Abstract
Background
Spontaneous premature ovarian failure presents most commonly with secondary amenorrhea. Young women with the disorder are infertile and experience the symptoms and sequelae of estrogen deficiency. The mechanisms that give rise to spontaneous premature ovarian failure are largely unknown, but many reports suggest a genetic mechanism in some cases. The small family size associated with infertility makes genetic linkage analysis studies extremely difficult. Another approach that has proven successful has been to examine candidate genes based on known genetic phenotypes in other species. Studies in mice have demonstrated that c-kit, a transmembrane tyrosine kinase receptor, plays a critical role in gametogenesis. Here we test the hypothesis that human KIT mutations might be a cause of spontaneous premature ovarian failure.
Methods and Results
We examined 42 women with spontaneous premature ovarian failure and found partial X monosomy in two of them. In the remaining 40 women with known 46,XX spontaneous premature ovarian failure we evaluated the entire coding region of the KIT gene. We did this using polymerase chain reaction based single-stranded conformational polymorphism analysis and DNA sequencing. We did not identify a single mutation that would alter the amino acid sequence of the c-KIT protein in any of 40 patients (upper 95% confidence limit is 7.2%). We found one silent mutation at codon 798 and two intronic polymorphisms.
Conclusion
Mutations in the coding regions of the KIT gene appear not to be a common cause of 46,XX spontaneous premature ovarian failure in North American women.
Background
Premature ovarian failure is a condition characterized by cessation of normal ovarian function before the age of 40. In most cases a precise mechanism for the ovarian failure is not identified; however, there is evidence suggesting a genetic mechanism in at least some cases. [1-4]
Normal ovarian function in reproductive-aged women is dependent upon the presence of a store of functional primordial follicles. This store develops from primordial germ cells, a small population of cells that differentiate from other cell lineages in very early embryonic life. [5] Premature depletion of primordial follicles is a known mechanism of premature ovarian failure.[6,7]
Studies in mice have demonstrated that c-kit, a transmembrane tyrosine kinase receptor, plays a critical role in hematopoeisis, melanogenesis, and gametogenesis. The c-kit receptor kinase and its ligand KL (kit ligand, stem cell factor) are encoded at the white spotting (W) and steel (Sl) loci of the mouse, respectively. Various mutations at the W and the Sl loci have been shown to cause anemia, pigmentation defects, and sterility.[8] Point mutations in the Kit receptor tyrosine kinase in mice can selectively impair fertility without inducing detectable abnormalities in hematopoesis or pigmentation.[9] Studies employing cultured mouse primordial germ cells have demonstrated that the Steel/Kit interaction is required for germ cell survival. [10-12]
These observations in mice suggest that mutations in the human KIT gene (MIM 164920) might be a cause of premature ovarian failure in women. The human KIT gene is located on chromosome 4 at map locus 4q12. Mutations in the human KIT gene have been identified as a cause of Piebaldism, a rare autosomal dominant disorder of melanogenesis characterized by patchy absence of pigmentation of the skin and overlying hair.[13,14] Here we test the hypothesis that specific human KIT mutations might be a cause of spontaneous premature ovarian failure even in the absence of detectable disorders of pigmentation or hematopoesis.
Methods
Subjects
Our institutional review board approved the study and all participants signed an informed consent and the procedures of the study were in compliance with the Helsinki Declaration. The study population comprised 42 women with premature ovarian failure and 10 normal healthy control women who had regular menses and proven fertility. Referring physicians made the diagnosis of premature ovarian failure based on the following criteria: development of at least 4 months of amenorrhea before age 40 associated with two serum FSH levels above 40 IU/L (drawn at least 1 month apart). Women with premature ovarian failure as a result of surgery, radiation, chemotherapy, or known karyotype abnormalities were not included in the study. Patients who had not previously had a karyotype analysis were included pending our analaysis. There were 34 Caucasian, 6 African American and two Hispanic women. The median age at the onset of menstrual irregularity was 25 years (range 14 to 39, 25th percentile 18.5, 75th percentile 34). Five women had a family history of premature ovarian failure (two patients had sisters with premature ovarian failure, one patient's mother had premature ovarian failure, one had a paternal aunt with premature ovarian failure, and one patient had a paternal aunt, a grandmother, and a great-grandmother who were thought to have the condition). All patients underwent a history and physical examination and laboratory screening to confirm the diagnosis of premature ovarian failure.
