Abstract
Objectives:
The rise of antibiotic-resistant strains of Staphylococcus aureus in dairy milk products is a global concern, compromising treatment efficacy and highlighting the need for innovative solutions. Therefore, a study was conducted to isolate S. aureus strains (N = 21) from raw milk samples of cows infected with subclinical bovine mastitis. Additionally, the resistance of these strains against 12 different antibiotics was examined.
Materials and Methods:
Sixty raw cow milk samples, 20 from each of three separate dairy farms in Lahore city, were collected and screened for the presence of S. aureus. It was discovered that 70% of these milk samples were contaminated with this bacterium, indicating a widespread presence across the farms. Different isolation tests were employed in this study, including gram staining, capsule staining, catalase, mannitol salt fermentation, DNase, coagulase, and oxidase.
Results:
The obtained results revealed that the isolated strains of S. aureus showed % of their resistance against different antibiotics in the order of amoxicillin (85%), penicillin (71%), gentamicin (CN) (42%), carbenicillin and trimethoprim-sulfamethoxazole (33%), streptomycin, ciprofloxacin, and oxytetracycline (28%), cefotaxime (10%), and chloramphenicol (4%) in decreasing order, respectively. However, these strains showed no resistance against Bacitracin and Ampicillin.
Conclusion:
The existence of resistant strains of S. aureus has been attributed to various factors, such as poor milk hygiene, delayed milk transportation, subclinical bovine mastitis among dairy cows, and antibiotic-resistant genes. Thus, our present study will provide useful information about the resistant strains of S. aureus, which may transfer through cows into milk and then produce serious food-borne diseases in human beings. This study will be helpful to improve and control the quality of dairy products in Pakistan.
Keywords: Staphylococcus aureus, antibiotic resistance, cow milk, dairy farms, Lahore
Introduction
Pakistan is the largest agriculture and livestock-producing country in the world because its dairy and livestock industries play a key role in its economy. Pakistan stands 5th with the production of 36.2 million tons of milk annually in the world. However, the milk or dairy industries of Pakistan sometimes face many problems, including unhealthy animals and infections due to the inattention of workers or milkmen that later may affect the quality of dairy products, which causes certain serious or minor infections in humans upon their consumption. Between 1996 and 2006, the distribution of livestock in Pakistan revealed that Punjab and Sindh provinces harbored a significant majority, with 72.4% of cattle and 92% of buffalo populations. Over the same period, milk production from cows and buffaloes increased by 35.6%, with an annual growth rate of 3.6%. This growth, particularly in cow milk production, resulted in cow milk’s share of total production rising from 33.1% to 34.7%. This increase can be attributed to the expansion of large cattle farms in Pakistan. In Punjab, the average yield of cow milk per animal/day is 6.31 L, and a total of 25,580,103 L of cow milk is produced per day [1].
Staphylococcus aureus, increasingly important as a major pathogen, is rising in significance due to its increasing resistance against antibiotics. It can affect most mammals, including humans. Staphylococcus aureus can be transmitted from animals as well as from one person to another through air droplets or aerosols suspended in the air by coughing or sneezing or direct contact with an object containing bacteria or through the bite of an infected animal. Approximately 30% of healthy humans harbor S. aureus in their noses, the back of their throats, and their skin [2]. It is obvious that in S. aureus, the most common antibiotic resistance was associated with its plasmids. The genetics of this bacterium are essentially similar to the genetics of Escherichia coli. Its resistance to antibiotics has increased physicians’ concern because 60% and 90% of S. aureus strains showed resistance against penicillin. Therefore, the use of antibiotics for its treatment often proves ineffective and exacerbates the issue of antibiotic resistance [3].
Milk, a vital staple in our diet, is consumed both fresh and as the base for various dairy products. Bovine mastitis, an inflammation of the mammary gland, occasionally results in systemic infection, significantly impacting animal welfare and milk quality. It is the most prevalent disease affecting high-yield dairy cows. The International Dairy Federation has recognized bovine mastitis as a critical issue in dairy animal pathology, emphasizing its significance. However, bovine mastitis, primarily caused by S. aureus, is a major challenge to the dairy sector. Staphylococcus aureus is responsible for 30%–40% of bovine mastitis cases globally, with Pakistan reporting a prevalence of 29%–57% in dairy cattle. Such a disease results in substantial economic losses, accounting for 17% of total animal disease-related costs in Pakistan and approximately $33B worldwide [4–7]. Contamination of milk due to mastitis can occur directly from infected udders or through improper handling, with bacterial loads reaching up to 10⁸ CFU/ml [8].
