Abstract
BioPROTACs consist of a target-binding unit and a component of the ubiquitin-proteasome system. However, the specific biophysical features influencing their effectiveness are poorly understood. We investigated the design principles defining the target-binding moiety of bioPROTACs. We used our recently developed assay for accurately measuring degradation rates, based on microinjection and live-cell microscopy, independent of other confounding factors like biosynthesis and transport. We used a very efficient UPS interaction domain from CHIP E3 ligase, and 9 different well-characterized DARPins to test degradation of the proof-of-principle target eGFP. All but two DARPins work well in this context, one sterically preventing E2 binding in the complex, the other overlapping with the target ubiquitination epitope. Affinity and thermodynamic stability of the binders had only a modest role. BioPROTACs constructed in this way were able to degrade eGFP catalytically. DARPins by themselves could also accelerate degradation of bound GFP, using other cellular E3 systems, but in a non-catalytic manner. The most important factor for efficient degradation by a bioPROTAC in trans is the correct orientation of the complex for target ubiquitination and presentation to the proteasome, still to be determined empirically. The strategies developed here show an efficient pathway to characterize and optimize such systems.
Subject terms: Ubiquitylated proteins, Proteasome, Fluorescence imaging, X-ray crystallography, Protein design
Systematic characterization of bioPROTACs reveals determinants of efficient target degradation by the ubiquitin-proteasome system.
Introduction
Selective protein degradation plays a crucial role in maintaining cellular homeostasis as well as protein quality control and is responsible for regulating a wide range of cellular processes. Most cellular proteins undergo degradation by the ubiquitin-proteasome-system (UPS)1. The proteasomal pathway involves the degradation of proteins by the proteasome, a large complex of enzymes that recognizes and degrades proteins that have been marked for degradation by the addition of polyubiquitin chains. Target ubiquitination is achieved by a cascade of E1, E2, and E3 enzymes, whereby ubiquitin is most often covalently attached to lysine residues on the target protein and on ubiquitin itself2. Also, serine, threonine as well as cysteine ubiquitination have been reported3. The specificity of substrate ubiquitination is conferred by an estimated number of 600–700 different E3 ligases4 that bind degron motifs and thereby select targets for degradation.
Degradation signals that induce regulated protein degradation are still poorly understood, but it is believed that successful degradation is dependent on three factors: an E3 binding site (degron), one or more ubiquitination sites in a defined surface region, thereby influencing proteasomal interaction, and an unstructured initiation site as the starting point for proteasomal unfolding in the proximity of the ubiquitination site5. Degradation signals that are separated onto different subunits within protein complexes can also act in trans to degrade specific subunits that carry a favorable initiation site6. Proteins lacking an initiation site or with an initiation site of the wrong amino acid composition, for example, have been shown to escape degradation despite ubiquitination7–9.
Targeted protein degradation is an emerging field of drug discovery which uses the ubiquitin proteasome system to specifically degrade a target protein of choice. It has potential therapeutic applications in a range of diseases, including cancer, neurodegeneration, and genetic disorders10–12. The approach functions by recruitment of an E3 ligase complex in close proximity of a target protein, which leads to target ubiquitination and subsequent proteasomal degradation. Targeted protein degradation offers several advantages over traditional modes of actions of drugs, such as competitive inhibition by small molecules. Molecules used for targeted protein degradation have been termed PROTACs (proteolysis-targeting chimeras) and contain a binding moiety for a pocket on the target protein as well as a binding moiety for an E3 ligase, attached by a linker. PROTACs have the potential to provide longer-lasting effects, because they induce the degradation of the target protein, which means that their effects might be sustained even after the drug has been cleared from the body. Moreover, they may allow application at lower dosage because of the potential catalytic character of the drug. Currently, numerous small-molecule PROTACs are undergoing pre-clinical and clinical development, directed to specific binding sites on the androgen and estrogen receptors, as well as Bruton’s kinase13.
Small-molecule PROTACs come with their own set of caveats. Finding small-molecule binders to the target can be challenging for many proteins, and so far, only a limited number of E3 ligases are available for targeting. Therefore, the engagement of “undruggable” targets without defined binding pockets may not be accessible. Also, the linker length between target moiety and E3 binding moiety needs to be carefully optimized since rigid small molecule PROTACs require a defined stereometry of the binding entities with defined linker length for assembly of the ternary complex, all while maintaining cell permeability and acceptable pharmacokinetics14.
To circumvent these problems, targeted protein degradation strategies have been expanded to protein-based bispecific degraders containing designed protein binders to engage with arbitrary target proteins15. Since protein binders mostly engage with larger target surfaces in folded or unstructured domains, no binding pockets are required. We collectively refer to such entities as “bioPROTACs”, regardless of the underlying binding domain, such as DARPins, antibodies, nanobodies or monobodies. By using protein binders, high specificity and versatility can be achieved. Several of these bioPROTACs were effective in degrading target proteins such as GFP-tagged PCNA and Ras, and combinations of different E3 adapter proteins and small-protein binders were tested15–18.
Despite these promising results, general design principles for efficient degradation by bioPROTACs with regard to binding affinity, epitope, geometry and perhaps unknown factors have remained elusive. It has remained unclear why some PROTACs fail to degrade a target despite binding to both the target and E3 efficiently19. Further, we do not fully understand the signals that control ubiquitination or how ubiquitin modifications are interpreted in the cell. In the present study, we aimed at expanding the knowledge of degradation signals in binders and complexes in general, as well as for the rational design of bioPROTACs by systematically characterizing target binders separately, and in combination with CHIP E3 ligase as fully functional bioPROTACs. To produce a fair comparison, we measured here, for the first time, directly the actual degradation rates of degrader complexes and the proof-of-principle target GS-eGFP by a recently developed method involving microinjection and live-cell fluorescence microscopy20. This single-cell approach avoids entangling the actual rates of degradation with the kinetics of biosynthesis or cellular uptake.
For the rational design of protein binders, we examined DARPins and focused on several properties, including their degradation rates themselves, structural stability, affinity, lysine content and the binding epitope. The latter has become of interest since it was found that an initiation site is required for effective degradation21. Ultimately, we found two different criteria that determine whether a designed degrader is effective. Using our degradation assay, we also showed that a functioning bioPROTAC with an eGFP-binding DARPin was able to degrade more than one GS-eGFP molecule, thereby confirming catalytic behavior in the living cell. Our findings provide new insights into degradation principles in general, as well as serve as a basis for designed bioPROTAC engineering.
Results
DARPin-based bioPROTAC efficiency shows high variation, despite high sequence similarities
To elucidate efficiency-defining principles for DARPin-based bioPROTAC design, we genetically fused 9 different eGFP-binders22 with the UPS-interacting domain of CHIP23 (C-terminus of Hsc70-interacting protein, Uniprot Q9UNE7) Ubox-type E3 ligase. Our aim was to elucidate the correlation between degradation efficiency of eGFP and characteristics of the selected DARPins.
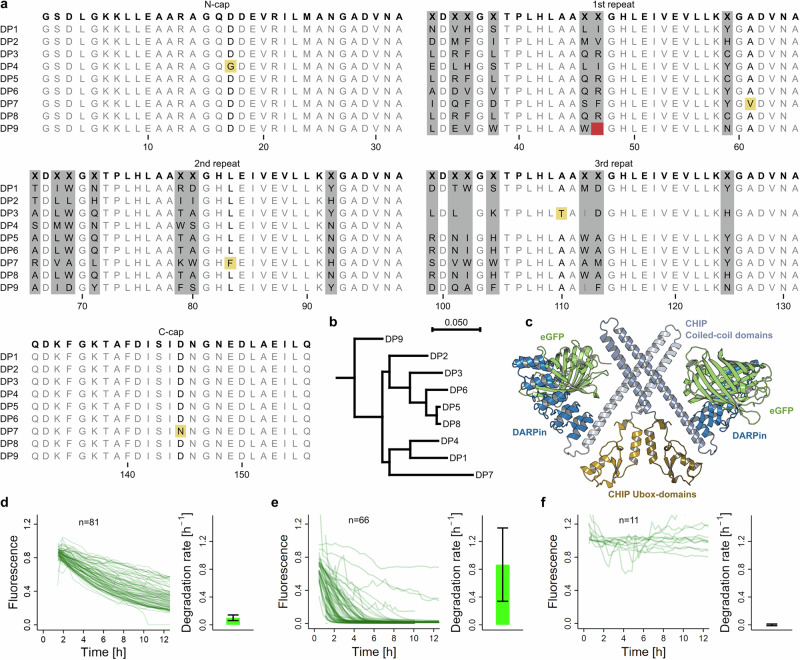
The DARPins used for bioPROTAC design represent the most promising binders selected by ribosome display24, which have previously been assessed for affinity and specificity to eGFP22. As indicated in the alignment shown in Fig. 1a, the selected DARPins (DP1-9) contain either two or three internal repeats, which are surrounded by constant N- and C-terminal capping repeats25. The relationship between the DARPin sequences is shown in a dendrogram (Fig. 1b). The UPS interaction domain, represented by the N-terminally truncated version of human E3 ligase CHIP (CHIPΔTPR), was previously reported for its potent function in bioPROTAC design26,27 and contains the Ubox-domain that interacts with E2 enzymes, and a coiled-coil domain facilitating dimerization, which is required for proteasome targeting ubiquitination activity in wtCHIP28,29. A structural model of the active bioPROTAC dimer is shown in Fig. 1c, based on the previously observed CHIP homo- instead of heterodimerization when TPR repeats are removed30. eGFP was selected as a proof-of-concept target due to its suitability for quantification using fluorescence-based readouts of our kinetic assay20. Importantly, in our experiments, we have used GS-eGFP, which comprises two additional N-terminal amino acids, Gly-Ser, compared to eGFP31 (resulting in the N-terminal sequence GSMVSK). We could previously show that proteasomal unfolding and degradation of eGFP without this N-terminal extension is hindered after ubiquitination, because it would require N-terminal engagement with the proteasome20. Protein sequences of all constructs used in this study are listed in Supplementary Data 1.
Fig. 1. DARPins used in Ubox-based bioPROTACs and degradation rate determination by fluorescence measurement.
a Sequence alignment of eGFP-binding DARPins. The library consensus is shown in bold letters on the top. Randomized positions are highlighted in gray and labeled with character “X”. Mutations outside of randomized regions, acquired by directed evolution, are colored in yellow. A deletion in the 1st repeat of DP9 is colored in red. b Dendrogram of DARPin sequences created in CLC with tree construction method of neighbor joining and Jukes-Cantor protein distance measure. c Model of a CHIP-based dimeric bioPROTAC/eGFP complex. The monomer bioPROTAC structures were produced using AlphaFold and consist of the N-terminal DARPin (blue), followed by the CHIP E3 ligase coiled-coil region (light and dark gray) and Ubox-domain (light and dark yellow) that interact with E2 enzymes (not shown). The orientation of the two monomeric CHIPΔTPR domains is taken from the structure PDB ID: 2F42. The DARPin:GFP orientation is derived from PDB ID: 5MAD. d–f Representative single cell fluorescence signals of cells injected with specific analyte (respective left graphs) and mean rate constant derived from exponential decay fit of the single cell fluorescence signals (respective right graphs); error bars represent standard deviations. d GS-eGFP injection from one exemplary experiment. e DP6-bioPROTAC+GS-eGFP. f DP6-bioPROTAC+GS-eGFP with added proteasome inhibitor.
To simultaneously assess degradation rates of bioPROTAC and target, we used an assay involving the cytosolic introduction of purified protein using microinjection and subsequent live-cell fluorescent microscopy20. While GS-eGFP degradation can be followed directly by its fluorescence, the bioPROTAC is fused to a fluorescent dye with very fast clearance from the cell such that bioPROTAC degradation is also rate-limiting for its fluorescence decrease20. The fluorescent TMR5-maleimide dye was coupled to intrinsic cysteines of the CHIPΔTPR domain (Supplementary Fig. S1), and labeling was verified by ESI-MS analysis (Supplementary Figs. S2–S10). Prior to microinjection into HEK293 cells, we formed 1:1 complexes of purified target and fluorescently labeled bioPROTAC-dimers (see cartoon in Fig. 2c and SDS-PAGEs and representative complex purification profiles in Supplementary Fig. S11), thereby mitigating the potential influence of pre-interaction degradation or off-target interactions on the observed degradation rates. Approximately 30 single-cell degradation rates were measured for each analyte to calculate the mean degradation rate. Specific numbers of injected cells are shown in Supplementary Tables ST1 and ST2. Single-cell and average degradation data can be found in Supplementary Data 2. The statistical significance of the differences between various constructs is presented in Supplementary Figs. S12–S16. Examples of single-cell fluorescence signals of specific analytes and resulting mean rate constants are shown in Fig. 1d–f. Relatively large standard deviations were observed for the degradation rates, consistent with previous reports on degradation rate intercellular variability from us and others20,32,33. This variation likely reflects differences in the cell-to-cell intracellular concentrations of limiting components within the degradation machinery or related regulatory factors.
