Abstract
Background
The time after hospital discharge carries high rates of mortality in neonates and young children in sub-Saharan Africa. Previous work using logistic regression to develop risk assessment tools to identify those at risk for postdischarge mortality has yielded fair discriminatory value. Our objective was to determine if machine learning models would have greater discriminatory value to identify neonates and young children at risk for postdischarge mortality.
Methods
We conducted a planned secondary analysis of a prospective observational cohort at Muhimbili National Hospital in Dar es Salaam, Tanzania and John F. Kennedy Medical Center in Monrovia, Liberia. We enrolled neonates and young children near the time of discharge. The outcome was 60-day postdischarge mortality. We collected socioeconomic, demographic, clinical, and anthropometric data during hospital admission and used machine learning (ie, eXtreme Gradient Boosting (XGBoost), Hist-Gradient Boost, Support Vector Machine, Neural Network, and Random Forest) to develop risk assessment tools to identify: (1) neonates and (2) young children at risk for postdischarge mortality.
Results
A total of 2310 neonates and 1933 young children enrolled. Of these, 71 (3.1%) neonates and 67 (3.5%) young children died after hospital discharge. XGBoost, Hist Gradient Boost, and Neural Network models yielded the greatest discriminatory value (area under the receiver operating characteristic curves range: 0.94–0.99) and fewest features, which included six features for neonates and five for young children. Discharge against medical advice, low birth weight, and supplemental oxygen requirement during hospitalisation were predictive of postdischarge mortality in neonates. For young children, discharge against medical advice, pallor, and chronic medical problems were predictive of postdischarge mortality.
Conclusions
Our parsimonious machine learning-based models had excellent discriminatory value to predict postdischarge mortality among neonates and young children. External validation of these tools is warranted to assist in the design of interventions to reduce postdischarge mortality in these vulnerable populations.
Keywords: Machine Learning, Infant, Neonatology, Mortality
WHAT IS ALREADY KNOWN ON THIS TOPIC
Postdischarge mortality is increasingly recognised as a contributor to persistently high rates of mortality among neonates and young children in sub-Saharan Africa.
Previous work using logistic regression to develop risk assessment tools to identify those at risk for postdischarge mortality has yielded fair discriminatory value.
WHAT THIS STUDY ADDS
From a prospective cohort study that included a total of 2310 neonates and 1933 young children, eXtreme Gradient Boosting, Hist-Gradient Boost, Neural Network, and Random Forest models with as few as six features demonstrated excellent discriminatory value (ie, area under the receiver operating characteristic curve 0.94–0.99) and outperformed traditional logistic regression models in both age groups.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
After external validation, machine learning models to identify neonates and young children at risk for postdischarge mortality may be used to direct resources to at-risk infants and young children.
Introduction
Postdischarge mortality rates among neonates and young children in sub-Saharan Africa are as high as 3–13%,1,3 far outpacing rates of mortality during hospital readmission in settings like the USA (ie, 0.1%).4 The accurate identification of young children at risk for postdischarge mortality is the first step towards developing interventions to reduce mortality during this vulnerable time for young children in sub-Saharan Africa.5 Clinical care providers, without the use of clinical decision aids, are unable to identify neonates or young children at risk for postdischarge mortality in the weeks following hospital discharge.6 Hospital discharge provides a valuable opportunity for clinicians to identify risk factors that confer greater risk for postdischarge mortality among young children in regions with high rates of postdischarge mortality.
Risk assessment tools are powerful instruments derived using statistical analyses of observational data and can be used at the bedside to reduce uncertainty and improve accuracy in medical decision making.7 They can be effective across a range of settings, including those with limited resources.8 9 Several studies have leveraged data available at the time of hospital discharge, including vital signs, anthropometry, and hospital clinical diagnoses, to develop logistic regression-based risk assessment tools to identify neonates, infants, and children at risk for postdischarge mortality in sub-Saharan African countries.10,13
However, risk assessment tools derived using logistic regression have demonstrated only fair-to-good discriminatory value (ie, area under the receiver operating characteristic curve (AUROC) from 0.77 to 0.82).10 11 13 14 Thus, more accurate risk assessment tools to identify young children at risk for postdischarge mortality are warranted. Machine learning is one approach to improve risk assessment as it augments the discriminatory value of these tools by identifying complex latent relationships between patient features not identifiable through standard linear or non-linear regression methods.15,17 However, the application of machine learning methods to identify young children at risk for postdischarge mortality has been limited.18
Machine learning model applications in clinical medicine are rapidly evolving and may have superior accuracy to logistic regression models. Thus, to develop more accurate risk assessment tools to identify (1) neonates and (2) young children at risk for postdischarge mortality, our objective was to determine if non-linear machine learning tools yield greater discriminatory value to identify young children at risk for postdischarge mortality compared with standard logistic regression models that we have previously developed.13 19 We hypothesised that machine learning-based risk assessment tools would have greater discriminatory capacity than our logistic regression models to identify young children at risk for postdischarge mortality. Furthermore, we hypothesised that machine learning models would have excellent discriminatory value (ie, AUROC ≥0.90).20
Methods
Study design
We conducted a planned secondary analysis of data collected in a prospective observational cohort study that included neonates (aged 0–28 days) and young children (aged 1–59 months) discharged from two national referral hospitals in sub-Saharan Africa and were followed for 60 days (2019–2022). The full protocol has been published previously.21
Patient and public involvement statement
The development of the research question was informed by the disease burden of postdischarge mortality among children in sub-Saharan Africa. Patients were not involved in the design, recruitment, or conduct of the study, nor were they advisers in this study. Results of this study will be made publicly available through publication.
