Abstract
Background
To support the positional cloning of the mouse mutation wobbler (wr) the corresponding regions on human Chr2p13-14 and mouse Chr11 were analyzed in detail and compared with respect to gene content, order, and orientation.
Results
The gene content of the investigated regions was highly conserved between the two species: 20 orthologous genes were identified on our BAC/YAC contig comprising 4.5 Mb between REL/Rel and RAB1A/Rab1a. Exceptions were pseudogenes ELP and PX19 whose mouse counterparts were not located within the analyzed region. Two independently isolated genomic clones indicate an inversion between man and mouse with the inverted segment being identical to the wobbler critical interval. We investigated the wobbler critical region by extensive STS/EST mapping and genomic sequencing. Additionally, the full-length cDNA sequences of four newly mapped genes as well as the previously mapped gene Otx1 were established and subjected to mutation analysis. Our data indicate that all genes in the wr critical region have been identified.
Conclusion
Unexpectedly, neither mutation analysis of cDNAs nor levels of mRNAs indicated which of the candidate genes might be affected by the wr mutation. The possibility arises that there might be hitherto unknown effects of mutations, in addition to structural changes of the mRNA or regulatory abnormalities.
Background
The conservation of genomic structure between two or more species is the basis of comparative genomics. During the process of evolution the distribution of genes on chromosomes is rearranged by illegitimate recombination and translocation. Species sharing a recent common ancestor tend to have fewer chromosomal rearrangements than distantly related species [1]. Orthologous genes serve as landmarks for the definition of related chromosomal regions. Orthologous segments are defined by at least one pair of orthologous genes; conserved synteny relates to at least two pairs of orthologous genes located on the same chromosomal segment in two different species [2].
With increasing sequence data comparative genomics is no longer restricted to genes, as evolutionary conserved intergenic sequences [3] can now also be taken into account. In addition, comparison of genomic sequence data of two different species facilitates the identification of novel genes and conserved regulatory elements.
We use comparative genomics for a positional cloning approach in order to identify candidate genes for the mouse mutation wobbler (wr) which causes muscle weakness due to motor neuron degeneration [4] and a spermiogenesis defect [5]. The wobbler mouse is used as an animal model for human spinal muscular atrophies (SMAs) although there is no human SMA known to be located within the wr homologous region.
The wr mutation is located on proximal mouse chromosome 11 [6] within a region homologous to human Chr2p13-14. Some genes of that region have previously been mapped to our Chr11 / Chr2p13-14 physical map: RAB1A/Rab1a [7], MDH1/Mor2, REL/Rel and OTX1/Otx1[8], PELI1/Peli1 [9], and CCT4/Cct4 [10].
Here we present a detailed transcript map of the orthologous chromosomal regions on proximal mouse Chr 11 and human Chr 2p13-14 with newly mapped genes and newly established mouse cDNA sequences serving as candidate genes for the wobbler mutation. The genomic regions analyzed are highly conserved in gene content, but two independent genomic clones indicate that the gene order is disrupted by an inversion.
Results
Our physical map of mouse proximal Chr 11 is based on 68 BAC and 11 YAC clones covering 3 Mb. The present mostly BAC-based contig has been established by STS/EST mapping improves the previously described YAC contig [8]. Our homologous human YAC contig (twelve clones, [8]) spans approximately 4.5 MB, exceeding the mouse contig on the distal side.
Ten loci defined by the following genes and cDNAs were newly mapped on mouse Chr 11 (Fig. 1): hepatocellular carcinoma antigen 8 (Hcc8), uridin-diphosphoglucose-pyrophosphorylase 2 (Ugp2), Kiaa0582, Kiaa0127, Flj20080, Flj21950, Hspc159, Kiaa0903, Homoloc2, Homoloc13 (homologous to human LOC51057) and Murr1 which had previously been physically mapped by Nabetani et al. [11] were found to be located on our Chr 11 contig.