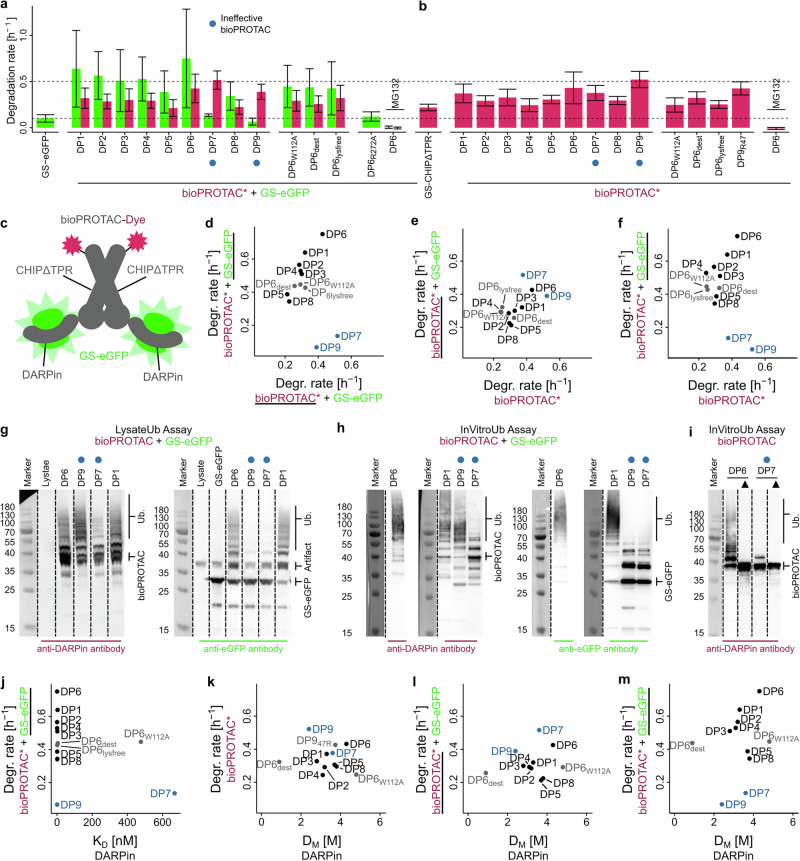
Fig. 2. Design and mechanism of bioPROTACs, based on genetic fusion of DARPins and CHIP E3 ligase.
Asterisks (*) indicate dye label. a, b Mean degradation rate constants of DARPin-CHIPΔTPR bioPROTACS, labeled with TMR dye (red bars), either alone (b) or in a one-to-one complex with GS-eGFP (green bars, a), determined by microinjection into HEK293 cells. Error bars represent standard deviations. Numbers of analyzed cells per analyte are shown in Supplementary Table ST1. Ineffective bioPROTACs are indicated by a blue circle. c Graphical representation of labeled DARPin-CHIPΔTPR bioPROTAC dimer in complex with GS-eGFP used for degradation determination. d–f Plots comparing the indicated degradation rate constants shown in (a, b), each axis shows the underlined compound. g–i Western blot images of ubiquitination reactions. Samples were detected with either anti-eGFP antibody or anti-DARPin polyclonal antibody to show GS-eGFP or bioPROTAC ubiquitination. Shown are images from single experiments. To image marker bands and antibody-stained bands, the same membranes were imaged in two separate channels. Uncropped membranes are shown in Supplementary Fig. S17. g Western blot images resulting from the LysateUb assay, based on HEK293 lysates and involving GS-eGFP/bioPROTAC complexes. h, i Western blot images of InVitroUb assays, an in vitro ubiquitination assay with purified E1, E2 and ubiquitin with bioPROTACs (i) or GS-eGFP/bioPROTAC complexes (h). Black triangle (▲) shows controls without added ubiquitination reaction components. j Graph comparing DARPin affinities determined by SPR with indicated degradation rate constant of GS-eGFP. k–m Comparison of DARPin denaturation midpoints, determined by guanidinium-induced unfolding, and degradation rate constants. Pearson’s correlation coefficients (r) are 0.86 for (d) when ignoring the two outliers in blue, 0.58 for (e), and 0.70 for (f) when ignoring the two outliers in blue. The r values for j–m are between 0.21 and −0.41, therefore not supporting a correlation. For a and b, the number of analyzed cells are shown in Supplementary Table ST1.
As displayed in Fig. 2a, GS-eGFP degradation rates were increased in the presence of most bioPROTACs, while the bioPROTACs themselves were generally degraded at lower rates. Comparisons of GS-eGFP and bioPROTAC degradation rates within those complexes showed a ratio of approximately 1.8 (Fig. 2d). This suggests a substoichiometric degradation mechanism for active bioPROTACs, which is discussed in a later section. The highest increase in GS-eGFP turnover was induced by DP6-bioPROTAC, which modulated GS-eGFP half-life from 7 h to approximately 55 min. Confirmation that this increase in GS-eGFP degradation is indeed promoted by the CHIP/Ubox mechanism was achieved by the introduction of the point mutation R272A in the E2-interacting CHIP domain, which resulted in degradation rates similar to GS-eGFP alone. A complete halt of degradation was achieved by the addition of the proteasome inhibitor MG132. In contrast to all other bioPROTAC constructs, those derived from DP7 and DP9 did not alter GS-eGFP degradation, thereby demonstrating that the exact nature of the binder is very important for degradation efficiency. We explore the nature of these differences in the subsequent sections.
We initially asked whether bioPROTAC efficiencies are influenced by degradation rates of the bioPROTACs themselves, independent of their target. Half-life differences, obtained from microinjection experiments, exhibited some variation ranging from 1.3 h (DP9-bioPROTAC) to 2.8 h (DP4-bioPROTAC), as shown in Fig. 2b. All constructs displayed a slight increase in degradation compared to CHIPΔTPR, which most likely stems from differences in DARPin ubiquitination, as discussed below. The degradation kinetics of bioPROTACs are unlikely to be affected by the binding of GS-eGFP, as shown by the similar degradation rates observed for bioPROTACs alone and when forming a complex with GS-eGFP (Fig. 2e). Consequently, the linear relationship between degradation rates within a single complex (Fig. 2d) remains consistent when comparing the degradation of GS-eGFP with that of the respective free bioPROTACs (Fig. 2f). One possible explanation for this linear relationship is that the performance of a bioPROTAC is influenced by its ubiquitination efficacy, impacting not only the target’s but also the bioPROTAC’s degradation through self-ubiquitination.
Ubiquitination is a qualitative predictor of bioPROTAC performance
The lack of GS-eGFP degradation induced by the DP7- and DP9-based bioPROTACs (Fig. 2a) was concomitant with a lack of GS-eGFP polyubiquitination, as assessed using an assay using HEK293 lysate, with results shown in Fig. 2g (raw images shown in Supplementary Fig. S17). These results could be reproduced in an orthogonal in vitro ubiquitination assay, based on a defined mixture of purified components of the UPS, as shown in Fig. 2h. A key difference between these approaches is the presence of the complete UPS in the HEK293 lysate-based ubiquitination assay (“LysateUb assay”), likely involving a spectrum of E3 ligases, additional E2 enzymes, and the accompanying complex regulatory system. This is in contrast to the in vitro ubiquitination assay (“InVitroUb assay”), which contains no other E3 ligases and only defined components of the UPS, thus reporting only on CHIP-based ubiquitination. Importantly, both assays allow for parallel detection of both bioPROTAC and target.
Ubiquitin chain formation on the bioPROTAC itself was detected for active bioPROTACs DP1 and DP6 in both assays (Figs. 2g–i and 3c). This was expected in light of the reported CHIP auto-ubiquitination34, which can serve as a reporter for the effectiveness of CHIP/E2 interaction35. Among the two non-functional bioPROTACs, DP9-bioPROTAC showed auto-ubiquitination, while no ubiquitination of DP7-bioPROTAC was observed using the InVitroUb assay, with results shown in Fig. 2h. The lack of DP7-bioPROTAC auto-ubiquitination, even in the absence of GS-eGFP, was also confirmed using InVitroUb assays, as shown in Fig. 2i. This observation strongly suggests the involvement of different and distinct mechanisms causing the inability of DP7- and DP9-based bioPROTACs to induce GS-eGFP degradation. Notably, the LysateUb assay (Fig. 2g) showed polyubiquitination for all bioPROTACs (although slightly reduced for DP7), indicating the engagement of alternative pathways in HEK293, either concurrently with CHIP, or exclusively in cases where CHIP functionality is compromised, as was observed for DP7-bioPROTAC. One possible explanation for the lack of auto-ubiquitination is hindrance of E2 engagement caused by competing non-specific interactions. However, thus far, fluorescence anisotropy measurements and SEC have failed to demonstrate any interaction between DP7 and either Ubox domain or E2. But it is noteworthy that DP7 generally showed some unspecific interactions to column material in SEC (Supplementary Fig. S18), as well as in SPR experiments.
Fig. 3. DARPin binding epitopes and GS-eGFP ubiquitination.
a Mass spectrometric analysis (MS-MS) of ubiquitination sites after in vitro ubiquitination of GS-eGFP, induced by DP6-based bioPROTAC. The sample was digested in the three indicated ways to improve coverage. Ubiquitinated lysines carry a covalent GG or LRGG adduct and are circled in different colors. For representative spectra, see Supplementary Fig. S27. b Structure of eGFP (gray, PDB ID: 5MAD) in complex with DP6 (yellow, PDB ID: 5MA6) and DP9 (blue, PDB ID: 5MAD) created in PyMOL. To create bioPROTACs, the CHIPΔTPR domain is genetically fused at the indicated C-terminus. GS-eGFP N- and C-termini are shown in black and purple, respectively. eGFP lysine ubiquitination sites upon DP6-bioPROTAC engagement are indicated in the same colors as shown in (a). The C-terminal K241 is not indicated, since the residue is not resolved in the structure, but close to the indicated C-terminus (M233, purple). c–e Assays involving GS-eGFP mutants. GS-eGFP_mut1: K104R_K159R, GS-eGFP_mut2: K6R_K104R_K159R, GS-eGFP_mut3: K104R_K159R_K241R, GS-eGFP_mut4: K6R_K104R_K159R_K241R. c Western-blot of InVitroUb ubiquitination assay of DP6-bioPROTAC in complex with indicated GS-eGFP lysine mutants. Samples were detected with either anti-eGFP antibody or anti-DARPin polyclonal antibody to show GS-eGFP or bioPROTAC ubiquitination. To image marker bands and antibody-stained bands, the same membranes were imaged in two separate channels. Uncropped membranes are shown in Supplementary Fig. S17. d Degradation rate constants determined by microinjection into HEK293 cells of GS-eGFP and GS-eGFP mutants alone (/) or in complex with DP6-bioPROTAC or inactive DP6-bioPROTACR272A mutant. Shown are average degradation rate constants of single cells. Error bars represent standard deviations. Numbers of analyzed cells per analyte are shown in Supplementary Table ST1. The four lysines or arginines within the GS-eGFP mutants are color-coded according to mutated lysine residues as shown in (a) and (b). e Western-blot of LysateUb ubiquitination assay of DP6-bioPROTAC or inactive DP6-bioPROTACR272A mutant alone or in complex with indicated GS-eGFP lysine mutants. Samples were detected by anti-eGFP antibody to show GS-eGFP ubiquitination. f–k Structures of indicated DARPins (blue) in complex with eGFP (gray) determined by x-ray crystallography. PDB IDs: DP1 9F22, DP2 9F23, DP3 6MWQ, DP4 9F24, DP6 5MA6 (differs in 5 mutations in the C-cap compared to our construct), DP9 5MAD. Models of DP5, DP7, and DP8 are shown in Supplementary Fig. S30. Crystallographic data are shown in Table 2. For d, the number of analyzed cells are shown in Supplementary Table ST1.
Affinity modulation has little impact on degradation efficiency
Target affinity would be a likely parameter to influence bioPROTAC degradation dynamics. On the one hand, excessive affinity may impede catalytic turnover by degrading the bioPROTAC quantitatively, or prevent degradation altogether, as was shown for a covalent PROTAC against Bruton’s Tyrosine Kinase36. Conversely, insufficient affinity might limit ubiquitination and thereby degradation by insufficient engagement time with the target.
We determined the affinity of the DARPins to GS-eGFP using surface plasmon resonance (SPR) and fluorescence anisotropy (Supplementary Figs. S19, S20 and Table 1) and found no direct correlation of affinity with GS-eGFP degradation rate (Fig. 2j). Most constructs exhibited high affinities in the low nano- to picomolar range. Despite this strong interaction, the measured higher degradation rates of the target compared to the bioPROTAC support a degradation mechanism with multiple turnovers, which constitutes promising prospects for drug design strategies using such high-affinity small protein binders. Importantly, the high affinity of the DP9-bioPROTAC for GS-eGFP suggests other factors as the determinant for the absence of target degradation. The only binder that showed markedly lower affinity was DP7. However, since several possible alternative explanations for the lack of ubiquitination by DP7-bioPROTAC could not be unambiguously verified, no definitive correlation regarding lower affinity and degradation could be inferred in this case.
Table 1.