Study setting
We enrolled neonates and young children who were discharged from Muhimbili National Hospital (MNH) in Dar es Salaam, Tanzania and John F. Kennedy Medical Center (JFKMC) in Monrovia, Liberia. These are both large national referral hospitals in their respective countries. Both hospitals are located in urban settings and are supported by the Ministry of Health in each country. Both are teaching hospitals and serve as major training sites for medical students and paediatrics residents in their respective countries. The neonatal and paediatric wards of each hospital are staffed by clinicians who provide clinical care according to each country’s national guidelines. All participants received standard clinical care provided by the clinical care teams and not by study staff. The decision to discharge a neonate or young child was not influenced by our study staff.
Study population
Participants were included if their caregivers consented to have their hospital admission data collected, had access to a phone, and agreed to receive follow-up phone calls following hospital discharge. We enrolled neonates who were admitted for an illness (ie, not routine newborn care) with no restriction based on reason for hospital admission. We enrolled young children who were admitted for an injury or illness with no restriction to admission or discharge diagnoses. We excluded neonates and young children who: (1) died during the initial hospital admission, (2) were older than 59 months of age at enrolment, or (3) had non-consenting caregivers.
Study procedures
Research staff was present on clinical rounds each day to identify potential participants and to conduct consecutive enrolment of discharged neonates and young children. Caregivers of neonates and young children were approached by research staff for potential enrolment near the time of hospital discharge. Caregivers provided written consent in Tanzania and, due to local preference, oral consent in Liberia.
Consenting caregivers agreed to have their child’s clinical data extracted, to respond to sociodemographic questions, and to receive phone calls to ascertain the child’s well-being following hospital discharge. Research staff at each site collected detailed demographic, socioeconomic, anthropometric, and clinical data that were collected during each participant’s hospitalisation. Caregivers were contacted by our research staff at each site by telephone 7 days, 14 days, 30 days, 45 days, and 60 days after discharge to assess the participants’ vital status. All data were stored in password-protected electronic data capture forms (Microsoft SQL in Tanzania and KoboToolbox in Liberia).
Outcome and candidate features
The outcome of postdischarge mortality within 60 days of hospital discharge was determined through caregiver report during follow-up telephone calls. All candidate features were selected prior to the enrolment of the first participant and prior to knowledge of the outcome. Candidate features were selected by the investigator team based on: (1) clinical experience, (2) frequency of assessment in routine clinical care, (3) results of prior studies on postdischarge mortality in the region,1 2 22 23 (4) a list of candidate features from a modified Delphi survey of public health experts, paediatricians and epidemiologists in sub-Saharan Africa,24 25 and (5) availability to clinicians at the time of hospital discharge. As previously published,13 19 a total of 115 features were considered for the neonatal models and 121 features were considered for the models for infants and children.
Statistical analyses
We developed traditional non-linear machine learning based risk assessment tools using eXtreme Gradient Boosting (XGBoost), Hist-Gradient Boosting, Support Vector Machines, Neural Networks, and Random Forest to separately identify features predictive of postdischarge mortality among (1) neonates and (2) young children. To develop these models, we first randomly assigned the (1) neonatal population and (2) the population of young children to derivation (80%) and validation (20%) cohorts with equal partitions to ensure equal numbers of participants who experienced postdischarge mortality in each group combining datasets from Tanzania and Liberia. We elected to combine datasets from both sites to minimise potential overfitting that may occur with non-random data splitting and to develop models that are generalisable beyond a single site. To mitigate class imbalance during the training phase of algorithm development, we performed data augmentation through the synthetic minority oversampling technique (SMOTE),26 which addresses the skewed distribution of data points between target classes by oversampling minority class data and assigning higher weights to misclassified minority populations. SMOTE was used to synthetically generate new examples of the minority class until its size reached 50% of the majority class, using five nearest neighbours to construct each synthetic sample. Subsequently, we applied random undersampling to reduce the majority class to 80% of the (post-SMOTE) minority class. Before SMOTE, 3.1% of the population in the training and validation cohort had postdischarge mortality. Following SMOTE, which was applied to the training cohort only, 44.4% of the training cohort had postdischarge mortality. To prepare the dataset for machine learning model development and to prevent potential overfitting, we performed data transformation, where categorical variables were one-hot encoded and continuous variables were scaled to a mean of zero and standard deviation of one.
To identify features predictive of postdischarge mortality, we used minimal redundancy/maximum relevance to find 30 non-redundant clinical features that were predictive of the outcome ranked from most to least predictive. However, as 30 features were thought to be beyond what could be clinically useful in the absence of electronic health records, using backwards selection and clinical relevance, we tested the discriminatory value (ie, AUROC) of models with each machine learning approach with as few as five features to find the minimum number of features that achieved AUROC ≥0.90 in derivation. As we used 10 features in our previously developed regression models that had fair discriminatory value (ie, AUROC= 0.77),13 19 we postulated that ≤10 features would allow for even greater discriminatory value with more advanced and precise machine learning approaches. Aligning with other machine learning studies,27 28 in order to visualise the relative predictive contribution of each feature, we plotted Shapley additive explanations for each model.
Post hoc, we identified the feature ‘discharge against medical advice’ was strongly predictive of the outcome of postdischarge mortality, which may have suggested that feature violated the causality constraint (ie, ‘discharge against medical advice’ was both a predictor of, and a proxy for, postdischarge mortality). Thus, in order to identify other potential features that would be present before the decision is made to ‘discharge against medical advice’ and to ensure that our models performed well without this feature, we sought additional covariates that were most strongly predictive of ‘discharge against medical advice’ through minimal redundancy/maximum relevance and using the same approach for feature selection for the main models and replaced ‘discharge against medical advice’ with the six most strongly predictive and clinically relevant features in the XGBoost, Support Vector Machine, and Random Forest models to predict postdischarge mortality as the outcome.
