Abstract
Background
Microbial production of conjugated linoleic acid (CLA) has garnered wide attention for the possibility to increase the CLA content in food products, therefore achieving higher concentrations of beneficial compounds for consumers. However, this approach has only been done using metabolically active cells, particularly in Bifidobacterium spp., thus being a major limitation given the anaerobic and fastidious nature of bifidobacteria. In this study, we aimed to investigate the capacity of Bifidobacterium breve JKL2022 (KACC81214BP) to convert free linoleic acid (LA) into CLA using growing cells and postbiotic preparations (washed cells and crude protein extracts) as catalysts.
Results
Bifidobacterium breve JKL2022 demonstrated high CLA production as early as 6 h and continued to increase until 12–15 h of incubation. Moreover, CLA production was observed in JKL2022 washed cells (97.42 ± 3.64%) and crude protein fractions (33.87 ± 4.05%– 103.65 ± 2.70%) obtained after cell lysis, highlighting its superior CLA-converting activity compared to the B. breve JCM strains. In vitro CLA reaction conditions were optimal at pH 7.0, following the first-order kinetics within the first 5 min of reaction, and the extraction efficiency of the isopropanol-hexane protocol increased after adjusting the pH to 5.0–5.5. Finally, RT-qPCR and in silico analysis revealed a strong correlation between the expression levels of lai (JKL2022_00014) and tetR (JKL2022_00217) genes, suggesting the potential role of TetR in upregulating the lai gene expression in JKL2022 that could explain the LA conversion in washed JKL2022 cells.
Conclusions
The ability of B. breve JKL2022 strain to convert free LA to CLA during growth, as well as using washed cells and crude protein extracts, suggests strain specificity and superior enzymatic activity. In addition to its potential application as a probiotic strain with CLA-enhancing properties, washed JKL2022 cells or crude protein extracts can be developed as postbiotic preparations for the same purpose.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12934-025-02766-1.
Keywords: Conjugated linoleic acid, Probiotics, Postbiotics, Linoleic acid isomerase, Transcriptional regulation, Catalytic activity
Background
Several dairy and meat products contain beneficial fatty acids, including monounsaturated fatty acids (MUFA) (e.g., oleic acid and palmitoleic acid) or polyunsaturated fatty acids (PUFA) such as omega-3 (e.g., α-linolenic acid, eicosapentaenoic acid, and docosahexaenoic acid) and omega-6 fatty acids (e.g., linoleic acid (LA) and arachidonic acid) [1–3]. Consumption of foods rich in MUFAs and PUFAs is believed to be beneficial for heart health, as they can help lower low-density lipoprotein cholesterol levels [4, 5], along with exerting anti-inflammatory properties [6, 7]. With the growing demand for functional foods and increasing interest in healthier lifestyles, research efforts have focused on adding value to various food products to achieve different health benefits [8]. Among them, conjugated linoleic acid (CLA)-enhanced dairy products and CLA-producing bacteria have garnered substantial interest [9, 10]. CLA is a group of geometric and positional dienoic isomers of LA, with the cis-9, trans-11 isomer being the most abundant and biologically active [11, 12]. Several benefits of CLA have been reported, including anti-inflammatory [13–15], anti-carcinogenic [16, 17], anti-obesity [18–20], and immunomodulatory properties [21, 22].
However, the application of CLA-producing bacteria has been limited to growing cells, particularly in the case of Bifidobacterium, wherein CLA production occurs only in metabolically active cells [23–25]. Andrade et al. highlighted that most studies on LA conversion in Bifidobacterium used growing cells, predominantly with B. breve strains [12]. Several independent studies have corroborated this observation, highlighting a major limitation in applying Bifidobacterium for CLA production [26, 27]. Furthermore, the toxicity of free LA as a substrate for microbial LA conversion presents another hurdle [28, 29]. As LA exposure slows the growth of the producing strain, it remains challenging to optimize the conditions for maximum CLA production using growing Bifidobacterium spp. cells.
Recently, the concept of postbiotics, defined as a “preparation of inanimate microorganisms and/or their components that confers a health benefit on the host,” [30] has gained increasing attention. Similar to probiotics, postbiotics may offer the similar health benefits to the host. The main difference lies in the viability of their components, which confers advantages to postbiotics under certain conditions, such as a longer shelf-life compared to that of probiotics, a lower risk of transmitting virulence factors and antimicrobial resistance genes through horizontal gene transfer, being safer to administer with a well-defined composition, and generally being more stable [31].
Bifidobacterium breve JKL2022 (KACC81214BP), a strain isolated from fecal samples of healthy infants, exhibited CLA-converting activity when washed cells, partially lysed cells, and crude cell-free protein extracts were used as catalysts [32]. The present study aimed to provide concrete evidence of the outstanding CLA-converting properties of the JKL2022 strain with efficient production of specific CLA isomers (cis-9, trans-11 isomer; 73.27 ± 3.80%) from free LA as compared to JCM strains, which failed to convert CLA when using their washed cells or crude protein extracts [32]. Additionally, this work investigated the potential regulatory role of TetR protein (tetR, JKL2022_00217) in the expression of the lai gene of B. breve using RT-qPCR, providing the foundational framework for understanding the CLA biosynthesis in bifidobacteria, which aligns with previous reports of TetR regulation of CLA biosynthesis in Lactobacillus plantarum [33]. The development of JKL2022 as pro- and postbiotic agents with strain-specific CLA biosynthesis capacity serves as a promising aspect for enhancing the CLA content of various food systems, particularly when the development of functional probiotics for food incorporation is challenging.
Materials and methods
Bacterial strains, growth conditions, and reagents
Bifidobacterium breve JKL2022 (KACC81214BP), isolated from fecal matter of healthy infants [34] and other Bifidobacterium strains (B. breve JCM7017, JCM7019, JCM1273, JCM1192, and B. animalis subsp. lactis BB12, obtained from culture collections) were routinely cultured in de Man, Rogosa, and Sharpe (MRS) broth (BD Difco, USA) supplemented with 0.05% (wt/vol) L-cysteine hydrochloride (cys-MRS) (Sigma, USA) at 37 °C under anaerobic conditions (GasPak™ EZ anaerobic chamber, BD Difco). All strains were kept in 10% skim milk + glycerol (3:1, vol: vol) at -80 °C unless stated otherwise. Stock solutions of LA (50 mg/mL, Sigma Aldrich) were prepared as emulsions in 50 mM sodium phosphate buffer (pH 7.0) containing 2% (vol/vol) Tween 80. Emulsification was achieved by vigorously vortexing the suspension prior to storage at -21 °C. The stock LA solutions were thawed on ice (4 °C) before use.
CLA production in growing cells and postbiotic preparations
CLA production in growing conditions
Bifidobacterium breve JKL2022 and other bifidobacterial strains were activated from glycerol stocks and inoculated (1%, vol/vol, approx. 1 × 107 CFU/mL) in 5 mL cys-MRS broth and cys-MRS broth supplemented with LA (0.50 mg/mL). Viable cell count, pH, and CLA concentration were measured every 3 h for 24 h. Cell viability was determined by plating on cys-MRS agar incubated anaerobically at 37 °C. The pH profile was measured using a BP3001 benchtop pH meter (Trans Instruments, Singapore). The CLA concentration was determined using the isopropanol-hexane extraction protocol with minor modifications [35]. Briefly, 400 µL of bacterial cell culture was transferred to a sterile 2.0 mL microfuge tube, followed by sequential addition of 800 µL of isopropanol (Sigma) and 600 µL of hexane (Sigma). The mixture was vortexed for 5 min, followed by centrifugation (980 × g, 5 min, 20 °C) to facilitate phase separation. The hexane (top) layer containing the conjugated fatty acids was diluted in methanol (Sigma) in a 100:900 ratio (vol:vol), and the absorbance was measured at 233 nm in a UV-transparent 96-well plate (UVMax, SPL, Korea). All optical density (OD) readings were performed with an INNO spectrophotometer (Korea). All experiments were performed thrice in triplicate.