Properties of eGFP-binding DARPins
| Name | Original Name | KD (Anisotropy) [nM] | KD (SPR) [nM] | kon (SPR)[M−1 s−1 106] | koff(SPR) [s−1 10−4] | DM (GdmCl) [M] n = 2 | Tm [°C] |
|---|---|---|---|---|---|---|---|
| DP1 | 3G190.24 | nd | 2.76 | 2.56 | 70.8 | 3.3 | 73 |
| DP2 | 2G156 | nd | 0.235 | 6.62 | 15.6 | 3.2 | 71 |
| DP3 | 3G86.32 | <5 (Ref. 22) | 1.38 | 0.68 | 9.4 | 2.8 | 69 |
| DP4 | 2G71 | nd | 0.108 | 48.4 | 52.4 | 3.1 | 65 |
| DP5 | 3G168 | <5 (Ref. 22) | 0.682 | 0.929 | 6.34 | 3.7 | >90 °C |
| DP6 | 3G124 | <5 (Ref. 22) | 0.271 | 2.09 | 5.67 | 4.3 | >90 °C |
| DP6dest | – | nd | 3.94 | 1.53 | 60.3 | 0.9 | nd |
| DP6W112A | – | 25,210 ± 10,000 | 477 | nd | nd | 4.8 | >90 °C |
| DP6lysfree | – | nd | 1.07 | 3.76 | 0.353 | nd | 87 |
| DP7 | 3G146 | 669 ± 156 | nd | nd | nd | 3.6 | 81 |
| DP8 | 3G86.1 | <5 (Ref. 22) | 1.34 | 0.767 | 10.2 | 3.8 | >90 °C |
| DP9 | 3G61 | 76 ± 21 (Ref. 22) | 0.376 | 3.42 | 12.9 | 2.4 | 57 |
Kinetics were determined by SPR spectroscopy (Supplementary Fig. S19). KD values of select DARPins were determined by fluorescence anisotropy (Supplementary Fig. S20). Denaturation midpoints (DM) were determined by guanidinium chloride unfolding measured with CD-spectroscopy (Supplementary Fig. S21). Melting temperatures as indicated were determined by SYPRO orange dye binding on a qPCR cycler (Supplementary Fig. S23).
nd not determined.
For this reason, we wished to directly test target affinity as a determinant of bioPROTAC performance. We therefore deliberately decreased the affinity of binder DP6, an excellent degrader (Fig. 2a), by introducing a Trp to Ala mutation in position 112 (DP6W112A-bioPROTAC, see structural representation in Fig. 4j). This resulted in a 1000-fold reduction of affinity to 477 nM, as determined in SPR measurements, and even lower affinity estimated from fluorescence anisotropy measurements (see Supplementary Figs. S19 and S20), while structural integrity was unchanged, as shown in a denaturant-based unfolding assay (Supplementary Fig. S21). Degradation of the GS-eGFP complex by DP6W112A-bioPROTAC was similar to the other active bioPROTACs and only moderately reduced from wt DP6-bioPROTAC (Fig. 2a, j), while also retaining polyubiquitination of the complex, as shown by LysateUb assay (Supplementary Fig. S22a). Taken together, our results suggest that target affinity in the ranges investigated alone does not determine degradation efficiency at the intracellular concentrations injected. It is conceivable that degradation performance could be reduced for very low-affinity binders or concentrations significantly below the median injection concentrations of 450 nM.
Fig. 4. Influence of DARPin binding on GS-eGFP degradation.
a–c Degradation rate constants of different DARPins or DARPin/GS-eGFP complexes. Shown are average degradation rate constants of single cells as determined upon microinjection of one-to-one complexes into the cytosol of living cells in combination with fluorescent live-cell imaging. Error bars indicate standard deviations. Numbers of analyzed cells per analyte are shown in Supplementary Table ST1. a Unlabeled DARPin/GS-eGFP complexes, GS-eGFP degradation rates shown in green. b DARPins C-terminally labeled with TMR dye (red bars) in complex with GS-eGFP (green bars). Asterisk (*) indicates dye label. c DARPins injected alone, C-terminally labeled with TMR dye (red bars). d, e Comparison of indicated degradation rate constants from (a–c). f Comparison of DARPin denaturation midpoints determined by GdmCl-induced equilibrium unfolding with DARPin degradation rates from (c). DP6 and DP9 and their variants with destabilizing and stabilizing mutations are shown in gold and cyan. g Degradation rate ratios within complexes of DARPin/GS-eGFP shown in (b) and bioPROTAC/GS-eGFP complexes from Fig. 2a. h Exemplary western blot of ubiquitination assays of DARPin/GS-eGFP complexes in HEK293 lysate performed as biological triplicates. Samples were split and loaded onto two separate SDS-PAGE gels for western-blotting with anti-eGFP antibody or anti-DARPin serum to show GS-eGFP or DARPin ubiquitination. To image marker bands and antibody-stained bands, the same membranes were imaged in two separate channels. Uncropped membranes are shown in Supplementary Fig. S17. i Quantification of GS-eGFP (green bars) and DARPin (pink bars) ubiquitination intensities from blots shown in (h) as well as two replicates shown in Supplementary Fig. S35. Error bars represent standard deviations derived from three biological replicates. Quantification was performed using the unedited blots shown in Supplementary Fig. S17. Ubiquitination signal intensities (upper rectangle) was normalized to the respective GS-eGFP or DARPin band (lower rectangle). A one-way ANOVA coupled with Tukey’s multiple comparison test was used to test for significance of differences within pairs of data. Significant differences are shown by an asterisk (*). j Crystal structure of DP6/eGFP complex (PDB ID: 5MA6). Mutated residues for DP6dest are shown in blue (L8A in each repeat) and red (L24A in each repeat) and W112 as critical interacting residue is shown in yellow. k–o Degradation rate constants of DP6 variants alone and in complex with GS-eGFP. Asterisks (*) indicate dye label. k DP6 variants alone, C-terminally labeled with TMR dye. l DP6 variants C-terminally labeled with TMR dye (red bars) in complex with GS-eGFP (green bars). m Degradation rates of unlabeled DP6 variants in complex with GS-eGFP. n Degradation rates of DP6 variants in complex with GS-eGFP with inhibitor controls and N-terminal acetylation. o Degradation rates of DP1 and DP4 variants in complex with GS-eGFP. Pearson’s correlation coefficients (r) are 0.68 for (e) and 0.97 for (g). In contrast, r values are 0.16 for (d) and −0.38 for (f), therefore not supporting a correlation. For a–c and k–o, the number of analyzed cells are shown in Supplementary Table ST1.
Structural stability is not a reliable predictor for bioPROTAC performance
In addition to the observed variations in ubiquitination, structural stability might be of relevance for the bioPROTACs’ performance. Low structural stability can potentially cause bioPROTAC unfolding and fast degradation of the bioPROTAC itself by attracting other E3 ligases and enhanced proteasomal unfolding efficiency. This, in turn, could impede target degradation.
Given that all constructs share the CHIPΔTPR domain, it was anticipated that differences in stability would arise primarily due to variations in the DARPins. The stability of DARPins alone, without fusion to other domains, was therefore determined by GdmCl-induced equilibrium unfolding, measured by circular dichroism (CD, Supplementary Fig. S21 and Table 1). For comparison of the DARPins among each other, the main transition midpoint was used, which probably represents the unfolding of the internal repeats together with the N-cap37. Denaturation midpoints in GdmCl assays ranged from 2.4 to 4.3 M GdmCl. It should be noted that most DARPins used here do not follow a two-state transition37, thereby precluding the extraction of ΔG values. We additionally used a thermal unfolding assay based on the binding of SYPRO Orange to hydrophobic regions, which become exposed upon partial unfolding (Supplementary Fig. S23 and Table 1). Both assays give a similar ranking of the DARPins, as shown in a comparison displayed in Supplementary Fig. S24.
No correlation of bioPROTAC degradation rates with the DARPins’ structural stability was observed (Fig. 2k, l). This might be attributed to the low variation amongst bioPROTAC degradation rates as well as denaturation midpoints. DARPins are generally very stable proteins due to their rigid structure and rapid refolding37. With unfolding at 2.4 M GdmCl or 57 °C, DP9 showed the lowest stability and fastest bioPROTAC degradation. To elucidate a possible causal link, the stability of DP9 was increased by re-introducing an arginine in position 47. This mutation significantly increased structural stability (Supplementary Fig. S21) but abrogated GS-eGFP binding ability in SEC purification. Degradation of DP9-bioPROTAC with and without stabilizing 47 R was similar (Fig. 2b), which indicates that their stability (within the ranges investigated here) is unlikely the main determinant of degradation differences. In summary, the degradation of DP9-bioPROTAC itself in complex with GS-eGFP is comparable to the other active bioPROTACs, implying the absence of a causal relationship between the bioPROTACs’ degradation rates and the rates of the target’s degradation.
DARPins with similar structural stability produced high variations in GS-eGFP degradation (Fig. 2m). To selectively determine the impact of low binder stability on GS-eGFP degradation, six Leu to Ala mutations (residues 41, 54, 74, 90, 107 and 123) in the non-binding backbone that were previously shown to destabilize DARPins38 were introduced into DP6 (DP6dest, structure shown in Fig. 4j). This did indeed lead to a pronounced destabilization, with unfolding of DP6dest at 0.9 M GdmCl (Supplementary Fig. S21 and Table 1). The destabilized DP6dest-bioPROTAC generated a moderately smaller increase in GS-eGFP degradation rates compared to wt DP6-bioPROTAC (Fig. 2a), albeit still being very similar to that observed with other bioPROTACs. Unexpectedly, the degradation of DP6dest-bioPROTAC in the absence of GS-eGFP was slightly lower compared to DP6-bioPROTAC, albeit showing pronounced self-polyubiquitination (Supplementary Fig. S22b), suggesting that factors other than the so far identified ones play a role. In summary, we found no convincing correlation of structural stability with bioPROTAC self-degradation or target degradation efficiency.
Lysine positioning in the target-binding moiety of bioPROTACs do not influence their degradation capabilities
Given the impact of the differences of the different bioPROTACS on ubiquitination and their predominant mechanism of conjugation, we wanted to test whether the removal of lysines as primary ubiquitination sites on the bioPROTAC itself could alter GS-eGFP degradation efficiency. We chose DP6, and we determined the most suitable replacements for the seven lysines in DP6 by Rosetta-based energy calculations (see Supplementary Fig. S25c–e for structure). The mutations had little effect on binding affinity as determined by SPR (Table 1). As expected, the DP6lysfree-bioPROTAC degradation rate was moderately lowered compared to DP6-bioPROTAC but still similar to the other constructs (Fig. 2b).
To determine whether a specific lysine ubiquitination site is responsible for the difference in rates, seven single-lysine mutants were produced. Little variation between constructs was observed, neither for GS-eGFP degradation, nor for their own degradation in the absence of target, as shown in Supplementary Fig. S26b. Nonetheless, the DP6single-lysine-bioPROTAC mutant, containing the most C-terminal lysine K137, was most quickly degraded on its own, which could indicate a possible involvement in bioPROTAC degradation. GS-eGFP degradation by DP6lysfree-bioPROTAC, as well as the seven DP6single-lysine-bioPROTACs were only moderately reduced compared to wt DP6-bioPROTAC/GS-eGFP degradation (Fig. 2a and Supplementary Fig. S26a). We conclude that the removal of lysines does not greatly alter bioPROTAC efficiency. Conversely, the presence of lysines, and thereby potential ubiquitination sites within a binder, does not hinder bioPROTAC performance, which is advantageous as it reduces constraints in the selection process of prospective binders.
Specific ubiquitination sites are involved in GS-eGFP degradation
For efficient degradation of a protein, ubiquitination not just anywhere, but of a residue in a specific distance to a proteasomal initiation site is required5. We could show previously20 that the N-terminus of GS-eGFP is the initiation site for proteasomal degradation within the CHIP/degrader pathway with DP6-bioPROTAC. It was therefore conceivable that ubiquitination at a specific lysine structurally close to the N-terminus is required for successful GS-eGFP degradation. Consequently, we investigated which of the large number of surface-exposed lysines of GS-eGFP are modified upon bioPROTAC engagement. For this purpose, we conducted experiments involving in vitro ubiquitination and subsequent tandem mass spectrometry, which allowed for the identification of four lysine ubiquitination sites at residues 6, 104, 159 and 241 of GS-eGFP, as indicated in Fig. 3a, b, for the degradation-inducing DP6-bioPROTAC (see Supplementary Fig. S27 for corresponding MS-MS spectra). Surprisingly, ubiquitination at three of the four lysines was also detected for DP7- and DP9-based bioPROTACs, although at a lower level. This suggests that their inability to induce degradation is likely caused by too low a level of ubiquitination or inefficient ubiquitin chain elongation, rather than a complete lack of ubiquitination. However, some of the recorded MS-MS spectra of ubiquitination by DP7- and DP9-bioPROTAC are not well resolved, indicating a lower likelihood of actual modification, and should be interpreted with caution (see Supplementary Figs. S28 and S29).
The obligatory presence of these residues for CHIP-based bioPROTAC degradation was confirmed by the introduction of Lys to Arg mutations in GS-eGFP. Mutation of lysines 104, 159 and 241 were sufficient to completely abolish GS-eGFP ubiquitination by DP6-bioPROTAC in an InVitroUb assay (Fig. 3c). bioPROTAC activity was also drastically reduced in vivo, and was also mirrored by slightly reduced ubiquitination of GS-eGFP in the LysateUb assay (Fig. 3e). However, in light of the still existing poly-ubiquitination in these mutants, we suspect that ubiquitination at other sites is generally possible, likely in combination with a different E2 or other additional factors, given the difference in ubiquitination observed between InVitroUb and LysateUb assays. Removal of ubiquitination sites at K104 and K159 rendered the bioPROTAC ineffective. Counterintuitively, additional mutation of the C-terminal K241 increased degradation, potentially by inducing ubiquitination at a new position. The observed variances in degradation rates (Fig. 3d) among the different GS-eGFP lysine mutants align well with prior research findings5, indicating that the ubiquitination of a specific site is determining the efficacy of protein degradation. In the case of GS-eGFP, K104 and K159 seem to be especially implicated in degradation.
