Abstract
Background
Twelve populations of the bacterium, Escherichia coli, adapted to a simple, glucose-limited, laboratory environment over 10,000 generations. As a consequence, these populations tended to lose functionality on alternative resources. I examined whether these populations in turn became inferior competitors in four alternative environments. These experiments are among the first to quantify and compare dimensions of the fundamental and realized niches.
Results
Three clones were isolated from each of the twelve populations after 10,000 generations of evolution. Direct competition between these clones and the ancestor in the selective environment revealed average fitness improvements of ~50%. When grown in the wells of Biolog plates, however, evolved clones grew 25% worse on average than the ancestor on a variety of different carbon sources. Next, I competed each evolved population versus the ancestor in four foreign environments (10-fold higher and lower glucose concentration, added bile salts, and dilute LB media). Surprisingly, nearly all populations were more fit than the ancestor in each foreign environment, though the margin of improvement was least in the most different environment. Most populations also evolved increased sensitivity to novobiocin.
Conclusions
Reduced functionality on numerous carbon sources suggested that the fundamental niche of twelve E. coli populations had narrowed after adapting to a specific laboratory environment. However, in spite of these results, the same populations were competitively superior in four novel environments. These findings suggest that adaptation to certain dimensions of the environment may compensate for other functional losses and apparently enhance the realized niche.
Background
"Part of the folk wisdom of evolutionary biology is that specialization leads to adaptive decay for environments outside the domain of specialization." – R. D. Holt ([1], p. 9)
As reflected in the epigraph, evolutionists generally assume that genetic adaptation to any particular environment is associated with the loss of fitness in dissimilar environments. Indeed, nearly all mathematical models of niche breadth – diet, physiological tolerance, life history features, and so on – assume the existence of tradeoffs which fulfill this assumption (e.g. [2-5]). Yet despite the central importance of this assumption for theories in evolutionary ecology, it remains largely untested [6]. Part of the problem lies in defining specialization in a manner that would allow this assumption to be rigorously tested. Futuyma and Moreno ([6], p. 208) recognized this problem and suggested "often specialization must lie in the eye of the beholder." I attempt to avoid this ambiguity and for this paper define specialization to be a reduction in niche breadth associated with adaptation to any particular environment.
The "niche" also poses its own challenges. Hutchinson's [7] classic definition of the niche as an "n-dimensional hypervolume" is a compelling theoretical construct that is, unfortunately, empirically overwhelming. Any two environmental variables that limit an organism's survival and ability to reproduce describe only one plane of this volume. Pragmatism demands that we limit research to a handful of variables while bearing in mind the existence of many others. To complicate matters further, the fundamental niche described by Hutchinson, which is bounded by the absolute functional capacities and tolerances of the organism, may be quite different from its actual habitat. The environmental space actually inhabited by the organism is known as the realized niche, and may be considerably smaller because of chance, biogeography, or constraints mounted by competitors or predators. The extent to which these two descriptions of the niche differ remains an open question in need of experimentation.
Nonetheless, empirical evaluation of evolutionary and ecological theory is limited less by semantics than by the complexity of most biological systems. One of the most conspicuous examples of niche specialization is the loss of vision and pigmentation among cave organisms [8]. Because the history of adaptation by cave creatures and the pressures of their new, dark environment are equally murky, however, the mechanisms by which natural selection produced blindness and colorlessness are uncertain. In other, more carefully controlled laboratory and field experiments, strong selection on particular traits often leads to adaptation, but trade-offs have proven surprisingly elusive (see, for example, [9-18]).
Given these challenges, I chose to study the niche of one of the simplest model organisms, the bacterium Escherichia coli. E. coli's rapid replication rate allow many generations to be followed, its genetics and metabolism are well characterized, and robust techniques for measuring relative fitness exist. Further, a wide variety of characters can be readily sampled to identify any losses of function that might alter niche dimensions. In this study, I quantify diet breadth during long-term evolution in a single-resource environment.
The experimental system
The design of the evolution experiment has been described elsewhere [19]. In short, 12 lines were derived from a single, strictly asexual clone of E. coli B that has been in the laboratory for several decades (see: http://myxo.css.msu.edu/ecoli/strainsource.html for more information). The strain is prototrophic (it can synthesize all components of a cell from a single energy source, such as glucose, and inorganic salts, nitrogen, etc.) and has undoubtedly undergone some general adaptation to the laboratory environment, but has not been selected under any specific conditions like the evolution protocol (prolonged serial transfer in minimal medium). A spontaneous mutant of the ancestor capable of using arabinose (Ara+) was used to found six of the replicates, whereas the other six were founded with the Ara- ancestor. This trait can be used to distinguish between populations on indicator plates and is neutral in the selective environment [19]. The populations are maintained by the daily transfer of 0.1 ml of culture into 9.9 ml of fresh Davis minimal media supplemented with 25 μg/ml of glucose (DM25). These conditions allow roughly 5 × 107 cells/ml at stationary phase. Every 500 generations (75 days), samples of each population were stored in a glycerol suspension at -80°C.