CLA production using postbiotic preparations
Different B. breve postbiotic preparations were investigated for their capacity to convert CLA, more specifically, heat-inactivated cells, washed cells, and crude cell-free preparations. The washed Bifidobacterium cells were prepared from overnight cultures (18–21 h) in cys-MRS (approx. 1 × 109 CFU/mL) by centrifuging (8,000 × g, 4 °C, 10 min) and resuspending the cell pellet twice with 1× PBS. The final resuspended cells were collected and referred to as washed cells (1× cell concentration, approx. 1 × 109 CFU/mL). Heat-inactivated cells were prepared by incubating the washed cells at 70 °C for 10, 20, and 30 min in a water bath and immediately transferring into a 4 °C ice bath [36]. The crude protein fractions were prepared through cell lysis by sonication (2 s on, 10 s off, 30 cycles) on ice using a Vibra cell sonicator VCX 500™. The resulting cell lysate was centrifuged (17,500 × g, 4 °C, 20 min) and filtered through a 0.45 μm Acrodisc® syringe filter (Pall Corporation, USA), hereby designated as the cell-free extract (CFE). The remaining pellet was resuspended in 1× PBS supplemented with 2% (vol/vol) Triton X-100, incubated at 4 °C for 2 h, and centrifuged and filtered under the same conditions previously described. The collected fraction was designated as the membrane extract fraction (T100). The protein concentrations of the CFE and T100 fractions were standardized to 1.00 mg/mL using 1× PBS. Protein concentration was measured using the Bradford assay with BSA as the protein standard [37].
The LA conversion test using the washed cells, heat-inactivated cells, CFE, and T100 fractions was performed as follows: First, 4 µL of 50 mg/mL LA was emulsified in the reaction buffer (50 mM sodium phosphate (Na-P) buffer + 0.5% Tween 80, pH 6.5–7.0). Subsequently, 40 µL of the washed cells, CFE, or T100 fractions was added, and the tubes were mixed by inversion before incubating at 37 °C for 20 min. The pH of the reaction mixture was adjusted to pH 5.0–5.5 by adding 10 µL of 1 N HCl, and the CLA was extracted and quantified through the isopropanol-hexane method described above. All experiments were performed in triplicate with three independent trials.
GC-FID analysis of LA conversion
The CLA production by B. breve JKL2022 was further validated by GC-FID, as previously described [32]. The CLA fractions, extracted through the isopropanol-hexane method from the in vitro LA conversion reactions using washed cells as catalysts, were used as the starting material for fatty acid methylation: T0, CLA fraction extracted immediately after the addition of cells; T20, CLA fraction extracted after 20 min of reaction. Heptadecenoic acid (C17:0) was added as the internal standard at a final concentration of 1.0 mg/mL. Then, the fatty acids were collected using the Folch method [38], and methyl esterification was performed using 14% BF3-MeOH (Sigma). The fatty acid methyl ester (FAME) samples were analyzed using an HP 6890 GC (Hewlett-Packard, USA) equipped with a flame ionization detector (FID) and HP-88 column (100 m × 0.25 mm i.d., 0.20 μm film thickness, Agilent, USA). The column oven temperature was maintained at 140 °C for 5 min, increased by 4 °C/min until reaching 240 °C, where it was maintained for 20 min. Approximately 1 µL of the samples were injected with a 1:50 split ratio with N2 used as the carrier gas (1 mL/min). The temperatures of the injection and detection ports were set to 260 °C and 270 °C, respectively. Results are expressed as the peak area ratio using the formula below:
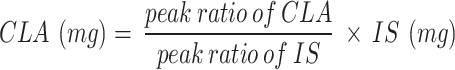 |
where IS represents the weight (mg) of the internal standard.
Optimization of in vitro LA conversion
Effect of pH on LA conversion
The effect of reaction pH on the final CLA yield and extraction efficiency was investigated. Briefly, buffers with different pH values were prepared as described in Table 1 and Table S3. Following this, LA was emulsified in the respective buffer at a final concentration of 0.50 mg/mL (1% vol/vol) and mixed with 40 µL of 2.0 mg/mL CFE obtained from a 24 h culture of JKL2022. The reaction was subsequently incubated at 37 °C for 20 min, and the pH was adjusted by adding either 1 N HCl or 1 N NaOH before CLA extraction using the isopropanol-hexane protocol, as previously described. A reaction mix without CFE was used as the negative control.
Table 1.
Reaction conditions and pH adjustments for determining the effect of pH on LA conversion activity and extraction efficiency
| Reaction Buffer | Extraction pH | pH adjustments |
|---|---|---|
| 50 mM Citric Acid- Sodium Phosphate Buffer, pH 4.5 | 4.5 | - |
| 5.0 | 2.53 µL NaOH | |
| 5.5 | 5.33 µL NaOH | |
| 7.0 | 13.20 µL NaOH | |
| 50 mM Citric Acid- Sodium Phosphate Buffer, pH 5.0 | 4.5 | 3.33 µL HCl |
| 5.0 | - | |
| 5.5 | 2.53 µL NaOH | |
| 7.0 | 10.67 µL NaOH | |
| 50 mM Citric Acid- Sodium Phosphate Buffer, pH 5.5 | 4.5 | 6.67 µL HCl |
| 5.0 | 3.60 µL HCl | |
| 5.5 | - | |
| 7.0 | 8.14 µL NaOH | |
| 50 mM Citric Acid- Sodium Phosphate Buffer, pH 6.0 | 4.5 | 10.05 µL HCl |
| 5.0 | 6.67 µL HCl | |
| 5.5 | 3.60 µL HCl | |
| 7.0 | 5.61 µL NaOH | |
|
50 mM Sodium Phosphate Buffer, pH 7.0 |
4.5 | 12.27 µL HCl |
| 5.0 | 11.73 µL HCl | |
| 5.5 | 11.06 µL HCl | |
| 7.0 | - |
Effect of tween 80 concentration in LA stock solution
The effect of the concentration of Tween 80 used to emulsify the LA stock solution on the final CLA yield and LA toxicity in growing cells was investigated [39]. Initially, the LA stock was prepared at a final concentration of 50 mg/mL in Na-P buffer (pH 7.0) supplemented with either 2.0% or 5.0% (v/v) Tween 80 and vortexed to facilitate emulsification. Then, Bifidobacterium strains were inoculated in (A) cys-MRS + 1% (v/v) LA in 2.0% Tween 80 or (B) cys-MRS (adjusted to contain 0.5% Tween 80) + 1% (v/v) LA in 5.0% Tween 80, respectively. The cultures were incubated anaerobically at 37 °C. Viable cell count, pH, and CLA production were measured after 12 and 24 h incubation as previously described. The reaction mix without the addition of CFE was used as a negative control.
Effect of emulsification method on LA availability
The effect of different emulsification methods on the availability of LA as a substrate for LA conversion was investigated. Different methods for preparing LA stocks in Tween 80-supplemented buffer were compared [40, 41]. First, 50 mg of LA was mixed with 50 mM sodium phosphate buffer (pH 7.0) containing 5% Tween 80 and emulsified by (1) vortexing for 1–2 min using a stand vortex (Scientific Industries, Vortex-Genie 2); (2) ultrasonic bath (Bransonic® Ultrasonic Bath 3510R-DTH) for 3 min; and (3) ultrasonic processing (Sonics & Materials, Vibra Cell™) for 1 min. The emulsified LA stocks were used for the LA conversion assay as previously described.
