Abstract
Background
In a variant of the standard PCR reaction termed bridging, or jumping, PCR the primer-bound sequences are originally on separate template molecules. Bridging can occur if, and only if, the templates contain a region of sequence similarity. A 3' end of synthesis in one round of synthesis that terminates in this region of similarity can prime on the other. In principle, Bridging PCR (BPCR) can detect a subpopulation of one template that terminates synthesis in the region of sequence shared by the other template. This study considers the sensitivity and noise of BPCR as a quantitative assay for backbone interruptions. Bridging synthesis is also important to some methods for computing with DNA.
Results
In this study, BPCR was tested over a 328 base pair segment of the E. coli lac operon and a signal to noise ratio (S/N) of approximately 10 was obtained under normal PCR conditions with Taq polymerase. With special precautions in the case of Taq or by using the Stoffel fragment the S/N was improved to 100, i.e. 1 part of cut input DNA yielded the same output as 100 parts of intact input DNA.
Conclusions
In the E. coli lac operator region studied here, depending on details of protocol, between 3 and 30% per kilobase of final PCR product resulted from bridging. Other systems are expected to differ in the proportion of product that is bridged consequent to PCR protocol and the sequence analyzed. In many cases physical bridging during PCR will have no informational consequence because the bridged templates are of identical sequence, but in a number of special cases bridging creates, or, destroys, information.
Background
Bridging, or jumping, PCR (BPCR) can convert DNA backbone interruptions anywhere in the region of shared sequence between primer binding sites into a single and unique amplified band [1]. The logic of BPCR is illustrated in Figure 1. In principle BPCR can detect backbone interruptions anywhere within a chosen sequence region. This manuscript reports a study in which BPCR is used to detect a minority of interrupted molecules in the lac operon of Escherichia coli and describes a system to measure noise in BPCR.
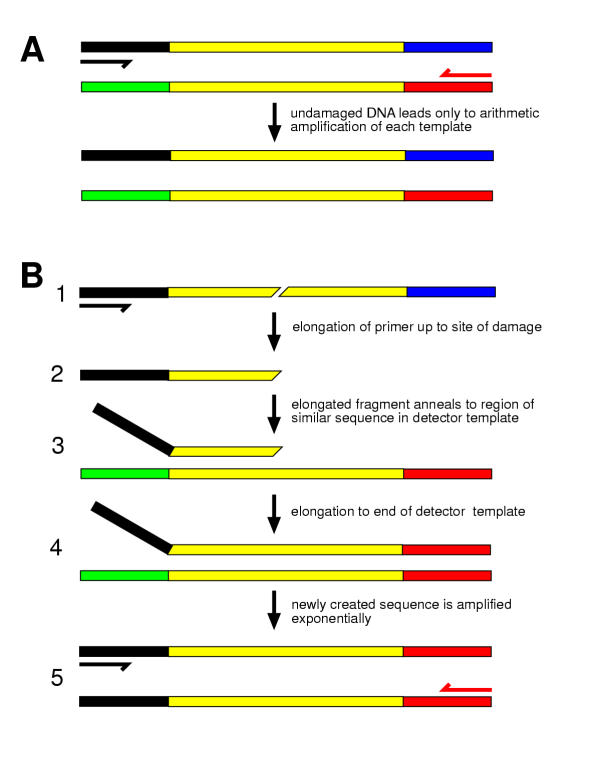
Figure 1.
The principle of Bridging PCR (BPCR) A. Only arithmetic amplification occurs when there are two templates and two primers but only one primer is complementary to each template. B. Consider the case in which two templates share a central region of similar sequence (shown in yellow), flanked by regions of different sequence (black, blue, green, red). 1) If one template is interrupted, then its primer can extend only up to the point of interruption. 2) Synthesis up to the break creates a new 3' end at the site of the interruption. 3) On the next round of PCR the new 3' end created by the interruption anneals to the second template is in the region of shared (yellow) sequence and, 4) primes synthesis to the end of the second template. 5) Extension to the end of the second template creates a recombinant molecule with both primer binding sites. This new molecule is exponentially amplified in subsequent rounds of normal PCR.
Results
Signal and noise in bridging PCR
Bridging PCR (BPCR) is a combination of two processes, a recombination between two template sequences and an amplification of the recombinant template. Two parental sequences share a homologous region (a region in which the sequence is identical) and diverge in the nonhomologous flanking sequences. One side of the nonhomologous flanking sequence of each of the two "parental" molecules contains a different PCR primer-binding site. Neither parental template has both primer-binding sites. Therefore a "chain reaction" cannot occur and only linear amplification is expected.
However, during the course of cycling, recombination can occur between the homologous regions via a copy choice mechanism (illustrated in Figure 1). A 3' end of one template sequence, if it is within the homologous region, can anneal to the other template. During the next extension cycle the invading strand will grow from its 3' end, and newly synthesized DNA will cross the region of sequence similarity and continue into sequences unique to the second template, including the second template's primer binding site. The result is a recombinant sequence, identical to the product of in vivo homologous recombination, with one flank from the original strand and the other flank from the second template. Because the two primer sites are now in cis, the recombinant sequence can be amplified exponentially.