Current treatment methods for bovine mastitis, which rely on antibiotics, are often ineffective and contribute to the development of antibiotic-resistant strains. Therefore, this study aimed to identify the prevalence of S. aureus in raw milk from mastitis-affected cows in Lahore. Additionally, it also analyzed the antibiotic resistance profiles of the isolated strains of this bacterium to assess potential health risks and foodborne diseases among milk consumers.
Materials and Methods
Ethical approval
The study protocol and ethical considerations for this research have received approval from the Ethical Committee of the University of Veterinary and Animal Sciences (UVAS) in Lahore, under approval number FBS-QAU-90. A consent form was signed by the owners of dairy farms before sampling to get their approval for milk sampling from infected cows.
Study area
Lahore is the second biggest city in Pakistan and is highly inhabited by 13 million people. It is the capital of Punjab province and is situated at 31°32’59” North and 74°20’37” East, and its total area is about 684 sq. meters. It is situated in the northeast of Pakistan, close to the border of India. The average temperature ranges from 21°C (December 2023) to 40°C (June 2023).
Study design
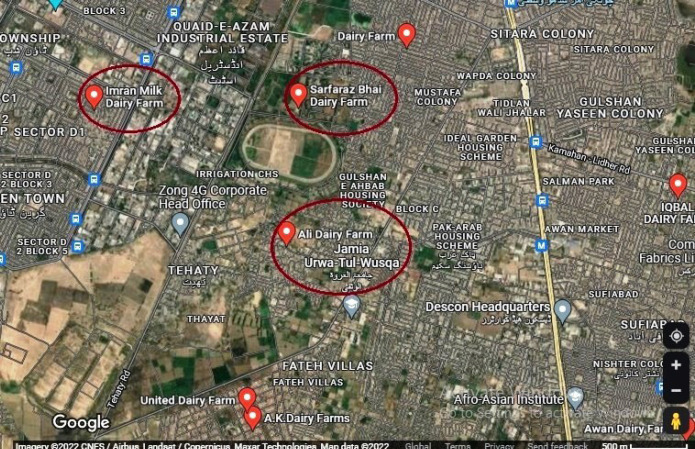
During the period from July to December 2021, a total of 60 raw milk samples (20 samples from each dairy farm) of cows particularly infected with subclinical bovine mastitis were collected from three dairy farms in Lahore city of Pakistan, as shown in Figure 1, including, i.e., Ali Dairy Farms (SA1 = 20), Imran Milk Dairy (SA2 = 20), and Sarfaraz Bhai Dairy Farm (SA3 = 20), Lahore, for isolating and identifying 21 strains of S. aureus present in raw milk gathered from bovine mastitis cows and estimating their resistance against selected antibiotics.
Figure 1. Google map of sampling sites in Lahore city of Pakistan.
Sample size and collection
Sixty raw milk samples (N = 60) were gathered from infected cows with subclinical bovine mastitis present in three different dairy farms in Lahore. 10 ml milk samples were taken from each infected cow, placed in sterilized bottles, and immediately transferred to the QOL/WTO (Quality Operation Laboratory) of UVAS, as shown in Figure 1.
Bacteriological isolation and identification
Milk samples were spread on the Mannitol salt agar and placed at 37°C for 24 h in an incubator to obtain isolated colonies. After incubation, size, shape, margins, pigmentation, elevation, and opacity are observed with the naked eye. The colonies were preliminarily identified through staining reactions with Gram stain and capsule staining. Colonies that grew on MSA were subcultured on nutrient media, and the cultures were preserved and maintained for isolates’ characterization. Oxidase tests, tube coagulase tests, and DNase tests were conducted on samples identified as positive for coagulase-positive S. aureus following the methods outlined by Girmay et al. [9].