Large-scale CLA production using washed cells
Increasing concentrations of washed JKL2022 cells were used to evaluate the strain’s capacity to convert increasing LA concentrations into CLA. First, different volumes (4, 8, 20, 40, and 80 µL) of 50 mg/mL LA were mixed with the reaction buffer (50 mM Na-P buffer + 0.5% Tween 80, pH 6.5–7.0) to reach a final reaction volume of 400 µL and emulsified by vortexing. Subsequently, 40 µL of concentrated washed cells was added to achieve final cell concentrations of 0.1×, 0.5×, 1.0×, 2.0×, and 5.0×, with 1.0× cell concentration equivalent to approx. 1 × 109 CFU/mL. The reaction mixtures were incubated as previously described. Then, the pH of the reaction mixture was adjusted by adding 10 µL of 1 N HCl, and CLA was extracted and quantified using the isopropanol-hexane method described above. The total CLA converted was calculated using the linear equation derived from the standard curve (Fig. S1) of CLA:
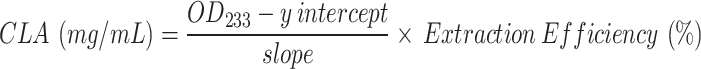 |
where OD233 is the net absorbance of the extracted hexane fraction measured at 233 nm, and extraction efficiency is the calculated efficiency of the isopropanol-hexane extraction (55–60%). For 100:900 dilution, the y intercept is 0.1493 and the slope is 4.9, while for 50:950 dilution, the y intercept is 0.017 and the slope is 2.7334. The conversion rate (%) was calculated using the following formula:
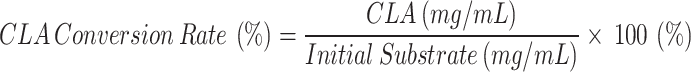 |
where CLA (mg/mL) is the concentration of CLA calculated from the reaction, and Initial Substrate (mg/mL) is the concentration of LA used for the reaction.
Timepoint LA conversion
A timepoint conversion experiment was performed to determine the rate of the enzyme reaction in vitro using LA or LNA as substrate. The reaction mixture was prepared by emulsifying 1.0 mg/mL of LA or LNA, as described previously. Enzyme reaction was initiated by adding 40 µL of JKL2022 CFE (0.288 mg/mL) and incubated in a 25 °C water bath. The reactions were terminated after 0.5-, 1-, 2-, 5-, 10-, and 20-min incubation by heat treatment at 80 °C for 5 min. The CLA (CLnA) was extracted using the isopropanol-hexane extraction method, and the concentration was measured spectrophotometrically at 233 nm. The catalytic activity of LAI was determined using the Beer-Lambert's Law:
 |
where ΔA/min is the change in absorbance over time, ε CLA at 233 nm is equal to 23,360 Lmol− 1 cm− 1, d is the pathlength of the cell (0.72 cm), V is the volume of the reaction (400 µL), v is the volume of the enzyme (40 µL), and c is the concentration of substance (CFE, 0.288 mg/mL). Purified recombinant LAI_sGFP (0.10 mg/mL) [32], was used to standardize the in vitro CLA reaction.
RT-qPCR and predictive analysis of lai transcription regulation by TetR
The expression profiles of lai and tetR across B. breve strains were evaluated. Total RNA was extracted from 15 h cultures grown in cys-MRS and cys-MRS supplemented with LA using the Invitrogen PureLink RNA extraction kit (USA) with on-column rDNase treatment. The total RNA was reverse transcribed using the PrimeScript First Strand cDNA synthesis kit (Takara, Japan) following the standard protocol. The resulting cDNA was diluted in RNase-free water (1:20), and the concentration was measured using nanodrop spectrophotometry, being adjusted to concentrations of 60–70 ng/µL for subsequent application as the template in the following reaction. The RT-qPCR analysis was performed using the Dyne Fast qPCR 2× Premix (SYBR Green with low ROX, DYNE BIO, Korea) using a standard PCR condition in a CFX Opus 96 Real-time PCR system (Bio-Rad). The primer sequences are listed in Table S1. Melting peaks, melting curves, and relevant CT values for each amplification reaction were recorded automatically. The gapdh gene was used for data normalization. The ΔΔCT method was used to quantify the expression levels of the target genes [42]. Three technical replicates of each amplification reaction were performed for each of the four biological replicates.
Prediction of functional protein-protein interaction networks was performed using the STRING ver. 12.0 [43] by searching multiple proteins by sequence. The amino acid sequences of the proteins used for the analysis are listed in Table S2. Integrated protein-DNA docking was performed using the HDOCK server ver. 2023-09-09 (http://hdock.phys.hust.edu.cn/) to determine the interaction between the TetR proteins and the lai promoter region [44]. The PDB file of TetR proteins and a 40-bp long DNA, containing the −10 and −35 ribosome recognition sites upstream of the lai gene, were used for this analysis.
Statistical analyses
Differences in the means for the effects of LA exposure on Bifidobacterium spp. growth and CLA production, CLA production using washed cells and protein extracts, the effect of Tween 80 concentration on LA toxicity, the effect of different emulsification methods on LA conversion, and large-scale LA conversion using increasing washed cell concentrations were measured using the ordinary two-way analysis of variance (ANOVA) (α, 0.05) with Šidák’s multiple comparison post-hoc test. The effect of pH on LA conversion and extraction efficiency, and the difference in gene expression levels of lai, tetR-217, and tetR-159 relative to B. breve JKL2022 were measured using the ordinary one-way ANOVA (α, 0.05) with Tukey’s multiple comparison post-hoc test. The differences in lai and tetR-217 expression levels as a function of LA exposure were measured using a two-way RM ANOVA (α, 0.05) with Šidák’s multiple comparison post-hoc test. The differences in lai and tetR-217 expression levels across the strains tested were measured using a two-way RM ANOVA (α, 0.05) with Tukey’s multiple comparison post-hoc test. All analyses were performed using GraphPad Prism v. 9.5.1.
Results
CLA production by Bifidobacterium spp
Bifidobacterium breve JKL2022, B. breve JCM strains, and B. animalis subsp. lactis Bb-12 were investigated for LA conversion under four conditions: growing cells, washed cells, heat-inactivated cells, and crude protein extracts. When cultured in cys-MRS supplemented with 0.50 mg/mL of free LA, the growth of strains was negatively affected (Fig. 1). In particular, JKL2022 and JCM7019 exhibited stunted growth and failed to reach 9.0 log(CFU/mL) after 18–24 h of incubation, whereas the other strains exhibited only a prolonged log phase (Fig. 1E). In terms of their pH profile, LA exposure resulted in slightly higher pH values after incubation. Nevertheless, all B. breve strains exhibited favorable CLA production within 6–9 h of incubation, with JKL2022 and JCM7017 producing the highest amount of CLA among the strains tested (Fig. 1F). In contrast, B. animalis subsp. lactis Bb-12 did not produce CLA during its growth.
Fig. 1.
Difference in growth curve, fermentation profile and CLA production of Bifidobacterium strains cultured under normal conditions (A-C) and exposed to 0.5 mg/mL LA (D-F). a−d Different letters denote significantly different results at each timepoint (p < 0.05)
In terms of CLA production using postbiotic preparations (i.e., washed cells or CFE), only JKL2022 exhibited significant CLA production, whereas the remaining strains failed to produce detectable amounts of CLA, regardless of cell concentration (Fig. 2). Additionally, the crude protein fraction derived from the T100 fraction showed significantly higher (66.61%) CLA production (OD233, 1.411, equivalent to 0.287–0.313 mg/mL CLA) compared to the CFE fraction (OD233, 0.471, equivalent to 0.091–0.100 mg/mL CLA). However, heat-inactivated JKL2022 cells lost all CLA-converting activities. GC-FID analysis confirmed the LA conversion activity of JKL2022, with cis-9, trans-11 CLA as the predominant isomer (69.48–77.07%) based on peak-area ratio. Other CLA isomers (trans-10, cis-12 CLA; cis-9, cis-11 CLA; and trans-9, trans-11 CLA) were present at considerably lower concentrations. A schematic diagram for the preparation of postbiotic agents derived from B. breve JKL2022 is presented in Figure S2.
Fig. 2.
