27b [261]. Although this cluster is generally found to be upregulated, several studies have reported a downregulation and a tumor suppressor role of its members in CRC. miR-27b acts as a tumor suppressor miRNA by targeting ARFGEF1 and the paxillin/c-Src circuit at focal adhesions [262]. miR-23b has pleiotropic functions; thus if dysregulated, it can lead to a variety of diseases, including cancer. In CRC, the downregulation of miR-23b modulates the expression levels of its target PDE7A, which is involved in the development of this tumor [263]. miR-27a plays a critical role in colon tumorigenesis, possibly influencing the anti-tumor immune response. Specifically, miR-27a modulates MHC surface exposure by targeting calreticulin, a highly conserved chaperone protein, important for the assembly and expression on the cell surface of MHC class I molecules and thus for the recognition of the presented tumor-associated antigen by CD8 T-cells [264].
The miR-10a/b, miR-99a/b, miR-100 and miR-125a/b, constituting the miR-10 family, possess tumor-suppressive properties. miR-100 targets RAP1B and modulates CRC cell growth and invasion phenotype [265]. miR-125b targets TP53 and other regulators of apoptosis, including PUMA, BAK1 and cyclin C, thus regulating cell cycle transition [266]. Moreover, miR-99, miR-100, and miR-125 genomic loci are physically clustered with the loci encoding for the let-7 miRNA family. Therefore, chromosomal deletions or transcriptional silencing of these genomic regions may influence both miR-10 and miR-let-7 families, although no validations of these hypotheses have been documented yet. Notably, let-7 family members play an important tumor-suppressor role due to their anti-proliferative function and pro-differentiation effects. LIN28A and LIN28B are specific and strong inhibitors of let-7 members by interfering with the biogenesis of the whole let-7 family [267]. Indeed, LIN28B is found overexpressed in several tumor types, including CRC, where it promotes colon cell malignant transformation through the suppression of let-7 [268].
The miR-34a, miR-34b and miR-34c family members regulate the expression of genes involved in the cell cycle, cell growth, DNA damage repair and apoptosis. miR-34a and miR-34b/c are transcribed from two different loci, both direct transcriptional targets of the tumor suppressor TP53 [269,270]. In turn, miR-34 directly represses MDM4 (HDM4 in humans), which encodes a RING-finger protein that binds p53 and blocks its ability to activate target genes. Thus, miR-34a may promote tumorigenesis, especially in the case of p53 haploinsufficiency [271]. A study performed by Gao J et al. on a cohort of 268 CRC patients showed that miR-34a-5p inhibits CRC metastasis by repressing cell growth, migration and invasion, inducing cell apoptosis and cell cycle arrest in a p53-dependent manner [272]. p53 transactivates miR-34, which represses the transcriptional activity TCF/LEF complexes by targeting genes encoding elements of the Wnt pathway. Thus, in CRC, loss of p53 or miR-34 promotes neoplastic progression, enhancing the Wnt signaling [273]. The expression of miR-34 may also depend on its methylation status. For instance, in FFPE colon cancer samples compared to normal colon mucosa, miR-34 was downregulated due to promoter hypermethylation [274]. Besides TP53, MYC can also promote the expression of miR-34. MK5 indirectly regulates MYC translation by activating the expression of miR-34b and miR-34c, which in turn bind the 3′UTR of MYC. Specifically, MK5 phosphorylates FoxO3a, thereby promoting its nuclear localization, inducing miR-34b/c expression and the inhibition of cancer cell proliferation [275]. The miR-34 family also plays a role in the regulation of tumor-initiating cancer stem cells (TICs). In CRC, TICs generally present intrinsic drug resistance mechanisms leading to chemotherapeutic failure. Such drug resistance mechanisms seem to be associated with miR-34a and miR-146 dysregulation [276]. miR-34a is a cell fate determinant in early-dividing colon cancer stem cells (CCSCs). Specifically, miR-34a targets Notch1 mRNA to generate a net threshold response in which a bimodal Notch signal specifies the choice between self-renewal and differentiation enabling cells to distinctly choose between maintaining a stem-like state or committing to differentiation [277]. These data suggest that miRNAs can indirectly promote asymmetric division, but it remains unclear whether and how miRNAs and proteins drive the cell fate. Another study showed that miR-34a targets Numb in early CCSCs and inhibits asymmetric division in cooperation with miR-146 [278]. In this scenario, it is known that the correct number of stem cells for self-renewal is maintained through asymmetric cell division (ACD). In cancer cells, the deregulation of ACD causes an alteration of the stem cell pool and promotes tumor growth. The EMT inducer Snail is responsible for the switch from ACD to symmetrical cell division (SCD) in CRC. Specifically, Snail induces the expression of miR-146a via the β-catenin-TCF4 complex. In turn, miR-146a targets Numb to stabilize β-catenin, which forms a feedback loop to maintain Wnt activity and directs SCD [279].
Another key relevant miRNA family is that of the miR-17/92 cluster, whose miRNAs actively cooperate with several oncogenic miRNAs, including miR-21-5p, miR-31, miR-135b and miR-145. All these miRNAs were investigated in clinically diagnosed early-stage CRC (24 colonic polyps containing early-stage adenocarcinoma). In particular, miR-17 showed increased expression in the transition zone from normal to adenomatous tissue, while miR-21-5p expression increased in the tumor-associated stroma, with an even more evident increase from adenoma to adenocarcinoma; in contrast, miR-145 expression decreased gradually during the normal-adenoma-adenocarcinoma progression. Therefore, these miRNAs may play a role in CRC development [280].
The miR-17/92 cluster, which includes miR-17, miR-18a, miR-19a, miR-20a, miR-19b, and miR-92a, is commonly upregulated in both hematological malignancies and solid tumors, including CRC [281,282]. Its overexpression is often associated with c-Myc activation and copy number gain of its locus on chromosome 13q31 [283]. Functionally, the members of the miR-17/92 family promote cell proliferation and angiogenesis, while inhibiting differentiation and apoptosis by modulating key oncogenic signaling pathways, such as JAK/STAT, PI3K/AKT/mTOR, and PTEN [280,284]. The expression of the miR-17/92 cluster is also modulated by the APC-β-catenin pathway; specifically, activated β-catenin resulting from APC loss can bind to and activate the miR-17/92 promoter region [285]. The expression of miR-18a correlates with APC mutations and is highly expressed in colon cancer [68]. In CRC, miR-20 influences the activation of the cyclin-dependent kinase inhibitor 1A/p21 (CDKN1A/p21), which negatively regulates TGFβ, thus preventing its antiproliferative effect [286]. miR-17/92 cluster is also associated with invasion, metastasis and decreased survival. Of the six members of the miR-17/92 cluster, miR-19a and miR-19b have been described as key promoters of cancer development and cancer cell proliferation. Even belonging to the miR-17/92 cluster, the miR-18a plays a conflicting role in CRC since it was found downregulated in CRC, suggesting that this miRNA may have tumor-suppressive effects compared to the other members that are often found overexpressed and associated with CRC cell proliferation [287]. Humphreys KJ et al., suggested that individual miR-17/92 cluster members have opposite effects on CRC cell proliferation. Specifically, miR-19a and miR-19b were primarily responsible for increased cell proliferation, while miR-18a showed the opposite effect by silencing the transcription of genes involved in cell proliferation, such as NEDD9 and CDK19 [288]. Thus, high miR-17/92 cluster activity without an increase in miR-18a can promote CRC progression. Indeed, while other members of the miR-17/92 cluster activate the PI3K pathway, thereby promoting cell growth, miR-18a can suppress growth by targeting CDC42 and CCND1 [287]. In addition, several post-transcriptional regulatory mechanisms influence the abundance of specific members of the miR-17/92 cluster. For instance, it was observed that miR-18a is the only member of the miR-17/92 cluster that requires the RNA-binding protein hnRNPA1 for its processing [289]. Furthermore, pri-miR-17-92 has a compact globular tertiary structure, which makes difficult the maturation of miRNAs [290].
Besides its role within the miR-17/92 cluster, miR-92a is also a member of a conserved miRNA family including miR-92a-1, miR-92a-2, miR-363 and miR-25. miR-92a is overexpressed in several tumors and its upregulation was associated with poor long-term prognosis in CRC [291]. In CRC, miR-92a exerts its tumorigenic role by influencing several mechanisms that lead to the downregulation of tumor suppressor and apoptotic genes and the upregulation of genes involved in cell proliferation [64]. Yamada N. et al., suggest that at the intracellular level, miR-92a targets DKK3, while when secreted through MVs this miRNA promotes angiogenesis [292]. It was also demonstrated that the expression levels of miR-92a are positively regulated by the pro-inflammatory IL-6/STAT3 pathway. As a result, miR-92a targets KLF4, GSK3β and DKK3 involved in the negative regulation of Wnt/β-catenin signaling [293]. In addition, miR-92a plays a crucial role in the regulation of apoptosis by targeting the anti-apoptotic molecule BCL-2-interacting mediator of cell death (BIM) [294].
Moreover, miR-17 belongs also to the miR-17 family consisting of miR-17, miR-18a/b, miR-20a/b, miR-93, and miR-106a/b. miR-17-5p is an oncogenic miRNA that regulates cancer development and progression by targeting P130 (RB transcriptional co-repressor like 2, RBL2) and subsequently activating the Wnt/β-catenin pathway [177]. Transfection of CRC cells with a miR-17 inhibitor reduced the proliferation of cancer cells by inducing G0/G1 arrest via RND3 targeting [181]. Ataxia telangiectasia mutated (ATM) gene encodes a key enzyme involved in DNA damage repair. ATM transcript is targeted by miR-18a that, when overexpressed in CRC, affects DNA damage repair [295,296]. miR-20a affects the cellular response to TGF-β and favors G1/S transition, promoting cell cycle progression [297]. miR-106b appears to have functions in the EMT of CRC. Indeed, miR-106b downregulation induces cytoskeletal reorganization and increases the expression of Rho GTPases (RAC1 and CDC42) and TIAM1. TGF-β1 can downregulate miR-106b and in turn, miR-106b also influences TGF-β1 expression, establishing a negative feedback loop that regulates the expression of PRRX1, a direct target of miR-106b [298].
The miR-135a/b family is often upregulated in CRC and targets APC, thus suppressing its expression and inducing the downstream activation of the Wnt pathway [299]. Other miR-135a/b targets associated with the Wnt signaling pathway are the secreted frizzled-related protein 4 (SFRP4), which binds and represses extracellular Wnt proteins [300] and ZNRF3, which is involved in the negative regulation of the Wnt pathway [301]. Valeri N et al., demonstrated that the overexpression of miR-135b is associated with APC loss, the deregulation of the PTEN/PI3K pathway and the overexpression of SRC. The upregulation of miR-135b also promotes malignant transformation and tumor progression, especially in sporadic and inflammatory bowel disease-associated human CRC. The overexpression of this miRNA also correlates with tumor stage and poor patients’ prognosis [302].
Besides those already mentioned families, several other miRNAs involved in the regulation of Wnt/β-catenin signaling have been identified. In particular, miR-552 is able to regulate the Wnt/β-catenin signaling pathway by targeting the cell fate-determining factor Dachshund family transcription factor 1 (DACH1) [303]. C-MYC can stimulate the expression of miR-552 by binding the miR-552 promoter. In turn, miR-522 targets TP53 exerting its oncogenic properties [304]. miR-590-3p targets WIF1 which inhibits WNT and DKK1 and in turn the LRP6 co-receptor inhibiting the β-catenin-dependent Wnt signaling [305]. miR-425-5p may promote tumorigenesis and metastasis by activating catenin-δ1 (CTNND1) mediated β-catenin pathway [306]. miR-29b, miR-29c and miR-93 are other inhibitors of Wnt ligands or β-catenin-associated factors. miR-29b targets BCL9L, a co-activator of β-catenin [307], miR-29c targets GNA13 and PTP4A [308]; miR-93 targets SMAD7, which promotes nuclear accumulation of β-catenin [309]; miRNA-29a targets the phosphoinositide 3-kinase, phosphorylated (p)-protein kinase B (AKT), p-glycogen synthase kinase 3β (GSK3β) [310].
Finally, one of the most widely studied oncomiRs involved in CRC pathogenesis is miR-21-5p, which is responsible for the regulation of multiple tumor-promoting mechanisms. A recent study detected high levels of miR-21-5p in CRC-derived exosomes. The authors demonstrated that the treatment of colon cells with isolated CRC-derived exosomes or miR-21-5p mimic leads to increased expression of genes involved in cell proliferation, invasion and extracellular matrix formation, including PDCD4, TPM1, and PTEN [311]. Particularly, PDCD4 is a pro-inflammatory factor that is activated by apoptotic stimuli and inhibits tumor proliferation by modulating NF-κB activity. In the case of low miR-21-5p expression, inflammatory infiltration decreased and fewer tumor-associated inflammatory cytokines, such as TNF-α, IL-6, IL-17A and IL-21, were produced. Thus, miR-21-5p seems to promote the development of colon cancer by promoting inflammation [312]. Furthermore, another study observed that miR-21-5p expression increased during the transition from precancerous colorectal adenoma to advanced carcinoma. In addition, the expression patterns of miR-21-5p and its target PDCD4 were mutually exclusive [313]. miR-21-5p may also potentiate TCF4/β-catenin-mediated transcriptional activation [314,315]. Lin PL et al., analyzed the APC mutation from 165 CRC samples and found that miR-21-5p was associated with β-catenin phosphorylation at Ser552 via the PTEN/AKT axis and played a critical role in β-catenin nuclear translocation in APC-mutated cells, but not in APC-wild-type cells [314].
All these studies indicate that CRC-associated oncogenic and tumor-suppressive signaling pathways and inflammatory pathways are finely regulated by specific miRNAs, such as miR-145, miR-34, and the miR-17/92 cluster, which play essential roles in modulating cell differentiation, proliferation, apoptosis, and immune response. Dysregulation of these miRNAs fosters CRC progression through mechanisms like stem cell pluripotency, Wnt signaling, and EMT, underscoring their potential as therapeutic targets (Table 2).
Table 2.
miRNA clusters and their targets involved in CRC development.
| miRNA/Cluster | Key Targets | Pathways Affected | Role in CRC | References |
|---|---|---|---|---|
| miR-145 | OCT4, SOX2, KLF4, EGFR, RASA1, MEKK, RREB1 | Wnt, Ras-MAPK, Pluripotency factors | Tumor suppressor, downregulated in early CRC, promotes cell differentiation | [[255], [256], [257]] |
| miR-143 | KRAS, ERK1/2, ELK1, MDM2, HKII, DNMT3A | Ras-MAPK, Cell cycle, Metabolism | Tumor suppressor, modulates KRAS pathway, downregulated early in CRC | [257][88,101,104] |
| miR-23b/27b/24 Cluster | FOXP2, ARFGEF1, PDE7A | Cell migration, c-Src circuit, Immune response | Tumor suppressor, downregulation affects cell migration, immune response modulation | [[261], [262], [263]] |
| miR-10 Family (miR-10a/b, miR-99a/b, miR-100, miR-125a/b) | RAP1B, p53, PUMA, BAK1, Cyclin C | Apoptosis, Cell cycle, Invasion | Tumor suppressor, regulates apoptosis and cell growth, downregulation in CRC | [265,266] |
| miR-let-7 | - | Proliferation, Differentiation | Tumor suppressor, let-7 regulates differentiation, suppressed by LIN28 A/B | [268] |
| miR-34 Family (miR-34a/b/c) | MDM4, β-catenin, Numb | p53, Wnt, DNA damage response | Tumor suppressor, regulates cell cycle and apoptosis, loss of miR-34 linked to p53 deficiency | [269,271,272] |
| miR-17–92 Cluster (miR-17, miR-18a, miR-19a/b, miR-20a, miR-92a) | NEDD9, CDK19, PTEN, Cyclin D1 | PI3K/AKT/mTOR, JAK/STAT, Cell proliferation, Apoptosis | Oncogenic, promotes proliferation, inhibits apoptosis, miR-18a tumor-suppressive, others oncogenic | [281,282,285,286] |
| miR-92a | DKK3, KLF4, GSK3β, BCL-2, BIM | Wnt/β-catenin, Apoptosis | Oncogenic, overexpression leads to poor prognosis, targets tumor suppressor genes | [291,293,294] |
| miR-135a/b | APC, SFRP4, ZNRF3 | Wnt, PTEN/PI3K | Oncogenic, suppresses APC and activates Wnt signaling | [299,301,302] |
| miR-522 | TP53, DACH1 | Wnt/β-catenin | Oncogenic, stimulated by c-Myc, regulates Wnt pathway via TP53 targeting | [303,304] |
| miR-590-3p | WIF1, DKK1 | Wnt | Inhibits Wnt/β-catenin signaling by targeting WIF1 and DKK1 | [305] |
| miR-425-5p | CTNND1 (catenin δ-1) | β-catenin pathway | Promotes tumorigenesis and metastasis | [306] |
| miR-29 Family (miR-29a/b/c) | BCL9L, GNA13, PTP4A | Wnt/β-catenin | Regulates β-catenin co-activators | [307,308] |
| miR-93 | SMAD7 | Wnt/β-catenin | Inhibits SMAD7 and promotes β-catenin accumulation | [309] |
| miR-21 | PDCD4, TPM1, PTEN | NF-κB, Inflammation, ECM formation | Oncogenic, promotes tumorigenesis by inducing inflammation, upregulated in CRC exosomes | [[311], [312], [313]] |
In this intricate genetic and epigenetic scenario, other ncRNAs have been found to influence miRNA expression, adding another layer to CRC pathogenesis. Indeed, miRNA dysregulation may be due to aberrant transcriptional activity, a change in epigenetics, altered miRNA biogenesis, as well as sponging of lncRNAs. For instance, miR-200a and miR-138, known to attenuate EMT, are modulated by the H19 lncRNA that is upregulated in CRC tissues [316]. Another lncRNA-miRNA interaction found in CRC is between LINC00152 and miR-139-5p, which results in increased cell proliferation, promotion of metastasis, and confers resistance to 5-FU [317]. NEAT1 lncRNA is up-regulated in CRC tissues and correlates with poor overall and disease-free survival. NEAT1, functioning as a ceRNA, modulates miRNA-34a expression, resulting in the repression of the miR-34a/SIRT1 axis and in the activation of the Wnt/β-catenin signaling pathway [318]. XIST is another lncRNA that negatively modulates miR-34a expression, leading to an increase of its target WNT1 [319]. ZEB1-AS1 lncRNA is significantly upregulated in CRC and promotes CRC cell proliferation, repressing apoptosis via the downregulation of miR-181a-5p and positively regulating the Wnt/β-catenin signaling [320]. The downregulation of miR-181a-5p can also be mediated by CRNDE lncRNA sponging, which results in the inhibition of cell proliferation and the reduction of chemoresistance [321]. The lncRNA MIR4435-2HG increases tumor growth and metastasis formation by sponging miR-206 that regulates the Yes-associated protein 1 (YAP1) transcription factor, a major effector and downstream regulator of the Hippo pathway [322]. YAP1 expression is also regulated by miR-139-5p, which in turn is regulated by the overexpressed oncogenic lncRNA RP11-757G1.5 [323]. The LINC00689 lncRNA can target miR-31-5p. In CRC, LINC00689 is downregulated, while miR-31-5p is upregulated. The target of miR-31-5p, Large Tumour Suppressor Kinase 2 (LATS2), phosphorylates YAP1, which regulates genes involved in cell proliferation, death, and migration. Furthermore, the activation of YAP1 could stimulate the activity of other transcription factors such as SMAD, trigger EMT and thus increase metastasis and invasiveness of cancer cells [324].
3.2. microRNAs in colorectal cancer progression
Besides their key role in CRC development, miRNAs also influence CRC progression and aggressiveness. Indeed, by targeting genes involved in EMT, apoptosis, cell growth and proliferation, miRNAs can promote angiogenesis, metastasis and tumor progression [325].
miR-155 regulates a variety of cellular functions, including EMT. The expression level of miR-155 is higher in primary CRC tissue than in adjacent normal mucosa. miR-155 has been shown to increase the migratory and invasive capacity of SW480 inducing claudin-1 expression [326]. Through RNA sequencing, another study revealed high levels of miR-146a-5p and miR-155-5p in CRC cells overexpressing the C-X-C motif chemokine receptor 7 (CXCR7). Specifically, CXCR7 binds the C-X-C motif chemokine ligand 12 (CXCL12), favoring the formation of CRC metastasis. In this process, CAFs are also involved in tumor progression through the secretion of both miR-146a-5p and miR-155-5p via exosome trafficking. In particular, CAFs may take up these miRNAs promoted by the JAK2-STAT3/NF-κB signaling. With a positive feedback loop, CAF-produced miR-146a-5p and miR-155-5p target the suppressor of cytokine signaling 1 (SOCS1) and the zinc finger and BTB domain containing 2 (ZBTB2), promoting the production of inflammatory cytokines, including IL-6, TNF-α, TGF-β and CXCL12 favoring tumor progression [327].
Tumor-associated macrophages (TAMs) can also promote CRC initiation and progression by influencing miRNA expression. Since exosomes derived from M2 macrophages (MDE) have high levels of miR-21-5p and miR-155-5p, they may contribute to migration and invasion in CRC [328]. Moreover, the tumor-promoting role of miR-155-5p was also observed in FAP patients vs non-FAP controls, where a significant downregulation of miR-155-5p expression was found in FAP patients and APC and β-catenin mutant colorectal cancer cell lines. Furthermore, miR-155-5p can regulate WNT/β-catenin signaling by targeting both AXIN1 and TCF4 [329].
miR-34 also seems to play a role in CRC progression. The expression of miR-34 in CRC is significantly downregulated. In SW480 cells, miR-34a attenuates migration and invasion by targeting Notch1 and Jagged1, suggesting a key role in suppressing CRC metastasis [330]. The ectopic expression of miR-34a in HCT-116 and RKO colon cancer cell lines caused complete suppression of cell proliferation and induced senescence-like phenotypes through the modulation of the E2F signaling pathway [331]. Other studies confirmed the role of miR-34 in the suppression of EMT. Specifically, ZNF281 is one of the putative targets of miR-34. Noteworthy, SNAIL induces EMT by activating ZNF281 transcription and repressing miR-34a/b/c, which cannot inhibit ZNF281 mRNA. Besides its role in EMT, ZNF281 overexpression also induces the stemness markers LGR5 and CD133 [332]. As described before, p53 transcriptionally activates miR-34a and, in turn, miR-34a downregulates the expression of silent information regulator 1 (SIRT1). By suppressing miR-34, SIRT1 promotes apoptosis in WT human colon cancer cells but not in those with deficient p53 [333]. miR-34 also plays a key role in inflammation as demonstrated by the active loop involving IL-6R/STAT3/miR-34a, which is required for CRC EMT, invasion and metastasis. This axis is also associated with lymph node and distant metastasis in CRC patients [334].
Communication between tumor cells and blood capillaries plays an important role in tumor growth, invasion, and spreading. A coculture assay showed that SW480 cells form functional gap junctions composed of connexin-43 (CX43) with human microvascular endothelial cells (HMECs). By overexpressing miR-145-5p in HMECs, the level of miR-145 also increases dramatically in SW480. In SW480 cells, miR-145 regulates the expression of CX43 and inhibits its pro-angiogenic capabilities. However, although miR-145 is transferred from SW480 cells to HMECs, the exact mechanisms underlying this transfer remain unclear. Notably, this transfer does not occur in non-contact co-cultures, thus excluding the involvement of soluble exosomes [335].
CRC progression and metastasis are prompted by constitutive and epigenetic RAS activation. In pancreatic cancer, it was demonstrated that RAS signaling leads to the repression of the miR-143/145 cluster. The downregulation of this cluster may be due to the repression of the cluster promoter operated by RREB1. Both KRAS and RREB1 are targets of miR-143/145, suggesting a feed-forward mechanism that enhances RAS signaling [336]. Certain circRNAs bind miRNAs and sequester them by inhibiting their functions. Hsa_circ_001569 promotes the proliferation and invasion of CRC cells by sponging miR-145 and induces the upregulation of miR-145 targets like E2F5, BAG4, and FMNL2 [337].
miR-20a and miR-92a belong to the miR-17/92 cluster and are overexpressed in CRC. miR-20a expression seems to correlate with lymph node metastasis and distant metastasis. Transfection of SW480 CRC cells with miR-20a promoted migration and invasion and induced EMT in CRC cells partly through the suppression of SMAD4 expression [203,338]. In SW480 cells, miR-92a induced EMT and regulated cell growth, migration and invasion via PTEN [339]. Moreover, miR-92a can promote CRC invasion and migration by targeting RECK [291].
The downregulated miR-124 has several biological functions and is involved in cell proliferation, autophagy and neuronal differentiation. miR-124 is abnormally expressed in inflammatory diseases and immune disorders by acting as an inhibitor of the inflammatory response [340]. A study analyzed the effects of methylation, overexpression and downregulation of miR-124 revealing how miR-124 suppresses CRC proliferation, migration and invasion by targeting DNMT3B [341]. Furthermore, miR-124 can modulate autophagy and apoptosis in CRC cells by inhibiting STAT3 [342,343] and the polypyrimidine tract-binding protein 1 (PTBP1) [344].
The miR-200 family, consisting of miR-200a/b/c, miR-141 and miR-429, located in two gene clusters, is often reported to be associated with epithelial differentiation and repression of EMT [345]. This miRNA family is also found downregulated in CRC due to promoter methylation [346]. As regards each family member, miR-200 was found to directly target the mRNA of the pro-mesenchymal transcription factors ZEB1, ZEB2 and PRRX1 [347]. Moreover, miR-200 suppresses EMT and metastasis and targets PD-L1, acting as a tumor suppressor miRNA. However, miR-200 is transcriptionally repressed by ZEB1, an activator of EMT, inducing the overexpression of PD-L1 and leading to immunosuppression of CD8(+) T-cells and metastasis [348]. As an example of the role of the miR-200 family in the onset of metastasis, miR-200c and miR-141 were found to be overexpressed in liver metastases compared to primary CRC tumors [349]. In agreement with these results, it was shown that serum levels of miR-200c are also high in patients with CRC metastases [350]. miR-141 by targeting SIP1 affects migration and invasion of CRC cells [211]. miR-141-3p negatively regulates proliferation, migration and invasion and sensitizes CRC cells to cetuximab through suppression of EGFR, thus serving as a potential predictive biomarker for response to cetuximab [212]. These miRNAs are also modulated by other ncRNAs, including H19 lncRNA, which activates the β-catenin pathway by sponging miR-141. Furthermore, H19 is highly expressed in CRC samples and has been associated with colorectal cancer stem cell (CSC). H19 has also been detected in CAF-derived exosomes, which in turn promote CSC stemness and chemoresistance of CRC cells [351].
The role of CAFs in the secretion of metastasis-inducing miRNAs was also observed for miR-31. Specifically, the expression of miR-31 was found to be increased in colorectal CAFs compared to normal colorectal fibroblasts (NFs). Overexpression of miR-31 in CAFs represses the expression of the autophagy-related genes BECN1, ATG, DRAM, and LC3, with negative effects on cell proliferation, invasion and apoptosis, and positive effects on CRC cells radiosensitivity [352].
MiR-200c, miR-17, and miR-192 were identified as major miRNAs involved in the regulation of genes crucial for extracellular matrix remodeling. Accordingly, expression of these miRNAs in human colon fibroblasts co-cultured with colon cancer cells significantly reduced cancer cell invasion [353]. A recent study compared the expression of selected miRNAs and EMT markers in biopsy samples from patients (n = 45) with primary CRC or metastatic CRC. The study indicated miR-17, miR-19b, miR-106a and miR-9 and the EMT-specific markers MMP2 and VEGFA as biomarkers with potential diagnostic, predictive and prognostic values in CRC progression and metastasis [354]. In addition, miR-106a was found to be highly expressed in metastatic CRC cells and seems to promote migration and invasion of tumor cells by targeting transforming growth factor receptor β (TGFBR2) [355].
RAN binding protein 1 (RANBP1) expression has been strongly associated with TNM stages and poor prognosis. RANBP1 could affect the nucleocytoplasmic transport of the pre-miRNAs of miR-18a, miR-183 and miR-106 and promotes YAP expression by influencing the Hippo pathway. YAP in turn functions as a transcriptional cofactor together with TEAD4 to activate RANBP1 transcription [356].