Determination of antibiotic vulnerability in S. aureus isolates
After confirming the presence of S. aureus, the effectiveness of antibiotics was assessed using the Kirby-Bauer (K-B) test, a standard method endorsed by the NCCLS (National Committee for Clinical Laboratory Standards) for antibiotic vulnerability testing, as described by Jackie-Reynolds [10] and Zhang et al. [11]. The K-B test, also known as the disc diffusion test, helps determine the resistance or sensitivity of bacteria to specific antibiotics, aiding in patient treatment decisions. The procedure involved swabbing the bacterium onto agar. The test procedure involves swabbing the bacterium onto agar, followed by placing antibiotic discs on top. As the antibiotics diffuse into the agar, their concentration decreases, creating varying levels of inhibition around the discs. If the organism is susceptible to the antibiotic, there will be a clear zone of inhibition where bacterial growth is inhibited. The size of these zones is measured and interpreted using standardized charts to determine bacterial sensitivity, resistance, or intermediate status. Many charts also provide the minimal inhibitory concentration (MIC) for each drug, offering more precise information on effective dosages. The Mueller-Hinton medium, a protein-rich agar, supports bacterial growth. Bacterial colonies are collected with sterile swabs and spread onto Muller Hinton plates to cover the entire surface. Antibiotic discs are then placed on the plates using sterile forceps, and the plates are incubated at 37°C for 24 h. Results were interpreted based on the MIC.
Statistical analysis
All statistical analysis of data was performed by using MS Excel 2010 software.
Results
Identifying Staphylococcus species in cow-milk samples through cultural characteristics analysis
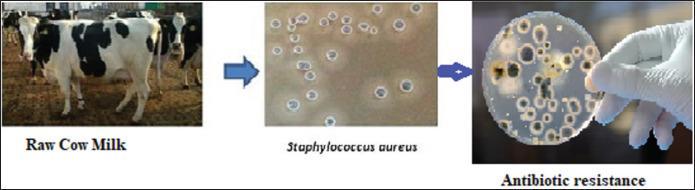
Sixty raw cow milk samples were obtained from infected cows with clinical and subclinical bovine mastitis present in three different dairy farms in Lahore. Then these milk samples were screened for identifying the occurrence of S. aureus. Out of these 60 raw milk samples, S. aureus strains were identified and present in only 42 raw milk samples (70%), while the remaining 18 samples (30%) were contaminated with some other bacterium species (Table 1). The presence of S. aureus in raw milk shows its poor quality, which can cause food-borne illnesses in consumers. Isolated S. aureus strains from milk samples were designated as SB1, SB2, SB3, SB4, SB5, SB6, SB7, SB8, SB9, SB10, SB11, SB12, SB13, SB14, SB15, SB16, SB17, SB18, SB19, SB20, and SB21. These isolates were differentiated into resistant, intermediate, and susceptible to antibiotics. The isolated strains of S. aureus showed their resistance against different antibiotics due to the presence of antibiotic-resistant genes (Fig. 2).
Table 1. Collection of milk samples from three different Lahore dairy farms.
| Sl No. | Sampling area | Sampling abbreviations/codes | No. of milk samples | No. of milk samples contaminated with bacteria isolates |
|---|---|---|---|---|
| 1 | Ali Dairy Farms Lahore | SA1 | 20 | 17a |
| 2 | Imran Milk Dairy Farm Lahore | SA2 | 20 | 13b |
| 3 | Sarfaraz Bhai Dairy Farm Lahore | SA3 | 20 | 12c |
| Total | N = 60 | 42 |
Note: Superscript shows the highest and lowest values.
Figure 2. Isolation of S. aureus from raw cow milk and disc diffusion test of the bacteria for assessing antibiotic resistance patterns.
Morphological and biochemical characterization
Golden yellow pigmented round colonies of convex shape appeared. Bacteria showed a purple color and are in a grape-like structure. Staphylococcus aureus is a gram-positive bacterium that showed a faint blue halo capsule around the purple cell as a result of capsule staining and showed positive results of catalase, mannitol salt fermentation, DNase, and coagulase but was oxidase negative.