Cytogenetics
Karyotypes were obtained on all patients. Peripheral blood specimens were cultured for 72–96 hours using standard methods and methotrexate-thymidine synchronization.[15] Harvesting and GTW-banding were also performed according to standard cytogenetic protocols.[16] Fifty metaphases were examined for each patient and scored for numerical or structural chromosome abnormalities. Band resolution was at the 600 band stage or higher.
Fluorescence in situ hybridization (FISH) was performed on metaphases from cultured lymphocytes of two patients with abnormal G-band findings. Chromosome and probe denaturation, hybridization, detection, and counterstaining were all done according to standard protocols as recommended by the manufacturer.
DNA extraction and polymerase chain reaction (PCR)
Genomic DNA was extracted from peripheral blood using standard procedures. Specific primers for PCR amplification of the exons were designed based on the human KIT genomic sequence (Genebank accession number AC006553, Human chromosome 4) (Table 1). In exon 21 the reverse primer is located in the 3' noncoding region, in all other cases the primers are located at introns flanking each exon. The PCR amplification was carried out in a total volume of 25 μl reaction mixture containing 1.5 mmol/l of MgCL2, 0.2 mmol/l of each dNTP, 50 ng of genomic DNA, 0.5 μm of each primer, and 1.25 IU of Taq DNA polymerase per manufacturer's instructions (Invitrogen, Carlsbad, CA). The PCR began with an initial denaturing at 95°C for 5 minutes. Thereafter the cycling profile consisted of 35 cycles of denaturing at 95°C for 30 seconds, annealing of the primer pair at the appropriate temperature for 30 seconds (Table 1), and then extension at 72°C for 60 seconds. This was followed by a final stabilization step of 10 minutes at 72°C. After PCR, 5 μl of the amplified product was examined by 1.5% agarose gel electrophoresis.
Table 1.
Primer sets and conditions for amplification of human KIT exons. Forward and reverse primers flanking each of the 21 exons are shown. The size of each exon-coding region, the size of each PCR product generated by each set of primers, and the respective annealing temperatures are indicated. Sequences marked by a * are derived from Spritz et al..[28]
| Exon No | Exon # Size (bp) | Primers | PCR product Size (bp) | Annealing Tm(°C) |
| Forward Reverse | ||||
| 1 | 88(67) | 5'GGACCAGAGCTCGGATC3' / 3'AGTCCTCTCTCCGGATG5' | 198 | 62 |
| 2 | 270 | 5'CTCAGTATTGGAAGAAGTGC3' / 3'TATCTATGGCTCAGTCATCC5' | 438 | 55 |
| 3 | 282 | 5'GCTTCTATAGATCCTGCC3' / 3'GATCAACGAGAAGAGAAGTC5' * | 367 | 55 |
| 4 | 137 | 5'TGTACACATTTGAGGAGAAA3' * / 3'CTGACAGACGCACTAGTCG5' * | 330 | 55 |
| 5 | 169 | 5'TGGAGAAGTTAATTGCTGCT3' * / 3'TGTGCTTTCATTGCAAGAGG5' | 435 | 55 |
| 6 | 190 | 5'TTGTAATTCCAAGATGAGG3' * / 3'TACTAGGATGAGGACATAGG5' | 422 | 55 |
| 7 | 116 | 5'TATGTGTGTGCGTGTTTATG3' * / 3'CAAGTTGAGTCCTTGCAGCTG5' * | 372 | 62 |
| 8 | 115 | 5'CTCCTTGGTTCAGATTCTGC3' / 3'GTGAATTGCAGTCCTTCC5' | 321 | 55 |
| 9 | 194 | 5'TATGCCACATCCCAAGTG3' / 3'GGTGTGATGCATGTATTACC5' | 390 | 55 |
| 10 | 107 | 5'ACATAGCTTTGCATCCTGC3' / 3'ATTGTCTCAGTCATTAGAGCAC5' * | 280 | 55 |
| 11 | 127 | 5'GAGTGCTCTAATGACTGAG3' / 3'CCACTGGAGTTCCTTAAAG5' | 266 | 56 |
| 12 | 105 | 5'ATGGTCCTTCAATTCCACC3' / 3'TTCTGTCAAATGGGCACTC5' | 270 | 55 |
| 13 | 111 | 5'GACATCAGTTTGCCAGTTG3' / 3'GCAAGAGAGAACAACAG5' | 296 | 58 |
| 14 | 151 | 5'GACTAAGTAGTCTGATCC3' / 3'ACCCCATGAACTGCCTGTC5' * | 327 | 55 |