A study evaluated the role of miR-181a in tumor angiogenesis. miR-181a targets SRC kinase signaling inhibitor 1 (SRCIN1), resulting in the activation of SRC and the subsequent secretion of VEGF, leading to increased angiogenesis [357]. Moreover, the expression of miR-181a is higher in CRC with liver metastases; indeed, high levels of miR-181a correlate with advanced-stage, distant metastases and serve as an independent prognostic factor of poor overall survival. The overexpression of miR-181a in CRC cells promotes cell motility and invasion partly due to the inhibition of expression of its target WIF1 [358].
The expression of miR-15 and miR-16 is regulated by the promoter of their host gene SMC4 [359]. miRNA 16-1 is frequently deleted or downregulated in several tumors, including CRC, where it plays a role in EMT, contributing to the capacity of CRC cells to metastasize [128]. Most of the targets of miR-15a-5p and miR-16-5p in CRC are genes involved in EMT regulation, such as CCNB1 [360] or transcription factor AP4 [128]. AP4 is a helix-loop-helix transcription factor encoded by c-MYC which is upregulated in CRC. A study identified hundreds of induced and repressed AP4 target genes. Other gene targeted by AP4 are the stemness markers LGR5 and CD44 as well as genes involved in EMT, such as SNAIL, E-cadherin/CDH1, OCLN, VIM, FN1 and claudins 1, 4 and 7. Hence, AP4 promotes EMT and increases the migration and invasion of CRC cells [361]. In clinical CRC samples, miR-15a levels are inversely correlated with AP4 protein levels, which in turn correlate with distant metastasis and poor survival [128].
miR-206 is frequently downregulated in many human malignancies, including CRC. miR-206 suppresses CRC cell proliferation by arresting CRC cells in the G1/G0 phase, accelerates apoptosis, and inhibits cell invasion by targeting FMNL2 and c-MET [362]. In HCT116 and Caco-2 cells treated with prostaglandin E2 (PGE2), the expression of miR-206 decreases while the expression of its target TM4SF1 increases, resulting in cell proliferation and repression of apoptosis [363]. NOTCH3 is an established target of miR-206, frequently expressed in human CRC samples and involved in CRC cell modulation and tumorigenic potential. Transient transfection of miR-206 mimic into SW480 and SW620 cells results in the inhibition of cell proliferation, cell cycle blockade and activation of apoptosis through downregulation of NOTCH3 and potential indirect inhibition of other signaling pathways involving CDH2 and MMP9 [113]. miR-206, together with miR-1 and miR-133a/b, belongs to the group of myo-miRNAs that are muscle-specific miRNAs [364]. Besides miR-206, another myo-miRNA generally down-regulated in CRC cell lines and tissue samples is miR-133a. Ectopic expression of miR-133a inhibited cell proliferation and migration. Stable overexpression of miR-133a was sufficient to suppress tumor growth and intrahepatic and pulmonary metastasis in vivo [108]. It was observed that in CRC, overexpression of CXCR4 promotes EMT and the infiltration of myeloid-derived suppressor cells (MDSCs) and macrophages into colonic tissue, accelerating APC mutation-associated colitis and CRC progression. In addition, it was observed that miR-133a-3p significantly decreased after XIST sponging, determining an increase of the target RhoA, which is involved in cytoskeletal reorganization and cell motility in HCT116 cells [365].
The expression levels of miR-320a in CRC cell lines and tumor tissues were found to be frequently downregulated. The restoration of miR-320a inhibited CRC cell proliferation and repressed its direct target β-catenin [366]. Similarly, a lentiviral-mediated re-expression of miR-320c inhibits the growth and migration of HCT116 and sensitizes CRC cells to 5-FU [367].
TET1, downregulated in CRC, is a miR-21-5p target that acts as a tumor suppressor and inhibits cell growth [368]. Moreover, miR-21-5p can enhance cell migration, intravasation, and metastasis by targeting programmed cell death 4 (PDCD4) [369].
Several immune factors contribute to the progression of CRC. For instance, myeloid-derived granulocyte suppressor cells (G-MDSCs) increase cancer growth. CRC tissues have been found to contain G-MDSC cells that secrete exosomes containing miR-166-5p. These exosomes accelerate cancer progression by promoting cell proliferation. miR-166-5p by targeting integral membrane protein 2B (ITM2B), which in turn activates the PI3K/Akt signaling pathway, promotes cell proliferation in CRC [370].
Overexpression of miR-195-5p in DLD1 and HCT116 cells represses cell growth, colony formation, invasion and migration by suppressing the Hippo-YAP pathway by targeting YAP [122].
The expression of miR-203 was quantified in primary CRC (pCRC) and corresponding liver metastasis (LM) and serum samples from CRC patients. The expression of miR-203 was significantly upregulated in LM compared to the corresponding pCRC tissues. Serum levels of miR-203 were elevated in a stage-dependent manner and high miR-203 expression was associated with poor survival in CRC patients in both patient cohorts [371].
miR-23b also appears to play a role in metastasis by interacting with BTBD7 [372]. miR-23a is overexpressed in CRC cell lines and tissues and regulates PDK4 expression by targeting its mRNA. PDK4 negatively regulates CRC proliferation through suppression of PDH activity. Accordingly, up-regulation of miR-23a promotes CRC cell proliferation by directly repressing PDK4 [373]. A study showed that miR-23a was significantly elevated in MSI CRC cells and tissues compared to CRC cells and tissues with stable microsatellite status (MSS). Ectopic expression of miR-23a increased the viability and survival of CRC MSS cells, while the downregulation of miR-23a reduced viability and promoted cell apoptosis in CRC MSI cells treated with 5-FU. In these models, ABCF1 was found as a direct target of miR-23a and its repression sensitizes CRC MSI cells to 5-FU [374].
Preliminary evidence was also obtained on the role of miR-221, miR-222, miR-let-7c, miR-638, miR-187 and miR-10b in CRC progression and metastasis formation. In particular, miR-221 and miR-222 regulate the activation of NF-κB and STAT3 in human CRC cell lines via RelA mRNA targeting; both factors are involved in the development and progression of CRC when constitutively activated [375].
The miRNA let-7c is downregulated in primary tumor tissues. Ectopic expression of let-7c in highly metastatic Lovo CRC cells significantly suppressed cell migration and invasion in vitro through the downregulation of KRAS, MMP11 and PBX3. In contrast, the inhibition of let-7c in poorly metastatic HT29 cells increased cell motility and invasion through increased gene expression of its targets KRAS, MMP11 and PBX3 [376].
Many other miRNAs affect EMT by targeting EMT-associated genes, such as miR-638, which targets SOX2 [377], miR-187 hinders SMAD-mediated EMT by directly suppressing the expression of SOX4, NT5E and PTK6 [378], and miR-10b targets HOXD10 [379]. The following table shows the main miRNAs involved in CRC progression, their respective target genes and altered molecular pathways (Table 3).
Table 3.
miRNAs and their targets involved in CRC progression and metastasis.
| Function | miRNA | Key Targets | Pathways Affected | Role in CRC | References |
|---|---|---|---|---|---|
| EMT and Metastasis | miR-20a | SMAD4 | EMT | Promotes migration, invasion, and EMT; upregulated in CRC | [203,338] |
| miR-92a | PTEN, RECK | EMT | Induces EMT and promotes CRC invasion, migration, and cell growth | [291,339] | |
| miR-106a | TGFBR2 | EMT | Promotes migration and invasion of tumor cells; upregulated in metastatic CRC | [354,355] | |
| miR-15a/16 | Cyclin B1, AP4 | EMT | Suppresses EMT, metastasis, and CRC progression; downregulated in CRC | [128,360] | |
| miR-10b | HOXD10 | EMT | Promotes EMT and invasion in CRC | [379] | |
| let-7c | KRAS, MMP11, PBX3 | EMT | Suppresses CRC migration and invasion; downregulated in CRC | [376] | |
| miR-638 | SOX2 | EMT | Suppresses EMT in CRC | [377] | |
| miR-187 | SOX4, NT5E, PTK6 | EMT | Suppresses EMT in CRC | [378] | |
| miR-133a | CXCR4, RhoA | EMT, cytoskeletal reorganization | Suppresses tumor growth and metastasis; downregulated in CRC | [108,365] | |
| miR-34 | Notch1, Jagged1, ZNF281, SIRT1 | EMT, E2F signaling, IL-6R/STAT3 | Suppresses migration, invasion, and EMT; inhibits metastasis and cell proliferation; downregulated in CRC; modulates p53 and inflammation | [[330], [331], [332], [333], [334]] | |
| EMT, Inflammation and Immune Regulation | miR-155 | Claudin-1, SOCS1, ZBTB2, AXIN1, TCF4 | EMT, JAK2-STAT3/NF-κB, WNT/β-catenin | Promotes migration, invasion, metastasis, and inflammation; upregulated in CRC; involved in tumor progression and metastasis | [[325], [326], [327], [328], [329]] |
| miR-200 family | ZEB1, ZEB2, PRRX1, PD-L1, SIP1 | EMT, immune suppression | Suppresses EMT and metastasis; downregulated in CRC; promotes immunosuppression via PD-L1 | [211,[347], [348], [349], [350]] | |
| Inflammation and Immune Regulation | miR-146a-5p | SOCS1, ZBTB2 | JAK2-STAT3/NF-κB | Promotes tumor progression and metastasis via inflammatory cytokines; involved in exosome trafficking from CAFs | [327] |
| miR-221/222 | RelA | NF-κB, STAT3 | Promotes CRC development and progression; regulates inflammatory signaling | [375] | |
| Inflammation Immune Regulation, Apoptosis, Autophagy, Cell Proliferation and Tumor Growth | miR-124 | DNMT3B, STAT3, PTB1 | Autophagy, apoptosis, inflammatory response | Suppresses CRC proliferation, migration, invasion, and STAT3 signaling; downregulated in CRC | [[340], [341], [342], [343], [344]] |
| Apoptosis, Autophagy, Cell Proliferation and Tumor Growth | miR-206 | FMNL2, c-MET, TM4SF1, NOTCH3 | Cell cycle, apoptosis | Suppresses CRC proliferation, invasion, and metastasis; downregulated in CRC | [113,362,363] |
| miR-21-5p | PDCD4, TET1 | Apoptosis, migration, metastasis | Enhances CRC migration, intravasation, and metastasis; upregulated in CRC | [368,369] | |
| miR-31 | Beclin-1, ATG, DRAM, LC3 | Autophagy | Promotes proliferation, invasion, and radiosensitivity in CRC; overexpressed in CAFs | [352] | |
| miR-145-5p | CX43, RAS, E2F5, BAG4, FMNL2 | Gap junctions, RAS signaling | Inhibits angiogenesis, proliferation, and invasion; regulates extracellular matrix remodeling | [[335], [336], [337],353] | |
| miR-195-5p | YAP | Hippo-YAP | Represses cell growth, colony formation, and invasion; downregulates the Hippo-YAP pathway | [122] | |
| Cell Proliferation and Tumor Growth | miR-181a | SRCIN1, WIF-1 | SRC, VEGF, Wnt | Promotes angiogenesis, motility, and invasion; correlates with advanced stage and metastasis | [357,358] |
| miR-320a/c | β-catenin | WNT/β-catenin | Inhibits CRC cell proliferation and migration; downregulated in CRC | [366,367] | |
| miR-166-5p | ITM3E | PI3K/Akt | Promotes cell proliferation; involved in G-MDSC-induced CRC progression | [370] | |
| Cell Proliferation, Tumor Growth, Drug Resistance, MSI, Prognostic Value | miR-203 | - | – | Associated with poor survival; upregulated in liver metastases | [371] |
| miR-23a | PDK4, ABCF1 | Microsatellite instability (MSI) | Promotes CRC cell proliferation; associated with drug resistance and MSI | [373,374] |
3.3. microRNAs in colorectal cancer chemo- and radioresistance
Chemoresistance refers to the ability of tumor cells to withstand the effects of chemotherapy, leading to reduced treatment effectiveness, treatment failure, and, ultimately, the progression of the disease. Tumor cells develop chemoresistance through various mechanisms, including overexpression of ABC transporters and efflux of chemotherapeutic drugs, the overexpression of thymidylate synthase, the overexpression of anti-apoptotic proteins and resistance to apoptosis; these mechanisms enable tumor cells to resist apoptosis and survive treatment [380]. Such mechanisms are particularly active in CCSCs, which show strong resistance to chemotherapy and are the main cause of CRC recurrence [381].
Several molecular pathways are particularly associated with CRC chemoresistance due to their roles in cell survival, proliferation, and drug response. The multidrug resistance (MDR) pathway is currently responsible for the low effectiveness of chemotherapeutic agents. One of the key characteristics of CRC cells exhibiting MDR is the overexpression of the insulin-like growth factor type I receptor (IGF-IR). Suppressing IGF-IR leads to the inhibition of the PI3K/Akt signaling pathway, which in turn downregulates Nrf2-ARE-dependent transcriptional activity. This leads to a reduced activity of the multidrug resistance-associated protein-2 (MRP-2) promoter, limiting MRP-2 expression and contributing to the reversal of chemoresistance [382]. Leptin is a pluripotent cytokine secreted by adipocytes and involved in the regulation of appetite and energy balance in the brain. Bartucci M. et al., found that obesity and increased leptin levels could counteract the cytotoxic effect of 5-FU promoting the growth and survival of CCSCs [383]. Apoptosis resistance is another strategy adopted by CRC cells for chemoresistance. For instance, the Human Ring-Finger homologous to Inhibitor of apoptosis protein type (hRFI) gene is involved in the inhibition of death receptor-mediated apoptosis in CRC cells. In a study by Konishi T et al., CRC cells were stably transfected with hRFI. The overexpression of hRFI resulted in cellular resistance to 5-FU through the inhibition of the mitochondrial apoptotic pathway, the upregulation of BCL-2 and BCL-X, and the activation of NF-kB [384]. Moreover, elevated expression of thiamine synthase, BCL-2, BCL-XL and Mcl-1 have been related to 5-FU resistance [385]. The tryptophan-aspartate repeat domain 43 (WDR43) is highly expressed in CRC tissues and its overexpression is associated with poor prognosis. WDR43 increases the ubiquitination of p53 by MDM2 through binding to RPL11. WDR43 suppression significantly inhibits cell growth and enhances the effect of oxaliplatin chemotherapy both in vitro and in vivo [386]. The sex-determining region Y-box2 (SOX2), a master regulator of embryonic and induced pluripotent stem cells, sustains CSCs and plays an important role in tumor initiation and aggressiveness. A study showed that SOX2 promotes chemoresistance through the transcriptional activation of ABCC2 expression. Specifically, SOX2 interacts with β-catenin and Beclin1 and increases their nuclear expression and transcriptional activity. Overexpression of β-catenin or Beclin1, in turn, promotes the expression of ABCC2, which, together with Beclin1 and SOX2, influences chemoresistance, stemness and EMT in CRC [387].
As miRNAs regulate signaling pathways involved in chemoresistance, their altered expression may affect cellular sensitivity to chemotherapeutic agents. Indeed, numerous studies have shown that miRNAs contribute to drug resistance by modulating mechanisms and pathways associated with cell survival [388]. Slattery ML et al. performed an analysis of miRNAs and apoptosis-related genes on 217 CRC and normal tissues. Several miRNAs were identified as involved in the regulation of BIRC5, CTSS and CSF2R, all genes associated with apoptosis. Specifically, the authors demonstrated that BIRC5 could be a potential target of miR-145-5p, miR-150-5p, miR-195-5p, and miR-650; CSF2RB a target of miR-92a-3p; CTSS a target of miR-20b-5p and miR-501-3p [389].
The miRNA-mediated dysregulation of genes involved in double-strand break (DSB) repair also contributes to the promotion of chemoresistance mechanisms [390]. For instance, a study investigating the co‐regulatory networks of tumor suppressor genes, oncogenes, and miRNAs occurring in CRC revealed that the overexpression of miR-17, miR-425 and miR-92 was significantly associated with up-regulation of BRCA1, counteracting the usually observed downregulation of genes involved in the mismatch repair pathway, including MLH1, MSH2 and MSH6 [391]. In line with these findings, other research groups have also investigated the role of antioxidant mechanisms in the occurrence of CRC chemoresistance. In this context, they demonstrated that the epigenetic regulation of glutathione (GSH) homeostasis is another mechanism that may induce the acquisition of drug resistance [392]. Specifically, miRNAs involved in the GSH homeostasis, such as miR-18a [287] or miR-214 [393] may influence the sensitivity of tumor cells to various therapeutic approaches.
miR-195-5p and miR-497-5p are downregulated in CRC tissues and have been widely studied in the context of drug resistance. HCT116 and RKO cells with MSI/P53 wild-type had increased sensitivity to oxaliplatin following transfection with miR-195-5p and miR-497-5p mimics [394]. Low miR-497 expression was strongly correlated with clinical stages and lymph node metastases. Furthermore, Ras suppressor kinase 1 (KSR1), a known oncogene overexpressed in human CRC samples, was identified as a direct target of miR-497. Overexpression of miR-497 in SW1116 CRC cells inhibited cell proliferation, migration and invasion and increased chemosensitivity to 5-FU, whereas forced expression of KSR1 had the opposite effect [120]. In addition, miR-497 by targeting IGF1-R promotes inhibition of cell proliferation and invasion and promotes apoptosis induced by several stimuli, including the chemotherapeutic drugs cisplatin and 5-FU [117]. miR-497 was also found to be downregulated in the multidrug-resistant human gastric cancer cell line SGC7901/vincristine (VCR) and in the multidrug-resistant human lung cancer cell line A549/cisplatin (CDDP). In these models, the downregulation of miR-497 correlates with the upregulation of BCL2 protein, one of its direct targets. Thus, miR-497 could play a role in MDR through modulation of apoptosis by targeting BCL2 [395]. Moreover, BCL2 is a direct target of miR-195 and the overexpression of this miRNA in HT29 and LoVo cells promotes cell apoptosis and suppresses tumorigenicity [123]. In Dox-resistant CRC lines HT29/DOX and LOVO/DOX, miR-195 was significantly downregulated. Knockdown of miR-195 in HT29 and LOVO-sensitive cells inhibited Dox cytotoxicity, whereas overexpression of miR-195 sensitized Dox-resistant cells by targeting BCL2L2 [124].
miR-125 is down-regulated in both colon cancer tissue and colon cancer cell lines demonstrating a tumor suppressor role; indeed, its overexpression inhibited cell proliferation and induced apoptosis in colon cancer cells. Overexpression of miR-125 leads to the repression of apoptosis, as the anti-apoptotic genes BCL2, BCL2L12 and Mcl-1 are direct targets of this miRNA [396]. Some studies have demonstrated the role of this miRNA in FOLFOX therapeutic efficacy. Notably, the FOLFOX regimen, consisting of the combination of 5-FU, leucovorin and oxaliplatin, is effective for the treatment of CRC [397]. However, the circRNA circ_0032833 was found significantly up-regulated in FOLFOX-resistant CRC and associated with drug resistance. Furthermore, circ_0032833 sequesters miR-125-5p, preventing its tumor-suppressing action. Among the targets of miR-125-5p, Musashi1 (MSI1) appears to be involved in 5-FU and oxaliplatin sensitization in FOLFOX-resistant CRC cells [398]. Another study demonstrated that the activation of the CXCL12/CXCR4 axis promotes EMT and the upregulation of miR-125b in CRC cells. Consequently, miR-125b promotes EMT, tumor invasion and CXCR4 expression, thus generating a positive feedback that also involves the Wnt/β-catenin signaling since APC appears to be targeted by miR-125b. miR-125b also appears to confer resistance to 5-FU in CRC, probably through increased autophagy [399]. All these data suggest the dual role of miR-125, with some subtypes acting as tumor suppressor miRNAs and others as tumor-promoting ones [400].
The cluster miR-143/145 is often downregulated in CRC cells compared to normal colon epithelia. Restoration of miR-143 and miR-145 in CRC cells reduced proliferation, migration and chemoresistance [86]. miR-145 seems to sensitize LS174T cells to 5-FU by repression of Fli-1 [401]. CBR3-AS1 lncRNA is upregulated in CRC tissues and cell lines and correlates with poor prognosis and adverse clinicopathological features of CRC patients. Furthermore, it was observed that CBR3-AS1 promotes resistance to oxaliplatin in CRC cells by sponging and inhibiting miR-145 [402]. Stable expression of miR-143 decreases viability and increases cell death in CRC cells treated with 5-FU, probably through the modulation of pathways regulated by the extracellular protein kinase 5/NF-kB [100]. Hexokinase 2 (HK II) encodes for a limiting enzyme of glutamine metabolism and is responsible for the dysregulation of glycolysis in tumors. HK II is overexpressed in CRC and positively correlates with 5-FU resistance. miR-143, which is significantly downregulated in 5-FU-resistant CRC patients and colon cancer cells, targets HK II. The overexpression of miR-143 inhibits the rate of glycolysis by directly targeting HK II, leading to the resensitization of 5-FU-resistant colon cancer cells [403]. By analyzing miRNA expression in both 5FU-sensitive and 5FU-resistant DLD-1 cell lines, as well as in their corresponding extracellular microvesicles (MVs) before and after 5-FU treatment, it was found that miR-34a and miR-145 were actively secreted via MVs in both cell types. This suggests that these miRNAs may play a role in cellular communication and possibly in the development of chemoresistance [404].
Besides its mutual action with miR-145, miR-34a is down-regulated in 5-FU-resistant DLD-1 cells when compared with sensitive parental DLD-1 clones. SIRT1, a miR-34a target, is associated with drug resistance and is up-regulated in 5-FU-resistant cells. Ectopic expression of miR-34a in resistant cells attenuates 5-FU resistance through the down-regulation of SIRT1 and E2F3 [405]. In addition, mutations affecting p53 are important determinants of chemoresistance in CRC. Leucine-rich pentatricopeptide repeat-containing protein (LRPPRC) is a key downstream functional factor of p53 that can bind mRNA of ATP-binding cassette subfamily B member 1 mRNA 1 (MDR1), increasing its stability and protein expression. In normal cells, miR-34a represses LRPPRC, reducing MDR1 expression. However, in p53 mutated cells, the accumulation of LRPPRC and MDR1 promotes drug resistance. To corroborate these findings, p53 mutated cells treated with gossypol-acetic acid (GAA), a specific inhibitor of LRPPRC, showed a reduced chemoresistance [406]. miR-34a was also found significantly downregulated in CRC clinical samples obtained from oxaliplatin-resistant patients and in multidrug-resistant CRC cells. Ectopic expression of miR-34a resensitized multidrug-resistant HCT-8/OR cells to oxaliplatin treatment, whereas miR-34a inhibition increased oxaliplatin resistance in chemoresistant HCT-8 cells. In these models, the mRNA of ornithine decarboxylase 2 (OAZ2) enzyme is targeted by miR-34a; therefore, the suppression of miR-34a/OAZ2 signal expression by chemotherapeutic agents increases the activation of MDR-associated ATP-binding cassette (ABC) transporters and anti-apoptosis pathways, thus leading to the development of MDR in CRC models [407].
As previously described, the miR-17/92 cluster is upregulated in CRC. miR-19b-3p expression was evaluated in 211 colon cancer patients, revealing its overexpression in patients with poor prognosis. Moreover, miR-19b-3p mediates resistance to oxaliplatin-based chemotherapy via SMAD4 [408]. Exosomal miR-19b has been identified as a key contributor to oxaliplatin resistance in cancer cells. Inhibition of its secretion using GW4869, a pharmacological agent known to block exosome release, enhances the sensitivity of SW480 cells to oxaliplatin. This suggests that targeting exosomal pathways, specifically miR-19b, could be a promising strategy for overcoming chemoresistance and improving therapeutic efficacy in oxaliplatin-resistant cancers. By disrupting exosomal signaling, the potential for re-sensitizing resistant cancer cells to treatment becomes a viable approach for enhancing the effectiveness of chemotherapy [409]. By evaluating miRNA expression profiles in CRC patients, comparing a cohort of 295 chemosensitive and chemoresistant patients, miRNA-17-5p expression was found to be increased in the chemoresistant group. In addition, overexpression of miR-17-5p promoted the invasiveness and MDR of COLO205 via PTEN targeting [180]. Although miR-20b is generally up-regulated in CRC, a study reported its downregulation in 5-FU-resistant compared to 5-FU-sensitive tissues and cells. Restoration of miR-20b resensitizes 5-FU-resistant HCT116 by inducing apoptosis and repressing the expression of its targets ADAM9 and EGFR [410].
The myo-miRNAs miR-206 and miR-133, often downregulated in CRC, also appear to play a role in chemoresistance. miR-206 was downregulated in 5-FU resistant CRC lines compared to their parental cell lines and this downregulation promotes drug resistance. The resistance conferred by the downregulation of miR-206 might depend on the increase of its target Bcl-2 [411]. As regards miR-133b, this is a tumor suppressor miRNA in CRC. Indeed, a study demonstrated that miR-133b is downregulated in CRC spheroids, which are enriched in CSCs and show stem cell-like characteristics and high chemoresistance. Overexpression of miR-133b reduced CRC stemness and abrogated chemoresistance to 5-FU and oxaliplatin. These effects may depend on the role of miR-133b in regulating its direct target disruptor of telomeric silencing 1-like (DOT1L), an exclusive H3K79 methyltransferase important for stem cell gene modification [412].
In three oxaliplatin-resistant CRC lines, HT29, RKO, and HCT116, miR-203 was found to be up-regulated. The downregulation of miR-203 sensitized chemoresistant CRC cells to oxaliplatin. Moreover, ATM, a primary mediator of DNA damage response, is targeted by miR-203 and stable knockdown of ATM is associated with oxaliplatin resistance in chemosensitive CRC cells [413]. As mentioned in other sections, the lncRNA HOTAIR is upregulated in CRC tissues compared to adjacent control tissues and downregulates miR-203a-3p in CRC in vitro models. HOTAIR promotes the proliferation and drug resistance of CRC cells and the overexpression of miR-203a-3p in CRC cell lines inhibits cell proliferation and reduces chemoresistance [58].
miR-192/miR-215 expression levels were decreased in clinical colon cancer specimens compared with adjacent normal tissues of the same patients [414]. In CRC cells, miR-192 and miR-215 bind TYMS, one of the specific targets of fluoropyrimidine-based chemotherapies. Cell proliferation and S-phase cells are reduced by overexpression of miR-192/215. Consequently, the effects of S-phase-specific drugs are attenuated. These results suggest that mechanisms other than TYMS overexpression are essential for directing 5-FU resistance [415]. In patients treated with fluoropyrimidine-based chemotherapies, the miR-200 family seems to influence survival. For instance, high levels of miR-200a, miR-200c, miR-141, or miR-429 were correlated with longer overall and disease-free survival. In particular, high miR-429 levels result in the inhibition of CRC cell invasion after 5-FU treatment [416]. Table 4 lists the miRNAs related to chemoresistance acquisition identified so far (Table 4).