If the 3' end of either template is not within the region of similar sequence, then no 3' end will hybridize to the other template and synthesis does not lead to recombination. The creation of recombinant sequences during PCR reflects the presence of templates whose 3' ends are within the region of sequence similarity. BPCR ought to be utilizable to assay the proportion of input templates with 3' ends in the region of sequence similarity. However, the reaction itself can produce 3' ends which then lead to recombinant templates. For maximum sensitivity and S/N ratio, the PCR reaction itself should not produce ends at any sites that did not exist in the input DNA. PCR itself generates new 3'ends [2,3]. In BPCR new ends yield recombinant products, even if the initial template was completely intact.
BPCR between plasmid pBS and Phagescript
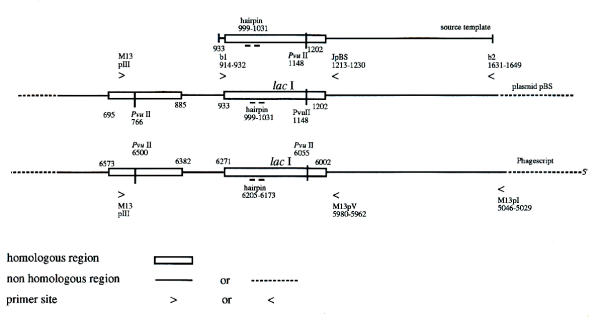
Figure 2 shows the map and sequence of templates and primer sites used in BPCR between a portion of plasmid pBS and Phagescript, a derivative of the ssDNA M13 phage. This pair of substrates meets the requirement for BPCR substrates because the region of shared sequence, the lac operator and the lac operon, are flanked by sequences that are different between the two vectors. The lac operon is one of the most frequently used genes for the study of in vivo mutagenesis [4-7].
Figure 2.
Maps and primer locations Maps of the templates and of the primer locations for bridging PCR between plasmid pBS(-) and phagescript particles. The base numbers (the numbers of on the maps) are from the published sequences by the supplier (Stratagene). The DNA of pBS(-) is double stranded. The sequences for the primers whose positions are shown in Figure 2 are given in Table 1.
Determination of signal to noise (S/N)
Two samples with known quantities of templates were prepared: one sample contained the pBS (-) sequence template with a cut in the homologous region created by PvuII digestion (cut template) and the intact ssDNA Phagescript DNA (reference template). In the other sample both the pBS (-) (intact template) and the Phagescript DNA (reference template) were intact. The molar ratio of reference template (ssDNA) to either intact template (dsDNA) or to cut template (dsDNA, considering only one of the two fragments) was about 10.
The concentration of pBS (-) sequence template, either cut or intact, was adjusted to be about 0.1 nM and to be equal in the two samples. The equality was confirmed by a 20 thermal cycle conventional PCR with primers b1 and M13 revI. The PCR outputs from serial dilutions (10 fold dilution between each step) were similar between the two samples (data not shown).
Each of the two samples was serially diluted into BPCR reactions, the dilution involving only the cut and intact template, with everything else unchanged. The highest concentration of template was about 1014 molecules/μl. Typically, dilution steps were 10 fold each and covered a million fold range. The PCRs were run in parallel (same concentrations, same volume and running on the same thermal block at the same time). After the BPCR, output of recombinant DNA was assayed by electrophoresis. For each dilution one can obtain the concentration of output [R] as a function of [Ts], the concentration of template (cut or intact depending on the sample). For each sample one defines [Ts]off as a cut-off concentration below which [R] is no longer saturated. The signal to noise is the ratio between [Ts]off in the two samples:
![]()
For example, if the cut-off concentration of the cut sample is 100 times less than the intact cut-off concentration then the S/N is 100.
Pre-annealing and pre-extension
A pre-annealing and pre-extension regime improved the S/N. In this regime, templates in the reaction tube with 1 × PCR buffer and dNTP's were heated to 90°C for 10 minutes then gradually cooled (by approximately 1°C/minute) to 72°C.
Pre-extension was carried out by holding the temperature at 72°C and adding polymerase to the pre-annealing tube. The temperature was held at 72°C for three more minutes before it was dropped to room temperature. The rationale for pre-extension [8] was that cut template molecules that are annealed to reference molecules should be extended by the polymerase to become full length. There are no primers present in the pre-extension cycle and therefore this cycle does not generate the class of noise due to incomplete extension from normal primers.
Immediately following pre-extension, i.e. without storage at 4°C, the samples were serially diluted and PCR amplified with the BPCR primers.