LA conversion of (A) washed cells and (B) crude protein extracts of Bifidobacterium strains. The LA conversion was confirmed by GC-FID analysis using T0 (C) and T20 (D) B. breve JKL2022 in vitro reactions. Heptadecanoic acid (C17:0) was used as internal standard (IS)
Effect of tween 80 emulsion on CLA production
Increasing the Tween 80 concentration from 2.0 to 5.0% during LA emulsification reduced its toxicity (Fig. 3). For JKL2022, cell viability increased by 13.92% and 31.22% after 12 and 24 h, respectively, accompanied by shifts in the fermentation profile consistent with improved metabolic activity. For the remaining strains, an increase in the Tween 80 concentration induced only a slight improvement in growth. In terms of LA conversion, all strains exhibited improved CLA production when LA was emulsified in 5% Tween 80. After 12 h of incubation, B. breve JCM7019 exhibited the largest increase in CLA concentration (69.32%), followed by JKL2022 (60.20%), JCM1273 (47.29%), JCM7017 (20.69%), and JCM1273 (4.52%). Additionally, the effect of different emulsification methods (e.g., vortexing, ultrasonic bath, and sonication) on substrate availability was examined. The results indicated that sonication of LA in Tween 80 yielded the highest CLA yield, followed by vortexing and ultrasonication (Fig. 4). However, the difference in CLA production between LA prepared by sonication and vortexing was not statistically significant (p > 0.05) regardless of the substrate concentration.
Fig. 3.
Effect of Tween 80 concentration on LA toxicity measured in terms of pH, cell viability, and CLA production of (A) B. breve JKL2022, (B) B. breve JCM7017, (C) B. breve JCM7019, (D) B. breve JCM1273, (E) B. breve JCM1192, and (F) B. animalis subsp. lactis Bb-12. * Indicate significantly different means (p < 0.05)
Fig. 4.
Effect of LA-emulsification method on LA conversion of Bifidobacterium breve JKL2022. a − b Different letters denote significantly different results (p < 0.05)
Optimization of reaction conditions and extraction protocol
Different pH conditions were tested to determine the effect of the pH of the reaction and the extraction method on the final CLA yield. Different acidity of reaction buffers yielded different CLA concentrations. Under normal extraction conditions, conversion reactions at pH 7.0 yielded the lowest CLA concentration (0.571 ± 0.09 OD233) while acidic reaction buffers resulted in higher CLA concentrations. Specifically, reactions at pH 5.5, pH 5.0, and pH 4.5 yielded 1.624 ± 0.278, 1.415 ± 0.198, and 1.528 ± 0.105 CLA (OD233), respectively. To determine the effect of pH on the extraction efficiency of the isopropanol-hexane protocol, the pH was adjusted after the LA conversion reaction. Generally, lowering the buffer pH immediately prior to isopropanol-hexane extraction resulted in a higher CLA yield compared to extractions performed without pH alterations. In contrast, neutralization of the acidic reaction buffers resulted in lower CLA yields. Overall, the highest CLA production was achieved when the reaction was catalyzed at pH 7.0 and extraction was performed at pH 5.0–5.5 (1.700 ± 0.142 to 1.756 ± 0.205 OD233), as shown in Fig. 5.
Fig. 5.
Effect of different reaction buffer pH on LA conversion and pH adjustments on extraction efficiency. a−c Different letters denote significantly different results (p < 0.05)
Increased CLA production in washed cells
To determine the capacity of JKL2022 to produce higher amounts of CLA compared to the standard reaction, LA conversion using increasing concentrations of washed cells and substrate was investigated (Fig. 6). Washed JKL2022 cells were inoculated into the reaction mixture at final concentrations of 0.1×, 0.5×, 1.0×, 2.0×, and 5.0×. Each cell concentration was incubated with 0.5, 1.0, 2.5, 5.0, and 10.0 mg/mL LA. Using 5.0× washed cells, the conversion rate reached 73.02 ± 2.32%, 86.31 ± 2.56%, 89.79 ± 4.80%, 91.70 ± 2.35%, and 98.67 ± 4.90% when reacted with 10.0, 5.0, 2.5, 1.0, and 0.5 mg/mL LA, respectively.
Fig. 6.
Large-scale LA conversion using increasing concentrations of Bifidobacterium breve JKL2022 washed cells expressed as (A) total CLA concentration and (B) conversion rate (%). Legend, concentration of LA used in each reaction (mg/mL). a − e Different letters denote significantly different results (p < 0.05)
Catalytic activity of LAI
Based on the in vitro timepoint LA conversion reactions, it was confirmed that the LAI obtained from JKL2022 CFE follows the first-order kinetics from 0 to 5 min of reaction (Fig. 7). The CLA concentration started to plateau after 10 min of reaction. Using the Beer-Lambert’s Law, the specific activity of JKL2022 CFE (0.288 mg/mL) for LA and LNA was calculated to be 11,664.08 µU/g and 56,790.12 µU/g, respectively, whereas purified LAI_sGFP has a specific activity of 33,770.93 µU/g for LA and 143,661.10 µU/g for LNA (Table 2).
Fig. 7.
Catalytic activity of linoleic acid isomerase derived from Bifidobacterium breve JKL2022 and recombinant LAI_sGFP based on timepoint LA conversion assay using (A) LA and (B) LNA as initial substrate
Table 2.
Catalytic activity of LAI derived from JKL2022 CFE and purified Recombinant lai_sgfp based on Beer-Lambert’s law
| Catalyst | Substrate | ΔA/min | ΔC/min | Enzyme Units (mol/min) |
Enzyme Activity (Units/L) | Specific Activity (µU/g) |
Linear Regression Equation |
|---|---|---|---|---|---|---|---|
|
JKL2022 CFE (0.288 mg/mL) |
LA | 0.057 | 3.36E-06 | 1.34E-08 | 3.36E-03 | 11664.08 | Y = (0.05652)X + 0.5295; R2 = 0.9263 |
| LNA | 0.294 | 1.64E-05 | 6.54E-08 | 1.64E-02 | 56790.12 | Y = (0.2944)X + 0.7336; R2 = 0.9557 | |
|
LAI_sGFP (0.10 mg/mL) |
LA | 0.057 | 3.38E-06 | 1.35E-08 | 3.38E-03 | 33770.93 | Y = (0.05679)X + 0.4618; R2 = 0.8392 |
| LNA | 0.259 | 1.44E-05 | 5.74E-08 | 1.44E-02 | 143611.10 | Y = (0.2585)X + 0.6994; R2 = 0.9246 |
ΔA/min is the change in absorbance over time, equivalent to the slope of the linear regression equation (absorbance at OD233)
ΔC/min is the substrate conversion per time unit (mol/s)
TetR regulation of Lai expression
The expression levels of lai (JKL2022_00014) and tetR (JKL2022_00217 and JKL2022_00159) were investigated by RT-qPCR analysis to examine the possible role of TetR in lai expression. Total RNA was extracted from B. breve strains cultured in cys-MRS and cys-MRS supplemented with LA to examine the expression patterns of lai, tetR-217, and tetR-159 genes and correlate them with the differences in CLA production activity among the strains. Based on in vitro CLA assays, all B. breve strains used in this study converted LA into CLA during growth, but only B. breve JKL2022 converted CLA when using washed cells or CFE.
When cultured under normal conditions (cys-MRS, 37 °C, anaerobic conditions), the lai expression level in B. breve JKL2022 was significantly higher compared to the JCM strains (13.80–61.88× higher). The tetR-217 gene follows the same expression pattern (33.88–62.71× higher) as seen in Fig. 8A, highlighting the higher transcription level of both lai and tetR-217 in B. breve JKL2022, consistent with the LAI activity in washed cells and CFE of JKL2022. In JCM strains, the low expression of lai and tetR-217 resulted in low concentrations of the LAI enzyme, rendering their washed cells and CFE incapable of converting LA into CLA. In contrast, tetR-159 only showed moderate correlation with the lai expression level and CLA-converting activity across the strains. Statistical analysis confirmed that the strains account for 91.51% of the total variance (p < 0.0001), and the effect is considered extremely significant. Therefore, despite encoding the same enzyme, differences in their transcription levels resulted in completely different capacities for CLA production.
