Abstract
The present review explores the influence of the gut microbiota on antibiotic resistance dynamics, particularly those associated with dysbiosis. The improper use of antibiotics can induce resistance in pathogens through various pathways, which is a topic of increasing interest within the scientific community. This review highlights the importance of microbial diversity, gut metabolism, and inflammatory responses against the dysbiosis due to the action of antibiotics. Additionally, it examines how secondary metabolites secreted by pathogens can serve as biomarkers for the early detection of antibiotic resistance. Although significant progress has been made in this field, key research gaps persist, including the need for a deeper understanding of the long-term effects of antibiotic-induced dysbiosis and the specific mechanisms driving the evolution of resistance in gut bacteria. Based on these considerations, this review systematically analyzed studies from PubMed, Web of Science, Embase, Cochrane Library, and Scopus up to July 2024. This study aimed to explore the dynamics of the interactions between gut microbiota and antibiotic resistance, specifically examining how microbial composition influences the development of resistance mechanisms. By elucidating these relationships, this review provides insights into management strategies for drug resistance and improves our understanding of microbial contributions to host health.
Keywords: Antibiotic resistance, Gut microbiota, Dysbiosis, Homeostasis, Metabolites
Graphical abstract
1. Introduction
The human gut microbiota, which comprises bacteria, archaea, viruses, and eukaryotes, plays a crucial role in human health and has attracted significant scientific attention [1]. Although the study of gut microbiota dates back centuries, recent advancements have dramatically improved our understanding of it. Enhanced culture techniques have also enabled precise isolation and characterization of bacterial species, establishing gut microbiology as a distinct field. The gut microbiota plays a pivotal role in human metabolism by contributing enzymes not encoded by the human genome, particularly in the breakdown of polysaccharides and polyphenols and the synthesis of essential vitamins. This has driven significant research to identify specific microorganisms and their metabolic pathways, particularly those involved in processing dietary components. Microbial enzymes such as glycoside hydrolases and polysaccharide lyases facilitate the fermentation of indigestible dietary fibers into short-chain fatty acids (SCFAs), including acetate, propionate, and butyrate [2]. SCFAs serve as energy sources for colonic epithelial cells and regulate various metabolic processes, including lipid metabolism, gluconeogenesis, and appetite control. The microbiota also synthesizes essential vitamins, such as vitamins K and B12, which are crucial for host metabolic functions [2,3]. The composition and metabolic activity of the gut microbiota significantly influence host dietary intake and growth, as microbial byproducts such as SCFAs and vitamins can have beneficial and detrimental effects on host health, depending on the context [2,3].
The symbiotic relationship between humans and their gut microbiota is characterized by mutualistic interactions, wherein the host provides a conducive environment for microbial growth, and the microbiota offers additional metabolic functions that benefit human health [4]. Several factors, including diet, age, and genetics, modulate the dynamic balance of the gut microbiota, influencing the immune, endocrine, and neurological systems and ultimately affecting overall health outcomes [5]. A disruption in the composition and function of the gut microbiota, known as dysbiosis, is associated with numerous diseases and health disorders [5]. The establishment and diversity of the gut microbiota are shaped by host genetics, early life exposure, diet, antibiotic use, and environmental factors [6,7].
The rise of antibiotic-resistant pathogens within the gut highlights the need to understand the complex interplay between gut microbiota and antibiotic resistance. Antibiotic resistance develops when bacteria acquire mechanisms to evade the effects of antibiotics, thereby rendering them less effective [8]. With its diverse microbial community, the gut microbiota plays a vital role in both health and disease [9]. This study reviews the impact of the gut microbiota on antibiotic resistance and provides a mechanistic overview of their interactions. Specifically, this study aimed to explore the dynamics of the interactions between the gut microbiota and antibiotic resistance, specifically examining how microbial composition influences the development of resistance mechanisms. By elucidating these relationships, this review provides insights into management strategies for drug resistance and improves our understanding of microbial contributions to host health.
1.1. Overview of the gut microbiota
Healthy gut microbiota primarily consists of several major phyla: Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria, Fusobacteria, and Verrucomicrobia. Firmicutes and Bacteroidetes account for approximately 90 % of the gut microbiota [9]. Firmicutes includes genera such as Lactobacillus, Bacillus, Clostridium, Enterococcus, and Ruminococcus, with Clostridium being the most prevalent, representing approximately 95 % of bacteria in this phylum. Bacteroidetes features notable genera, such as Bacteroides and Prevotella. Although less abundant, the phylum Actinobacteria is predominantly represented by the genus Bifidobacterium [9].
Commensal bacteria in the human gut, collectively referred to as the gut microbiota, reside in a complex ecosystem in which glycans are the primary nutrient source [10]. The host immune system regulates the presence and activity of these bacteria are regulated by the host's immune system [11]. These bacteria possess genes encoding enzymes capable of degrading various substrates, many of which originate in the host [12]. Some bacteria utilize and break down carbohydrate chains from the host mucosal glycans to provide nutrients [10]. Although the microbial communities vary among individuals, certain bacterial genes are consistently found in humans and influence the colonization of gut commensals [13,14].
The gut microbiota modulates the expression of numerous host genes. According to recent studies, the intestines of germ-free mice show the underexpression of certain genes compared to conventionally raised mice, and this effect can persist after the introduction of gut commensals, affecting susceptibility to enteric infections [15,16]. Such studies indicate that mice are common organisms for studying gut microbiota and provide insights relevant to human gut commensals [17]. Understanding the influence of the gut microbiota on host gene expression is crucial for concepts such as colonization resistance, which refers to the ability of the gut microbiota to prevent the growth of pathogenic microorganisms [18]. Colonization resistance can be due to nutrient competition, the production of inhibitory compounds such as SCFAs from microbial fermentation, or immune system stimulation [19]. Insights into these interactions can provide a scientific platform for therapies that can regulate the immune system and microbial composition to enhance colonization resistance against pathogens [19].
1.2. Antibiotic resistance
The emergence and global dissemination of new antibiotic resistance mechanisms pose a significant threat to the effectiveness of treatments for common infectious diseases, leading to prolonged illness, disability, and increased mortality [20]. Despite advances in microbiology, the development of new antibiotics has not kept pace with the rise in antibiotic resistance, leading to a critical situation in which existing antibiotics are becoming increasingly ineffective [21]. This scenario, which is often referred to as the post-antibiotic era, has profound implications for global public health. The difficulty in treating even minor infections may increase, and the failure of antibiotic therapy to provide a clinical cure could result in higher morbidity and mortality rates [22]. The spread of drug-resistant bacteria is exacerbated by antibiotic misuse and socioeconomic factors, diminishing the effectiveness of current treatments [23]. Currently, antibiotic resistance is responsible for more than 700,000 deaths annually worldwide, with projections suggesting that this number could escalate to 10 million by 2050 if new therapeutic strategies are not developed [24]. The mortality rate due to antimicrobial resistance could potentially surpass other leading causes of mortality by 2050, underlining the urgency of tackling this issue on a global scale [25]. This can be cited with various studies where increasing prevalence of multidrug-resistant (MDR) and carbapenem-resistant bacteria, such as Klebsiella pneumoniae, is particularly alarming as they are responsible for numerous infections, especially in hospital settings [26]. Its reported that the molecular epidemiology of carbapenem-resistant classic (CR-cKp) and hypervirulent (CR-hvKp) Klebsiella pneumoniae are being detected in various countries [27]. The findings revealed a high prevalence of MDR CR-hvKp isolates, with significant beta-lactamase gene occurrences, particularly bla NDM and bla GES. The study emphasized the need for long-term surveillance and effective treatment strategies to combat the spread of these resistant strains [27].
Similarly, the spread of antimicrobial resistance is not limited to Klebsiella pneumoniae. In similar terms its reported that Pseudomonas aeruginosa have revealed high levels of quinolone resistance mediated by plasmid-borne resistance genes like qnrB [28]. Further, resistance mechanism is also evident in Shigella species, where ESBL production is widespread, and resistance to third-generation cephalosporins has reached alarming levels [29].
Apart from these pathogens methicillin-resistant Staphylococcus aureus (MRSA) and coagulase-negative staphylococci (CoNS) are also contributing to antimicrobial resistance, which are found in the oral cavity, demonstrate significant multidrug resistance, exacerbating treatment challenges [30].
Apart from hospital-acquired infections, antimicrobial resistance also impacts maternal health which was reported in northern Ethiopia [31]. Hence, understanding these mechanisms will envision targeting specific genes for effective antimicrobial therapies. Furthermore, resistance arises due to the selective pressure exerted by the inappropriate use of antibiotics, which kill or inhibit most bacteria but allow resistant strains to survive and proliferate, thereby passing on their genetic resistance [32,33].
2. Influence of the gut microbiota on antibiotic resistance dynamics
The interaction between gut microbiota and antibiotic resistance is a critical aspect of human health [33]. Antibiotic administration can profoundly alter the composition and diversity of the gut microbiota, disrupt homeostasis, and create selective pressure that promotes the proliferation of antibiotic-resistant bacteria [34]. This disruption impairs the protective functions of the microbiota, leading to increased susceptibility to resistant pathogens, reduced microbial diversity, and changes in the abundance and metabolic functions of specific bacterial populations [35].
Antibiotics directly contribute to the emergence and dissemination of resistance genes and can substantially affect the structure and function of gut microbiota. For instance, a study that analyzed gut microbial communities in patients undergoing β-lactam therapy revealed a significant reduction in microbial diversity and metabolic capabilities, including bile acid, cholesterol, hormone, and vitamin metabolism [36]. This alteration disrupts the microbial balance, creating niches that favor opportunistic pathogens and resistant strains. Research has shown that antibiotic treatment can decrease the diversity of detectable bacterial species by up to 20-fold and increase the prevalence of opportunistic pathogens such as Acinetobacter calcoaceticus/baumannii complex, Chlamydia abortus, Bacteroides fragilis, and Bacteroides thetaiotaomicron [37].
Additionally, antibiotic-induced dysbiosis compromises the metabolic and immune functions of the gut microbiota, leading to increased susceptibility to infections and inflammatory disorders. Case studies have indicated that antibiotic-induced dysbiosis reduces the abundance of beneficial bacteria, such as Lachnospiraceae, Muribaculaceae, and Ruminococcaceae, while increasing the abundance of harmful taxa, such as Enterococcaceae and Clostridiales [38]. This imbalance is associated with decreased levels of SCFAs and tryptophan and increased levels of purines, which are linked to food allergies. Dysbiosis also results in elevated specific IgE and IgG levels, increased inflammation, and severe allergic symptoms, including damage to the intestinal villi and decreased levels of tight junction proteins [38]. Moreover, disruptions in microbial communities also affect nutrient metabolism, immune development, and neurological function [39].
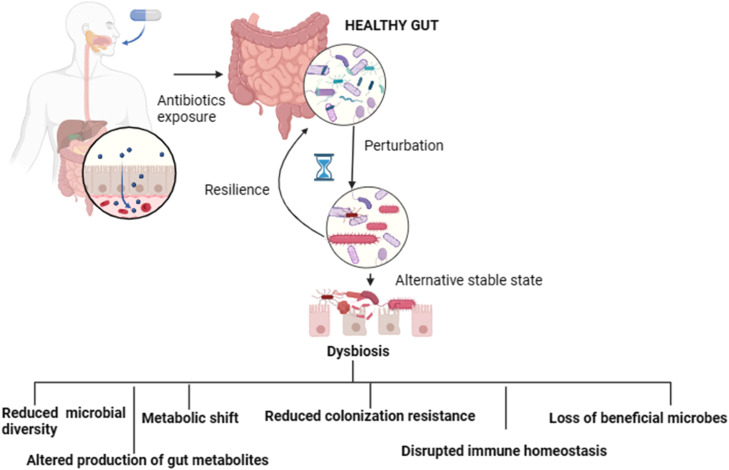
The administration of antibiotics exerts multifaceted effects beyond the facilitation of antibiotic resistance, emphasizing the intricate interactions between antimicrobial treatments and gut microbiota. As shown in Fig. 1, antibiotic exposure can lead to significant disruptions in the gut microbiota, causing persistent dysbiosis characterized by diminished microbial diversity, loss of essential microbial taxa, and notable metabolic changes. This state of dysbiosis compromises the inherent resistance of the gut to pathogenic colonization, thereby enhancing the risk of disease development.
Fig. 1.
Antibiotic-induced Dysbiosis in the Gut Microbiota.
3. Antibiotic usage and resistance mechanisms
The widespread use of antibiotics, including aminoglycosides, β-lactams, fluoroquinolones, macrolides, and tetracyclines, is well documented. However, the improper and excessive use of these antibiotics has been a pivotal factor in the swift emergence of antibiotic resistance mechanisms, posing a significant challenge to public health.