We used five-fold cross-validation to perform parameter tuning for each model in the derivation sets (online supplemental table 1). For validation of the models, we evaluated the AUROC and area under the precision-recall curve (AUPRC) in the 20% validation sets. We internally validated our risk assessment tools using bootstrapping methodology with 100 repetitions and calculated the 95% CIs for the AUROC and AUPRC curve for each model.29,32 We compared the developed machine learning models for effective predictions of young children at risk for postdischarge mortality to our previously developed logistic regression models using the DeLong test. We used multiple imputation for variables with data missing at random by using the chained equations R package to create 10 imputed datasets based on fully conditional specifications.33 Individuals were weighted by one minus their fraction of missing data divided by the number of imputed datasets to account for multiple imputations.34 35 Multiple imputation was performed for variables with >70% complete data prior to splitting each dataset into derivation and validation sets. All analyses were conducted using Python.
Results
A total of 4243 participants enrolled and had 60-day survival status available (2310 (54.4%) neonates and 1933 (45.6%) young children; table 1). Just over half (n=2249, 53.0%) of the enrolled participants were discharged from JFKMC and 47% (n=2014) were enrolled from MNH. Of enrolled participants, there were 138 (3.3%) who died within 60 days of hospital discharge (71 [51.4%] neonates and 67 [48.6%] young children). Among the 71 neonatal deaths that occurred within 60 days of discharge, 26.8% occurred in the first 7 days and 50.7% of the 67 deaths among infants and children occurred within 30 days of discharge.
Table 1. Characteristics of enrolled participants who were discharged from referral hospitals in Dar es Salaam, Tanzania and Monrovia, Liberia.
| Characteristic | Overall, N=4243, n (%) | Neonates, N=2310, n (%) | Young children, N=1933, n (%) |
|---|---|---|---|
| Postdischarge mortality | 138 (3.3) | 71 (3.1) | 67 (3.5) |
| Site | |||
| Liberia | 2249 (53) | 1165 (50) | 1084 (56) |
| Tanzania | 1994 (47) | 1145 (50) | 849 (44) |
| Age at discharge, median (IQR) | –* | 8 (4, 15) | 11 (4, 23) |
| (Missing) | 189 | 50 | 139 |
| Sex | |||
| Female | 1859 (44) | 1068 (46) | 791 (41) |
| Male | 2373 (56) | 1238 (54) | 1135 (59) |
| (Missing) | 11 | 4 | 7 |
| Disposition from hospital | |||
| Discharge | 4093 (97) | 2224 (96) | 1869 (97) |
| Against medical advice | 145 (3.4) | 85 (3.7) | 60 (3.1) |
| Transfer | 3 (<0.1) | 1 (<0.1) | 2 (0.1) |
| (Missing) | 2 | 0 | 2 |
| Number of discharge diagnoses, median (IQR) | 2 (1, 3) | 2 (1, 2) | 2 (1, 3) |
| Presence of any chronic medical conditions | 403 (9.5) | 68 (2.9) | 335 (17) |
| (Missing) | 2 | 0 | 2 |
Neonates' age was measured in days, infants and children were measured in months.
Risk assessment tools to identify neonates at risk for postdischarge mortality
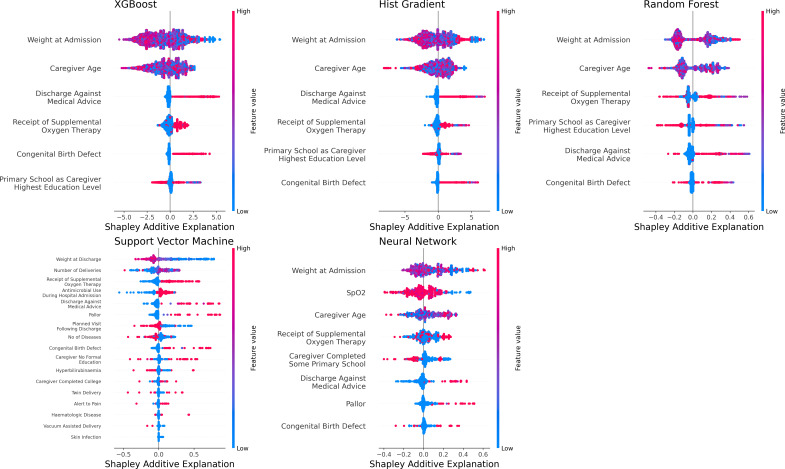
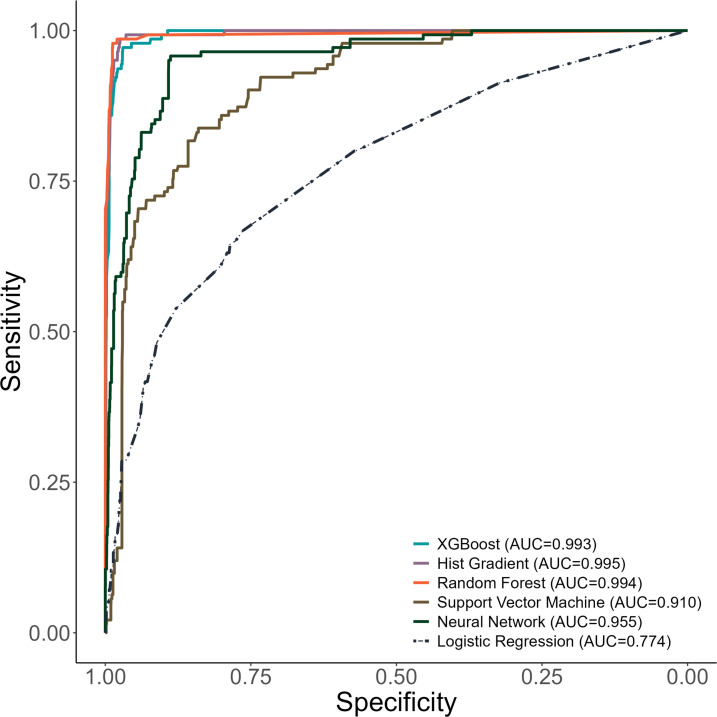
The XGBoost model to identify neonates at risk for postdischarge mortality selected discharge against medical advice, caregiver age, the receipt of oxygen therapy, caregiver education level, the presence of congenital birth defects, and admission weight of the neonate as features predictive of postdischarge mortality (figure 1). The XGBoost model for neonates demonstrated an AUROC of 0.99 (95% CI 0.99, 0.99) in the derivation and an AUROC of 0.99 (95% CI 0.97, 0.99) in the validation set (figure 2). The AUPRCs of the XGBoost model for neonates were 0.99 (95% CI 0.99, 0.99) in the derivation and 0.82 (95% CI 0.73, 0.87) in the validation set (online supplemental figure 1).