BPCR with Taq polymerase yielded S/N of approximately 10
Templates were prepared by 25 cycles of conventional PCR with Taq polymerase [9] using primer b1 and Jp on template plasmid pBS(-). Phagescript phage particles, rather than purified DNA prepared from the particles by phenol extraction, were used as the reference template [10]. The BPCR primer pair was b1 and M13 pI (see figure 2 for primer locations) which would give a 1261 bp recombination product. The S/N of BPCR with Taq polymerase was approximately 10 (see figure 3, lanes 1–5). This result is consistent with the report that Taq polymerase's nuclease activity [2,11] cuts in the lac operator [3]. The lac operator cut site is within the homologous region in this system. BPCR was conducted for 30 cycles with Taq polymerase.
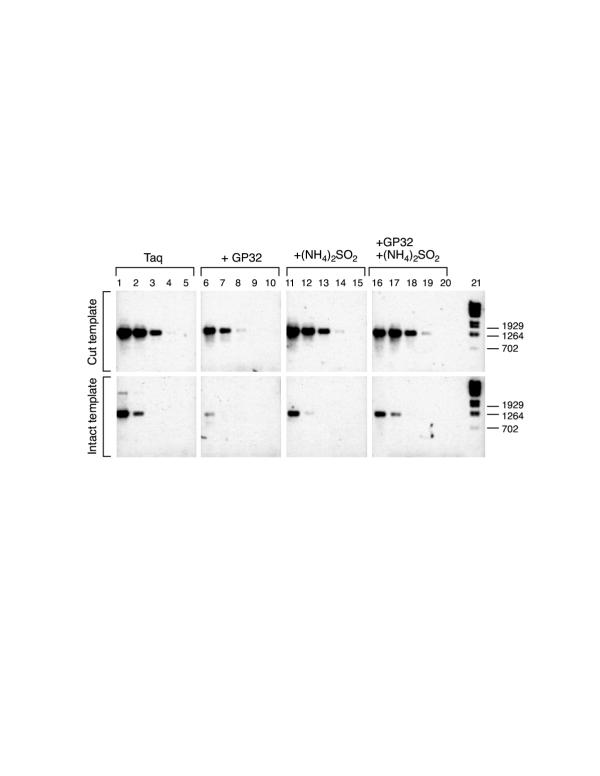
Figure 3.
Taq polymerase alone yields a S/N of 10 but can be improved Bridging PCR with Taq polymerase and the improvement of the S/N by addition of gp32 and (NH4)2SO4. The cut or intact templates was prepared by PCR with Taq polymerase and primers b1 and Jp on double-stranded plasmid pBS(-). BPCR was carried out with Taq polymerase and primers b1 and M13 pI. (See Figure 2 for location of primers and Table 1 for their sequence.) The other template was Phagescript phage particles. The 1 × PCR buffer for lanes 1–10 was 50 mM KCl, 10 mM Tris-HCl (pH 8.8 at 25°C), 1.5 mM MgCl2. The 1 × PCR buffer with (NH4)2SO4, (lanes 11 to 20) was 10 mM KCl, 20 mM Tris-HCl (pH 8.8 at 25°C), 2.0 mM MgSO4, 10 mM (NH4)2SO4, 0.1% Triton X-100 and 0.1 mg/ml bovine serum albumin. The ethidium bromide fluorescence image of the 1.0% agarose electrophoresis gel was photographed. The photograph was digitized and processed with Scion Image Adobe Photoshop software on a Macintosh computer. The image intensity was inverted.
Each lane has two rows which are labeled as cut, i.e. PvuII digested (upper row), or intact (lower row). The BPCR product bands are about 1261 bp, as expected from the template sequences (see figure 2). Lanes 1 to 5 show BPCR with Taq polymerase and no other additions. Lanes 6 to 10 show the enhancement of the S/N of BPCR by the addition of 0.01% gp32. Lanes 11 to 15, show the enhancement of BPCR by the addition of 10 mM (NH4)2SO4. Lanes 16 to 20, show BPCR with gp32 and (NH4)2SO4. The addition of both gp32 and (NH4)2SO4 did not improve the S/N. Each lane has two samples separately loaded into the two separate loading wells, one on each row. In the figure, the upper row is of source template cut with PvuII, and the lower DNA sample is of the same source template intact (without PvuII cut). The source template concentrations in the two samples were equal. Lanes 2, 3, 4 and 5 are serial template dilutions of lane 1. Lanes 7, 8, 9 and 10 are serial template dilutions of lane 6. Lanes 12, 13, 14 and 15 are serial template dilutions of lane 11. Lanes 17, 18, 19 and 20 are serial template dilutions of lane 16. Lane 21 contains DNA size markers made from Lambda DNA, BstE II digested, 125 ng. The source template concentrations of lanes 1, 6, 11 and 17 are adjusted to equal to 1 × 10-17 mol/μl PCR solution.
Several variations of BPCR with Taq polymerase were carried out (data not shown), including: 1) cut and intact templates were prepared with primers b1 and b2, 2) purified ssDNA instead of phage particles was used as a reference template and 3) cut and intact templates were prepared with the Stoffel fragment of Taq polymerase. The S/N was about 10 in these variations. Increase in the number of PCR cycles and/or a longer extension time (5 minutes) were also tested and found to be not acceptable because non-specific output, in the form of multiple gel bands or smearing gel lanes, frequently appeared. Combining Taq polymerase with a small quantity of Pfu polymerase [12] had little effect on the S/N.
