Abstract
Three new ent-kaurane diterpenoids, isodosins E–G (1–3), along with 20 known ones (4–23), were obtained from the aerial parts of Isodon serra (Maxim.) Hara. The structures of the new compounds were elucidated using 1D/2D NMR spectra and HREIMS data, and their absolute configurations were determined by electronic circular dichroism (ECD) calculations. The in vitro anti-hepatocarcinoma activities of compounds 2, 3, 5, 8, 13, 19, and 23 were evaluated against HepG2 and Huh7 cell lines using the CCK-8 assay. Among them, compounds 3, 8, and 23 exhibited high inhibitory effects on HepG2 cells, with IC50 values of 6.94 ± 9.10 μM, 71.66 ± 10.81 μM, and 43.26 ± 9.07 μM, respectively. In a Hepa1-6 xenograft mouse model, compound 8 significantly inhibited tumor growth at doses of 50 and 100 mg kg−1, demonstrating its potent anti-hepatocarcinoma activity.
Three new ent-kaurane diterpenoids, isodosins E–G (1–3), along with 20 known ones (4–23), were obtained from the aerial parts of Isodon serra (Maxim.) Hara.
1. Introduction
The genus Isodon (Lamiaceae), comprising approximately 150 species worldwide, is mainly distributed in tropical and subtropical Asia.1 Plants of this genus are a prolific source of diverse diterpenoids with extensive biological activities, and more than 1200 diterpenoids have been isolated.1–4 Among these diterpenoids, ent-kaurane diterpenoids have attracted much attention due to their promising anti-tumor activities.5–7 For example, oridonin, a prominent ent-kaurane diterpenoid, has garnered widespread attention for its significant anti-tumor activity, demonstrating dose-dependent cytotoxicity against HepG2 cells with an IC50 value of 37.90 μM.8,9 Numerous studies have further revealed that oridonin inhibited tumorigenesis and progression through multiple mechanisms, including induction of tumor apoptosis and autophagy, suppression of the cell cycle progression, and inhibition of angiogenesis.8,10 In terms of molecular mechanisms, it has been demonstrated to exert its anti-tumor effects through the regulation of ROS, MAPK, PI3K/Akt, Bax/Bcl-2, and NF-κB pathways.9,11 The findings support the potential of ent-kaurane diterpenoids as anti-cancer lead compounds and highlight the need for further research and development in this field.
Isodon serra (Maxim.) Hara, which belongs to the genus Isodon, is predominantly distributed in Guangdong, Fujian, and Jiangxi provinces, China.12 Traditionally, it has been used to treat various ailments, such as hepatitis, cholecystitis, jaundice, and enteritis.13 Previous phytochemical studies of this species have led to the isolation of abundant bioactive ent-kaurane diterpenes, including enmein, spirolactone, C-20 non-oxygenated, and C-20 oxygenated types.1,14–16 These compounds have demonstrated significant pharmacological activities, including cytotoxic, antibacterial, and anti-inflammatory activities.16,17 In our previous research, a series of ent-kaurane diterpenoids were isolated from the petroleum ether fraction of I. serra, and some of them exhibited selective cytotoxicity against HepG2 cells in the MTT assay.18 In particular, odonicin significantly inhibited the proliferation of HepG2 cells, and this effect was achieved by inducing apoptosis.18
To further search for potential bioactive diterpenoids, the chemical composition of the ethyl acetate fraction from I. serra was investigated in the present study. As a result, three new diterpenoids (1–3) and 20 known ones (4–23) were isolated and characterized (Fig. 1). Compounds 3, 8, and 23 demonstrated potent inhibitory activity against HepG2 cells. Furthermore, compound 8 exhibited significant tumor growth inhibition in a Hepa1-6 xenograft mouse model.
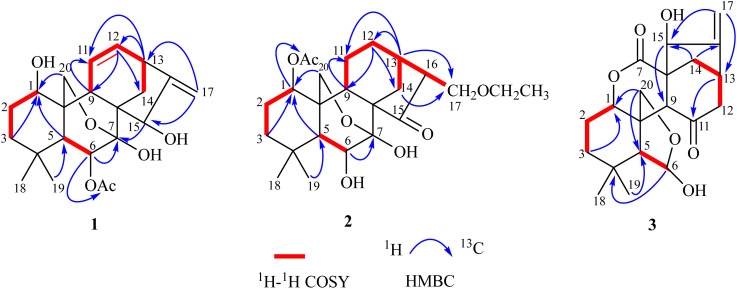
Fig. 1. Chemical structures of compounds 1–23.
2. Results and discussion
The aerial parts of I. serra were extracted three times and partitioned with petroleum ether (PE), ethyl acetate (EA), and aqueous solution (AS). The EA fraction was subjected to silica gel column chromatography, and further purified using MPLC and semi-preparative HPLC. Ultimately, three new diterpenoids (1–3) and 20 known compounds (4–23) were obtained.