Figure 1. Features predictive of postdischarge mortality among neonates in each machine learning model.
Figure 2. Receiver operating characteristic curves for machine learning models to identify neonates at risk for postdischarge mortality. AUC, area under curve; XGBoost, eXtreme Gradient Boosting.
The Hist Gradient Boost model for neonates selected the same six features as the XGBoost model (figure 1). This model also yielded excellent discriminatory value in the derivation (AUROC 0.99, 95% CI 0.99, 0.99) and validation sets (AUROC 0.99, 95% CI 0.99, 0.98; figure 2) and an AUPRC above the prevalence of postdischarge mortality in the derivation (AUPRC 0.99, 95% CI 0.99, 0.99) and validation sets (0.86, 95% CI 0.83, 0.91; online supplemental figure 1).
The Random Forest model for neonates included the same six features as both boosted models (figure 1) and demonstrated excellent discriminatory value in derivation (AUROC 0.99, 95% CI 0.99, 0.99) and validation (AUROC 0.99, 0.98, 0.99; figure 2). The Random Forest model demonstrated an AUPRC above the prevalence of postdischarge mortality in both derivation (AUPRC 0.99, 95% CI 0.99, 0.99) and validation (AUPRC 0.92, 95% CI 0.89, 0.94; online supplemental figure 1).
The Support Vector Machine model for neonates required the inclusion of 17 features (figure 1) to yield good discriminatory value in the derivation set (ie, AUROC 0.88, 95% CI 0.86, 0.91) and validation set (ie, AUROC 0.89, 95% CI 0.89, 0.90; figure 2). Despite its lower discriminatory value, the AUPRC of the Support Vector Machine model for neonates had an AUPRC above the prevalence of postdischarge mortality in both the derivation (AUPRC 0.21, 95% CI 0.17, 0.25) and validation sets (0.84, 95% CI 0.83, 0.85; online supplemental figure 1).
The Neural Network model to identify neonates at risk for postdischarge mortality included eight variables (figure 1) and had excellent discriminatory value in derivation (AUROC 0.95, 95% CI 0.92, 0.97) and validation (AUROC 0.97, 95% CI 0.96, 0.98; figure 2). The AUPRC for the Neural Network model was above the prevalence of postdischarge mortality in derivation (AUPRC 0.48, 95% CI 0.39, 0.56) and validation (AUPRC 0.96, 95% CI 0.95, 0.97; online supplemental figure 1).
Each of the machine-learning models for neonates had greater discriminatory value than the logistic regression model for predicting postdischarge mortality (all p<0.001 by DeLong test). With the exception of the Support Vector Machine model, all machine-learning models for neonates had F-1 scores >0.90 and had excellent sensitivity and specificity at 50% probability of postdischarge mortality (online supplemental table 2).
Risk assessment tools to identify young children at risk for postdischarge mortality
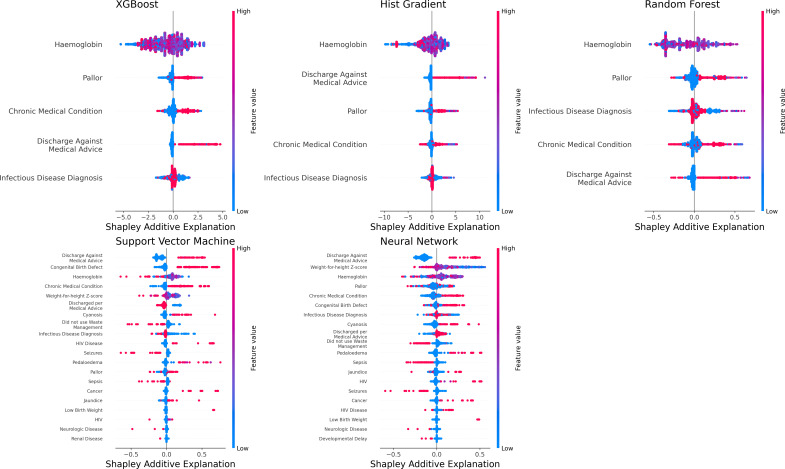
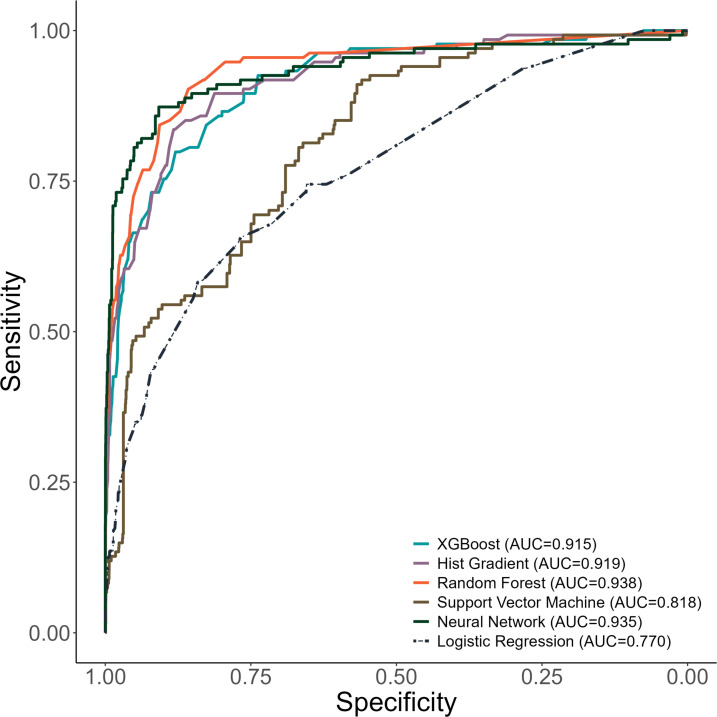
Our XGBoost model for young children selected these five features as most predictive of postdischarge mortality: discharge against medical advice, presence of pallor, haemoglobin level, presence of an infectious condition, and presence of chronic medical problems (figure 3). This model resulted in an AUROC of 0.94 (95% CI 0.94, 0.95) in derivation and AUROC of 0.91 (95% CI 0.88, 0.93) in validation (figure 4). The AUPRC for the XGBoost model for young children was above the prevalence of postdischarge mortality among young children in derivation (AUPRC 0.94, 95% CI 0.93, 0.94) and validation (AUPRC 0.49, 95% CI 0.46, 0.59; online supplemental figure 2).
Figure 3. Features predictive of postdischarge mortality among infants and children in each machine learning model. *Infectious disease diagnosis includes any of the following: pneumonia, malaria, sepsis, tuberculosis, infectious diarrhoea, meningitis, or skin and soft tissue infection.