The bacterial populations that are the focus of this study have been well studied [19-26]. A brief review of the relevant observations follows. First, each of these lines has adapted to growth in a serially diluted, or seasonal, environment of glucose-limited minimal media [19-22]. The dynamics of adaptation are well known: after a period of rapid adaptation during the first 1,000 generations, the rate of improvement has subsequently slowed to almost one-thirtieth of the initial rate [20,27]. Moreover, relatively few mutations generated this early, rapid adaptation [19,20,24]. Thus, these populations have experienced two types of dynamics: one of rapid adaptation followed by one of much slower improvement. Nonetheless, this gradual adaptation contributed significantly to the mean fitness relative to the ancestor of ~1.7 by generation 20,000 [27].
Travisano and Lenski [22,23] demonstrated that adaptations specific to growth on glucose were partly responsible for the initial fitness improvements. They found that evolved populations tended to outcompete the ancestor on substrates that share membrane transport pathways with glucose, but were inferior on substrates that did not. To assess the longer-term consequences of these adaptations, Cooper and Lenski [27] employed Biolog™ plates, which are microtiter plates containing 95 different carbon sources and an indicator dye that reflects the amount of respiration on each substrate. Sampling three clones per population after generations 2,000, 10,000, and 20,000, the authors found that average performance on the 64 informative, foreign substrates decayed significantly over time. The primary objective of this study was to deduce which of two population genetic mechanisms, antagonistic pleiotropy and mutation accumulation, was more responsible for specialization (results were strongly in favor of antagonistic pleiotropy). However, this experiment also approximated the evolution of one aspect of the fundamental niche, diet breadth, in these populations.
Cooper et al. [28] continued this line of research by studying the evolution of thermal niche in these populations, which are maintained at a constant 37°C. Maximum growth rate, which has been shown to be the primary determinant of fitness in this system [21], was measured across a range of temperatures (20–42°C) over evolutionary time. The authors found a surprising amount of correlated adaptation to "moderate" temperatures surrounding 37°C, suggesting that selection at this temperature had favored mutations that also improved performance at (presumably) physiologically similar temperatures. However, mean performance at "extreme" temperatures (20°C, >40°C) decreased over evolutionary time, with several evolved clones failing to grow at these temperatures. Judging the effect of this laboratory evolution on the thermal niche of these bacterial populations therefore depends on one's perspective. Mean growth at extreme temperatures tended to decrease, but mean growth rates across a range of moderate temperatures actually increased over time.
In summary, the functionality of these evolved E. coli populations under certain conditions has been compromised by long-term adaptation to a single-resource, single-temperature environment. Yet most of these experiments have approximated the fundamental niche by examining each clone in isolation from its ancestor and other members of the population. By comparison, measuring the realized niche requires a head-to-head test of fitness with the ancestor. After a survey of diet breadth using Biolog plates, this study focuses on a series of direct competitions between evolved isolates from 10,000 generations and the ancestor in four foreign environments. Assays of individual functions and whole-organism fitness are clearly quite different, but it is the latter, poorly measured property that ultimately determines both evolutionary success and the tangible dimensions of the niche.
Experimental design
These experiments focus only on isolates from the populations at 10,000 generations and their common ancestors. Three clones from each population were picked randomly from platings of these cultures and frozen separately; each experiment therefore compares 36 clones (12 populations × 3 clones) with the two ancestors (Ara- and Ara+). I studied multiple clones from each population because there are within-population polymorphisms that may be associated with functional differences [25,28-30].
The environments are described as follows. First, I both increased and decreased the concentration of glucose (25 μg/ml of glucose is standard) in the evolutionary medium in order to test the specificity of adaptation to the environment. I hypothesized that adaptation to a particular concentration of glucose in the medium might reduce competitive ability at either lower (2.5 μg/ml) or higher (250 μg/ml) concentrations, perhaps because of tradeoffs between resource affinity and maximum growth rate. Next, I added bile salts, which are organic detergents that are lethal to most non-enteric bacteria, to the standard experimental medium. Tolerance to bile salts is a characteristic that has been used to define enteric bacteria over the years, which is shared by the ancestor in this experiment. In E. coli, it has been established that bile acids may traverse the outer membrane through the OmpF porin, which is the same porin through which glucose passes [31]. If selection for improved glucose transport resulted in a greater number of OmpF porins relative to the more bile-resistant, smaller-channel OmpC porin [31,32], for example, it is possible that tolerance to bile salts was compromised as the populations evolved. As a second test of the effect of selection on the outer membrane, I measured the resistance to the antibiotic novobiocin of each population. While novobiocin acts primarily as a DNA gyrase inhibitor, resistance is mediated by the concentration of phospholipids in the outer membrane, for which the cls gene (encoding cardiolipin synthase) is partly responsible [33]. Again, if selection had increased membrane permeability, sensitivity to novobiocin might have increased relative to the partially resistant ancestor.