2.1. Structural elucidation
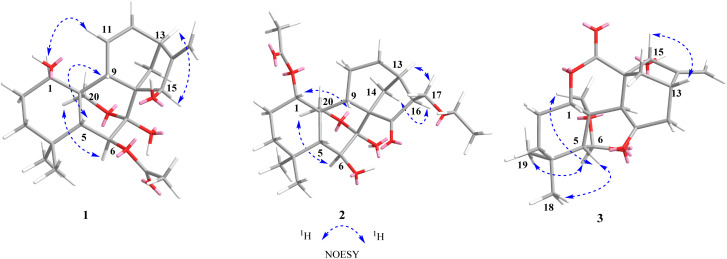
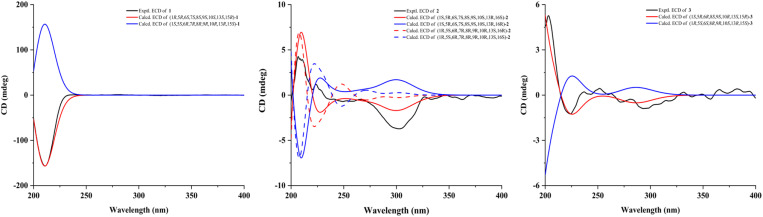
Compound 1, isolated as a light yellow powder, displayed an ion peak at m/z 413.1950 [M + Na]+ (calcd for C22H30O6Na, 413.1935) by its HR-ESI-MS, which suggested a molecular formula of C22H30O6 with eight degrees of unsaturation. The IR spectrum displayed the characteristic absorptions for hydroxyl (2935 cm−1) and carbonyl (1738 cm−1) groups (Fig. S8†). In the 1H NMR spectrum (Table 1), three methyl singlets [δH 0.89 (3H, s, H-19), 1.13 (3H, s, H-18), and 2.18 (3H, s, OAc)], five oxygenated methylene or methine protons [δH 3.79 (1H, d, J = 9.0 Hz, H-20b) and 3.81 (1H, dd, J = 9.0, 2.5 Hz, H-20a), δH 3.74 (1H, m, H-1), 4.60 (1H, s, H-15), and 5.20 (1H, d, J = 4.2 Hz, H-6)], and four olefinic protons [δH 5.00 (1H, d, J = 2.4 Hz, H-17b), 5.05 (1H, s, H-17a), 5.76 (1H, dd, J = 9.4, 2.4 Hz, H-11), and 6.36 (1H, ddd, J = 10.0, 7.1, 2.9 Hz, H-12)] were observed. The 13C NMR data revealed that 1 contained 20 carbon atoms, which were classified into three methyls, five methylenes (one olefinic and one oxygenated), eight methines (two olefinic and three oxygenated), and six non-protonated carbons (one olefinic, one ester carbonyl, and one oxygenated) by DEPT and HSQC spectra (Table 1). These data were consistent with the skeleton of a 7,20-epoxy-ent-kaur-16-en-1-one and were similar to those of 1β,6β,7β-trihydroxy-15β-acetoxy-7,20-epoxy-ent-kaur-16-ene (4).19 The major difference between the two except that the two methylenes at C-11/C-12 (δC 26.4 and 31.7) in 4 were replaced by a double bond (δC 123.3 and 138.6) in 1. This hypothesis was confirmed by the 1H–1H COSY correlations of H-9/H-11/H-12/H-13, as well as the HMBC correlations of H-9, H-13/C-11 (δC 123.3) and H-13, H-14a/C-12 (δC 138.6) (Fig. 2). In the NOESY spectrum, the configurations of H-1/H-11 and H-13/H-15 indicated that the hydroxyl groups at C-1 and C-15 were assigned as β-orientation. The acetoxy group at C-6 was determined to be β-oriented based on the interactions of H-6/H-20a (Fig. 3). The calculated ECD spectrum of 1 displayed Cotton Effects (CEs) analogous to the experimental ECD data, which demonstrated the absolute configuration was 1R,5R,6S,7S,8S,9S,10S,13S,15R (Fig. 4). Thus, the structure of 1 was elucidated and named isodosin E.
Table 1. 1H NMR (600 MHz) and 13C NMR (150 MHz) spectroscopic data of compounds 1–3 in CDCl3 (δ in ppm, J in Hz).
| Position | 1 | 2 | 3 | |||
|---|---|---|---|---|---|---|
| δ C (type) | δ H (mult., J in Hz) | δ C (type) | δ H (mult., J in Hz) | δ C (type) | δ H (mult., J in Hz) | |
| 1 | 65.5 (CH) | 3.74 (m) | 68.1 (CH) | 5.19 (t, 4.8) | 79.1 (CH) | 4.62 (dd, 11.4, 5.9) |
| 2a | 26.7 (CH2) | 1.64 (m) | 41.3 (CH2) | 1.46 (m) | 23.5 (CH2) | 1.88 (m) |
| 2b | 1.25 (m) | 1.13 (m) | 1.78 (d, 12.5) | |||
| 3a | 33.4(CH2) | 1.78 (d, 10.3) | 28.2(CH2) | 2.20 (m) | 36.7 (CH2) | 1.50 (m) |
| 3b | 1.28 (m) | 1.91 (dd, 16.7, 5.0) | 1.54 (d, 5.3) | |||
| 4 | 33.7 (C) | 33.9 (C) | 31.1 (C) | |||
| 5 | 51.3 (CH) | 1.86 (d, 4.2) | 59.8 (CH) | 1.21 (d, 7.0) | 54.6 (CH) | 2.97 (s) |
| 6 | 75.2 (CH) | 5.20 (d, 4.2) | 74.5 (CH) | 3.78 (dd, 11.9, 7.0) | 102.1 (CH) | 5.34 (s) |
| 7 | 97.2 (C) | 94.7 (C) | 173.1 (C) | |||
| 8 | 51.6 (C) | 59.3 (C) | 54.3 (C) | |||
| 9 | 39.8 (CH) | 2.89 (d, 2.4) | 52.9 (CH) | 1.54 (m) | 51.2 (CH) | 3.79 (s) |
| 10 | 41.7 (C) | 36.8 (C) | 47.7 (C) | |||
| 11a | 123.3 (CH) | 5.76 (dd, 9.4, 2.4) | 18.3 (CH) | 1.44 (m) | 210.3 (C) | |
| 11b | 1.36 (m) | |||||
| 12a | 138.6 (CH) | 6.36 (t, 8.2) | 30.5 (CH2) | 1.34 (m) | 52.5 (CH2) | 2.68 (dd, 16.0, 7.9) |
| 12b | 1.18 (m) | 2.46 (d, 16.0) | ||||
| 13 | 38.6 (CH) | 2.97 (dd, 7.4, 4.5) | 28.6 (CH) | 3.44 (m) | 36.8 (CH) | 3.07 (m) |
| 14a | 31.8 (CH2) | 1.95 (dd, 11.6, 4.5) | 28.4 (CH2) | 2.73 (m) | 36.5 (CH2) | 1.96 (dd, 12.4, 4.1) |
| 14b | 1.55 (d, 11.6) | 1.95 (dd, 16.5, 9.5) | 1.74 (d, 12.4) | |||
| 15 | 74.3 (CH) | 4.60 (s) | 221.7 (C) | 79.4 (CH) | 5.24 (s) | |
| 16 | 155.6 (C) | 56.3 (CH) | 2.74 (m) | 153.9 (C) | ||
| 17a | 107.5 (CH2) | 5.05 (s) | 66.4 (CH2) | 4.08 (d, 9.2) | 111.5 (CH2) | 5.41 (s) |
| 17b | 5.00 (d, 2.4) | 5.37 (s) | ||||
| 18 | 31.7 (CH3) | 1.13 (s) | 33.7 (CH3) | 1.12 (s) | 32.9 (CH3) | 1.08 (s) |
| 19 | 21.6 (CH3) | 0.89 (s) | 22.4 (CH3) | 1.10 (s) | 23.7 (CH3) | 0.97 (s) |
| 20a | 66.4 (CH2) | 3.81 (dd, 9.0, 2.5) | 68.8 (CH2) | 4.17 (d, 9.5) | 72.1 (CH2) | 4.04 (d, 9.2) |
| 20b | 3.79 (d, 9.0) | 3.70 (dd, 9.5, 3.0) | 3.93 (d, 9.2) | |||
| OAc-6 | 170.9 | |||||
| 21.4 (CH3) | 2.18 (s) | |||||
| OAc-1 | 169.8 (C) | |||||
| 21.9 (CH3) | 2.10 (s) | |||||
| OEt–CH2 | 66.7 (CH2) | 3.49 (m) | ||||
| OEt–CH3 | 15.1 (CH3) | 1.17 (t, 7.0) | ||||
Fig. 2. 1H–1H COSY and key HMBC correlations of compounds 1–3.