Figure 4. Receiver operating characteristic curves for machine learning models to identify infants and children at risk for postdischarge mortality. AUC: area under curve; XGBoost: eXtreme Gradient Boosting.
The Hist-Gradient Boost model for young children also demonstrated excellent discriminatory value in derivation (AUROC 0.95, 0.94, 0.95) and validation (0.91, 95% CI 0.88, 0.94; figure 4) with the selection of the same five variables as the XGBoost model for young children. The Hist-Gradient Boost model yielded an AUPRC above the prevalence of postdischarge mortality in derivation (AUPRC 0.94, 95% CI 0.94, 0.95) and validation (0.52, 95% CI 0.48, 0.61; online supplemental figure 2).
The Random Forest model for infants and children included the same five features as the boosted models and had excellent discriminatory value in derivation (AUROC 0.98, 0.97, 0.98) and validation (0.93, 95% CI 0.90, 0.95; figure 4). The Random Forest model also had an AUPRC far above the prevalence of postdischarge mortality among young children (ie, AUPRC 0.97, 95% CI 0.97, 0.97 in derivation and 0.61, 95% CI 0.54, 0.67 in validation; online supplemental figure 2).
The Support Vector Machine model selected 25 features (figure 3) to create a model with good discriminatory value in derivation (AUROC 0.84, 95% CI 0.80, 0.86) and in validation (AUROC 0.86, 95% CI 0.85, 0.86; figure 4). This model also demonstrated an AUPRC above the prevalence of postdischarge mortality among young children (0.21, 95% CI 0.16, 0.28 in derivation and 0.81, 95% CI 0.80, in validation; online supplemental figure 2).
The Neural Network model for young children selected 25 features (figure 3) and demonstrated an AUROC of 0.95 (95% CI 0.93, 0.97) in derivation and AUROC 0.98 (95% CI 0.97, 0.99) in validation to identify young children at risk for postdischarge mortality (figure 4). The Neural Network model also demonstrated an AUPRC above the prevalence of postdischarge mortality among young children (AUPRC 0.73, 0.66, 0.78 in derivation and 0.98, 95% CI 0.97, 0.98 in validation; online supplemental figure 2).
Each of the machine-learning models for young children had greater discriminatory value compared with the logistic regression model for predicting postdischarge mortality (all p<0.001 using the DeLong test). All machine-learning models for infants and children had F-1 scores >0.90 (online supplemental table 2). The XGBoost, Hist-Gradient Boost, and Random Forest models had excellent specificity at 50% probability of postdischarge mortality.
Our post-hoc analyses on the prediction of postdischarge mortality supplanting the feature ‘discharge against medical advice’ with features predictive of ‘discharge against medical advice’ demonstrated similar discriminatory value to our main models for both neonates and infants and children (online supplemental table 3).
Discussion
We developed several machine learning models to identify neonates and young children at risk for all-cause 60-day postdischarge mortality at two sites in sub-Saharan Africa. Each of the developed models had greater discriminatory value than previously published risk assessment tools for postdischarge mortality among children in sub-Saharan Africa,1014 36,38 including the tools we developed from the same dataset using logistic regression.13 19 The XGBoost, Hist-Gradient Boost, and Random Forest models may be best suited for clinical use due to their inclusion of fewer features while also demonstrating excellent discriminatory value in internal validation.