Analysis of zone of inhibition (in mm) by using the Kirby-Bauer method
The Zone of Inhibition (in mm) in the Kirby-Bauer antibiotic susceptibility test indicates the effectiveness of an antibiotic against a particular bacterial strain. A larger zone suggests that the antibiotic is more effective in inhibiting the growth of the bacteria being tested. The MIC (minimum inhibitory concentration) that inhibits bacterial growth was also determined by using the Kirby-Bauer (K-B) method. This procedure determines the concentration of an antibiotic required to inhibit pathogen growth, providing insight into the effective dosage for controlling the infection in patients. For each antibiotic, 21 strains of S. aureus were differentiated into three categories, i.e., resistant, intermediate, and susceptible, according to their antibiotic potential, as shown in Table 2. The antimicrobial effect of each antibiotic depends on different factors such as (1) the severity of the infection of the animal from which milk was obtained and (2) the concentration of the antibiotic’s discs. The range of diameter of the zone of inhibition (ZOI) ranged from 8 to 20 mm. Some S. aureus strains showed resistance, while others had more susceptibility to antibiotics. The concentration of the antibiotic disc also had a great effect on MIC. A high concentration of dose had a significant effect on microbial growth, while a low dose had a low effect. The average value of MIC for the selected 12 antibiotics was also shown by 21 strains of S. aureus ranging from 11 to 25 mm, as shown in Table 3. Staphylococcus aureus strains indicated the highest resistance to both Amoxicillin and penicillin, which was 85% and 71%, respectively (Fig. 3), while showing no resistant effect against ciprofloxacin and gentamicin but revealing high susceptibility to these two antibiotics. Staphylococcus aureus resistivity for other antibiotics was found in order of Bacitracin 42%, Carbenicillin 33%, Trimethoprim-Sulphamethoxazole 33%, Ampicillin 28%, Oxytetracycline 28%, Streptomycin 28%, Cefotaxime 25%, and for Chloramphenicol 4%. Among them, penicillin shows the lowest diameter of MIC, which is 11 mm, whereas, cefotaxime shows the highest diameter of MIC, which is 25 mm.
Table 2. Determination of the diameter of the ZOI of Staphylococcus aureus isolated from cow milk.
| S. No. | Antibiotics | Abbreviations | Disc Conc. | Diameter of Zone of inhibition (ZOI) in mm | ||
|---|---|---|---|---|---|---|
| Resistant | Intermediate | Susceptible | ||||
| 1. | Penicillin | P | 10IU | ≤ 20a | 21–28a | ≥ 29a |
| 2. | Oxytetracycline | OT | 30 µg | ≤ 14e | 15–18d | ≥19e |
| 3. | Chloramphenicol | C | 30 µg | ≤ 12f | 13–17e | ≥ 18f |
| 4. | Gentamycin | CN | 10 µg | ≤ 12f | 13–14f | ≥ 15h |
| 5. | Bacitracin | B | 10U | ≤ 8i | 9–11j | ≥ 13j |
| 6. | Ampicillin | AM | 10 mcg | ≤ 11g | 12–13h | ≥ 14i |
| 7. | Streptomycin | S | 10 µg | ≤ 11g | 12–14g | ≥ 15h |
| 8. | Amoxicillin | AX | 25 mcg | ≤ 19b | NIL | ≥ 20d |
| 9. | Carbenicillin | CAR | 100 µg | ≤ 17c | 18–22b | ≥ 23b |
| 10. | Ciprofloxacin | CIP | 5 µg | ≤ 15d | 16–20c | ≥ 21c |
| 11. | Trimethoprim-Sulphamethoxazole | SXT | 25 µg | ≤ 10h | 11–15i | ≥ 16g |
| 12. | Cefotaxime | CTX | 30 µg | ≤ 14e | 15–17d | ≥ 18f |
Note: Superscript shows the highest and lowest ZOI values.
Table 3. Shows of antibiotic resistance potential of Staphylococcus aureus strains against twelve selected antibiotics.