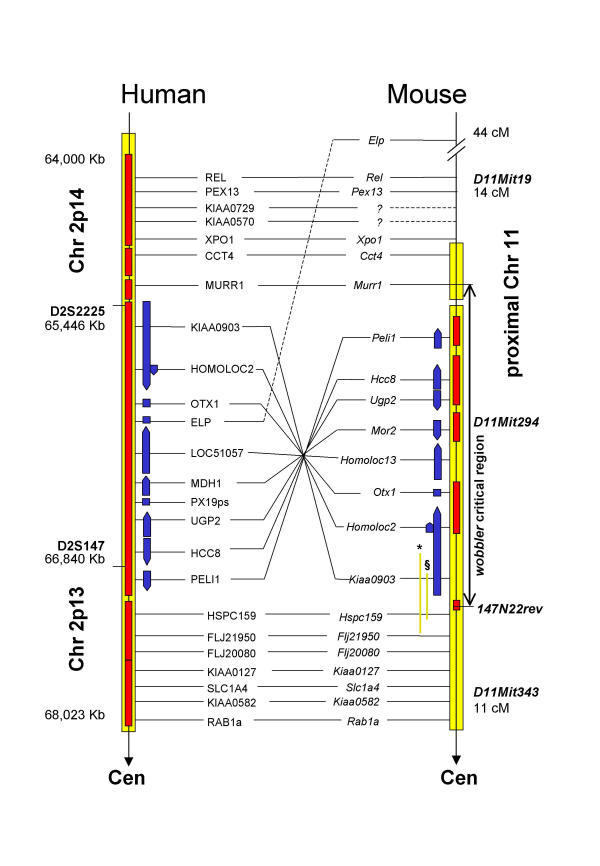
Figure 1.
Region of conserved synteny of human Chr2-13-14 and mouse prox Chr11 Orthologous genes of human and mouse and their positions on the respective physical map are shown except for Kiaa0570 and Kiaa0729, which could not be mapped with radiation hybrids. Mouse orthologs for ELP and PX19 are not located within the investigated region. Gene order of HCC8 and UGP2 is based on our STS/EST mapping and sequence analysis. Gene order between human and mouse is conserved in three sections; the proximal and distal sections show the same orientation whereas the central section is inverted between the two species. The central part of the figure including the inverted section is shown slightly enlarged. For orientation, positions of microsatellites D2S2225 and D2S147 (human) and D11Mit19, D11Mit294 and D11Mit343 (mouse) are indicated. Blue arrows show orientation and relative genomic sizes of genes, the arrowhead points to the 3' end. Yellow bars represent chromosomal regions covered by YAC or BAC contigs. Red overlays show sequenced sections (human: public draft sequence, mouse: BACs sequenced by our group, AC091422, AC091423, AC091424, AC091428, AC091419, AC091420, AC091421). Green bars indicate the genomic clones (YAC ymWIBR100H6, *; BAC 135B4, §), which cover Kiaa0903 and Hspc159. Approximate distances are given in Kb for human Chr2p13-14, in cM for mouse prox Chr11.
All human orthologs of these genes and cDNAs were mapped to the human contig of the homologous region which contains, in addition, peroxisome biogenesis factor 13 (PEX13) [12], exportin 1 (XPO1) [13] and cDNAs KIAA0729 [14] and KIAA0570 [15]. The murine orthologs Pex13 and Xpo1 were radiation hybrid mapped (T31 radiation hybrid Panel RH04.05, Research Genetics) between Rel and Cct4 but this technique failed for Kiaa0570 and Kiaa0729.
The human pseudogenes endozepine like peptide (ELP) [16] and px19-like protein (PX19) [17] are localized in the human genome sequence of the interval under study and their positions were confirmed by STS-mapping on our human physical map. However, their counterparts were not detected on the mouse contig. Murine Elp was radiation hybrid mapped but located far distally (at 40 cM) on Chr11 (Fig 1). The product of Elp is a testis specific isoform of the ubiquitous acyl-CoA binding protein (ACBP) which is highly expressed in the haploid stages of male germ cell development [16]. PX19 was first identified in chicken as a cDNA with a LEA (late embryogenesis abundant) motif [17]. By searching the human draft genome sequence we detected more than 5 intronless copies of this gene, and also in the limited data of the mouse genome several copies were present (data not shown).