Table 4.
miRNAs and their targets involved in CRC chemoresistance.
| miRNA | Key Targets | Pathways Affected | Role in CRC | References |
|---|---|---|---|---|
| miR-145-5p | BIRC5, Fli-1 | Apoptosis, Drug resistance | Sensitizes CRC cells to 5-FU by repression of Fli-1; potential target of BIRC5; downregulated in CRC and associated with oxaliplatin resistance | [389,401,402] |
| miR-150-5p | BIRC5 | Apoptosis | Regulates apoptosis by targeting BIRC5; downregulation linked to CRC progression | [389] |
| miR-195-5p | BIRC5, BCL2, BCL2L2, YAP | Apoptosis, Drug resistance, Hippo-YAP | Downregulated in CRC; sensitizes cells to oxaliplatin and 5-FU by targeting BCL2 and BCL2L2; promotes apoptosis; overexpression inhibits tumorigenicity; associated with 5-FU resistance in HT29/DOX and LOVO/DOX cells | [123,124,389,394] |
| miR-650 | BIRC5 | Apoptosis | Potential regulator of apoptosis by targeting BIRC5 | [389] |
| miR-92a-3p | CSF2RB | Apoptosis | Targets CSF2RB; involved in apoptosis regulation | [389] |
| miR-20b-5p | CTSS, ADAM9, EGFR | Apoptosis, EMT | Downregulated in 5-FU-resistant CRC cells; resensitizes CRC cells to 5-FU by inducing apoptosis and repressing ADAM9 and EGFR | [389,410] |
| miR-501-3p | CTSS | Apoptosis | Potential regulator of CTSS and apoptosis in CRC | [389] |
| miR-497-5p | KSR1, BCL2, IGF1-R | Apoptosis, Drug resistance, EMT | Downregulated in CRC; sensitizes cells to oxaliplatin and 5-FU; promotes apoptosis by targeting IGF1-R and BCL2; inhibits EMT and cell proliferation; enhances chemosensitivity | [117,120,394,395] |
| miR-125-5p | MSI1, BCL2, Mcl-1, CXCR4 | Apoptosis, EMT, Autophagy | Tumor suppressor in CRC; downregulation leads to resistance to FOLFOX; targets BCL2, BCL2L12, and Mcl-1; modulates CXCR4 and APC signaling; also involved in drug resistance and autophagy | [396,[398], [399], [400]] |
| miR-34a | SIRT1, E2F3, OAZ2 | Apoptosis, Drug resistance, EMT | Downregulated in 5-FU-resistant CRC cells; targets SIRT1, E2F3, and OAZ2; sensitizes multidrug-resistant CRC cells to 5-FU and oxaliplatin treatment | [[405], [406], [407]] |
| miR-19b-3p | SMAD4 | Apoptosis, Drug resistance | Mediates oxaliplatin resistance by targeting SMAD4; overexpressed in patients with poor prognosis; inhibition of its exosomal secretion enhances oxaliplatin sensitivity | [408,409] |
| miR-17-5p | PTEN | Apoptosis, Drug resistance | Upregulated in chemoresistant CRC; promotes invasiveness and multidrug resistance (MDR) by targeting PTEN | [180] |
| miR-143 | HK II | Glycolysis, Apoptosis, Drug resistance | Downregulated in CRC and 5-FU-resistant CRC cells; inhibits glycolysis by targeting HK II, resensitizes cells to 5-FU; inhibits proliferation and migration | [86,100,403] |
| miR-206 | Bcl-2 | Apoptosis, Drug resistance | Downregulated in CRC; promotes drug resistance by targeting Bcl-2 | [411] |
| miR-133b | DOT1L | Stemness, Chemoresistance | Tumor suppressor miRNA; downregulated in CRC; reduces CRC stemness and abrogates chemoresistance to 5-FU and oxaliplatin by targeting DOT1L | [412] |
| miR-203 | ATM, HOTAIR | DNA damage response, Drug resistance | Upregulated in oxaliplatin-resistant CRC lines; downregulation sensitizes cells to oxaliplatin by targeting ATM | [375,413] |
| miR-192/215 | TYMS | S-phase, Drug resistance | Downregulated in CRC; modulates TYMS expression, which affects the efficacy of fluoropyrimidine-based chemotherapy | [414,415] |
| miR-200 family | - | EMT, Drug resistance | Associated with better prognosis in CRC patients treated with fluoropyrimidine-based chemotherapy; high levels correlate with longer survival | [416] |
Besides chemotherapy, radiotherapy is another major treatment for unresectable or drug-resistant tumors, especially CRC. However, neoplastic cells can also acquire resistance to radiation exposure by developing a radioresistant phenotype through the modulation of various mechanisms, including autophagy, apoptosis, cell cycle control, ROS pathways, cancer stem cells (CSCs) and epithelial-mesenchymal transition (EMT) [417]. Similarly to what was described for the acquisition of resistance to chemotherapeutics, miRNAs may also act as modulators in cell signaling pathways that confer radioresistance [418].
For instance, it has been shown that miR-7-5p, which targets KLF4, is reduced in cancerous tissues of CRC patients radiotherapy resistant and that the miR-7-5p/KLF4 axis can induce radiosensitivity [419]. Sun T and colleagues suggested that miR-19b inhibition could enhance the efficacy of radiotherapy in CRC cells [194]. miR-195 can increase the radiosensitivity of CRC cells by targeting CARM1 [420]. miR-185 can enhance radiosensitivity in CRC by targeting IGF1R and IGF2 [421]. Circ-ACAP2 may promote CRC progression and radioresistance, in part by sponging miR-143-3p, which in turn modulates Wnt/β-catenin signaling [422]. miR-106b could induce cell radioresistance by directly targeting PTEN and p21 [423]. Long noncoding RNA SP100-AS1 induces radioresistance in CRC by sponging of miR-622 which targets ATG3 and influences autophagy activity [424]. A study suggested that the circulating miRNAs miR-506-3p and miR-140-5p may have roles as biomarkers of radiosensitivity as they have higher expression levels in radiosensitive patients than in radioresistant patients [425]. The restoration of miR-1 promotes the expression of Bax and E-cadherin and decreases the expression of BCL2, MMP2 and MMP9, apparently impairing the invasion and migration of CRC cells in synergy with radiotherapy [426]. miR-222 and miR-155 could promote radioresistance in CRC by targeting PTEN and FOXO3a, respectively [427]. miR-29a may regulate the radiosensitivity of CRC cells by targeting PTEN [428]. miR-124 can radiosensitize CRC cells by targeting PRRX, an EMT inducer and regulator of stemness [429]. miR-378a-5p could resensitize CRC cells to radiotherapy by modulating the LRP8/β-catenin axis [430]. miR-1226-5p is involved in CRC radioresistance and through IRF1 suppression activates M2 macrophages and induces TGF-β secretion [431]. ATG12 and LC3 are overexpressed in radioresistant CRC samples and miR-214 can promote radiosensitivity by inhibiting ATG12-mediated autophagy [393]. ATG12 is also a target of miR-93, which in turn is sponged by the long non-coding RNA HOTAIR. Knockdown of HOTAIR increases radiosensitivity by modulating the miR-93/ATG12 axis [432]. Exosome-mediated transfer of miR-93-5p from CAFs to CRC cells can confer radioresistance through downregulation of FOXA1 and upregulation of TGFB3 [433]. Similarly, miR-590-3p transfer via CAFs-derived exosomes was found to enhance radioresistance in CRC through positive regulation of the PI3K/Akt signaling pathway [434]. Table 5 lists the miRNAs related to radioresistance acquisition identified so far (Table 5).
Table 5.
miRNAs and their targets involved in CRC radioresistance.
| miRNA | Key Targets | Pathways Affected | Role in CRC | References |
|---|---|---|---|---|
| miR-7-5p | KLF4 | Stemness and radioresistance | Antitumor function in the regulation of CSC properties and radiosensitivity | [419] |
| miR-19b | FBXW7 | Stemness and radioresistance | Modulation of the FBXW7/Wnt/β-catenin axis | [194] |
| miR-195 | CARM1 | Apoptosis and radioresistance | Downregulated in CRC, inhibits the expression of CARM1 which in turn regulates the expression of p53 and NF-κB involved in radiosensitivity | [420] |
| miR-185 | IGF1R and IGF2 | Apoptosis and radioresistance | Upregulation enhances radiosensitivity by targeting IGF1R and IGF2 | [421] |
| miR-143-3p | FZD4 | Progression and radioresistance | Modulation of the Wnt/β-catenin signaling by circ-ACAP2/miR-143-3p/FZD4 axis | [422] |
| miR-106b | PTEN and p21 | Cell proliferation, tumour growth and radioresistance | Upregulation downregulates PTEN and p21 and subsequently enhances radioresistance. | [423] |
| miR-622 | ATG3 | Autophagy and radioresistance | Downregulated as sponged by SP100-AS1, affects autophagic activity by targeting ATG3 and contributes to radioresistance | [424] |
| miR-506-3p and miR-140-5p | – | – | Circulating biomarkers of radiosensitivity | [425] |
| miR-1 | BCL2, MMP2 and MMP9 | Apoptosis and radioresistance | Downregulated in CRC, enhances radiosensitivity by inducing cell apoptosis | [426] |
| miR-222 | PTEN | Cell proliferation, apoptosis inhibition, cell invasion and radioresistance | Upregulated, mediates radioresistance via PI3/Akt pathway | [427] |
| miR-155 | FOXO3a | Cell proliferation, apoptosis inhibition, cell invasion and radioresistance | Upregulated, mediates radioresistance via PI3/Akt pathway | [427] |
| miR-29a | PTEN | Cell proliferation, tumour growth and radioresistance | Radiosensitivity regulation by targeting the PTEN gene | [428] |
| miR-124 | PRRX | EMT, stemness regulation and radioresistance | Downregulated in CRC, enhances radiosensitivity by targeting PRRX | [429] |
| miR-378a-5p | LRP8 | Cancer development and progression, radioresistance | Downregulated in CRC, regulates radioresistance via modulation of the Wnt/β-catenin pathway | [430] |
| miR-1226-5p | IRF1 | EMT, migration, invasion, and tumor growth | In radioresistant CRC promoted EMT by targeting IRF1 | [431] |
| miR-214 | ATG12 | Autophagy and radioresistance | Modulation of radioresistance by targeting ATG12 | [393] |
| miR-93 | ATG12 | Apoptosis, autophagy and radiosensitivity | Modulation of radioresistance by targeting ATG12 | [432] |
| miR-93-5p | FOXA1 | Apoptosis, cell proliferation and radioresistance | Modulation of TGF-β signaling pathway and of radioresistance by targeting FOXA1. | [433] |
| miR-590-3p | CLCA4 | Tumor growth | Enhances radioresistance through positive regulation of the CLCA4-dependent PI3K/Akt signaling pathway. | [434] |
4. Role of circulating microRNAs in colorectal cancer
Notably, miRNAs are widely used as effective biomarkers for different diseases, including cancer. Several studies have demonstrated the diagnostic and prognostic value of differentially expressed miRNAs detected in both tissue and liquid biopsy samples obtained from CRC patients and healthy controls [435,436]. More recently, circulating miRNAs were proposed as non-invasive and reliable biomarkers for tumor diagnosis and patients’ follow-up due to their stability, the low costs of the analysis and the possibility of repeat sampling multiple times during the treatments [437].
Several studies have investigated the diagnostic accuracy of miRNAs and circulating miRNAs in CRC in terms of sensitivity, specificity, odds ratio (OR) and area under the ROC curve (AUC). Examples are miR-21-5p [[438], [439], [440], [441], [442]], miR-1290 [443], miR-210 [438,441], miR-378e [444], miR-1246 [445], miR-92a-1 [446], miR-320d [443], miR-15b [442] or miR-150-5p [447] which showed AUC values ranging from 0.7 to > 0.95 thus demonstrating significant potential as diagnostic biomarkers for the detection of CRC. Moreover, the comparison between protein biomarkers and the circulating levels of miR-133a, miR-574-3p and miR-27a has demonstrated a better sensitivity of these latter biomarkers (AUC = 0.736 (0.639–0.834) for CA19.9 and 0.88 (0.814–0.946 for CEA), both when analyzed alone or in combination (AUC = 0.974 (0.948–1.000) for miR-133a, 0.975 (0.948–1.000) for miR-574-3p and 0.904 (0.849–0.958) for miR-27a) [448]. Other studies have also investigated the potential prognostic role of circulating miRNAs. In particular, the serum levels of miR-93-5p could play a prognostic role for early disease recurrence (p = 0.035) in CRC patients with liver metastases who showed higher levels in metastatic vs non-metastatic tumors (p < 0.001) [449]. Also circulating miR-618 has been suggested as a possible prognostic biomarker in metastatic colon cancer since its up-regulation is associated with a better prognosis (overall survival (OS) of 21 months) compared to patients with low miR-618 expression (OS of 16 months; HR = 0.51, 95 % CI: 0.30–0.86, p = 0.012) [450]. The overexpression of miR-326, miR-27b and miR-148a was associated with low PFS, while miR-326 was associated with low OS [451]. Circulating miRNAs may also serve as predictive biomarkers for treatment response, offering a non-invasive tool to anticipate the efficacy of specific therapeutic strategies. For instance, Zhang J and colleagues proposed a profile of five serum miRNAs (miR-20a, miR-130, miR-145, miR-216 and miR-372) as a biomarker to predict CRC chemosensitivity [452]. High expression of miR-345 was associated with a non-response to treatment with irinotecan and cetuximab [453]. It has been observed that an increased serum level of miR-155 after surgery and chemotherapy is a sign of chemoresistance in CRC, and elevated levels of miR-155, miR-200c and miR-210 imply local recurrence and distant metastases as well as a poor prognosis [454]. Circulating miR-20b-5p, miR-29b-3p and miR-155-5p were significantly associated with PFS and OS as well as with response to bevacizumab in patients with metastatic CRC [455].
Moreover, circulating miRNAs may play a significant role in the development of CRC chemoresistance by modulating gene expression and influencing various cellular processes related to drug response. A study investigated circulating miRNAs as biomarkers of chemoresistance for oxaliplatin therapy in CRC patients. In particular, six miRNAs, miR-100, miR-92a, miR-16, miR-30e, miR-144-5p and let-7i, were verified as significantly and consistently downregulated (>1.5-fold, P < 0.05) in oxaliplatin-resistant patients. GO and KEGG pathway analysis showed that these miRNAs were able to modulate the RNA polymerase II transcription and the PI3K-AKT signaling pathway, AMPK signaling pathway and FoxO signaling pathway [456]. Jin G and colleagues selected 30 miRNAs that are aberrantly expressed during CRC progression based on previous microarray analyses. Subsequently, the expression levels of these miRNAs were assessed in oxaliplatin/5-FU-resistant CRC cell lines and in the corresponding secreted exosomes. Notably, miR-21-5p, miR-1246, miR-1229-5p, miR-135b, miR-425 and miR-96-5p were found up-regulated in exosomes obtained from the supernatant of resistant cells. Through GO and pathway prediction analysis, it was found that these miRNAs are involved in the PI3K-Akt, FoxO and autophagy signaling pathways. Therefore, targeting these miRNAs could promote chemosensitivity to oxaliplatin and 5-FU, representing a promising strategy for the treatment of resistant CRC [457]. Another study assessed the modulation of circulating miRNA levels in peripheral blood samples obtained from 77 5-FU-treated CRC patients. Differential expression of circulating miRNA levels was evaluated at three different time points: baseline, after 3 and after 6 months of treatment. Specifically, the expression levels of five miRNAs, miR-223-3p, miR-20a-5p, miR-17-5p, miR-19a-3p and miR-7-5p, and the expression of three proteins, PTEN, ERK and EGFR, were assessed. At baseline, CRC patients had significantly higher levels of circulating miRNAs than healthy controls. These levels decreased during 5-FU therapy and then increased significantly only in responder patients after 6 months. In particular, miR-19a-3p demonstrated a marked change in patients with elevated ERK, EGFR, and PTEN protein levels, showing a significant correlation with increased risk of disease recurrence and progression at the 6-month evaluation. This pattern suggests that miR-19a-3p could serve as a potential biomarker for early detection of aggressive disease behavior, particularly in patients with these specific molecular profiles [458]. In a study by Chen Q et al., the differential expression of circulating miRNAs from the serum of drug-responsive and drug-resistant patients was analyzed by microarray. Among the most significantly differentially expressed miRNAs between responders and non-responders, miR-221, miR-222, miR-122, miR-19a and miR-144 were selected for further validations in an independent cohort (N = 72). Notably, serum miR-19a levels were found to predict both intrinsic and acquired drug resistance [459]. miR-21-5p was frequently up-regulated in solid tumors, including CRC. The expression of miR-21-5p was found to be significantly up-regulated in the exosomes of CRC cells compared to normal human colon epithelial cells. Treatment of CRC cells with isolated exosomes or miR-21-5p mimic resulted in increased expression of genes involved in cell proliferation, invasion, and extracellular matrix degradation. These effects depended on miR-21-5p-mediated downregulation of its targets PDCD4, TPM1 and PTEN. In particular, miR-21-mediated PDCD4 silencing increases CRC resistance to 5-FU [311].
5. Conclusions
CRC is the fourth-deadliest cancer in the world and its incidence is constantly increasing worldwide. As highlighted in this review, miRNA expression profiles differ between normal mucosa and CRC tissue. The data here reported strongly support the role of miRNAs in CRC development and progression since miRNAs regulate cancer cell proliferation, migration, and invasion by modulating several molecular pathways, including Wnt/β-catenin, PI3K-AKT, RAS, MAPK, TGF-β and p53 signaling.
Several studies have also proposed miRNAs both as markers and therapeutic targets or for the development of novel RNA-based antitumor treatments. In this context, a better understanding of the role of miRNAs in CRC tumorigenesis and progression may provide new insights for non-invasive diagnostic tools for CRC screening and personalized therapy [460]. The present comprehensive review also highlights the role of miRNAs in mediating CRC drug resistance. These findings suggest the need for innovative in vitro and in vivo studies aimed at investigating the potential therapeutic application of miRNAs.
Despite these promising updates on miRNA research, several critical limitations remain in our understanding of the miRNA–CRC axis, which must be addressed to translate current knowledge into clinical impact. First, the context-dependent nature of miRNA activity remains a major challenge. A single miRNA can bind different targets and may act as a tumor suppressor in one setting and as an oncogene in another, depending on the cellular environment, the presence of specific cofactors, or even the cancer stage. This plasticity complicates therapeutic targeting and calls for more refined models that can account for tumor heterogeneity and dynamic miRNA–target interactions [461].
Moreover, as widely discussed in this review article, while many studies have identified dysregulated miRNAs in CRC through high-throughput profiling, relatively few have functionally validated these findings in relevant in vivo models. There is a significant gap between correlative studies and mechanistic investigations that clarify the downstream pathways affected by miRNAs, their upstream regulators, and the crosstalk with other molecular networks such as epigenetic modifications, immune response, and microbiota-host interactions.
Additionally, the therapeutic potential of miRNA-based interventions, though promising, is still constrained by delivery challenges, off-target effects, and the lack of tumor-specific targeting strategies. Most delivery systems used in preclinical models are not yet clinically feasible, and systemic administration of miRNA mimics or inhibitors may lead to unintended modulation of non-target tissues.
In the context of circulating miRNAs as non-invasive biomarkers, several limitations in the standardization of sample processing, the definition of normalization strategies, and the use of analytical platforms still exist. Specifically, circulating miRNA profiles are usually analyzed using RNA sequencing and microarray platforms and then validated through reverse transcription quantitative polymerase chain reaction (RT-PCR) or digital droplet PCR (ddPCR), with profound differences among these techniques. As regards miRNA profiling, microarray technology guarantees high throughput and multiplexing. However, conventional microarray technologies have a limited dynamic range and sensitivity [462]. SmallRNA sequencing by NGS is the most adopted method as it requires less starting material, allows the identification of miRNA isoforms and has the highest throughput [463]. Among the validation methods, despite RT-qPCR having low throughput compared to other techniques, it has advantages in terms of cost-effectiveness and speed [464]. When using RT-qPCR, it is important to normalize miRNA expression in order to reproduce data between studies, however, no stable endogenous controls have been identified yet. In contrast, ddPCR exhibits a higher tolerance to inhibitors than conventional RT-qPCR and allows an absolute quantification of miRNA expression [465,466]. Other technical issues related to miRNA quantification are due to pre-analytical factors, including appropriate sample volumes, sample handling, RNA extraction methods, quantification and normalization methods. The conflicting data on the expression levels of miRNAs in the different studies are partly due to the differences in these variables.
Finally, miRNAs involved in CRC chemo- and radioresistance are often studied in isolation, ignoring the complex interplay within the tumor microenvironment and the compensatory pathways that may undermine therapeutic efficacy. The dynamic response of miRNA expression to treatment further complicates their use as predictive markers, underscoring the need for longitudinal and integrative studies that combine transcriptomic, proteomic, and functional data.
Overall, all the findings here discussed highlight critical gaps in the current knowledge of miRNA-CRC axis; therefore, future research should prioritize the functional validation of miRNA–target interactions in clinically relevant models, the development of robust, specific delivery platforms for therapeutic use, the multi-omics integration to map miRNA-mediated networks and the development of consensus protocols for the clinical evaluation of circulating miRNAs as biomarkers.
CRediT authorship contribution statement
Federica Longo: Writing – review & editing, Writing – original draft, Data curation, Conceptualization. Giuseppe Gattuso: Writing – original draft, Investigation, Formal analysis. Graziana Spoto: Writing – original draft, Investigation, Formal analysis. Daria Ricci: Writing – original draft, Investigation, Formal analysis. Anastasia Cristina Venera Vitale: Writing – original draft, Investigation, Formal analysis. Alessandro Lavoro: Investigation, Formal analysis. Saverio Candido: Investigation, Formal analysis. Massimo Libra: Writing – review & editing, Visualization, Supervision, Funding acquisition. Luca Falzone: Writing – review & editing, Writing – original draft, Supervision, Funding acquisition, Data curation, Conceptualization.
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Funding
This work was supported in part by: European Union - NextGenerationEU through the Italian Ministry of University and Research under PNRR M4C2—Action 1.4—Call “Potenziamento strutture di ricerca e creazione di “campioni nazionali di R&S”—Project “National Center for Gene Therapy and Drugs based on RNA Technology” (CN00000041) to Professor Massimo Libra (CUP: E63C22000950006). The views and opinions expressed are those of the authors only and do not necessarily reflect those of the European Union or the European Commission. Neither the European Union nor the European Commission can be held responsible for them.
Prof. Luca Falzone was supported by the PIAno di inCEntivi per la RIcerca di Ateneo 2024/2026 - Linea di Intervento 1 “Progetti di ricerca collaborativa" (Project Code: BioEpiRes), and the Piano di incentivi per la ricerca di Ateneo 2024/2026 (Pia.ce.ri.), Linea di intervento 3 - Starting Grant (Project Code: ResCOr), University of Catania (Catania, Italy).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the Italian League Against Cancer (LILT) of Catania.
Footnotes
Peer review under the responsibility of Editorial Board of Non-coding RNA Research.
Contributor Information
Massimo Libra, Email: m.libra@unict.it.