Fig. 3. Key NOESY correlations of compounds 1–3.
Fig. 4. Experimental and calculated ECD spectra of compounds 1–3.
Compound 2 was obtained as a white amorphous powder. The molecular formula was determined as C24H36O7 based on the HR-ESI-MS ion peak at m/z 437.2569 [M + H]+ (calcd for C24H36O7H, 437.2534), indicating seven degrees of unsaturation. The IR absorption bands at 2924 and 1718 cm−1 suggested the existence of hydroxyl and carbonyl groups (Fig. S18†). The 1H NMR spectrum of 2 (Table 1) showed signals of three methyls [δH 1.10 (3H, s, H-19), 1.12 (3H, s, H-18), and 2.10 (3H, s, OAc)]. The 13C NMR, DEPT 135, and HSQC spectra (Table 1) revealed 24 carbon signals, including four methyls, seven methylenes, seven methines, and six non-protonated carbons (one carbonyl, one ester carbonyl, and six oxygenated). The above information indicated that the structure of 2 was similar to that of adenolin E,20 except for two notable differences. Specifically, the methoxy group at C-17 (δC 68.9) in adenolin E was absent in 2 and an ethoxy group was present in 2. Furthermore, the oxygenated methine at C-11 (δC 62.9) in adenolin E was replaced by a methylene group (δC 56.3) in 2. These observations were also supported by 1H–1H COSY correlations of H-9/H-11/H-12 and the HMBC correlation of H-17 with OEt–CH2 (δC 66.7) (Fig. 2). In the NOESY spectrum (Fig. 3), the correlation of H-1/H-9 indicated the acetoxy group at C-1 was α-oriented. The hydroxyl group at C-6 was β-oriented by the correlation of H-6/H-20a. The ECD spectrum of 2 was recorded in CHCl3, and its absolute configuration was assigned as 1S,5R,6S,7S,8S,9S,10S,13R,16S by comparing the experimental data with calculated ECD curves of four possible stereoisomers (Fig. 4). Thus, the structure of 2 was elucidated and named isodosin F.
Compound 3 was obtained as a white amorphous powder, and its molecular formula was determined to be C20H26O6 based on the HR-ESI-MS ion at m/z 385.1630 [M + Na]+ (calcd for C20H26O6Na, 385.1622), requiring eight degrees of unsaturation. Analysis of the 1H NMR data of 3 (Table 1) indicated two singlet methyl groups [δH 0.97 (3H, s, H-19) and 1.08 (3H, s, H-18)] and two olefinic protons [δH 5.37 (1H, s, H-17b) and 5.41 (1H, s, H-17a)]. The 13C NMR spectra of 3 showed 20 carbons classified into two methyls (δC 32.9, 23.7), six methylenes (δC 111.5, 72.1, 52.5, 36.7, 36.5, 23.5), six methines (δC 102.1, 79.4, 79.1, 54.6, 51.2, 36.8), and six non-protonated carbons (δC 210.3, 173.1, 153.9, 54.3, 47.7, 31.1), which were supported by DEPT and HSQC spectra. Analysis of its NMR data suggested many similarities to those of 6β,15α-dihydroxy-6α,20-epoxy-6,7-seco-ent-kaur-16-en-1α,7-olide (20),19 with the major difference was that the methylene at C-11 (δC 18.6) in 20 was replaced by the ketone carbonyl group at C-11 (δC 210.3) in 3. This hypothesis was confirmed by the 1H–1H COSY correlations of H-12/H-13/H-14 and the HMBC correlations of H-12/H-13 with C-11 (Fig. 2). The hydroxyl group at C-6 was assigned as β-oriented based on the NOESY correlations of H-1/H-5, H-5/H-18, and H-6/H-19. The α-orientation of the hydroxyl group at C-15 was deduced by the H-13/H-15 correlation (Fig. 3). The ECD curve of compound 3 showed a positive Cotton effect at 204 nm and 249 nm, which was consistent with the calculated ECD spectrum of (1S,5R,6R,8S,9S,10R,13S,15R)-3 (Fig. 4). Thus, the structure of 3 was confirmed and named isodosin G.