| Sr. no. | Antibiotics | Disc. Conc. | No. of strains of S. aureus | Antibiotic resistance (%) | Average value of MIC (mm) | ||
|---|---|---|---|---|---|---|---|
| Resistant no. (%) | Intermediate no. (%) | Susceptible no. (%) | |||||
| 1. | Penicillin | 10IU | 15 (71.4) | 2 (9.5) | 4 (19.04) | 71.0b | 11.0i |
| 2. | Oxytetracycline | 30 µg | 6 (28.6) | 0 | 15 (71.4) | 28.0e | 19.0e |
| 3. | Chloramphenicol | 30 µg | 1 (4.8) | 5 (23.8) | 15 (71.4) | 4.0g | 22.0c |
| 4. | Gentamycin | 10 µg | 0 | 2 (9.5) | 19 (90.5) | 0.0h | 19.0e |
| 5. | Bacitracin | 10U | 9 (4.29) | 3 (14.3) | 9 (42.9) | 42.0c | 11.0i |
| 6. | Ampicillin | 10 mcg | 6 (28.6) | 3 (14.3) | 12 (57.1) | 28.0e | 16.0f |
| 7. | Streptomycin | 10 µg | 6 (28.6) | 4 (19.0) | 11 (52.4) | 28.0e | 16.0f |
| 8. | Amoxicillin | 25 mcg | 18 (85.7) | 0 | 3 (14.3) | 85.0a | 14.0h |
| 9. | Carbenicillin | 100 µg | 7 (33.3) | 6 (28.6) | 8 (38.1) | 33.0d | 20.0d |
| 10. | Ciprofloxacin | 5 µg | 0 | 1 (4.8) | 20 (95.2) | 0.0h | 24.0b |
| 11. | Trimethoprim- Sulphamethoxazole | 25 µg | 7 (33.3) | 2 (9.5) | 12 (57.1) | 33.0d | 15.0g |
| 12. | Cefotaxime | 30 µg | 2 (9.5) | 1 (4.8) | 18 (85.7) | 10.0f | 25.0a |
Note: MIC = minimum inhibitory concentration. Superscript shows the highest and lowest values.
Figure 3. Antibiotic resistance of S. aureus against different antibiotics.
Discussion
Bacteria play a crucial role in developing certain medicines used in clinical practice. Trabectedin, an anti-cancer drug derived from tunicates, is produced by their symbiotic bacteria. Similarly, many antibiotics, immunosuppressive drugs, and anticancer medications are derived from bacterial products. However, isolating antibiotics from the same environment where pathogenic strains reside can lead to resistance to naturally produced antibiotics [12,13]. Staphylococcus aureus, a major bacterium causing bovine mastitis, leads to significant economic losses in Pakistan’s dairy industry due to limited knowledge of its genetic diversity and antimicrobial resistance genes. This study highlights that initially, milk drawn from healthy animals may be bacteria-free but later becomes contaminated, either from the hands of milkmen or the udders of animals harboring microorganisms such as Corynebacteria, Coliforms, Klebsiella, Streptococci, Staphylococci, and Salmonellae. Dirty teats contaminated with dung and mud serve as sources of bacteria for milk contamination. Additionally, utensils used for milk storage and transportation also contribute to bacterial presence. However, the primary source of contamination often stems from adding contaminated water to milk to increase its volume. These findings indicate that raw milk undergoes unhygienic conditions during transportation, leading to significant contamination over time due to high temperatures, which promote bacterial proliferation. Montelongo et al. [14] noted that the genome of Staphylococcus contains a diverse array of genes, a significant portion of which are obtained through lateral gene transfer. Many antibiotic resistance genes are found on plasmids or other mobile genetic elements. The impressive capacity of S. aureus to acquire advantageous genes from diverse sources is underscored by the intricate nature of its genome and the clear evidence of lateral gene transfer.
Several studies, including those by Onanuga et al. [15], have noted significant resistance in S. aureus against various antibiotics. They found high resistance rates to ampicillin (91.7%), clindamycin (78.3%), cephalexin (75%), methicillin (71.7%), and vancomycin (68.3%) while showing very low resistance to gentamicin (3.3%), ciprofloxacin (3.3%), ofloxacin (3.3%), sparfloxacin (3.3%), and pefloxacin (10.0%). Another study by Ikeagwu et al. [16] indicated that S. aureus exhibited the highest sensitivity to Ofloxacin (65%) and the least to Co-trimoxazole (6%). Additionally, Gali et al. [17] discovered vancomycin-resistant S. aureus (VRSA) strains in milk samples in Nigeria, with 5.4% showing resistance. Notably, pasteurization and fermentation seemed to reduce the pathogen’s occurrence, as VRSA was absent in yogurt and less prevalent in pasteurized milk compared to raw milk. The VRSA strains exhibited high resistance to penicillin (100%), tetracycline (85%), amoxicillin (65%), methicillin (40%), and oxacillin (40%), but were sensitive to amikacin (5%) and sulfamethoxazole/trimethoprim (10%). Similarly, a study by Jahan et al. [5] revealed high resistance of S. aureus in raw milk of cows to amoxicillin (100%), penicillin (100%), and erythromycin (75%), while being sensitive to oxacillin (100%), cloxacillin (100%), neomycin (100%), and ciprofloxacin (83.33%). In our current work, S. aureus strains demonstrated the highest resistance to amoxicillin (85%) and penicillin (71%), whereas ciprofloxacin and gentamicin showed no resistance. Tanzin et al. [18] observed that both S. aureus and Escherichia coli are antibiotic-resistant strains found in milk samples of cattle and buffaloes in Bangladesh and can be controlled in the case of bovine mastitis by using antibiotics like ciprofloxacin and levofloxacin that showed low resistance rates. Additionally, Naushad et al. [19] studies highlighted the importance of antibiotic choice in controlling antibiotic-resistant strains in milk samples, with antibiotics like ciprofloxacin and levofloxacin showing low resistance rates. Furthermore, genetic diversity among S. aureus isolates from bovine milk revealed high resistance against beta-lactams and sulfonamides but low resistance against other antibiotics. Finally, Hassani et al. [20] investigations in Iran’s northwest regions found antibiotic resistance genes, such as blaTEM and blaSHV in E. coli and Salmonella species, and mecA and blaZ in S. aureus isolates, contributing to high rates of antibiotic resistance against amoxicillin, penicillin, and cephalexin, respectively.