Binding to a particular epitope can prevent target degradation
Given the importance of site-specific ubiquitination for GS-eGFP degradation, we turned towards discerning the localization of the DARPin-binding epitopes. We obtained crystal structures of six of the nine DARPins in complex with eGFP (Fig. 3f–k). Three were taken from previous publications22, while those of DP1, DP2 and DP4 are presented here for the first time (see crystallographic data in Table 2). DP5 and DP8 differ only minimally in sequence from each other and from DP6 and DP3 (Fig. 1a, b), and are therefore expected to share the same epitope, which is also supported by AlphaFold predictions (Supplementary Fig. S30). For DP7, fluorescence-anisotropy-based epitope binning in combination with energy calculations (Supplementary methods) supports an epitope similar to DP6 (Supplementary Fig. S31). Most DARPins, therefore, have a similar binding epitope with only small variations in distance to the eGFP termini, with only DP9 showing a distinctly different epitope from all other DARPins.
Table 2.
X-ray crystallography data collection and refinement statistics of DARPin/eGFP complexes
| Short names, selection names | DP1, 3G190.24 | DP2, 2G156 | DP4, 2G71 |
|---|---|---|---|
| PDB ID | 9F22 | 9F23 | 9F24 |
| Data statistics | |||
| Resolution range | 40.43–2.20 (2.27–2.20) | 48.08–1.59 (1.63–1.59) | 43.02–2.06 (2.18–2.06) |
| Space group | P3221 | P21 | I4 |
| Unit cell | 77.04 77.04 152.47 90.0 90.0 120.0 | 61.87 95.16 69.47 90.0 115.76 90.0 | 136.03 136.03 75.21 90.0 90.0 90.0 |
| Total reflections | 266,569 (24,113) | 641,964 (37,974) | 360,597 (57,114) |
| Unique reflections | 27,210 (2676) | 95,818 (3412) | 42,465 (6598) |
| Multiplicity | 9.7 (10.4) | 6.7 (11.1) | 8.5 (8.7) |
| Completeness (%) | 99.62 (99.41) | 98.63 (92.94) | 98.90 (96.60) |
| Mean I/sigma(I) | 7.3 (1.1) | 14.12 (2.20) | 10.75 (0.96) |
| Wilson B-factor | 48.45 | 15.06 | 50.77 |
| R-merge | 0.196 (2.815) | 0.086 (0.859) | 0.074 (1.362) |
| R-meas | 0.207 (2.960) | 0.093 (0.947) | 0.086 (1.574) |
| R-pim | 0.066 (0.910) | n.d. | n.d. |
| CC1/2 | 0.997 (0.415) | 0.999 (0.836) | 0.998 (0.421) |
| Refinement statistics | |||
| Resolution range | 40.43–2.20 (2.27–2.20) | 48.08–1.59 (1.61–1.59) | 43.02–2.06 (2.11–2.06) |
| Reflections used in refinement | 27,210 (2676) | 95,818 (2976) | 42,465 (2794) |
| Reflections used for R-free | 1397 (146) | 4667 (160) | 2124 (140) |
| R-work | 0.1741 | 0.1736 | 0.1822 |
| R-free | 0.2229 | 0.2117 | 0.2216 |
| Number of non-hydrogen atoms | 3189 | 6867 | 3054 |
| Macromolecules | 3002 | 5902 | 2771 |
| Ligands | 31 | 221 | 22 |
| Solvent | 156 | 744 | 261 |
| Protein residues | 383 | 702 | 354 |
| RMS(bonds) | 0.009 | 0.011 | 0.012 |
| RMS(angles) | 1.44 | 1.51 | 1.65 |
| Ramachandran favored (%) | 98.67 | 98.41 | 98.56 |
| Ramachandran allowed (%) | 1.06 | 1.59 | 1.44 |
| Ramachandran outliers (%) | 0.27 | 0.00 | 0.00 |
| Rotamer outliers (%) | 1.23 | 1.55 | 2.35 |
| Clashscore | 5.34 | 7.61 | 7.07 |
| Average B-factor | 58.28 | 20.63 | 53.87 |
| Macromolecules | 58.44 | 18.76 | 53.39 |
| Ligands | 50.58 | 31.82 | 41.76 |
| Solvent | 56.70 | 32.11 | 59.93 |
DP9 is unable to polyubiquitinate GS-eGFP as a bioPROTAC, even in an in vitro assay. We have excluded affinity and stability of the DARPin as influential factors and could also demonstrate that DP9-bioPROTAC is able to effectively interact with E2 proteins, since it is able to auto-ubiquitinate in vitro. Consequently, one remaining possible explanation for the failure to ubiquitinate and thereby degrade GS-eGFP is the different epitope. We determined three lysine ubiquitination sites that are necessary for efficient GS-eGFP degradation. The epitope of DP9 is located in close proximity of those lysines, whereas the other DARPins bind more closely to the N-terminus (Fig. 3b). The binding epitopes of most DARPins allow unobstructed access of the C-terminally located active Ubox-domain to those relevant lysines, which we expect to enable rapid polyubiquitination. However, DP9 itself binds in proximity of the relevant lysine residues, which implies that the C-terminally fused Ubox domain is located further away (Fig. 3b). It also suggests that E2 proteins encounter restricted access to the ubiquitination sites, with those on DP9 itself potentially competing with neighboring sites on GS-eGFP. We therefore suggest that DP9-bioPROTAC does not degrade GS-eGFP because the binding epitope itself obstructs access to, sterically hinders, or outcompetes polyubiquitination at the relevant lysine residues.
bioPROTACs are able to engage in multiple rounds of target degradation
Having elucidated the factors responsible for efficient degrader function, we next wanted to assess whether they are capable of multiple turnovers. In contrast to other drugs, small-molecule PROTACs derive their potency from promoting target degradation in a catalytic manner. As discussed before, the observed ratio of active bioPROTAC and GS-eGFP degradation rates already suggested a degradation mechanism with multiple turnovers (Fig. 2a, d). Our aim was to demonstrate that bioPROTACs, just as small-molecule PROTACs, possess this capability for catalytic target degradation, which would be advantageous for applicability in therapeutic contexts. To test this, we performed microinjection with the DP6-bioPROTAC and an increasing excess of GS-eGFP. The measured rates of GS-eGFP degradation were very similar for 1:1, 2:1 and 3:1 ratios of GS-eGFP and bioPROTAC (Supplementary Fig. S32). Moreover, the rates differed substantially from the predicted rate of a non-catalytic mechanism, thereby strongly implying a catalytic mode of degradation. The precise stoichiometry governing the degradation mechanism of CHIP-based bioPROTACs remains to be elucidated. When using a 16-fold excess of GS-eGFP, however, a considerably lower overall degradation rate was observed, potentially due to saturation of the DP6-bioPROTAC pathway, so that independent degradation pathways then have to take on the bulk of GS-eGFP degradation.
Binders without UPS interaction domain can promote target degradation
Initially conceived as a control for bioPROTAC degradation, we also tested degradation rates of DARPin/GS-eGFP complexes lacking any intentional UPS interaction domain. Unexpectedly, we also observed significant variations in their promotion of target degradation. Microinjection of one-to-one complexes of DARPins with GS-eGFP revealed a surprisingly high degradation rate of GS-eGFP by DP1 and DP6lysfree, but not DP6 (Fig. 4a). These two DARPins reduced GS-eGFP half-life from 7 h to 1.9 h and 2.5 h, respectively, whereas the other DARPins exhibited only marginal changes, averaging to a GS-eGFP half-life of 5.3 h. Importantly, we observed no correlation between the target degradation rates facilitated by DARPin-bioPROTACs and unfused DARPins, thus highlighting the potency of the CHIP E3 ligase to outperform a competing degradation mechanism targeting the fused DARPins (Fig. 4d). Nevertheless, the induction of GS-eGFP degradation by certain binders, nearly reaching levels comparable to those achieved by CHIP-based bioPROTACs, prompted us to explore the underlying principles further.
Given the catalytic activity in degradation observed for bioPROTACs, we were interested in elucidating the stoichiometry of degradation rates within the DARPin/GS-eGFP complexes as well. C-terminal labeling of the DARPins20 reduced the degradation rate of the GS-eGFP slightly (cf. Fig. 4a, b), but allows the assessment of relative rates of both components within the complexes. A 1:1 ratio in degradation rates was observed for most DARPin/GS-eGFP complexes, indicating a non-catalytic degradation mechanism (Fig. 4b). DP7 and 9 show distinctly different degradation behavior than the other DARPins, and we will discuss this also in the context of their different behavior as bioPROTACS in the next section. As expected, a slight correlation in degradation of DARPins without GS-eGFP and within GS-eGFP-complexes was observed (Fig. 4e). However, the constructs DP1 and DP6lysfree, characterized by their degradation-inducing properties, exhibit an increased degradation rate only when engaged in a complex with GS-eGFP, not as single proteins. The possibility of binding-induced GS-eGFP destabilization could be excluded as an explanation for increased complex degradation using a GdmCl unfolding assay (Supplementary Fig. S33). Degradation of the complex was found to be both proteasome- and ubiquitin-dependent, as shown by controls with E1 and proteasome inhibitors (Fig. 4n and Supplementary Fig. S34).
Given the involvement of ubiquitination, we suspected that DP1 and DP6lysfree potentially contain degron sequences which are recognized by E3 ligases to selectively ubiquitinate GS-eGFP. This would explain degradation induction selectively within the complex but not for the DARPins themselves. Indeed, an increase in GS-eGFP ubiquitination by those two DARPins was found in the LysateUb assay (Fig. 4h, I and Supplementary Fig. S35). However, DARPin ubiquitination was increased slightly as well, although not to a significant extent. This was surprising for DP6lysfree, given the deliberate removal of primary amines as ubiquitination sites. Ubiquitination on the N-terminal primary amine could be excluded by blockage through acetylation (Fig. 4n). Ubiquitination on the DARPin, therefore, does not appear to be critical for degradation, as is evidenced by the comparable degradation rates observed for DP6lysfree and DP6 in their unfused state. To determine the specific ubiquitination sites, MS-MS analysis after a pulldown from HEK293 lysate was performed (Supplementary Fig. S25). A single ubiquitination site on the GS-eGFP target within the DP6lysfree/GS-eGFP complex was identified. This C-terminal lysine has been previously implicated as a ubiquitination site specifically targeted by bioPROTACs for initiating GS-eGFP degradation, suggesting its potential for mediating degradation within this context. For the DP1/GS-eGFP complex, no ubiquitination sites on GS-eGFP were identified. Instead, two sites were found on the DARPin in proximity to its termini (Supplementary Fig. S36). Nonetheless, GS-eGFP ubiquitination cannot be rigorously excluded, as complete sequence coverage of the identified fragments was not achieved. An alternative mechanism for degradation could involve site-specific ubiquitination of the DARPin, positioning GS-eGFP in a favorable conformation for proteasomal unfolding. However, it remains unclear why DP1 is not degraded more rapidly in the absence of GS-eGFP, unless ubiquitination is facilitated only when DP1 is bound to GS-eGFP under conditions that have yet to be elucidated.
To elucidate the amino acids potentially conferring degron quality to a yet unidentified E3 ligase, we tested a set of DP1 and DP6lysfree mutants. Four amino acids unique to DP1, relative to the other eight eGFP-binding DARPins, were substituted with those present in DP6. Indeed, all mutants reduced GS-eGFP degradation within the complex, which could implicate those specific amino acids in a degron (Supplementary Fig. S36c). For DP6lysfree, the degron sequence or epitope is likely contained by the introduced lysine mutations, since wt DP6 itself does not show induced GS-eGFP degradation. Note that all lysines had been mutated to Arg, except at position 7 (to Met) and at position 137 (to His). This is supported by an increase in degradation of complexes of two other DARPins with the same lysfree mutations (Fig. 4o).
Single-lysine restoring mutants were produced to test which of the seven mutated amino acids are responsible (Fig. 4m). Mutation of DP6lysfree Met7 back to a lysine drastically reduced complex degradation, indicating its involvement in E3 recognition (Fig. 4m). Notably, an additional lysine ubiquitination site on the DARPin in combination with a potential degron sequence is able to induce even faster DARPin as well as GS-eGFP degradation, depending on the position (Fig. 4k–m). This highlights the importance of ubiquitin chain positioning for proteasomal degradation.
In summary, we found that protein binders without E3 fusion can induce fast non-catalytic degradation of target complexes. This is most likely due to E3 recognition sequences, naturally occurring in some binders, or produced by site-specific mutation.
DARPin degradation behavior as indicator of bioPROTAC performance
Our investigation revealed an absence of correlation between the degradation rates of DARPin/GS-eGFP and bioPROTAC/GS-eGFP complexes. However, a notable correlation was observed concerning the rate ratios within these complexes (Fig. 4g). DP7 and 9 are degraded independently from GS-GFP, irrespective of the presence or absence of the CHIPΔTPR domain. While the precise degradation mechanism of the DARPins has remained unclear in the absence of a fused E3 domain, the correlation observed in the degradation rate ratios suggests a potential common underlying factor.
We have observed not only a distinctive ratio in degradation rates within the two complexes but also a variation in binder degradation when comparing them isolated versus when within the complex. While DP7 degradation is slightly increased within the GS-eGFP complex, it is somewhat reduced for DP9 (Fig. 4e). This underscores the influence of GS-eGFP binding on the DARPin degradation and suggests distinct degradation pathways for the two DARPins. Through SPR and fluorescence anisotropy analyses, we determined that DP7 exhibits markedly lower affinity for the target, compared to the other DARPins (Table 1). This discrepancy in affinity may explain its distinct behavior and its degradation independent of its target. However, with mutant DP6W112A, which demonstrates an equally reduced affinity in the same range, degradation occurs at a 1:1 stoichiometry with GS-eGFP, indicating that low affinity alone is not sufficient to produce substoichiometric ratios. Conceivably, low affinity, coupled with a propensity for nonspecific binding as observed during protein preparation, could contribute to the accelerated degradation of DP7 within the GS-eGFP complex, possibly constituting another degron. Nonetheless, the precise mechanism underlying these observations remains to be elucidated.