3.1. Aminoglycosides
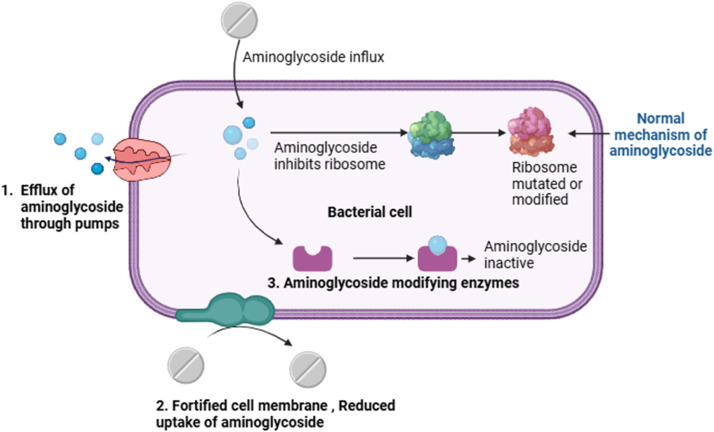
Aminoglycosides are commonly used to treat bacterial infections. Common aminoglycosides such as gentamicin, streptomycin, amikacin, and tobramycin are widely used, and the inappropriate use of these antibiotics can contribute substantially to the threat of antibiotic resistance due to their widespread use and the tendency of bacteria to develop resistance mechanisms against them [40]. These resistance mechanisms include fortified cell walls, efflux pumps that expel aminoglycosides, mutations in ribosomal targets, and ribosome methyltransferases. The most prevalent resistance mechanism involves the inactivation of aminoglycosides via aminoglycoside-modifying enzymes [40]. The mechanism underlying aminoglycoside resistance is shown in Fig. 2. A study that analyzed 160,000 publicly available genomes across 27 clusters of genes encoding aminoglycoside-modifying enzymes found that approximately 25 % of the sequenced bacteria sampled from various continents and terrestrial biomes carried these genes [41]. Antibiotic-associated diarrhea is a common adverse event of antibiotic therapy. It is characterized by diarrhea that occurs without another clear cause during antibiotic treatment. Scientific reports have highlighted that the cephalosporins gentamicin and cefradine can cause diarrhea in mice. This might be due to the disruption of the microbiota during antibiotic-associated diarrhea with dominant species includes Firmucutes and Proteobacteria. One study reported a decrease in bacterial diversity after antibiotic administration, with notable increases in the abundance of opportunistic pathogens, such as Enterococcus and Clostridium. These findings suggest a link between antibiotic-induced microbiota alterations and antibiotic-associated diarrhea, highlighting the potential biomarkers of these conditions [42]. High-level gentamicin resistance in Enterococcus faecalis poses a significant challenge for treatment, especially in severe infections, where it undermines the effectiveness of aminoglycoside-based therapies. The widespread presence of the aac(6′)-Ie-aph(2″)-Ia gene in E. faecalis strains from both human and animal sources exacerbates the issue by conferring resistance to most aminoglycosides except streptomycin [43]. The ability of this gene to transfer horizontally among different Enterococcus species raises concerns regarding its spread. The overuse of antibiotics in veterinary medicine can also fuel the rise of drug-resistant bacteria, necessitating innovative approaches, including the development of new antibacterial drugs and the exploration of alternative therapies, such as semi-purified bacteriocins and probiotics. Understanding high-level gentamicin resistance and its implications for public health emphasizes the importance of addressing its spread and potential transfer to other bacteria [43].
Fig. 2.
Aminoglycoside mode of action and its resistance mechanism.
3.2. β-lactams
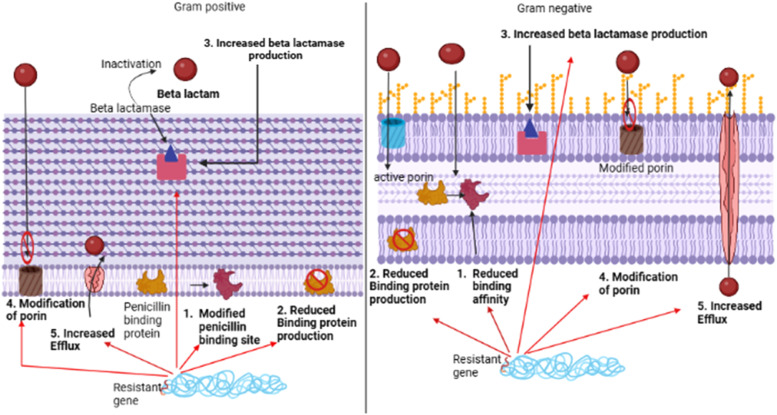
Increased antibiotic resistance due to β-lactam antibiotics in the gut microbiota poses a significant health challenge at the individual and population levels [44]. This family of antibiotics, including penicillins, cephalosporins, and carbapenems, are widely and commonly used to combat bacterial infections [45]. However, their widespread use has spurred the emergence and dissemination of resistance mechanisms within the gut microbial community, leading to profound alterations in its composition and functionality. Resistance to β-lactams primarily arises from the production of β-lactamase enzymes by gut bacteria, which dismantle the crucial β-lactam ring of these antibiotics, rendering them ineffective [46]. Originally, β-lactamase enzymes were primarily of the serine β-lactamase type. However, the recent emergence of highly resistant Gram-negative strains has introduced metallo-β-lactamase enzymes as a source of resistance. Although both types of enzymes have the same function in bacteria, they differ structurally and mechanistically. The serine β-lactamase type employs an active serine group to initiate the hydrolysis of the β-lactam ring, forming a covalent intermediate that is subsequently cleaved. In contrast, metallo-β-lactamase enzymes use a zinc ion to activate the β-lactam, facilitating nucleophilic attack by a hydroxide anion positioned between two zinc ions [46]. Furthermore, mutations in penicillin-binding proteins, which are the primary targets, such as acquiring an extra low-affinity penicillin-binding protein, overexpressing an existing low-affinity penicillin-binding protein, and modifying endogenous penicillin-binding proteins through point mutations or homologous recombination, individually or in combination, can confer resistance [47]. Such mechanisms undermine the efficacy of β-lactam antibiotics and disrupt the delicate balance of the gut microbiota. This disruption often results in dysbiosis, marked by shifts in microbial diversity and composition, potentially fostering the colonization of antibiotic-resistant pathogens [48]. The β-lactam resistance mechanism is illustrated in Fig. 3.
Fig. 3.
β-lactam resistance mechanism.
Amoxicillin is another widely used β-lactam antibiotic. Exposure to amoxicillin leads to notable shifts in the gut microbiota composition, including the increased abundance of Klebsiella and Bacteroides uniformis, decreased levels of Parabacteroides, Bifidobacterium, and Phascolarctobacterium, as well as altered functional pathways [49]. Additionally, this exposure increases the number of β-lactam resistance genes and metabolic and immune system disease genes, with bloomed pathogens being strongly correlated to these changes [49]. These effects are particularly pronounced in the ascending colon and persist even after the discontinuation of amoxicillin treatment. Overall, these findings highlight the potential negative consequences of amoxicillin and emphasize the importance of careful consideration when prescribing this antibiotic [49].
In a study on mice prone to type 1 diabetes, mice administered pulsed therapeutic antibiotics when young were more likely to develop type 1 diabetes in comparison with the control mice without treatment of antibiotics. The pulsed therapeutic antibiotic group had different gut bacteria, fewer immune cells in their intestines, and changes in genes related to inflammation and cholesterol. This shows that antibiotics can affect the gut bacteria, body processes, and immune functions, thereby accelerating the onset of type 1 diabetes [50]. Collectively, these results suggest that understanding the dynamics of β-lactam antibiotic resistance in the gut microbiota and its consequences is imperative for devising strategies to combat antibiotic resistance while preserving the beneficial functions of the microbiota.
3.3. Fluoroquinolones
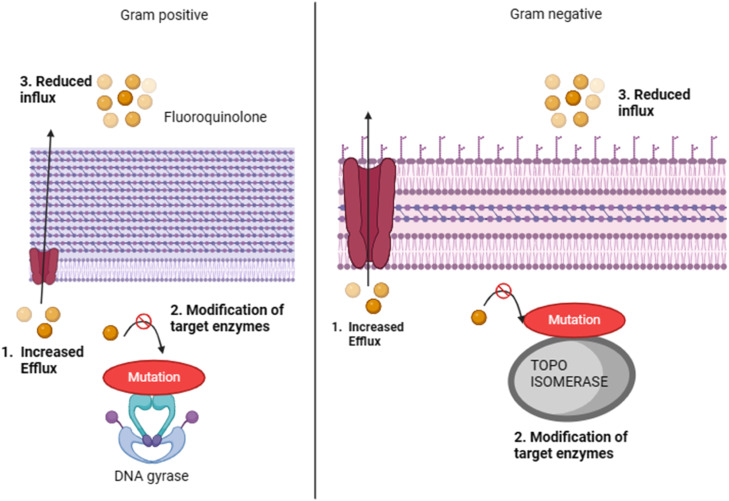
The surge in antibiotic resistance in the gut microbiota attributed to fluoroquinolones poses a considerable health challenge [51]. These antibiotics, which are widely used to treat bacterial infections, disrupt the gut microbial balance [52]. Fluoroquinolones inhibit bacterial DNA gyrase and topoisomerase IV, which are crucial enzymes for DNA replication and repair. Resistance typically arises from mutations in the genes encoding these enzymes, diminishing antibiotic binding and efficacy [51]. Additionally, bacteria can resist fluoroquinolones through efflux pumps and expel antibiotics from within [51]. Fluoroquinolone resistance primarily develops through two mechanisms: mutations in target enzymes, such as DNA gyrase in Gram-negative bacteria and topoisomerase IV in Gram-positive bacteria, and enhanced efflux of antibiotics through specific systems. The increased expression of chromosomal genes can lead to an elevated efflux of fluoroquinolones, reducing their accumulation within bacterial cells. This dual mechanism enables bacteria to withstand the effects of fluoroquinolones, posing a challenge for effective antibiotic therapies [53]. This selective pressure fosters the growth of resistant strains, leading to dysbiosis and an increased chance of colonization by resistant pathogens. The mechanism of fluoroquinolone resistance is illustrated in Fig. 4.
Fig. 4.
Fluoroquinolone resistance mechanism.
Understanding fluoroquinolone resistance in gut microbiota is vital for guiding the use of antibiotics and preserving their efficacy. Ciprofloxacin is commonly used to treat Crohn's disease. In a study of patients with Crohn's disease who required abscess drainage, approximately 38.5 % had no bacteria detected, whereas 61.5 % had various microorganisms isolated. Most of these microorganisms are bacteria, including Gram-positive and Gram-negative aerobes, Gram-negative anaerobes, and fungi [54]. The most common bacteria found were Streptococci spp. and Escherichia coli. However, a significant proportion of Gram-negative aerobes, including E. coli, are resistant to ciprofloxacin. Patients infected with ciprofloxacin-resistant bacteria had a longer disease duration and hospitalization than those infected with ciprofloxacin-sensitive bacteria[54]. In another case study, high doses of ciprofloxacin (1 mg/mL) caused weight loss, nervousness, decreased appetite, and increased gut microbial cell death; lower doses (0.2 mg/mL) had less impact [55]. Ciprofloxacin treatment further reduced the levels of tight junction proteins and antibacterial genes while increasing the secretion of the inflammatory cytokine IL-1β [48]. It also alters the gut bacterial diversity and richness. The synthesis of important metabolites such as indole, butyric acid, and valeric acid were significantly reduced due to disruptions in gut bacterial diversity and composition. Thus, high-dose ciprofloxacin treatment impaired gut barrier function, highlighting the need for cautious antibiotic use in disease treatment [55]. In a case study, moxifloxacin reduced the abundance of Enterococci and Enterobacteria, whereas clarithromycin decreased the abundance of Escherichia coli but increased that of Enterococci, Enterobacter, Citrobacter, Klebsiella, and Pseudomonas. Both antibiotics had minimal impact on other microbial populations, while clarithromycin suppressed the beneficial gut microbiota Bifidobacteria, Lactobacilli, and Clostridia in anaerobic microflora. These findings highlight the distinct effects on the intestinal microbiota, suggesting potential implications for therapeutic efficacy and microbiota recovery [56].