Human gene content and order on our contig was compared with the annotated draft sequence of the human genome (UCSC genome browser, December 2001). Nearly all genes/cDNAs are present on both and the gene order is identical with one exception (HCC8 and UGP2) which is due to gaps in the human draft sequence. On the basis of the human draft sequence and our YAC contig it was possible to determine the orientation of most human genes (PELI1 [9], MDH1 [18], HCC8 [19], UGP2 [20], LOC51057 and KIAA0903 [21]). In the mouse, genes were ordered and oriented by our high resolution BAC contig combined with sequence analysis of seven selected mouse BACs (AC091422, AC091423, AC091424, AC091428, AC091419, AC091420, AC091421).
Murine genomic clones, YAC clone ymWIBR100H6 (500 Kb) and BAC clone 135B4 (200 kb), comprised sequences from both, Kiaa0903 and Hspc159; this indicated a different gene order of mouse and man in the wobbler critical region (Fig. 1). We did not identify any YAC or BAC clones that would contain both Murr1 and Peli1 which match the distal border of the presumed inversion. On the basis of close neighbourhood of Kiaa0903 and Hspc159, three segments were defined. Within each segment order and orientation of genes are conserved between man and mouse, but the central one is inverted. The distal segment is defined by Rel and Murr1, the proximal one by Hspc159 and Rab1a and the central one by Kiaa0903 and Peli1.
Segregation analysis of the wr candidate interval
The candidate interval of the wobbler mutation was refined in comparison with [8] by using the interspecies backcross Mus musculus C57BL/6J-wr x Mus musculus castaneus.
New polymorphisms between the two strains were established all over the candidate region using BAC-end or intron sequences. Two recombinations were detected between BAC-end 147N22rev and wr; and four recombinations were detected between anonymous cDNA Murr1 and wr. Thus the candidate region of the wobbler mutation was narrowed down to the interval between the two loci 147N22rev (located distally to Kiaa0903) and Murr1.
Interestingly, the apparently inverted region seems to be identical to the wobbler critical interval. It contains the newly mapped genes Hcc8, Ugp2, Kiaa0903, Homoloc13 and the previously mapped genes Otx1 [8], Mor2 [8], and Peli1 [9]. Our high resolution physical maps in addition to the human draft sequence suggests that all orthologous genes have been identified. Because no additional genes were found by BLAST search using genomic sequences or by analysis of mouse sequences with exon prediction programs we believe that the set of candidate genes for the wobbler mutation is complete.
Full length cDNA sequences for the newly mapped genes Hcc8, Ugp2, Kiaa0903, Homoloc13 and the previously mapped gene Otx1 were established according to Materials and Methods. All genes were subjected to mutation analysis by comparative sequencing of the coding regions, and parts of 3' and 5' UTRs, but no mutation was detected so far. Using RNAs from testis and spinal cord (the affected organs) of adult wobbler (wr/wr) and wildtype (+/+) mice, transcript levels of the canididate genes were analyzed by Northern blotting and RT-PCR, but no difference between wobbler and wildtype mice was found (data not shown).
A summary of nucleotide and amino acid features for all genes within the wobbler critical region including accession numbers, genomic structure and mouse-man homologies is displayed in Table 1.
Table 1.