| 15 | 92 | 5'TAGAGCATGACCCATGAG3' / 3'ACCCACTTGCAACCCTAACT5' * | 325 | 60 |
| 16 | 128 | 5'GGTATGTCATTGCCACTG3' / 3'GGCTCTAAAATGCTCTGTTCT5' * | 296 | 55 |
| 17 | 123 | 5'GTGAACATCATTCAAGGCG3' / 3'TTACATTATGAAAGTCACAGG5' * | 390 | 55 |
| 18 | 112 | 5'CACATTTCAGCAACAGC3' / 3'CCTTCCTTGATCATCTTGT5' * | 382 | 53 |
| 19 | 100 | 5'CTCAGAGCATCTTCTTGAAG3' / 3'ACATCTGGGTTTCTGTCTC5' | 252 | 58 |
| 20 | 106 | 5'CCATATGTCCAGTTGCATAG3' / 3'TACCTGAAGCCCAATTTGC5' | 257 | 60 |
| 21 | 2407(129) | 5'TGGCCACAAAGTTCTTGG3' / 3'TATCCTGGAGTTGGATGC5' | 369 | 55 |
Single-stranded conformational polymorphism (SSCP) analysis
Analysis of DNA polymorphism was conducted using a method of "Cold SSCP" as described previously.[17] Briefly, the PCR products (4 μl) were mixed with SSCP sample buffer (6 μl, formamide with 0.05% bromophenol blue and xylene cyanol). After denaturing at 95°C for 5 minutes, the DNA samples were placed on ice immediately and loaded onto precast 20% TBE acrylamide gels (Tris Base, Boric Acid, EDTA, 4% glycerol, and 20% acrylamide, Invitrogen, Carlsbad, CA). Gel elecrophoresis was run in 1 X TBE buffer at 200 V with the circulator set at 4°C (using a Penquin Water-cooled Dual-Gel Electrophoresis system attached to a thermostatically controlled refrigerated circulator, Amersham & Pharmacia, Uppsala, Sweden). Gels were stained with ethidium bromide (1 μg/ml) for 20 minutes at room temperature. DNA bands were visualized and photographed under UV light (340 nm).
Direct DNA sequencing
PCR products were either purified using ExoSAP-IT (USB Co, Cleveland, OH) or subcloned into a TA cloning vector (Invitrogen, Carlsbad, CA) per the manufacturer's instructions. DNA sequencing was conducted by PCR (26 cycles of 96°C, 10 seconds; 50°C 5 seconds; 60°C 4 minutes) using a dRhodamine terminator cycle sequencing kit (PE Applied Biosystems, Foster, CA) and 10 μl of purified PCR product or 1 μg of purified plasmid DNA. The sequencing samples were separated by 5% Long Ranger gel electrophoresis on the ABI Automatic 310 Sequencer (Applied Biosystems, Foster, CA)
Results
Clinical
None of the 42 women were found to have clinical findings to suggest Turner syndrome. None of the patients had clinical or laboratory findings to suggest a stem cell deficiency in melanogenesis or hematopoiesis. Two women were found to have partial monosomy of the X chromosome that was felt to be related to the presentation of spontaneous premature ovarian failure. For this reason they were excluded from further analysis. These findings highlight the usefulness of careful cytogenetic screening of women with spontaneous premature ovarian failure. The remaining 40 women had a normal 46,XX karyotype analysis.
One woman of normal stature had a karyotype analysis showing: 46,X, ?rec(X) dup(Xp) inv(X)(p11.4 q24). ish rec(X) dup(Xp) inv(X)(p11.4 q24) (KAL++). In all cells examined there was one normal X chromosome and one X chromosome that had most of Xp (Xp11.4→Xpter) replacing a portion of Xq (Xq24→Xqter). This rearrangement was confirmed with a FISH study. A probe to the Kallman syndrome region (Oncor Inc., Gaithersburg, MD) at Xp22.3 was hybridized to patient's metaphases and showed signal on both ends of the abnormal X chromosome (as well as the normal X chromosome) confirming the G-band impression that both ends of this chromosome was comprised of Xp. In summary, the patient had partial trisomy Xp and partial monosomy Xq as demostrated by G-banding and FISH.