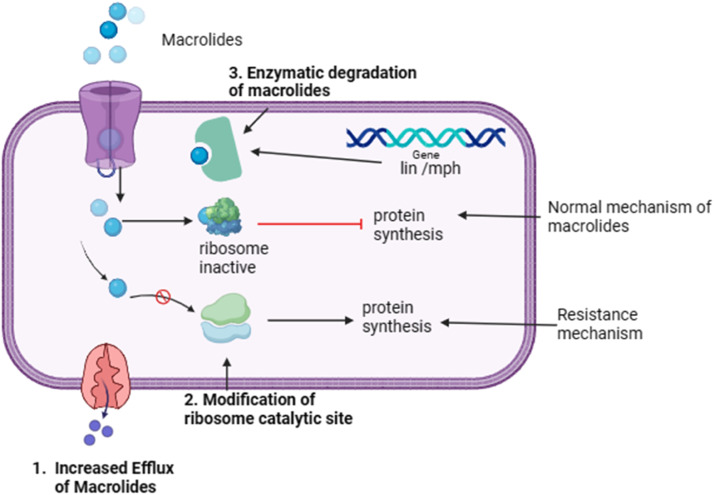
3.4. Macrolides
Macrolide antibiotics function by binding to the 50S ribosomal subunit of bacteria, hindering protein synthesis and impeding bacterial growth where macrolide molecules, along with particular nascent peptides within the ribosomal tunnel, induce allosteric changes in the functional characteristics of the ribosome's catalytic center [57] Resistance to these antibiotics arises through several mechanisms, including modifications to the drug's target site on the ribosome, efflux pumps that expel antibiotics from bacterial cells, and the enzymatic inactivation of the antibiotic [58]. This mechanism is illustrated in Fig. 5. These mechanisms allow bacteria to evade the effects of macrolides and survive exposure to antibiotics [58]. Furthermore, in resistant cells, the presence of uncoupling agents, such as carbonylcyanide-m-chlorophenylhydrazone, 2,4-dinitrophenol, and arsenate, increases the accumulation of drugs to the same extent as in susceptible cells [59]. Macrolide-resistant cells have transporter proteins resembling the 12-transmembrane domain found in efflux proteins; these proteins are driven by proton-motive forces. In contrast, macrolide-streptogramin type B or partial macrolide- and streptogramin-type resistant cells have transporter proteins similar to ATP-binding segments in ATP-driven efflux proteins [59]. The two primary mechanisms of macrolide antibiotic inactivation include lactone ring degradation by an esterase encoded by the ere gene and modification through phosphorylation mediated by mph, and nucleotidylation mediated by the lin gene. However, this resistance is due to the inappropriate use of macrolides, which exert selective pressure on the gut microbiota and favor the proliferation of resistant bacterial strains [59].
Fig. 5.
Macrolides resistance mechanism.
Consequently, there was a shift in the composition of the gut microbiota, with a decline in the abundance of susceptible species and an increase in the abundance of resistant species. For instance, in one study, the mass drug administration of azithromycin led to an increase in the prevalence of the gastrointestinal carriage of macrolide-resistant bacteria. Fecal metagenomics of 60 children before treatment and 122 children after four rounds of mass drug administration (half receiving azithromycin and half receiving placebo) revealed this trend. The abundance of several species, including Escherichia albertii, a potential human enteropathogen, increased after treatment. This study suggests that the mass administration of azithromycin may have a limited impact on clinically relevant bacteria; however, the increase in the abundance of enteropathogenic Escherichia species warrants further investigation [60].
One study analyzed erythromycin-resistant lactic acid bacteria in the feces of healthy individuals following different diets, such as vegans, ovo-lacto vegetarians, and omnivores. They found that out of 155 lactic acid bacterial isolates, 97 were resistant to erythromycin, with Enterococcus faecium being the most common species across all dietary groups. Among the resistant isolates, 19 carried the Erm(B) gene, and omnivores had the highest number of carriers. Interestingly, Enterococcus avium from omnivores contained both the Erm(B) and Erm(A) genes [61]. This study also investigated the transferability of erythromycin resistance genes and found that four out of six tested isolates were capable of transferring the Erm(B) gene [61]. Overall, isolates from omnivores exhibited greater resistance to antibiotics and carried a greater number of antibiotic resistance genes than those from ovo-lacto vegetarians and vegans. These findings suggest that diet does not significantly influence the occurrence of erythromycin-resistant bacteria; however, commensal strains can serve as reservoirs and sources for the spread of antibiotic resistance genes [61]. Furthermore, such alterations in the gut microbiota can have far-reaching consequences on human health, as the microbiota plays a crucial role in various physiological processes, including digestion, metabolism, and immune function[62]. In one study, the impact of two commonly used antibiotics, azithromycin (macrolide) and amoxicillin (β-lactam), on the human gut microbiota was investigated. Metagenomic sequencing was then used to analyze the gut microbiota of individuals who received these antibiotics. The results revealed the significant and sustained effects of antibiotics, particularly azithromycin, on the gut microbial community. Specifically, clear alterations were observed with decrease in Bifidobacterium species, along with an increased prevalence of erm genes associated with macrolide resistance [63].
Disruption of the gut microbiota owing to macrolide resistance may contribute to the development of gastrointestinal disorders, immune system dysregulation, and increased susceptibility to infections[64]. Healthy adults exposed to four weeks of low-dose erythromycin or azithromycin, which are normally used clinically, showed significant shifts in gut microbiota composition. These shifts included a reduction in the microbial capacity related to carbohydrate metabolism and SCFA biosynthesis. Simultaneously, alterations were noted in systemic biomarkers associated with immunity, such as interleukin-5, interleukin-10, and monocyte chemoattractant protein-1, and metabolic biomarkers, such as serotonin 5-HT and C-peptide homeostasis. Furthermore, transplanting erythromycin-exposed murine microbiota into germ-free mice revealed changes in metabolic homeostasis and gastrointestinal motility, but not in systemic immune regulation [64]. This suggests that the long-term use of low-dose macrolide antibiotics could affect the function of our bodies by changing the gut bacteria [64]. Thus, addressing the challenge of macrolide resistance requires not only wise antibiotic use, but also the exploration of alternative therapeutic approaches to mitigate the impact on both microbial ecology and human health.
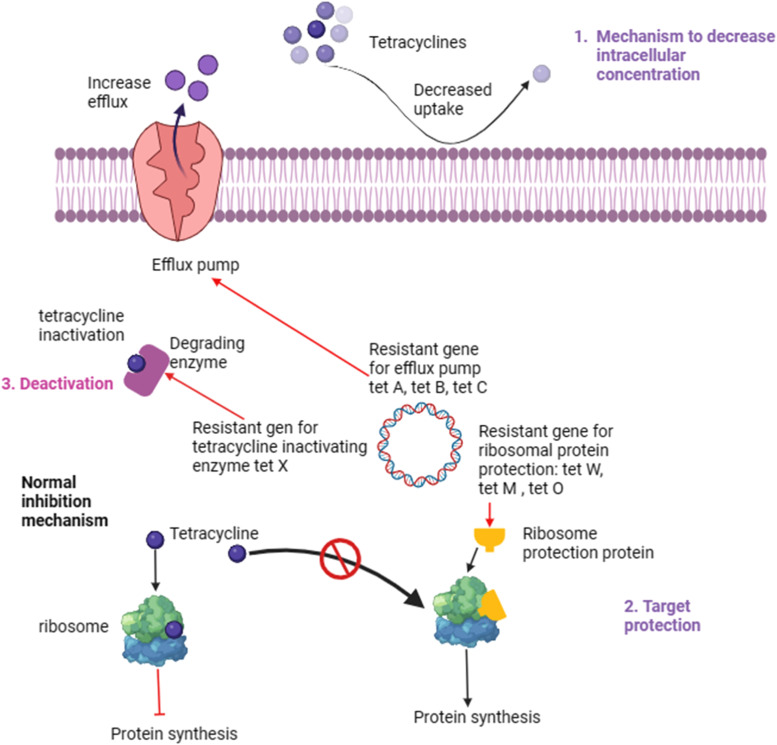
3.5. Tetracyclines
Tetracyclines exhibit a broad spectrum of activity against diverse bacterial species, including Gram-positive and Gram-negative bacteria. Their antimicrobial efficacy is achieved through binding to the 30S ribosomal subunit, which inhibits the attachment of aminoacyl-tRNA to ribosomes. This mechanism effectively halts bacterial protein synthesis and enhances the proliferation of pathogenic organisms [65]. Tetracycline resistance in the gut microbiota typically arises through multiple mechanisms involving the acquisition of specific resistance genes and alterations in bacterial physiology[66]. One primary mechanism involves the production of efflux pumps such as Tet(A), Tet(B), and Tet(C), which actively pump tetracycline antibiotics out of bacterial cells, preventing them from reaching effective concentrations [65,[67], [68], [69]]. Another mechanism involves the protection of the bacterial ribosome target site through the action of ribosomal protection proteins encoded by the Tet(M), Tet(O), and Tet(W) genes, which prevent tetracyclines from binding and inhibit protein synthesis [70]. Additionally, the enzymatic inactivation of tetracyclines by tetracycline-inactivating enzymes, such as Tet(X), can also confer resistance [71]. These mechanisms collectively contribute to the ability of the gut bacteria to withstand tetracycline exposure. The mechanism underlying tetracycline resistance is shown in Fig. 6.
Fig. 6.
Tetracycline resistance mechanism.
The burden of tetracycline resistance on gut microbiota is multifaceted and can have significant implications for both individual and public health [72,73]. Tetracycline resistance genes can spread horizontally among gut bacterial populations, thereby facilitating the dissemination of resistance to other antibiotics and potentially pathogenic bacteria [73]. A previous study revealed the potential for the horizontal transfer of antibiotic resistance genes within the gastrointestinal tract. Through metagenomic analysis of a healthy mother-infant pair, it was found that tetracycline resistance genes were present in both individuals, but were encoded by different genes and organisms. Interestingly, identical tetracycline resistance gene sequences were identified in diverse bacterial families and even phyla within the mother's gut, suggesting horizontal transfer. Furthermore, in the infant gut, tetracycline resistance genes were exclusively found in Streptococci carrying a composite transposon, indicating the potential for the joint spread of tetracycline and erythromycin resistance within the infant gut environment [73]. These findings highlight the complexity of antibiotic resistance gene transmission within the gut and emphasize the importance of understanding horizontal gene transfer dynamics in microbial communities [73]. This dissemination can occur through mobile genetic elements such as plasmids and transposons, leading to the transfer of resistance genes between different bacterial species [74]. The identification of new conjugative transposons harboring various ribosomal protective Tet genes indicates a significant development in the mechanism of resistance through the horizontal transfer and dissemination of tetracycline resistance. [74].
Moreover, the overuse and misuse of tetracycline antibiotics in humans, food, and veterinary medicine contribute to the selection pressure for tetracycline-resistant bacteria in the gut microbiota [75]. The presence of tetracycline-resistant bacteria in the gut microbiota can compromise the effectiveness of tetracycline antibiotics when used to treat infections, leading to treatment failures and the need for alternative, often more potent, antibiotics [76]. In a case study of 128 Swedish infants, tetracycline resistance genes were found in 12 % of E. coli strains despite no direct tetracycline exposure. These strains carried either the Tet(A) or Tet(B) genes. Surprisingly, the resistance observed was unrelated to antibiotic treatment. Resistant strains frequently harbor virulence factor genes. Although resistant and susceptible strains showed similar persistence and numbers in the gut, their coexistence led to lower counts of resistant strains. Some initially resistant strains lose their Tet genes over time. These findings suggest that there is limited pressure against Tet gene carriage in the human infant gut, emphasizing the need to understand the dynamics of tetracycline-resistant bacteria in the gut microbiota, which can compromise the effectiveness of tetracycline antibiotics [77].
Additionally, disruptions in the composition and diversity of the gut microbiota caused by tetracycline resistance can have broader implications for host health, including increased susceptibility to colonization by pathogenic bacteria, dysregulation of immune function, and alterations in metabolic processes [78]. In a case study involving mice, the administration of tetracycline led to significant changes in the composition and function of the gut microbiota. Specifically, an increase in Firmicutes abundance and a decrease in Bacteroidetes abundance were observed. Metagenomic analyses revealed alterations in microbial functions related to carbohydrate metabolism, ribosomal activity, and cell wall processes. Tetracycline treatment also resulted in an increase in the number of antibiotic resistance genes, plasmids, and integrons in the mouse intestinal microbiota. These findings suggest that fecal discharge spreads antibiotic resistance genes and mobile genetic elements into the environment[78]. Therefore, addressing tetracycline resistance in gut microbiota is crucial for preserving the efficacy of tetracycline antibiotics and maintaining overall gut health.