To our knowledge, ours is the first study to use machine learning approaches to develop parsimonious risk assessment tools to identify neonates and young children at risk for postdischarge mortality. Investigators in the multicountry Childhood Acute Illness and Nutrition Network used XGBoost to identify 25 factors predictive of postdischarge mortality among children aged 2–24 months in six countries (including four sub-Saharan African countries).18 However, their objective was not to develop tools that could be used in clinical practice to accurately identify individual young children at risk for postdischarge mortality. Other studies have used traditional logistic regression to develop risk assessment tools for postdischarge mortality.10 13 14 19 36 37 Although some studies suggest that logistic regression-based models may outperform machine learning models,38 our study suggests that, with recent advances in prediction modelling, more precise approaches for identifying children at risk for postdischarge mortality are possible through machine learning.
Although each model for neonates and young children demonstrated superior discriminatory value to traditional logistic regression-based models, some of our models were more parsimonious than others. Specifically, we found that XGBoost, Hist Gradient Boost, and Random Forest models for both neonates and young children had the greatest discriminatory value, the fewest features, and were less computationally taxing than Support Vector Machine and Neural Network approaches. We elected to test several approaches to identify models that may balance discriminatory value with feasibility for use in clinical practice in busy settings with limited resources. Prior studies have demonstrated that the optimal machine learning approach may vary by disease process and outcome.39,41 In our study, the Support Vector Machine approach was suboptimal as it required the inclusion of 17 features for neonates and 25 for young children to achieve good discriminatory value, which may not be feasible for risk stratification in clinical practice in the absence of electronic health records as is currently the case in many settings in sub-Saharan Africa.
The features selected by our XGBoost, Hist Gradient Boost, and Random Forest models align with those identified in prior studies,10 37 which suggests that both social and biological phenomena drive postdischarge mortality. For example, discharge against medical advice, having lower haemoglobin levels or pallor, and having lower birth weight were predictive of postdischarge mortality, as observed in previous studies.10 13 19 37 Despite the use of sophisticated modelling, all the identified features can be collected in routine clinical practice, which makes the future use of these models potentially feasible. Discharge against medical advice can be measured at discharge and, although strongly associated with postdischarge mortality, could be supplanted by other features in our models and retain excellent discriminatory value.
Although the machine learning risk assessment tools we developed require sophisticated digitised health data, which is not widely available in sub-Saharan Africa, their clinical applicability at the bedside will only be possible with a simple calculator, which may be developed in the form of a smartphone application. Such applications have demonstrated feasibility in sub-Saharan Africa,42 but have yet to be implemented widely. The use of such tools has the potential to more precisely direct resources to the highest risk populations, which warrants further study. However, prior to the development of such mobile applications for our machine learning models, geographical external validation is necessary to ensure the reliability of our findings. To this end, we will conduct an external validation study of these models at two sites in Western Kenya with enrolment beginning in 2025.
Limitations
Our findings should be interpreted in the context of their limitations. First, although we used a list of >110 candidate features to develop our models, there may be other unmeasured confounding variables not assessed in our study that could contribute to postdischarge mortality. Due to the number of events of postdischarge mortality, we were unable to assess our models’ confidence at each follow-up time point. Additional studies with larger sample sizes may allow for further development of models that can add precise prediction of clinical parameters predictive of postdischarge mortality at specific time points following discharge. This study was also conducted at national referral hospitals in Tanzania and Liberia and thus may not be representative of other settings in sub-Saharan Africa, such as district or lower-level hospitals. Thus, external validation studies are warranted prior to the clinical use of our machine learning models for postdischarge mortality prediction.
Conclusions
Our parsimonious machine learning models had excellent discriminatory value to predict all-cause, 60-day postdischarge mortality among neonates and young children in two large hospitals in sub-Saharan Africa. These models required fewer features than previous models and demonstrated superior discriminatory value. However, before their implementation, external validation studies of these tools are warranted. This may assist in the design of appropriate interventions to reduce postdischarge mortality among the most vulnerable populations.
Supplementary material
Acknowledgements
We would like to acknowledge Reza Sameni and Tony Pan in the Emory Biomedical Informatics Core, and Michael Proctor, Ryan Birmingham and Sheida Habibi also in the Emory Biomedical Informatics Core for their assistance with the computation of this study. The Emory Biomedical Informatics Core is supported by CTSA Grant UL1TR002378 from the National Institutes of Health. This work was also supported by the Emory University Pediatric Biostatistics Core (RRID:SCR_025834). CR and ALW had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Footnotes
Funding: The authors wish to acknowledge funding for this work from the National Institutes of Health (K24 DK104676 and P30 DK040561 to CPD, K24 AT009893 to CRM, and K23HL173694 to CAR), the Boston Children’s Hospital Global Health Program (Grant Number N/A) to CAR, the Palfrey Fund for Child Health Advocacy (Grant Number N/A) to CAR and the Emory Pediatric Research Alliance Junior Faculty Focused Award (Grant Number N/A) to CAR. The funders had no role in the study design or in the collection, analysis, or interpretation of the data. The funders did not write the report and had no role in the decision to submit the paper for publication.
Provenance and peer review: Not commissioned; externally peer reviewed.
Patient consent for publication: Consent obtained from parent(s)/guardian(s)
Ethics approval: This study involves human participants and was approved by The study received ethical clearance from the Tanzania National Institute of Medical Research (#NIMR/HQ/R8a/Vol.IX/3494), the Muhimbili University of Health and Allied Sciences Research and Ethics Committee (#307/323/01), the John F. Kennedy Medical Center Institutional Review Board (#08062019), the Boston Children’s Hospital Institutional Review Board (#P00033242), and the use of de-identified data was exempted from review by the Emory University Institutional Review Board (no number provided for exempted studies). Participants gave informed consent to participate in the study before taking part.
Data availability free text: Data may be made available upon reasonable request to the corresponding author.