Additives that improved Taq polymerase S/N to approximately 100
Some additives were found to enhance the S/N ratio. Among the additives tested, gp32 protein [13] or (NH4)2SO4 improved the S/N most efficiently and the S/N reached 100 (See figure 3. Compare cut vs intact rows in lanes 6–10 and 11–15). Combining gp32 and (NH4)2SO4 did not further improve S/N (See figure 3. Compare cut vs intact rows in lanes 16–20.).
The Stoffel fragment of Taq polymerase gave a S/N of approximately 100
The Stoffel fragment of Taq polymerase is a truncated portion of the enzyme which retains polymerase activity but lacks the 5'->3' exonuclease activity of Taq polymerase [14]. The Stoffel fragment does not cut at the lac operator as does Taq polymerase [3]. It might therefore be a good choice of polymerase for jumping PCR. Consistent with the expectation, results of using the Stoffel fragment, without additives such as gp32, (NH4)2SO4 or detergent, gave a S/N of 100 (See figure 4, compare intact lanes 3, 4, 5, 6 to cut lanes 7,8,9,10). Addition of any of the above additives did not further improve the S/N. The source templates used were also prepared by standard PCR with the Stoffel fragment and the primer pair b1 and Jp on pBS (see figure 2).
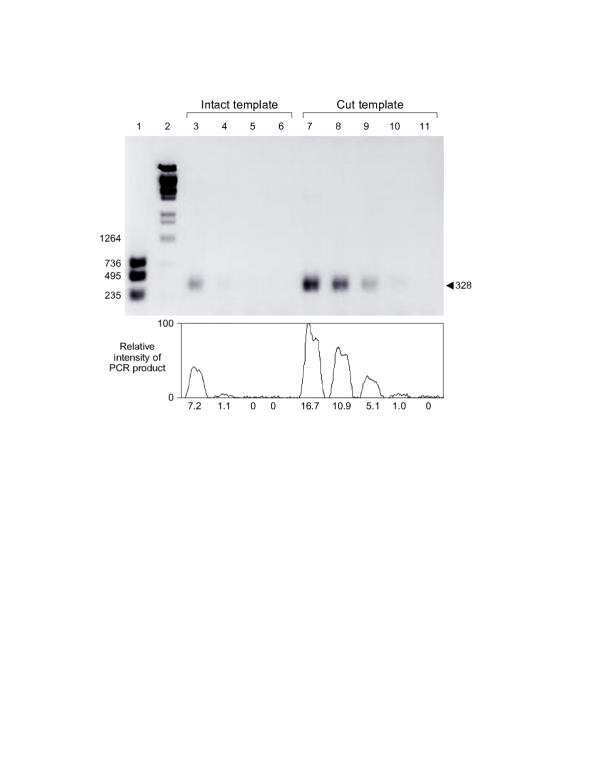
Figure 4.
Stoffel fragment of Taq yields a S/N of 100 The Stoffel fragment of Taq polymerase was used in BPCR. The gel was 1% agarose containing ethidium bromide. The fluorescent image of the gel was acquired and processed with a Macintosh computer, a video camera and the software Scion Image 1.59 (from NIH). The gel image intensity was inverted, and the background was subtracted (horizontal 1D). The band intensity of the 328 bp PCR product was recorded. The intensity profile curve of the 328 bp band is aligned and plotted below the gel lanes. The relative area under each peak is given below the peak. Area values are equalized to Lane 10, whose area value is set as 1.0. Lane 1, DNA size markers: a mixture of three separate PCR products: 235 bp (with primers b1 and M13 revI and template pBS(-)), 495 bp (primers M13 pIII and M13 revI and template Phagescript) and 736 bp (primers b1 and b2 and template pBS (-)). Lanes 2, size markers of Lambda DNA, BstE II-digested, 75 ng/lane. Lanes 3, 4, 5 and 6, PCR with intact source template. Each lane had 5 μl PCR product. Lanes 7, 8, 9, 10 and 11, PCR with source template cut with PvuII. Each lane had 5 μl PCR product. The concentrations of source template of lanes 3 and 7 are adjusted to be equal, 1 × 10-16 mol/μl PCR solution. Lanes 4, 5 and 6 are serial template dilutions of lane 3. Lanes 8, 9, 10 and 11 are serial template dilutions of lane 7.
BPCR was carried out with the Stoffel fragment of Taq polymerase and primers b1 and M13 pV. (See Figure 2 for location of primers and Table 1 for their sequence.) The cut or intact template was made by PCR with the Stoffel fragment, primers b1 and Jp and a plasmid preparation of pBS(-). The other template (reference template) was ssDNA from Phagescript particles. The 1 × PCR buffer contains 10 mM KCl, 10 mM Tris-HCl (pH 8.3 at 25°C) and 2.5 mM MgCl2.