DP9 exhibits a unique characteristic, demonstrating a half-life of 1.5 h compared to the typical half-lives of approximately 3 h observed for most DARPins (Fig. 4c). We established that structural stability did not exert an influence on bioPROTAC behavior (see above). However, it is noteworthy that these bioPROTACs contain the large CHIPΔTPR domain, which could potentially mask the underlying effects of the binding protein. Given that unfolding by the proteasome can potentially serve as a rate-determining step in degradation39,40, we sought to ascertain whether the DARPin’s low stability contributes to its rapid degradation. However, no correlation between DARPin denaturation midpoints and degradation rates was found (Fig. 4f). Although destabilization had some impact, as DP6dest exhibited increased degradation, with its half-life decreasing from 14.3 to 5.9 h, while DP9 and the DP947R mutant featuring enhanced stability, were degraded at substantially faster rates. This suggests the involvement of other factors in driving degradation, most likely mediated by the targeting of an, as yet, unidentified E3 ligase. Indeed, increased DP9 ubiquitination was detected in western blot analysis after LysateUb assays (Fig. 4h, i). This is in agreement with 5 lysine ubiquitination sites found by MS-MS analysis (Supplementary Fig. S37). Nonetheless, our attempts to identify the specific degron have thus far been unsuccessful, since mutating single amino acids differing from DP6 failed to reduce DP9 degradation (Supplementary Fig. S37k).
The question then arises as to why ubiquitination at DP9 is insufficient to degrade GS-eGFP concurrently, a phenomenon seemingly effective in other binder complexes such as DP1/GS-eGFP. It has been proposed that proteins lacking ubiquitination can be targeted to the proteasome by a ubiquitinated binding partner that serves as a proteasome adapter6 and the proteasome can unravel up to three proteins at the same time41, given that they contain sufficient initiation sites. Based on our findings from bioPROTAC studies, the most plausible explanation for GS-eGFP evading degradation within the DP9 complex can be attributed to the different epitope of DP9. We hypothesize that the specific binding position of this DARPin might either inhibit ubiquitination and/or impede favorable initiation site interactions of the target at the proteasome, thereby enabling GS-eGFP to escape degradation, while DP9 undergoes successful unfolding. Further experiments are needed to elucidate this specific mode of proteasomal initiation within the DARPin complexes. Although evaluating degradation principles without knowledge of the involved E3 ligases presents challenges, the correlation of ratios of degradation rates between target and DARPin- or bioPROTAC-complexes may serve as an early indication of the suitability of certain binders for use as bioPROTACs.
Discussion
PROTACs can be highly effective drugs that have successfully exploited targeted protein degradation42. Extensive research has focused on refining their design principles, particularly concerning linker length, binding affinity, and E3 ubiquitin ligase engagement. Nonetheless, they remain limited to those target proteins with binding pockets for small molecules. BioPROTACs, based on protein binders and peptide or protein-based UPS interaction domains, have been less intensively studied. Nonetheless, bioPROTACs hold promise for surpassing conventional PROTACs in several respects, including facile selection of target binders for challenging proteins and potentially higher affinities with less intensive linker optimization. While small-molecule PROTACS need to be empirically optimized to maintain cell permeability and acceptable pharmacokinetics14, bioPROTACS need to rely on successful cytoplasmic delivery43, e.g. by fusion with domains forming a complex with cationic and ionizable lipids via electrostatic interactions44, or by using a gene delivery technology45,46. In the present study, we only concentrated on the design principles to analyze what is needed to optimize their efficiency as degraders.
In our study to improve the understanding of principles behind the design of efficient bioPROTACs, we investigated the target-binding domains in the presence and absence of an UPS recruitment domain and focused on three properties: affinity, stability and binding epitope. To test target binder properties, bioPROTACs were constructed as fusions of DARPins that recognize GS-eGFP to the N-terminally truncated E3 ligase CHIP. This produced powerful degraders that drastically reduced the GS-eGFP half-life. Degradation rate ratios within DARPin-CHIPΔTPR /GS-eGFP complexes and further experiments with excess target indicated that the degraders are able to degrade GS-eGFP in a catalytic manner with multiple turnovers in the living cell. Considering the catalytic nature of small-molecule PROTACs, this could position bioPROTACs that are based on fusions with CHIP E3 ligase as compelling candidates for therapeutic advancement.
We found that the main determining factor for the potency of bioPROTACs is the binding epitope, which influences the engagement of the target’s ubiquitination and proteasomal initiation site5. It is widely understood that a ubiquitination site within a specific distance close to an initiation site for proteasomal unfolding and degradation is required16,47. We showed previously that the small 2-amino-acid N-terminal extension, Gly-Ser, of the proof-of-principle target eGFP is necessary and sufficient to provide such an initiation site20, converting the otherwise inert GFP into a convenient model target. Here, we could show that ubiquitination at specific lysines determines the degradation rate of GS-eGFP and that this ubiquitination is likely inhibited when an incompatible binding epitope is engaged. This underscores the importance for a comprehensive characterization of each potential target and target binder for their susceptibility to degradation. For a new potential therapeutic target, it is therefore advisable to generate a range of binders with diverse epitopes to create effective bioPROTACs.
We were surprised to find that one DARPin DP7 produced no self-ubiquitination as a CHIPΔTPR-fusion. This observation suggests a potential hindrance to the interaction between the Ubox-domain and E2 enzymes, which are crucial for effective ubiquitination. This underscores that specific properties of binders can impede effective ubiquitination, emphasizing the critical need for rigorous assessment of suitable binders during the selection of new targets. Depending on the particular E3 pathway involved, this can be evaluated by assessing autoubiquitination efficiency in vitro. Importantly, despite its inability to undergo self-ubiquitination, DP7-bioPROTAC is ubiquitinated and undergoes rapid degradation within cells. This degradation may result from interactions with another E3 ligase, potentially recognizing a specific surface feature of the protein. However, ubiquitination and rapid degradation of bioPROTAC alone are not sufficient to induce target degradation, as demonstrated by both DP7- and DP9-bioPROTAC/GS-eGFP complexes. This observation suggests the possibility of an independent mechanism, which warrants further investigation.
We found only a modest influence of target affinity on degradation rate, using a mutant with drastically decreased affinity for GS-eGFP. It is plausible that a brief, yet impactful interaction with the target could still enable highly efficient ubiquitination mediated by E3 domains such as CHIP/Ubox. Consequently, even binders with lower affinities may present compelling prospects for targeted protein degradation. However, it is probable that there exists a lower limit of necessary affinity. Binders very close to this limit may be influenced by cellular factors that shift the binding equilibrium, such that the in vitro measurements of affinity do not allow a determination of a strict cut-off. Furthermore, since preassembled complexes were injected into cells to study degradation in this study, it remains to be determined whether differences in affinity play a role in an endogenous cellular context. Our findings contradict those reported limits by Lim et al.16, who asserted that target affinity constituted the primary determinant for bioPROTAC activity, as demonstrated with SPOP-based bioPROTACs. Lim et al.16 reported that two monobody-SPOP bioPROTACs, featuring an affinity of approximately 250 nM, failed to induce degradation of H2B-GFP in transiently transfected HEK293 cells. In contrast, our study revealed that a bioPROTAC constructed with a DARPin mutant possessing an even lower affinity efficiently degraded GS-eGFP in HEK293 cells. Considering that intracellular concentrations achieved via transfection and microinjection are expected to be comparable, as indicated by fluorescence intensities observed in our transfection experiments with HEK293 cells, it is plausible that performance disparities based on affinity may be contingent upon the specific E3 pathway involved. Nonetheless, our findings suggest that qualitative differences in degradation, as observed in this instance, may stem from mechanisms beyond mere differences in binding affinity.
In our investigation of the structural stability of binders, we did not observe a correlation with the performance of bioPROTACs. We investigated DP6, a very powerful degrader when fused to CHIPΔTPR, and compared it with its destabilized version, DP6dest, fused in the same manner. Notably, destabilization exerted only a modest impact on the degradation rate of the target. Generally, degradation rates were largely comparable between DARPin-based-bioPROTACs themselves, especially in when in complex with GS-eGFP. Discrepancies observed in degradation rates were likely attributable to sequence differences resulting in varying levels of ubiquitination rather than differences in structural stability. Notably, the elimination of ubiquitination sites within DP6 fused to CHIPΔTPR did not enhance GS-eGFP degradation. This highlights the importance of effective CHIP E2 interaction and the ubiquitination mechanisms themselves as the principal determinants of bioPROTAC efficacy, rather than the premature degradation of the bioPROTAC itself.
Finally, we examined the impact of the binder in the absence of a deliberately fused degron or E3-interacting component. Our findings indicate that binders possess the capability to facilitate target degradation even in the absence of fusion to an E3 interaction domain. However, this degradation occurred through a non-catalytic mechanism. Degradation of complexes with the target is presumably facilitated by a degron sequence inherent within the binder, which may arise naturally following selection or be induced through mutations, as exemplified by the lysine removal within the DARPins. This is important to consider even when working with thermodynamically highly stable proteins such as DARPins. In fact, we found that sequence differences, presumably constituting interaction sites with a yet unidentified set of E3 enzymes, had a much more pronounced effect on DARPin degradation than structural stability. Although deliberate destabilization of a DARPin was correlated with faster degradation, one eGFP-binding DARPin (DP9) was degraded in the cytosol of HEK293 exceptionally fast, even when stabilized by a mutation (DP9R47) which corrects a single amino acid deletion that had occurred during directed evolution. This observation raises the possibility that the DARPin may harbor a binding site for an E3 ligase. So far, we were unable to find this specific degron site by introducing point mutations in DP9. Since not all E3 ligases bind to peptide degrons and have linear binding motifs5, it is possible that this specific E3 binding site is a conformational epitope, and single mutations are not sufficient to prevent E3 binding.
Lastly, we could show that, while induced degradation of complexes mediated by DARPin binding lacks correlation with bioPROTAC performance, a high ratio of DARPin to GS-eGFP degradation within the complex was associated with poor bioPROTAC efficacy. This correlation can potentially be used to select binders for bioPROTAC design before any E3 fusion.
In summary, we showed important design principles for bioPROTACs. In the context of binders, the main criterion appears to the suitability of the binding to epitopes, avoiding disruption of effective E2 interaction, while factors such as affinity and structural stability exert only minor to medium influences. The conclusions we could draw may serve as promising initial steps towards the rational design of bioPROTACs targeting therapeutically significant molecular targets.
Methods
Design and cloning of expression constructs
Proteins containing an N-terminal MRGS-His tag, followed by an Avi-tag and TEV cleavage site, were expressed from the pQIq vector48 (a lacIq-encoding derivative of pQE30; Qiagen, Hilden, Germany). The N-terminal fragments were inserted via EcoRI/BamHI sites. Fragments coding for DARPins were inserted via BamHI/PstI sites. C-terminal fragments (Gly-Ser linkers with single cysteines, linker followed by CHIPΔTPR domain, HA-tags) were inserted by PstI/HindIII sites. Fragments were synthesized by Twist Bioscience and Integrated DNA Technologies (IDT).
Protein expression and purification
The E. coli strain BL21 was used for expression. Constructs containing a C-terminal single cysteine for dye labeling were expressed by induction with 400 µM IPTG in 1 L TB-medium. Cells were grown at 37 °C until an OD600 of approximately 1.5 was reached, and the temperature was reduced to 25 °C for single cysteine constructs, and expression was performed for 16 h. Other constructs were expressed by autoinduction in 1 L autoinduction medium49 at 25 °C for 24 h. Cells were harvested by centrifugation at 4 °C. Lysis buffer (50 mM Tris, 500 mM NaCl, 10% w/v glycerol, 0.5 mM EDTA, 0.2 mM 4-(2-aminoethyl) benzene sulfonyl fluoride hydrochloride (AEBSF), 1.4 µM pepstatin-A, 1 µM leupeptin and 1 mg/ml lysozyme, pH 8 at 4 °C) was added and cells were lysed with a French press. The supernatant was cleared by ultracentrifugation at 20,000 × g at 4 °C. Bench-top IMAC was performed with 5 ml Ni-NTA bed volume in running buffer (50 mM Tris, 500 mM NaCl, pH 8 at 4 °C). Bound protein was washed with running buffer and then with wash buffer (50 mM Tris, 500 mM NaCl, 10 mM imidazole, pH 8 at 4 °C) before elution with elution buffer (50 mM Tris, 500 mM NaCl, 250 mM imidazole, pH 8 at 4 °C). Proteins were buffer-exchanged by overnight dialysis at 4 °C against dialysis buffer (20 mM Tris, 100 mM NaCl, pH 8 at 4 °C) with simultaneous TEV cleavage at a molar ratio of 1:10. Single-cysteine containing proteins were additionally reduced before dialysis with 10 mM DTT for 30 min, and 1 mM DTT was added to the dialysis buffer. Uncleaved protein and His-TEV were removed by bench-top reverse IMAC with dialysis buffer, where the proteins of interest are in the flow-through. Constructs containing CHIP/Ubox domains were eluted from the reverse IMAC by 37.5 mM imidazole, since they bind to the Ni-NTA column even without a His-tag. Protein samples were diluted 1:1 with double-distilled H2O for subsequent anion exchange chromatography using a MonoQ 5/50 GL column on an ÄKTA PURE system. The protein was loaded using AEX running buffer (10 mM Tris, 50 mM NaCl, pH 8 at 4 °C) and eluted by gradient elution from 0–100% elution buffer (10 mM Tris, 500 mM NaCl, pH 8 at 4 °C) in 40 CV. Samples were pooled and flash-frozen in liquid nitrogen for storage at −80 °C.