The known compounds were identified as 1β,6β,7β-trihydroxy-15β-acetoxy-7,20-epoxy-ent-kaur-16-ene (4),19 maoecrystal S (5),21 neorabdosin (6),22 parvifoline G (7),23 odonicin (8),24 effusanin A (9),25 (6β,7α,15β)-6,15-bis(acetyloxy)-7,20-epoxy-7,13-dihydroxykaura-2,16-dien-1-one (10),26 rabdolasional (11),27 shikokianin (12),28 isodoternifolin B (13),29 (6β,7α,11α)-7,20-epoxy-6,7,11-trihydroxykaur-16-en-15-one (14),30 maoecrystal X (15),31 nodosin (16),32 isodocarpin (17),33 15β-hydroxy-6,7-seco-6,11β:6,20-diepoxy-1α,7-olide-ent-kaur-16-ene (18),34 6-acetylepinodosinol (19),35 6β,15α-dihydroxy-6α,20-epoxy-6,7-seco-ent-kaur-16-en-1α,7-olide (20),19 maoyecrystal E (21),36 lushanrubescensin H (22),37 and isorubesin C (23).38 Their structures were confirmed by comparing the spectroscopic data with those previously reported, in which compounds 4–6, 10–15, and 19–23 were isolated from I. serra for the first time.
2.2. Anti-hepatocarcinoma activities in vitro
Hepatocellular carcinoma (HCC), as the third leading cause of cancer-related death worldwide, presents substantial therapeutic challenges due to the high rate of late diagnosis, severe side effects, and frequent drug resistance.39–42 Diterpenoids, a structurally diverse class of natural products, have attracted considerable interest owing to their potent anti-cancer effects.8,9
Given the anti-cancer activity of diterpenoids from I. serra, the main isolated compounds 2, 3, 5, 8, 13, 19, and 23 were evaluated for their cytotoxic effects on HepG2 and Huh7 cell lines using the CCK-8 assay, with cisplatin as the positive control. As shown in Fig. 5 and Table 2, compounds 2, 5, 8, and 23 reduced the viability of HepG2 cells in a dose-dependent manner at concentrations of 0, 12.5, 25, 50, 100, and 200 μM. Among them, compound 3 exhibited strong inhibition effects with an IC50 of 6.94 ± 9.10 μM, followed by compounds 23 (IC50 = 43.26 ± 9.07 μM) and 8 (IC50 = 71.66 ± 10.81 μM). By contrast, compounds 13 and 19 showed relatively weak cytotoxicity against HepG2 cells under the same conditions. In comparison, the IC50 value of cisplatin against HepG2 cells was 20.73 ± 5.71 μM. Notably, at a concentration of 100 μM, the inhibitory effect of compound 23 against HepG2 cells was comparable to that of cisplatin, and at 200 μM, its inhibitory effect even exceeded that of the reference drug. These results suggested that certain diterpenoids from I. serra exhibited promising anti-hepatocarcinoma activity.
Fig. 5. Anti-hepatocarcinoma activities of compounds 2, 3, 5, 8, 13, 19, and 23, with cisplatin as the positive control. Data were presented as mean ± SD of three parallel measurements. *p < 0.05, **p < 0.01, ***p < 0.001 versus the control group.
Table 2. Effects of compounds 2, 3, 5, 8, 13, 19, and 23 on the proliferation of HepG2 and Huh7 cells.
| Compound | IC50a (μM) | |
|---|---|---|
| HepG2 | Huh7 | |
| 2 | 105.00 ± 6.49 | >200 |
| 3 | 6.94 ± 9.10 | 23.34 ± 17.83 |
| 5 | 95.22 ± 18.86 | >200 |
| 8 | 71.66 ± 10.81 | >200 |
| 13 | >200 | >200 |
| 19 | >200 | >200 |
| 23 | 43.26 ± 9.07 | 109.60 ± 12.18 |
| Cisplatinb | 20.73 ± 5.71 | 96.15 ± 9.29 |
IC50 values were means ± SD (n = 3).
Positive control.
2.3. Anti-hepatocarcinoma activity of compound 8in vivo
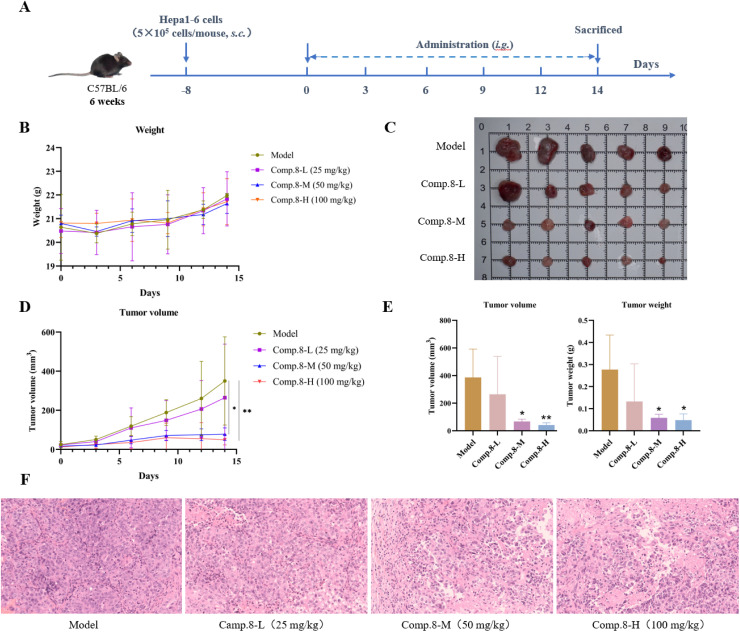
Given its potent in vitro anti-hepatocarcinoma activity against HepG2 cells and sufficient yield from isolation, compound 8 was selected for further in vivo evaluation using a subcutaneous Hepa1-6 xenograft mouse model, as depicted in Fig. 6A. Compared to the model group, there was no significant change in the body weight of mice treated with 8 (p > 0.05), suggesting that the mice exhibited a favorable tolerance to 8 (Fig. 6B). The result was important as it demonstrated the safety profile of 8 with no apparent toxicity or adverse effects on overall health. As shown in Fig. 6C–E, tumor growth inhibition was observed in the medium and high dose groups, with significant reductions in both tumor volume and weight compared to the model group. At the doses of 25, 50, and 100 mg kg−1, the tumor inhibition rates were 52.13%, 78.88%, and 82.70%, respectively, which demonstrated a dose-dependent anti-hepatocarcinoma effect of 8in vivo. Histopathological examination using H&E staining (Fig. 6F) provided further insights into the cellular effects of 8 on tumor tissues. In the model group, the tumor cell structure remained intact with deeply stained nuclei. In contrast, tumor tissues from the medium and high dose groups showed vacuole-like degeneration, unclear cell membrane boundaries, and irregular nuclear enlargement, with the presence of necrosis and nuclear deformation.