BPCR primers b1 and M13 pV, produced the expected 328 bp recombinant sequence. In this size range the Stoffel fragment of Taq polymerase yielded a better S/N than Taq polymerase. However, the Stoffel fragment had low output in BPCR between primers b1 and M13 pI. This is consistent with the manufacturer's description that the Stoffel fragment is not optimal for the amplification of sequences longer than 1 kb.
Vent polymerase lacking nuclease activity [15] is also expected to give a low noise level. However, it yielded insufficient output (data not shown).
Discussion
Bridging, or jumping, PCR (BPCR) was discovered during the analysis of ancient DNA [1]. In reconstruction experiments new alleles of lysozyme genes were generated in vitro, during a normal PCR reaction, and, since damage to DNA promotes bridging, it was suggested that BPCR could be developed as an assay for DNA damage. Since its fortuitous discovery, BPCR has been used to make pools as part of algorithms for the generation and selection of new alleles [16,17], as well for the assembly of genes [18,19] and replicons [19] from oligonucleotides. BPCR has the potential to be developed into a method for the detection of nucleotide backbone interruptions or other factors that impede 3' extension by polymerase.
Certain algorithms for "Computing with DNA"[20] utilize bridging synthesis[21,22]. Noise in the context of computing with DNA could follow bridging between unintended sequences via the 3' end of a partially-extended primer. Bridging errors in the computation could limit the size of problem that can be solved experimentally using algorithms that involve bridging synthesis. Forensic DNA fingerprinting can be carried out via PCR and analysis of tandem repeats (e.g. [23]). Forensic samples may contain damaged DNA [24] and it is seems likely that they are thereby prone to BPCR between repeated sequences [8] leading to artifacts.
Sources of noise in BPCR
PCR itself creates DNA damage and aberrant molecules. Taq polymerase harbors nuclease activities [2,3]. Sites of stem-loop structure are particularly vulnerable to both degradation and to polymerase stalling [25-27] both of which generate noise in BPCR. Gp32, an additive that increased S/N for Taq, is a single stranded binding protein that reduces the stem-loop structures [13] associated with both cleavage and stalling by polymerases [25][2,3,11,28]. Templates are damaged by thermal cycling [29] even in the absence of enzyme.
The buffer for Taq polymerase including (NH4)2SO2 and for the Stoffel fragment had a low KCl concentration (10 mM). Since the stem-loop structure of the template is less likely to form at low KCl, this low concentration could also contribute to the increase of S/N. Because the Taq and Stoffel polymerase both reached the same optimum S/N of 100, it seems most likely that template damage via heating-cooling cycles as found by Gustafson et al. [29] is limiting S/N in this system.
Applications and limitations of bridging PCR as an assay for DNA damage
It is a basic aspect of materials science in any system to study discontinuities. This is especially the case in materials that store information. Several methods are available to assay discontinuities in DNA, and for many applications, one of the several current methods may suffice [30][31-36]. DNA backbone interruptions are key intermediates in genetic metabolism [37][38][39][40][41,42][43]. Chemotherapy [44], ionizing radiation [45] and dietary mutagens [46] also cause distributed DNA strand interruptions [47]. Chemical modifications [48-50][51] need not break the DNA backbone to impede the progress of DNA polymerase. Other modifications can be converted into backbone interruptions. For example, it has been proposed that alkali-sensitive ribonucleotides in DNA are a source of mutations [10].
It is likely that the presently attained S/N of 100 for BPCR is barely adequate for in vivo studies. Further improvement of the S/N by another factor of 10 or 100 would give greater assurance. Even with the achieved S/N of 100, we are faced with a dilemma because of the lack of other techniques that can detect a defect in less than 1% of the input DNA over a long region, 328 bp in this study. Background breaks at any site in the region contribute to noise, therefore the background breakage at any given site is apparently less than one in 30,000.
Conclusions
Break-copy recombination [52][53] almost inevitably occurs during PCR; it has been called variously: bridging PCR, jumping PCR, recombinogenic PCR, PCR sewing, gene shuffling, and sexual PCR. Sometimes this process is a 'bug' in the system leading to artifacts; sometimes it is a 'feature' leading to diverse libraries of products. Bridging PCR is to a certain extent- ca 3%/kb- found in this work, unavoidable in PCR. With a proper configuration of markers, BPCR can be used as a sensitive method for detecting discontinuities in DNA. In another context, fidelity of bridging synthesis is important for computing with DNA. For assay and computing applications it is necessary to consider noise in BPCR. In this work, BPCR was studied over a 328 base segment of the lac operon, and the noise level was measured as approximately 10% when no special precautions were taken with Taq polymerase. With buffer modifications or the addition of T4 gp32 the S/N of Taq was improved to 1%. The Stoffel fragment yielded a S/N of 1% without modifications of the standard protocol but was not further improved by buffer modifications or gp32. In this system with normal PCR using Taq polymerase, the level of bridging that occured was ca 30% per kilobase of amplifed sequence. It must be cautioned that our results have been obtained in only one system with respect to the test sequence and the primers. Differences of the sequence and of reaction conditions are expected to alter the proportion of bridging that occurs in any particular system. With respect to sequence, it is anticipated that regions of potential secondary structure or other sites that are targets for the nuclease activity of Taq may become hotspots for "crossing over" during PCR.