A second woman of normal stature had a karyotype analysis showing: 46,X, der(X) t (X;13) (q22.3;q14.1). All cells examined in this patient had extra material on Xq which was 13q in origin. FISH was performed using whole chromosome probes to the X and 13 (Oncor, Gaithersburg, MD). The X probe showed signal covering the entire normal X as well as Xp and the proximal half of Xq on the derivative chromosome. Whole chromosome probe 13 showed signal on the normal 13 as well as on the derivative X chromosome. These results are consistant with partial monosomy Xq from q22→q terminus and partial trisomy 13 from q 14.1→q terminus. Given the lack of symptoms/characteristics associated with trisomy 13 (multiple congenital anomolies with severe cognitive delays) we hypothesize that the derivative X chromosome is the inactivated X, thus sparing the patient the expected phenotype.
Screening of the KIT gene
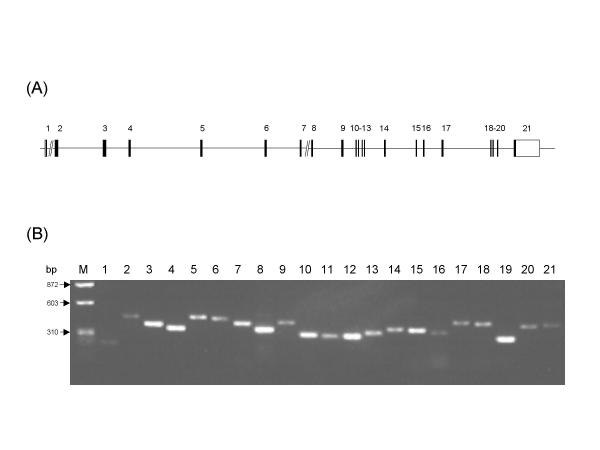
All the exons of the gene (Figure 1A) were successfully amplified by PCR in all 10 controls and all 40 patients with 46,XX spontaneous premature ovarian failure. Except for product 21, which carried only the coding region, each PCR product contained one complete exon. All PCR products were of the expected size. As shown in Figure 1B, the size of the 21 PCR products ranged from 198 to 438 bp and all appeared as a single clear band.
Figure 1.
Human KIT organization and PCR amplification of its exons. A. Schematic representation of exon-intron map of human KIT. It is composed of 21 exons and 20 introns. Vertical bars and horizontal solid lines represent the exons and introns, respectively. The // indicates that these introns are not in scale. Arabic numbers above each of the vertical bars indicate the exon number. Modified from Giebel et al.[27]. B. Analysis of PCR products of the human KIT gene. Human genomic DNA spanning each of the human KIT exons were amplified using specific primer pairs. PCR products for each of the exons were separated by 1.5% agarose-electrophoresis and stained with ethidium-bromide. The Arabic numbers above each of the PCR products represent the exon number. DNA size markers (φ X174 RF DNA/HaeIII) are shown at the left of the panel (M).