Fig. 8.
Regulatory role of TetR on lai gene expression based on (1) RT-qPCR analysis: (A) gene expression of lai, tetR-217, and tetR-159 under normal growing conditions relative to B. breve JKL2022; (B) gene expression lai, tetR-217, and tetR-159 under LA-exposed conditions relative to B. breve JKL2022; and (C) relative gene expression of lai, tetR-217, and tetR-159 as function of LA-exposure; (2) in silico predictions of (D) functional protein association network using STRING, and (3) recognition of lai promoter region by (E) TetR-217 and (F) TetR-159 dimers using HDOCK analysis. a-d Different letters denote significantly different results (p < 0.05)
When cultured in the presence of LA, the expression levels of lai, tetR-217, and tetR-159 genes were upregulated (Figure S3). All JCM strains showed strong upregulation of lai (3.51–30.05-fold) genes due to LA exposure, while B. breve JKL2022 was weakly downregulated by 0.52-fold. The tetR-217 gene of JKL2022 (2.24-fold), JCM7017 (1.87-fold), and JCM7019 (1.92-fold) was upregulated, whereas those of JCM1192 (0.49-fold) and JCM1273 (0.76-fold) were downregulated. In contrast, the tet-159 gene of all strains was upregulated (1.20–2.74-fold). Nevertheless, the expression levels of lai and tetR-217 of JCM strains under LA-exposed conditions remain lower than that of JKL2022, except for the tetR-217 gene of JCM7019 (Fig. 8B). In this regard, strong upregulation of lai and tetR-217 was observed in the strains with higher CLA-converting activity (e.g., JCM7017 and JCM7019).
Discussion
Bifidobacterium breve has been widely utilized for its high CLA production activity compared to other bifidobacterial species [45, 46]. As a result, increasing research has focused on utilizing this species for product development with various health benefits, primarily CLA-enhanced or probiotic dairy products [47, 48]. However, the LA conversion mechanism has mostly been elucidated only for members of Lactobacillus, while that for Bifidobacterium remains unclear [49, 50]. The CLA pathway in lactobacilli involves at least three key enzymes: myosin cross-reactive antigen, short-chain dehydrogenase, and acetoacetate decarboxylase [51, 52]. Although homologs of these enzymes are also present in bifidobacteria, they are not involved in the bifidobacterial LA conversion pathway [53]. Particularly, the previously hypothesized MCRA-homolog in bifidobacteria was experimentally proven to be unrelated to the CLA biosynthesis pathway [54]. Instead, Bifidobacterium either employs a specific linoleic acid isomerase (LAI) that converts LA into CLA without any intermediates, or the conversion mechanism is too rapid for the intermediates to be easily detected [32, 46, 55]. Continuous research to elucidate the LA conversion pathway in different lactic acid bacteria remains crucial for the development of novel probiotics with enhanced health benefits.
In this study, B. breve JKL2022 (KACC81214BP) demonstrated excellent CLA-converting activity under growing conditions, in washed cells, and in crude CFE obtained from lysed cells. Under growth conditions, JKL2022, JCM7017, and JCM7019 showed significant CLA production after 9 h of incubation, which continued to increase until 12–15 h of incubation. Minimal changes in CLA concentration were observed after 24 h incubation. The highest LA conversion efficiencies were observed for JKL2022 and JCM7017, followed by JCM7019, JCM1273, and JCM1192. However, B. animalis Bb-12 did not produce detectable levels of CLA during growth. Several bifidobacteria exhibit CLA-converting activity, including B. bifidum LMG 10,645 (35% conversion) [56], B. dentium NCFB 2243 (29.05%) [23], B. dentium FhuBCZ5M2 (> 5.0%) [57], B. longum DPC6315 and DPC6320 (14.67–70.86%) [23], and B. pseudocatenulatum NCIMB8811 and FGSYC4M2 (3.89–5.0%) [23, 57]. However, B. breve strains demonstrate significantly higher CLA production capacity, reaching up to > 90.0% under optimal conditions [23, 24, 45]. Phylogenetic analysis of bifidobacterial LAI demonstrates that B. longum LAI shares the highest sequence homology (% identity) with B. breve LAI (91.84–92.04%), followed by B. pseudocatenulatum (84.37–84.99%) and B. dentium (83.88–83.97%) LAI. Notably, non-bifidobacterial GIT bacteria also possess a portion of the LAI gene sequence (186–395 bp), suggesting a strong conservation of this region, which may indicate its potential role in the active site of the enzyme. A recent predictive analysis confirmed that the binding site for LA is located in the conserved domain of bifidobacterial LAI [32], highlighting the strong conservation pressure of this region across different taxa.
In this study, we confirmed that the addition of free LA to the culture medium had a negative effect on the growth and fermentation profiles of the strains, which is consistent with previous reports. LA toxicity occurs because free fatty acids induce metabolic stress in the bacterial cells, manifesting as reduced growth and slower fermentation [58]. The LA conversion activity of certain bacterial genera (i.e., Lactobacillus and Bifidobacterium) is regarded as a detoxification mechanism that converts a toxic substrate into a less toxic form [59, 60]. Notably, emulsification of the LA with a higher Tween 80 concentration reduced the observed toxicity of free LA towards B. breve cells, manifested by higher cell counts and CLA production. This effect may be due to excess Tween 80 competing for oxidation or forming mixed micelles with LA [61]. Increasing the concentration of the emulsifier also causes the droplet size to decrease, which may improve the interaction between the enzyme and substrate, thereby improving the efficiency of the LA to CLA conversion [40, 61].
Despite demonstrating efficient LA conversion during growth, suggesting their potential use for increasing the CLA content of dairy products, one of the main challenges in applying bifidobacteria as probiotics or adjunct cultures in dairy products is their sensitivity to oxygen stress [62, 63]. Exposure to oxygen substantially affects survival in various media, resulting in slowed growth and impaired metabolic activity. However, the ability of JKL2022 to mediate LA conversion independent of its growth might be advantageous under conditions where the cell viability is not essential. Consistent with previous reports, all other tested strains failed to convert CLA using washed cells or CFE [64, 65], suggesting that this trait may be strain-specific [56]. Consequently, the potential of JKL2022 as a source of postbiotics with CLA-enhancing properties is being explored. Accordingly, the CLA production activity of washed JKL2022 cells and crude protein extracts was further investigated, focusing on increasing the substrate concentration to determine the maximum efficiency of potential postbiotic preparations.
In vitro LA conversion reactions demonstrated that only the washed cells and crude protein extracts derived from JKL2022 can mediate CLA production. The findings of this study demonstrate that postbiotic preparations, particularly cell membrane-bound protein extracts, exhibited satisfactory CLA production, achieving up to 100% conversion rates. The putative LAI of JKL2022 was recently identified and characterized as a membrane-spanning protein. Although B. breve strains share significant amino acid sequence homology with JKL2022 LAI, washed cells and crude protein extracts derived from JCM strains failed to produce CLA when used as catalysts for the reaction, which may be attributed to inter-strain differences in lai expression levels [32].