Fig. 6. Anti-hepatocarcinoma activity of Comp. 8in vivo. (A) Schematic design of 8in vivo anti-tumor effect study. (B) Changes in body weight of mice. (C) Images of the tumors from all mice at the endpoint. (D) Changes in the tumor volume of mice. (E) Tumor volume and weight changes after 8 treatment. (F) H&E staining of tumor tissue (scale bar = 100 μm). All data are shown as mean ± SD (n = 5). *p < 0.05, **p < 0.01, versus the model group.
Combined with the in vitro results, these findings provided evidence that 8 exerted significant anti-hepatocarcinoma effects. Further elucidation of the underlying molecular mechanisms and signaling pathways will contribute to a deeper understanding of its therapeutic mechanisms.
3. Experimental section
3.1. General experimental procedures
IR spectra were measured on a Bruker TENSOR37 infrared spectrometer with KBr disks. UV spectra were acquired on a Yokechina double-beam ultraviolet-visible spectrophotometer. Optical rotations were recorded on an Autopol l automatic polarimeter (Rudolph Research Analytical). 1D and 2D NMR spectra were obtained on a Bruker Ascend 600 NMR spectrometer with TMS as an internal reference. HR-ESI-MS were obtained on an Agilent 1290 Infinity LC and an Agilent 6530 Q-TOF mass spectrometer (Agilent, Palo Alto, CA, USA). Column chromatography (CC) was performed with silica gel (80–100 mesh, Huanghai, China). Medium-pressure liquid chromatography (MPLC) was performed using an RP-18 column (YMC C18, 46 × 600 mm, 40–60 μm particle size) on a RUlHE LC-2100 liquid chromatography instrument. Semi-preparative HPLC purifications were conducted on a RUlHE LC-2010 liquid chromatography instrument with a Waters Xbridge Prep C18 column (10 × 250 mm, 5 μm) or an XSelect Prep C18 column (10 × 250 mm, 5 μm). Ultra Performance Liquid Chromatography (UPLC) was performed on the Waters XSelect Premier HSS T3 column (100 mm × 2.0 mm, 2.2 μm).
The HPLC-grade solvents (methanol and acetonitrile) were obtained from Thermo Fisher Scientific (Suwanee, GA, USA). Petroleum ether, ethyl acetate, methanol, and acetonitrile were acquired from Ghtech (Guangzhou, China). Deuterated chloroform (CDCl3) was purchased from Macklin (Shanghai, China). The Cell Count Kit-8 (CCK-8) was purchased from Beyotime (Shanghai, China). Dulbecco's modified Eagle medium (DMEM), RPML-1640, 0.25% trypsin-ethylene diamine tetraacetic acid (EDTA) solution, penicillin–streptomycin solution, and phosphate buffer saline (PBS) were purchased from Servicebio (Wuhan, China). Fetal bovine serum (FBS) was obtained from Gibco (WalthamMA, USA).
3.2. Plant materials
The collection and identification of the aerial parts of I. serra were consistent with our previous study.18
3.3. Extraction and isolation
As recorded in our earlier study,18 80 kg of the aerial parts of I. serra were powdered and extracted with 80% EtOH (320 L) under reflux three times. The extracts were concentrated and then partitioned successively with petroleum ether (PE), ethyl acetate (EA), and aqueous solution (AS). The EA fraction (672 g) was subjected to silica gel CC using a gradient of PE–EtOAc–MeOH (1 : 0 : 0–0 : 0 : 1, v/v/v) to obtain eight fractions (Fr.A–H). Fr.C (62.7 g) was further separated by MPLC and eluted with a stepwise gradient of ACN–H2O (10 : 90–100 : 0, v/v) to yield eight subfractions (Fr.C1–C8). Fr.C4 was purified by semi-preparative HPLC eluted with MeOH–H2O (40 : 60) to yield compounds 1 (10.40 mg) and 4 (51.82 mg). Similarly, compound 5 (60.23 mg) was purified from Fr.C5 using semi-preparative HPLC (MeOH–H2O, 49 : 51). Fr.C6 was separated via semi-preparative HPLC eluted with MeOH–H2O (58 : 42) to afford compounds 6 (30.09 mg) and 7 (30.76 mg). Compound 8 (2.24 g) was obtained from Fr.C7 by repeated recrystallization using MeOH. Fr.E (53.0 g) was chromatographed on a C18 MPLC column eluted with a gradient of ACN–H2O (15 : 85–100 : 0, v/v) to give ten subfractions, Fr.E1-E10. Compound 16 (20.36 mg) was acquired using recrystallization from Fr.E2. The residue of Fr.E2 was purified using semi-preparative HPLC eluted with MeOH–H2O (41 : 59) to afford compounds 17 (7.38 mg). Following, Fr.E3 was isolated by MPLC and eluted with MeOH–H2O (35 : 65–54 : 46, v/v) to obtain two subfractions (Fr.E3-a and E3-b). Fr.E3-a and Fr.E3-b were respectively subjected to semi-preparative HPLC (MeOH–H2O, 43 : 57 and 50 : 50) to obtain compounds 21 (10.27 mg) and 3 (35.81 mg). Compound 9 (107.24 mg) was obtained from Fr.E4 by repeated recrystallization using MeOH. The residue of Fr.E4 was subjected to sim-PHPLC (MeOH–H2O, 48 : 52) to yield compound 10 (8.26 mg). Fr.E5 was purified by semi-preparative HPLC (MeOH–H2O, 54 : 46) to give compounds 18 (10.32 mg) and 19 (28.65 mg). Compound 11 (98.04 mg) was afforded from Fr.E6 by sim-PHPLC (MeOH–H2O, 58 : 42). Fr.E7 (1.88 g) was further separated by MPLC and eluted with a stepwise gradient of 54–78% MeOH in H2O to provide seven subfractions, Fr.E7-a–E7-g. Fr.E7-b was further purified by HPLC eluted with MeOH–H2O (58 : 42) to yield compound 22 (18.23 mg). Fr.E7-c was separated via semi-preparative HPLC (MeOH–H2O, 61 : 39) to afford compound 23 (19.26 mg). Similarly, compounds 12 (15.17 mg) and 20 (25.02 mg) were isolated from Fr.E7-d by semi-preparative HPLC (MeOH–H2O, 65 : 35). Compound 13 (162.37 mg) was isolated from Fr.E7-f by repeated recrystallization using MeOH. Fr.E7-g was further purified by HPLC eluted with MeOH–H2O (72 : 28) to provide compound 14 (55.66 mg). Fr.E9 was separated by an MPLC column and eluted with MeOH–H2O (70 : 30–80 : 20, v/v) to afford four fractions (Fr.E9-a–E9-d). Fr.E9-c was separated via semi-preparative HPLC (MeOH–H2O, 72 : 28) to afford compound 15 (21.61 mg). Fr.E0-d was purified by semi-preparative HPLC (MeOH–H2O, 75 : 25) to give compound 2 (48.2 mg).