4. Antibiotic-induced dysbiosis and related diseases
Antibiotics, while effective against pathogens, can significantly disrupt the symbiotic relationship between the host and resident gut microbiota, often leading to antibiotic resistance [35]. This disruption causes dysbiosis, in which beneficial gut microbiota are replaced by opportunistic pathogens, paving the way for a cascade of inflammatory processes within the intestinal milieu [35]. The presence of pathogenic bacterial components in the intestinal mucosa due to antibiotic-induced dysbiosis triggers an immune response [79].
Immune cells such as macrophages and dendritic cells recognize microbial components through pattern recognition receptors such as Toll-like receptors. Activation of these receptors triggers the production of pro-inflammatory cytokines like interleukin-1β, interleukin-6, tumor necrosis factor, and colony-stimulating factor [80]. These cytokines activate the inflammatory cascade and contribute to tissue damage and systemic inflammation. Abnormal metabolites released by an imbalanced gut microbiota can also facilitate inflammatory responses by altering the phenotype and function of immune cells and activating inflammation-related pathways [80].
SCFAs such as indoleacetic acid, gamma-aminobutyric acid, and trimethylamine N-oxide (TMAO) play crucial roles in immune regulation [81]. For instance, SCFAs have anti-inflammatory properties and can promote the differentiation of regulatory T cells (Tregs), which helps suppress excessive inflammation [82]. In contrast, dysbiosis may lead to decreased SCFA production and increased inflammation [82]. SCFAs, including acetate, propionate, and butyrate, are generated through the fermentation of dietary fibers by gut bacteria [83]. These SCFAs exert diverse physiological effects, such as the regulation of immune function, maintenance of gut barrier integrity, and modulation of energy metabolism.
In a case study involving 25 patients with irritable bowel syndrome (IBS), stool samples were analyzed, indicating a decrease in the concentration of SCFAs, such as acetic acid, butyric acid, propionic acid, valeric acid, and isovaleric acid, which serve as reliable biomarkers for IBS [84]. Further studies have suggested that a decrease in SCFA concentration is indicative of antibiotic-associated diarrhea, pouchitis caused by ulcerative colitis, and diversion colitis [85]. Moreover, an analysis of fecal SCFA concentrations in patients with celiac disease, adenomatous polyposis, and colorectal cancer suggested a promising noninvasive diagnostic approach. Another study suggested that the concentration of fecal SCFAs could potentially serve as markers for detecting colorectal cancer and adenomatous polyposis [86].
TMAO, another important gut metabolite, is derived from the microbial metabolism of dietary choline, phosphatidylcholine, and carnitine [87]. During antibiotic-induced dysbiosis, elevated levels of TMAO are associated with an increased risk of neurological and cardiovascular diseases such as atherosclerosis, hypertension, and coronary artery disease [87,88]. TMAO has been implicated in promoting inflammation by activating pathways, such as nuclear factor kappa B, and altering the polarization of macrophages towards the a pro-inflammatory M1 phenotype [87]. Dysbiosis-mediated inflammation contributes to endothelial dysfunction, arterial plaque formation, and systemic inflammation, which are the hallmarks of cardiovascular disorders [89]. Pathogenesis mediated by TMAO involves the activation of inflammatory signaling pathways, including nuclear factor kappa B, nucleotide-binding oligomerization domain, leucine-rich repeat, pyrin domain-containing protein 3 inflammasome, and mitogen-activated protein kinase/c-Jun N-terminal kinase, both in the periphery and brain. These mechanisms elucidate inflammatory processes underlying TMAO-induced pathogenic effects in various tissues and provide insights into potential therapeutic targets for mitigating TMAO-related diseases [87].
Bile acids, which are synthesized in the liver and metabolized by gut microbes, play critical roles in lipid digestion and absorption [90]. Antibiotics significantly affect bile acid metabolism and, consequently, gut microbiota composition and function [91]. By disrupting the balance of microbial communities in the gut, antibiotics can impair the transformation of primary bile acids into secondary or tertiary bile acids, leading to alterations in bile acid profiles [92]. These changes can result in an imbalance in specific bile acids that affect the physiological functions of various microbes. Reduced microbial diversity due to antibiotic use can exacerbate these issues by diminishing the ability of the gut to effectively regulate bile acid metabolism [91]. This disruption increases the abundance of harmful pathogens and reduces the abundance of beneficial microbes, potentially leading to inflammation and compromised intestinal barrier function. Thus, antibiotics influence bile acid metabolism by affecting the gut microbiota, ultimately disrupting gut health and homeostasis.
Dysregulation of bile acid metabolism has also been linked to gastrointestinal disorders such as cholestasis and colorectal cancer, as well as metabolic conditions such as obesity and diabetes [93]. Bile acid dysregulation and gut dysbiosis are implicated in pre-carcinogenic effects such as inflammation, oxidative DNA damage, and heightened cell proliferation [94]. Mechanistically, bile acid dysregulation, mediated by key receptors such as the farnesoid X receptor, apical sodium-dependent bile acid transporter, and TGR5, is linked to enterohepatic carcinogenesis. This highlights the potential of bile acids as cancer markers and sheds light on their mechanistic roles in gastrointestinal carcinogenesis [94].
Similarly, the gut microflora seriously impacts the gut-brain axis, which forms a network connecting the gastrointestinal tract with the brain and involves the gut microbiota, enteric nervous system (ENS), and central nervous system [95]. This system allows the exchange of information through various biochemical signaling pathways [96]. The gut microbiota, comprising trillions of microorganisms, produces metabolites and neurotransmitters, such as serotonin and gamma-aminobutyric acid, which influence brain function and mood [96]. The ENS, a network of neurons within the gut walls, communicates directly with the brain via the vagus nerve, affecting gastrointestinal motility and sensation [97]. Additionally, biochemical signaling involves neurotransmitters, hormones, and immune mediators that affect mood, stress responses, and cognitive functions [96]. Dysregulation of this axis has been linked to mental health disorders such as depression and anxiety, as well as gastrointestinal conditions such as IBS and inflammatory bowel disease (IBD) [98]. Emerging therapeutic approaches, including dietary interventions, probiotics, and prebiotics, aim to restore balance in the gut-brain axis, offering potential benefits for both mental and gut health. Understanding individual variations in this axis can lead to personalized treatments and enhance therapeutic outcomes for patients with gut-related and neurological conditions.
In addition, the overuse of antibiotics can compromise the gut barrier, making it more permeable and allowing pathogens to secrete toxins [99]. These toxins enter the bloodstream and trigger systemic inflammation and immune responses [99]. Alterations in the gut microbiota also affect the metabolism of dietary components, leading to deficiencies and disrupted metabolic functions. Furthermore, a compromised microbial environment heightens susceptibility to infections, including those caused by antibiotic-resistant bacteria and Clostridium difficile [92,100].
Thus, the impact of antibiotic-induced dysbiosis extends far beyond the gastrointestinal tract and encompasses a large network of metabolic and immune-mediated disorders, as shown in Fig. 7 and Table 1. Monitoring the levels of these gut metabolites provides valuable insights into the gut microbiota composition and function and holds promise for the development of diagnostic and therapeutic strategies for a wide range of health conditions. Studies have highlighteds the influence of antibiotics can reduce beneficial bacteria, increase pathogenic strains, and affect SCFA levels, contributing to conditions like obesity, inflammatory bowel disease, and cardiovascular disorders. For instance, the use of ceftriaxone conditionally increases the abundance of pathogenic bacteria such as E. coli and decreases SCFA levels, leading to inflammation and long-term dysbiosis [101]. Conversely, the combination of cefoperazone/sulbactam with SCFAs reduced antibiotic resistance, suggesting a synergistic effect enhancing bacterial susceptibility [102]. Meropenem disrupts beneficial bacterial populations and increases the expression of inflammatory markers, although the microbial composition tends to revert to baseline within 60 d [103]. Aztreonam leads to significant gut disruption with persistent changes even after treatment [103]. Fluoroquinolones such as levofloxacin cause transient shifts in microbial composition, whereas aminoglycosides reduce beneficial taxa and metabolites, resulting in metabolic disturbances and prolonged inflammatory responses [104]. Finally, macrolides diminish the capacity of microbes for carbohydrate metabolism and SCFA synthesis, affecting gastrointestinal function and systemic homeostasis [105].
Fig. 7.
Antibiotic-induced dysbiosis, its inflammatory response, and diseases.
Table 1.
Impact of antibiotics on gut microbiota, metabolism, and systemic homeostasis.
| Antibiotic | Gut Microbiota Composition | Effect on gut metabolite | Effect on Homeostasis | Inflammation makers | Withdrawal Impact | References |
|---|---|---|---|---|---|---|
| Ceftriaxone (β-lactam) | Increase in conditionally pathogenic bacteria (E. coli, Clostridium, Staphylococcus spp.) | Decreased SCFA levels, impaired receptor (FFA2, FFA3) function, altered transporter activity (SMCT1, MCT1, MCT4) | Disrupted oxidant-antioxidant balance, increased epithelial permeability, tissue remodelling | Elevated TNF-α, IL-10 | Long-term dysbiosis, lasting changes in SCFAs, increased susceptibility to colitis even 56 days after withdrawal. | [101] |
| Cefoperazone/Sulbactam, Ceftazidime/Avibactam, Cefepime/Enmetazobactam (β-lactam/β-lactamase inhibitors) | Significant reduction in resistance rates when combined with SCFAs | MIC of SCFAs was 3750 μg/mL (60 mM) against E. coli; SCFAs decreased MIC values for β-lactams | Colonic SCFA concentrations (65–123 mM) significantly suppressed E. coli growth and virulence | Reduction in expression of virulence genes (fliC, ipaH, FimH, BssS) at colonic SCFA concentrations | Withdrawal not discussed; study focused on in vitro effects and immediate bacterial response to SCFAs along with β-lactam | [102] |
| Meropenem (β-lactam) | Significant disruption; increased opportunistic Enterococcaceae | Decrease in SCFA-producing bacteria (e.g., Roseburia, Lachnospiraceae, Ruminococcaceae) | Disruption of gut homeostasis; impaired colonization resistance | Increased IL-1β, IL-6, IL-12, IL-17, TNF-α | Microbial composition returned to pretreatment levels within 60 days; persistent elevated cytokines | [103] |
| Cefoperazone/Sulbactam (β-lactam) | Significant disruption; depletion of beneficial taxa, increased pathogens | Decrease in SCFA production; reduced butyrate-producing taxa | Disruption of gut homeostasis; increased carbohydrate availability | Increased IL-1β, IL-6, IL-12, IL-17, TNF-α | Similar to Meropenem; microbial composition tended to return but cytokine levels elevated | [103] |
| Aztreonam (β-lactam) | Probiotic strains resistant; significant disruption in gut microbiota | No specific effects on SCFA reported | Disruption of gut homeostasis; potential long-term effects | Increased IL-1β, IL-6, IL-12, IL-17, TNF-α | Microbial composition changes persisted long after treatment | [103] |
| Levofloxacin (Fluoroquinolone) | Transient increase in Firmicutes; decrease in Bacteroidetes | No significant effect reported | Return to baseline levels on Day 8 and Day 60 | Similar to control group | Microbial composition returned to baseline levels on Day 8 and Day 60 | [103] |
| Neomycin, Gentamicin (Aminoglycoside) | Significant disruption; reduced beneficial taxa; loss of diversity | 17 metabolites decreased; notably, indole-3-propionic acid and hippuric acid remarkably decreased | Disruption observed; energy metabolism changes | Increased IL-1β, IL-6, IL-12, IL-17, TNF-α | Changes persisted long after treatment | [104] |
| Moxifloxacin, Levofloxacin (Fluoroquinolone) | Increased opportunistic taxa; decreased Firmicutes | 15 metabolites decreased; increased complex lipids; decreased hippuric acid and indole-3-acetic acid | Noted disruption; complex lipids increased | Increased IL-1β, IL-6, IL-12, IL-17, TNF-α | Microbial changes seen; some metabolites elevated post-treatment | [104] |
| Doxycycline, Tetracycline (Tetracycline) | Significant disruption; loss of beneficial taxa | 13 metabolites decreased; 9 increased; decreased hippuric acid and indole-3-acetic acid noted | Disruption observed; changes in energy metabolism | Increased IL-1β, IL-6, IL-12, IL-17, TNF-α | Microbial composition changes persisted | [104] |
| Erythromycin, Azithromycin (Macrolide) | Reduced microbial capacity for carbohydrate metabolism and SCFA biosynthesis | Depletion of keystone bacteria; reduced SCFA biosynthesis; alterations in serotonin (5-HT) and C-peptide | Disruption in metabolic homeostasis; changes in gastrointestinal motility | Reduction in MCP-1, IL-5, and IL-10 (significant with azithromycin); trends for IL-4, IL-6, IL-7, TNF-α, IFN-γ (not significant) | Long-term changes in gut microbiota composition, impacting systemic homeostasis | [105] |
Although TMAO and SCFAs are promising diagnostic biomarkers of gut health and metabolic status, several challenges hinder their clinical application. For instance, variability in production poses a significant obstacle. TMAO synthesis is influenced by dietary precursors such as choline and carnitine, which are predominantly found in animal products, leading to considerable differences in levels among individuals based on their dietary habits[106]. Additionally, gut microbiota diversity affects the production of both TMAO and SCFAs, complicating the interpretation of biomarker levels concerning health status [107]. Clinical interpretation is further complicated by the complexity of dysbiosis, which can elevate TMAO levels associated with cardiovascular diseases and metabolic disorders but is influenced by factors such as inflammation and renal function [108]. Moreover, TMAO and SCFAs lack specificity, as elevated levels may be found in individuals without specific diseases [107]. Analytical challenges also arise in quantifying these biomarkers, as their measurement often requires sophisticated techniques, such as mass spectrometry or high-performance liquid chromatography. Moreover, variabilities in sample collection, processing, and storage can affect the stability and accuracy of biomarkers, highlighting the need for consensus guidelines for measurement protocols[109]. Furthermore, inter-individual variability in metabolic responses adds another layer of complexity because genetic differences and underlying health conditions can lead to varying concentrations of these metabolites [109]. These challenges necessitate a comprehensive understanding of the role of the gut microbiome in health and disease, emphasizing the importance of continued research on the interactions between diet, microbiota composition, and metabolic output. Establishing clear protocols and a consensus on measurement techniques is vital for effectively utilizing TMAO and SCFAs as diagnostic biomarkers in clinical settings, ultimately enhancing disease prevention and management strategies.