Luca Falzone, Email: luca.falzone@unict.it.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Brenner H., Heisser T., Cardoso R., Hoffmeister M. Reduction in colorectal cancer incidence by screening endoscopy. Nat. Rev. Gastroenterol. Hepatol. 2024;21:125–133. doi: 10.1038/s41575-023-00847-3. [DOI] [PubMed] [Google Scholar]
- 3.Fearon E.R., Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 4.Gharib E., Robichaud G.A. From crypts to cancer: a holistic perspective on colorectal carcinogenesis and therapeutic strategies. Int. J. Mol. Sci. 2024;25:9463. doi: 10.3390/ijms25179463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kasi A., Handa S., Bhatti S., Umar S., Bansal A., Sun W. Molecular pathogenesis and classification of colorectal carcinoma. Curr. Colorectal Cancer Rep. 2020;16:97–106. doi: 10.1007/s11888-020-00458-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byers T., Levin B., Rothenberger D., Dodd G.D., Smith R.A. American cancer society guidelines for screening and surveillance for early detection of colorectal polyps and cancer: update 1997. American cancer society detection and treatment advisory group on colorectal cancer. CA Cancer J. Clin. 1997;47:154–160. doi: 10.3322/canjclin.47.3.154. [DOI] [PubMed] [Google Scholar]
- 7.Zhao R., Xia D., Chen Y., Kai Z., Ruan F., Xia C., Gong J., Wu J., Wang X. Improved diagnosis of colorectal cancer using combined biomarkers including Fusobacterium nucleatum, fecal occult blood, transferrin, CEA, CA19-9, gender, and age. Cancer Med. 2023;12:14636–14645. doi: 10.1002/cam4.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuipers E.J., Grady W.M., Lieberman D., Seufferlein T., Sung J.J., Boelens P.G., van de Velde C.J.H., Watanabe T. Colorectal cancer. Nat. Rev. Dis. Primers. 2015;1 doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keum N., Giovannucci E. Global burden of colorectal cancer: emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019;16:713–732. doi: 10.1038/s41575-019-0189-8. [DOI] [PubMed] [Google Scholar]
- 10.Douillard J.-Y., Rong A., Sidhu R. RAS mutations in colorectal cancer. N. Engl. J. Med. 2013;369:2159–2160. doi: 10.1056/NEJMc1312697. [DOI] [PubMed] [Google Scholar]
- 11.Jonker D.J., O'Callaghan C.J., Karapetis C.S., Zalcberg J.R., Tu D., Au H.-J., Berry S.R., Krahn M., Price T., Simes R.J., Tebbutt N.C., van Hazel G., Wierzbicki R., Langer C., Moore M.J. Cetuximab for the treatment of colorectal cancer. N. Engl. J. Med. 2007;357:2040–2048. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- 12.André T., Shiu K.-K., Kim T.W., Jensen B.V., Jensen L.H., Punt C., Smith D., Garcia-Carbonero R., Benavides M., Gibbs P., de la Fouchardiere C., Rivera F., Elez E., Bendell J., Le D.T., Yoshino T., Van Cutsem E., Yang P., Farooqui M.Z.H., Marinello P., Diaz L.A. KEYNOTE-177 investigators, pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N. Engl. J. Med. 2020;383:2207–2218. doi: 10.1056/NEJMoa2017699. [DOI] [PubMed] [Google Scholar]
- 13.Ma S.-C., Zhang J.-Q., Yan T.-H., Miao M.-X., Cao Y.-M., Cao Y.-B., Zhang L.-C., Li L. Novel strategies to reverse chemoresistance in colorectal cancer. Cancer Med. 2023;12:11073–11096. doi: 10.1002/cam4.5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu W.K.K., Law P.T.Y., Lee C.W., Cho C.H., Fan D., Wu K., Yu J., Sung J.J.Y. MicroRNA in colorectal cancer: from benchtop to bedside. Carcinogenesis. 2011;32:247–253. doi: 10.1093/carcin/bgq243. [DOI] [PubMed] [Google Scholar]
- 15.Wei L., Wang X., Lv L., Zheng Y., Zhang N., Yang M. The emerging role of noncoding RNAs in colorectal cancer chemoresistance. Cell. Oncol. 2019;42:757–768. doi: 10.1007/s13402-019-00466-8. [DOI] [PubMed] [Google Scholar]
- 16.Annese T., Tamma R., De Giorgis M., Ribatti D. microRNAs biogenesis, functions and role in tumor angiogenesis. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denli A.M., Tops B.B.J., Plasterk R.H.A., Ketting R.F., Hannon G.J. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 18.Schwarz D.S., Hutvágner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 19.Helwak A., Kudla G., Dudnakova T., Tollervey D. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell. 2013;153:654–665. doi: 10.1016/j.cell.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alizadeh M., Ghasemi H., Bazhan D., Mohammadi Bolbanabad N., Rahdan F., Arianfar N., Vahedi F., Khatami S.H., Taheri-Anganeh M., Aiiashi S., Armand N. MicroRNAs in disease states. Clin. Chim. Acta. 2025;569 doi: 10.1016/j.cca.2025.120187. [DOI] [PubMed] [Google Scholar]
- 21.Tâlvan C.-D., Tâlvan E.-T., Mohor C.I., Budişan L., Grecu V., Mihalache M., Neagoe I.B., Zănoagă O., Oprinca G.C., Cristian A.N. The impact of miRNA expression on colon cancer severity, invasiveness, and localization. Cancers (Basel) 2025;17:1091. doi: 10.3390/cancers17071091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pettorossi F., Gasparotto M., Ghirardello A., Franco C., Ceolotto G., Giannella A., Iaccarino L., Zanatta E., Doria A., Gatto M. MicroRNAs in idiopathic inflammatory myopathies: state-of-the-art and future perspectives. Curr. Opin. Rheumatol. 2023;35:374–382. doi: 10.1097/BOR.0000000000000960. [DOI] [PubMed] [Google Scholar]
- 23.Macvanin M., Obradovic M., Zafirovic S., Stanimirovic J., Isenovic E.R. The role of miRNAs in metabolic diseases. Curr. Med. Chem. 2023;30:1922–1944. doi: 10.2174/0929867329666220801161536. [DOI] [PubMed] [Google Scholar]
- 24.Colpaert R.M.W., Calore M. MicroRNAs in cardiac diseases. Cells. 2019;8:737. doi: 10.3390/cells8070737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavoro A., Gattuso G., Grillo C., Spandidos D., Salmeri M., Lombardo C., Candido S., Falzone L. Role of microRNAs as novel diagnostic biomarkers and potential therapeutic targets for hearing disorders (Review) Int. J. Epigen. 2022;2:3. doi: 10.3892/ije.2022.12. [DOI] [Google Scholar]
- 26.Oglesby I.K., McElvaney N.G., Greene C.M. MicroRNAs in inflammatory lung disease--master regulators or target practice? Respir. Res. 2010;11:148. doi: 10.1186/1465-9921-11-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., Rassenti L., Kipps T., Negrini M., Bullrich F., Croce C.M. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jurj A., Fontana B., Varani G., Calin G.A. Small molecules targeting microRNAs: new opportunities and challenges in precision cancer therapy. Trends Cancer. 2024;10:809–824. doi: 10.1016/j.trecan.2024.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Melo S., Villanueva A., Moutinho C., Davalos V., Spizzo R., Ivan C., Rossi S., Setien F., Casanovas O., Simo-Riudalbas L., Carmona J., Carrere J., Vidal A., Aytes A., Puertas S., Ropero S., Kalluri R., Croce C.M., Calin G.A., Esteller M. Small molecule enoxacin is a cancer-specific growth inhibitor that acts by enhancing TAR RNA-binding protein 2-mediated microRNA processing. Proc. Natl. Acad. Sci. U. S. A. 2011;108:4394–4399. doi: 10.1073/pnas.1014720108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gumireddy K., Young D.D., Xiong X., Hogenesch J.B., Huang Q., Deiters A. Small-molecule inhibitors of microrna miR-21 function. Angew Chem. Int. Ed. Engl. 2008;47:7482–7484. doi: 10.1002/anie.200801555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Velagapudi S.P., Cameron M.D., Haga C.L., Rosenberg L.H., Lafitte M., Duckett D.R., Phinney D.G., Disney M.D. Design of a small molecule against an oncogenic noncoding RNA. Proc. Natl. Acad. Sci. U. S. A. 2016;113:5898–5903. doi: 10.1073/pnas.1523975113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Velagapudi S.P., Gallo S.M., Disney M.D. Sequence-based design of bioactive small molecules that target precursor microRNAs. Nat. Chem. Biol. 2014;10:291–297. doi: 10.1038/nchembio.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costales M.G., Haga C.L., Velagapudi S.P., Childs-Disney J.L., Phinney D.G., Disney M.D. Small molecule inhibition of microRNA-210 reprograms an oncogenic hypoxic circuit. J. Am. Chem. Soc. 2017;139:3446–3455. doi: 10.1021/jacs.6b11273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi Z., Zhang J., Qian X., Han L., Zhang K., Chen L., Liu J., Ren Y., Yang M., Zhang A., Pu P., Kang C. AC1MMYR2, an inhibitor of dicer-mediated biogenesis of Oncomir miR-21, reverses epithelial-mesenchymal transition and suppresses tumor growth and progression. Cancer Res. 2013;73:5519–5531. doi: 10.1158/0008-5472.CAN-13-0280. [DOI] [PubMed] [Google Scholar]
- 35.Li Y., Disney M.D. Precise small molecule degradation of a noncoding RNA identifies cellular binding sites and modulates an oncogenic phenotype. ACS Chem. Biol. 2018;13:3065–3071. doi: 10.1021/acschembio.8b00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winkle M., El-Daly S.M., Fabbri M., Calin G.A. Noncoding RNA therapeutics - challenges and potential solutions. Nat. Rev. Drug Discov. 2021;20:629–651. doi: 10.1038/s41573-021-00219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bader A.G., Brown D., Stoudemire J., Lammers P. Developing therapeutic microRNAs for cancer. Gene Ther. 2011;18:1121–1126. doi: 10.1038/gt.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seyhan A.A. Trials and tribulations of MicroRNA therapeutics. Int. J. Mol. Sci. 2024;25:1469. doi: 10.3390/ijms25031469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lima J.F., Cerqueira L., Figueiredo C., Oliveira C., Azevedo N.F. Anti-miRNA oligonucleotides: a comprehensive guide for design. RNA Biol. 2018;15:338–352. doi: 10.1080/15476286.2018.1445959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kara G., Calin G.A., Ozpolat B. RNAi-based therapeutics and tumor targeted delivery in cancer. Adv. Drug Deliv. Rev. 2022;182 doi: 10.1016/j.addr.2022.114113. [DOI] [PubMed] [Google Scholar]
- 41.Brillante S., Volpe M., Indrieri A. Advances in MicroRNA therapeutics: from preclinical to clinical studies. Hum. Gene Ther. 2024;35:628–648. doi: 10.1089/hum.2024.113. [DOI] [PubMed] [Google Scholar]
- 42.Krützfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M., Stoffel M. Silencing of microRNAs in vivo with “antagomirs”. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 43.Lennox K.A., Behlke M.A. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011;18:1111–1120. doi: 10.1038/gt.2011.100. [DOI] [PubMed] [Google Scholar]
- 44.Tassone P., Di Martino M.T., Arbitrio M., Fiorillo L., Staropoli N., Ciliberto D., Cordua A., Scionti F., Bertucci B., Salvino A., Lopreiato M., Thunarf F., Cuomo O., Zito M.C., De Fina M.R., Brescia A., Gualtieri S., Riillo C., Manti F., Caracciolo D., Barbieri V., Di Paola E.D., Di Francesco A.E., Tagliaferri P. Safety and activity of the first-in-class locked nucleic acid (LNA) miR-221 selective inhibitor in refractory advanced cancer patients: a first-in-human, phase 1, open-label, dose-escalation study. J. Hematol. Oncol. 2023;16:68. doi: 10.1186/s13045-023-01468-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ebert M.S., Neilson J.R., Sharp P.A. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat. Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Krützfeldt J., Kuwajima S., Braich R., Rajeev K.G., Pena J., Tuschl T., Manoharan M., Stoffel M. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bader A.G., Brown D., Winkler M. The promise of microRNA replacement therapy. Cancer Res. 2010;70:7027–7030. doi: 10.1158/0008-5472.CAN-10-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiriboga C.A., Swoboda K.J., Darras B.T., Iannaccone S.T., Montes J., De Vivo D.C., Norris D.A., Bennett C.F., Bishop K.M. Results from a phase 1 study of nusinersen (ISIS-SMN(Rx)) in children with spinal muscular atrophy. Neurology. 2016;86:890–897. doi: 10.1212/WNL.0000000000002445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garber K. Big win possible for Ionis/Biogen antisense drug in muscular atrophy. Nat. Biotechnol. 2016;34:1002–1003. doi: 10.1038/nbt1016-1002. [DOI] [PubMed] [Google Scholar]
- 50.Garber K. Alnylam launches era of RNAi drugs. Nat. Biotechnol. 2018;36:777–778. doi: 10.1038/nbt0918-777. [DOI] [PubMed] [Google Scholar]
- 51.Scott L.J. Givosiran: first approval. Drugs (Basel) 2020;80:335–339. doi: 10.1007/s40265-020-01269-0. [DOI] [PubMed] [Google Scholar]
- 52.Smolarz B., Durczyński A., Romanowicz H., Szyłło K., Hogendorf P. miRNAs in cancer (review of literature) Int. J. Mol. Sci. 2022;23:2805. doi: 10.3390/ijms23052805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Omar A., Govan D., Penny C. Epigenetic regulation in colorectal cancer: the susceptibility of microRNAs 145, 143 and 133b to DNA demethylation and histone deacetylase inhibitors. PLoS One. 2023;18 doi: 10.1371/journal.pone.0289800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee Y.S., Dutta A. MicroRNAs in cancer. Annu. Rev. Pathol. 2009;4:199–227. doi: 10.1146/annurev.pathol.4.110807.092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma B., Wang S., Wu W., Shan P., Chen Y., Meng J., Xing L., Yun J., Hao L., Wang X., Li S., Guo Y. Mechanisms of circRNA/lncRNA-miRNA interactions and applications in disease and drug research. Biomed. Pharmacother. 2023;162 doi: 10.1016/j.biopha.2023.114672. [DOI] [PubMed] [Google Scholar]
- 57.Chen L.-L., Kim V.N. Small and long non-coding RNAs: past, present, and future. Cell. 2024;187:6451–6485. doi: 10.1016/j.cell.2024.10.024. [DOI] [PubMed] [Google Scholar]
- 58.Xiao Z., Qu Z., Chen Z., Fang Z., Zhou K., Huang Z., Guo X., Zhang Y. LncRNA HOTAIR is a prognostic biomarker for the proliferation and chemoresistance of colorectal cancer via MiR-203a-3p-mediated wnt/ß-catenin signaling pathway. Cell. Physiol. Biochem. 2018;46:1275–1285. doi: 10.1159/000489110. [DOI] [PubMed] [Google Scholar]
- 59.Huang X., Lu S. MicroR-545 mediates colorectal cancer cells proliferation through up-regulating epidermal growth factor receptor expression in HOTAIR long non-coding RNA dependent. Mol. Cell. Biochem. 2017;431:45–54. doi: 10.1007/s11010-017-2974-4. [DOI] [PubMed] [Google Scholar]
- 60.Li P., Zhang X., Wang L., Du L., Yang Y., Liu T., Li C., Wang C. lncRNA HOTAIR contributes to 5FU resistance through suppressing miR-218 and activating NF-κB/TS signaling in colorectal cancer. Mol. Ther. Nucleic Acids. 2017;8:356–369. doi: 10.1016/j.omtn.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tang X.J., Wang W., Hann S.S. Interactions among lncRNAs, miRNAs and mRNA in colorectal cancer. Biochimie. 2019;163:58–72. doi: 10.1016/j.biochi.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 62.Tarasov V.A., Matishov D.G., Shin E.F., Boiko N.V., Timoshkina N.N., Makhotkin M.A., Lomonosov A.M., Kirpii A.A., Kit O.I., Maksimov A.Y. [Coordinated aberranit expression of miRNAs in colon cancer] Genetika. 2014;50:1232–1244. [PubMed] [Google Scholar]
- 63.Hernández R., Sánchez-Jiménez E., Melguizo C., Prados J., Rama A.R. Downregulated microRNAs in the colorectal cancer: diagnostic and therapeutic perspectives. BMB Rep. 2018;51:563–571. doi: 10.5483/BMBRep.2018.51.11.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stark V.A., Facey C.O.B., Viswanathan V., Boman B.M. The role of miRNAs, miRNA clusters, and isomiRs in development of cancer stem cell populations in colorectal cancer. Int. J. Mol. Sci. 2021;22:1424. doi: 10.3390/ijms22031424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pritchard C.C., Cheng H.H., Tewari M. MicroRNA profiling: approaches and considerations. Nat. Rev. Genet. 2012;13:358–369. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sato A., Fujita Y., Otsuka K., Sasaki A., Suzuki H., Matsumoto T., Sugai T. Differential expression of microRNAs in colorectal cancer: different patterns between isolated cancer gland and stromal cells. Pathol. Int. 2020;70:21–30. doi: 10.1111/pin.12872. [DOI] [PubMed] [Google Scholar]
- 67.Sun D., Yu F., Ma Y., Zhao R., Chen X., Zhu J., Zhang C.-Y., Chen J., Zhang J. MicroRNA-31 activates the RAS pathway and functions as an oncogenic MicroRNA in human colorectal cancer by repressing RAS p21 GTPase activating protein 1 (RASA1) J. Biol. Chem. 2013;288:9508–9518. doi: 10.1074/jbc.M112.367763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reid J.F., Sokolova V., Zoni E., Lampis A., Pizzamiglio S., Bertan C., Zanutto S., Perrone F., Camerini T., Gallino G., Verderio P., Leo E., Pilotti S., Gariboldi M., Pierotti M.A. miRNA profiling in colorectal cancer highlights miR-1 involvement in MET-dependent proliferation. Mol. Cancer Res. 2012;10:504–515. doi: 10.1158/1541-7786.MCR-11-0342. [DOI] [PubMed] [Google Scholar]
- 69.Shaath H., Toor S.M., Nada M.A., Elkord E., Alajez N.M. Integrated whole transcriptome and small RNA analysis revealed multiple regulatory networks in colorectal cancer. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-93531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Almeida M.I., Nicoloso M.S., Zeng L., Ivan C., Spizzo R., Gafà R., Xiao L., Zhang X., Vannini I., Fanini F., Fabbri M., Lanza G., Reis R.M., Zweidler-McKay P.A., Calin G.A. Strand-specific miR-28-5p and miR-28-3p have distinct effects in colorectal cancer cells. Gastroenterology. 2012;142:886–896.e9. doi: 10.1053/j.gastro.2011.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ling H., Pickard K., Ivan C., Isella C., Ikuo M., Mitter R., Spizzo R., Bullock M., Braicu C., Pileczki V., Vincent K., Pichler M., Stiegelbauer V., Hoefler G., Almeida M.I., Hsiao A., Zhang X., Primrose J., Packham G., Liu K., Bojja K., Gafà R., Xiao L., Rossi S., Song J.H., Vannini I., Fanini F., Kopetz S., Zweidler-McKay P., Wang X., Ionescu C., Irimie A., Fabbri M., Lanza G., Hamilton S.R., Berindan-Neagoe I., Medico E., Mirnezami A., Calin G.A., Nicoloso M.S. The clinical and biological significance of MIR-224 expression in colorectal cancer metastasis. Gut. 2016;65:977–989. doi: 10.1136/gutjnl-2015-309372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shen K., Liang Q., Xu K., Cui D., Jiang L., Yin P., Lu Y., Li Q., Liu J. MiR-139 inhibits invasion and metastasis of colorectal cancer by targeting the type I insulin-like growth factor receptor. Biochem. Pharmacol. 2012;84:320–330. doi: 10.1016/j.bcp.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 73.Jevšinek Skok D., Hauptman N., Boštjančič E., Zidar N. The integrative knowledge base for miRNA-mRNA expression in colorectal cancer. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-54358-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Falzone L., Scola L., Zanghì A., Biondi A., Di Cataldo A., Libra M., Candido S. Integrated analysis of colorectal cancer microRNA datasets: identification of microRNAs associated with tumor development. Aging (Albany NY) 2018;10:1000–1014. doi: 10.18632/aging.101444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kandhavelu J., Subramanian K., Khan A., Omar A., Ruff P., Penny C. Computational analysis of miRNA and their gene targets significantly involved in colorectal cancer progression. MicroRNA. 2019;8:68–75. doi: 10.2174/2211536607666180803100246. [DOI] [PubMed] [Google Scholar]
- 76.He M.-Q., Wan J.-F., Zeng H.-F., Tang Y.-Y., He M.-Q. miR-133a-5p suppresses gastric cancer through TCF4 down-regulation. J. Gastrointest. Oncol. 2021;12:1007–1019. doi: 10.21037/jgo-20-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Crimi S., Falzone L., Gattuso G., Grillo C.M., Candido S., Bianchi A., Libra M. Droplet digital PCR analysis of liquid biopsy samples unveils the diagnostic role of hsa-miR-133a-3p and hsa-miR-375-3p in oral cancer. Biology (Basel) 2020;9:379. doi: 10.3390/biology9110379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yu J., Wang F., Yang G.-H., Wang F.-L., Ma Y.-N., Du Z.-W., Zhang J.-W. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochem. Biophys. Res. Commun. 2006;349:59–68. doi: 10.1016/j.bbrc.2006.07.207. [DOI] [PubMed] [Google Scholar]
- 79.Pidíková P., Herichová I. miRNA clusters with up-regulated expression in colorectal cancer. Cancers (Basel) 2021;13:2979. doi: 10.3390/cancers13122979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pidíkova P., Reis R., Herichova I. miRNA clusters with down-regulated expression in human colorectal cancer and their regulation. Int. J. Mol. Sci. 2020;21:4633. doi: 10.3390/ijms21134633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dong Y., Wu W.K.K., Wu C.W., Sung J.J.Y., Yu J., Ng S.S.M. MicroRNA dysregulation in colorectal cancer: a clinical perspective. Br. J. Cancer. 2011;104:893–898. doi: 10.1038/bjc.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schetter A.J., Leung S.Y., Sohn J.J., Zanetti K.A., Bowman E.D., Yanaihara N., Yuen S.T., Chan T.L., Kwong D.L.W., Au G.K.H., Liu C.-G., Calin G.A., Croce C.M., Harris C.C. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Earle J.S.L., Luthra R., Romans A., Abraham R., Ensor J., Yao H., Hamilton S.R. Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J. Mol. Diagn. 2010;12:433–440. doi: 10.2353/jmoldx.2010.090154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shi B., Sepp-Lorenzino L., Prisco M., Linsley P., deAngelis T., Baserga R. Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J. Biol. Chem. 2007;282:32582–32590. doi: 10.1074/jbc.M702806200. [DOI] [PubMed] [Google Scholar]
- 85.Sachdeva M., Mo Y.-Y. MicroRNA-145 suppresses cell invasion and metastasis by directly targeting mucin 1. Cancer Res. 2010;70:378–387. doi: 10.1158/0008-5472.CAN-09-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pagliuca A., Valvo C., Fabrizi E., di Martino S., Biffoni M., Runci D., Forte S., De Maria R., Ricci-Vitiani L. Analysis of the combined action of miR-143 and miR-145 on oncogenic pathways in colorectal cancer cells reveals a coordinate program of gene repression. Oncogene. 2013;32:4806–4813. doi: 10.1038/onc.2012.495. [DOI] [PubMed] [Google Scholar]
- 87.La Rocca G., Badin M., Shi B., Xu S.-Q., Deangelis T., Sepp-Lorenzinoi L., Baserga R. Mechanism of growth inhibition by MicroRNA 145: the role of the IGF-I receptor signaling pathway. J. Cell. Physiol. 2009;220:485–491. doi: 10.1002/jcp.21796. [DOI] [PubMed] [Google Scholar]
- 88.Zhang J., Sun Q., Zhang Z., Ge S., Han Z.-G., Chen W.-T. Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene. 2013;32:61–69. doi: 10.1038/onc.2012.28. [DOI] [PubMed] [Google Scholar]
- 89.Strippoli A., Cocomazzi A., Basso M., Cenci T., Ricci R., Pierconti F., Cassano A., Fiorentino V., Barone C., Bria E., Ricci-Vitiani L., Tortora G., Larocca L.M., Martini M. c-MYC expression is a possible keystone in the colorectal cancer resistance to EGFR inhibitors. Cancers (Basel) 2020;12:638. doi: 10.3390/cancers12030638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yin Y., Yan Z.-P., Lu N.-N., Xu Q., He J., Qian X., Yu J., Guan X., Jiang B.-H., Liu L.-Z. Downregulation of miR-145 associated with cancer progression and VEGF transcriptional activation by targeting N-RAS and IRS1. Biochim. Biophys. Acta. 2013;1829:239–247. doi: 10.1016/j.bbagrm.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 91.Feng Y., Zhu J., Ou C., Deng Z., Chen M., Huang W., Li L. MicroRNA-145 inhibits tumour growth and metastasis in colorectal cancer by targeting fascin-1. Br. J. Cancer. 2014;110:2300–2309. doi: 10.1038/bjc.2014.122. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 92.Chen Q., Zhou L., Ye X., Tao M., Wu J. miR-145-5p suppresses proliferation, metastasis and EMT of colorectal cancer by targeting CDCA3. Pathol. Res. Pract. 2020;216 doi: 10.1016/j.prp.2020.152872. [DOI] [PubMed] [Google Scholar]
- 93.Yang Y., Li X.-J., Li P., Guo X.-T. MicroRNA-145 regulates the proliferation, migration and invasion of human primary colon adenocarcinoma cells by targeting MAPK1. Int. J. Mol. Med. 2018;42:3171–3180. doi: 10.3892/ijmm.2018.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sathyanarayanan A., Chandrasekaran K.S., Karunagaran D. microRNA-145 downregulates SIP1-expression but differentially regulates proliferation, migration, invasion and Wnt signaling in SW480 and SW620 cells. J. Cell. Biochem. 2018;119:2022–2035. doi: 10.1002/jcb.26365. [DOI] [PubMed] [Google Scholar]
- 95.Yamada N., Noguchi S., Mori T., Naoe T., Maruo K., Akao Y. Tumor-suppressive microRNA-145 targets catenin δ-1 to regulate Wnt/β-catenin signaling in human colon cancer cells. Cancer Lett. 2013;335:332–342. doi: 10.1016/j.canlet.2013.02.060. [DOI] [PubMed] [Google Scholar]
- 96.Gregersen L.H., Jacobsen A.B., Frankel L.B., Wen J., Krogh A., Lund A.H. MicroRNA-145 targets YES and STAT1 in colon cancer cells. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang Z., Zhang X., Yang Z., Du H., Wu Z., Gong J., Yan J., Zheng Q. MiR-145 regulates PAK4 via the MAPK pathway and exhibits an antitumor effect in human colon cells. Biochem. Biophys. Res. Commun. 2012;427:444–449. doi: 10.1016/j.bbrc.2012.06.123. [DOI] [PubMed] [Google Scholar]
- 98.Li S., Wu X., Xu Y., Wu S., Li Z., Chen R., Huang N., Zhu Z., Xu X. miR-145 suppresses colorectal cancer cell migration and invasion by targeting an ETS-related gene. Oncol. Rep. 2016;36:1917–1926. doi: 10.3892/or.2016.5042. [DOI] [PubMed] [Google Scholar]
- 99.Yu Y., Nangia-Makker P., Farhana L., G Rajendra S., Levi E., Majumdar A.P.N. miR-21 and miR-145 cooperation in regulation of colon cancer stem cells. Mol. Cancer. 2015;14:98. doi: 10.1186/s12943-015-0372-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Borralho P.M., Kren B.T., Castro R.E., da Silva I.B.M., Steer C.J., Rodrigues C.M.P. MicroRNA-143 reduces viability and increases sensitivity to 5-fluorouracil in HCT116 human colorectal cancer cells. FEBS J. 2009;276:6689–6700. doi: 10.1111/j.1742-4658.2009.07383.x. [DOI] [PubMed] [Google Scholar]
- 101.Chen X., Guo X., Zhang H., Xiang Y., Chen J., Yin Y., Cai X., Wang K., Wang G., Ba Y., Zhu L., Wang J., Yang R., Zhang Y., Ren Z., Zen K., Zhang J., Zhang C.-Y. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–1392. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- 102.Akao Y., Nakagawa Y., Naoe T. MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 2007;26:311–320. doi: 10.1089/dna.2006.0550. [DOI] [PubMed] [Google Scholar]