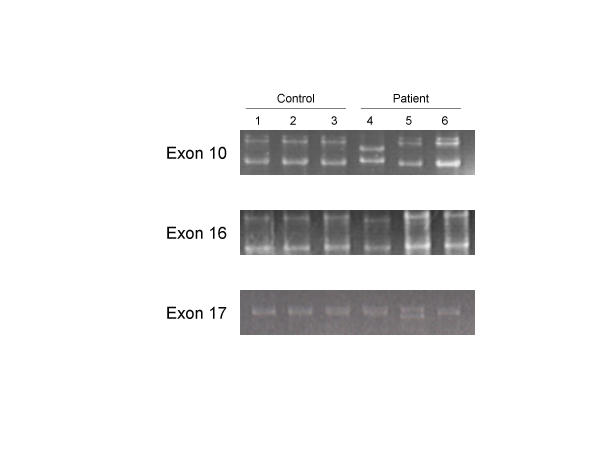
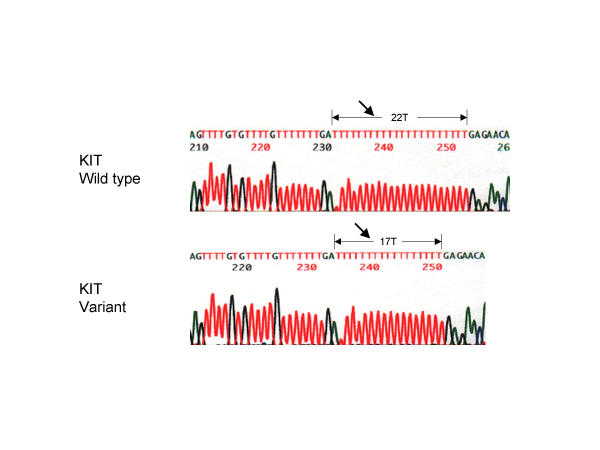
The PCR products were screened for mutations by SSCP analysis. We found mobility shifts in the PCR products from two of the patients and none of the controls. As shown in Figure 2, patient 4 had a mobility shift in the products from two exons, 10 and 16; patient 5 had a mobility shift in the product from exon 17. In patient 4 both the polymorphisms were determined by sequencing to be within intron regions, and would thus not change the sequence of the c-KIT protein. In the polymorphism 71 nucleotides upstream of exon 10, substitution at nucleotide 71 replaced the normal T with an A [IVS9-71T>A]. In the polymorphism downstream of exon 16 there was a deletion of 5 T nucleotides from within a normal series of 22 consecutive T nucleotides [IVS16 +66_+70delTTTTT] (Figure 3). In patient 5 sequencing uncovered a mutation at codon 798; however, the mutation C798T was silent with no change in amino acid sequence (Isoleucine).
Figure 2.
SSCP analysis of KIT PCR products The PCR products were separated on 20% TBE-acrylamide gel electrophoresis and stained with ethidium bromide. SSCP analysis of samples from three normal control women and three patients with spontaneous premature ovarian failure are shown for exon 10,16, and 17. DNA mobility of PCR products from exons 10 and 16 was altered in one patient (Lane 4). One patient showed an alternation of DNA mobility in the PCR product from exon 17 (Lane 5). Also shown is the DNA mobility of samples from normal women (Lanes 1–3) and from a representative patient with no changes in DNA mobility (Lane 6).
Figure 3.
DNA sequence analysis of a KIT intronic region variant downstream of exon 16 Electropherogram displaying the sequence of the KIT variant of the intronic sequence downstream of exon 16 compared to the wild-type sequence. Arrows indicate a deletion of 5T nucleotides in the variant as compared to a normal series of 22 consecutive T nucleotides in the wild-type.
Restriction Fragment Length Polymorphism Analysis
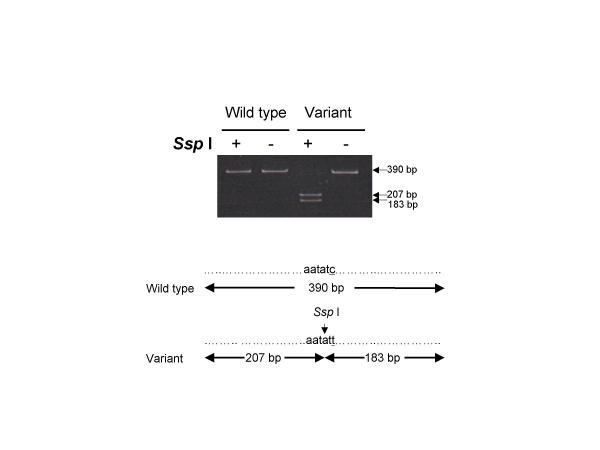
The C798T substitution in exon 17 of patient 5 generates a new restriction site for SspI (AAT ATT). To determine if this mutation was homozygous or heterozygous we analysed restriction fragment length polymorphism (RFLP) on PCR products. As shown in Figure 4, the mutant PCR product (390 bp) for exon 17 was digested with SspI to yield two shorter DNA fragments only (207 and 183 bp). These data suggest that this mutation is homozygous. We did not perform RFLP analysis on the mutant PCR products from exons 10 and 16 because no new restriction sites were generated by the mutations. Other methodologies such as forced RFLP and sequencing of multiple cloned products could be applied to determine if these mutations are homozygous or heterozygous.