Nucleic acid and polypeptide features of mouse genes of the central section of the investigated region on Chr11.
| Gene | Protein | ||||||||
| mouse gene / cDNA | Structure | % nucleotide identity | Size | % sequence identity | |||||
| designation | function | cDNA length (bp) | genomic length (kb) | number of exons | human | other species | length (aa) | human | other species |
| Ugp2 | phosphorylation of UDP-glucose, glycogen metabolism | 2017 AF424698 | 60 | 10 | 89% (NM_006759) | Bos taurus 89% (L14019) Sus scrofa 88% (X99312) | 508 | 92% (NP_006750) | Bos taurus 92% (AAA30801) Sus scrofa 92% (CAA67690) |
| Peli1 | putative member of cytoplasmic signalling pathway (IRAK) | 3710 AF302503 | 14 | 6 | 85% (AF302505) | n.a. | 418 (AAG15391) | 99% (AAG15393) | Ciona intestinalis 68% (BAB00628.1) Drosophila m. 74% (AAC96298) |
| Otx1 | homeoboxgene, transcription factor during brain development | 1068 AF424700 | 4 | 3 | 85% (XM_049268) | Rattus norvegicus 96% (NM_013109.1) | 355 (AAB27579.1) | 97% (P32242) | Rattus norvegicus 98% (NP_037241.1) Danio rerio 74% (BAA05158.1) |
| Hcc8 | hepatocellular carcinoma antigen | 4328 AF424699 | ~ 64 | ≥ 22 | 90% (AF102177) | Rattus norvegicus 92% (AJ010392.2) | 977 | 90% (AAF37319) | Rattus norvegicus 92% (CAB96885.1) |
| Mor2 | cytoplasmic malate dehydrogenase | 1288 (NM_008618) | 15 | 9 | 90% (XM_002358) | Rattus norvegicus 94% (AF093773) Sus scrofa 87% (SSU44846) | 334 (NP_032644) | 97% (XP_002358) | Rattus norvegicus 98% (AAC64180.1) Sus scrofa 94% (AAC48610.1) |
| Homoloc13 | unknown | 2151 AF424701 | ~ 150 | 11–12 | 84% (NM_015910) | n.a. | 643 | 80% (NP_056994) | n.a. |
| Kiaa0903 | unknown, contains actinin binding domain, CAAX box | 4820 AF4246987 | ~ 350 | 25 | 88 % (AX030068) | n.a. | 1207 | 77% (BAA74926.1) | n.a. |
| Homoloc2 | unknown | 3045 | >3 | n.a. | 85% (AA305160) | n.a. | no ORF | n.a. | n.a. |
Accession numbers in brackets are previously published results. n.a. = not available
Discussion
Knowledge of gene order and content is crucial for the identification of disease genes. During the draft phase of genome projects sequence information has to be used with caution since multiple gaps lead to incorrect orientation and order of the sequence fragments. High-resolution maps based on contigs of genomic clones as presented in this work help to overcome this problem. For example, the positions of genes UGP2 and HCC8, whose 5' ends are less than 2 kb apart, were shown to be inverted in the human draft version relative to the correct orientation, and most of the UGP2 gene was not available. The correct gene order was resolved with the high resolution map presented here. Furthermore, genes are missing from the chromosome maps if their transcripts have not been described although the protein is known. For example, OTX1 was not annotated in the draft sequence because its cDNA sequence was not present in the database.
By the identification of 20 orthologous gene pairs we were able to show that the gene content of proximal mouse Chr11 and human Chr2p13-14 is highly conserved. On the other hand, the gene order appears disrupted by an inversion which comprises 1.5 Mb. Despite the fact that the present mouse draft sequence suggests an orientation as in the human genome our results indicate an inversion. The exact inversion breakpoints are not known, and despite extensive screens no mouse BACs spanning the distal breakpoint were detected. Low complexity and reiterative sequences may be responsible for both, the chromosomal rearrangement between mouse and man and the difficulty to clone these regions. A comparable inversion of about 1 Mb has been reported for human Chr19p13.3 and mouse Chr10 [22]. Carver and Stubbs [1] suggest that continuous regions of conserved syntenies without insertions, deletions and inversions are rather the exception than the rule. Further small-scale rearrangements within regions of conserved synteny are expected to be discovered with high-resolution maps and availability of sequence data.