Materials and Methods
The sources of materials used in this study: Phagescript and pBS(-) plasmid were from Stratagene; Taq polymerase and the Stoffel fragment of Taq were from Perkin Elmer Cetus; gp32 was from Pharmacia; BstE II-digested Lambda DNA was from New England Biolabs. PvuII digestion was carried out according to the manufacturer's directions (New England Biolabs), and completeness was assayed by gel electrophoresis. Single stranded phage DNA was prepared from phage particle suspension with phenol extraction [54]. Phenol was tris-equilibrated at pH 8.0 (Sigma). The Bridging PCR reaction solution contained 0.5 mM dNTP's, 70 nM of each primer, 2.5 units/100 μl of polymerase in 1× buffer. (NH4)2SO4 at 10 mM was used in some reactions (see Figure 3) as was Triton X-100 at 0.1%. When used (see Figure 3), gp32 was 0.01% (wt/vol).
The PCR thermal cycles were as follows: An initial 3 minutes at 94°C followed by 30 cycles, each cycle consisting of 30 seconds at 94°C, 30 seconds at 55°C and 2 minutes at 72°C.
PCR was "hot start", i.e., the polymerase mixed in 10% of the final reaction volume in 1 × buffer was added to the rest of the mix during the initial denaturing step when the sample temperature was already at 94°C. Hot start and cycling was carried out on a Perkin Elmer Gene Amp 9600. PCR products were visualized by ethidium bromide staining and fluorescence after electrophoresis through 1.0% agarose gel.
Primer sequences used in this study are given in Table 1 and primer locations are shown in Figure 2.
Table 1.
Primer sequences
| Primer name | 5'- sequence -3' |
| Jp | GCGAGTCAGTGAGCGAGG |
| b1 | GTCGACCTGCAGGCATGCA |
| b2 | CCCGACACACGTGCTTGG |
| M13 pI | GTCTGTGCCACGTATTCT |
| M13 pV | GCTGTTGCCCGTCTCGCTG |
| M13revI | CTGGCACGACAGGTTTCCCGA |
| PIII | GCAACTGTTGGGAAGGGCGATCG |
Authors' contributions
Author 1, SL, carried out the experiments and drafted part of the manuscript. Author 2, DST, conceived of the study, with author 1 created the experimental design, and wrote parts of the manuscript. Author 3, AJL, made intellectual contributions.
Acknowledgments
Acknowledgements
Thanks to Fiona Doetsch, Joshua Lederberg, Aguan Wei, Peter Kaplan and Noel Goddard, for insight and encouragement. FD immensely improved presentation of the figures and Michael Badmeav helped with primer analysis. This work was partially supported by a grant from the Sloan Foundation to Marcelo Magnasco and David Thaler. DT is a Senior Scholar of the Ellison Medical Foundation.
Contributor Information
Shumo Liu, Email: shumo@research.nj.nec.com.
David S Thaler, Email: thalerd@mail.rockefeller.edu.
Albert Libchaber, Email: libchbr@mail.rockefeller.edu.
References
- Paabo S, Irwin DM, Wilson AC. DNA damage promotes jumping between templates during enzymatic amplification. J Biol Chem. 1990;265:4718–4721. [PubMed] [Google Scholar]
- Lyamichev V, Brow MD, Dahlberg JE. Structure-specific endonucleolytic cleavage of nucleic acids by eubacterial DNA polymerases. Science. 1993;260:778–783. doi: 10.1126/science.7683443. [DOI] [PubMed] [Google Scholar]
- Tombline G, Bellizzi D, Sgaramella V. Heterogeneity of primer extension products in asymmetric PCR is due both to cleavage by a structure-specific exo/endonuclease activity of DNA polymerases and to premature stops. Proc Natl Acad Sci. 1996;93:2724–2728. doi: 10.1073/pnas.93.7.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook A, Lederberg J. Recombination studies of lactose nonfermenting mutants in Escherichia coli K-12. Genetics. 1962;47:1335–1353. doi: 10.1093/genetics/47.10.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen J, Vijg J. A selective system for lacZ- phage using a galactose-sensitive E. coli host. Biotechniques. 1993;14:326–7. [PubMed] [Google Scholar]
- Cairns J, Overbaugh J, Miller S. The origin of mutants. Nature. 1988;335:142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- Cupples CG, et al. A set of lacZ mutations in Escherichia coli that allow rapid detection of specific frameshift mutations. Genetics. 1990;125:275–280. doi: 10.1093/genetics/125.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan DJ. DNA regeneration in the polymerase chain reaction. J Theor Biol. 1993;162:289–307. doi: 10.1006/jtbi.1993.1088. [DOI] [PubMed] [Google Scholar]
- Abramson RD. Thermostable DNA polymerases, In: J.J. Sninsky, editor. in PCR Strategies. Academic Press: New York; 1995. pp. 39–57. [Google Scholar]