Figure 4.
Restriction fragment length polymorphism analysis of a KIT variant in exon 17 using Ssp I Undigested DNA and wild-type DNA digested with Ssp I gives a single band of 390 bp. Homozygosity for the variant yields two fragments of 207 and 183 bp. A heterozygote carrier would have demonstrated all three of the fragment lengths. Therefore, this patient is homozygous for this variant.
Discussion
Primordial germs cells originate from a small number of progenitor cells that arise in very early development. Thereafter, complex factors that are not well understood direct their proliferation, survival, migration to the genital ridge, differentiation into oocytes, and the subsequent expenditure of oocytes via ovulation or apoptotic death.[5,7][18] Mutations or deletions in any of the genes involved in this complex process could theoretically be a cause of premature ovarian failure on the basis of germ cell deficiency, either due to a deficient initial endowment of primordial follicles or an accelerated expenditure of this endowment.
Evidence that genetic factors play a role in some cases of premature ovarian failure has been around for a long time [19,20]. However, the small pedigrees necessarily associated with this condition make it extremely difficult to conduct genetic linkage analysis that can be used to identify candidate genes.[4] One approach to this dilemma has been to evaluate candidate genes based on existing knowledge of ovarian physiology, as has been reported in the case of inhibin.[21] Other candidate genes are those that when mutated are known to cause germ cell deficiency in other species, for example the diaphanous gene of drosophila.[22] Numerous growth factors have been associated with primordial germ cell behavior during migration in mice using in vitro studies, but in most cases it is unknown whether these factors function in vivo.[5] One exception is the Steel/Kit interaction. Steel and W are genetic loci identified in strains of mice that exhibit sterility on the basis of germ cell deficiency. The products of these two loci are now known to be the tyrosine kinase receptor Kit (W) and its ligand Steel factor (Steel). It was on this basis that we set out to examine patients with premature ovarian failure for mutations in KIT.
The SSCP technique has been used successfully for mutation searching in other genes. [23-26] Using the PCR-SSCP technique we were able to detect only one polymorphism in the entire coding regions of the KIT gene, and this polymorphism did not change the amino acid sequence at codon 798. Therefore, these results suggest that mutations in the coding regions of the KIT gene are unlikely to be a common cause of premature ovarian failure. The upper 95% confidence limit of the proportion 0/40 is 7.2% (no mutations found in 40 women with 46,XX spontaneous premature ovarian failure). The DNA polymorphisms that we identified within intron regions upstream of exon 10 and downstream of exon 16 would not change the amino acid sequence of the c-KIT protein. However, we cannot exclude the possibility that these changes in nucleotides that are close to exons might alter c-KIT transcription, or that mutations in the regulatory regions of the KIT gene might be a cause of spontaneous premature ovarian failure. Furthermore, it should be noted that SSCP methodology does not detect all mutations of PCR products. Although in this report using this technique we were able to detect a single nucleotide mutation in a PCR product of 390 bp, most laboratories would suggest that SSCP detects approximately 70 to 80% of mutations. Screening a larger group of patients with a more sensitive technique such as denaturing high-performance liquid chromatography might detect more mutations.
Conclusions
We were unable to detect any significant mutations in the entire coding region of the KIT gene in 40 patients with 46,XX spontaneous premature ovarian failure. Therefore, mutations in the coding regions of the KIT gene do not appear to be a common cause of 46,XX spontaneous premature ovarian failure in North American women.
Competing interests
None declared.
Authors' contributions
K.S., Z-B.T, and K.V. participated in the design of the study and carried out the molecular genetic laboratory studies. K.S. and Z-B.T. drafted the manuscript. V.H.V. recruited the patients and helped characterized them clinically. L.M.N. conceived the study, participated in its design, coordination, analysis and manuscript preparation. All authors read and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Acknowledgments
Acknowledgements
We thank Jeanne Meck, Ph.D. and the staff of the Georgetown University Hospital Cytogenetics Laboratory for classical and molecular cytogenetics expertise. We thank Constantine Stratakis, M.D., D.Sc. for reviewing our manuscript prior to submission.