- 103.Ng E.K.O., Tsang W.P., Ng S.S.M., Jin H.C., Yu J., Li J.J., Röcken C., Ebert M.P.A., Kwok T.T., Sung J.J.Y. MicroRNA-143 targets DNA methyltransferases 3A in colorectal cancer. Br. J. Cancer. 2009;101:699–706. doi: 10.1038/sj.bjc.6605195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gregersen L.H., Jacobsen A., Frankel L.B., Wen J., Krogh A., Lund A.H. MicroRNA-143 down-regulates Hexokinase 2 in colon cancer cells. BMC Cancer. 2012;12:232. doi: 10.1186/1471-2407-12-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang Y., Wang Z., Chen M., Peng L., Wang X., Ma Q., Ma F., Jiang B. MicroRNA-143 targets MACC1 to inhibit cell invasion and migration in colorectal cancer. Mol. Cancer. 2012;11:23. doi: 10.1186/1476-4598-11-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Law I.K.M., Bakirtzi K., Polytarchou C., Oikonomopoulos A., Hommes D., Iliopoulos D., Pothoulakis C. Neurotensin--regulated miR-133α is involved in proinflammatory signalling in human colonic epithelial cells and in experimental colitis. Gut. 2015;64:1095–1104. doi: 10.1136/gutjnl-2014-307329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zheng K., Liu W., Liu Y., Jiang C., Qian Q. MicroRNA-133a suppresses colorectal cancer cell invasion by targeting Fascin1. Oncol. Lett. 2021;22:644. doi: 10.3892/ol.2021.12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang H., An H., Wang B., Liao Q., Li W., Jin X., Cui S., Zhang Y., Ding Y., Zhao L. miR-133a represses tumour growth and metastasis in colorectal cancer by targeting LIM and SH3 protein 1 and inhibiting the MAPK pathway. Eur. J. Cancer. 2013;49:3924–3935. doi: 10.1016/j.ejca.2013.07.149. [DOI] [PubMed] [Google Scholar]
- 109.Dong Y., Zhao J., Wu C.-W., Zhang L., Liu X., Kang W., Leung W.-W., Zhang N., Chan F.K.L., Sung J.J.Y., Ng S.S.M., Yu J. Tumor suppressor functions of miR-133a in colorectal cancer. Mol. Cancer Res. 2013;11:1051–1060. doi: 10.1158/1541-7786.MCR-13-0061. [DOI] [PubMed] [Google Scholar]
- 110.Taniguchi K., Sakai M., Sugito N., Kumazaki M., Shinohara H., Yamada N., Nakayama T., Ueda H., Nakagawa Y., Ito Y., Futamura M., Uno B., Otsuki Y., Yoshida K., Uchiyama K., Akao Y. PTBP1-associated microRNA-1 and -133b suppress the Warburg effect in colorectal tumors. Oncotarget. 2016;7:18940–18952. doi: 10.18632/oncotarget.8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hu G., Chen D., Li X., Yang K., Wang H., Wu W. miR-133b regulates the MET proto-oncogene and inhibits the growth of colorectal cancer cells in vitro and in vivo. Cancer Biol. Ther. 2010;10:190–197. doi: 10.4161/cbt.10.2.12186. [DOI] [PubMed] [Google Scholar]
- 112.Duan F.-T., Qian F., Fang K., Lin K.-Y., Wang W.-T., Chen Y.-Q. miR-133b, a muscle-specific microRNA, is a novel prognostic marker that participates in the progression of human colorectal cancer via regulation of CXCR4 expression. Mol. Cancer. 2013;12:164. doi: 10.1186/1476-4598-12-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wang X.-W., Xi X.-Q., Wu J., Wan Y.-Y., Hui H.-X., Cao X.-F. MicroRNA-206 attenuates tumor proliferation and migration involving the downregulation of NOTCH3 in colorectal cancer. Oncol. Rep. 2015;33:1402–1410. doi: 10.3892/or.2015.3731. [DOI] [PubMed] [Google Scholar]
- 114.Lyu J., Sun Y., Li X., Ma H. MicroRNA-206 inhibits the proliferation, migration and invasion of colorectal cancer cells by regulating the c-Met/AKT/GSK-3β pathway. Oncol. Lett. 2021;21:147. doi: 10.3892/ol.2020.12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Furukawa S., Kawasaki Y., Miyamoto M., Hiyoshi M., Kitayama J., Akiyama T. The miR-1-NOTCH3-Asef pathway is important for colorectal tumor cell migration. PLoS One. 2013;8 doi: 10.1371/journal.pone.0080609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xu W., Zhang Z., Zou K., Cheng Y., Yang M., Chen H., Wang H., Zhao J., Chen P., He L., Chen X., Geng L., Gong S. MiR-1 suppresses tumor cell proliferation in colorectal cancer by inhibition of Smad3-mediated tumor glycolysis. Cell Death Dis. 2017;8 doi: 10.1038/cddis.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Guo S.T., Jiang C.C., Wang G.P., Li Y.P., Wang C.Y., Guo X.Y., Yang R.H., Feng Y., Wang F.H., Tseng H.-Y., Thorne R.F., Jin L., Zhang X.D. MicroRNA-497 targets insulin-like growth factor 1 receptor and has a tumour suppressive role in human colorectal cancer. Oncogene. 2013;32:1910–1920. doi: 10.1038/onc.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang N., Shen Q., Zhang P. miR-497 suppresses epithelial-mesenchymal transition and metastasis in colorectal cancer cells by targeting fos-related antigen-1. OncoTargets Ther. 2016;9:6597–6604. doi: 10.2147/OTT.S114609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xu Y., Chen J., Gao C., Zhu D., Xu X., Wu C., Jiang J. MicroRNA-497 inhibits tumor growth through targeting insulin receptor substrate 1 in colorectal cancer. Oncol. Lett. 2017;14:6379–6386. doi: 10.3892/ol.2017.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Wang L., Jiang C., Li D., Ge X., Shi Z., Li C., Liu X., Yin Y., Zhen L., Liu L.-Z., Jiang B.-H. MicroRNA-497 inhibits tumor growth and increases chemosensitivity to 5-fluorouracil treatment by targeting KSR1. Oncotarget. 2016;7:2660–2671. doi: 10.18632/oncotarget.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Luo Q., Zhang Z., Dai Z., Basnet S., Li S., Xu B., Ge H. Tumor-suppressive microRNA-195-5p regulates cell growth and inhibits cell cycle by targeting cyclin dependent kinase 8 in colon cancer. Am J Transl Res. 2016;8:2088–2096. [PMC free article] [PubMed] [Google Scholar]
- 122.Sun M., Song H., Wang S., Zhang C., Zheng L., Chen F., Shi D., Chen Y., Yang C., Xiang Z., Liu Q., Wei C., Xiong B. Integrated analysis identifies microRNA-195 as a suppressor of Hippo-YAP pathway in colorectal cancer. J. Hematol. Oncol. 2017;10:79. doi: 10.1186/s13045-017-0445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liu L., Chen L., Xu Y., Li R., Du X. microRNA-195 promotes apoptosis and suppresses tumorigenicity of human colorectal cancer cells. Biochem. Biophys. Res. Commun. 2010;400:236–240. doi: 10.1016/j.bbrc.2010.08.046. [DOI] [PubMed] [Google Scholar]
- 124.Qu J., Zhao L., Zhang P., Wang J., Xu N., Mi W., Jiang X., Zhang C., Qu J. MicroRNA-195 chemosensitizes colon cancer cells to the chemotherapeutic drug doxorubicin by targeting the first binding site of BCL2L2 mRNA. J. Cell. Physiol. 2015;230:535–545. doi: 10.1002/jcp.24366. [DOI] [PubMed] [Google Scholar]
- 125.Piccinno E., Scalavino V., Labarile N., Bianco G., Savino M.T., Armentano R., Giannelli G., Serino G. Downregulation of γ-catenin by miR-195-5p inhibits colon cancer progression, regulating desmosome function. Int. J. Mol. Sci. 2023;25:494. doi: 10.3390/ijms25010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ye Q., Liu S., Lin S., Xie W. Circular RNA circSEMA5A facilitates colorectal cancer development by regulating microRNA-195-5p to target CCNE1 axis. Cell. Signal. 2023;107 doi: 10.1016/j.cellsig.2023.110649. [DOI] [PubMed] [Google Scholar]
- 127.Gu H., Xu Z., Zhang J., Wei Y., Cheng L., Wang J. circ_0038718 promotes colon cancer cell malignant progression via the miR-195-5p/Axin2 signaling axis and also effect Wnt/β-catenin signal pathway. BMC Genom. 2021;22:768. doi: 10.1186/s12864-021-07880-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Shi L., Jackstadt R., Siemens H., Li H., Kirchner T., Hermeking H. p53-induced miR-15a/16-1 and AP4 form a double-negative feedback loop to regulate epithelial-mesenchymal transition and metastasis in colorectal cancer. Cancer Res. 2014;74:532–542. doi: 10.1158/0008-5472.CAN-13-2203. [DOI] [PubMed] [Google Scholar]
- 129.Liu L., Yao H., Zhou X., Chen J., Chen G., Shi X., Wu G., Zhou G., He S. MiR-15a-3p regulates ferroptosis via targeting glutathione peroxidase GPX4 in colorectal cancer. Mol. Carcinog. 2022;61:301–310. doi: 10.1002/mc.23367. [DOI] [PubMed] [Google Scholar]
- 130.Deng J., Wang H., Liang Y., Zhao L., Li Y., Yan Y., Zhao H., Zhang X., Zou F. miR-15a-5p enhances the malignant phenotypes of colorectal cancer cells through the STAT3/TWIST1 and PTEN/AKT signaling pathways by targeting SIRT4. Cell. Signal. 2023;101 doi: 10.1016/j.cellsig.2022.110517. [DOI] [PubMed] [Google Scholar]
- 131.Liu L., Yu T., Jin Y., Mai W., Zhou J., Zhao C. MicroRNA-15a carried by mesenchymal stem cell-derived extracellular vesicles inhibits the immune evasion of colorectal cancer cells by regulating the KDM4B/HOXC4/PD-L1 Axis. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.629893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ji D., Zhan T., Li M., Yao Y., Jia J., Yi H., Qiao M., Xia J., Zhang Z., Ding H., Song C., Han Y., Gu J. Enhancement of sensitivity to chemo/radiation therapy by using miR-15b against DCLK1 in colorectal cancer. Stem Cell Rep. 2018;11:1506–1522. doi: 10.1016/j.stemcr.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhao C., Zhao Q., Zhang C., Wang G., Yao Y., Huang X., Zhan F., Zhu Y., Shi J., Chen J., Yan F., Zhang Y. miR-15b-5p resensitizes colon cancer cells to 5-fluorouracil by promoting apoptosis via the NF-κB/XIAP axis. Sci. Rep. 2017;7:4194. doi: 10.1038/s41598-017-04172-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Sun L.-N., Zhi Z., Chen L.-Y., Zhou Q., Li X.-M., Gan W.-J., Chen S., Yang M., Liu Y., Shen T., Xu Y., Li J.-M. SIRT1 suppresses colorectal cancer metastasis by transcriptional repression of miR-15b-5p. Cancer Lett. 2017;409:104–115. doi: 10.1016/j.canlet.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 135.Ma Q., Wang X., Li Z., Li B., Ma F., Peng L., Zhang Y., Xu A., Jiang B. microRNA-16 represses colorectal cancer cell growth in vitro by regulating the p53/survivin signaling pathway. Oncol. Rep. 2013;29:1652–1658. doi: 10.3892/or.2013.2262. [DOI] [PubMed] [Google Scholar]
- 136.Cai K., Yang Y., Guo Z.-J., Cai R.-L., Hashida H., Li H.-X. Amentoflavone inhibits colorectal cancer epithelial-mesenchymal transition via the miR-16-5p/HMGA2/β-catenin pathway. Ann. Transl. Med. 2022;10:1009. doi: 10.21037/atm-22-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huang X., Hou Y., Weng X., Pang W., Hou L., Liang Y., Wang Y., Du L., Wu T., Yao M., Wang J., Meng X. Diethyldithiocarbamate-copper complex (CuET) inhibits colorectal cancer progression via miR-16-5p and 15b-5p/ALDH1A3/PKM2 axis-mediated aerobic glycolysis pathway. Oncogenesis. 2021;10:4. doi: 10.1038/s41389-020-00295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Young L.E., Moore A.E., Sokol L., Meisner-Kober N., Dixon D.A. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol. Cancer Res. 2012;10:167–180. doi: 10.1158/1541-7786.MCR-11-0337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xu Y., Shen L., Li F., Yang J., Wan X., Ouyang M. microRNA-16-5p-containing exosomes derived from bone marrow-derived mesenchymal stem cells inhibit proliferation, migration, and invasion, while promoting apoptosis of colorectal cancer cells by downregulating ITGA2. J. Cell. Physiol. 2019;234:21380–21394. doi: 10.1002/jcp.28747. [DOI] [PubMed] [Google Scholar]
- 140.Gil-Zamorano J., Martin R., Daimiel L., Richardson K., Giordano E., Nicod N., García-Carrasco B., Soares S.M.A., Iglesias-Gutiérrez E., Lasunción M.A., Sala-Vila A., Ros E., Ordovás J.M., Visioli F., Dávalos A. Docosahexaenoic acid modulates the enterocyte Caco-2 cell expression of microRNAs involved in lipid metabolism. J. Nutr. 2014;144:575–585. doi: 10.3945/jn.113.189050. [DOI] [PubMed] [Google Scholar]
- 141.Zhao K., Ye Z., Li Y., Li C., Yang X., Chen Q., Xing C. LncRNA FTX contributes to the progression of colorectal cancer through regulating miR-192-5p/eif5a2 Axis. OncoTargets Ther. 2020;13:2677–2688. doi: 10.2147/OTT.S241011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chang H.-Y., Ye S.-P., Pan S.-L., Kuo T.-T., Liu B.C., Chen Y.-L., Huang T.-C. Overexpression of miR-194 reverses HMGA2-driven signatures in colorectal cancer. Theranostics. 2017;7:3889–3900. doi: 10.7150/thno.20041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu T., Fang Y. MiR-194-3p modulates the progression of colorectal cancer by targeting KLK10. Histol. Histopathol. 2022;37:301–309. doi: 10.14670/HH-18-413. [DOI] [PubMed] [Google Scholar]
- 144.Wang B., Shen Z., Gao Z., Zhao G., Wang C., Yang Y., Zhang J., Yan Y., Shen C., Jiang K., Ye Y., Wang S. MiR-194, commonly repressed in colorectal cancer, suppresses tumor growth by regulating the MAP4K4/c-Jun/MDM2 signaling pathway. Cell Cycle. 2015;14:1046–1058. doi: 10.1080/15384101.2015.1007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sundaram P., Hultine S., Smith L.M., Dews M., Fox J.L., Biyashev D., Schelter J.M., Huang Q., Cleary M.A., Volpert O.V., Thomas-Tikhonenko A. p53-responsive miR-194 inhibits thrombospondin-1 and promotes angiogenesis in colon cancers. Cancer Res. 2011;71:7490–7501. doi: 10.1158/0008-5472.CAN-11-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhao H.-J., Ren L.-L., Wang Z.-H., Sun T.-T., Yu Y.-N., Wang Y.-C., Yan T.-T., Zou W., He J., Zhang Y., Hong J., Fang J.-Y. MiR-194 deregulation contributes to colorectal carcinogenesis via targeting AKT2 pathway. Theranostics. 2014;4:1193–1208. doi: 10.7150/thno.8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sun B., Fang Y.-T., Jin D.-J., Chen Z.-Y., Li Z.-Y., Gu X.-D., Xiang J.-B. miR-194 inhibits the proliferation of SW620 colon cancer stem cells through downregulation of SSH2 expression. Cancer Manag. Res. 2019;11:10229–10238. doi: 10.2147/CMAR.S221150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Feng Y., Xu Y., Gao Y., Chen Y., Wang X., Chen Z. A novel lncRNA SOX2OT promotes the malignancy of human colorectal cancer by interacting with miR-194-5p/SOX5 axis. Cell Death Dis. 2021;12:499. doi: 10.1038/s41419-021-03756-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wang M., Han D., Yuan Z., Hu H., Zhao Z., Yang R., Jin Y., Zou C., Chen Y., Wang G., Gao X., Wang X. Long non-coding RNA H19 confers 5-Fu resistance in colorectal cancer by promoting SIRT1-mediated autophagy. Cell Death Dis. 2018;9:1149. doi: 10.1038/s41419-018-1187-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Vychytilova-Faltejskova P., Merhautova J., Machackova T., Gutierrez-Garcia I., Garcia-Solano J., Radova L., Brchnelova D., Slaba K., Svoboda M., Halamkova J., Demlova R., Kiss I., Vyzula R., Conesa-Zamora P., Slaby O. MiR-215-5p is a tumor suppressor in colorectal cancer targeting EGFR ligand epiregulin and its transcriptional inducer HOXB9. Oncogenesis. 2017;6:399. doi: 10.1038/s41389-017-0006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li J., Han Y., Zhou M., Liu N., Li H., Huang G., Yu Z., Luo D., Zhang H., Zheng X., Liang F., Chen R. Electroacupuncture ameliorates AOM/DSS-induced mice colorectal cancer by inhibiting inflammation and promoting autophagy via the SIRT1/miR-215/Atg14 axis. Aging (Albany NY) 2023;15:13194–13212. doi: 10.18632/aging.205236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Jones M.F., Hara T., Francis P., Li X.L., Bilke S., Zhu Y., Pineda M., Subramanian M., Bodmer W.F., Lal A. The CDX1-microRNA-215 axis regulates colorectal cancer stem cell differentiation. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E1550–E1558. doi: 10.1073/pnas.1503370112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zheng S., Zhong Y.-F., Tan D.-M., Xu Y., Chen H.-X., Wang D. miR-183-5p enhances the radioresistance of colorectal cancer by directly targeting ATG5. J. Biosci. 2019;44:92. [PubMed] [Google Scholar]
- 154.Shang A., Wang X., Gu C., Liu W., Sun J., Zeng B., Chen C., Ji P., Wu J., Quan W., Yao Y., Wang W., Sun Z., Li D. Exosomal miR-183-5p promotes angiogenesis in colorectal cancer by regulation of FOXO1. Aging (Albany NY) 2020;12:8352–8371. doi: 10.18632/aging.103145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yu J.-H., Tan J.-N., Zhong G.-Y., Zhong L., Hou D., Ma S., Wang P.-L., Zhang Z.-H., Lu X.-Q., Yang B., Zhou S.-N., Han F.-H. Hsa_circ_0020134 promotes liver metastasis of colorectal cancer through the miR-183-5p-PFN2-TGF-β/Smad axis. Transl. Oncol. 2024;39 doi: 10.1016/j.tranon.2023.101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pan D., Hao J., Wu T., Shen T., Yu K., Li Q., Hu R., Yang Z., Li Y. Sodium butyrate inhibits the malignant proliferation of colon cancer cells via the miR-183/DNAJB4 Axis. Biochem. Genet. 2024;62:4174–4190. doi: 10.1007/s10528-023-10599-z. [DOI] [PubMed] [Google Scholar]
- 157.Wang G., Zhou J., Lu F., Qiu L., Xu L., Yang X., Miao Y. Downregulation of microRNA-183-5p inhibits the proliferation and invasion of colorectal cancer cells by inactivating the reticulocalbin-2/Wnt/β-catenin signaling pathway. Mol. Med. Rep. 2019;19:4475–4483. doi: 10.3892/mmr.2019.10059. [DOI] [PubMed] [Google Scholar]
- 158.Ge T., Xiang P., Mao H., Tang S., Zhou J., Zhang Y. Inhibition of miR-96 enhances the sensitivity of colorectal cancer cells to oxaliplatin by targeting TPM1. Exp. Ther. Med. 2020;20:2134–2140. doi: 10.3892/etm.2020.8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang X., Liu F., Cui Z., Li Z., Lv Y. Carboxypeptidase A6 suppresses the proliferation and invasion of colorectal cancer cells and is negatively regulated by miR-96-3p. Arch. Biochem. Biophys. 2023;740 doi: 10.1016/j.abb.2023.109595. [DOI] [PubMed] [Google Scholar]
- 160.Yue C., Chen J., Li Z., Li L., Chen J., Guo Y. microRNA-96 promotes occurrence and progression of colorectal cancer via regulation of the AMPKα2-FTO-m6A/MYC axis. J. Exp. Clin. Cancer Res. 2020;39:240. doi: 10.1186/s13046-020-01731-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Gao F., Wang W. MicroRNA-96 promotes the proliferation of colorectal cancer cells and targets tumor protein p53 inducible nuclear protein 1, forkhead box protein O1 (FOXO1) and FOXO3a. Mol. Med. Rep. 2015;11:1200–1206. doi: 10.3892/mmr.2014.2854. [DOI] [PubMed] [Google Scholar]
- 162.Zhou L., Liu H., Chen Z., Chen S., Lu J., Liu C., Liao S., He S., Chen S., Zhou Z. Downregulation of miR-182-5p by NFIB promotes NAD+ salvage synthesis in colorectal cancer by targeting NAMPT. Commun. Biol. 2023;6:775. doi: 10.1038/s42003-023-05143-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Guo Z., Liu X., Shao H. E2F4-induced AGAP2-AS1 up-regulation accelerates the progression of colorectal cancer via miR-182-5p/CFL1 axis. Dig. Liver Dis. 2022;54:878–889. doi: 10.1016/j.dld.2021.08.002. [DOI] [PubMed] [Google Scholar]
- 164.Li F., Zhou Y.-D., Liu J., Cai J.-D., Liao Z.-M., Chen G.-Q. RBP-J promotes cell growth and metastasis through regulating miR-182-5p-mediated Tiam1/Rac1/p38 MAPK axis in colorectal cancer. Cell. Signal. 2021;87 doi: 10.1016/j.cellsig.2021.110103. [DOI] [PubMed] [Google Scholar]
- 165.Li X., Zhang X., Zhang Q., Lin R. miR-182 contributes to cell proliferation, invasion and tumor growth in colorectal cancer by targeting DAB2IP. Int. J. Biochem. Cell Biol. 2019;111:27–36. doi: 10.1016/j.biocel.2019.04.002. [DOI] [PubMed] [Google Scholar]
- 166.Brocato J., Costa M. SATB1 and 2 in colorectal cancer. Carcinogenesis. 2015;36:186–191. doi: 10.1093/carcin/bgu322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Jia L., Luo S., Ren X., Li Y., Hu J., Liu B., Zhao L., Shan Y., Zhou H. miR-182 and miR-135b mediate the tumorigenesis and invasiveness of colorectal cancer cells via targeting ST6GALNAC2 and PI3K/AKT pathway. Dig. Dis. Sci. 2017;62:3447–3459. doi: 10.1007/s10620-017-4755-z. [DOI] [PubMed] [Google Scholar]
- 168.Zhang Y., Wang X., Wang Z., Tang H., Fan H., Guo Q. miR-182 promotes cell growth and invasion by targeting forkhead box F2 transcription factor in colorectal cancer. Oncol. Rep. 2015;33:2592–2598. doi: 10.3892/or.2015.3833. [DOI] [PubMed] [Google Scholar]
- 169.Amodeo V., Bazan V., Fanale D., Insalaco L., Caruso S., Cicero G., Bronte G., Rolfo C., Santini D., Russo A. Effects of anti-miR-182 on TSP-1 expression in human colon cancer cells: there is a sense in antisense? Expert Opin. Ther. Targets. 2013;17:1249–1261. doi: 10.1517/14728222.2013.832206. [DOI] [PubMed] [Google Scholar]
- 170.Yu W., Wang J., Li C., Xuan M., Han S., Zhang Y., Liu P., Zhao Z. miR-17-5p promotes the invasion and migration of colorectal cancer by regulating HSPB2. J. Cancer. 2022;13:918–931. doi: 10.7150/jca.65614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sun K., Chen L., Li Y., Huang B., Yan Q., Wu C., Lai Q., Fang Y., Cai J., Liu Y., Chen J., Wang X., Zhu Y., Dong S., Tan J., Li A., Liu S., Zhang Y. METTL14-dependent maturation of pri-miR-17 regulates mitochondrial homeostasis and induces chemoresistance in colorectal cancer. Cell Death Dis. 2023;14:148. doi: 10.1038/s41419-023-05670-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Xu J., Meng Q., Li X., Yang H., Xu J., Gao N., Sun H., Wu S., Familiari G., Relucenti M., Zhu H., Wu J., Chen R. Long noncoding RNA MIR17HG promotes colorectal cancer progression via miR-17-5p. Cancer Res. 2019;79:4882–4895. doi: 10.1158/0008-5472.CAN-18-3880. [DOI] [PubMed] [Google Scholar]
- 173.Zhang Y., Wang S., Lai Q., Fang Y., Wu C., Liu Y., Li Q., Wang X., Gu C., Chen J., Cai J., Li A., Liu S. Cancer-associated fibroblasts-derived exosomal miR-17-5p promotes colorectal cancer aggressive phenotype by initiating a RUNX3/MYC/TGF-β1 positive feedback loop. Cancer Lett. 2020;491:22–35. doi: 10.1016/j.canlet.2020.07.023. [DOI] [PubMed] [Google Scholar]
- 174.Ji J., Fu J. MiR-17-3p facilitates aggressive cell phenotypes in colon cancer by targeting PLCD1 through affecting KIF14. Appl. Biochem. Biotechnol. 2023;195:1723–1735. doi: 10.1007/s12010-022-04218-7. [DOI] [PubMed] [Google Scholar]
- 175.Wang Y., Zhao J., Wang Y., Gao J., Yang H., Li H. MiR-17-5p targets and downregulates CADM2, activating the malignant phenotypes of colon cancer cells. Mol. Biotechnol. 2022;64:1388–1400. doi: 10.1007/s12033-022-00515-y. [DOI] [PubMed] [Google Scholar]
- 176.Ma H., Pan J.-S., Jin L.-X., Wu J., Ren Y.-D., Chen P., Xiao C., Han J. MicroRNA-17∼92 inhibits colorectal cancer progression by targeting angiogenesis. Cancer Lett. 2016;376:293–302. doi: 10.1016/j.canlet.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 177.Ma Y., Zhang P., Wang F., Zhang H., Yang Y., Shi C., Xia Y., Peng J., Liu W., Yang Z., Qin H. Elevated oncofoetal miR-17-5p expression regulates colorectal cancer progression by repressing its target gene P130. Nat. Commun. 2012;3:1291. doi: 10.1038/ncomms2276. [DOI] [PubMed] [Google Scholar]
- 178.Kim T.W., Lee Y.S., Yun N.H., Shin C.H., Hong H.K., Kim H.H., Cho Y.B. MicroRNA-17-5p regulates EMT by targeting vimentin in colorectal cancer. Br. J. Cancer. 2020;123:1123–1130. doi: 10.1038/s41416-020-0940-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Sun W., Cui J., Ge Y., Wang J., Yu Y., Han B., Liu B. Tumor stem cell-derived exosomal microRNA-17-5p inhibits anti-tumor immunity in colorectal cancer via targeting SPOP and overexpressing PD-L1. Cell Death Discov. 2022;8:223. doi: 10.1038/s41420-022-00919-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Fang L., Li H., Wang L., Hu J., Jin T., Wang J., Yang B.B. MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget. 2014;5:2974–2987. doi: 10.18632/oncotarget.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Luo H., Zou J., Dong Z., Zeng Q., Wu D., Liu L. Up-regulated miR-17 promotes cell proliferation, tumour growth and cell cycle progression by targeting the RND3 tumour suppressor gene in colorectal carcinoma. Biochem. J. 2012;442:311–321. doi: 10.1042/BJ20111517. [DOI] [PubMed] [Google Scholar]
- 182.Boces-Pascual C., Mata-Ventosa A., Martín-Satué M., Boix L., Gironella M., Pastor-Anglada M., Pérez-Torras S. OncomiRs miR-106a and miR-17 negatively regulate the nucleoside-derived drug transporter hCNT1. Cell. Mol. Life Sci. 2021;78:7505–7518. doi: 10.1007/s00018-021-03959-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Yin S.L., Xiao F., Liu Y.F., Chen H., Guo G.C. Long non-coding RNA FENDRR restrains the aggressiveness of CRC via regulating miR-18a-5p/ING4 axis. J. Cell. Biochem. 2020;121:3973–3985. doi: 10.1002/jcb.29555. [DOI] [PubMed] [Google Scholar]
- 184.Kang L., Sun J., Liu J., Xu F., Zhu Q., Shi X. Long non-coding RNA CASC2 functions as A tumor suppressor in colorectal cancer via modulating the miR-18a-5p/BTG3 pathway. Cell J. 2022;24:665–672. doi: 10.22074/cellj.2022.8036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Huang G., Wu X., Li S., Xu X., Zhu H., Chen X. The long noncoding RNA CASC2 functions as a competing endogenous RNA by sponging miR-18a in colorectal cancer. Sci. Rep. 2016;6 doi: 10.1038/srep26524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Liu G., Liu Y., Yang Z., Wang J., Li D., Zhang X. Tumor suppressor microRNA-18a regulates tumor proliferation and invasion by targeting TBPL1 in colorectal cancer cells. Mol. Med. Rep. 2015;12:7643–7648. doi: 10.3892/mmr.2015.4335. [DOI] [PubMed] [Google Scholar]
- 187.Zhang Y., Liu X., Zhang J., Xu Y., Shao J., Hu Y., Shu P., Cheng H. Inhibition of miR-19a partially reversed the resistance of colorectal cancer to oxaliplatin via PTEN/PI3K/AKT pathway. Aging (Albany NY) 2020;12:5640–5650. doi: 10.18632/aging.102929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Fan H., Ai R., Mu S., Niu X., Guo Z., Liu L. MiR-19a suppresses ferroptosis of colorectal cancer cells by targeting IREB2. Bioengineered. 2022;13:12021–12029. doi: 10.1080/21655979.2022.2054194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Li H., Huang B. <em>miR-19a</em> targeting <em>CLCA4</em> to regulate the proliferation, migration, and invasion of colorectal cancer cells. Eur. J. Histochem. 2022;66:3381. doi: 10.4081/ejh.2022.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Yin Q., Wang P.-P., Peng R., Zhou H. MiR-19a enhances cell proliferation, migration, and invasiveness through enhancing lymphangiogenesis by targeting thrombospondin-1 in colorectal cancer. Biochem. Cell. Biol. 2019;97:731–739. doi: 10.1139/bcb-2018-0302. [DOI] [PubMed] [Google Scholar]
- 191.Chen M., Lin M., Wang X. Overexpression of miR-19a inhibits colorectal cancer angiogenesis by suppressing KRAS expression. Oncol. Rep. 2018;39:619–626. doi: 10.3892/or.2017.6110. [DOI] [PubMed] [Google Scholar]