Over the past two decades, 75% of infectious diseases in humans have been traced back to animals, either directly or indirectly. Staphylococcus aureus, a global pathogenic strain, poses significant health risks, particularly through dairy products. While milk offers essential nutrients, improper handling during production and distribution can lead to food-borne illnesses, as it provides an ideal environment for harmful pathogens to thrive. Staphylococcus bacteria, including S. aureus, are commonly found in animals and humans, causing a range of ailments from mild skin infections to severe toxic shock syndrome and food poisoning. The emergence of antibiotic-resistant strains, notably methicillin-resistant S. aureus, presents a considerable challenge for healthcare and dairy industries. These resistant strains, including vancomycin-resistant S. aureus, pose therapeutic challenges and health risks, with milk being reported as a potential source of contamination [21]. Proper sanitation and hygiene practices are crucial in mitigating the spread of antibiotic-resistant bacteria through dairy products, safeguarding public health [9].
According to Schneider [22], antibiotic resistance has emerged as a significant challenge for public health and society at large. Despite the introduction of new antibiotics to the market, they too eventually encounter the rise of pathogenic resistance. The rapid development of resistance appears to be closely tied to the microbial origin and function of these antibiotics in nature. Most clinically used antibiotics are derived from natural microbial products, but the presence of antimicrobial resistance genes in their environment makes it easy for pathogens to acquire them through horizontal gene transfer. Janeoo et al. [23] have observed that drug-resistant bacterial pathogens are now causing major health issues affecting a significant portion of the global population. Therefore, there is an urgent need to discover effective antibacterial drugs against specific drug-resistant bacterial strains for healthcare systems worldwide.
Conclusion
The raw cow milk used in Lahore City was contaminated with certain types of toxigenic or multidrug-resistant S. aureus strains that cause serious infections in consumers. Milk is a major source of food taken by human beings, and contaminated milk can cause various problems. Improving hygienic conditions and careful handling of cows during milking can effectively treat these issues. Thus, this study contributes to implementing suitable control measures to reduce contaminants and the spread of pathogenic bacterial strains in humans. We could further investigate the molecular characterization of the milk-isolated bacteria and the bacterial count per milliliter of milk.
Acknowledgments
We are highly indebted to my respected colleague Dr. Imran Altaf from the University of Veterinary and Animal Sciences (UVAS), who provided me with full support in milk sample analysis and techniques for the isolation of bacterial strains. The authors would like to acknowledge the Deanship of Graduate Studies and Scientific Research, Taif University of Saudi Arabia, for funding this work.
List of abbreviations
C, celsius; CFU, colony-forming unit; DNase, Deoxyribonuclease; h, hour; K-B, Kirby-Bauer; ml, milliliter; MIC, Minimal Inhibitory Concentration; mm, millimeter; NCCLS, National Committee for Clinical Laboratory Standards; VRSA, Vancomycin-resistant Staphylococcus aureus.
Conflict of interest
The authors declared that they have no conflict of interest that is directly or indirectly related to this work submitted for publication.