Contributor Information
Kyoko Shibanuma, Email: ShibanumaK@aol.com.
Zhi-Bin Tong, Email: TongZ@cc1.nichd.nih.gov.
Vien H Vanderhoof, Email: VanderhV@mail.nih.gov.
Konstantina Vanevski, Email: Vanevski@mail.nih.gov.
Lawrence M Nelson, Email: Lawrence_Nelson@nih.gov.
References
- Bondy CA, Nelson LM, Kalantaridou SN. The genetic origins of ovarian failure. J Womens Health. 1998;7:1225–1229. doi: 10.1089/jwh.1998.7.1225. [DOI] [PubMed] [Google Scholar]
- Kalantaridou SN, Davis SR, Nelson LM. Premature ovarian failure. Endocrinology and Metabolism Clinics of North America. 1998;27:989–1005. doi: 10.1016/s0889-8529(05)70051-7. [DOI] [PubMed] [Google Scholar]
- van Kasteren YM, Hundscheid RD, Smits AP, Cremers FP, van Zonneveld P, Braat DD. Familial idiopathic premature ovarian failure: an overrated and underestimated genetic disease? Hum Reprod. 1999;14:2455–2459. doi: 10.1093/humrep/14.10.2455. [DOI] [PubMed] [Google Scholar]
- Davis CJ, Davison RM, Payne NN, Rodeck CH, Conway GS. Female sex preponderance for idiopathic familial premature ovarian failure suggests an X chromosome defect: opinion. Hum Reprod. 2000;15:2418–2422. doi: 10.1093/humrep/15.11.2418. [DOI] [PubMed] [Google Scholar]
- Wylie C. Germ cells. Cell. 1999;96:165–174. doi: 10.1016/s0092-8674(00)80557-7. [DOI] [PubMed] [Google Scholar]
- Gosden RG, Faddy MJ. Biological bases of premature ovarian failure. Reprod Fertil Dev. 1998;10:73–78. doi: 10.1071/R98043. [DOI] [PubMed] [Google Scholar]
- Tilly JL. Commuting the death sentence: how oocytes strive to survive. Nat Rev Mol Cell Biol. 2001;2:838–848. doi: 10.1038/35099086. [DOI] [PubMed] [Google Scholar]
- Besmer P, Manova K, Duttlinger R, Huang EJ, Packer A, Gyssler C, et al. The kit-ligand (steel factor) and its receptor c-kit/W: pleiotropic roles in gametogenesis and melanogenesis. Dev Suppl. 1993:125–137. [PubMed] [Google Scholar]
- Kissel H, Timokhina I, Hardy MP, Rothschild G, Tajima Y, Soares V, et al. Point mutation in kit receptor tyrosine kinase reveals essential roles for kit signaling in spermatogenesis and oogenesis without affecting other kit responses. EMBO J. 2000;19:1312–1326. doi: 10.1093/emboj/19.6.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin I, Deed R, Cooke J, Zsebo K, Dexter M, Wylie CC. Effects of the steel gene product on mouse primordial germ cells in culture. Nature. 1991;352:807–809. doi: 10.1038/352807a0. [DOI] [PubMed] [Google Scholar]
- Dolci S, Williams DE, Ernst MK, Resnick JL, Brannan CI, Lock LF, et al. Requirement for mast cell growth factor for primordial germ cell survival in culture. Nature. 1991;352:809–811. doi: 10.1038/352809a0. [DOI] [PubMed] [Google Scholar]
- Matsui Y, Toksoz D, Nishikawa S, Nishikawa S, Williams D, Zsebo K, et al. Effect of Steel factor and leukaemia inhibitory factor on murine primordial germ cells in culture. Nature. 1991;353:750–752. doi: 10.1038/353750a0. [DOI] [PubMed] [Google Scholar]
- Spritz RA, Beighton P. Piebaldism with deafness: molecular evidence for an expanded syndrome. Am J Med Genet. 1998;75:101–103. doi: 10.1002/(SICI)1096-8628(19980106)75:1<101::AID-AJMG20>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Richards KA, Fukai K, Oiso N, Paller AS. A novel KIT mutation results in piebaldism with progressive depigmentation. J Am Acad Dermatol. 2001;44:288–292. doi: 10.1067/mjd.2001.112221. [DOI] [PubMed] [Google Scholar]