The only exceptions from the conservation of gene content are PX19 and ELP which are probably pseudogenes that have integrated on human Chr2p after the divergence of rodents and primates. Although some cDNA clones are listed in the databases, no expression of ELP could be detected and the sequences contain several mutations [23]. We conclude that the human ELP is a pseudogene. Murine Elp probably arose by retroposon-mediated gene duplication and secondarily acquired a testis-specific promotor and thus a limited expression. The Chr2p copy of PX19 is truncated compared to the functional PX19 cDNA, with the 3' UTR partially missing. No human EST corresponding to the Chr2p copy was detected, implying it to be a pseudogene. The polyA tail in the genomic sequence suggests that it may have arisen by the integration of an alternatively polyadenylated PX19 transcript. Although first identified as a human cDNA clone, Homoloc2 neither has an open reading frame nor could a transcript be detected by RT-PCR. The high conservation of its genomic sequence between man and mouse suggests a regulatory function.
Establishing full-length cDNA sequences for all genes within the wobbler critical region was the basis for an extensive comparative sequencing effort aimed to detect a mutation. So far, no mutation and no difference of transcript levels was found.
Conclusions
The comparative mouse-man genomics approach has proven a valuable tool to find new candidate genes for disease mutations [24]. However, inversions and insertions have to be taken into account if the human genome is to be used as a framework for the sequence assembly of the mouse. Besides providing new candidates comparative genomics leads to the identification of conserved noncoding sequences, which might have important biological functions. Mutations such as wobbler for which all candidate genes have been excluded might affect a conserved noncoding sequence with a cis-acting effect on genes that might be located outside the critical region as defined by recombination.
Methods
BAC and YAC clones
For the mouse BAC and YAC clones and the human YAC clones see Resch et al. [8]. The contigs were established by extensive STS/EST mapping and genomic sequencing.
STS/EST mapping
Standard PCR reactions were: 12.5 μl reaction volume of Qiagen (Hilden, Germany) Taq Mastermix Kit with 12.5 pmoles of each primer. PCR was performed on all BACs and YACs using 50 ng of template DNA.
Search for orthologous ESTs
Orthologous mouse ESTs for mapped human ESTs were identified via BLAST search. For ESTs with greater than 80% homology to ESTs of other species, PCR primers were generated. Subsequently, physical mapping was carried out by STS and radiation hybrid mapping [8].
Preparation of RNA and RT-PCR
Mouse RNA from wildtype and wobbler (wr/wr)was prepared from various tissues of C57Bl/6J mice by standard procedures [23]. Human RNA was purchased from Clontech (Palo Alto, USA). One μg of total RNA was reverse transcribed using the Omniscript RT-Kit from Qiagen (Hilden, Germany).
Expansion of ESTs to full length cDNAs
In order to expand EST sequences (length usually < 500 bp), overlapping sequences of cDNA clones belonging to a single EST were assembled in silico. To verify the assembled sequences, primers were generated for each end, and the transcript was amplified by RT-PCR, cloned and sequenced.
Genomic sequence analysis
BACs were sequenced using a combination of shotgun and directed approaches as described previously [24]. Assembled sequence contigs were analyzed by the automated annotation system RUMMAGE [25].
Authors' Contributions
Fuchs S. and Resch K. and Thiel C. carried out the the STS/EST-mapping and identified the novel genes and Ulbrich M. narrowed down the wobbler-critical interval. Platzer M. has subcloned and sequenced the mentioned mouse BACs. Jockusch H. and Schmitt-John T. instructed and coordinated the experimental work and drafted the manuscript.
Acknowledgments
Acknowledgments
We thank Melanie Ronsiek (Bielefeld), Beate Fischer, Hella Ludewig, and Evelin Michaelis (Jena) for expert technical assistance. This work was supported by a grant from Deutsche Forschungsgemeinschaft (SCHM1276/2), Volkswagenstiftung (I/72 444) and the German BMBF (BEO 0311108/0).
Contributor Information
Sonja Fuchs, Email: sonja.fuchs@uni-bielefeld.de.
Karin Resch, Email: resch@lavisionbiotec.com.
Cora Thiel, Email: cora.thiel@uni-bielefeld.de.