4.1. Clinical integration of TMAO and SCFAs as diagnostic biomarkers
The clinical integration of TMAO and SCFAs as diagnostic biomarkers presents several challenges. One major challenge is the variability in the levels of these metabolites influenced by diet, genetics, microbiome composition, and medication use. For instance, TMAO concentrations can fluctuate depending on the dietary intake of choline- and carnitine-rich foods, which affects the standardization of diagnostic thresholds across different populations [110]. Similarly, SCFA levels are influenced by dietary fiber intake, making it challenging to establish universal reference ranges for diagnostic purposes [111]. Furthermore, the complex and dynamic nature of the gut microbiota, which involves many interacting microbial species, complicates the isolation of specific contributions of TMAO and SCFAs to diseases such as cardiovascular disorders and inflammatory conditions [112].
Current methodologies for measuring TMAO and SCFAs, such as liquid chromatography-mass spectrometry, offer high precision and sensitivity; however, they are costly and require specialized laboratory environments, which limit their application in routine clinical diagnostics [113]. To address this challenge, there is a growing need to develop rapid point-of-care diagnostic tools that are both cost-effective and user-friendly while maintaining high accuracy. Recent advancements in biosensor technologies, such as electrochemical sensors, have shown promise for the detection of these metabolites. For instance, electrochemical sensors enhanced with nanomaterials have been explored to quantify indole derivatives and TMAO, showing the potential for more accessible clinical applications owing to their portability and affordability [114]. Additionally, enzyme-linked immunosorbent assays (ELISAs) have been considered an adaptation to detect TMAO, leveraging their established use in clinical laboratories for other biomarkers [115]. Integrating technologies such as electrochemical sensors and ELISA with microfluidic platforms can enhance the miniaturization and automation of SCFA and TMAO detection, improving their clinical utility; however, methods such as fluorescence in situ hybridization and microfluidics face challenges in high-density sample processing. Thus, large-scale studies are needed to validate these approaches in clinical settings to accurately detect metabolites in low-density microbial habitats [116]. Such technological advancements aim to bridge the gap between research settings and real-world clinical applications, making monitoring gut-derived metabolites more feasible for personalized healthcare.
Variability in blood levels of TMAO poses challenges for its use as a biomarker. Multiple factors, including diet, gut microbiota composition, gut-blood barrier permeability, liver enzyme activity, and methylamine excretion rates, influence TMAO levels. These complexities make it challenging to establish a direct correlation between TMAO levels and specific health conditions [106]. For instance, TMAO levels rise rapidly after choline, betaine, and l-carnitine consumption as the gut bacteria metabolize these compounds to trimethylamine (TMA). TMA is then absorbed into the bloodstream and oxidized to TMAO by the liver enzyme flavin-containing monooxygenase-3 (FMO-3). Other FMO forms play little to no role in TMA metabolism in humans [106]. The final elimination of TMAO occurs through urination, sweating, and exhalation, adding another layer of variability. Consequently, these factors must be considered when evaluating TMAO as a reliable biomarker for health assessments [106]. Further challenges in the clinical application of TMAO and SCFAs as biomarkers include their interactions with other metabolic indicators. For instance, TMAO has been implicated in the risk of cardiovascular diseases by promoting inflammation and enhancing platelet reactivity [108]. This suggests that TMAO could serve as a valuable adjunct to traditional cardiovascular risk markers, such as inflammatory cytokines and lipid profiles, offering a more holistic assessment of cardiovascular health [110,117]. Additionally, while SCFAs have demonstrated anti-inflammatory properties, dysbiosis often results in decreased SCFA production, which may indicate underlying health issues such as IBS or metabolic disorders. Therefore, understanding the interplay between SCFAs and other metabolic pathways may enhance their potential as biomarkers in clinical practice [118].
5. Mechanism for the transfer of resistance in gut microbiota
In the intricate gut microbiota ecosystem, antibiotic resistance mechanisms are multifaceted and dynamic, driven by evolutionary pressures and microbial interactions [119]. Horizontal gene transfer is a pivotal mechanism that facilitates the exchange of genetic material, including antibiotic resistance genes, among diverse microbial species [120]. A case study analyzed gut microbiota datasets from mother to child in 283 samples and longitudinal IBD in 148 patient samples, focusing on horizontal gene transfer. That study showed that horizontal gene transfer networks expand in complexity during early life, indicating microbiota transmission from mother to child. The group-specific network edges and communities were identified as potential biomarkers. The intestinal bowel disease patient networks contained more horizontal gene transfer edges in pathogenic genera such as Mycobacterium, Sutterella, and Pseudomonas, and the microbiota of children contained more genera of Bifidobacterium and Escherichia [121]. Through processes such as conjugation, transduction, and transformation, bacteria can acquire resistance genes from their environment or neighboring bacteria, rapidly spreading resistance traits within the microbiota [122].
Mutation and selection play fundamental roles in shaping antibiotic resistance profiles [116]. The evolution of antibiotic resistance is influenced by the mutation supply rate, efficacy of the resistance mechanism, fitness of resistant mutants at varying drug concentrations, and the intensity of selective pressures [123]. Bacterial populations continuously undergo genetic mutations, some of which confer antibiotic resistance. In one case study, pathogenic bacteria such as E. coli, Salmonella, Pseudomonas aeruginosa, Streptococcus pneumoniae, Neisseria meningitidis, and Helicobacter pylori acquired mutators with increased mutation rates, often due to defects in the methyl-directed mismatch repair system. These strains developed antibiotic resistance mutations at significantly higher rates, leading to enhanced resistance levels. Mutators also facilitate horizontal gene transfer, which aids the spread of antibiotic resistance. It has also been observed that a permanent mutator status can reduce fitness due to the accumulation of harmful mutations. Mutators also pose a risk during infection treatment, as they promote the selection of antibiotic-resistant mutants and can refine existing resistance mechanisms [124]. Thus, resistant mutants have a selective advantage under antibiotic pressure, allowing them to proliferate and dominate the microbial community.
Biofilm formation is a sophisticated strategy employed by bacteria to escape the action of antibiotics [125]. Within biofilms, microbial cells are encased in a protective matrix that shields them from antibiotics and immune responses. This protective barrier enhances bacterial survival and promotes the development of antibiotic resistance [126]. A previous study investigated biofilm-forming bacteria and antibiotic resistance in hospital effluent isolates. The results showed the prevalence of antibiotic-resistant and heavy metal-tolerant bacteria, with some producing extended-spectrum β-lactamase enzymes. Toxic metals, such as cadmium and copper, were selected for antibiotic resistance. Certain bacteria exhibited β-hemolysis and other virulence traits [127].
There was a positive correlation between age and the abundance of antibiotic resistance genes within the gut microbiota, indicating the age-related accumulation and complexity of these genes. A study on the relationship between age, gut microbiota, and antibiotic resistance genes was conducted through a metagenomic analysis of fecal samples from 246 individuals. The abundance and complexity of resistance genes in the gut microbiota increased with age, with the older age groups exhibiting the highest levels of these genes. Among the identified genes, Tet(Q), which is associated with tetracycline resistance, was the most prevalent and had the highest occurrence in Bacteroides [128]. Another study investigated the relationship between age and antibiotic resistance in human gut microbiota using DNA microarray analysis. The results indicate the progressive accumulation and higher complexity of antibiotic resistance genes from childhood through adulthood, highlighting the link between age and antibiotic resistance development in the gut microbiota [129]. By uncovering these age-related patterns, that study provided valuable insights for understanding and potentially mitigating antibiotic resistance in the context of the gut microbiota across different age groups.
5.1. Bacterial translocation
The migration of pathogens from the intestinal region to other parts of the body, referred to as bacterial translocation, can occur due to intestinal barrier dysfunction, inflammation, or immune suppression [130]. This process is facilitated by compromised mucosal integrity, changes in the microbial balance induced by antibiotic resistance, and weakened immune responses. Once translocated, pathogens can spread throughout the body, causing infections in distant organs or tissues and potentially leading to conditions such as sepsis [131]. Bacterial translocation highlights the complex relationship among gut health, antibiotic resistance, immune function, and systemic illnesses [132]. Bacterial translocation and its role in systemic diseases can be influenced by antibiotic resistance [133]. In this scenario, motile phagocytes may ingest intestinal bacteria that, if resistant to antibiotics, can survive within these cells during transport to extraintestinal sites. However, failure to effectively eliminate these bacteria intracellularly could release them into the extraintestinal environment [124]. This hypothesis aligns with various observations in the literature. First, antibiotic-resistant bacteria, often classified as facultative intracellular pathogens, are prone to translocate out of the intestinal tract. Second, non-motile intestinal particles, such as antibiotic-resistant strains, can also rapidly translocate from the intestinal lumen. Finally, the bacterial translocation rate may be influenced by antibiotic usage, which modulates immunity, including phagocytic functions [134]. Furthermore, antibiotics can cause gut bacteria to cross the colonic epithelium, triggering inflammation and increasing vulnerability to post-injury diseases [134]. This migration results from reduced microbial signals to colonic goblet cells, which form antigen passages for antibiotic-induced bacterial translocation. This study also suggests that the depletion of these cells, including CX3CR1+ dendritic cells, halts migration, indicating their importance in bacterial translocation [135]. Most antibiotics induce this process after a single dose, with heightened inflammation specifically linked to antibiotics that promote bacterial movement[135]. Collectively, these mechanisms emphasize the adaptability and resilience of the gut microbiota in response to antibiotic exposure, highlighting the urgent need for comprehensive strategies to combat antibiotic resistance and preserve the microbial balance in the gut [135].
7. Future perspectives
Future possibilities regarding the complexity of antibiotic resistance and its impact on gut microbiota dynamics are multifaceted and promise significant advancements in research and clinical practice. Enhancing our understanding of resistance mechanisms will be pivotal, involving investigations into the genetic and metabolic pathways contributing to antibiotic resistance and exploring horizontal gene transfer mechanisms among gut bacteria. This knowledge could pave the way for developing targeted antibiotics designed to specifically combat resistant strains without disrupting beneficial microbiota, potentially in combination with prebiotics or probiotics, to mitigate dysbiosis. Additionally, emerging evidence highlights the efficiency of breast milk in combating antimicrobial resistance, as it contains bioactive components like lactoferrin, oligosaccharides, and antimicrobial peptides, which can inhibit the growth of resistant pathogens and support the development of a healthy microbiome [136].