Patient and public involvement: Patients and/or the public were not involved in the design, conduct, reporting or dissemination plans of this research.
Author note: This manuscript is an honest and accurate account of the study being reported. No aspects of this study have been omitted or withheld.
Data availability statement
Data are available upon reasonable request.
References
- 1.Rees CA, Flick RJ, Sullivan D, et al. An Analysis of the Last Clinical Encounter before Outpatient Mortality among Children with HIV Infection and Exposure in Lilongwe, Malawi. PLoS One . 2017;12:e0169057. doi: 10.1371/journal.pone.0169057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiens MO, Pawluk S, Kissoon N, et al. Pediatric post-discharge mortality in resource poor countries: a systematic review. PLoS One. 2013;8:e66698. doi: 10.1371/journal.pone.0066698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nemetchek B, English L, Kissoon N, et al. Paediatric postdischarge mortality in developing countries: a systematic review. BMJ Open. 2018;8:e023445. doi: 10.1136/bmjopen-2018-023445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rees CA, Neuman MI, Monuteaux MC, et al. Mortality During Readmission Among Children in United States Children’s Hospitals. J Pediatr. 2022;246:161–9. doi: 10.1016/j.jpeds.2022.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiens MO, Kissoon N, Kabakyenga J. Smart Hospital Discharges to Address a Neglected Epidemic in Sepsis in Low- and Middle-Income Countries. JAMA Pediatr. 2018;172:213–4. doi: 10.1001/jamapediatrics.2017.4519. [DOI] [PubMed] [Google Scholar]
- 6.Rees CA, Kisenge R, Ideh RC, et al. Predictive value of clinician impression for readmission and postdischarge mortality among neonates and young children in Dar es Salaam, Tanzania and Monrovia, Liberia. BMJ Paediatr Open . 2023;7:e001972. doi: 10.1136/bmjpo-2023-001972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maguire JL, Kulik DM, Laupacis A, et al. Clinical prediction rules for children: a systematic review. Pediatrics. 2011;128:e666–77. doi: 10.1542/peds.2011-0043. [DOI] [PubMed] [Google Scholar]
- 8.Rees CA, Hooli S, King C, et al. External validation of the RISC, RISC-Malawi, and PERCH clinical prediction rules to identify risk of death in children hospitalized with pneumonia. J Glob Health . 2021;11:04062. doi: 10.7189/jogh.11.04062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rees CA, Colbourn T, Hooli S, et al. Derivation and validation of a novel risk assessment tool to identify children aged 2–59 months at risk of hospitalised pneumonia-related mortality in 20 countries. BMJ Glob Health . 2022;7:e008143. doi: 10.1136/bmjgh-2021-008143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiens MO, Kumbakumba E, Larson CP, et al. Postdischarge mortality in children with acute infectious diseases: derivation of postdischarge mortality prediction models. BMJ Open. 2015;5:e009449. doi: 10.1136/bmjopen-2015-009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Madrid L, Casellas A, Sacoor C, et al. Postdischarge Mortality Prediction in Sub-Saharan Africa. Pediatrics. 2019;143:e20180606. doi: 10.1542/peds.2018-0606. [DOI] [PubMed] [Google Scholar]
- 12.Diallo AH, Sayeem Bin Shahid ASM, Khan AF, et al. Childhood mortality during and after acute illness in Africa and south Asia: a prospective cohort study. Lancet Glob Health. 2022;10:e673–84. doi: 10.1016/S2214-109X(22)00118-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rees CA, Ideh RC, Kisenge R, et al. Identifying neonates at risk for post-discharge mortality in Dar es Salaam, Tanzania, and Monrovia, Liberia: Derivation and internal validation of a novel risk assessment tool. BMJ Open. 2024;14:e079389. doi: 10.1136/bmjopen-2023-079389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiens MO, Nguyen V, Bone JN, et al. Prediction models for post-discharge mortality among under-five children with suspected sepsis in Uganda: A multicohort analysis. PLOS Glob Public Health . 2024;4:e0003050. doi: 10.1371/journal.pgph.0003050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramgopal S, Horvat CM, Yanamala N, et al. Machine Learning To Predict Serious Bacterial Infections in Young Febrile Infants. Pediatrics. 2020;146:e20194096. doi: 10.1542/peds.2019-4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le S, Hoffman J, Barton C, et al. Pediatric Severe Sepsis Prediction Using Machine Learning. Front Pediatr. 2019;7:413. doi: 10.3389/fped.2019.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaswani A, Shazeer N, Parmar N, et al. Attention Is All You Need. Arxiv. 2023 https://arxiv.org/abs/1706.03762v7 Available. [Google Scholar]
- 18.Diallo AH, Sayeem Bin Shahid ASM, Khan AF, et al. Characterising paediatric mortality during and after acute illness in Sub-Saharan Africa and South Asia: a secondary analysis of the CHAIN cohort using a machine learning approach. eClinicalMedicine. 2023;57:101838. doi: 10.1016/j.eclinm.2023.101838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rees CA, Kisenge R, Godfrey E, et al. Derivation and Internal Validation of a Novel Risk Assessment Tool to Identify Infants and Young Children at Risk for Post-Discharge Mortality in Dar es Salaam, Tanzania and Monrovia, Liberia. J Pediatr. 2024;273:114147. doi: 10.1016/j.jpeds.2024.114147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol. 2010;5:1315–6. doi: 10.1097/JTO.0b013e3181ec173d. [DOI] [PubMed] [Google Scholar]