Michael Ulbrich, Email: michael.ulbrich@uni-bielefeld.de.
Matthias Platzer, Email: mplatzer@imb-jena.de.
Harald Jockusch, Email: h.jockusch@uni-bielefeld.de.
Thomas Schmitt-John, Email: thomas.john@uni-bielefeld.de.
References
- Carver EA, Stubbs L. Zooming in on the human-mouse comparative map: genome conservation re-examined on a high-resolution scale. Genome Res. 1997;7:1123–1137. doi: 10.1101/gr.7.12.1123. [DOI] [PubMed] [Google Scholar]
- Nadeau JH, Sankoff D. Counting on comparative maps. Trends Genet. 1998;14:495–501. doi: 10.1016/S0168-9525(98)01607-2. [DOI] [PubMed] [Google Scholar]
- Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- Duchen LW, Strich SJ. An hereditary motor neurone disease with progressive denervation of muscle in the mouse: the mutant 'wobbler'. J Neurol Neurosurg Psychiatry. 1968;31:535–542. doi: 10.1136/jnnp.31.6.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leestma JE, Sepsenwol S. Sperm tail axoneme alterations in the Wobbler mouse. J Reprod Fertil. 1980;58:267–270. doi: 10.1530/jrf.0.0580267. [DOI] [PubMed] [Google Scholar]
- Kaupmann K, Simon-Chazottes D, Guenet JL, Jockusch H. Wobbler, a mutation affecting motoneuron survival and gonadal functions in the mouse, maps to proximal chromosome 11. Genomics. 1992;13:39–43. doi: 10.1016/0888-7543(92)90199-3. [DOI] [PubMed] [Google Scholar]
- Wedemeyer N, Lengeling A, Ronsiek M, Korthaus D, Baer K, Wuttke M, Jockusch H. YAC contigs of the Rab1 and wobbler (wr) spinal muscular atrophy gene region on proximal mouse chromosome 11 and of the homologous region on human chromosome 2p. Genomics. 1996;32:447–454. doi: 10.1006/geno.1996.0140. [DOI] [PubMed] [Google Scholar]
- Resch K, Korthaus D, Wedemeyer N, Lengeling A, Ronsiek M, Thiel C, Baer K, Jockusch H, Schmitt-John T. Homology between human chromosome 2p13.3 and the wobbler critical region on mouse chromosome 11: comparative high-resolution mapping of STS and EST loci on YAC/BAC contigs. Mamm Genome. 1998;9:893–898. doi: 10.1007/s003359900890. [DOI] [PubMed] [Google Scholar]
- Resch K, Jockusch H, Schmitt-John T. Assignment of homologous genes, Peli1/PELI1 and Peli2/PELI2, for the Pelle adaptor protein Pellino to mouse chromosomes 11 and 14 and human chromosomes 2p13.3 and 14q21, respectively, by physical and radiation hybrid mapping. Cytogenet Cell Genet. 2001;92:172–174. doi: 10.1159/000056895. [DOI] [PubMed] [Google Scholar]
- Rathke-Hartlieb S, Schmidt VC, Jockusch H, Schmitt-John T, Bartsch JW. Spatiotemporal progression of neurodegeneration and glia activation in the wobbler neuropathy of the mouse. NeuroReport. 1999;10:3411–3416. doi: 10.1097/00001756-199911080-00028. [DOI] [PubMed] [Google Scholar]
- Nabetani A, Hatada I, Morisaki H, Oshimura M, Mukai T. Mouse U2af1-rs1 is a neomorphic imprinted gene. Mol Cell Biol. 1997;17:789–798. doi: 10.1128/mcb.17.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Kalish JE, Morrell JC, Bjorkman J, Urquhart AJ, Crane DI. Pex13p is an SH3 protein of the peroxisome membrane and a docking factor for the predominantly cytoplasmic PTs1 receptor. J Cell Biol. 1996;135:85–95. doi: 10.1083/jcb.135.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo N, Khochbin S, Nishi K, Kitano K, Yanagida M, Yoshida M, Horinouchi S. Molecular cloning and cell cycle-dependent expression of mammalian CRM1, a protein involved in nuclear export of proteins. J Biol Chem. 1997;272:29742–29751. doi: 10.1074/jbc.272.47.29742. [DOI] [PubMed] [Google Scholar]