Isodosin E (1): light yellow powder; [α]21.4D −360.72 (c = 0.5, MeOH); UV (MeOH) λmax (log ε) 210 (3.54), 295 (2.64), 320 (2.64) nm; IR (KBr) νmax 3479, 2935, 1738, 1371, 1248, 1021 cm−1; 1H and 13C NMR, see Table 1; HR-ESI-MS m/z 413.1950 [M + Na]+ (calcd for C22H30O6Na, 413.1935).
Isodosin F (2): white amorphous powder; [α]21.8D −29.88 (c = 0.5, CH2Cl2); UV (MeOH) λmax (log ε) 210 (3.64), 281 (3.00) nm; IR (KBr) νmax 3263, 2924, 1718, 1241, 1162 cm−1; 1H and 13C NMR, see Table 1; HR-ESI-MS m/z 437.2569 [M + H]+ (calcd for C24H36O7H, 437.2534).
Isodosin G (3): white amorphous powder; [α]21.8D −86.96 (c = 0.5, MeOH); UV (MeOH) λmax (log ε) 211 (3.54) nm; IR (KBr) νmax 2906, 1696, 1232, 1121, 1042 cm−1; 1H and 13C NMR, see Table 1; HR-ESI-MS m/z 385.1630 [M + Na]+ (calcd for C20H26O6Na, 385.1622).
3.4. ECD calculations
ECD calculations were carried out as previously reported, with appropriate modifications.18 The conformational analyses were performed using random searches in Spartan 16 (Wavenfunction, lrvine, CA, USA, 2016) with the MMFF94 force field. The conformers were then re-optimized using DFT at the B3LYP/6-31G(d,p) level using Gaussian 09 with the SMD model. For all conformers with Boltzmann populations exceeding 1%, energies, oscillator strengths, and rotational strengths (velocity) were subsequently computed via TDDFT at the B3LYP/6-311+g(d,p) level in MeOH. The ECD spectra were simulated by Gaussian convolution (σ = 0.30) before being averaged according to their Boltzmann distribution theory and relative Gibbs free energy (ΔG) values.
3.5. Cell lines and cell culture
Human hepatocellular carcinoma HepG2 and Huh7 cell lines, as well as the murine hepatoma Hepa1-6 cell line, were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). They were cultured in DMEM medium supplemented with 10% FBS and 1% antibiotics (penicillin and streptomycin) at 37 °C in a humidified 5% CO2 incubator.
3.6. Anti-hepatocarcinoma assay in vitro
The anti-hepatocarcinoma activities of the main compounds 2, 3, 5, 8, 13, 19, and 23, along with the positive control cisplatin, were evaluated using the CCK-8 assay. Briefly, HepG2 and Huh7 cells were seeded in 96-well plates at a density of 5 × 103 cells per well and incubated for 24 h. The cells were then treated with various concentrations (0, 12.5, 25, 50, 100, and 200 μM) of selected compounds and cisplatin. After a further 24 h incubation, 10 μL of CCK-8 solution was added and the cells were incubated at 37 °C for 4 h. Finally, the absorbance at 450 nm was measured using a Multiskan SkyHigh Spectrum (Thermo Scientific™, Waltham, MA, USA).
3.7. Animals and experimental protocols
All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Guangdong Pharmaceutical University and approved by the Animal Ethics Committee of Guangdong Pharmaceutical University (No. gdpulacspf2022559). Six-week-old male C57BL/6 mice were purchased from the Experimental Animals Center of Guangdong Province, China. Animals were housed at a temperature of 23–25 °C and relative humidity of (50 ± 10)% with a 12 hours of light and dark cycle. After a week of acclimation, the mice were subcutaneously injected with 5 × 105 Hepa1-6 cells (0.1 mL per mouse) in the right axillary. The mice were randomly divided into four groups (n = 5): model, Comp. 8-L group (i.g., 25 mg kg−1), Comp. 8-M group (i.g., 50 mg kg−1), and Comp. 8-H group (i.g., 100 mg kg−1). Mice in Comp. 8 treated groups were administered different doses of Comp. 8 (dissolved in 0.5% CMC-Na) by i.g. administration once daily for 14 days, and the model mice received the same volume of 0.5% CMC-Na. The size of the tumors was measured every 3 days with calipers, and the tumor volume was calculated using the formula, volume (mm3) = (length × width2)/2. At the end of the administration, mice were anesthetized and euthanized by cervical dislocation, and tumors were immediately removed, measured, and weighed.