- 21.Rees CA, Kisenge R, Ideh RC, et al. A Prospective, observational cohort study to identify neonates and children at risk of postdischarge mortality in Dar es Salaam, Tanzania and Monrovia, Liberia: the PPDM study protocol. BMJ Paediatr Open . 2022;6:e001379. doi: 10.1136/bmjpo-2021-001379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chhibber AV, Hill PC, Jafali J, et al. Child Mortality after Discharge from a Health Facility following Suspected Pneumonia, Meningitis or Septicaemia in Rural Gambia: A Cohort Study. PLoS One. 2015;10:e0137095. doi: 10.1371/journal.pone.0137095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ngari MM, Fegan G, Mwangome MK, et al. Mortality after Inpatient Treatment for Severe Pneumonia in Children: a Cohort Study. Paediatr Perinat Epidemiol. 2017;31:233–42. doi: 10.1111/ppe.12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiens MO, Kissoon N, Kumbakumba E, et al. Selecting candidate predictor variables for the modelling of post-discharge mortality from sepsis: a protocol development project. Afr Health Sci. 2016;16:162–9. doi: 10.4314/ahs.v16i1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nemetchek BR, Liang L, Kissoon N, et al. Predictor variables for post-discharge mortality modelling in infants: a protocol development project. Afr H Sci. 2018;18:1214. doi: 10.4314/ahs.v18i4.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chawla NV, Bowyer KW, Hall LO, et al. SMOTE: Synthetic Minority Over-sampling Technique. Jair. 2002;16:321–57. doi: 10.1613/jair.953. [DOI] [Google Scholar]
- 27.Li R, Shinde A, Liu A, et al. Machine Learning–Based Interpretation and Visualization of Nonlinear Interactions in Prostate Cancer Survival. JCO Clinical Cancer Informatics . 2020;4:637–46. doi: 10.1200/CCI.20.00002. [DOI] [PubMed] [Google Scholar]
- 28.Mahajan A, Esper S, Oo TH, et al. Development and Validation of a Machine Learning Model to Identify Patients Before Surgery at High Risk for Postoperative Adverse Events. JAMA Netw Open . 2023;6:e2322285. doi: 10.1001/jamanetworkopen.2023.22285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steyerberg EW, Harrell FE., Jr Prediction models need appropriate internal, internal-external, and external validation. J Clin Epidemiol. 2016;69:245–7. doi: 10.1016/j.jclinepi.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steyerberg EW, Harrell FE, Jr, Borsboom GJ, et al. Internal validation of predictive models: efficiency of some procedures for logistic regression analysis. J Clin Epidemiol. 2001;54:774–81. doi: 10.1016/s0895-4356(01)00341-9. [DOI] [PubMed] [Google Scholar]
- 31.Riley RD, Ensor J, Snell KIE, et al. Calculating the sample size required for developing a clinical prediction model. BMJ. 2020;368:m441. doi: 10.1136/bmj.m441. [DOI] [PubMed] [Google Scholar]
- 32.Iba K, Shinozaki T, Maruo K, et al. Re-evaluation of the comparative effectiveness of bootstrap-based optimism correction methods in the development of multivariable clinical prediction models. BMC Med Res Methodol. 2021;21:9. doi: 10.1186/s12874-020-01201-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Buuren S, Groothuis-Oudshoorn K. MICE: multivariate imputation by chained equations in R. J Stat Softw. 2011;45:1–67. doi: 10.18637/jss.v045.i03. [DOI] [Google Scholar]
- 34.Wood AM, White IR, Royston P. How should variable selection be performed with multiply imputed data? Stat Med. 2008;27:3227–46. doi: 10.1002/sim.3177. [DOI] [PubMed] [Google Scholar]
- 35.Austin PC, Lee DS, Ko DT, et al. Effect of Variable Selection Strategy on the Performance of Prognostic Models When Using Multiple Imputation. Circ Cardiovasc Qual Outcomes. 2019;12:e005927. doi: 10.1161/CIRCOUTCOMES.119.005927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmed SM, Brintz BJ, Talbert A, et al. Derivation and external validation of a clinical prognostic model identifying children at risk of death following presentation for diarrheal care. PLOS Glob Public Health . 2023;3:e0001937. doi: 10.1371/journal.pgph.0001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talbert A, Ngari M, Bauni E, et al. Mortality after inpatient treatment for diarrhea in children: a cohort study. BMC Med. 2019;17:20. doi: 10.1186/s12916-019-1258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finlayson SG, Beam AL, van Smeden M. Machine Learning and Statistics in Clinical Research Articles-Moving Past the False Dichotomy. JAMA Pediatr. 2023;177:448–50. doi: 10.1001/jamapediatrics.2023.0034. [DOI] [PubMed] [Google Scholar]
- 39.Gao C, Sun H, Wang T, et al. Model-based and Model-free Machine Learning Techniques for Diagnostic Prediction and Classification of Clinical Outcomes in Parkinson’s Disease. Sci Rep. 2018;8:7129. doi: 10.1038/s41598-018-24783-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volkova A, Ruggles KV. Predictive Metagenomic Analysis of Autoimmune Disease Identifies Robust Autoimmunity and Disease Specific Microbial Signatures. Front Microbiol. 2021;12:621310. doi: 10.3389/fmicb.2021.621310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Burns SM, Woodworth TS, Icten Z, et al. A Machine Learning Approach to Identify Predictors of Severe COVID-19 Outcome in Patients With Rheumatoid Arthritis. Pain Physician. 2022;25:593–602. [PubMed] [Google Scholar]
- 42.English LL, Dunsmuir D, Kumbakumba E, et al. The PAediatric Risk Assessment (PARA) Mobile App to Reduce Postdischarge Child Mortality: Design, Usability, and Feasibility for Health Care Workers in Uganda. JMIR Mhealth Uhealth. 2016;4:e16. doi: 10.2196/mhealth.5167. [DOI] [PMC free article] [PubMed] [Google Scholar]