- 192.Dai W., Zeng W., Lee D. lncRNA MCM3AP-AS1 inhibits the progression of colorectal cancer via the miR-19a-3p/FOXF2 axis. J. Gene Med. 2021;23 doi: 10.1002/jgm.3306. [DOI] [PubMed] [Google Scholar]
- 193.Shen P., Qu L., Wang J., Ding Q., Zhou C., Xie R., Wang H., Ji G. LncRNA LINC00342 contributes to the growth and metastasis of colorectal cancer via targeting miR-19a-3p/NPEPL1 axis. Cancer Cell Int. 2021;21:105. doi: 10.1186/s12935-020-01705-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Sun T., Yin Y.-F., Jin H.-G., Liu H.-R., Tian W.-C. Exosomal microRNA-19b targets FBXW7 to promote colorectal cancer stem cell stemness and induce resistance to radiotherapy. Kaohsiung J. Med. Sci. 2022;38:108–119. doi: 10.1002/kjm2.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Santos A., Cristóbal I., Caramés C., Luque M., Sanz-Álvarez M., Madoz-Gúrpide J., Rojo F., García-Foncillas J. Deregulation of the miR-19b/PPP2R5E signaling Axis shows high functional impact in colorectal cancer cells. Int. J. Mol. Sci. 2023;24:7779. doi: 10.3390/ijms24097779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Cruz-Gil S., Sanchez-Martinez R., Gomez de Cedron M., Martin-Hernandez R., Vargas T., Molina S., Herranz J., Davalos A., Reglero G., Ramirez de Molina A. Targeting the lipid metabolic axis ACSL/SCD in colorectal cancer progression by therapeutic miRNAs: miR-19b-1 role. J. Lipid Res. 2018;59:14–24. doi: 10.1194/jlr.M076752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Che J., Wang W., Huang Y., Zhang L., Zhao J., Zhang P., Yuan X. miR-20a inhibits hypoxia-induced autophagy by targeting ATG5/FIP200 in colorectal cancer. Mol. Carcinog. 2019;58:1234–1247. doi: 10.1002/mc.23006. [DOI] [PubMed] [Google Scholar]
- 198.Signs S.A., Fisher R.C., Tran U., Chakrabarti S., Sarvestani S.K., Xiang S., Liska D., Roche V., Lai W., Gittleman H.R., Wessely O., Huang E.H. Stromal miR-20a controls paracrine CXCL8 secretion in colitis and colon cancer. Oncotarget. 2018;9:13048–13059. doi: 10.18632/oncotarget.24495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Jiang Z., Li L., Hou Z., Liu W., Wang H., Zhou T., Li Y., Chen S. LncRNA HAND2-AS1 inhibits 5-fluorouracil resistance by modulating miR-20a/PDCD4 axis in colorectal cancer. Cell. Signal. 2020;66 doi: 10.1016/j.cellsig.2019.109483. [DOI] [PubMed] [Google Scholar]
- 200.Tang S., Fu H., Xu Q., Zhou Y. miR-20a regulates sensitivity of colorectal cancer cells to NK cells by targeting MICA. Biosci. Rep. 2019;39 doi: 10.1042/BSR20180695. BSR20180695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Qiang Y., Feng L., Wang G., Liu J., Zhang J., Xiang L., Su C., Zhang S., Xie X., Chen E. miR-20a/Foxj2 Axis mediates growth and metastasis of colorectal cancer cells as identified by integrated analysis. Med. Sci. Monit. 2020;26 doi: 10.12659/MSM.923559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ghofrani-Shahpar M., Pakravan K., Razmara E., Amooie F., Mahmoudian M., Heshmati M., Babashah S. Cancer-associated fibroblasts drive colorectal cancer cell progression through exosomal miR-20a-5p-mediated targeting of PTEN and stimulating interleukin-6 production. BMC Cancer. 2024;24:400. doi: 10.1186/s12885-024-12190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Cheng D., Zhao S., Tang H., Zhang D., Sun H., Yu F., Jiang W., Yue B., Wang J., Zhang M., Yu Y., Liu X., Sun X., Zhou Z., Qin X., Zhang X., Yan D., Wen Y., Peng Z. MicroRNA-20a-5p promotes colorectal cancer invasion and metastasis by downregulating Smad4. Oncotarget. 2016;7:45199–45213. doi: 10.18632/oncotarget.9900. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 204.Sehgal P., Lanauze C., Wang X., Hayer K.E., Torres-Diz M., Leu N.A., Sela Y., Stanger B.Z., Lengner C.J., Thomas-Tikhonenko A. MYC hyperactivates Wnt signaling in APC/CTNNB1-Mutated colorectal cancer cells through miR-92a-dependent repression of DKK3. Mol. Cancer Res. 2021;19:2003–2014. doi: 10.1158/1541-7786.MCR-21-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Li L., Zhang J., Peng H., Jiang X., Liu Z., Tian H., Hou S., Xie X., Peng Q., Zhou T. Knockdown of miR-92a suppresses the stemness of colorectal cancer cells via mediating SOCS3. Bioengineered. 2022;13:5613–5624. doi: 10.1080/21655979.2021.2022267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Alcantara K.M.M., Garcia R.L. MicroRNA-92a promotes cell proliferation, migration and survival by directly targeting the tumor suppressor gene NF2 in colorectal and lung cancer cells. Oncol. Rep. 2019;41:2103–2116. doi: 10.3892/or.2019.7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Lv H., Zhang Z., Wang Y., Li C., Gong W., Wang X. MicroRNA-92a promotes colorectal cancer cell growth and migration by inhibiting KLF4. Oncol. Res. 2016;23:283–290. doi: 10.3727/096504016X14562725373833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kim B.-K., Yoo H.-I., Lee A.-R., Choi K., Yoon S.K. Decreased expression of VLDLR is inversely correlated with miR-200c in human colorectal cancer. Mol. Carcinog. 2017;56:1620–1629. doi: 10.1002/mc.22618. [DOI] [PubMed] [Google Scholar]
- 209.Wang Z.-Z., Yang J., Jiang B.-H., Di J.-B., Gao P., Peng L., Su X.-Q. KIF14 promotes cell proliferation via activation of Akt and is directly targeted by miR-200c in colorectal cancer. Int. J. Oncol. 2018;53:1939–1952. doi: 10.3892/ijo.2018.4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Sui H., Cai G.-X., Pan S.-F., Deng W.-L., Wang Y.-W., Chen Z.-S., Cai S.-J., Zhu H.-R., Li Q. miR200c attenuates P-gp-mediated MDR and metastasis by targeting JNK2/c-Jun signaling pathway in colorectal cancer. Mol. Cancer Therapeut. 2014;13:3137–3151. doi: 10.1158/1535-7163.MCT-14-0167. [DOI] [PubMed] [Google Scholar]
- 211.Hu M., Xia M., Chen X., Lin Z., Xu Y., Ma Y., Su L. MicroRNA-141 regulates Smad interacting protein 1 (SIP1) and inhibits migration and invasion of colorectal cancer cells. Dig. Dis. Sci. 2010;55:2365–2372. doi: 10.1007/s10620-009-1008-9. [DOI] [PubMed] [Google Scholar]
- 212.Xing Y., Jing H., Zhang Y., Suo J., Qian M. MicroRNA-141-3p affected proliferation, chemosensitivity, migration and invasion of colorectal cancer cells by targeting EGFR. Int. J. Biochem. Cell Biol. 2020;118 doi: 10.1016/j.biocel.2019.105643. [DOI] [PubMed] [Google Scholar]
- 213.Sun Z., Shao B., Liu Z., Dang Q., Guo Y., Chen C., Guo Y., Chen Z., Liu J., Hu S., Yuan W., Zhou Q. LINC01296/miR-141-3p/ZEB1-ZEB2 axis promotes tumor metastasis via enhancing epithelial-mesenchymal transition process. J. Cancer. 2021;12:2723–2734. doi: 10.7150/jca.55626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Wang F., Zhao L., Zhang J., Meng Z., Zhou C., Wang G., Liu Y., Li M., Xi J., Niu W., Wang G. Chemotherapy-induced miR-141/MAP4K4 signaling suppresses progression of colorectal cancer. Biosci. Rep. 2018;38 doi: 10.1042/BSR20180978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Wu P.P., Zhu H.Y., Sun X.F., Chen L.X., Zhou Q., Chen J. MicroRNA-141 regulates the tumour suppressor DLC1 in colorectal cancer. Neoplasma. 2015;62:705–712. doi: 10.4149/neo_2015_084. [DOI] [PubMed] [Google Scholar]
- 216.Pei W., Wei K., Wu Y., Qiu Q., Zhu H., Mao L., Shi X., Zhang S., Shi Y., Tao S., Mao H., Pang S., Wang J., Liu M., Wang W., Yang Q., Chen C. Colorectal cancer tumor cell-derived exosomal miR-203a-3p promotes CRC metastasis by targeting PTEN-induced macrophage polarization. Gene (Amst.) 2023;885 doi: 10.1016/j.gene.2023.147692. [DOI] [PubMed] [Google Scholar]
- 217.Miao J., Hou N., Yang W., Jiang Q., Xue W., Wang X., Zhang H., Xiong X., Wang L., Zhao L., Huang C. miR-203a suppresses cell proliferation by targeting RING-finger protein 6 in colorectal cancer. Anti Cancer Drugs. 2020;31:583–591. doi: 10.1097/CAD.0000000000000874. [DOI] [PubMed] [Google Scholar]
- 218.Chen L., Gao H., Liang J., Qiao J., Duan J., Shi H., Zhen T., Li H., Zhang F., Zhu Z., Han A. miR-203a-3p promotes colorectal cancer proliferation and migration by targeting PDE4D. Am. J. Cancer Res. 2018;8:2387–2401. [PMC free article] [PubMed] [Google Scholar]
- 219.Qian Z., Gong L., Mou Y., Han Y., Zheng S. MicroRNA-203a-3p is a candidate tumor suppressor that targets thrombospondin 2 in colorectal carcinoma. Oncol. Rep. 2019;42:1825–1832. doi: 10.3892/or.2019.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Lu X., Yu Y., Liao F., Tan S. Homo sapiens circular RNA 0079993 (hsa_circ_0079993) of the POLR2J4 gene acts as an oncogene in colorectal cancer through the microRNA-203a-3p.1 and CREB1 Axis. Med. Sci. Monit. 2019;25:6872–6883. doi: 10.12659/MSM.916064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Yang Z., An Y., Wang N., Dong X., Kang H. LINC02595 promotes tumor progression in colorectal cancer by inhibiting miR-203b-3p activity and facilitating BCL2L1 expression. J. Cell. Physiol. 2020;235:7449–7464. doi: 10.1002/jcp.29650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Tian F., Wang P., Lin D., Dai J., Liu Q., Guan Y., Zhan Y., Yang Y., Wang W., Wang J., Liu J., Zheng L., Zhuang Y., Hu J., Wang J., Kong D., Zhu K. Exosome-delivered miR-221/222 exacerbates tumor liver metastasis by targeting SPINT1 in colorectal cancer. Cancer Sci. 2021;112:3744–3755. doi: 10.1111/cas.15028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Hu J., Zhang J., Yu M., Liu Z., Zou Y., Hong L., Zhang T., Sun J., Zheng M., Zhu X., Wang Z., Liu S. Circulating miR-221/222 reduces CD4+ T cells by inhibiting CD4 expression in colorectal cancer. Acta Biochim. Biophys. Sin. 2021;53:1367–1376. doi: 10.1093/abbs/gmab106. [DOI] [PubMed] [Google Scholar]
- 224.Xu K., Liang X., Shen K., Sun L., Cui D., Zhao Y., Tian J., Ni L., Liu J. MiR-222 modulates multidrug resistance in human colorectal carcinoma by down-regulating ADAM-17. Exp. Cell Res. 2012;318:2168–2177. doi: 10.1016/j.yexcr.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 225.Gao H., Cong X., Zhou J., Guan M. MicroRNA-222 influences migration and invasion through MIA3 in colorectal cancer. Cancer Cell Int. 2017;17:78. doi: 10.1186/s12935-017-0447-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Li S., Yan G., Yue M., Wang L. Extracellular vesicles-derived microRNA-222 promotes immune escape via interacting with ATF3 to regulate AKT1 transcription in colorectal cancer. BMC Cancer. 2021;21:349. doi: 10.1186/s12885-021-08063-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang X.-J., Zhang D., Yang Y.-T., Li X.-Y., Li H.-N., Zhang X.-P., Long J.-Y., Lu Y.-Q., Liu L., Yang G., Liu J., Hong J., Wu H.-G., Ma X.-P. Suppression of microRNA-222-3p ameliorates ulcerative colitis and colitis-associated colorectal cancer to protect against oxidative stress via targeting BRG1 to activate Nrf2/HO-1 signaling pathway. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1089809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Luo F., Zhou J., Wang S., Sun Z., Han Q., Bai C. microRNA-222 promotes colorectal cancer cell migration and invasion by targeting MST3. FEBS Open Bio. 2019;9:901–913. doi: 10.1002/2211-5463.12623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Liao D., Li T., Ye C., Zeng L., Li H., Pu X., Ding C., He Z., Huang G.-L. miR-221 inhibits autophagy and targets TP53INP1 in colorectal cancer cells. Exp. Ther. Med. 2018;15:1712–1717. doi: 10.3892/etm.2017.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sun K., Wang W., Zeng J., Wu C., Lei S., Li G. MicroRNA-221 inhibits CDKN1C/p57 expression in human colorectal carcinoma. Acta Pharmacol. Sin. 2011;32:375–384. doi: 10.1038/aps.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Dokhanchi M., Pakravan K., Zareian S., Hussen B.M., Farid M., Razmara E., Mossahebi-Mohammadi M., Cho W.C., Babashah S. Colorectal cancer cell-derived extracellular vesicles transfer miR-221-3p to promote endothelial cell angiogenesis via targeting suppressor of cytokine signaling 3. Life Sci. 2021;285 doi: 10.1016/j.lfs.2021.119937. [DOI] [PubMed] [Google Scholar]
- 232.Qin J., Luo M. MicroRNA-221 promotes colorectal cancer cell invasion and metastasis by targeting RECK. FEBS Lett. 2014;588:99–104. doi: 10.1016/j.febslet.2013.11.014. [DOI] [PubMed] [Google Scholar]
- 233.Lee Y., Kim S.J., Choo J., Heo G., Yoo J.-W., Jung Y., Rhee S.H., Im E. miR-23a-3p is a key regulator of IL-17C-induced tumor angiogenesis in colorectal cancer. Cells. 2020;9:1363. doi: 10.3390/cells9061363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Yong F.L., Wang C.W., Roslani A.C., Law C.W. The involvement of miR-23a/APAF1 regulation axis in colorectal cancer. Int. J. Mol. Sci. 2014;15:11713–11729. doi: 10.3390/ijms150711713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.You J., Li J., Ke C., Xiao Y., Lu C., Huang F., Mi Y., Xia R., Li Q. Oncogenic long intervening noncoding RNA Linc00284 promotes c-Met expression by sponging miR-27a in colorectal cancer. Oncogene. 2021;40:4151–4166. doi: 10.1038/s41388-021-01839-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Liu W., Zhang Z., Luo X., Qian K., Huang B., Deng J., Yang C. M6A promotes colorectal cancer progression via regulating the miR-27a-3p/BTG2 pathway. J. Oncol. 2023;2023 doi: 10.1155/2023/7097909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Chen S., Ji L., Wang Y., Zhang L., Xu M., Su Y., Zhang X. lncRNA RMST suppresses the progression of colorectal cancer by competitively binding to miR-27a-3p/RXRα axis and inactivating Wnt signaling pathway. Acta Biochim. Biophys. Sin. 2023;55:726–735. doi: 10.3724/abbs.2023065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Yin H., Gu S., Li G., Yu H., Zhang X., Zuo Y. Long noncoding RNA PVT1 predicts poor prognosis and promotes the progression of colorectal cancer through the miR-24-3p/NRP1 axis in zebrafish xenografts. Neoplasma. 2023;70:500–513. doi: 10.4149/neo_2023_221208N1169. [DOI] [PubMed] [Google Scholar]
- 239.Javanmard A.-R., Dokanehiifard S., Bohlooli M., Soltani B.M. LOC646329 long non-coding RNA sponges miR-29b-1 and regulates TGFβ signaling in colorectal cancer. J. Cancer Res. Clin. Oncol. 2020;146:1205–1215. doi: 10.1007/s00432-020-03145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zheng Z., Cui H., Wang Y., Yao W. Downregulation of RPS15A by miR-29a-3p attenuates cell proliferation in colorectal carcinoma. Biosci. Biotechnol. Biochem. 2019;83:2057–2064. doi: 10.1080/09168451.2019.1637712. [DOI] [PubMed] [Google Scholar]
- 241.Liu K., Yao H., Wen Y., Zhao H., Zhou N., Lei S., Xiong L. Functional role of a long non-coding RNA LIFR-AS1/miR-29a/TNFAIP3 axis in colorectal cancer resistance to pohotodynamic therapy. Biochim. Biophys. Acta Mol. Basis Dis. 2018;1864:2871–2880. doi: 10.1016/j.bbadis.2018.05.020. [DOI] [PubMed] [Google Scholar]
- 242.Tang W., Zhu Y., Gao J., Fu J., Liu C., Liu Y., Song C., Zhu S., Leng Y., Wang G., Chen W., Du P., Huang S., Zhou X., Kang J., Cui L. MicroRNA-29a promotes colorectal cancer metastasis by regulating matrix metalloproteinase 2 and E-cadherin via KLF4. Br. J. Cancer. 2014;110:450–458. doi: 10.1038/bjc.2013.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Xiong J., Zhang L., Tang R., Zhu Z. MicroRNA-301b-3p facilitates cell proliferation and migration in colorectal cancer by targeting HOXB1. Bioengineered. 2021;12:5839–5849. doi: 10.1080/21655979.2021.1962483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Song D., Zhang Q., Zhang H., Zhan L., Sun X. MiR-130b-3p promotes colorectal cancer progression by targeting CHD9. Cell Cycle. 2022;21:585–601. doi: 10.1080/15384101.2022.2029240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Yi R., Li Y., Wang F., Gu J., Isaji T., Li J., Qi R., Zhu X., Zhao Y. Transforming growth factor (TGF) β1 acted through miR-130b to increase integrin α5 to promote migration of colorectal cancer cells. Tumour Biol. 2016;37:10763–10773. doi: 10.1007/s13277-016-4965-6. [DOI] [PubMed] [Google Scholar]
- 246.Chen S., Wang Y., Li D., Wang H., Zhao X., Yang J., Chen L., Guo M., Zhao J., Chen C., Zhou Y., Liang G., Xu L. Mechanisms controlling MicroRNA expression in tumor. Cells. 2022;11:2852. doi: 10.3390/cells11182852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Suzuki H.I., Katsura A., Matsuyama H., Miyazono K. MicroRNA regulons in tumor microenvironment. Oncogene. 2015;34:3085–3094. doi: 10.1038/onc.2014.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Tito C., De Falco E., Rosa P., Iaiza A., Fazi F., Petrozza V., Calogero A. Circulating microRNAs from the molecular mechanisms to clinical biomarkers: a focus on the clear cell renal cell carcinoma. Genes. 2021;12:1154. doi: 10.3390/genes12081154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Müller M.F., Ibrahim A.E.K., Arends M.J. Molecular pathological classification of colorectal cancer. Virchows Arch. 2016;469:125–134. doi: 10.1007/s00428-016-1956-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Guo Y., Bao Y., Yang W. Regulatory miRNAs in colorectal carcinogenesis and metastasis. Int. J. Mol. Sci. 2017;18:890. doi: 10.3390/ijms18040890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Vaghari-Tabari M., Targhazeh N., Moein S., Qujeq D., Alemi F., Majidina M., Younesi S., Asemi Z., Yousefi B. From inflammatory bowel disease to colorectal cancer: what's the role of miRNAs? Cancer Cell Int. 2022;22:146. doi: 10.1186/s12935-022-02557-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Bopanna S., Ananthakrishnan A.N., Kedia S., Yajnik V., Ahuja V. Risk of colorectal cancer in Asian patients with ulcerative colitis: a systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2017;2:269–276. doi: 10.1016/S2468-1253(17)30004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Canavan C., Abrams K.R., Mayberry J. Meta-analysis: colorectal and small bowel cancer risk in patients with Crohn's disease. Aliment. Pharmacol. Ther. 2006;23:1097–1104. doi: 10.1111/j.1365-2036.2006.02854.x. [DOI] [PubMed] [Google Scholar]
- 254.Kent O.A., McCall M.N., Cornish T.C., Halushka M.K. Lessons from miR-143/145: the importance of cell-type localization of miRNAs. Nucleic Acids Res. 2014;42:7528–7538. doi: 10.1093/nar/gku461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Xu N., Papagiannakopoulos T., Pan G., Thomson J.A., Kosik K.S. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137:647–658. doi: 10.1016/j.cell.2009.02.038. [DOI] [PubMed] [Google Scholar]
- 256.Le Rolle A.-F., Chiu T.K., Zeng Z., Shia J., Weiser M.R., Paty P.B., Chiu V.K. Oncogenic KRAS activates an embryonic stem cell-like program in human colon cancer initiation. Oncotarget. 2016;7:2159–2174. doi: 10.18632/oncotarget.6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Kamatani A., Nakagawa Y., Akao Y., Maruyama N., Nagasaka M., Shibata T., Tahara T., Hirata I. Downregulation of anti-oncomirs miR-143/145 cluster occurs before APC gene aberration in the development of colorectal tumors. Med. Mol. Morphol. 2013;46:166–171. doi: 10.1007/s00795-013-0020-5. [DOI] [PubMed] [Google Scholar]
- 258.Kent O.A., Fox-Talbot K., Halushka M.K. RREB1 repressed miR-143/145 modulates KRAS signaling through downregulation of multiple targets. Oncogene. 2013;32:2576–2585. doi: 10.1038/onc.2012.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Ibrahim A.F., Weirauch U., Thomas M., Grünweller A., Hartmann R.K., Aigner A. MicroRNA replacement therapy for miR-145 and miR-33a is efficacious in a model of colon carcinoma. Cancer Res. 2011;71:5214–5224. doi: 10.1158/0008-5472.CAN-10-4645. [DOI] [PubMed] [Google Scholar]
- 260.Qin J., Wang F., Jiang H., Xu J., Jiang Y., Wang Z. MicroRNA-145 suppresses cell migration and invasion by targeting paxillin in human colorectal cancer cells. Int. J. Clin. Exp. Pathol. 2015;8:1328–1340. [PMC free article] [PubMed] [Google Scholar]
- 261.Nishida K., Kuwano Y., Rokutan K. The MicroRNA-23b/27b/24 cluster facilitates colon cancer cell migration by targeting FOXP2. Cancers (Basel) 2020;12:174. doi: 10.3390/cancers12010174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Matsuyama R., Okuzaki D., Okada M., Oneyama C. MicroRNA-27b suppresses tumor progression by regulating ARFGEF1 and focal adhesion signaling. Cancer Sci. 2016;107:28–35. doi: 10.1111/cas.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Hao L., Yu H. MiR-23b inhibits cell migration and invasion through targeting PDE7A in colon cancer cells. Int. J. Clin. Exp. Pathol. 2017;10:9436–9443. [PMC free article] [PubMed] [Google Scholar]
- 264.Colangelo T., Polcaro G., Ziccardi P., Pucci B., Muccillo L., Galgani M., Fucci A., Milone M.R., Budillon A., Santopaolo M., Votino C., Pancione M., Piepoli A., Mazzoccoli G., Binaschi M., Bigioni M., Maggi C.A., Fassan M., Laudanna C., Matarese G., Sabatino L., Colantuoni V. Proteomic screening identifies calreticulin as a miR-27a direct target repressing MHC class I cell surface exposure in colorectal cancer. Cell Death Dis. 2016;7 doi: 10.1038/cddis.2016.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Peng H., Luo J., Hao H., Hu J., Xie S.-K., Ren D., Rao B. MicroRNA-100 regulates SW620 colorectal cancer cell proliferation and invasion by targeting RAP1B. Oncol. Rep. 2014;31:2055–2062. doi: 10.3892/or.2014.3075. [DOI] [PubMed] [Google Scholar]
- 266.Fiala O., Sorejs O., Hosek P., Liska V., Vycital O., Bruha J., Kucera R., Topolcan O., Finek J., Maceckova D., Pitule P. Association of miR-125b, miR-17 and let-7c dysregulations with response to anti-epidermal growth factor receptor monoclonal antibodies in patients with metastatic colorectal cancer. Cancer Genomics Proteomics. 2020;17:605–613. doi: 10.21873/cgp.20217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Wang T., Wang G., Hao D., Liu X., Wang D., Ning N., Li X. Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer. Mol. Cancer. 2015;14:125. doi: 10.1186/s12943-015-0402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.King C.E., Wang L., Winograd R., Madison B.B., Mongroo P.S., Johnstone C.N., Rustgi A.K. LIN28B fosters colon cancer migration, invasion and transformation through let-7-dependent and -independent mechanisms. Oncogene. 2011;30:4185–4193. doi: 10.1038/onc.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Chang T.-C., Wentzel E.A., Kent O.A., Ramachandran K., Mullendore M., Lee K.H., Feldmann G., Yamakuchi M., Ferlito M., Lowenstein C.J., Arking D.E., Beer M.A., Maitra A., Mendell J.T. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.He L., He X., Lim L.P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., Jackson A.L., Linsley P.S., Chen C., Lowe S.W., Cleary M.A., Hannon G.J. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Okada N., Lin C.-P., Ribeiro M.C., Biton A., Lai G., He X., Bu P., Vogel H., Jablons D.M., Keller A.C., Wilkinson J.E., He B., Speed T.P., He L. A positive feedback between p53 and miR-34 miRNAs mediates tumor suppression. Genes Dev. 2014;28:438–450. doi: 10.1101/gad.233585.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Gao J., Li N., Dong Y., Li S., Xu L., Li X., Li Y., Li Z., Ng S.S., Sung J.J., Shen L., Yu J. miR-34a-5p suppresses colorectal cancer metastasis and predicts recurrence in patients with stage II/III colorectal cancer. Oncogene. 2015;34:4142–4152. doi: 10.1038/onc.2014.348. [DOI] [PubMed] [Google Scholar]
- 273.Kim N.H., Kim H.S., Kim N.-G., Lee I., Choi H.-S., Li X.-Y., Kang S.E., Cha S.Y., Ryu J.K., Na J.M., Park C., Kim K., Lee S., Gumbiner B.M., Yook J.I., Weiss S.J. p53 and microRNA-34 are suppressors of canonical Wnt signaling. Sci. Signal. 2011;4 doi: 10.1126/scisignal.2001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Roy S., Levi E., Majumdar A.P.N., Sarkar F.H. Expression of miR-34 is lost in colon cancer which can be re-expressed by a novel agent CDF. J. Hematol. Oncol. 2012;5:58. doi: 10.1186/1756-8722-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Kress T.R., Cannell I.G., Brenkman A.B., Samans B., Gaestel M., Roepman P., Burgering B.M., Bushell M., Rosenwald A., Eilers M. The MK5/PRAK kinase and Myc form a negative feedback loop that is disrupted during colorectal tumorigenesis. Mol. Cell. 2011;41:445–457. doi: 10.1016/j.molcel.2011.01.023. [DOI] [PubMed] [Google Scholar]
- 276.Strubberg A.M., Madison B.B. MicroRNAs in the etiology of colorectal cancer: pathways and clinical implications. Dis. Model. Mech. 2017;10:197–214. doi: 10.1242/dmm.027441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Bu P., Chen K.-Y., Chen J.H., Wang L., Walters J., Shin Y.J., Goerger J.P., Sun J., Witherspoon M., Rakhilin N., Li J., Yang H., Milsom J., Lee S., Zipfel W., Jin M.M., Gümüş Z.H., Lipkin S.M., Shen X. A microRNA miR-34a-regulated bimodal switch targets Notch in colon cancer stem cells. Cell Stem Cell. 2013;12:602–615. doi: 10.1016/j.stem.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Bu P., Wang L., Chen K.-Y., Srinivasan T., Murthy P.K.L., Tung K.-L., Varanko A.K., Chen H.J., Ai Y., King S., Lipkin S.M., Shen X. A miR-34a-numb feedforward loop triggered by inflammation regulates asymmetric stem cell division in intestine and colon cancer. Cell Stem Cell. 2016;18:189–202. doi: 10.1016/j.stem.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Hwang W.-L., Jiang J.-K., Yang S.-H., Huang T.-S., Lan H.-Y., Teng H.-W., Yang C.-Y., Tsai Y.-P., Lin C.-H., Wang H.-W., Yang M.-H. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat. Cell Biol. 2014;16:268–280. doi: 10.1038/ncb2910. [DOI] [PubMed] [Google Scholar]
- 280.Knudsen K.N., Nielsen B.S., Lindebjerg J., Hansen T.F., Holst R., Sørensen F.B. microRNA-17 is the most up-regulated member of the miR-17-92 cluster during early colon cancer evolution. PLoS One. 2015;10 doi: 10.1371/journal.pone.0140503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Hayashita Y., Osada H., Tatematsu Y., Yamada H., Yanagisawa K., Tomida S., Yatabe Y., Kawahara K., Sekido Y., Takahashi T. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 282.Yu G., Tang J.-Q., Tian M.-L., Li H., Wang X., Wu T., Zhu J., Huang S.-J., Wan Y.-L. Prognostic values of the miR-17-92 cluster and its paralogs in colon cancer. J. Surg. Oncol. 2012;106:232–237. doi: 10.1002/jso.22138. [DOI] [PubMed] [Google Scholar]