The superior LA conversion of JKL2022 was further investigated on the transcription level. The TetR family of transcriptional factors participates in the control of various biological processes. They recognize specific DNA sequences related to gene expression [66]. Although TetR-family transcription factors play key roles in fatty acid biosynthesis and lipid metabolism [67, 68], their specific role in bifidobacterial CLA biosynthesis has not yet been elucidated. In this study, RT-qPCR analysis revealed the potential transcription regulatory role of TetR (JKL2022_00217) in lai expression of JKL2022. The changes in transcription pattern of lai and tetR-217 due to LA exposure are consistent with the established hypothesis that conjugation of LA into CLA is part of a detoxification mechanism for bacteria, demanding the strains to produce the key enzymes to facilitate this mechanism [28]. Under normal culture conditions, the lai and tetR-217 genes are expressed at significantly higher levels in JKL2022, verifying that JKL2022 produces larger amounts of LAI than JCM strains. Consistently, based on transcriptome analysis, JKL2022 exhibited 20.06–39.29× and 3.02–3.12× higher expression levels for lai and tetR-217 genes, respectively, compared to JCM7017 and JCM7019. Therefore, LAI enzymes are already present in the washed cells and crude protein extracts of strain JKL2022, rendering these fractions capable of converting CLA from free LA. In contrast, under LA-exposed conditions, all strains exhibited upregulation of both lai and tetR-217. The degree of upregulation also correlates with the strength of CLA production, wherein strong CLA converters have stronger upregulation of lai and tetR-217. However, in the case of JKL2022, exposure to LA resulted only in the upregulation of tetR-217. Nonetheless, the lai expression level in JKL2022 was higher relative to those of the JCM strains even in LA-exposed conditions. The JCM strains with significantly lower lai and tetR-217 expression levels under normal conditions produce inactive washed cells and protein fractions. Thus, the superior CLA production of JKL2022 can be attributed to the higher base expression level of both tetR-217 and lai.
Moreover, predictive in silico analysis using protein-protein interaction networks revealed a strong relationship between tetR and lai genes on the basis of co-occurrence and neighborhood. In addition, TetR-217 exhibited a strong binding affinity to the promoter region of lai (docking score, -205.12; confidence, 0.7505) based on HDOCK analysis, consistent with a previous report that TetR regulates lipid biosynthesis through the activation of CLA synthesis gene transcription [33]. A recent study evaluated the role of TetR in the CLA activity of Lactiplantibacillus plantarum AR195, in which overexpression of TetR led to a 7% increase in CLA activity. In contrast, tetR gene knockout led to the downregulation of cla-hy, cla-dh, and cla-dc genes, which encode the enzymes for LA conversion in lactobacilli [33]. The results of this study provide the first evidence suggesting that TetR-217 influences the CLA activity of B. breve by upregulating lai transcription, similar to that reported in Lpb. plantarum AR195.
Increased concentrations of washed cells effectively converted increased amounts of substrate, substantiating their potential for postbiotic applications. Further optimization of the reaction conditions and extraction methods for in vitro CLA production revealed that optimal LA conversion occurs at a neutral pH. Acidification of the reaction mix before the addition of isopropanol and hexane improved the extraction of CLA. In particular, adjusting the reaction mixture from pH 7.0 to pH 5.0–5.5 resulted in the highest CLA yield. In contrast, adjusting the pH after the reaction from acidic to neutral resulted in lower CLA yield. Finally, the effect of different emulsification methods on LA conversion indicated that the method used in this study (vortexing) was as effective as emulsification by ultrasonication, providing a simple and rapid method of preparing LA stock for in vitro experiments [69]. Based on the timepoint LA conversion experiments, it was confirmed that LAI follows the first-order kinetics until 5 min of reaction, after which the concentration of the conjugated fatty acids started to plateau. In addition, LAI exhibited higher affinity for LNA than LA, manifested by higher specific activity (LNA, 56,790.12 µU/g; LA, 11,664.08 µU/g). Based on these results, in vitro reaction of 0.5–1.0 mg LA can be completed within 10–20 min of reaction.
Conclusions
The reported results in this study show that B. breve JKL2022 possesses superior CLA production capacity under growing conditions compared to that of other bifidobacteria. Additionally, only JKL2022 produced CLA when washed cells or protein extracts were used as catalysts, suggesting an inter-strain difference in lai expression. The ability of JKL2022 to convert CLA independent of growth suggests that the LA-converting enzyme is present at elevated levels and remains active after extraction from the cell membrane, attributed to the potential transcription regulatory role of TetR protein in LAI synthesis. This capability is advantageous for developing postbiotic preparations for CLA production or to enhance the CLA content of various dairy products. Currently, studies are being conducted to improve the CLA production of JKL2022 strains using in vitro digestion models and to disseminate this strain as a probiotic that can efficiently modify the MUFA and PUFA profiles of dairy products, improving their health benefits for consumers.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to give thanks to the BT Research Center of Chung-Ang University (Anseong, Korea) for letting us use their facilities.
Abbreviations
- LA
Linoleic Acid
- CLA
Conjugated Linoleic Acid
- LAI
Linoleic Acid Isomerase
- RT-qPCR
Quantitative Reverse Transcription Polymerase Chain Reaction
- CFE
Cell-Free Extract
- cDNA
Complementary Deoxyribonucleic Acid
- GC-FID
Gas Chromatography with Downstream Flame Ionization Detector
- MUFA
Monounsaturated Fatty Acid
- PUFA
Polyunsaturated Fatty Acid
- MRS
De Mann, Rogosa, Sharpe medium
- Cys-MRS
MRS media supplemented with 0.05% L-cysteine
- Na-P buffer
Sodium Phosphate buffer
- PBS
Phosphate-Buffered Saline
- GAPDH
Glyceraldehyde 3-Phosphate Dehydrogenase
- OD
Optical Density
- BSA
Bovine Serum Albumin
- MCRA
Myosin Cross-Reactive Antigen Protein
- DH
Short-Chain Dehydrogenase/Oxidoreductase
- DC
Acetoacetate Decarboxylase
Author contributions
AGE and G-BK designed and did the formal analysis of the experimental data of the study. AGE performed all statistical analyses and wrote the initial draft of the manuscript. All authors contributed with editing and approved the final manuscript.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation (NRF) funded by the Ministry of Science and ICT (2021R1A2C1093838).
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Glick NR, Fischer MH. The role of essential fatty acids in human health. Evid-Based Complement Altern Med. 2013;18:268–89. [Google Scholar]
- 2.Vissers LET, Rijksen J, Boer JMA, Verschuren WMM, van der Schouw YT, Sluijs I. Fatty acids from dairy and meat and their association with risk of coronary heart disease. Eur J Nutr. 2019;58:2639–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amores G, Virto M. Total and free fatty acids analysis in milk and dairy fat. Separations 2019, 6.
- 4.St-Onge MP, Farnworth ER, Jones PJ. Consumption of fermented and nonfermented dairy products: effects on cholesterol concentrations and metabolism. Am J Clin Nutr. 2000;71:674–81. [DOI] [PubMed] [Google Scholar]
- 5.Albano C, Morandi S, Silvetti T, Casiraghi MC, Manini F, Brasca M. Lactic acid bacteria with cholesterol-lowering properties for dairy applications: In vitro and in situ activity. J Dairy Sci. 2018;101:10807–18. [DOI] [PubMed] [Google Scholar]
- 6.Lordan R, Zabetakis I. Invited review: the anti-inflammatory properties of dairy lipids. J Dairy Sci. 2017;100:4197–212. [DOI] [PubMed] [Google Scholar]
- 7.Nagao K, Yanagita T. Conjugated fatty acids in food and their health benefits. J Biosci Bioeng. 2005;100:152–7. [DOI] [PubMed] [Google Scholar]
- 8.Jang Y, Elnar AG, Hur SJ, Kim G-B. Factors influencing conjugated Linoleic acid content of dairy products: challenges and strategies. Crit Rev Food Sci Nutr 2024:1–17. [DOI] [PubMed]
- 9.Gorissen L, Leroy F, De Vuyst L, De Smet S, Raes K. Bacterial production of conjugated Linoleic and linolenic acid in foods: a technological challenge. Crit Rev Food Sci Nutr. 2015;55:1561–74. [DOI] [PubMed] [Google Scholar]
- 10.Jang Y, Elnar AG, Kang MH, Kim G-B. Application of conjugated Linoleic acid-producing strain, Bifidobacterium breve JKL2022, in the development of probiotic dairy products. Food Sci Anim Resour 2024:1–25.