TMR labeling
To be able to quantify degradation of the bioPROTAC, two native cysteines in the coiled-coil region of CHIP were labeled with TMR-maleimide. Fluorophore conjugation to residues within the CHIP domain, instead of introduction of a single cysteine at the C-terminus, was chosen because mutation of either of the internal cysteines within CHIP was found to cause a decrease in degrader efficiency (Supplementary Fig. 1). DARPins without E3 domain were labeled at an introduced C-terminal cysteine behind a short GGGGSGGG linker.
A total of 200 µl protein was incubated with 10 mM DTT for 30 min at RT. The buffer was exchanged to degassed PBS, pH 7.5. and 1 mM EDTA using a PD25 MiniTrap (Cytiva) in a nitrogen-flooded cabinet to prevent air oxidation. Tetramethylrhodamine-5-maleimide (TMR5-maleimide) (94506, Sigma) at 20 mM in DMSO was added in a 1:1 molar ratio and incubated for 3 h at RT with shaking at 550 rpm. The reaction was quenched with 100 mM DTT for 10 min. The labeled proteins were separated from excess dye either by a PD25 MiniTrap and buffer exchange to PBS pH 7.5, or by anion exchange chromatography (AEX) with the same buffers and conditions as described above and a gradient of 80 CV. After AEX, the labeled DARPins were buffer-exchanged to PBS using a SD 75 10/300 GL column (Cytiva) on an ÄKTA PURE system (Cytiva). Labeling was confirmed by ESI-MS.
N-terminal acetylation
A total of 200 µl of protein was buffer-exchanged to phosphate buffer (0.1 M, pH 6.6) using a PD25 Minitrap column (Cytiva). Acetic anhydride (320102-100 ML, Sigma) was added at a molar ratio of 20:1. The reaction was incubated for 3 h at RT with shaking at 550 rpm. The acetylated protein was transferred into PBS through buffer exchange using a Superdex 75 10/300 GL column on an ÄKTA Pure system. The acetylation was verified by ESI-MS and LC-MS/MS analysis.
eGFP complexation
GS-eGFP complexes were formed by mixing the respective DARPin or bioPROTAC with GS-eGFP in a molar ratio of 1.5:1, with an excess of the smaller over the higher molecular weight species. 1:1 complexes were purified by SEC in PBS, pH 7.5, using either an SD 75 10/300 GL or an SD 200 10/300 GL column on an ÄKTA Pure system. Samples were pooled and flash-frozen in liquid nitrogen for storage at –80 °C.
ESI-MS
Mass spectrometry analyses were carried out at the Functional Genomics Center Zürich. For ESI-MS analysis, the samples were diluted threefold with 1% TFA and transferred into autosampler vials for LC/MS. A 7 µl aliquot was injected into an ACQUITY UPLC® BioResolve-RP-mAb 2.7 µm, 2.1 × 150 mm, 450 Å column (Waters, USA). Separation and elution were carried out using an ACQUITY UPLC system with a gradient of buffer A (0.1% DFA in water) and buffer B (0.1% DFA in acetonitrile/75% 2-propanol) at a flow rate of 200 µl/min and a temperature of 60 °C over 10 min. The analysis was conducted on a Synapt G2 mass spectrometer coupled directly to the UPLC system. Mass spectra were acquired in positive-ion mode by scanning the m/z range from 400 to 5000 Da, with a scan time of 1 s and an interscan delay of 0.1 s. The spray voltage was set to 3 kV, the cone voltage to 35 V, and the source temperature to 100 °C. Data acquisition was performed using the MassLynx 4.2 software (Waters, UK). When possible, individual peak m/z data were deconvoluted into mass spectra using the MaxEnt1 algorithm in MassLynx, applying a resolution of 0.5 Da/channel and a Uniform Gaussian Damage Model with a half-height of 0.5 Da.
Protein crystallization and structure determination
Protein complexes were isolated by SEC using a HiLoad 16/600 SD 75 pg or a HiLoad 16/600 SD 200 pg column (GE Healthcare) in 8 mM HEPES, 10 mM NaCl pH 7.5. The protein was concentrated to approximately 10 mg/ml. Sitting-drop vapor-diffusion experiments were performed using a Phoenix crystallization robot (Art Robbins Instruments). Screening for crystallization conditions was performed with sparse-matrix screens from Hampton Research and Molecular Dimensions in 3-Drop Intelli-Plates 96-3 LVR (Hampton) at 4 °C. Protein solutions were mixed with reservoir solutions at 1:1 and 2:1 ratios (200–300 nl final volume) and equilibrated against 70 μl of reservoir solution. A crystal structure for the DP1/eGFP complex was obtained in 0.1 M Na acetate pH 5.5, 10% w/v PEG 8 K, 10% PEG 1 K, 0.2 M KBr. A crystal structure for the DP2/eGFP complex was obtained in 50% v/v pentaerythritol propoxylate (5/4 PO/OH), 0.1 M Tris, pH 8. A crystal structure for the DP2/eGFP complex was obtained in 50% v/v pentaerythritol propoxylate (5/4 PO/OH), 0.1 M Tris, pH 8. A crystal structure for the DP4/eGFP complex was obtained in 0.1 M Tris, pH 8.5, 8% w/v PEG 20 K, 8% v/v PEG MME 550, 0.2 M KBr.
Crystals were mounted in cryo-loops from Hampton Research and flash-frozen (liquid N2) in mother liquor supplemented with 35% ethylene glycol. X-ray diffraction data were collected at a wavelength of 1.0 Å on beamlines X06SA and X06DA (Swiss Light Source, Paul Scherrer Institute, Villigen, Switzerland) equipped with an Eiger 16 M detector (Dectris, Baden-Wättwil, Switzerland). Data were processed with XDS, Aimless, and autoPROC with 5% of data set aside for calculating the Rfree value50–52. Initial phases were obtained by molecular replacement using Phaser53 with the structure of the full consensus N3C DARPin (PDB ID 2QYJ) as search model. Refinement was done using Refmac554, followed by model building in COOT55.
Modeling DARPin eGFP complexes
Since the DP7/GS-eGFP complex could not be crystallized, despite numerous attempts with different constructs, two modeling approaches were pursued. First, AlphaFold Multimer was employed, where 88 or the 100 complex structures obtained (including the 51 top-ranked) cluster with DP6. Second, ColabDock was used with default 1vN distance restraints, using the center of the DARPin binding site and each amino acid of each β-strand of GFP in turn in individual dockings. This results in a generation of clusters of all reasonable docking poses. The top 5 poses of each cluster were ranked by Rosetta interface score, and the cluster similar to DP6 again gave the best scores. Since this is also consistent with the mutational analysis (Supplementary Fig. S31), we conclude that DP7 binds in a similar manner to DP6.
Cell culture
HEK293 (ATCC CRL-1573) cells were cultured in DMEM containing heat-inactivated FCS (10%) and penicillin/streptomycin (1% v/v) at 37 °C with 5% CO2 in 75 cm2 plates. Every 4–5 days cells were detached by trypsin treatment and diluted 1:20.
Microinjection and live-cell imaging
Microinjection experiments were performed as described previously20. In short, microscopy dishes with HEK293 cells were seeded two days prior to injection to achieve 80–90% confluency on the injection day. The medium for microinjection and subsequent live-cell imaging was replaced 4 h before injection with Live Cell Imaging Solution (Invitrogen) containing 10% FCS. Analyte samples were prepared at 25 µM, supplemented with a 0.1 mM injection marker (Alexa Fluor 647-labeled dextran, average molecular weight 10 kDa; #D22914, Invitrogen). Microinjections were performed using an InjectMan4 and FemtoJet4i system (Eppendorf). Glass microinjection cuvettes (Femtotip II, Eppendorf) were used for manual injections of ~30–100 cells per analyte, with continuous flow controlled by manually inserting and withdrawing the needle from each cell, leading to variability in injected volume and concentration. Notably, degradation rates were shown to be independent of injected concentrations. For inhibition experiments, 10 µM MG132 (M8699, Sigma) or 100 nM E1 inhibitor TAK-243 (MLN7243, S8341-5MG, Selleck Chemicals) was added to the injection medium 1 h before injection. Live-cell imaging was conducted at room temperature for injection and at 37 °C for imaging, using a Visitron CSU-W1 microscope with up to four epifluorescent channels (405 nm, 488 nm, 561 nm and 640 nm excitation). Images were captured every 20 min for 12 h. Approximately 75% of injected cells (most often around 30 cells) were available for analysis, while cells undergoing mitosis, those with altered motility or shape, and cells detaching from the dish were excluded. Following flat- and dark-field corrections, cell outlines were automatically determined using the injection marker signal in CellProfiler. Total cellular fluorescence was measured at each time point. Single-cell degradation rates (k) were determined by fitting an exponential decay model (Eq. (1)) to the data using the NLS function in R56.
| 1 |
Outliers of single-cell degradation rates of each of the four channel signals (405, 488, 561 and 640) were removed using the interquartile range (IQR) method, where rates were only obtained if they lay within [Q1–1.5 × IQR, Q3 + 1.5 × IQR]. For the total number of analyzed cells and retained rates see Supplementary Data 2.
Spectral analysis
Measurements of the absorbance of GS-eGFP and GS-eGFP/binder-complexes were performed on a NanoDrop™One (Thermo) instrument at approximately 1 µM in a range of 200 nm to 850 nm. Measurements of GS-eGFP and GS-eGFP/binder-complex fluorescence were performed on a FluoroMax-4 spectrofluorometer (HORIBA Scientific). Protein samples were measured at a concentration of 150 nM in PBS pH 7.4 with an ultra-micro cuvette (Hellma®, 105-251-15-40) with 3 × 3 mm path length. Excitation was measured from 350–540 nm, emission 400–600 nm.
Denaturant-induced equilibrium unfolding measured by circular dichroism
Circular dichroism (CD) measurements were performed on Chirascan V100 (Applied Photophysics) instrument with an ultra-micro cuvette (Hellma®, 105-251-15-40) with 3 × 3 mm path length. Thirty stock solutions were prepared with guanidinium chloride concentrations ranging from 0 to 7.25 M in 0.25 M steps in PBS pH 7.5 Protein solutions were prepared in duplicates. Protein was added to a final concentration of 10 µM in a volume of 50 µl for each sample and incubated overnight at 4 °C. CD was monitored at 222 nm at 20 °C with a bandwidth of 1 nm and a time-per-point measurement of 5 s. To subtract the background of guanidinium chloride on the CD signal, a regression line was obtained by measurement of buffer stocks without added protein.
Data analysis of the collected CD signals was performed using R Studio. To determine the values for ∆G0 and the midpoint denaturant concentration D = (∆G0/m), Eq. (2) was fitted to the data using the nonlinear weighted least square function. However, since the DARPins do not follow a two-state transition (see main text), we do not report the ΔG0 values.
| 2 |
Here, yobs is the measured CD signal after background subtraction, yN is the CD signal at zero denaturant, yU is the CD signal of unfolded protein extrapolated to zero denaturant, m is the slope of the dependence of ∆G on denaturant concentration, mN is the slope of the pre-transition baseline, mU is the slope of the post-transition baseline, ∆G0 is the Gibbs free energy of unfolding, R is the gas constant, and T is the absolute temperature57.
Differential scanning fluorimetry (Thermofluor assay)
Twenty µL protein solution at 10 µM was prepared in 30 mM MOPS, pH 7, with 5× Sypro Orange (stock 5000×, New England Biolabs). Heat denaturation curves were obtained on a qPCR Agilent Mx3005p (Agilent Technologies) instrument by measurement of the fluorescence at 610 nm after excitation at 498 nm in the temperature range of 25–96 °C by increasing the temperature by 2 °C every minute.
GS-eGFP unfolding assays
Buffer stocks were prepared with 0.2 M MOPS, pH 7, plus added guanidinium chloride with concentrations from 0 to 5.5 M in 0.5 M steps. Final samples were prepared from the stock in 50 µL volumes and GS-eGFP or POI/GS-eGFP complexes were added at 150 nM final concentration. Fluorescence was recorded at 535 nm after excitation at 400 nm and 485 nm with 10 nm bandwidth each and 30 flashes using a Tecan Spark® multimode microplate reader instrument. The signals of buffer samples without added protein were recorded and subtracted from the raw data. Since this is not an equilibrium reaction, data analysis was performed by estimating the best fit using the three-parameter IRT model (3-parameter logistic model, 3PL) with non-linear least squares fitting using R. The first four points were omitted from the fit.
SPR
Expression and purification of biotinylated GS-eGFP
For subsequent immobilization on the SPR chip, GS-eGFP and BirA were co-expressed in E. coli BL21 cells by autoinduction at 25 °C for 27 h. E. coli BL21 cells were transformed with the same pQIq vector as described above and with the vector pBirAcm (avidity.com) for expression of BirA48. Half an hour after inoculation, the autoinduction medium was supplemented with a final concentration of 20 µM D-biotin. Cells were harvested and lysed as described above and purified by Ni-NTA purification and subsequent dialysis as described above, but without TEV cleavage. Successful biotinylation was shown by a higher molecular weight shift on an SDS-PAGE after incubation with PEG-streptavidin.