Regarding therapeutic strategies, advancing fecal microbiota transplantation techniques could restore the microbial balance in individuals affected by antibiotic-induced dysbiosis, while precision probiotics tailored to individual microbiota profiles may further support gut health post-treatment. Metabolomic profiling is another promising avenue that allows researchers to identify biomarkers associated with antibiotic-induced dysbiosis and investigate the role of metabolites in modulating resistance mechanisms. Using bioinformatics tools, artificial intelligence, and machine learning applications could also revolutionize our ability to analyze the complex interactions between antibiotic exposure, gut microbiota composition, and resistance patterns, leading to the development of predictive models for antibiotic resistance based on microbiota profiles. Furthermore, alternative therapeutic strategies, such as bacteriophage therapy and antimicrobial peptides, warrant exploration to treat infections while preserving the gut microbiota diversity. Employing advanced methods such as ERIC-PCR for quickly and affordably determining the clonal relationships of MDR isolates holds great potential. This approach could improve antimicrobial resistance tracking, especially in low-resource regions, and support the development of precise strategies to combat its spread globally [27]. Public health initiatives emphasizing prudent antibiotic use and establishing surveillance systems to monitor resistance patterns in the population are essential to combat the antibiotic resistance crisis. Longitudinal studies are needed to assess the long-term effects of antibiotic use on gut microbiota dynamics and resistance, along with the development of regulatory frameworks to ensure the safe use of antibiotics in clinical and agricultural settings. Finally, personalized medical approaches, including tailoring antibiotic regimens based on individual microbiota composition and incorporating pharmacogenomics, have the potential to minimize the emergence of disruption and resistance. Collectively, these possibilities highlight the need for a multidisciplinary approach to address the intricate relationship between antibiotic resistance and gut microbiota dynamics to ultimately enhance health outcomes and effectively manage antibiotic resistance. In conclusion, the relationship between antibiotic resistance and gut microbiota dynamics poses healthcare challenges. Understanding how resistance mechanisms affect beneficial microbes is essential for developing targeted treatments that minimize disruption of the gut microbiome. Future research should focus on uncovering the genetic and metabolic pathways of resistance and exploring strategies, such as precision probiotics and fecal microbiota transplantation, to restore microbial balance.
CRediT authorship contribution statement
H. Shayista: Writing – original draft. M.N. Nagendra Prasad: Writing – review & editing, Project administration, Formal analysis. S. Niranjan Raj: Writing – review & editing, Formal analysis. Ashwini Prasad: Visualization. S. Lakshmi: Resources. H.K. Ranjini: Data curation. K. Manju: Data curation. Ravikumara: Resources. Raghuraj Singh Chouhan: Methodology. Olga Y. Khohlova: Methodology. Olga V. Perianova: Resources. Syed Baker: Writing – review & editing, Visualization, Project administration, Methodology, Investigation, Conceptualization.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
All authors express their gratitude to Karnataka State Open University for providing the necessary facilities to carry out this work. The authors also extend their heartfelt thanks to the Editor and reviewers for their constructive feedback, which has significantly contributed to uplifting the manuscript's standards and aligning it with the journal's guidelines.
The Authors would like to thank Karnataka State Open University, Mysore for providing the infrastructure to carry out the present study.
Data Availability Statement
The data used to support the findings of this study have been included in the article.
References
- 1.Hugon P., Lagier J.-C., Colson P., Bittar F., Raoult D. Repertoire of human gut microbes. Microb. Pathog. 2017;106:103–112. doi: 10.1016/j.micpath.2016.06.020. [DOI] [PubMed] [Google Scholar]
- 2.Flint H.J., Scott K.P., Duncan S.H., Louis P., Forano E. Microbial degradation of complex carbohydrates in the gut. Gut. Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.den Besten G., van Eunen K., Groen A.K., Venema K., Reijngoud D.-J., Bakker B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013;54:2325–2340. doi: 10.1194/jlr.r036012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mondot S., De Wouters T., Doré J., Lepage P. The human gut microbiota and its dysfunctions. Digest. Dis. 2013;31:278–285. doi: 10.1159/000354678. [DOI] [PubMed] [Google Scholar]
- 5.Sirisinha S. The potential impact of gut on your health: Current status and future challenges. Asian Pac. J. Allergy Immunol. 2016 doi: 10.12932/ap0803. [DOI] [PubMed] [Google Scholar]
- 6.A. Muthaiyan, Determinants of the gut microbiota, gut microbiota and its impact on health and diseases. (2020) 19–62. 10.1007/978-3-030-47384-6_2.
- 7.Carding S., Verbeke K., Vipond D.T., Corfe B.M., Owen L.J. Dysbiosis of the gut microbiota in disease. Microbial. Ecol. Health Dis. 2015;26 doi: 10.3402/mehd.v26.26191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Modi S.R., Collins J.J., Relman D.A. Antibiotics and the gut microbiota. J. Clin. Investig. 2014;124:4212–4218. doi: 10.1172/jci72333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R., et al. Enterotypes of the human gut microbiota. Nat. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koropatkin N.M., Cameron E.A., Martens E.C. How glycan metabolism shapes the human gut microbiota. Nat. Rev. Microbiol. 2012;10(5):323–335. doi: 10.1038/nrmicro2746. 10 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shanahan F. The host–microbe interface within the gut. Best Pract. Res. Clin. Gastroenterol. 2002;16:915–931. doi: 10.1053/bega.2002.0342. [DOI] [PubMed] [Google Scholar]
- 12.S.A. Nasseri, A.C. Lazarski, I.L. Lemmer, C.Y. Zhang, E. Brencher, H.-M. Chen, et al., Functional metagenomics reveals an alternative, broad-specificity pathway for metabolism of carbohydrates in human gut commensal bacteria. (2024). 10.1101/2024.03.25.586180.
- 13.Schloissnig S., Arumugam M., Sunagawa S., Mitreva M., Tap J., Zhu A., Waller A., Mende D.R., Kultima J.R., Martin J., Kota K., Sunyaev S.R., Weinstock G.M., Bork P. Genomic variation landscape of the human gut microbiota. Nat. 2012;493(7430):45–50. doi: 10.1038/nature11711. 493 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sommer F., Anderson J.M., Bharti R., Raes J., Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 2017;15(10):630–638. doi: 10.1038/nrmicro.2017.58. 15 (2017) [DOI] [PubMed] [Google Scholar]
- 15.Rooks M.G., Garrett W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016;16(6):341–352. doi: 10.1038/nri.2016.42. 16 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janeckova L., Kostovcikova K., Svec J., Stastna M., Strnad H., Kolar M., et al. Unique gene expression signatures in the intestinal mucosa and organoids derived from germ-free and monoassociated mice. Int. J. Mol. Sci. 2019;20:1581. doi: 10.3390/ijms20071581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krych L., Hansen C.H., Hansen A.K., van den Berg F.W., Nielsen D.S. Quantitatively different, yet qualitatively alike: A meta-analysis of the mouse core gut microbiota with a view towards the human gut microbiota. PLoS. One. 2013;8 doi: 10.1371/journal.pone.0062578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan I., Bai Y., Zha L., Ullah N., Ullah H., Shah S.R., et al. Mechanism of the gut microbiota colonization resistance and enteric pathogen infection. Front. Cell Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.716299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawley T.D., Walker A.W. Intestinal colonization resistance. Immunol. 2013;138:1–11. doi: 10.1111/J.1365-2567.2012.03616.X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salam M.A., Al-Amin M.Y., Salam M.T., Pawar J.S., Akhter N., Rabaan A.A., Alqumber M.A.A. Antimicrobial resistance: a growing serious threat for global public health. Healthcare. 2023;11:1946. doi: 10.3390/HEALTHCARE11131946. 11 (2023) 1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansson K., Brenthel A. Imagining a post-antibiotic era: A cultural analysis of crisis and antibiotic resistance. Med. Humanit. 2022;48:381–388. doi: 10.1136/medhum-2022-012409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murray C.J., Ikuta K.S., Sharara F., Swetschinski L., Aguilar G.Robles, Gray A., et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet. 2022;399:629–655. doi: 10.1016/s0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chinemerem Nwobodo D., Ugwu M.C., Oliseloke Anie C., Al-Ouqaili M.T., Chinedu Ikem J., Victor Chigozie U., et al. Antibiotic resistance: The challenges and some emerging strategies for tackling a Global Menace. J. Clin. Lab. Anal. 2022;36 doi: 10.1002/jcla.24655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Antimicrobial resistance, (n.d.). https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed April 27, 2024).
- 25.Ahmed S.K., Hussein S., Qurbani K., Ibrahim R.H., Fareeq A., Mahmood K.A., Mohamed M.G. Antimicrobial resistance: Impacts, challenges, and future prospects. J. Med. Surg. Public Health. 2024;2 doi: 10.1016/J.GLMEDI.2024.100081. [DOI] [Google Scholar]
- 26.Raouf F.E.A., Benyagoub E., Alkhudhairy M.K., Akrami S., Saki M. Extended-spectrum beta-lactamases among Klebsiella pneumoniae from Iraqi patients with community-acquired pneumonia. Rev. Assoc. Med. Bras. 1992;68(2022):833–837. doi: 10.1590/1806-9282.20220222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saki M., Amin M., Savari M., Hashemzadeh M., Seyedian S.S. Beta-lactamase determinants and molecular typing of carbapenem-resistant classic and hypervirulent Klebsiella pneumoniae clinical isolates from southwest of Iran. Front. Microbiol. 2022;13 doi: 10.3389/FMICB.2022.1029686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saki M., Farajzadeh Sheikh A., Seyed-Mohammadi S., Asareh Zadegan Dezfuli A., Shahin M., Tabasi M., Veisi H., Keshavarzi R., Khani P. Occurrence of plasmid-mediated quinolone resistance genes in Pseudomonas aeruginosa strains isolated from clinical specimens in southwest Iran: a multicentral study. Sci. Rep. 2022;12(1):1–8. doi: 10.1038/s41598-022-06128-4. 12 (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farajzadeh Sheikh A., Moradi Bandbal M., Saki M. Emergence of multidrug-resistant Shigella species harboring extended-spectrum beta-lactamase genes in pediatric patients with diarrhea from southwest of Iran. Mol. Biol. Rep. 2020;47:7097–7106. doi: 10.1007/S11033-020-05776-X. [DOI] [PubMed] [Google Scholar]
- 30.Garbacz K., Wierzbowska M., Kwapisz E., Kosecka-Strojek M., Bronk M., Saki M., Międzobrodzki J. Distribution and antibiotic-resistance of different Staphylococcus species identified by matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) isolated from the oral cavity. J. Oral Microbiol. 2021;13 doi: 10.1080/20002297.2021.1983322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yalew G.T., Muthupandian S., Hagos K., Negash L., Venkatraman G., Hagos Y.M., Meles H.N., Weldehaweriat H.H., Al-Dahmoshi H.O.M., Saki M. Prevalence of bacterial vaginosis and aerobic vaginitis and their associated risk factors among pregnant women from northern Ethiopia: A cross-sectional study. PLoS One. 2022;17 doi: 10.1371/JOURNAL.PONE.0262692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levy S. Microbial resistance to antibiotics. Lancet. 1982;320:83–88. doi: 10.1016/s0140-6736(82)91701-9. [DOI] [PubMed] [Google Scholar]