3.8. H&E staining
To evaluate histological alterations, tumor tissues were fixed with 4% paraformaldehyde, embedded in paraffin, and cut into 5 μm sections. Sections were then stained with hematoxylin and eosin (H&E). Finally, the stained sections were examined under a light microscope.
3.9. Statistical analysis
All data were expressed as mean ± standard deviation (mean ± SD). Statistical analysis was performed using GraphPad Prism, Version 8.2.1 (GraphPad Software, Inc., San Diego, CA, USA) with a one-way analysis of variance (ANOVA) test. A value of p < 0.05 was considered statistically significant.
4. Conclusions
In summary, three new diterpenoids, including two 7,20-epoxy-ent-kaurane type diterpenoids (1–2) and one enmein-type ent-kaurane diterpenoid (3), together with 20 known diterpenoids (4–23), were isolated from the aerial parts of I. serra. The anti-hepatocarcinoma activities of the main compounds were evaluated against HepG2 and Huh7 cells, with compounds 3, 8, and 23 demonstrating significant inhibition effects on HepG2 cells. Moreover, compound 8 exhibited significant inhibition of tumor growth in vivo. These findings not only enriched the chemical composition of I. serra, but also supported the potential application of these diterpenoids in the therapy of HCC.
Author contributions
J. Yang conceived and designed the experiments; H. L. Wu contributed to the isolation, structure elucidation, and writing. C. Liu, S. Q. Li, and S. X. Wang contributed to the biological activity test; Z. Y. Huang and Y. H. Xu prepared the ESI;† X. He, C. C Zhang, and J. Ma revised the manuscript; X. He and G. Yang contributed to project management and funding acquisitions. All authors have read and agreed to the published version of the manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Supplementary Material
Acknowledgments
This research was funded by the Scientific Research Projects of Guangdong Provincial Bureau of Traditional Chinese Medicine (Grant No. 20241168, 20231206), the Science and Technology Planning Project of Guangzhou (Grant No. SL2022A04J00771, SL2023A04J02377), the Medical Scientific Research Foundation of Guangdong Province (Grant No. A2024479), the Guangdong Basic and Applied Basic Research Foundation (Grant No. 2022A1515110678), the National Natural Science Foundation of China (Grant No. 82304859), the Key Research and Development Program of Shanxi (Grant No. 2021ZDLSF04-03), and the CACMS Innovation Fund (Grant No. CI2021A03907).
Electronic supplementary information (ESI) available: HR-ESI-MS, 1D/2D NMR, IR, UV, and CD data of compounds 1–3, as well as NMR data of compounds 4–23. See DOI: https://doi.org/10.1039/d5ra02720a
Data availability
The raw data supporting the conclusions will be made available by the authors upon request.
Notes and references
- Liu M. Wang W. G. Sun H. D. Pu J. X. Nat. Prod. Rep. 2017;34:1090–1140. doi: 10.1039/c7np00027h. [DOI] [PubMed] [Google Scholar]
- Sun H. D. Huang S. X. Han Q. B. Nat. Prod. Rep. 2006;23:673–698. doi: 10.1039/b604174d. [DOI] [PubMed] [Google Scholar]
- Qiu C. L. Ye Z. N. Yan B. C. Hu K. Yang J. Yang X. Z. Li H. M. Li X. N. Sun H. D. Puno P. T. Bioorg. Chem. 2022;124:105811. doi: 10.1016/j.bioorg.2022.105811. [DOI] [PubMed] [Google Scholar]
- Li Y. X. Chi J. Zhang L. X. Wang F. Zhang W. J. Wang Z. M. Dai L. P. Phytochemistry. 2024;228:114247. doi: 10.1016/j.phytochem.2024.114247. [DOI] [PubMed] [Google Scholar]
- Ali M. A. Khan N. Ali A. Akram H. Zafar N. Imran K. Khan T. Khan K. Armaghan M. Palma-Morales M. Rodríguez-Pérez C. Caunii A. Butnariu M. Habtemariam S. Sharifi-Rad J. Food Sci. Nutr. 2024;12:3046–3067. doi: 10.1002/fsn3.3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X. Liao H. Hu S. Luo K. Zhu H. Mol. Cell. Probes. 2021;59:101759. doi: 10.1016/j.mcp.2021.101759. [DOI] [PubMed] [Google Scholar]
- Gou L. Yue G. G. Lee J. K. Puno P. T. Lau C. B. Biochem. Pharmacol. 2023;210:115491. doi: 10.1016/j.bcp.2023.115491. [DOI] [PubMed] [Google Scholar]
- Sobral P. J. M. Vicente A. T. S. Salvador J. A. R. Front. Chem. 2023;11:1066280. doi: 10.3389/fchem.2023.1066280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. Ye Y. Yu Z. L. Oncol. Rep. 2014;31:2165–2172. doi: 10.3892/or.2014.3081. [DOI] [PubMed] [Google Scholar]
- Hu X. Huang S. Ye S. Jiang J. Curr. Pharm. Biotechnol. 2024;25:655–664. doi: 10.2174/1389201024666230821110116. [DOI] [PubMed] [Google Scholar]