SPR experiments
SPR experiments were conducted on a ProteOn (Bio-Rad) apparatus at room temperature. A precoated ProteOn™NLC Sensor Chip (GE) was conditioned, and biotinylated GS-eGFP was subsequently immobilized to give approximately 300 RU, exploiting the neutravidin/biotin interaction, using TBS-T as running buffer. For immobilization, the biotinylated protein was injected at a concentration of 10 nM using a constant flow rate of 100 µL/min. The analyte concentration was measured in quintuplicates, and the mean concentration was calculated. Analytes were subsequently injected at 8 different concentrations, ranging from 0.1 nM to 200 nM with an internal dilution factor of 1.7, without regeneration. For analysis, only part of the curves were used that showed a discernible signal over the background and only until Rmax was reached.
Data analysis
Interspot data curves obtained from measurement of running buffer TBS-T were directly subtracted from raw data. Dissociation curves were isolated, and the data were fitted globally to a first-order exponential decay model to obtain the dissociation rate constant kd. To calculate the association rate constant ka, Eq. (3) was used to simulate an entire curve with arbitrary values for ka and Rmax and the fixed kd value separately determined using the Euler method in R.
| 3 |
where R is the signal, t is time, ka is the association rate constant, [A] is the concentration of analyte, Rmax is the signal reached at saturation, and kd is the dissociation rate constant. The Root Mean Square Error (RMSE) was calculated for the association phases between data and simulation, and the parameters were changed until a minimum was found (Nelder-Mead method58).
For DP6W112A Eq. (4) was used to determine Rmax. For this purpose, the mean signal at the plateaus was plotted against the injected concentration and then Eq. (4) was used to determine KD using GraphPad prism.
| 4 |
where R is the signal, [A] is the concentration of analyte, Rmax is the signal reached at saturation, and KD is the equilibrium dissociation constant.
Ubiquitination assays
Preparation of lysates
To prepare the lysate for lysate ubiquitination assays, HEK293 cells were grown in T150 plates until 80–90% confluency. Cells were washed once with warm PBS, then detached by trypsin treatment. Cells were then washed twice with ice-cold PBS. Cell extraction was performed by resuspending cells in double-distilled ice-cold H2O at 1 μl/105 cells plus protease inhibitor cocktail (Pierce). Cells were kept on ice for 20–30 min, followed by three cycles of freeze-thawing, using liquid nitrogen and a 37 °C water bath. The lysate was centrifuged for 30 min at 16,000 × g (4 °C) and extracted proteins were recovered by transferring the supernatant into a new reaction tube. The protein concentration of the cell extract was determined using the BCA assay (Pierce), performed according to the manufacturer’s instructions. Aliquots of the lysate were prepared with 300 μg total protein each (around 30 μl) and flash-frozen with liquid nitrogen for storage at −80 °C.
Ubiquitination assays and readout
Ubiquitination assays were either performed with the POI in HEK293 lysate (LysateUb assays) already containing all required enzymes or fully in vitro for the CHIP-in-vitro ubiquitination assay (InVitroUb assay) with added purified E1 and E2 proteins.
For LysateUb assays, the reaction was performed in a total volume of 60 μl. Reactants were added to the prepared lysate aliquots to yield a final concentration of 50 mM Tris-HCl (pH 8.3), 5 mM MgCl2, 2 mM DTT, 5 mM ATP, 10 mM creatine phosphate, 0.25 U/ml creatine kinase, 0.25 mM MG-132 (in DMSO), 50 μg/μl ubiquitin aldehyde (BML-UW8450-0050, Enzo Life Sciences), 1 μg/μl ubiquitin (U-100H, Bio-Techne) and 0.03 nmol of the POI or its one-to-one complex with GS-eGFP (final concentration 500 nM). LysateUb reactions were incubated for 3 h for DARPins and 2 h for bioPROTAC complexes at 37 °C in a PCR cycler and afterwards 20 μl of 4× reducing Laemmli buffer was added.
For InVitroUb assays, the procedure was adapted from that described previously59. Ubiquitination was performed in 20 μl reactions. The reaction mixture consisted of double-distilled H2O with 50 mM Tris (pH 7.5), 50 mM KCl, 0.2 mM DTT, 0.1 μM Ube1 (SRP6147, Sigma), 2.5 mM ATP, 2.5 mM MgCl2, 100 μM ubiquitin (U-100H, Bio-Techne), 1 μM UbcH5c (662098, Sigma) and 1 μM POI or POI/GS-eGFP complex. For InVitroUb reactions were incubated for 1 h at 37 °C in a PCR cycler and afterwards 20 μl of 4× reducing Laemmli buffer was added.
For both assays, samples were boiled for 10 min at 95 °C, then centrifuged using a tabletop centrifuge. SDS-PAGE was performed using a Mini Gel Tank (Thermo) at 120 V for 70 min with 12% BoltTM Bis-Tris Plus gels (Thermo) with 1x MOPS running buffer. For LysateUb assays 5 μl of each sample and for InVitroUb assays 10 μl of each sample were loaded onto two separate gels for detection of two different proteins by western blot. Gels were blotted onto an LF-PVDF membrane (Bio-Rad), using the Bio-Rad Trans-Blot Turbo system as per the manufacturer’s instructions. The “Mixed-MW” blotting protocol was selected. Blocking was performed for 1 h in 1× casein blocking buffer (Sigma). Incubation with primary antibody was performed overnight in PBS-T (0.05% Tween) and 1× casein blocking buffer at 4 °C on a roller shaker. Membrane duplicates were incubated with either polyclonal rabbit anti-DARPin serum (produced in house) or polyclonal rabbit anti-GFP antibody (600-401-215, Rockland Immunochemicals) at a dilution of 1:10,000 and 1:3000, respectively. Blots were washed 3× for 5 min with PBS-T. The secondary antibody, HRP-coupled goat anti-rabbit IgG (7074, Cell Signaling Technology), was diluted 1:2000 in PBS-T (0.05% Tween) and 1× casein blocking buffer and incubated with the membranes for 1 h at RT on a roller shaker. Blots were washed 3× for 5 min with PBS-T. Five minutes after HRP substrate (051730, Merck) was added to the membranes, chemiluminescence imaging was performed on Viber Fusion FX7 Imager instrument.
Ubiquitination and HA-pulldown
C-terminally HA-tagged DARPins were purified as described above. Ubiquitination of the proteins in HEK293 lysates was performed as described above for the LysateUb assays, but the volume was increased to 200 µL HEK293 lysate (1200 µg total protein, prepared as described above) per ubiquitination reaction with 0.48 nmol total DARPin-HA/eGFP complex for a total volume of 480 µl. The reaction was incubated for 4 h at 37 °C. Subsequently, 8.61 µl (20 µl/50 µg protein) anti-HA magnetic beads (Sigma, SAE0197) were added to the reaction and incubated for 2 h at RT on a rotating platform. Washing was performed with 5 × 200 μl PBS according to the manufacturer’s instructions.
MS-MS analyses of ubiquitination sites
Mass spectrometry analyses were carried out at the Functional Genomics Center Zürich.
LC-MS/MS sample processing
The washed anti-GFP or anti-HA-tag pulldown beads were re-suspended in 45 µl digestion buffer (10 mM Tris, 2 mM CaCl2, pH 8.2) and digested overnight using 2 µl of sequencing grade trypsin or chymotrypsin (0.1 ng/µl in digestion buffer) at 37 °C. Samples digested with chymotrypsin were further digested with 200 ng trypsin for another 4 h the next day. The supernatants were transferred into new tubes, and the beads were washed with 150 µl trifluoroacetic acid (TFA) (0.1% TFA, 50% acetonitrile) and combined with the first supernatant. The samples were dried to completeness. Samples from the in vitro ubiquitination reaction (InVitroUb assays) were denatured in a final concentration of 4% SDS and boiled for 10 min at 95 °C. Single-pot, solid-phase-enhanced sample preparation (SP3-assisted) protein capture and clean-up was performed. Proteins were reduced and alkylated by adding tris(2-carboxyethyl)phosphine (TCEP) and 2-chloroacetamide to a final concentration of 5 mM and 15 mM, respectively, followed by incubation for 30 min at 30 °C protected from light. The samples were diluted with ethanol to a final concentration of 60% EtOH (v/v) before adding the corresponding amount of hydrophilic and hydrophobic carboxylated magnetic beads (GE Life Sciences). The beads were washed three times with 80% EtOH. For the enzymatic digestion, trypsin, chymotrypsin or GluC in 50 mM tetraethylammonium bromide (TEAB) was added to the beads and, if needed, the pH was adjusted to pH 8. Samples were digested overnight at 37 °C. Samples first digested with GluC and chymotrypsin were incubated with trypsin for another 4 h at 37 °C. The supernatants were saved, and the remaining peptides were extracted with H2O. The two elutions were combined and dried to completeness. For LC-MS/MS analysis, samples were re-solubilized in 20 µL of MS sample buffer (3% acetonitrile, 0.1% formic acid) and peptide concentrations were determined using the Lunatic UV/Vis polychromatic spectrophotometer (Unchained Labs).
LC-MS/MS data acquisition
LC-MS/MS analysis was performed on an Orbitrap Fusion Lumos (Thermo Scientific) equipped with a Digital PicoView source (New Objective) and coupled to an M-Class UPLC (Waters). The solvent composition was 0.1% formic acid for channel A and 99.9% acetonitrile in 0.1% formic acid for channel B. The column temperature was 50 °C. For each sample, the respective volume equivalent to A280 absorbance of 0.1 was loaded onto a commercial ACQUITY UPLC M-Class Symmetry C18 Trap Column (100 Å, 5 µm, 180 µm × 20 mm, Waters) connected to a ACQUITY UPLC M-Class HSS T3 Column (100 Å, 1.8 µm, 75 µm × 250 mm, Waters). The peptides were eluted at a flow rate of 300 nL/min. After an initial hold for three min at 5% B, a gradient from 5 to 25% B in 35 min/50 min (for pulldown/in vitro reaction samples) and 25% to 35% B in an additional 10 min was applied. The column was cleaned after the run by increasing to 95% B within 5 min prior to re-establishing loading conditions. The mass spectrometer was operated in data-dependent mode with a maximum cycle time of 3 s, with spray voltage set to 2.3 kV, funnel RF level at 40% and heated capillary temperature at 275 °C. Full-scan MS spectra (350–2000 m/z) were acquired at a resolution of 120,000 at 200 m/z after accumulation to an automated gain control (AGC) target value of 400,000 or for a maximum injection time of 50 ms. Precursors with an intensity above 5000 were selected for MS/MS. Ions were isolated using a quadrupole mass filter with 1.6 m/z isolation window and fragmented by electron-transfer/higher-energy collision dissociation (EThcD). Fragments were detected in the Orbitrap with the resolution set to 120,000, an AGC target of 200,000 and maximum injection time mode set to 246 ms. Charge state screening was enabled, and singly, unassigned charge states and charge states higher than six were excluded. Precursor masses previously selected for MS/MS measurement were excluded from further selection for 20 s, applying a mass tolerance of 10 ppm. The samples were acquired using internal lock mass calibration on m/z 371.1012 and 445.1200.
LC-MS/MS data analysis
The acquired data were processed for identification and quantification using Proteome Discoverer 2.5.0.400 (Thermo Fisher Scientific). Spectra were searched against custom sequences covering the proteins of interest and common protein contaminants concatenated to its reversed decoyed FASTA database using Sequest HT with FDR calculation done using Percolator default settings. The following modifications were set as variable modifications for all samples: oxidation (M), acetylation (protein N-terminus), Met-loss (protein N-terminus), Met-loss+acetyl (protein N-terminus), GG (K, S) LRGG (K, S), and fixed modification for samples that were subjected to reduction and alkylation was set to carbamidomethyl (C). Enzyme specificity was set to trypsin/P only or chymotrypsin and trypsin or GluC and trypsin allowing a minimal peptide length of 6 amino acids and a maximum of two missed cleavages. Precursor tolerance was set to 10 ppm and fragment ion tolerance was set to 20 mmu.
The protein identification results were imported into Scaffold (Proteome Software) with 1% protein FDR, 0.1% peptide FDR and a minimum number of two peptides per protein. For PTM localization evaluation, Scaffold results were exported as mzIdentML files and imported into ScaffoldPTM.
The mass spectrometry proteomics data were handled using the local laboratory information management system (LIMS)60, and all relevant data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository61 with the data set identifier PXD063877.
KD determination with fluorescence anisotropy
20 nM GS-eGFP was titrated with DARPin with 24 dilutions starting at a concentration of approximately 100 µM and a dilution factor of 1.666. All samples were prepared in duplicates in black Greiner 96-Well Flat Bottom plates. Fluorescence was measured using a Tecan Spark plate reader equipped with a fluorescence polarization module. The monochromator excitation wavelength was 485 nm with a bandwidth of 10 nm, and the monochromator emission wavelength was 525 nm with a bandwidth of 25 nm. The G-factor was 1.082 with 30 flashes and the settle time 100 ms. The averages of two replicates were subtracted with the anisotropy of the lowest DARPin concentration and were fit, as previously described22, by the following equation:
| 5 |
where FDG is the anisotropy of GS-eGFP at the given concentrations, which is proportional to the fraction of the bound GS-eGFP, cD is the concentration of DARPin, cG is the fixed concentration of GS-eGFP, KD is the dissociation constant, and m is the anisotropy amplitude between the unbound and bound GS-eGFP.
Statistics and reproducibility
For degradation rate comparison, statistical analyses were performed using unpaired two-sided Student’s t tests to compare mean degradation rate constants between different analytes. Degradation rate constants were determined at the single-cell level, and results are reported as mean ± standard deviation (for numerical values see Supplementary Data). p values from these comparisons are provided in the Supplementary Information. Experimental replicates were defined as individual, healthy single cells injected with the same analyte, regardless of the date of injection. For each analyte, data were typically collected from 10 to 100 cells, as detailed in Supplementary Tables ST1 and ST2. We found degradation rates to be reproducible across multiple injection days for the proof-of-principle analyte eGFP, as described in our previous work20.