- 33.Zhao Q., Chen Y., Huang W., Zhou H., Zhang W. Drug-microbiota interactions: an emerging priority for precision medicine. Signal. Transduct. Target. Ther. 2023;8(1):1–27. doi: 10.1038/s41392-023-01619-w. 8 (2023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu L., Surathu A., Raplee I., Chockalingam A., Stewart S., Walker L., et al. The effect of antibiotics on the gut microbiota: a metagenomics analysis of microbial shift and gut antibiotic resistance in antibiotic treated mice. BMC Genomics. 2020;21 doi: 10.1186/s12864-020-6665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Francino M.P. Antibiotics and the human gut microbiota: dysbioses and accumulation of resistances. Front. Microbiol. 2016;6 doi: 10.3389/fmicb.2015.01543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pérez-Cobas A.E., Gosalbes M.J., Friedrichs A., Knecht H., Artacho A., Eismann K., et al. Gut microbiota disturbance during antibiotic therapy: A multi-omic approach. Gut. 2012;62:1591–1601. doi: 10.1136/gutjnl-2012-303184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raplee I., Walker L., Xu L., Surathu A., Chockalingam A., Stewart S., et al. Emergence of nosocomial associated opportunistic pathogens in the gut microbiota after antibiotic treatment. Antimicrob. Resist. Infect. Control. 2021;10 doi: 10.1186/s13756-021-00903-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Q., Cheng L., Wang J., Hao M., Che H. Antibiotic-induced gut microbiota dysbiosis damages the intestinal barrier, increasing food allergy in adult mice. Nutrients. 2021;13:3315. doi: 10.3390/nu13103315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dahiya D., Nigam P.S. Antibiotic-therapy-induced gut dysbiosis affecting gut microbiota—brain axis and cognition: Restoration by intake of probiotics and Synbiotics. Int. J. Mol. Sci. 2023;24:3074. doi: 10.3390/ijms24043074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Garneau-Tsodikova S., Labby K.J. Mechanisms of resistance to aminoglycoside antibiotics: Overview and perspectives. Medchemcomm. 2016;7:11–27. doi: 10.1039/c5md00344j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pradier L., Bedhomme S. Ecology, more than antibiotics consumption, is the major predictor for the global distribution of aminoglycoside-modifying enzymes. Elife. 2023:12. doi: 10.7554/elife.77015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shao H., Zhang C., Xiao N., Tan Z. Gut microbiota characteristics in mice with antibiotic-associated diarrhea. BMC Microbiol. 2020;20 doi: 10.1186/s12866-020-01999-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sparo M., Delpech G., García Allende N. Impact on public health of the spread of high-level resistance to gentamicin and vancomycin in enterococci. Front. Microbiol. 2018;9 doi: 10.3389/fmicb.2018.03073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang S.S., Apisarnthanarak A., Hsu L.Y. Mechanisms of β-lactam antimicrobial resistance and epidemiology of major community- and healthcare-associated multidrug-resistant bacteria. Adv. Drug Deliv. Rev. 2014;78:3–13. doi: 10.1016/j.addr.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 45.Lima L.M., da Silva B.N.M., Barbosa G., Barreiro E.J. β-lactam antibiotics: An overview from a medicinal chemistry perspective. Eur. J. Med. Chem. 2020;208 doi: 10.1016/J.EJMECH.2020.112829. [DOI] [PubMed] [Google Scholar]
- 46.Davies D.T., Everett M. Designing inhibitors of β-lactamase enzymes to overcome carbapenem resistance in gram-negative bacteria. Acc. Chem. Res. 2021;54:2055–2064. doi: 10.1021/acs.accounts.0c00863. [DOI] [PubMed] [Google Scholar]
- 47.Zapun A., Contreras-Martel C., Vernet T. Penicillin-binding proteins and β-lactam resistance. FEMS Microbiol. Rev. 2008;32:361–385. doi: 10.1111/j.1574-6976.2007.00095.x. [DOI] [PubMed] [Google Scholar]
- 48.Lu S., Huang Q., Wei B., Chen Y. Effects of β-lactam antibiotics on gut microbiota colonization and metabolites in late preterm infants. Curr. Microbiol. 2020;77:3888–3896. doi: 10.1007/s00284-020-02198-7. [DOI] [PubMed] [Google Scholar]
- 49.Liu L., Wang Q., Lin H., Das R., Wang S., Qi H., et al. Amoxicillin increased functional pathway genes and beta-lactam resistance genes by pathogens bloomed in intestinal microbiota using a simulator of the human intestinal microbial ecosystem. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.01213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Livanos A.E., Greiner T.U., Vangay P., Pathmasiri W., Stewart D., McRitchie S., et al. Antibiotic-mediated gut microbiota perturbation accelerates development of type 1 diabetes in mice. Nat. Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Lastours V., Fantin B. Impact of fluoroquinolones on human microbiota. Focus on the emergence of antibiotic resistance. Future Microbiol. 2015;10:1241–1255. doi: 10.2217/fmb.15.40. [DOI] [PubMed] [Google Scholar]
- 52.Jeong S.-H., Song Y.-K., Cho J.-H. Risk assessment of ciprofloxacin, flavomycin, olaquindox and colistin sulfate based on microbiological impact on human gut biota. Regul. Toxicol. Pharmacol. 2009;53:209–216. doi: 10.1016/j.yrtph.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 53.Brar R.K., Jyoti U., Patil R.K., Patil H.C. Fluoroquinolone antibiotics: An overview. Adesh Univ. J. Med. Sci. Res. 2020;2:26–30. doi: 10.25259/aujmsr_12_2020. [DOI] [Google Scholar]
- 54.Park S.-K., Kim K.-J., Lee S.-O., Yang D.-H., Jung K.W., Ye B.D., et al. Ciprofloxacin usage and bacterial resistance patterns in Crohn's disease patients with abscesses. J. Clin. Gastroenterol. 2014;48:703–707. doi: 10.1097/mcg.0000000000000024. [DOI] [PubMed] [Google Scholar]
- 55.Zhu S., Li H., Liang J., Lv C., Zhao K., Niu M., et al. Assessment of oral ciprofloxacin impaired gut barrier integrity on gut bacteria in mice. Int. Immunopharmacol. 2020;83 doi: 10.1016/j.intimp.2020.106460. [DOI] [PubMed] [Google Scholar]
- 56.Edlund Charlotta, Beyer G., Hiem M. Comparative effects of moxifloxacin and clarithromycin on the normal intestinal microflora. Scand. J. Infect. Dis. 2000;32:81–85. doi: 10.1080/00365540050164272. [DOI] [PubMed] [Google Scholar]
- 57.Vázquez-Laslop N., Mankin A.S. How macrolide antibiotics work. Trends. Biochem. Sci. 2018;43:668–684. doi: 10.1016/j.tibs.2018.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leclercq R. Mechanisms of resistance to macrolides and lincosamides: Nature of the resistance elements and their clinical implications. Clin. Infect. Dis. 2002;34:482–492. doi: 10.1086/324626. [DOI] [PubMed] [Google Scholar]
- 59.Nakajima Y. Mechanisms of bacterial resistance to macrolide antibiotics. J. Infect. Chemotherapy. 1999;5:61–74. doi: 10.1007/s101560050011. [DOI] [PubMed] [Google Scholar]
- 60.Pickering H., Hart J.D., Burr S., Stabler R., Maleta K., Kalua K., et al. Impact of azithromycin mass drug administration on the antibiotic-resistant gut microbiota in children: A randomized, controlled trial. Gut. Pathog. 2022;14 doi: 10.1186/s13099-021-00478-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Milanović V., Osimani A., Cardinali F., Litta-Mulondo A., Vignaroli C., Citterio B., et al. Erythromycin-resistant lactic acid bacteria in the healthy gut of vegans, ovo-lacto vegetarians and omnivores. PLoS One. 2019;14 doi: 10.1371/journal.pone.0220549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lathakumari R.H., Vajravelu L.K., Satheesan A., Ravi S., Thulukanam J. Antibiotics and the gut microbiota: Understanding the impact on human health. Med. Microecol. 2024;20 doi: 10.1016/j.medmic.2024.100106. [DOI] [Google Scholar]
- 63.Chopyk J., Cobián Güemes A.G., Ramirez-Sanchez C., Attai H., Ly M., Jones M.B., et al. Common antibiotics, azithromycin and amoxicillin, affect gut metagenomics within a household. BMC Microbiol. 2023;23 doi: 10.1186/s12866-023-02949-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Choo J.M., Martin A.M., Taylor S.L., Sun E., Mobegi F.M., Kanno T., et al. The impact of long-term macrolide exposure on the gut microbiota and its implications for metabolic control. Microbiol. Spectr. 2023;11 doi: 10.1128/spectrum.00831-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jaroszewski J., Mamun N., Czaja K. Bidirectional interaction between tetracyclines and gut microbiome. Antibiotics. 2023;12:1438. doi: 10.3390/antibiotics12091438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Speer B.S., Shoemaker N.B., Salyers A.A. Bacterial resistance to tetracycline: Mechanisms, transfer, and clinical significance. Clin. Microbiol. Rev. 1992;5:387–399. doi: 10.1128/cmr.5.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Cristobal R.E. Multidrug resistance pump acrAB-tolC is required for high-level, tet(A)-mediated tetracycline resistance in Escherichia coli. J. Antimicrobial Chemotherapy. 2006;58:31–36. doi: 10.1093/jac/dkl172. [DOI] [PubMed] [Google Scholar]
- 68.Beheshti M., Ardebili A., Beheshti F., Lari A.R., Siyadatpanah A., Pournajaf A., et al. Tetracycline resistance mediated by Tet efflux pumps in clinical isolates of Acinetobacter baumannii. Rev. Do Instituto Medicina Tropical São Paulo. 2020;62 doi: 10.1590/s1678-9946202062088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levy S.B. Active efflux, a common mechanism for biocide and antibiotic resistance. J. Appl. Microbiol. 2002;92 doi: 10.1046/j.1365-2672.92.5s1.4.x. [DOI] [PubMed] [Google Scholar]
- 70.Aires J., Doucet-Populaire F., Butel M.J. Tetracycline resistance mediated by tet (W), tet (M), and tet (O) genes of Bifidobacterium isolates from humans. Appl. Environ. Microbiol. 2007;73:2751–2754. doi: 10.1128/aem.02459-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang W., Moore I.F., Koteva K.P., Bareich D.C., Hughes D.W., Wright G.D. TetX is a flavin-dependent monooxygenase conferring resistance to tetracycline antibiotics. J. Biol. Chem. 2004;279:52346–52352. doi: 10.1074/jbc.m409573200. [DOI] [PubMed] [Google Scholar]
- 72.Akinbowale O.L., Peng H., Barton M.D. Diversity of tetracycline resistance genes in bacteria from aquaculture sources in Australia. J. Appl. Microbiol. 2007;103:2016–2025. doi: 10.1111/j.1365-2672.2007.03445.x. [DOI] [PubMed] [Google Scholar]
- 73.de Vries L.E., Vallès Y., Agersø Y., Vaishampayan P.A., García-Montaner A., Kuehl J.V., et al. The gut as reservoir of antibiotic resistance: Microbial diversity of tetracycline resistance in mother and infant. PLoS. One. 2011;6 doi: 10.1371/journal.pone.0021644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roberts M.C. Update on acquired tetracycline resistance genes. FEMS Microbiol. Lett. 2005;245:195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 75.Granados-Chinchilla F., Rodríguez C. Tetracyclines in food and feedingstuffs: From regulation to analytical methods, bacterial resistance, and environmental and Health Implications. J. Anal. Methods Chem. 2017:1–24. doi: 10.1155/2017/1315497. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jetty P., Gaddam S., Padi S.S. Emergence of third generation tetracyclines: New magic bullets to tackle antibiotic resistance in the post-antibiotic era. Asian J. Res. Infect. Dis. 2023;13:1–14. doi: 10.9734/ajrid/2023/v13i4271. [DOI] [Google Scholar]
- 77.Karami N., Nowrouzian F., Adlerberth I., Wold A.E. Tetracycline resistance in Escherichia coli and persistence in the infantile colonic microbiota. Antimicrob. Agents Chemother. 2006;50:156–161. doi: 10.1128/aac.50.1.156-161.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yin J., Zhang X.-X., Wu B., Xian Q. Metagenomic insights into tetracycline effects on microbial community and antibiotic resistance of Mouse Gut. Ecotoxicology. 2015;24:2125–2132. doi: 10.1007/s10646-015-1540-7. [DOI] [PubMed] [Google Scholar]
- 79.Aguilera M., Cerdà-Cuéllar M., Martínez V. Antibiotic-induced dysbiosis alters host-bacterial interactions and leads to colonic sensory and motor changes in mice. Gut Microbes. 2015;6:10–23. doi: 10.4161/19490976.2014.990790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Swer N.M., Venkidesh B.S., Murali T.S., Mumbrekar K.D. Gut microbiota-derived metabolites and their importance in neurological disorders. Mol. Biol. Rep. 2022;50:1663–1675. doi: 10.1007/s11033-022-08038-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li M., van Esch B.C.A.M., Wagenaar G.T.M., Garssen J., Folkerts G., Henricks P.A.J. Pro- and anti-inflammatory effects of short chain fatty acids on immune and endothelial cells. Eur. J. Pharmacol. 2018;831:52–59. doi: 10.1016/j.ejphar.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 83.Scott K.P., Duncan S.H., Flint H.J. Dietary fibre and the gut microbiota. Nutr. Bull. 2008;33:201–211. doi: 10.1111/j.1467-3010.2008.00706.x. [DOI] [Google Scholar]
- 84.Farup P.G., Rudi K., Hestad K. Faecal short-chain fatty acids - A diagnostic biomarker for irritable bowel syndrome? BMC Gastroenterol. 2016;16 doi: 10.1186/s12876-016-0446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mortensen P.B., Clausen M.R. Short-chain fatty acids in the human colon: Relation to gastrointestinal health and disease. Scand. J. Gastroenterol. 1996;31:132–148. doi: 10.3109/00365529609094568. [DOI] [PubMed] [Google Scholar]