Author’s contributions
SB, ZM, AU, and WK collected milk samples, designed the study, isolated bacterial strains, and tested their antibiotic resistance in the laboratory. SB, ZM, WK, and AU wrote the manuscript, and MBS, HB, AB Ismael and AAS edited its revised version. Funding Acquisitions made by HB, AB Isamel, and AAS. All the authors read and approved the final version.
References
- [1].Burki A, Khan M. Department of Economics, Lahore University of Management Sciences, Tetra Pak Pakistan. Lahore, Pakistan: 2016. Pakistan’s dairy sector: Lessons from the past to build a resilient dairy industry. [Google Scholar]
- [2].Song JH, Hsueh PR, Chung DR, Ko KS, Kang CI, Peck KR, et al. Spread of methicillin-resistant Staphylococcus aureus between the community and the hospitals in Asian countries: an ANSORP study. J Antimicrob Chemother. 2011;66(5):1061–69. doi: 10.1093/jac/dkr024. https://doi.org/10.1093/jac/dkr024. [DOI] [PubMed] [Google Scholar]
- [3].Ifediora AC, Enya E, Mbajiuka CS. Comparative evaluation of the antibiotic resistance profile of staphylococcus aureus isolated from breeders and livestock. Int J Public Health. 2024;69:1607603. doi: 10.3389/ijph.2024.1607603. https://doi.org/10.3389/ijph.2024.1607603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Le Maréchal C, Seyffert N, Jardin J, Hernandez D, Jan G, Rault L, et al. Molecular basis of virulence in Staphylococcus aureus mastitis. PLoS One. 2011;6(11):e27354. doi: 10.1371/journal.pone.0027354. https://doi.org/10.1371/journal.pone.0027354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jahan M, Parvej MS, Chowdhury SMZH, Haque ME, Talukder MAK, Ahmed S. Isolation and characterization of Staphylococcus aureus from raw cow milk in Bangladesh. J Adv Vet Anim Res. 2014;2(1):49–55. https://doi.org/10.5455/javar.2015.b47. [Google Scholar]
- [6].Aprodu I, Walcher G, Schelin J, Hein I, Norling B, Rådström P, et al. Advanced sample preparation for the molecular quantification of Staphylococcus aureus in artificially and naturally contaminated milk. Int J Food Microbiol. 2011;145:S61–5. doi: 10.1016/j.ijfoodmicro.2010.09.018. https://doi.org/10.1016/j.ijfoodmicro.2010.09.018. [DOI] [PubMed] [Google Scholar]
- [7].Shahzad MA, Yousaf A, Ahsan A, Irshad H, Riaz A, Khan A, et al. Virulence and resistance profiling of Staphylococcus aureus isolated from subclinical bovine mastitis in the Pakistani Pothohar region. Sci Rep. 2024;14:14569. doi: 10.1038/s41598-024-65448-9. https://doi.org/10.1038/s41598-024-65448-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lemma F, Alemayehu H, Stringer A, Eguale T. Prevalence and antimicrobial susceptibility profile of staphylococcus aureus in milk and traditionally processed dairy products in Addis Ababa, Ethiopia. Biomed Res Int. 2021;2021:5576873. doi: 10.1155/2021/5576873. https://doi.org/10.1155/2021/5576873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Girmay W, Gugsa G, Taddele H, Tsegaye Y, Awol N, Ahmed M, et al. Isolation and identification of methicillin-resistant Staphylococcus aureus (mrsa) from milk in shire dairy farms, Tigray, Ethiopia. Vet Med Int. 2020;2020:8833973. doi: 10.1155/2020/8833973. https://doi.org/10.1155/2020/8833973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Jackie-Reynolds RC. Dornbirn, Austria: 2011. Kirby-Bauer test for antibiotic susceptibility, YUMMY Publishing GmbH, Lustenauer Straße 66, 6850. Available via http://delrio.dcccd.edu/jreynolds/microbiology/2421/lab_manual/KB_antibiotic.pdf . [Google Scholar]
- [11].Zhang P, Wu XH, Su L, Wang HQ, Lin TF, Fang YP, et al. Rapid, label-free prediction of antibiotic resistance in Salmonella typhimurium by surface-enhanced raman spectroscopy. Int J Mol Sci. 2022;23(3):1356. doi: 10.3390/ijms23031356. https://doi.org/10.3390/ijms23031356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wolska-Gębarzewska M, Międzobrodzki J, Kosecka-Strojek M. Current types of staphylococcal cassette chromosome mec (SCCmec) in clinically relevant coagulase-negative staphylococcal (CoNS) species. Crit Rev Microbiol. 2024;50(6):1020–36. doi: 10.1080/1040841X.2023.2274841. https://doi.org/10.1080/1040841X.2023.2274841. [DOI] [PubMed] [Google Scholar]