For western blot-based comparison of ubiquitination of protein complexes statistical significance was assessed using one-way ANOVA followed by Tukey’s multiple comparison test. Results are reported as mean ± standard deviation (for numerical values see Supplementary Data) derived from three biological replicates, each performed independently by mixing and reacting all components on separate days. We found that differences in ubiquitination levels between complexes were reproducible across replicates.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Materials
Acknowledgements
We thank the Center for Microscopy and Image Analysis (ZMB), particularly Dr. Jana Döhner and Dr. Urs Ziegler, for microscope access and valuable technical advice. We are grateful to the Functional Genomics Center Zurich for proteomics analyses, and in particular Dr. Antje Dittmann and Laura Kunz for LC-MS-MS analysis of ubiquitination assays, and Dr. Serge Chesnov, for ESI-MS analysis of purified proteins. We also thank Cedric Kiss, Janosch Ehrenmann and Gwendal Loarer for help with X-ray crystallography. Finally, we thank Sven Furler for performing analytical HPLC and SEC-MALS experiments. Funding was obtained from the Schweizerische Nationalfonds, grant 310030_192689 (to A.P.).
Author contributions
D.V. and D.W. jointly designed experiments. D.W. performed ubiquitination assays and N-terminal acetylation. D.V. conducted statistical analyses and data fitting. D.V. and D.W. performed and analyzed microinjection experiments. A.H., L.R., A.U., and D.W. fluorescently labeled proteins. A.H., L.R., A.U., D.V., and D.W. expressed and purified proteins. A.H., A.U., D.V., and D.W. carried out CD spectroscopy experiments. A.H. and D.W. executed thermofluor assays. L.R., D.V., and D.W. performed SPR. D.W. conducted fluorescence anisotropy experiments. S.H., P.M., and D.W. worked on X-ray crystallography. D.V. and D.W. performed protein unfolding assays. E.M. performed protein energy calculations using Rosetta software for epitope binning. A.Ho. performed energy calculations for lysine mutations. J.K. established LysateUb assay. A.P. performed modeling of DARPin/GFP complexes. A.P., D.V., and D.W. wrote the manuscript. D.V. and D.W. jointly designed figures. A.P. supervised the work and provided input, critical feedback, and guidance.
Peer review
Peer review information
Communications Biology thanks Jonathan Taylor and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Laura Rodríguez Pérez. A peer review file is available.
Data availability
All protein sequences are included within this paper’s Supplementary Data 1 file. All uncropped gels are included within this paper’s Supplementary Information file, Supplementary Fig. S17. Source data for the graphs shown in this paper and underlying single-cell degradation rates are included in Supplementary Data file 2. Raw data on the single timepoint- and cell-level are available at https://github.com/theplueckthunlab/2024_paper_microinjection. The mass spectrometry proteomics have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository61 with the dataset identifier PXD063877. Molecular structures have been deposited to the PDB with the identifiers 9F22, 9F23 and 9F24.
Code availability
Code for single-cell degradation rate analysis has been deposited on GitHub and can be accessed at https://github.com/theplueckthunlab/2024_paper_microinjection.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Dorothea Winkelvoß, David Vukovic.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-025-08352-w.
References
- 1.Yen, H.-C. S., Xu, Q., Chou, D. M., Zhao, Z. & Elledge, S. J. Global protein stability profiling in mammalian cells. Science322, 918–923 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Komander, D. & Rape, M. The ubiquitin code. Ann. Rev. Biochem.81, 203–229 (2012). [DOI] [PubMed] [Google Scholar]
- 3.McClellan, A. J., Laugesen, S. H. & Ellgaard, L. Cellular functions and molecular mechanisms of non-lysine ubiquitination. Open Biol.9, 190147 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.George, A. J., Hoffiz, Y. C., Charles, A. J., Zhu, Y. & Mabb, A. M. A comprehensive atlas of E3 ubiquitin ligase mutations in neurological disorders. Front. Genet.9, 29 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guharoy, M., Bhowmick, P., Sallam, M. & Tompa, P. Tripartite degrons confer diversity and specificity on regulated protein degradation in the ubiquitin-proteasome system. Nat. Commun.7, 10239 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prakash, S., Inobe, T., Hatch, A. J. & Matouschek, A. Substrate selection by the proteasome during degradation of protein complexes. Nat. Chem. Biol.5, 29–36 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fishbain, S. et al. Sequence composition of disordered regions fine-tunes protein half-life. Nat. Struct. Mol. Biol.22, 214–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fishbain, S., Prakash, S., Herrig, A., Elsasser, S. & Matouschek, A. Rad23 escapes degradation because it lacks a proteasome initiation region. Nat. Commun.2, 192 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu, H. et al. Conserved sequence preferences contribute to substrate recognition by the proteasome. J. Biol. Chem.291, 14526–14539 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dale, B. et al. Advancing targeted protein degradation for cancer therapy. Nat. Rev. Cancer21, 638–654 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang, Y. et al. Progress and challenges in targeted protein degradation for neurodegenerative disease therapy. J. Med. Chem.65, 11454–11477 (2022). [DOI] [PubMed] [Google Scholar]
- 12.Swartzel, J. C. et al. Targeted degradation of mRNA decapping enzyme DcpS by a VHL-recruiting PROTAC. ACS Chem. Biol.17, 1789–1798 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang, X. et al. Annual review of PROTAC degraders as anticancer agents in 2022. Eur. J. Med. Chem.267, 116166 (2024). [DOI] [PubMed] [Google Scholar]
- 14.Troup, R. I., Fallan, C. & Baud, M. G. Current strategies for the design of PROTAC linkers: a critical review. Explor. Target. Antitumor Ther.1, 273–312 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.VanDyke, D., Taylor, J. D., Kaeo, K. J., Hunt, J. & Spangler, J. B. Biologics-based degraders—an expanding toolkit for targeted-protein degradation. Curr. Opin. Biotechnol.78, 102807 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim, S. et al. bioPROTACs as versatile modulators of intracellular therapeutic targets including proliferating cell nuclear antigen (PCNA). Proc. Natl. Acad. Sci. USA117, 5791–5800 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim, S. et al. Exquisitely specific anti-KRAS biodegraders inform on the cellular prevalence of nucleotide-loaded states. ACS Cent. Sci.7, 274–291 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon, G. et al. BIO-PROTAC: new treatment modality to treat undruggable KRAS mutants by application of cell penetrating scFv. Cancer Res.83, 569–569 (2023). [Google Scholar]
- 19.Bondeson, D. P. et al. Lessons in PROTAC design from selective degradation with a promiscuous warhead. Cell. Chem. Biol.25, 78–87.e75 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vukovic, D. et al. Protein degradation kinetics measured by microinjection and live cell fluorescence microscopy. Sci. Rep.14, 27153 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prakash, S., Tian, L., Ratliff, K. S., Lehotzky, R. E. & Matouschek, A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Biol.11, 830–837 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Brauchle, M. et al. Protein interference applications in cellular and developmental biology using DARPins that recognize GFP and mCherry. Biol. Open3, 1252–1261 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ballinger, C. A. et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol.19, 4535–4545 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plückthun, A. Ribosome display: a perspective. Methods Mol. Biol.805, 3–28 (2012). [DOI] [PubMed] [Google Scholar]
- 25.Binz, H. K., Stumpp, M. T., Forrer, P., Amstutz, P. & Plückthun, A. Designing repeat proteins: well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J. Mol. Biol.332, 489–503 (2003). [DOI] [PubMed] [Google Scholar]
- 26.DeLisa, M., Varner, J. & Portnoff, A. Targeted protein silencing using chimeras between antibodies and ubiquitination enzymes. US Patent Office US20140112922A1 (2014).
- 27.Portnoff, A. D., Stephens, E. A., Varner, J. D. & DeLisa, M. P. Ubiquibodies, synthetic E3 ubiquitin ligases endowed with unnatural substrate specificity for targeted protein silencing. J. Biol. Chem.289, 7844–7855 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikolay, R. et al. Dimerization of the human E3 ligase CHIP via a coiled-coil domain is essential for its activity. J. Biol. Chem.279, 2673–2678 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Balaji, V. et al. A dimer-monomer switch controls CHIP-dependent substrate ubiquitylation and processing. Mol. Cell82, 3239–3254 (2022). [DOI] [PubMed] [Google Scholar]
- 30.Ye, Z. et al. Symmetry breaking during homodimeric assembly activates an E3 ubiquitin ligase. Sci. Rep.7, 1789 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, T. T., Cheng, L. & Kain, S. R. Optimized codon usage and chromophore mutations provide enhanced sensitivity with the green fluorescent protein. Nucleic Acids Res.24, 4592–4593 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alber, A. B., Paquet, E. R., Biserni, M., Naef, F. & Suter, D. M. Single live cell monitoring of protein turnover reveals intercellular variability and cell-cycle dependence of degradation rates. Mol. Cell71, 1079–1091.e1079 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Das, S., Singh, A. & Shah, P. Evaluating single-cell variability in proteasomal decay. bioRxiv, 10.1101/2023.1108.1122.554358 (2023).
- 34.Jiang, J. et al. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem.276, 42938–42944 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Xu, Z. et al. Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Struct. Biol.8, 1–13 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tinworth, C. P. et al. PROTAC-mediated degradation of Bruton’s tyrosine kinase is inhibited by covalent binding. ACS Chem. Biol.14, 342–347 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Wetzel, S. K., Settanni, G., Kenig, M., Binz, H. K. & Plückthun, A. Folding and unfolding mechanism of highly stable full-consensus ankyrin repeat proteins. J. Mol. Biol.376, 241–257 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Verdurmen, W. P., Luginbühl, M., Honegger, A. & Plückthun, A. Efficient cell-specific uptake of binding proteins into the cytoplasm through engineered modular transport systems. J. Control. Release200, 13–22 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Bard, J. A. M., Bashore, C., Dong, K. C. & Martin, A. The 26S proteasome utilizes a kinetic gateway to prioritize substrate degradation. Cell177, 286–298.e215 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martinez-Fonts, K. et al. The proteasome 19S cap and its ubiquitin receptors provide a versatile recognition platform for substrates. Nat. Commun.11, 477 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee, C., Prakash, S. & Matouschek, A. Concurrent translocation of multiple polypeptide chains through the proteasomal degradation channel. J. Biol. Chem.277, 34760–34765 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Chen, Y. et al. Proteolysis-targeting chimera (PROTAC) delivery system: advancing protein degraders towards clinical translation. Chem. Soc. Rev.51, 5330–5350 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deprey, K., Becker, L., Kritzer, J. & Plückthun, A. Trapped! A critical evaluation of methods for measuring total cellular uptake versus cytosolic localization. Bioconjug. Chem.30, 1006–1027 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan, A. et al. Lipid-mediated intracellular delivery of recombinant bioPROTACs for the rapid degradation of undruggable proteins. Nat. Commun.15, 5808 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sung, Y. K. & Kim, S. Recent advances in the development of gene delivery systems. Biomater. Res.23, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang, C. et al. Emerging non-viral vectors for gene delivery. J. Nanobiotechnol.21, 272 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caussinus, E., Kanca, O. & Affolter, M. Fluorescent fusion protein knockout mediated by anti-GFP nanobody. Nat. Struct. Mol. Biol.19, 117–121 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Simon, M., Frey, R., Zangemeister-Wittke, U. & Plückthun, A. Orthogonal assembly of a designed ankyrin repeat protein-cytotoxin conjugate with a clickable serum albumin module for half-life extension. Bioconjug. Chem.24, 1955–1966 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Studier, F. W. Stable expression clones and auto-induction for protein production in E. coli. Methods Mol. Biol.1091, 17–32 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Vonrhein, C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. D Biol. Crystallogr.67, 293–302 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kabsch, W. Xds. Acta Crystallogr. D Biol. Crystallogr.66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Evans, P. R. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr. D Biol. Crystallogr.67, 282–292 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McCoy, A. J. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr.63, 32–41 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murshudov, G. N. et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr.67, 355–367 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr.60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 56.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/ (2021).
- 57.Pace, C. N. Measuring and increasing protein stability. Trends Biotechnol.8, 93–98 (1990). [DOI] [PubMed] [Google Scholar]
- 58.Nelder, J. A. & Mead, R. A simplex-method for function minimization. Comput. J.7, 308–313 (1965). [Google Scholar]
- 59.Scaglione, K. M. et al. Ube2w and ataxin-3 coordinately regulate the ubiquitin ligase CHIP. Mol. Cell43, 599–612 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Türker et al. B-Fabric: the Swiss Army Knife for life sciences. In EDBT ‘10: Proceedings of the 13th International Conference on Extending Database Technology. 10.1145/1739041.1739135 (2010).
- 61.Perez-Riverol, Y. et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res.50, D543–D552 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Materials
Data Availability Statement
All protein sequences are included within this paper’s Supplementary Data 1 file. All uncropped gels are included within this paper’s Supplementary Information file, Supplementary Fig. S17. Source data for the graphs shown in this paper and underlying single-cell degradation rates are included in Supplementary Data file 2. Raw data on the single timepoint- and cell-level are available at https://github.com/theplueckthunlab/2024_paper_microinjection. The mass spectrometry proteomics have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository61 with the dataset identifier PXD063877. Molecular structures have been deposited to the PDB with the identifiers 9F22, 9F23 and 9F24.
Code for single-cell degradation rate analysis has been deposited on GitHub and can be accessed at https://github.com/theplueckthunlab/2024_paper_microinjection.