- 11.Nasrollahzadeh A, Mollaei Tavani S, Arjeh E, Jafari SM. Production of conjugated Linoleic acid by lactic acid bacteria; important factors and optimum conditions. Food Chem: X. 2023;20:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrade JC, Ascencao K, Gullon P, Henriques SMS, Pinto JMS, Rocha-Santos TAP, Freitas AC, Gomes AM. Production of conjugated Linoleic acid by food‐grade bacteria: A review. Int J Dairy Technol. 2012;65:467–81. [Google Scholar]
- 13.Valenzuela C, Baker E, Miles E, Calder P. Conjugated Linoleic acids have anti-inflammatory effects in cultured endothelial cells. Int J Mol Sci. 2023;24:847–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reynolds CM, Roche HM. Conjugated Linoleic acid and inflammatory cell signalling. Prostaglandins Leukot Essent Fat Acids. 2010;82:199–204. [DOI] [PubMed] [Google Scholar]
- 15.Bassaganya-Riera J, Hontecillas R, Beitz DC. Colonic anti-inflammatory mechanisms of conjugated Linoleic acid. Clin Nutr. 2002;21:451–9. [DOI] [PubMed] [Google Scholar]
- 16.Islam MA, Kim YS, Oh TW, Kim GS, Won CK, Kim HG, Choi MS, Kim JO, Ha YL. Superior anticarcinogenic activity of trans,trans-conjugated Linoleic acid in N-methyl-N-nitrosourea-induced rat mammary tumorigenesis. J Agric Food Chem. 2010;58:5670–8. [DOI] [PubMed] [Google Scholar]
- 17.Dachev M, Bryndová J, Jakubek M, Moučka Z, Urban M. The effects of conjugated Linoleic acids on cancer. Processes. 2021;9:454–71. [Google Scholar]
- 18.Kondo S, Xiao JZ, Satoh T, Odamaki T, Takahashi S, Sugahara H, Yaeshima T, Iwatsuki K, Kamei A, Abe K. Antiobesity effects of Bifidobacterium breve strain B-3 supplementation in a mouse model with high-fat diet-induced obesity. Biosci Biotechnol Biochem. 2010;74:1656–61. [DOI] [PubMed] [Google Scholar]
- 19.Li JJ, Huang CJ, Xie D. Anti-obesity effects of conjugated Linoleic acid, docosahexaenoic acid, and eicosapentaenoic acid. Mol Nutr Food Res. 2008;52:631–45. [DOI] [PubMed] [Google Scholar]
- 20.Ibrahim KS, El-Sayed EM. Dietary conjugated Linoleic acid and medium-chain triglycerides for obesity management. J Biosci (Bangalore). 2021;46:1–14. [PubMed] [Google Scholar]
- 21.Chen Y, Jin Y, Stanton C, Ross RP, Wang Z, Zhao J, Zhang H, Yang B, Chen W. Dose-response efficacy and mechanisms of orally administered CLA-producing Bifidobacterium breve CCFM683 on DSS-induced colitis in mice. J Funct Foods 2020, 75.
- 22.Bassaganya-Riera J, Hontecillas R, Horne WT, Sandridge M, Herfarth HH, Bloomfeld R, Isaacs KL. Conjugated Linoleic acid modulates immune responses in patients with mild to moderately active crohn’s disease. Clin Nutr. 2012;31:721–7. [DOI] [PubMed] [Google Scholar]
- 23.Coakley M, Ross R, Nordgren M, Fitzgerald g, Devery R, Stanton C. Conjugated Linoleic acid biosynthesis by human-derived Bifidobacterium species. J Appl Microbiol. 2003;94:138–45. [DOI] [PubMed] [Google Scholar]
- 24.Park HG, Cho SD, Kim JH, Lee H, Chung SH, Kim SB, Kim HS, Kim T, Choi NJ, Kim YJ. Characterization of conjugated Linoleic acid production by Bifidobacterium breve LMC 520. J Agric Food Chem. 2009;57:7571–5. [DOI] [PubMed] [Google Scholar]
- 25.Choi SH, Lee KM, Kim KH, Kim GB. Development of a rapid method for the screening of conjugated Linoleic acid (CLA)-producing strains of Bifidobacterium breve. Food Sci Anim Resour. 2018;38:806–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao H, Yang B, Stanton C, Ross RP, Zhang H, Liu Z, Chen H, Chen W. Characteristics of bifidobacterial conjugated fatty acid and hydroxy fatty acid production and its potential application in fermented milk. LWT 2020, 120.
- 27.Song Y-S, Kang S-W, Oh D-K, Rho Y-T, Hong S-I, Kim S-W. Bioconversion of Linoleic acid to conjugated Linoleic acid by Bifidobacterium breve. Biotechnol Bioprocess Eng. 2005;10:357–61. [Google Scholar]
- 28.Fontes AL, Pimentel LL, Calzada J, Salsinha AS, Rodríguez JM, Gomes AM, Arqués JL, Rodríguez-Alcalá LM. Study of the association between genotypic potential and Linoleic acid tolerance with microbial production of conjugated Linoleic acid. J Agric Food Chem. 2023;3:2199–207. [Google Scholar]
- 29.Maia MR, Chaudhary LC, Figueres L, Wallace RJ. Metabolism of polyunsaturated fatty acids and their toxicity to the microflora of the rumen. Antonie Van Leeuwenhoek. 2007;91:303–14. [DOI] [PubMed] [Google Scholar]
- 30.Hill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S, et al. Expert consensus document. The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol. 2014;11:506–14. [DOI] [PubMed] [Google Scholar]
- 31.Nataraj BH, Ali SA, Behare PV, Yadav H. Postbiotics-parabiotics: the new horizons in microbial biotherapy and functional foods. Microb Cell Fact. 2020;19:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elnar AG, Jang Y, Kim G-B. Heterologous expression and polyphasic analysis of CLA-Converting Linoleic acid isomerase from Bifidobacterium breve JKL2022. J Agric Food Chem. 2025;73:1425–40. [DOI] [PubMed] [Google Scholar]
- 33.Liu XX, Dou H, Liu L, Wang GQ, Xiong ZQ, Ai LZ. Regulatory effect of transcriptional regulator TetR on the synthesis of conjugated Linoleic acid in Lactiplantibacillus plantarum AR195. J Agric Food Chem. 2024;72:25827–35. [DOI] [PubMed] [Google Scholar]
- 34.Elnar AG, Eum B, Kim GB. Genomic characterization and probiotic assessment of Bifidobacterium breve JKL2022 with strain-specific CLA-converting properties. Sci Rep. 2025;15:15419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung M, Kim G-B, Jang E, Jung Y, Park S, Lee B. Technical note: improved extraction method with hexane for gas chromatographic analysis of conjugated Linoleic acids. J Dairy Sci. 2006;89:90–4. [DOI] [PubMed] [Google Scholar]
- 36.Pimentel TC, Cruz AG, Pereira E, Almeida da Costa WK, da Silva Rocha R, Targino de Souza Pedrosa G, Rocha CS, Alves JM, Alvarenga VO, Sant’Ana AS, Magnani M. Postbiotics: an overview of concepts, inactivation technologies, health effects, and driver trends. Trends Food Sci Technol. 2023;138:199–214. [Google Scholar]
- 37.Elnar AG, Kim G-B. In vitro and in Silico characterization of N-formylated two-peptide bacteriocin from Enterococcus faecalis CAUM157 with anti-Listeria activity. Probiotics Antimicrob Proteins. 2024;16:1130–47. [DOI] [PubMed] [Google Scholar]
- 38.Folch J, Lees M, Stanley GHS. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 39.Panghyová E, Kačenová D, Matulová M, Kiss E. Composition of conjugated Linoleic acid isomers formed by Lactobacillus and Bifidobacterium spp. In conversion media. J Food Nutr Res. 2009;48:163–70. [Google Scholar]
- 40.Khiaosa-ard R, Leiber F, Soliva CR. Methods of emulsifying Linoleic acid in biohydrogenation studies in vitro May bias the resulting fatty acid profiles. Lipids. 2010;45:651–7. [DOI] [PubMed] [Google Scholar]