- Brown MG, Lawce HJ. Peripheral Blood Cytogenetic Methods. In: Barch MJ, Knutsen T, Spurbeck JL, editor. In The AGT Cytogenetics Laboratory Manual. Philadelphia: Lippincott-Raven; 1997. pp. 99–100. [Google Scholar]
- Gustashaw KM. Chromosome Stains. In: Barch MJ, Knutsen T, Spurbeck JL, editor. In The AGT Cytogenetics Laboratory Manual. Philadelphia: Lippincott-Raven;; 1997. p. 279. [Google Scholar]
- Hongyo T, Buzard GS, Calvert RJ, Weghorst CM. 'Cold SSCP': a simple, rapid and non-radioactive method for optimized single-strand conformation polymorphism analyses. Nucleic Acids Res. 1993;21:3637–3642. doi: 10.1093/nar/21.16.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehr M. The primordial germ cells of mammals: some current perspectives. Exp Cell Res. 1997;232:194–207. doi: 10.1006/excr.1997.3508. [DOI] [PubMed] [Google Scholar]
- Coulam CB, Stringfellow S, Hoefnagel D. Evidence for a genetic factor in the etiology of premature ovarian failure. Fertil Steril. 1983;40:693–695. doi: 10.1016/s0015-0282(16)47433-9. [DOI] [PubMed] [Google Scholar]
- Mattison DR, Evans MI, Schwimmer WB, White BJ, Jensen B, Schulman JD. Familial premature ovarian failure. Am J Hum Genet. 1984;36:1341–1348. [PMC free article] [PubMed] [Google Scholar]
- Shelling AN, Burton KA, Chand AL, van Ee CC, France JT, Farquhar CM, et al. Inhibin: a candidate gene for premature ovarian failure. Hum Reprod. 2000;15:2644–2649. doi: 10.1093/humrep/15.12.2644. [DOI] [PubMed] [Google Scholar]
- Bione S, Sala C, Manzini C, Arrigo G, Zuffardi O, Banfi S, et al. A human homologue of the Drosophila melanogaster diaphanous gene is disrupted in a patient with premature ovarian failure: evidence for conserved function in oogenesis and implications for human sterility. Am J Hum Genet. 1998;62:533–541. doi: 10.1086/301761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boute N, Gribouval O, Roselli S, Benessy F, Lee H, Fuchshuber A, et al. NPHS2, encoding the glomerular protein podocin, is mutated in autosomal recessive steroid-resistant nephrotic syndrome. Nat Genet. 2000;24:349–354. doi: 10.1038/74166. [DOI] [PubMed] [Google Scholar]
- Lee MH, Lu K, Hazard S, Yu H, Shulenin S, Hidaka H, et al. Identification of a gene, ABCG5, important in the regulation of dietary cholesterol absorption. Nat Genet. 2001;27:79–83. doi: 10.1038/87170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semina EV, Ferrell RE, Mintz-Hittner HA, Bitoun P, Alward WL, Reiter RS, et al. A novel homeobox gene PITX3 is mutated in families with autosomal- dominant cataracts and ASMD. Nat Genet. 1998;19:167–170. doi: 10.1038/527. [DOI] [PubMed] [Google Scholar]
- Azuma N, Hirakiyama A, Inoue T, Asaka A, Yamada M. Mutations of a human homologue of the Drosophila eyes absent gene (EYA1) detected in patients with congenital cataracts and ocular anterior segment anomalies. Hum Mol Genet. 2000;9:363–366. doi: 10.1093/hmg/9.3.363. [DOI] [PubMed] [Google Scholar]
- Giebel LB, Strunk KM, Holmes SA, Spritz RA. Organization and nucleotide sequence of the human KIT (mast/stem cell growth factor receptor) proto-oncogene. Oncogene. 1992;7:2207–2217. [PubMed] [Google Scholar]
- Spritz RA, Giebel LB, Holmes SA. Dominant negative and loss of function mutations of the c-kit (mast/stem cell growth factor receptor) proto-oncogene in human piebaldism. Am J Hum Genet. 1992;50:261–269. [PMC free article] [PubMed] [Google Scholar]