- Thaler DS, Tombline G, Zahn K. Short patch reverse transcription in Escherichia coli. Genetics. 1995;140:909–915. doi: 10.1093/genetics/140.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland PM, et al. Detection of specific polymerase chain reaction product by utilizing the 5'–3' exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes WM. PCR amplification of up to 35-kb DNA with high fidelity and high yield from lambda bacteriophage templates. Proc Natl Acad Sci USA. 1994;91:2216–2220. doi: 10.1073/pnas.91.6.2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapley R. Enhancing PCR amplification and sequencing using DNA-binding proteins. Mol Biotechnol. 1994;2:295–298. doi: 10.1007/BF02745882. [DOI] [PubMed] [Google Scholar]
- Lawyer FC, et al. High-level expression, purification, and enzymatic characterization of full-length Thermus aquaticus DNA polymerase and a truncated form deficient in 5' to 3' exonuclease activity. PCR Methods & Applic. 1993;2:275–287. doi: 10.1101/gr.2.4.275. [DOI] [PubMed] [Google Scholar]
- Kong H, Kucera RB, Jack WE. Characterization of a DNA polymerase from the hyperthermophile archaea Thermococcus litoralis. Vent DNA polymerase, steady state kinetics, thermal stability, processivity, strand displacement, and exonuclease activities. J Biol Chem. 1993;268:1965–1975. [PubMed] [Google Scholar]
- Horton RM, et al. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- Stemmer WP. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc Natl Acad Sci USA. 1994;91:10747–1075. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman K, et al. Polymerase chain reaction-mediated gene synthesis: synthesis of a gene coding for isozyme c of horseradish peroxidase. Proc Natl Acad Sci USA. 1991;88:4084–4088. doi: 10.1073/pnas.88.10.4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer WP, et al. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49–53. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- Adelman LM. Molecular computation of solutions to combinatorial problems. Science. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- Kaplan PD, et al. Parallel overlap extension for construction of computational DNA libraries. J Theor Biol. 1997;188:333–341. doi: 10.1006/jtbi.1997.0475. [DOI] [PubMed] [Google Scholar]
- Ouyang Q, et al. DNA solution of the maximal clique problem. Science. 1997;278:446–9. doi: 10.1126/science.278.5337.446. [DOI] [PubMed] [Google Scholar]
- Nellemann LJ, Moller A, Morling N. PCR typing of DNA fragments of the short tandem repeat (STR) system HUMTH01 in Danes and Greenland Eskimos. Forensic Science International. 1994;68:45–51. doi: 10.1016/0379-0738(94)90378-6. [DOI] [PubMed] [Google Scholar]
- Edwards H. More on DNA typing dispute. Nature. 1994;373:p 98.. doi: 10.1038/373098a0. [DOI] [PubMed] [Google Scholar]
- Gefter ML, Sherman LA. The role of DNA structure in DNA replication, In: K. Javaherian, editor. in The organization and expression of the eukaryotic genome. Academic Press: London; 1977. pp. 233–248. [Google Scholar]
- Olsen DB, Eckstein F. Incomplete primer extension during in vitro DNA amplification catalyzed by Taq polymerase; exploitation for DNA sequencing. Nucleic Acids Res. 1989;17:9613–9620. doi: 10.1093/nar/17.23.9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ. When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell. 1988;53:679–686. doi: 10.1016/0092-8674(88)90086-4. [DOI] [PubMed] [Google Scholar]
- Longley MJ, Bennett SE, Mosbaugh DW. Characterization of the 5' to 3' exonuclease associated with Thermus aquaticus DNA polymerase. Nucleic Acids Res. 1990;18:7317–1722. doi: 10.1093/nar/18.24.7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson CE, Alm RA, Trust TJ. Effect of heat denaturation of target DNA on the PCR amplification. Gene. 1993;123:241–4. doi: 10.1016/0378-1119(93)90130-U. [DOI] [PubMed] [Google Scholar]
- Xu F, Petes TD. Fine-structure mapping of meisosis-specific double-strand DNA breaks at a recombination hotspot associated with an insertion of telomeric sequences upstream of the HIS4 locus in yeast. Genetics. 1996;143:1115–1125. doi: 10.1093/genetics/143.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlissel M, et al. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5'-phosphorylated, RAG-dependent, and cell cycle regulated. Genes & Devel. 1993;7:2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- Yakes FM, Y Chen, Van Houten B. PCR-based assays for the detection and quantitation of DNA damage and repair, In: G.P. Pfeifer, editor. in Technologies for detection of DNA damage and mutations. Plenum Press: New York; 1996. pp. 171–184. [Google Scholar]