- 283.Diosdado B., van de Wiel M.A., Terhaar Sive Droste J.S., Mongera S., Postma C., Meijerink W.J.H.J., Carvalho B., Meijer G.A. MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br. J. Cancer. 2009;101:707–714. doi: 10.1038/sj.bjc.6605037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Lanza G., Ferracin M., Gafà R., Veronese A., Spizzo R., Pichiorri F., Liu C., Calin G.A., Croce C.M., Negrini M. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol. Cancer. 2007;6:54. doi: 10.1186/1476-4598-6-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Li Y., Lauriola M., Kim D., Francesconi M., D'Uva G., Shibata D., Malafa M.P., Yeatman T.J., Coppola D., Solmi R., Cheng J.Q. Adenomatous polyposis coli (APC) regulates miR17-92 cluster through β-catenin pathway in colorectal cancer. Oncogene. 2016;35:4558–4568. doi: 10.1038/onc.2015.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Bai X., Hua S., Zhang J., Xu S. The MicroRNA family both in normal development and in different diseases: the miR-17-92 cluster. BioMed Res. Int. 2019;2019 doi: 10.1155/2019/9450240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Humphreys K.J., McKinnon R.A., Michael M.Z. miR-18a inhibits CDC42 and plays a tumour suppressor role in colorectal cancer cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0112288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Humphreys K.J., Cobiac L., Le Leu R.K., Van der Hoek M.B., Michael M.Z. Histone deacetylase inhibition in colorectal cancer cells reveals competing roles for members of the oncogenic miR-17-92 cluster. Mol. Carcinog. 2013;52:459–474. doi: 10.1002/mc.21879. [DOI] [PubMed] [Google Scholar]
- 289.Guil S., Cáceres J.F. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 290.Chakraborty S., Mehtab S., Patwardhan A., Krishnan Y. Pri-miR-17-92a transcript folds into a tertiary structure and autoregulates its processing. RNA. 2012;18:1014–1028. doi: 10.1261/rna.031039.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Wei Q.-D., Zheng W.-B., Sun K., Xue Q., Yang C.-Z., Li G.-X. MiR-92a promotes the invasion and migration of colorectal cancer by targeting RECK. Int. J. Clin. Exp. Pathol. 2019;12:1565–1577. [PMC free article] [PubMed] [Google Scholar]
- 292.Yamada N., Nakagawa Y., Tsujimura N., Kumazaki M., Noguchi S., Mori T., Hirata I., Maruo K., Akao Y. Role of intracellular and extracellular MicroRNA-92a in colorectal cancer. Transl. Oncol. 2013;6:482–492. doi: 10.1593/tlo.13280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Zhang G.-J., Li L.-F., Yang G.-D., Xia S.-S., Wang R., Leng Z.-W., Liu Z.-L., Tian H.-P., He Y., Meng C.-Y., Liu D.-Z., Hou S.-L., Tang X.-G., Zhou T. MiR-92a promotes stem cell-like properties by activating Wnt/β-catenin signaling in colorectal cancer. Oncotarget. 2017;8:101760–101770. doi: 10.18632/oncotarget.21667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Tsuchida A., Ohno S., Wu W., Borjigin N., Fujita K., Aoki T., Ueda S., Takanashi M., Kuroda M. miR-92 is a key oncogenic component of the miR-17-92 cluster in colon cancer. Cancer Sci. 2011;102:2264–2271. doi: 10.1111/j.1349-7006.2011.02081.x. [DOI] [PubMed] [Google Scholar]
- 295.Song L., Lin C., Wu Z., Gong H., Zeng Y., Wu J., Li M., Li J. miR-18a impairs DNA damage response through downregulation of ataxia telangiectasia mutated (ATM) kinase. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Wu C.-W., Dong Y.-J., Liang Q.-Y., He X.-Q., Ng S.S.M., Chan F.K.L., Sung J.J.Y., Yu J. MicroRNA-18a attenuates DNA damage repair through suppressing the expression of ataxia telangiectasia mutated in colorectal cancer. PLoS One. 2013;8 doi: 10.1371/journal.pone.0057036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Sokolova V., Fiorino A., Zoni E., Crippa E., Reid J.F., Gariboldi M., Pierotti M.A. The effects of miR-20a on p21: two mechanisms blocking growth arrest in TGF-β-responsive colon carcinoma. J. Cell. Physiol. 2015;230:3105–3114. doi: 10.1002/jcp.25051. [DOI] [PubMed] [Google Scholar]
- 298.Zheng L., Zhang Y., Lin S., Sun A., Chen R., Ding Y., Ding Y. Down-regualtion of miR-106b induces epithelial-mesenchymal transition but suppresses metastatic colonization by targeting Prrx1 in colorectal cancer. Int. J. Clin. Exp. Pathol. 2015;8:10534–10544. [PMC free article] [PubMed] [Google Scholar]
- 299.Nagel R., le Sage C., Diosdado B., van der Waal M., Oude Vrielink J.A.F., Bolijn A., Meijer G.A., Agami R. Regulation of the adenomatous polyposis coli gene by the miR-135 family in colorectal cancer. Cancer Res. 2008;68:5795–5802. doi: 10.1158/0008-5472.CAN-08-0951. [DOI] [PubMed] [Google Scholar]
- 300.Jacobsen A., Silber J., Harinath G., Huse J.T., Schultz N., Sander C. Analysis of microRNA-target interactions across diverse cancer types. Nat. Struct. Mol. Biol. 2013;20:1325–1332. doi: 10.1038/nsmb.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Li L., Wang A., Cai M., Tong M., Chen F., Huang L. Identification of stool miR-135b-5p as a non-invasive diaognostic biomarker in later tumor stage of colorectal cancer. Life Sci. 2020;260 doi: 10.1016/j.lfs.2020.118417. [DOI] [PubMed] [Google Scholar]
- 302.Valeri N., Braconi C., Gasparini P., Murgia C., Lampis A., Paulus-Hock V., Hart J.R., Ueno L., Grivennikov S.I., Lovat F., Paone A., Cascione L., Sumani K.M., Veronese A., Fabbri M., Carasi S., Alder H., Lanza G., Gafa R., Moyer M.P., Ridgway R.A., Cordero J., Nuovo G.J., Frankel W.L., Rugge M., Fassan M., Groden J., Vogt P.K., Karin M., Sansom O.J., Croce C.M. MicroRNA-135b promotes cancer progression by acting as a downstream effector of oncogenic pathways in colon cancer. Cancer Cell. 2014;25:469–483. doi: 10.1016/j.ccr.2014.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Cao J., Yan X.-R., Liu T., Han X.-B., Yu J.-J., Liu S.-H., Wang L.-B. MicroRNA-552 promotes tumor cell proliferation and migration by directly targeting DACH1 via the Wnt/β-catenin signaling pathway in colorectal cancer. Oncol. Lett. 2017;14:3795–3802. doi: 10.3892/ol.2017.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Kwak B., Kim D.U., Kim T.O., Kim H.-S., Kim S.-W. MicroRNA-552 links Wnt signaling to p53 tumor suppressor in colorectal cancer. Int. J. Oncol. 2018;53:1800–1808. doi: 10.3892/ijo.2018.4505. [DOI] [PubMed] [Google Scholar]
- 305.Feng Z.-Y., Xu X.-H., Cen D.-Z., Luo C.-Y., Wu S.-B. miR-590-3p promotes colon cancer cell proliferation via Wnt/β-catenin signaling pathway by inhibiting WIF1 and DKK1. Eur. Rev. Med. Pharmacol. Sci. 2017;21:4844–4852. [PubMed] [Google Scholar]
- 306.Liu D., Zhang H., Cui M., Chen C., Feng Y. Hsa-miR-425-5p promotes tumor growth and metastasis by activating the CTNND1-mediated β-catenin pathway and EMT in colorectal cancer. Cell Cycle. 2020;19:1917–1927. doi: 10.1080/15384101.2020.1783058. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 307.Subramanian M., Rao S.R., Thacker P., Chatterjee S., Karunagaran D. MiR-29b downregulates canonical Wnt signaling by suppressing coactivators of β-catenin in human colorectal cancer cells. J. Cell. Biochem. 2014;115:1974–1984. doi: 10.1002/jcb.24869. [DOI] [PubMed] [Google Scholar]
- 308.Zhang J.X., Mai S.J., Huang X.X., Wang F.W., Liao Y.J., Lin M.C., Kung H.F., Zeng Y.X., Xie D. MiR-29c mediates epithelial-to-mesenchymal transition in human colorectal carcinoma metastasis via PTP4A and GNA13 regulation of β-catenin signaling. Ann. Oncol. 2014;25:2196–2204. doi: 10.1093/annonc/mdu439. [DOI] [PubMed] [Google Scholar]
- 309.Tang Q., Zou Z., Zou C., Zhang Q., Huang R., Guan X., Li Q., Han Z., Wang D., Wei H., Gao X., Wang X. MicroRNA-93 suppress colorectal cancer development via Wnt/β-catenin pathway downregulating. Tumour Biol. 2015;36:1701–1710. doi: 10.1007/s13277-014-2771-6. [DOI] [PubMed] [Google Scholar]
- 310.Han X., Zheng J., Wang Y., Gao Z. miRNA-29a inhibits colon cancer growth by regulation of the PTEN/Akt/GSK3β and Wnt/β-catenin signaling pathways. Oncol. Lett. 2018;16:2638–2644. doi: 10.3892/ol.2018.8905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Sun L.-H., Tian D., Yang Z.-C., Li J.-L. Exosomal miR-21 promotes proliferation, invasion and therapy resistance of colon adenocarcinoma cells through its target PDCD4. Sci. Rep. 2020;10:8271. doi: 10.1038/s41598-020-65207-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Shi C., Yang Y., Xia Y., Okugawa Y., Yang J., Liang Y., Chen H., Zhang P., Wang F., Han H., Wu W., Gao R., Gasche C., Qin H., Ma Y., Goel A. Novel evidence for an oncogenic role of microRNA-21 in colitis-associated colorectal cancer. Gut. 2016;65:1470–1481. doi: 10.1136/gutjnl-2014-308455. [DOI] [PubMed] [Google Scholar]
- 313.Yamamichi N., Shimomura R., Inada K., Sakurai K., Haraguchi T., Ozaki Y., Fujita S., Mizutani T., Furukawa C., Fujishiro M., Ichinose M., Shiogama K., Tsutsumi Y., Omata M., Iba H. Locked nucleic acid in situ hybridization analysis of miR-21 expression during colorectal cancer development. Clin. Cancer Res. 2009;15:4009–4016. doi: 10.1158/1078-0432.CCR-08-3257. [DOI] [PubMed] [Google Scholar]
- 314.Lin P.-L., Wu D.-W., Huang C.-C., He T.-Y., Chou M.-C., Sheu G.-T., Lee H. MicroRNA-21 promotes tumour malignancy via increased nuclear translocation of β-catenin and predicts poor outcome in APC-mutated but not in APC-wild-type colorectal cancer. Carcinogenesis. 2014;35:2175–2182. doi: 10.1093/carcin/bgu110. [DOI] [PubMed] [Google Scholar]
- 315.Lan F., Yue X., Han L., Shi Z., Yang Y., Pu P., Yao Z., Kang C. Genome-wide identification of TCF7L2/TCF4 target miRNAs reveals a role for miR-21 in Wnt-driven epithelial cancer. Int. J. Oncol. 2012;40:519–526. doi: 10.3892/ijo.2011.1215. [DOI] [PubMed] [Google Scholar]
- 316.Liang W.-C., Fu W.-M., Wong C.-W., Wang Y., Wang W.-M., Hu G.-X., Zhang L., Xiao L.-J., Wan D.C.-C., Zhang J.-F., Waye M.M.-Y. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6:22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Bian Z., Zhang J., Li M., Feng Y., Yao S., Song M., Qi X., Fei B., Yin Y., Hua D., Huang Z. Long non-coding RNA LINC00152 promotes cell proliferation, metastasis, and confers 5-FU resistance in colorectal cancer by inhibiting miR-139-5p. Oncogenesis. 2017;6:395. doi: 10.1038/s41389-017-0008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Luo Y., Chen J.-J., Lv Q., Qin J., Huang Y.-Z., Yu M.-H., Zhong M. Long non-coding RNA NEAT1 promotes colorectal cancer progression by competitively binding miR-34a with SIRT1 and enhancing the Wnt/β-catenin signaling pathway. Cancer Lett. 2019;440–441:11–22. doi: 10.1016/j.canlet.2018.10.002. [DOI] [PubMed] [Google Scholar]
- 319.Sun N., Zhang G., Liu Y. Long non-coding RNA XIST sponges miR-34a to promotes colon cancer progression via Wnt/β-catenin signaling pathway. Gene (Amst.) 2018;665:141–148. doi: 10.1016/j.gene.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 320.Lv S.-Y., Shan T.-D., Pan X.-T., Tian Z.-B., Liu X.-S., Liu F.-G., Sun X.-G., Xue H.-G., Li X.-H., Han Y., Sun L.-J., Chen L., Zhang L.-Y. The lncRNA ZEB1-AS1 sponges miR-181a-5p to promote colorectal cancer cell proliferation by regulating Wnt/β-catenin signaling. Cell Cycle. 2018;17:1245–1254. doi: 10.1080/15384101.2018.1471317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Han P., Li J.-W., Zhang B.-M., Lv J.-C., Li Y.-M., Gu X.-Y., Yu Z.-W., Jia Y.-H., Bai X.-F., Li L., Liu Y.-L., Cui B.-B. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol. Cancer. 2017;16:9. doi: 10.1186/s12943-017-0583-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Dong X., Yang Z., Yang H., Li D., Qiu X. Long non-coding RNA mir4435-2HG promotes colorectal cancer proliferation and metastasis through miR-206/YAP1 Axis. Front. Oncol. 2020;10:160. doi: 10.3389/fonc.2020.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Zhu X., Bu F., Tan T., Luo Q., Zhu J., Lin K., Huang J., Luo C., Zhu Z. Long noncoding RNA RP11-757G1.5 sponges miR-139-5p and upregulates YAP1 thereby promoting the proliferation and liver, spleen metastasis of colorectal cancer. J. Exp. Clin. Cancer Res. 2020;39:207. doi: 10.1186/s13046-020-01717-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Du Y.-L., Liang Y., Shi G.-Q., Cao Y., Qiu J., Yuan L., Yong Z., Liu L., Li J. LINC00689 participates in proliferation, chemoresistance and metastasis via miR-31-5p/YAP/β-catenin axis in colorectal cancer. Exp. Cell Res. 2020;395 doi: 10.1016/j.yexcr.2020.112176. [DOI] [PubMed] [Google Scholar]
- 325.Zhang N., Hu X., Du Y., Du J. The role of miRNAs in colorectal cancer progression and chemoradiotherapy. Biomed. Pharmacother. 2021;134 doi: 10.1016/j.biopha.2020.111099. [DOI] [PubMed] [Google Scholar]
- 326.Zhang G.-J., Xiao H.-X., Tian H.-P., Liu Z.-L., Xia S.-S., Zhou T. Upregulation of microRNA-155 promotes the migration and invasion of colorectal cancer cells through the regulation of claudin-1 expression. Int. J. Mol. Med. 2013;31:1375–1380. doi: 10.3892/ijmm.2013.1348. [DOI] [PubMed] [Google Scholar]
- 327.Wang D., Wang X., Song Y., Si M., Sun Y., Liu X., Cui S., Qu X., Yu X. Exosomal miR-146a-5p and miR-155-5p promote CXCL12/CXCR7-induced metastasis of colorectal cancer by crosstalk with cancer-associated fibroblasts. Cell Death Dis. 2022;13:380. doi: 10.1038/s41419-022-04825-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Lan J., Sun L., Xu F., Liu L., Hu F., Song D., Hou Z., Wu W., Luo X., Wang J., Yuan X., Hu J., Wang G. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. 2019;79:146–158. doi: 10.1158/0008-5472.CAN-18-0014. [DOI] [PubMed] [Google Scholar]
- 329.Prossomariti A., Piazzi G., D'Angelo L., Miccoli S., Turchetti D., Alquati C., Montagna C., Bazzoli F., Ricciardiello L. miR-155 is downregulated in familial adenomatous polyposis and modulates WNT signaling by targeting AXIN1 and TCF4. Mol. Cancer Res. 2018;16:1965–1976. doi: 10.1158/1541-7786.MCR-18-0115. [DOI] [PubMed] [Google Scholar]
- 330.Zhang X., Ai F., Li X., Tian L., Wang X., Shen S., Liu F. MicroRNA-34a suppresses colorectal cancer metastasis by regulating Notch signaling. Oncol. Lett. 2017;14:2325–2333. doi: 10.3892/ol.2017.6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Tazawa H., Tsuchiya N., Izumiya M., Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Hahn S., Jackstadt R., Siemens H., Hünten S., Hermeking H. SNAIL and miR-34a feed-forward regulation of ZNF281/ZBP99 promotes epithelial-mesenchymal transition. EMBO J. 2013;32:3079–3095. doi: 10.1038/emboj.2013.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Yamakuchi M., Ferlito M., Lowenstein C.J. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. U. S. A. 2008;105:13421–13426. doi: 10.1073/pnas.0801613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Rokavec M., Öner M.G., Li H., Jackstadt R., Jiang L., Lodygin D., Kaller M., Horst D., Ziegler P.K., Schwitalla S., Slotta-Huspenina J., Bader F.G., Greten F.R., Hermeking H. IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated colorectal cancer invasion and metastasis. J. Clin. Investig. 2014;124:1853–1867. doi: 10.1172/JCI73531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Thuringer D., Jego G., Berthenet K., Hammann A., Solary E., Garrido C. Gap junction-mediated transfer of miR-145-5p from microvascular endothelial cells to colon cancer cells inhibits angiogenesis. Oncotarget. 2016;7:28160–28168. doi: 10.18632/oncotarget.8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Kent O.A., Chivukula R.R., Mullendore M., Wentzel E.A., Feldmann G., Lee K.H., Liu S., Leach S.D., Maitra A., Mendell J.T. Repression of the miR-143/145 cluster by oncogenic Ras initiates a tumor-promoting feed-forward pathway. Genes Dev. 2010;24:2754–2759. doi: 10.1101/gad.1950610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Xie H., Ren X., Xin S., Lan X., Lu G., Lin Y., Yang S., Zeng Z., Liao W., Ding Y.-Q., Liang L. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Zhang G.-J., Li Y., Zhou H., Xiao H.-X., Zhou T. miR-20a is an independent prognostic factor in colorectal cancer and is involved in cell metastasis. Mol. Med. Rep. 2014;10:283–291. doi: 10.3892/mmr.2014.2144. [DOI] [PubMed] [Google Scholar]
- 339.Zhang G., Zhou H., Xiao H., Liu Z., Tian H., Zhou T. MicroRNA-92a functions as an oncogene in colorectal cancer by targeting PTEN. Dig. Dis. Sci. 2014;59:98–107. doi: 10.1007/s10620-013-2858-8. [DOI] [PubMed] [Google Scholar]
- 340.Jia X., Wang X., Guo X., Ji J., Lou G., Zhao J., Zhou W., Guo M., Zhang M., Li C., Tai S., Yu S. MicroRNA-124: an emerging therapeutic target in cancer. Cancer Med. 2019;8:5638–5650. doi: 10.1002/cam4.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Shahmohamadnejad S., Nouri Ghonbalani Z., Tahbazlahafi B., Panahi G., Meshkani R., Emami Razavi A., Shokri Afra H., Khalili E. Aberrant methylation of miR-124 upregulates DNMT3B in colorectal cancer to accelerate invasion and migration. Arch. Physiol. Biochem. 2022;128:1503–1509. doi: 10.1080/13813455.2020.1779311. [DOI] [PubMed] [Google Scholar]
- 342.Zhang J., Lu Y., Yue X., Li H., Luo X., Wang Y., Wang K., Wan J. MiR-124 suppresses growth of human colorectal cancer by inhibiting STAT3. PLoS One. 2013;8 doi: 10.1371/journal.pone.0070300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Roshani Asl E., Rasmi Y., Baradaran B. MicroRNA-124-3p suppresses PD-L1 expression and inhibits tumorigenesis of colorectal cancer cells via modulating STAT3 signaling. J. Cell. Physiol. 2021;236:7071–7087. doi: 10.1002/jcp.30378. [DOI] [PubMed] [Google Scholar]
- 344.Taniguchi K., Sugito N., Kumazaki M., Shinohara H., Yamada N., Nakagawa Y., Ito Y., Otsuki Y., Uno B., Uchiyama K., Akao Y. MicroRNA-124 inhibits cancer cell growth through PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer Lett. 2015;363:17–27. doi: 10.1016/j.canlet.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 345.Zaravinos A. The regulatory role of MicroRNAs in EMT and cancer. J. Oncol. 2015;2015 doi: 10.1155/2015/865816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Davalos V., Moutinho C., Villanueva A., Boque R., Silva P., Carneiro F., Esteller M. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene. 2012;31:2062–2074. doi: 10.1038/onc.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Park S.-M., Gaur A.B., Lengyel E., Peter M.E. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Chen L., Gibbons D.L., Goswami S., Cortez M.A., Ahn Y.-H., Byers L.A., Zhang X., Yi X., Dwyer D., Lin W., Diao L., Wang J., Roybal J., Patel M., Ungewiss C., Peng D., Antonia S., Mediavilla-Varela M., Robertson G., Suraokar M., Welsh J.W., Erez B., Wistuba I.I., Chen L., Peng D., Wang S., Ullrich S.E., Heymach J.V., Kurie J.M., Qin F.X.-F. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Hur K., Toiyama Y., Takahashi M., Balaguer F., Nagasaka T., Koike J., Hemmi H., Koi M., Boland C.R., Goel A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut. 2013;62:1315–1326. doi: 10.1136/gutjnl-2011-301846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Toiyama Y., Hur K., Tanaka K., Inoue Y., Kusunoki M., Boland C.R., Goel A. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann. Surg. 2014;259:735–743. doi: 10.1097/SLA.0b013e3182a6909d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Ren J., Ding L., Zhang D., Shi G., Xu Q., Shen S., Wang Y., Wang T., Hou Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8:3932–3948. doi: 10.7150/thno.25541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Yang X., Xu X., Zhu J., Zhang S., Wu Y., Wu Y., Zhao K., Xing C., Cao J., Zhu H., Li M., Ye Z., Peng W. miR-31 affects colorectal cancer cells by inhibiting autophagy in cancer-associated fibroblasts. Oncotarget. 2016;7:79617–79628. doi: 10.18632/oncotarget.12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Ast V., Kordaß T., Oswald M., Kolte A., Eisel D., Osen W., Eichmüller S.B., Berndt A., König R. MiR-192, miR-200c and miR-17 are fibroblast-mediated inhibitors of colorectal cancer invasion. Oncotarget. 2018;9:35559–35580. doi: 10.18632/oncotarget.26263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Hanusova V., Matouskova P., Manethova M., Soukup J., John S., Zofka M., Vošmikova H., Krbal L., Rudolf E. Comparative analysis of miRNA and EMT markers in metastatic colorectal cancer. Cancer Invest. 2023;41:837–847. doi: 10.1080/07357907.2023.2283495. [DOI] [PubMed] [Google Scholar]
- 355.Feng B., Dong T.T., Wang L.L., Zhou H.M., Zhao H.C., Dong F., Zheng M.H. Colorectal cancer migration and invasion initiated by microRNA-106a. PLoS One. 2012;7 doi: 10.1371/journal.pone.0043452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Zheng D., Cao M., Zuo S., Xia X., Zhi C., Lin Y., Deng S., Yuan X. Correction: RANBP1 promotes colorectal cancer progression by regulating pre-miRNA nuclear export via a positive feedback loop with YAP. Oncogene. 2022;41:1070. doi: 10.1038/s41388-021-02152-2. [DOI] [PubMed] [Google Scholar]
- 357.Sun W., Wang X., Li J., You C., Lu P., Feng H., Kong Y., Zhang H., Liu Y., Jiao R., Chen X., Ba Y. MicroRNA-181a promotes angiogenesis in colorectal cancer by targeting SRCIN1 to promote the SRC/VEGF signaling pathway. Cell Death Dis. 2018;9:438. doi: 10.1038/s41419-018-0490-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Ji D., Chen Z., Li M., Zhan T., Yao Y., Zhang Z., Xi J., Yan L., Gu J. MicroRNA-181a promotes tumor growth and liver metastasis in colorectal cancer by targeting the tumor suppressor WIF-1. Mol. Cancer. 2014;13:86. doi: 10.1186/1476-4598-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Ozsolak F., Poling L.L., Wang Z., Liu H., Liu X.S., Roeder R.G., Zhang X., Song J.S., Fisher D.E. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Dai L., Wang W., Zhang S., Jiang Q., Wang R., Dai L., Cheng L., Yang Y., Wei Y.-Q., Deng H.-X. Vector-based miR-15a/16-1 plasmid inhibits colon cancer growth in vivo. Cell Biol. Int. 2012;36:765–770. doi: 10.1042/CBI20110404. [DOI] [PubMed] [Google Scholar]
- 361.Jackstadt R., Röh S., Neumann J., Jung P., Hoffmann R., Horst D., Berens C., Bornkamm G.W., Kirchner T., Menssen A., Hermeking H. AP4 is a mediator of epithelial-mesenchymal transition and metastasis in colorectal cancer. J. Exp. Med. 2013;210:1331–1350. doi: 10.1084/jem.20120812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Ren X.L., He G.Y., Li X.M., Men H., Yi L.Z., Lu G.F., Xin S.N., Wu P.X., Li Y.L., Liao W.T., Ding Y.Q., Liang L. MicroRNA-206 functions as a tumor suppressor in colorectal cancer by targeting FMNL2. J. Cancer Res. Clin. Oncol. 2016;142:581–592. doi: 10.1007/s00432-015-2053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Park Y.R., Seo S.Y., Kim S.L., Zhu S.M., Chun S., Oh J.-M., Lee M.R., Kim S.H., Kim I.H., Lee S.O., Lee S.T., Kim S.W. MiRNA-206 suppresses PGE2-induced colorectal cancer cell proliferation, migration, and invasion by targetting TM4SF1. Biosci. Rep. 2018;38 doi: 10.1042/BSR20180664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.McCarthy J.J. MicroRNA-206: the skeletal muscle-specific myomiR. Biochim. Biophys. Acta. 2008;1779:682–691. doi: 10.1016/j.bbagrm.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Yu X., Wang D., Wang X., Sun S., Zhang Y., Wang S., Miao R., Xu X., Qu X. CXCL12/CXCR4 promotes inflammation-driven colorectal cancer progression through activation of RhoA signaling by sponging miR-133a-3p. J. Exp. Clin. Cancer Res. 2019;38:32. doi: 10.1186/s13046-018-1014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Sun J.-Y., Huang Y., Li J.-P., Zhang X., Wang L., Meng Y.-L., Yan B., Bian Y.-Q., Zhao J., Wang W.-Z., Yang A.-G., Zhang R. MicroRNA-320a suppresses human colon cancer cell proliferation by directly targeting β-catenin. Biochem. Biophys. Res. Commun. 2012;420:787–792. doi: 10.1016/j.bbrc.2012.03.075. [DOI] [PubMed] [Google Scholar]
- 367.Vishnubalaji R., Hamam R., Yue S., Al-Obeed O., Kassem M., Liu F.-F., Aldahmash A., Alajez N.M. MicroRNA-320 suppresses colorectal cancer by targeting SOX4, FOXM1, and FOXQ1. Oncotarget. 2016;7:35789–35802. doi: 10.18632/oncotarget.8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Ma Y., Qin C., Li L., Miao R., Jing C., Cui X. MicroRNA-21 promotes cell proliferation by targeting tumor suppressor TET1 in colorectal cancer. Int. J. Clin. Exp. Pathol. 2018;11:1439–1445. [PMC free article] [PubMed] [Google Scholar]
- 369.Asangani I.A., Rasheed S.a.K., Nikolova D.A., Leupold J.H., Colburn N.H., Post S., Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 370.Ma T., Jiang J., Shi M., Xu H. Exosomal miRNA-166-5p derived from G-MDSCs promotes proliferation by targeting ITM3E in colorectal cancer. Environ. Toxicol. 2024;39:803–814. doi: 10.1002/tox.23980. [DOI] [PubMed] [Google Scholar]
- 371.Hur K., Toiyama Y., Okugawa Y., Ide S., Imaoka H., Boland C.R., Goel A. Circulating microRNA-203 predicts prognosis and metastasis in human colorectal cancer. Gut. 2017;66:654–665. doi: 10.1136/gutjnl-2014-308737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Grisard E., Coan M., Cesaratto L., Rigo I., Zandonà L., Paulitti A., Andreuzzi E., Rampioni Vinciguerra G.L., Poletto E., Del Ben F., Brisotto G., Biscontin E., Turetta M., Dassi E., Mirnezami A., Canzonieri V., Vecchione A., Baldassarre G., Mongiat M., Spizzo R., Nicoloso M.S. Sleeping beauty genetic screen identifies miR-23b::BTBD7 gene interaction as crucial for colorectal cancer metastasis. EBioMedicine. 2019;46:79–93. doi: 10.1016/j.ebiom.2019.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Deng Y.H., Deng Z.H., Hao H., Wu X.L., Gao H., Tang S.H., Tang H. MicroRNA-23a promotes colorectal cancer cell survival by targeting PDK4. Exp. Cell Res. 2018;373:171–179. doi: 10.1016/j.yexcr.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 374.Li X., Li X., Liao D., Wang X., Wu Z., Nie J., Bai M., Fu X., Mei Q., Han W. Elevated microRNA-23a expression enhances the chemoresistance of colorectal cancer cells with microsatellite instability to 5-fluorouracil by directly targeting ABCF1. Curr. Protein Pept. Sci. 2015;16:301–309. doi: 10.2174/138920371604150429153309. [DOI] [PubMed] [Google Scholar]