- Nagase T, Ishikawa K, Suyama M, Kikuno R, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. XI. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1998;5:277–286. doi: 10.1093/dnares/5.5.277. [DOI] [PubMed] [Google Scholar]
- Nagase T, Ishikawa K, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. IX. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res, 1998;5:31–39. doi: 10.1093/dnares/5.1.31. [DOI] [PubMed] [Google Scholar]
- Valentin M, Balvers M, Pusch W, Weinbauer GF, Knudsen J, Ivell R. Structure and expression of the mouse gene encoding the endozepine-like peptide from haploid male germ cells. Eur J Biochem. 2000;267:5438–5449. doi: 10.1046/j.1432-1327.2000.01603.x. [DOI] [PubMed] [Google Scholar]
- Guzman-Rojas L, Sims JC, Rangel R, Guret C, Sun Y, Alcocer JM, Martinez-Valdez H. PRELI, the human homologue of the avian px19, is expressed by germinal center B lymphocytes. Int Immunol. 2000;12:607–612. doi: 10.1093/intimm/12.5.607. [DOI] [PubMed] [Google Scholar]
- Larson LM, Bruce AW, Saumur JH, Wasdahl WA. Further evidence by gene dosage for the regional assignment of erythrocyte acid phosphatase (ACP1) and malate dehydrogenase (MDH1) loci on chromosome 2p. Clin Genet. 1982;22:220–225. doi: 10.1111/j.1399-0004.1982.tb01437.x. [DOI] [PubMed] [Google Scholar]
- Stenner-Liewen F, Luo G, Sahin U, Tureci O, Koslovski M, Kautz I, Liewen H, Pfreundschuh M. Definition of tumor-associated antigens in hepatocellular carcinoma. Cancer Epidemiol Biomarkers Prev. 2000;9:285–290. [PubMed] [Google Scholar]
- Cheng SD, Peng HL, Chang HY. Localization of the human UGP2 gene encoding the muscle isoform of UDPglucose pyrophosphorylase to 2p13-p14 by fluorescence in situ hybridization. Genomics. 1997;39:414–416. doi: 10.1006/geno.1996.4426. [DOI] [PubMed] [Google Scholar]
- Nagase T, Ishikawa K, Suyama M, Kikuno R, Hirosawa M, Miyajima N, Tanaka A, Kotani H, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. XII. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1998;5:355–364. doi: 10.1093/dnares/5.6.355. [DOI] [PubMed] [Google Scholar]
- Puttagunta R, Gordon LA, Meyer GE, Kapfhamer D, Lamerdin JE, Kantheti P, Portman KM, Chung WK, Jenne DE, Olsen AS, Burmeister M. Comparative maps of human 19p13.3 and mouse chromosome 10 allow identification of sequences at evolutionary breakpoints. Genome Res. 2000;10:1369–1380. doi: 10.1101/gr.145200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Platzer M, Rotman G, Bauer D, Uziel T, Savitsky K, Bar-Shira A, Gilad S, Shiloh Y, Rosenthal A. Ataxia-telangiectasia locus: sequence analysis of 184 kb of human genomic DNA containing the entire ATM gene. Genome Res. 1997;7:592–605. doi: 10.1101/gr.7.6.592. [DOI] [PubMed] [Google Scholar]
- Taudien S, Rump A, Platzer M, Drescher B, Schattevoy R, Gloeckner G, Dette M, Baumgart C, Weber J, Menzel U, Rosenthal A. RUMMAGE – a high-throughput sequence annotation system. Trends Genet. 2000;16:519–520. doi: 10.1016/S0168-9525(00)02127-2. [DOI] [PubMed] [Google Scholar]