- Ding Y. Ding C. Ye N. Liu Z. Wold E. A. Chen H. Wild C. Shen Q. Zhou J. Eur. J. Med. Chem. 2016;122:102–117. doi: 10.1016/j.ejmech.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J. Liu M. Jiang H. Y. Yang J. Du X. Li X. N. Wang W. G. Li Y. Pu J. X. Sun H. D. Phytochemistry. 2016;130:244–251. doi: 10.1016/j.phytochem.2016.05.014. [DOI] [PubMed] [Google Scholar]
- Xing H. An L. Song Z. Li S. Wang H. Wang C. Zhang J. Tuerhong M. Abudukeremu M. Li D. Lee D. Xu J. Lall N. Guo Y. J. Nat. Prod. 2020;83:2844–2853. doi: 10.1021/acs.jnatprod.9b01281. [DOI] [PubMed] [Google Scholar]
- Xie R. J. Yan F. L. Hai G. F. Hou R. J. Ding M. M. Bai Y. X. Fitoterapia. 2011;82:726–730. doi: 10.1016/j.fitote.2011.03.003. [DOI] [PubMed] [Google Scholar]
- Zheng Q. Cui J. Fu H. Zhongguo Zhongyao Zazhi. 2011;36:2203–2206. [PubMed] [Google Scholar]
- Liu X. Bian Z. Tian Y. Li H. Hu S. Li C. Pandey P. Ferreira D. Chittiboyina A. G. Hamann M. T. Ma X. Wang S. Wang X. Fitoterapia. 2024;176:106019. doi: 10.1016/j.fitote.2024.106019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. J. Li B. L. Xie J. D. Liang H. J. Li Q. R. Yuan J. Wu J. W. Nat. Prod. Res. 2022;36:2021–2027. doi: 10.1080/14786419.2020.1841189. [DOI] [PubMed] [Google Scholar]
- Li S. Liang F. Huang D. Wu H. Tan X. Ma J. Wei C. Wang S. Huang Z. Yang G. He X. Yang J. Molecules. 2024;29:2733. doi: 10.3390/molecules29122733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita E. Fujita T. Shibuya M. Shingu T. Tetrahedron. 1969;25:2517–2530. [Google Scholar]
- Ping Z. R. Hongjie Z. Zhongwen L. Yulin Z. Handong S. Phytochemistry. 1992;31:4237–4240. [Google Scholar]
- Niu X. M. Li S. H. Li M. L. Zhao Q. S. Mei S. X. Na Z. Wang S. J. Lin Z. W. Sun H. D. Planta Med. 2002;68:528–533. doi: 10.1055/s-2002-32551. [DOI] [PubMed] [Google Scholar]
- Tao C. Li Q. J. Ye L. H. Zhang J. X. Yang X. S. Nat. Prod. Res. Dev. 2015;27:26–30. [Google Scholar]
- Li L. M. Li G. Y. Huang S. X. Li S. H. Zhou Y. Xiao W. L. Lou L. G. Ding L. S. Sun H. D. J. Nat. Prod. 2006;69:645–649. doi: 10.1021/np0600200. [DOI] [PubMed] [Google Scholar]
- Li B. L. Li J. Tong L. Pan Y. J. Yu K. B. Bull. Korean Chem. Soc. 2004;25:304–306. [Google Scholar]
- Fujita T. Takeda Y. Shingu T. Ueno A. Chem. Lett. 1980;9:1635–1638. [Google Scholar]
- Wang W.-G. Yang J. Wu H.-y. Kong L.-M. Su J. Li X.-N. Xue D. Zhan R. Min Z. Li Y. Pu J. Sun H.-D. Tetrahedron. 2015;71:9161–9171. [Google Scholar]
- Takeda Y. Fujita T. Sun H.-d. Minami Y. Ochi M. Chen C.-C. Chem. Pharm. Bull. 1990;38:1877–1880. [Google Scholar]
- Li B. Pan Y.-J. J. Pan W. Helv. Chim. Acta. 2001;84:3418–3422. [Google Scholar]
- Wang Z. M. Feng H. Liang X. T. Yuan S. T. Xu M. J. Acta Pharm. Sin. 1996:764–769. [PubMed] [Google Scholar]
- Gou L. Hu K. Yang Q.-F. Li X.-N. Sun H.-D. Xiang C. L. Puno P. T. Tetrahedron. 2019;75:2797–2806. [Google Scholar]
- Shen Y. H. Wen Z. Y. Xu G. Xiao W. L. Peng L. Y. Lin Z. W. Sun H. D. Chem. Biodiversity. 2005;2:1665–1672. doi: 10.1002/cbdv.200590136. [DOI] [PubMed] [Google Scholar]
- Kakinuma K. Okada Y. Ikekawa N. Kada T. Nomoto M. Agric. Biol. Chem. 1984;48:1647–1648. [Google Scholar]
- Yan F. L. Guo L. Bai S. Sun H.-D. J. Chin. Chem. Soc. 2008;55:933–936. [Google Scholar]
- Yan F. L. Guo L. Q. Zhang J. X. Bai S. P. Sun H. D. Chin. Chem. Lett. 2008;19:441–443. [Google Scholar]
- Chen S. N. Lin Z. W. Qin G. W. Sun H. D. Chen Y. Z. Planta Med. 1999;65:472–474. doi: 10.1055/s-2006-960818. [DOI] [PubMed] [Google Scholar]
- Wu Y. X. Ye F. Zhang W. Li J. C. Liu N. China J. Chin. Mater. Med. 2011;36:1772–1775. [PubMed] [Google Scholar]
- Han Q. B. Li M. L. Li S. H. Mou Y. K. Lin Z. W. Sun H. D. Chem. Pharm. Bull. 2003;51:790–793. doi: 10.1248/cpb.51.790. [DOI] [PubMed] [Google Scholar]
- Gao X. M. Luo X. Pu J. X. Wu Y. L. Zhao Y. Yang L. B. He F. Li X. N. Xiao W. L. Chen G. Q. Sun H. D. Planta Med. 2011;77:169–174. doi: 10.1055/s-0030-1250165. [DOI] [PubMed] [Google Scholar]
- Bray F. Laversanne M. Sung H. Ferlay J. Siegel R. L. Soerjomataram I. Jemal A. Ca-Cancer J. Clin. 2024;74:229–263. doi: 10.3322/caac.21834. [DOI] [PubMed] [Google Scholar]
- Chidambaranathan-Reghupaty S. Fisher P. B. Sarkar D. Adv. Cancer Res. 2021;149:1–61. doi: 10.1016/bs.acr.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W. Chen Z. Zhang W. Cheng Y. Zhang B. Wu F. Wang Q. Wang S. Rong D. Reiter F. P. De Toni E. N. Wang X. Signal Transduction Targeted Ther. 2020;5:87. doi: 10.1038/s41392-020-0187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwanwan D. Singh S. K. Singh S. Saikam V. Singh R. Biochim. Biophys. Acta, Rev. Cancer. 2020;1873:188314. doi: 10.1016/j.bbcan.2019.188314. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions will be made available by the authors upon request.