- 375.Liu S., Sun X., Wang M., Hou Y., Zhan Y., Jiang Y., Liu Z., Cao X., Chen P., Liu Z., Chen X., Tao Y., Xu C., Mao J., Cheng C., Li C., Hu Y., Wang L., Chin Y.E., Shi Y., Siebenlist U., Zhang X. A microRNA 221- and 222-mediated feedback loop maintains constitutive activation of NFκB and STAT3 in colorectal cancer cells. Gastroenterology. 2014;147:847–859.e11. doi: 10.1053/j.gastro.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Han H.-B., Gu J., Zuo H.-J., Chen Z.-G., Zhao W., Li M., Ji D.-B., Lu Y.-Y., Zhang Z.-Q. Let-7c functions as a metastasis suppressor by targeting MMP11 and PBX3 in colorectal cancer. J. Pathol. 2012;226:544–555. doi: 10.1002/path.3014. [DOI] [PubMed] [Google Scholar]
- 377.Ma K., Pan X., Fan P., He Y., Gu J., Wang W., Zhang T., Li Z., Luo X. Loss of miR-638 in vitro promotes cell invasion and a mesenchymal-like transition by influencing SOX2 expression in colorectal carcinoma cells. Mol. Cancer. 2014;13:118. doi: 10.1186/1476-4598-13-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Zhang F., Luo Y., Shao Z., Xu L., Liu X., Niu Y., Shi J., Sun X., Liu Y., Ding Y., Zhao L. MicroRNA-187, a downstream effector of TGFβ pathway, suppresses Smad-mediated epithelial-mesenchymal transition in colorectal cancer. Cancer Lett. 2016;373:203–213. doi: 10.1016/j.canlet.2016.01.037. [DOI] [PubMed] [Google Scholar]
- 379.Wang Y., Li Z., Zhao X., Zuo X., Peng Z. miR-10b promotes invasion by targeting HOXD10 in colorectal cancer. Oncol. Lett. 2016;12:488–494. doi: 10.3892/ol.2016.4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Dhanyamraju P.K. Drug resistance mechanisms in cancers: execution of pro-survival strategies. J. Biomed. Res. 2024;38:95–121. doi: 10.7555/JBR.37.20230248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Zhong C., Wang G., Guo M., Zhu N., Chen X., Yan Y., Li N., Yu W. The role of tumor stem cells in colorectal cancer drug resistance. Cancer Control. 2024;31 doi: 10.1177/10732748241274196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Shen K., Cui D., Sun L., Lu Y., Han M., Liu J. Inhibition of IGF-IR increases chemosensitivity in human colorectal cancer cells through MRP-2 promoter suppression. J. Cell. Biochem. 2012;113:2086–2097. doi: 10.1002/jcb.24080. [DOI] [PubMed] [Google Scholar]
- 383.Bartucci M., Svensson S., Ricci-Vitiani L., Dattilo R., Biffoni M., Signore M., Ferla R., De Maria R., Surmacz E. Obesity hormone leptin induces growth and interferes with the cytotoxic effects of 5-fluorouracil in colorectal tumor stem cells. Endocr. Relat. Cancer. 2010;17:823–833. doi: 10.1677/ERC-10-0083. [DOI] [PubMed] [Google Scholar]
- 384.Konishi T., Sasaki S., Watanabe T., Kitayama J., Nagawa H. Overexpression of hRFI inhibits 5-fluorouracil-induced apoptosis in colorectal cancer cells via activation of NF-kappaB and upregulation of BCL-2 and BCL-XL. Oncogene. 2006;25:3160–3169. doi: 10.1038/sj.onc.1209342. [DOI] [PubMed] [Google Scholar]
- 385.Zhang N., Yin Y., Xu S.-J., Chen W.-S. 5-Fluorouracil: mechanisms of resistance and reversal strategies. Molecules. 2008;13:1551–1569. doi: 10.3390/molecules13081551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Di Y., Jing X., Hu K., Wen X., Ye L., Zhang X., Qin J., Ye J., Lin R., Wang Z., He W. The c-MYC-WDR43 signalling axis promotes chemoresistance and tumour growth in colorectal cancer by inhibiting p53 activity. Drug Resist. Updates. 2023;66 doi: 10.1016/j.drup.2022.100909. [DOI] [PubMed] [Google Scholar]
- 387.Zhu Y., Huang S., Chen S., Chen J., Wang Z., Wang Y., Zheng H. SOX2 promotes chemoresistance, cancer stem cells properties, and epithelial-mesenchymal transition by β-catenin and Beclin1/autophagy signaling in colorectal cancer. Cell Death Dis. 2021;12:449. doi: 10.1038/s41419-021-03733-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Vaghari-Tabari M., Majidinia M., Moein S., Qujeq D., Asemi Z., Alemi F., Mohamadzadeh R., Targhazeh N., Safa A., Yousefi B. MicroRNAs and colorectal cancer chemoresistance: new solution for old problem. Life Sci. 2020;259 doi: 10.1016/j.lfs.2020.118255. [DOI] [PubMed] [Google Scholar]
- 389.Slattery M.L., Mullany L.E., Sakoda L.C., Wolff R.K., Samowitz W.S., Herrick J.S. Dysregulated genes and miRNAs in the apoptosis pathway in colorectal cancer patients. Apoptosis. 2018;23:237–250. doi: 10.1007/s10495-018-1451-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Peraza-Vega R.I., Valverde M., Rojas E. Interactions between miRNAs and double-strand breaks DNA repair genes, pursuing a fine-tuning of repair. Int. J. Mol. Sci. 2022;23:3231. doi: 10.3390/ijms23063231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Slattery M.L., Herrick J.S., Mullany L.E., Samowitz W.S., Sevens J.R., Sakoda L., Wolff R.K. The co-regulatory networks of tumor suppressor genes, oncogenes, and miRNAs in colorectal cancer. Genes Chromosomes Cancer. 2017;56:769–787. doi: 10.1002/gcc.22481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Marengo B., Pulliero A., Izzotti A., Domenicotti C. miRNA regulation of glutathione homeostasis in cancer initiation, progression and therapy resistance. MicroRNA. 2020;9:187–197. doi: 10.2174/2211536609666191218103220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Hu J.L., He G.Y., Lan X.L., Zeng Z.C., Guan J., Ding Y., Qian X.L., Liao W.T., Ding Y.Q., Liang L. Inhibition of ATG12-mediated autophagy by miR-214 enhances radiosensitivity in colorectal cancer. Oncogenesis. 2018;7:16. doi: 10.1038/s41389-018-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Poel D., Boyd L.N.C., Beekhof R., Schelfhorst T., Pham T.V., Piersma S.R., Knol J.C., Jimenez C.R., Verheul H.M.W., Buffart T.E. Proteomic analysis of miR-195 and miR-497 replacement reveals potential candidates that increase sensitivity to oxaliplatin in MSI/P53wt colorectal cancer cells. Cells. 2019;8:1111. doi: 10.3390/cells8091111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Zhu W., Zhu D., Lu S., Wang T., Wang J., Jiang B., Shu Y., Liu P. miR-497 modulates multidrug resistance of human cancer cell lines by targeting BCL2. Med. Oncol. 2012;29:384–391. doi: 10.1007/s12032-010-9797-4. [DOI] [PubMed] [Google Scholar]
- 396.Tong Z., Liu N., Lin L., Guo X., Yang D., Zhang Q. miR-125a-5p inhibits cell proliferation and induces apoptosis in colon cancer via targeting BCL2, BCL2L12 and MCL1. Biomed. Pharmacother. 2015;75:129–136. doi: 10.1016/j.biopha.2015.07.036. [DOI] [PubMed] [Google Scholar]
- 397.Falzone L., Bordonaro R., Libra M. SnapShot: cancer chemotherapy. Cell. 2023;186:1816–1816.e1. doi: 10.1016/j.cell.2023.02.038. [DOI] [Google Scholar]
- 398.Li S., Zheng S. Down-regulation of Circ_0032833 sensitizes colorectal cancer to 5-fluorouracil and oxaliplatin partly depending on the regulation of miR-125-5p and MSI1. Cancer Manag. Res. 2020;12:11257–11269. doi: 10.2147/CMAR.S270123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Yu X., Shi W., Zhang Y., Wang X., Sun S., Song Z., Liu M., Zeng Q., Cui S., Qu X. CXCL12/CXCR4 axis induced miR-125b promotes invasion and confers 5-fluorouracil resistance through enhancing autophagy in colorectal cancer. Sci. Rep. 2017;7 doi: 10.1038/srep42226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Banzhaf-Strathmann J., Edbauer D. Good guy or bad guy: the opposing roles of microRNA 125b in cancer. Cell Commun. Signal. 2014;12:30. doi: 10.1186/1478-811X-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Zhang J., Guo H., Zhang H., Wang H., Qian G., Fan X., Hoffman A.R., Hu J.-F., Ge S. Putative tumor suppressor miR-145 inhibits colon cancer cell growth by targeting oncogene Friend leukemia virus integration 1 gene. Cancer. 2011;117:86–95. doi: 10.1002/cncr.25522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Xie L., Cui G., Li T. Long noncoding RNA CBR3-AS1 promotes stem-like properties and oxaliplatin resistance of colorectal cancer by sponging miR-145-5p. J. Oncol. 2022;2022 doi: 10.1155/2022/2260211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Chen W., Chen Y., Hui T. microRNA-143 interferes the EGFR-stimulated glucose metabolism to re-sensitize 5-FU resistant colon cancer cells via targeting hexokinase 2. J. Chemother. 2023;35:539–549. doi: 10.1080/1120009X.2022.2157617. [DOI] [PubMed] [Google Scholar]
- 404.Akao Y., Khoo F., Kumazaki M., Shinohara H., Miki K., Yamada N. Extracellular disposal of tumor-suppressor miRs-145 and -34a via microvesicles and 5-FU resistance of human colon cancer cells. Int. J. Mol. Sci. 2014;15:1392–1401. doi: 10.3390/ijms15011392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Akao Y., Noguchi S., Iio A., Kojima K., Takagi T., Naoe T. Dysregulation of microRNA-34a expression causes drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer Lett. 2011;300:197–204. doi: 10.1016/j.canlet.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 406.Yang Y., Yuan H., Zhao L., Guo S., Hu S., Tian M., Nie Y., Yu J., Zhou C., Niu J., Wang G., Song Y. Targeting the miR-34a/LRPPRC/MDR1 axis collapse the chemoresistance in P53 inactive colorectal cancer. Cell Death Differ. 2022;29:2177–2189. doi: 10.1038/s41418-022-01007-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Li Y., Gong P., Hou J.-X., Huang W., Ma X.-P., Wang Y.-L., Li J., Cui X.-B., Li N. miR-34a regulates multidrug resistance via positively modulating OAZ2 signaling in colon cancer cells. J. Immunol. Res. 2018;2018 doi: 10.1155/2018/7498514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Jiang T., Ye L., Han Z., Liu Y., Yang Y., Peng Z., Fan J. miR-19b-3p promotes colon cancer proliferation and oxaliplatin-based chemoresistance by targeting SMAD4: validation by bioinformatics and experimental analyses. J. Exp. Clin. Cancer Res. 2017;36:131. doi: 10.1186/s13046-017-0602-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Gu Y.Y., Yu J., Zhang J.F., Wang C. Suppressing the secretion of exosomal miR-19b by gw4869 could regulate oxaliplatin sensitivity in colorectal cancer. Neoplasma. 2019;66:39–45. doi: 10.4149/neo_2018_180306N155. [DOI] [PubMed] [Google Scholar]
- 410.Fu Q., Cheng J., Zhang J., Zhang Y., Chen X., Luo S., Xie J. miR-20b reduces 5-FU resistance by suppressing the ADAM9/EGFR signaling pathway in colon cancer. Oncol. Rep. 2017;37:123–130. doi: 10.3892/or.2016.5259. [DOI] [PubMed] [Google Scholar]
- 411.Meng X., Fu R. miR-206 regulates 5-FU resistance by targeting Bcl-2 in colon cancer cells. OncoTargets Ther. 2018;11:1757–1765. doi: 10.2147/OTT.S159093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Lv L., Li Q., Chen S., Zhang X., Tao X., Tang X., Wang S., Che G., Yu Y., He L. miR-133b suppresses colorectal cancer cell stemness and chemoresistance by targeting methyltransferase DOT1L. Exp. Cell Res. 2019;385 doi: 10.1016/j.yexcr.2019.111597. [DOI] [PubMed] [Google Scholar]
- 413.Zhou Y., Wan G., Spizzo R., Ivan C., Mathur R., Hu X., Ye X., Lu J., Fan F., Xia L., Calin G.A., Ellis L.M., Lu X. miR-203 induces oxaliplatin resistance in colorectal cancer cells by negatively regulating ATM kinase. Mol. Oncol. 2014;8:83–92. doi: 10.1016/j.molonc.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Braun C.J., Zhang X., Savelyeva I., Wolff S., Moll U.M., Schepeler T., Ørntoft T.F., Andersen C.L., Dobbelstein M. p53-Responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res. 2008;68:10094–10104. doi: 10.1158/0008-5472.CAN-08-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Boni V., Bitarte N., Cristobal I., Zarate R., Rodriguez J., Maiello E., Garcia-Foncillas J., Bandres E. miR-192/miR-215 influence 5-fluorouracil resistance through cell cycle-mediated mechanisms complementary to its post-transcriptional thymidilate synthase regulation. Mol. Cancer Therapeut. 2010;9:2265–2275. doi: 10.1158/1535-7163.MCT-10-0061. [DOI] [PubMed] [Google Scholar]
- 416.Diaz T., Tejero R., Moreno I., Ferrer G., Cordeiro A., Artells R., Navarro A., Hernandez R., Tapia G., Monzo M. Role of miR-200 family members in survival of colorectal cancer patients treated with fluoropyrimidines. J. Surg. Oncol. 2014;109:676–683. doi: 10.1002/jso.23572. [DOI] [PubMed] [Google Scholar]
- 417.Wu Y., Song Y., Wang R., Wang T. Molecular mechanisms of tumor resistance to radiotherapy. Mol. Cancer. 2023;22:96. doi: 10.1186/s12943-023-01801-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Pedroza-Torres A., Romero-Córdoba S.L., Montaño S., Peralta-Zaragoza O., Vélez-Uriza D.E., Arriaga-Canon C., Guajardo-Barreto X., Bautista-Sánchez D., Sosa-León R., Hernández-González O., Díaz-Chávez J., Alvarez-Gómez R.M., Herrera L.A. Radio-miRs: a comprehensive view of radioresistance-related microRNAs. Genetics (Austin, Tex.) 2024;227 doi: 10.1093/genetics/iyae097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Shang Y., Zhu Z., Zhang Y., Ji F., Zhu L., Liu M., Deng Y., Lv G., Li D., Zhou Z., Lu B., Fu C.-G. MiR-7-5p/KLF4 signaling inhibits stemness and radioresistance in colorectal cancer. Cell Death Discov. 2023;9:42. doi: 10.1038/s41420-023-01339-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Zheng L., Chen J., Zhou Z., He Z. miR-195 enhances the radiosensitivity of colorectal cancer cells by suppressing CARM1. OncoTargets Ther. 2017;10:1027–1038. doi: 10.2147/OTT.S125067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Afshar S., Najafi R., Sedighi Pashaki A., Sharifi M., Nikzad S., Gholami M.H., Khoshghadam A., Amini R., Karimi J., Saidijam M. MiR-185 enhances radiosensitivity of colorectal cancer cells by targeting IGF1R and IGF2. Biomed. Pharmacother. 2018;106:763–769. doi: 10.1016/j.biopha.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 422.Zhang G., Liu Z., Zhong J., Lin L. Circ-ACAP2 facilitates the progression of colorectal cancer through mediating miR-143-3p/FZD4 axis. Eur. J. Clin. Invest. 2021;51 doi: 10.1111/eci.13607. [DOI] [PubMed] [Google Scholar]
- 423.Zheng L., Zhang Y., Liu Y., Zhou M., Lu Y., Yuan L., Zhang C., Hong M., Wang S., Li X. MiR-106b induces cell radioresistance via the PTEN/PI3K/AKT pathways and p21 in colorectal cancer. J. Transl. Med. 2015;13:252. doi: 10.1186/s12967-015-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Zhou Y., Shao Y., Hu W., Zhang J., Shi Y., Kong X., Jiang J. A novel long noncoding RNA SP100-AS1 induces radioresistance of colorectal cancer via sponging miR-622 and stabilizing ATG3. Cell Death Differ. 2023;30:111–124. doi: 10.1038/s41418-022-01049-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Liao F., Chen X., Peng P., Dong W. RWR-algorithm-based dissection of microRNA-506-3p and microRNA-140-5p as radiosensitive biomarkers in colorectal cancer. Aging (Albany NY) 2020;12:20512–20522. doi: 10.18632/aging.103907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.Wu Y., Pu N., Su W., Yang X., Xing C. Downregulation of miR-1 in colorectal cancer promotes radioresistance and aggressive phenotypes. J. Cancer. 2020;11:4832–4840. doi: 10.7150/jca.44753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 427.Khoshinani H.M., Afshar S., Pashaki A.S., Mahdavinezhad A., Nikzad S., Najafi R., Amini R., Gholami M.H., Khoshghadam A., Saidijam M. Involvement of miR-155/FOXO3a and miR-222/PTEN in acquired radioresistance of colorectal cancer cell line. Jpn. J. Radiol. 2017;35:664–672. doi: 10.1007/s11604-017-0679-y. [DOI] [PubMed] [Google Scholar]
- 428.Wang J., Xu J., Fu J., Yuan D., Guo F., Zhou C., Shao C. MiR-29a regulates radiosensitivity in human intestinal cells by targeting PTEN gene. Radiat. Res. 2016;186:292–301. doi: 10.1667/RR14428.1. [DOI] [PubMed] [Google Scholar]
- 429.Zhang Y., Zheng L., Huang J., Gao F., Lin X., He L., Li D., Li Z., Ding Y., Chen L. MiR-124 Radiosensitizes human colorectal cancer cells by targeting PRRX1. PLoS One. 2014;9 doi: 10.1371/journal.pone.0093917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Hu G., Che P., Deng L., Liu L., Liao J., Liu Q. MiR-378a-5p exerts a radiosensitizing effect on CRC through LRP8/β-catenin axis. Cancer Biol. Ther. 2024;25 doi: 10.1080/15384047.2024.2308165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Choi J.Y., Seok H.J., Lee D.H., Kwon J., Shin U.S., Shin I., Bae I.H. miR-1226-5p is involved in radioresistance of colorectal cancer by activating M2 macrophages through suppressing IRF1. J. Transl. Med. 2024;22:980. doi: 10.1186/s12967-024-05797-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Liu Y., Chen X., Chen X., Liu J., Gu H., Fan R., Ge H. Long non-coding RNA HOTAIR knockdown enhances radiosensitivity through regulating microRNA-93/ATG12 axis in colorectal cancer. Cell Death Dis. 2020;11:175. doi: 10.1038/s41419-020-2268-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Chen X., Liu J., Zhang Q., Liu B., Cheng Y., Zhang Y., Sun Y., Ge H., Liu Y. Exosome-mediated transfer of miR-93-5p from cancer-associated fibroblasts confer radioresistance in colorectal cancer cells by downregulating FOXA1 and upregulating TGFB3. J. Exp. Clin. Cancer Res. 2020;39:65. doi: 10.1186/s13046-019-1507-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Chen X., Liu Y., Zhang Q., Liu B., Cheng Y., Zhang Y., Sun Y., Liu J. Exosomal miR-590-3p derived from cancer-associated fibroblasts confers radioresistance in colorectal cancer. Mol. Ther. Nucleic Acids. 2021;24:113–126. doi: 10.1016/j.omtn.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Nagy Z.B., Barták B.K., Kalmár A., Galamb O., Wichmann B., Dank M., Igaz P., Tulassay Z., Molnár B. Comparison of circulating miRNAs expression alterations in matched tissue and plasma samples during colorectal cancer progression. Pathol. Oncol. Res. 2019;25:97–105. doi: 10.1007/s12253-017-0308-1. [DOI] [PubMed] [Google Scholar]
- 436.Gattuso G., Longo F., Spoto G., Ricci D., Lavoro A., Candido S., Di Cataldo A., Broggi G., Salvatorelli L., Magro G., Libra M., Falzone L. Diagnostic and prognostic significance of a four-miRNA signature in colorectal cancer. Int. J. Mol. Sci. 2025;26:1219. doi: 10.3390/ijms26031219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.El-Daly S.M., Gouhar S.A., Abd Elmageed Z.Y. Circulating microRNAs as reliable tumor biomarkers: opportunities and challenges facing clinical application. J. Pharmacol. Exp. Therapeut. 2023;384:35–51. doi: 10.1124/jpet.121.000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Sabry D., El-Deek S.E.M., Maher M., El-Baz M.A.H., El-Bader H.M., Amer E., Hassan E.A., Fathy W., El-Deek H.E.M. Role of miRNA-210, miRNA-21 and miRNA-126 as diagnostic biomarkers in colorectal carcinoma: impact of HIF-1α-VEGF signaling pathway. Mol. Cell. Biochem. 2019;454:177–189. doi: 10.1007/s11010-018-3462-1. [DOI] [PubMed] [Google Scholar]
- 439.Ghareib A.F., Mohamed R.H., Abd El-Fatah A.R., Saadawy S.F. Assessment of serum MicroRNA-21 gene expression for diagnosis and prognosis of colorectal cancer. J. Gastrointest. Cancer. 2020;51:818–823. doi: 10.1007/s12029-019-00306-w. [DOI] [PubMed] [Google Scholar]
- 440.Elaguizy M., Sheta M., Ibrahim N., Eltaweel A., Mostafa A. Serum microRNA-18a, MicroRNA-21 and microRNA-92a as diagnostic markers in colorectal cancer patients. J. BUON. 2020;25:1443–1448. [PubMed] [Google Scholar]
- 441.Li G., Wang Q., Li Z., Shen Y. Serum miR-21 and miR-210 as promising non-invasive biomarkers for the diagnosis and prognosis of colorectal cancer. Rev. Esp. Enferm. Dig. 2020;112:832–837. doi: 10.17235/reed.2020.6801/2019. [DOI] [PubMed] [Google Scholar]
- 442.Han L., Shi W.-J., Xie Y.-B., Zhang Z.-G. Diagnostic value of four serum exosome microRNAs panel for the detection of colorectal cancer. World J. Gastrointest. Oncol. 2021;13:970–979. doi: 10.4251/wjgo.v13.i8.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.Liu X., Xu X., Pan B., He B., Chen X., Zeng K., Xu M., Pan Y., Sun H., Xu T., Hu X., Wang S. Circulating miR-1290 and miR-320d as novel diagnostic biomarkers of human colorectal cancer. J. Cancer. 2019;10:43–50. doi: 10.7150/jca.26723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Wang X., Li Z., Fu J., Xu W., Li Z. Diagnostic value and prognostic significance of LI-cadherin and miR-378e in colorectal cancer. Oncol. Lett. 2020;20:2456–2464. doi: 10.3892/ol.2020.11755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Salah M., Shaheen I., El-Shanawany P., Saad N.E., Saad R., El Guibaly M., Momen N. Detection of miR-1246, miR-23a and miR-451 in sera of colorectal carcinoma patients: a case-control study in Cairo University hospital. Afr. Health Sci. 2020;20:1283–1291. doi: 10.4314/ahs.v20i3.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Shi Y., Liu Z. Serum miR-92a-1 is a novel diagnostic biomarker for colorectal cancer. J. Cell Mol. Med. 2020;24:8363–8367. doi: 10.1111/jcmm.15282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Zhao Y.J., Song X., Niu L., Tang Y., Song X., Xie L. Circulating exosomal miR-150-5p and miR-99b-5p as diagnostic biomarkers for colorectal cancer. Front. Oncol. 2019;9:1129. doi: 10.3389/fonc.2019.01129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 448.Abo-Elela D.A., Salem A.M., Swellam M., Hegazy M.G. Potential diagnostic role of circulating MiRNAs in colorectal cancer. Int. J. Immunopathol. Pharmacol. 2023;37 doi: 10.1177/03946320221144565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 449.Despotović J., Bogdanović A., Dragičević S., Galun D., Krivokapić Z., Nikolić A. Prognostic potential of circulating miR-93-5p in patients with colorectal cancer liver metastases. Neoplasma. 2022;69:430–442. doi: 10.4149/neo_2021_210603N749. [DOI] [PubMed] [Google Scholar]
- 450.Radanova M., Mihaylova G., Mihaylova Z., Ivanova D., Tasinov O., Nazifova-Tasinova N., Pavlov P., Mirchev M., Conev N., Donev I. Circulating miR-618 has prognostic significance in patients with metastatic colon cancer. Curr. Oncol. 2021;28:1204–1215. doi: 10.3390/curroncol28020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Kjersem J.B., Ikdahl T., Lingjaerde O.C., Guren T., Tveit K.M., Kure E.H. Plasma microRNAs predicting clinical outcome in metastatic colorectal cancer patients receiving first-line oxaliplatin-based treatment. Mol. Oncol. 2014;8:59–67. doi: 10.1016/j.molonc.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Zhang J., Zhang K., Bi M., Jiao X., Zhang D., Dong Q. Circulating microRNA expressions in colorectal cancer as predictors of response to chemotherapy. Anti Cancer Drugs. 2014;25:346–352. doi: 10.1097/CAD.0000000000000049. [DOI] [PubMed] [Google Scholar]
- 453.Schou J.V., Rossi S., Jensen B.V., Nielsen D.L., Pfeiffer P., Høgdall E., Yilmaz M., Tejpar S., Delorenzi M., Kruhøffer M., Johansen J.S. miR-345 in metastatic colorectal cancer: a non-invasive biomarker for clinical outcome in non-KRAS mutant patients treated with 3rd line cetuximab and irinotecan. PLoS One. 2014;9 doi: 10.1371/journal.pone.0099886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 454.Chen J., Wang W., Zhang Y., Chen Y., Hu T. Predicting distant metastasis and chemoresistance using plasma miRNAs. Med. Oncol. 2014;31:799. doi: 10.1007/s12032-013-0799-x. [DOI] [PubMed] [Google Scholar]
- 455.Ulivi P., Canale M., Passardi A., Marisi G., Valgiusti M., Frassineti G.L., Calistri D., Amadori D., Scarpi E. Circulating plasma levels of miR-20b, miR-29b and miR-155 as predictors of bevacizumab efficacy in patients with metastatic colorectal cancer. Int. J. Mol. Sci. 2018;19:307. doi: 10.3390/ijms19010307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Han J., Sun W., Liu R., Zhou Z., Zhang H., Chen X., Ba Y. Plasma exosomal miRNA expression profile as oxaliplatin-based chemoresistant biomarkers in colorectal adenocarcinoma. Front. Oncol. 2020;10:1495. doi: 10.3389/fonc.2020.01495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Jin G., Liu Y., Zhang J., Bian Z., Yao S., Fei B., Zhou L., Yin Y., Huang Z. A panel of serum exosomal microRNAs as predictive markers for chemoresistance in advanced colorectal cancer. Cancer Chemother. Pharmacol. 2019;84:315–325. doi: 10.1007/s00280-019-03867-6. [DOI] [PubMed] [Google Scholar]
- 458.Badr D., Fouad M.A., Hussein M., Salem S., Zekri A., Shouman S. Rebound increase in microRNA levels at the end of 5-FU-based therapy in colorectal cancer patients. Sci. Rep. 2023;13 doi: 10.1038/s41598-023-41030-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Chen Q., Xia H.-W., Ge X.-J., Zhang Y.-C., Tang Q.-L., Bi F. Serum miR-19a predicts resistance to FOLFOX chemotherapy in advanced colorectal cancer cases. Asian Pac. J. Cancer Prev. 2013;14:7421–7426. doi: 10.7314/apjcp.2013.14.12.7421. [DOI] [PubMed] [Google Scholar]
- 460.Marletta S., Rizzo A., Spoto G., Falzone L. Predictive and prognostic biomarkers in cancer: towards the precision medicine era. Explor. Target Antitumor Ther. 2024;5:1321–1325. doi: 10.37349/etat.2024.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Svoronos A.A., Engelman D.M., Slack F.J. OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Res. 2016;76:3666–3670. doi: 10.1158/0008-5472.CAN-16-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 462.Ekiz Kanik F., Celebi I., Sevenler D., Tanriverdi K., Lortlar Ünlü N., Freedman J.E., Ünlü M.S. Attomolar sensitivity microRNA detection using real-time digital microarrays. Sci. Rep. 2022;12 doi: 10.1038/s41598-022-19912-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Coenen-Stass A.M.L., Magen I., Brooks T., Ben-Dov I.Z., Greensmith L., Hornstein E., Fratta P. Evaluation of methodologies for microRNA biomarker detection by next generation sequencing. RNA Biol. 2018;15:1133–1145. doi: 10.1080/15476286.2018.1514236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Hardikar A.A., Farr R.J., Joglekar M.V. Circulating microRNAs: understanding the limits for quantitative measurement by real-time PCR. J. Am. Heart Assoc. 2014;3 doi: 10.1161/JAHA.113.000792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Dingle T.C., Sedlak R.H., Cook L., Jerome K.R. Tolerance of droplet-digital PCR vs real-time quantitative PCR to inhibitory substances. Clin. Chem. 2013;59:1670–1672. doi: 10.1373/clinchem.2013.211045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 466.Hindson C.M., Chevillet J.R., Briggs H.A., Gallichotte E.N., Ruf I.K., Hindson B.J., Vessella R.L., Tewari M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods. 2013;10:1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.