- 41.Hwangbo SA, Lee SY, Kim BA, Moon CK. Preparation of surfactant-free nano oil particles in water using ultrasonic system and the mechanism of emulsion stability. Nanomaterials (Basel) 2022, 12. [DOI] [PMC free article] [PubMed]
- 42.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. [DOI] [PubMed] [Google Scholar]
- 43.Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, Gable AL, Fang T, Doncheva NT, Pyysalo S, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51:D638–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan Y, Tao H, He J, Huang SY. The HDOCK server for integrated protein-protein Docking. Nat Protoc. 2020;15:1829–52. [DOI] [PubMed] [Google Scholar]
- 45.Chung S, Kim I, Park H, Kang H, Yoon C, Jeong H, Choi N, Eung G, Kim Y. Synthesis of conjugated linoelic acid by human-derived Bifidobacterium breve LMC 017: utilization as a functional starter culture for milk fermentation. J Agric Food Chem. 2008;56:3311–6. [DOI] [PubMed] [Google Scholar]
- 46.Mei Y, Chen H, Yang B, Zhao J, Zhang H, Chen W. Computational analysis and heterologous expression of BBI-like proteins from food-grade Bifidobacterium species reveal possibly a key factor in conjugated Linoleic acid bioconversion. J Agric Food Chem. 2023;71:8093–103. [DOI] [PubMed] [Google Scholar]
- 47.Jena R, Choudhury PK. Bifidobacteria in fermented dairy foods: a health beneficial outlook. Probiotics Antimicrob Proteins 2023, Ahead of print:1–22. [DOI] [PubMed]
- 48.Jankovic I, Sybesma W, Phothirath P, Ananta E, Mercenier A. Application of probiotics in food products - challenges and new approaches. Curr Opin Biotechnol. 2010;21:175–81. [DOI] [PubMed] [Google Scholar]
- 49.Lee HY, Park JH, Seok SH, Baek MW, Kim DJ, Lee KE, Paek KS, Lee Y, Park JH. Human originated bacteria, Lactobacillus rhamnosus PL60, produce conjugated Linoleic acid and show anti-obesity effects in diet-induced obese mice. Biochim Biophys Acta. 2006;1761:736–44. [DOI] [PubMed] [Google Scholar]
- 50.Liu XX, Zhang HY, Song X, Yang Y, Xiong ZQ, Xia YJ, Ai LZ. Reasons for the differences in biotransformation of conjugated Linoleic acid by Lactobacillus plantarum. J Dairy Sci. 2021;104:11466–73. [DOI] [PubMed] [Google Scholar]
- 51.Yang B, Qi H, Gu Z, Zhang H, Chen W, Chen H, Chen YQ. Characterization of the triple-component Linoleic acid isomerase in Lactobacillus plantarum ZS2058 by genetic manipulation. J Appl Microbiol. 2017;123:1263–73. [DOI] [PubMed] [Google Scholar]
- 52.Salsinha AS, Pimentel LL, Fontes AL, Gomes AM, Rodriguez-Alcala LM. Microbial production of conjugated Linoleic acid and conjugated linolenic acid relies on a multienzymatic system. Microbiol Mol Biol Rev. 2018;82:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rosberg-Cody E, Liavonchanka A, Göbel C, Ross R, O’Sullivan O, Fitzgerald GF, Feussner I, Stanton C. Myosin-cross-reactive antigen (MCRA) protein from Bifidobacterium breve is a FAD-dependent fatty acid hydratase which has a function in stress protection. BMC Bioinformatics. 2011;12:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.O’Connell KJ, Motherway MOC, Hennessey AA, Brodhun F, Ross RP, Feussner I, Stanton C, Fitzgerald GF, van Sinderen D. Identification and characterization of an oleate hydratase-encoding gene from Bifidobacterium breve. Bioengineered. 2014;4:313–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mei Y, Li X, Yang B, Zhao J, Zhang H, Chen H, Chen W. Heterologous expression of a novel Linoleic acid isomerase BBI, and effect of fusion tags on its performance. Curr Res Food Sci. 2022;5:2053–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gorissen L, De Vuyst L, Raes K, De Smet S, Leroy F. Conjugated Linoleic and linolenic acid production kinetics by bifidobacteria differ among strains. Int J Food Microbiol. 2012;155:234–40. [DOI] [PubMed] [Google Scholar]
- 57.Mei Y, Chen H, Yang B, Zhao J, Zhang H, Chen W. Linoleic acid triggered a metabolomic stress condition in three species of bifidobacteria characterized by different conjugated Linoleic acid-producing abilities. J Agric Food Chem. 2021;69:11311–21. [DOI] [PubMed] [Google Scholar]
- 58.Senizza A, Rocchetti G, Callegari ML, Lucini L, Morelli L. Linoleic acid induces metabolic stress in the intestinal microorganism Bifidobacterium breve DSM 20213. Sci Rep. 2020;10:5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu C, Chen H, Mei Y, Yang B, Zhao J, Stanton C, Chen W. Advances in research on microbial conjugated Linoleic acid bioconversion. Prog Lipid Res. 2024;93:1–15. [DOI] [PubMed] [Google Scholar]
- 60.Mei Y, Chen H, Yang B, Zhao J, Zhang H, Chen W. Research progress on conjugated Linoleic acid bio-conversion in Bifidobacterium. Int J Food Microbiol 2022, 369. [DOI] [PubMed]
- 61.Ponginebbi L, Nawar W, Chinachoti P. Oxidation of Linoleic acid in emulsions: effect of substrate, emulsifier, and sugar concentration. J Am Oil Chem Soc 1999, 76.
- 62.Talwalkar A, Kailasapathy K. A review of oxygen toxicity in probiotic yogurts: influence on the survival of probiotic bacteria and protective techniques. Compr Rev Food Sci Food Saf. 2004;3:117–24. [DOI] [PubMed] [Google Scholar]
- 63.Bolduc M-P, Raymond Y, Fustier P, Champagne CP, Vuillemard J-C. Sensitivity of bifidobacteria to oxygen and redox potential in non-fermented pasteurized milk. Int Dairy J. 2006;16:1038–48. [Google Scholar]
- 64.Terán V, Pizarro PL, Zacarías MF, Vinderola G, Medina R, Van Nieuwenhove C. Production of conjugated dienoic and trienoic fatty acids by lactic acid bacteria and bifidobacteria. J Funct Foods. 2015;19:417–25. [Google Scholar]
- 65.Oh D-K, Hong G-H, Lee Y, Min S, Sin H-S, Cho SK. Production of conjugated Linoleic acid by isolated Bifidobacterium strains. World J Microbiol Biotechnol. 2003;19:907–12. [Google Scholar]
- 66.Cuthbertson L, Nodwell JR. The TetR family of regulators. Microbiol Mol Biol Rev. 2013;77:440–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Singh P, Jain A, Chhabra R, Kaur J. TetR family transcriptional regulators: lipid metabolism and drug resistance in mycobacteria. Gene Rep 2024, 36.
- 68.Wang K, Sybers D, Maklad HR, Lemmens L, Lewyllie C, Zhou X, Schult F, Brasen C, Siebers B, Valegard K, et al. A TetR-family transcription factor regulates fatty acid metabolism in the archaeal model organism Sulfolobus acidocaldarius. Nat Commun. 2019;10:1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gammill W, Proctor A, Jain V. Comparative study of high-linoleic acid vegetable oils for the production of conjugated Linoleic acid. J Agric Food Chem. 2010;58:2952–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.