- 86.Niccolai E., Baldi S., Ricci F., Russo E., Nannini G., Menicatti M., et al. Evaluation and comparison of short chain fatty acids composition in gut diseases. World J. Gastroenterol. 2019;25:5543–5558. doi: 10.3748/wjg.v25.i36.5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Praveenraj S.S., Sonali S., Anand N., Tousif H.A., Vichitra C., Kalyan M., et al. The role of a gut microbial-derived metabolite, trimethylamine N-oxide (TMAO), in neurological disorders. Mol. Neurobiol. 2022;59:6684–6700. doi: 10.1007/s12035-022-02990-5. [DOI] [PubMed] [Google Scholar]
- 88.Mutengo K.H., Masenga S.K., Mweemba A., Mutale W., Kirabo A. Gut microbiota dependant trimethylamine N-oxide and hypertension. Front. Physiol. 2023;14 doi: 10.3389/fphys.2023.1075641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Livshits G., Kalinkovich A. Inflammaging as a common ground for the development and maintenance of sarcopenia, obesity, cardiomyopathy and dysbiosis. Ageing Res. Rev. 2019;56 doi: 10.1016/j.arr.2019.100980. [DOI] [PubMed] [Google Scholar]
- 90.Chiang J.Y. Bile acid metabolism and signaling. Compr. Physiol. 2013:1191–1212. doi: 10.1002/cphy.c120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Larabi A.B., Masson H.L.P., Bäumler A.J. Bile acids as modulators of gut microbiota composition and function. Gut. Microbes. 2023;15 doi: 10.1080/19490976.2023.2172671. PMID: 36740850; PMCID: PMC9904317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Andreescu M. Molecular insights into the role of gut microbiota in antibiotic therapy selection and resistance mitigation. Cureus. 2023;15 doi: 10.7759/cureus.50318. PMID: 38089944; PMCID: PMC10714069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Joyce S.A., Gahan C.G.M. Disease-associated changes in bile acid profiles and links to altered gut microbiota. Digest. Dis. 2017;35:169–177. doi: 10.1159/000450907. [DOI] [PubMed] [Google Scholar]
- 94.Tsuei J., Chau T., Mills D., Wan Y.-J.Y. Bile acid dysregulation, gut dysbiosis, and gastrointestinal cancer. Exp. Biol. Med. 2014;239:1489–1504. doi: 10.1177/1535370214538743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Carabotti M., Scirocco A., Maselli M.A., Severi C. The gut-brain axis: interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015;28:203–209. PMID: 25830558; PMCID: PMC4367209. [PMC free article] [PubMed] [Google Scholar]
- 96.Chen Y., Xu J., Chen Y. Regulation of neurotransmitters by the gut microbiota and effects on cognition in neurological disorders. Nutrients. 2021;13:2099. doi: 10.3390/nu13062099. PMID: 34205336; PMCID: PMC8234057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Geng Z.H., Zhu Y., Li Q.L., Zhao C., Zhou P.H. Enteric nervous system: The bridge between the gut microbiota and neurological disorders. Front. Aging Neurosci. 2022;14 doi: 10.3389/fnagi.2022.810483. PMID: 35517052; PMCID: PMC9063565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bartocci B., Dal Buono A., Gabbiadini R., Busacca A., Quadarella A., Repici A., Mencaglia E., Gasparini L., Armuzzi A. Mental illnesses in inflammatory bowel diseases: mens sana in corpore sano. Medicina (Kaunas) 2023;59:682. doi: 10.3390/medicina59040682. PMID: 37109640; PMCID: PMC10145199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shah T., Baloch Z., Shah Z., Cui X., Xia X. The intestinal microbiota: Impacts of antibiotics therapy, colonization resistance, and diseases. Int. J. Mol. Sci. 2021;22:6597. doi: 10.3390/ijms22126597. PMID: 34202945; PMCID: PMC8235228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hou K., Wu Z.X., Chen X.Y., et al. Microbiota in health and diseases. Signal. Transd. Targeted Therapy. 2022;7:135. doi: 10.1038/s41392-022-00974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Holota Y., Dovbynchuk T., Kaji I., Vareniuk I., Dzyubenko N., Chervinska T., et al. The long-term consequences of antibiotic therapy: Role of colonic short-chain fatty acids (SCFA) system and intestinal barrier integrity. PLoS One. 2019;14 doi: 10.1371/journal.pone.0220642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kadry A.A., El-Antrawy M.A., El-Ganiny A.M. Impact of short chain fatty acids (scfas) on antimicrobial activity of new β-lactam/β-lactamase inhibitor combinations and on virulence of escherichia coli isolates. J. Antibiot. (Tokyo) 2023;76:225–235. doi: 10.1038/s41429-023-00595-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gu S.-L., Gong Y., Zhang J., Chen Y., Wu Z., Xu Q., et al. Effect of the short-term use of fluoroquinolone and β-lactam antibiotics on mouse gut microbiota. Infect. Drug Resist. 2020;13:4547–4558. doi: 10.2147/idr.s281274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Behr C., Kamp H., Fabian E., Krennrich G., Mellert W., Peter E., et al. Gut microbiome-related metabolic changes in plasma of antibiotic-treated rats. Arch. Toxicol. 2017;91:3439–3454. doi: 10.1007/s00204-017-1949-2. [DOI] [PubMed] [Google Scholar]
- 105.Choo J.M., Martin A.M., Taylor S.L., Sun E., Mobegi F.M., Kanno T., et al. The impact of long-term macrolide exposure on the gut microbiome and its implications for metabolic control. Microbiol. Spectr. 2023;11 doi: 10.1128/spectrum.00831-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Nowiński A., Ufnal M. Trimethylamine N -oxide: A harmful, protective or diagnostic marker in lifestyle diseases? Nutrition. 2018;46:7–12. doi: 10.1016/j.nut.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 107.Rüb A.M., Tsakmaklis A., Gräfe S.K., Simon M.-C., Vehreschild M.J., Wuethrich I. Biomarkers of human gut microbiota diversity and dysbiosis. Biomark. Med. 2021;15:139–150. doi: 10.2217/bmm-2020-0353. [DOI] [PubMed] [Google Scholar]
- 108.Shayista H., Prasad M.N.N., Raj S.N., Ranjini H.K., Manju K., Baker S. Mechanistic overview of gut microbiota and mucosal pathogens with respect to cardiovascular diseases. Microbiology. 2024;5 doi: 10.1016/j.microb.2024.100160. [DOI] [Google Scholar]
- 109.Kruger K., Myeonghyun Y., van der Wielen N., Kok D.E., Hooiveld G.J., Keshtkar S., et al. Evaluation of inter- and intra-variability in gut health markers in healthy adults using an optimised faecal sampling and processing method. Sci. Rep. 2024;14 doi: 10.1038/s41598-024-75477-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tang W.H.W., Wang Z., Levison B.S., Koeth R.A., Britt E.B., Fu X., et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 2013;368:1575–1584. doi: 10.1056/nejmoa1109400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Louis P., Flint H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2016;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 112.Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., et al. Host-gut microbiota metabolic interactions. Science (1979) 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 113.Zhu W., Gregory J.C., Org E., Buffa J.A., Gupta N., Wang Z., et al. Gut microbial metabolite TMAO enhances platelet hyperreactivity and thrombosis risk. Cell. 2016;165:111–124. doi: 10.1016/j.cell.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yadav A.K., Verma D., Sajwan R.K., Poddar M., Yadav S.K., Verma A.K., et al. Nanomaterial-based electrochemical Nanodiagnostics for human and gut metabolites diagnostics: Recent advances and challenges. Biosensors. 2022;12:733. doi: 10.3390/bios12090733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang L., Xie F., Tang H., Zhang X., Hu J., Zhong X., et al. Gut microbial metabolite tmao increases peritoneal inflammation and peritonitis risk in peritoneal dialysis patients. Transl. Res. 2022;240:50–63. doi: 10.1016/j.trsl.2021.10.001. [DOI] [PubMed] [Google Scholar]
- 116.Lloyd-Price J., Abu-Ali G., Huttenhower C. The healthy human microbiome. Genome Med. 2016;8 doi: 10.1186/s13073-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang Z., Klipfell E., Bennett B.J., Koeth R., Levison B.S., DuGar B., et al. Gut Flora Metabolism of phosphatidylcholine promotes cardiovascular disease. Nature. 2011;472:57–63. doi: 10.1038/nature09922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Koh A., De Vadder F., Kovatcheva-Datchary P., Bäckhed F. From dietary fiber to host physiology: Short-chain fatty acids as key bacterial metabolites. Cell. 2016;165:1332–1345. doi: 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- 119.Lerner A., Matthias T., Aminov R. Potential effects of horizontal gene exchange in the human gut. Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.01630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zeng X., Lin J. Factors influencing horizontal gene transfer in the intestine. Anim. Health Res. Rev. 2017;18:153–159. doi: 10.1017/s1466252317000159. [DOI] [PubMed] [Google Scholar]
- 121.Li C., Chen J., Li S.C. Understanding horizontal gene transfer network in human gut microbiota. Gut. Pathog. 2020;12 doi: 10.1186/s13099-020-00370-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.McInnes R.S., McCallum G.E., Lamberte L.E., van Schaik W. Horizontal transfer of antibiotic resistance genes in the human gut microbiota. Curr. Opin. Microbiol. 2020;53:35–43. doi: 10.1016/j.mib.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 123.Hughes D., Andersson D.I. Evolutionary trajectories to antibiotic resistance. Annu. Rev. Microbiol. 2017;71:579–596. doi: 10.1146/annurev-micro-090816-093813. [DOI] [PubMed] [Google Scholar]
- 124.Chopra I., O'Neill A.J., Miller K. The role of mutators in the emergence of antibiotic-resistant bacteria. Drug Resist. Updates. 2003;6:137–145. doi: 10.1016/s1368-7646(03)00041-4. [DOI] [PubMed] [Google Scholar]
- 125.Lin D., Chen K., Guo J., Ye L., Li R., Chan E.W., et al. Contribution of biofilm formation genetic locus, pgaabcd, to antibiotic resistance development in gut microbiota. Gut. Microbes. 2020;12 doi: 10.1080/19490976.2020.1842992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lewis K. Multidrug tolerance of biofilms and Persister cells. Curr. Top. Microbiol. Immunol. 2008:107–131. doi: 10.1007/978-3-540-75418-3_6. [DOI] [PubMed] [Google Scholar]
- 127.Nath S., Sinha A., Singha Y.S., Dey A., Bhattacharjee N., Deb B. Prevalence of antibiotic-resistant, toxic metal-tolerant and biofilm-forming bacteria in hospital surroundings. Environ. Anal. Health Toxicol. 2020;35 doi: 10.5620/eaht.2020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wu L., Xie X., Li Y., Liang T., Zhong H., Ma J., et al. Metagenomics-based analysis of the age-related cumulative effect of antibiotic resistance genes in gut microbiota. Antibiotics. 2021;10:1006. doi: 10.3390/antibiotics10081006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lu N., Hu Y., Zhu L., Yang X., Yin Y., Lei F., et al. DNA microarray analysis reveals that antibiotic resistance-gene diversity in human gut microbiota is age related. Sci. Rep. 2014;4 doi: 10.1038/srep04302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wiest R. Bacterial translocation. Biosci. Microflora. 2005;24:61–90. doi: 10.12938/bifidus.24.61. [DOI] [Google Scholar]
- 131.Wang C., Li Q., Ren J. Microbiota-immune interaction in the pathogenesis of gut-derived infection. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.01873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jin S., Wetzel D., Schirmer M. Deciphering mechanisms and implications of bacterial translocation in human health and disease. Curr. Opin. Microbiol. 2022;67 doi: 10.1016/j.mib.2022.102147. [DOI] [PubMed] [Google Scholar]
- 133.Yu L.C.-H., Shih Y.-A., Wu L.-L., Lin Y.-D., Kuo W.-T., Peng W.-H., et al. Enteric dysbiosis promotes antibiotic-resistant bacterial infection: Systemic dissemination of resistant and commensal bacteria through epithelial transcytosis. Am. J. Physiol.-Gastrointestinal Liver Physiol. 2014;307 doi: 10.1152/ajpgi.00070.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wells C.L., Maddaus M.A., Simmons R.L. Proposed mechanisms for the translocation of intestinal bacteria. Clin. Infect. Dis. 1988;10:958–979. doi: 10.1093/clinids/10.5.958. [DOI] [PubMed] [Google Scholar]
- 135.Knoop K.A., McDonald K.G., Kulkarni D.H., Newberry R.D. Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut. 2015;65:1100–1109. doi: 10.1136/gutjnl-2014-309059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Baker S., Niranjan Raj.S., Manju K., Ranjini H.K., Shayista H. Efficacy of breast milk components against microbial pathogens to combat drug-resistance. Microbe. 2023;1 doi: 10.1016/J.MICROB.2023.100010. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study have been included in the article.