- Vinograd J, Lebowitz J, Watson R. Early and late helix-coil transitions in closed circular DNA The number of superhelical turns in polymoma DNA. J Mol Biol. 1968;33:173–197. doi: 10.1016/0022-2836(68)90287-8. [DOI] [PubMed] [Google Scholar]
- Grossman LI, Watson R, Vinograd J. The presence of ribonucleotides in mature closed-circular mitochondrial DNA. Proc Natl Acad Sci USA. 1973;70:3339–3343. doi: 10.1073/pnas.70.12.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Sakai T, Hotchi M. Histochemical demonstration of DNA double strand breaks by in situ 3'- tailing reaction in apoptotic endometrium. Biotech Histochem. 1995;70:33–9. doi: 10.3109/10520299509108314. [DOI] [PubMed] [Google Scholar]
- Komura J, Riggs AD. Terminal transferase-dependent PCR: a versatile and sensitive method for in vivo footprinting and detection of DNA adducts. Nucleic Acids Res. 1998;26:1807–11. doi: 10.1093/nar/26.7.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler DS, Stahl FW. DNA double-chain breaks in recombination of phage lambda and of yeast. Ann Rev Genet. 1988;22:169–197. doi: 10.1146/annurev.ge.22.120188.001125. [DOI] [PubMed] [Google Scholar]
- Burgin ABJ, Nash HA. Suicide substrates reveal properties of the homology-dependent steps during integrative recombination of bacteriophage lambda. Curr Biol. 1995;5:1312–1321. doi: 10.1016/s0960-9822(95)00258-2. [DOI] [PubMed] [Google Scholar]
- Ramsden DA, Gellert M. Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes & Devel. 1995;9:2409–2420. doi: 10.1101/gad.9.19.2409. [DOI] [PubMed] [Google Scholar]
- Cooper DL, Lahue RS, Modrich P. Methyl-directed mismatch repair is bidirectional. J Biol Chem. 1993;268:11823–11829. [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W. DNA Repair and Mutagenesis. Washington DC: Am Sci Microbiol Press. 1995. p. 698.
- Mosig G, et al. Multiple initiation mechanisms adapt phage T4 DNA replication to physiological changes during T4's development. FEMS Microbiol Revs. 1995;17:83–98. doi: 10.1016/0168-6445(94)00069-7. [DOI] [PubMed] [Google Scholar]
- Zechiedrich EL, Cozzarelli NR. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes & Devel. 1995;9:2859–69. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]
- Oshita F, et al. Correlation of therapeutic outcome in non-small cell lung cancer and DNA damage assayed by polymerase chain reaction in leukocytes damaged in vitro. Cancer Res. 1995;55:2334–2337. [PubMed] [Google Scholar]
- James SJ, Enger SM, Makinodan T. DNA strand breaks and DNA repair response in lymphocytes after chronic in vivo exposure to very low doses of ionizing radiation in mice. Mutat Res. 1991;249:255–263. doi: 10.1016/0027-5107(91)90152-E. [DOI] [PubMed] [Google Scholar]
- Thompson LH. Properties and Applications of Human DNA Repair Genes. Mutat Res. 1991;247:213–219. doi: 10.1016/0027-5107(91)90017-I. [DOI] [PubMed] [Google Scholar]
- Ames BN, Gold LS, Willett WC. The causes and prevention of cancer. Proc Natl Acad Sci USA. 1995;92:5258–5265. doi: 10.1073/pnas.92.12.5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holler E, Bauer R, Bernges F. Monofunctional DNA-platinum(II) adducts block frequently DNA polymerases. Nucleic Acids Res. 1992;20:2307–2312. doi: 10.1093/nar/20.9.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu AK, et al. Effect of site-specifically located mitomycin C-DNA monoadducts on in vitro DNA synthesis by DNA polymerases. Biochemistry. 1993;32:4708–4718. doi: 10.1021/bi00069a004. [DOI] [PubMed] [Google Scholar]
- Lindsley JE, Fuchs RP. Use of single-turnover kinetics to study bulky adduct bypass by T7 DNA polymerase. Biochemistry. 1994;33:764–772. doi: 10.1021/bi00169a018. [DOI] [PubMed] [Google Scholar]
- Raizis AM, Schmitt F, Jost JP. A bisulfite method of 5-methylcytosine mapping that minimizes template degradation. Analyt Biochem. 1995;226:161–166. doi: 10.1006/abio.1995.1204. [DOI] [PubMed] [Google Scholar]
- Meselson M. On the mechanism of genetic recombination between DNA molecules. J Mol Biol. 1964;9:734–745. doi: 10.1016/s0022-2836(64)80178-9. [DOI] [PubMed] [Google Scholar]
- Mosig G. Relationship of T4 replication and recombination, In: P.B. Berget, editor. in Bacteriophage T4. American Society for Microbiology: Washington D.C.; 1983. pp. 120–130. [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning a laboratory manuel. Plainview, NY: Cold Spring Harbor Press. 1989.