- [13].Li H, Liu X, Li S, Rong J, Xie S, Gao Y, et al. KleTy: integrated typing scheme for core genome and plasmids reveals repeated emergence of multi-drug resistant epidemic lineages in Klebsiella worldwide. Genome Med. 2024;16(1):130. doi: 10.1186/s13073-024-01399-0. https://doi.org/10.1186/s13073-024-01399-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Montelongo C, Mores CR, Putonti C, Wolfe AJ, Abouelfetouh A. Whole-genome sequencing of Staphylococcus aureus and Staphylococcus haemolyticus clinical isolates from Egypt. Microbiol Spectr. 2022;10(4):e0241321. doi: 10.1128/spectrum.02413-21. https://doi.org/10.1128/spectrum.02413-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Onanuga A, Oyi AR, Olayinka BO, Onaolapo JA. Prevalence of community-associated multi-resistant Staphylococcus aureus among healthy women in Abuja, Nigeria. Afr J Biotechnol. 2005;4(9):942–5. http://dx.doi.org/10.4314/ajb.v4i9.71202. [Google Scholar]
- [16].Ikeagwu IJ, Amadi ES, Iroha IR. Antibiotic sensitivity pattern of Staphylococcus aureus in Abakaliki, Nigeria. [01 January 2025];Pak J Med Sci. 2008 24(2):231–5. Available via http://www.embase.com/search/results?subaction = viewrecord&from = export&id = L351798604 . [Google Scholar]
- [17].Gali AU, Ibrahim ST, Abdulhamid HBS, Zubairu A, Sale AA, Olumuyiwa AB, et al. Staphylococcus aureus in soup samples from restaurants/food canteens and some families in Jalingo Metropolis, North-Eastern Nigeria. Afr J Microbiol Res. 2014;8(31):2964–9. https://doi.org/10.5897/ajmr2014.6913. [Google Scholar]
- [18].Tanzin T, Nazir KHMNH, Zahan MN, Parvej MS, Zesmin K, Rahman MT. Antibiotic resistance profile of bacteria isolated from raw milk samples of cattle and buffaloes. J Adv Vet Anim Res. 2016;3(1):62–7. https://doi.org/10.5455/javar.2016.c133. [Google Scholar]
- [19].Naushad S, Nobrega DB, Naqvi SA, Barkema HW, De Buck J. Genomic analysis of bovine Staphylococcus aureus isolates from milk to elucidate diversity and determine the distributions of antimicrobial and virulence genes and their association with mastitis. mSystems. 2020;5(4):e00063–20. doi: 10.1128/mSystems.00063-20. https://doi.org/10.1128/mSystems.00063-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Hassani S, Moosavy MH, Gharajalar SN, Khatibi SA, Hajibemani A, Barabadi Z. High prevalence of antibiotic resistance in pathogenic foodborne bacteria isolated from bovine milk. Sci Rep. 2022;12(1):3878. doi: 10.1038/s41598-022-07845-6. https://doi.org/10.1038/s41598-022-07845-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Qolbaini EN, Khoeri MM, Safari D. Identification and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus-associated subclinical mastitis isolated from dairy cows in Bogor, Indonesia. Vet World. 2021;14:1180–4. doi: 10.14202/vetworld.2021.1180-1184. https://doi.org/10.14202/vetworld.2021.1180-1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Schneider YK. Bacterial natural product drug discovery for new antibiotics: strategies for tackling the problem of antibiotic resistance by efficient bioprospecting. Antibiotics. 2021;10(7):842. doi: 10.3390/antibiotics10070842. https://doi.org/10.3390/antibiotics10070842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Janeoo S, Kaur H, Kaul G, Akhir A, Chopra S, Banerjee S, et al. Fluorine-containing 2, 3-diaryl quinolines as potent inhibitors of methicillin and vancomycin-resistant Staphylococcus aureus: synthesis, antibacterial activity and molecular docking studies. J Mol Struct. 2021;1244:130924. https://doi.org/10.1016/j.molstruc.2021.130924. [Google Scholar]