I chose the LB (Luria broth) environment because its nutrients are so different from the selection medium, but other conditions remain the same (e.g. culture vessel, temperature, and serial transfer regime). LB media consists of tryptone, yeast extract (which is the water-soluble portion of autolyzed yeast) and salt. In short, it is a complex of nutrients largely devoid of simple carbohydrates like glucose. The LB environment was diluted with distilled water to generate comparable cell density with the standard selective environment. For each competition in a foreign environment, fitness was also simultaneously measured in the standard medium for a direct reference.
Results
Diet breadth narrows
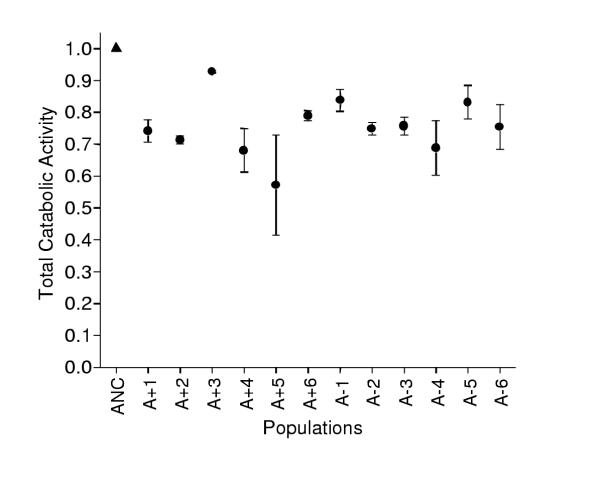
Consistent with a previous study [27], I found a significant decline in diet breadth of an average of 25% for all evolved populations (Figure 1, one sampled t-test, t = 9.49, df = 11, p < .0001). This decline in diet breadth proved to be general and not limited to a few substrates; on average, evolved populations tended to perform worse than the ancestor. There was no significant variation among populations in the sum of all catabolic functions, but there was significant variation among clones within populations (Table 1). A single "specialist" clone in population A-5 that grew less than 50% as well as the ancestor on 47 of 68 substrates caused much of this effect, but this clone was otherwise normal on glucose. Another "specialist" clone that was abnormally deficient on fewer substrates was also found in population A+4. These clones were not better or worse competitors than other clones in the evolution environment because of their reduced functionality, however (data not shown). If these two outlying clones are omitted from the nested ANOVA of diet breadth, then significant variation exists among populations (F11, 22 = 2.34, p = .043) but no longer among clones within populations (F22,34 = .957, p = .534).
Figure 1.
Effect of 10,000 generations of evolution on total catabolic function. Each point is the weighted average of performance on 68 substrates; 1.0 is the ancestral value. The evolved populations (circles) had significantly lower diet breadth (t = 9.49, df = 11, p < .0001, one-tailed) than the mean of the common ancestors (ANC, triangle). Error bars are standard errors.
Table 1.
Nested ANOVA for total catabolic activity of the 12 evolved populations.
| Source | df | MS | F | P |
| Population | 11 | 61.66 | 1.203 | .339 |
| Clone (Population) | 23 | 51.26 | 3.125 | .001 |
| Error | 35 | 16.40 |
Total catabolic activity is the sum of absorbance scores for 93 different substrates. Population and clone are random effects.
Fitness improved in foreign environments
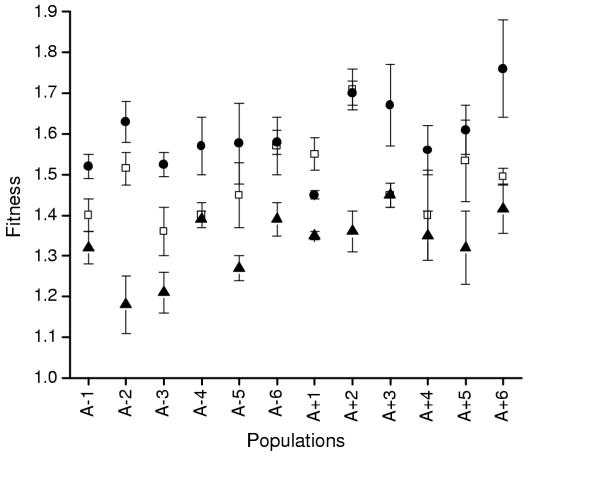
I evaluated diet breadth in a second, contrasting manner by competing clones from evolved populations directly against the ancestor in four foreign environments. In the first experiment, I measured competitive fitness versus the ancestor over a 100-fold range of glucose concentrations in the otherwise standard experimental environment. Clones were competed in two "test" environments (DM2.5 and DM250) as well as in the control environment (DM25). I found a significant effect of glucose concentration on population mean fitness (Table 2). Populations (and clones within populations) also varied significantly in their performances at different glucose concentrations, as evidenced by the significant interaction terms in the ANOVA in Table 3. Not surprisingly, populations were most fit in the DM25 environment in which they evolved (Figure 2; t-tests for paired comparisons: DM2.5 vs. DM25, ts = 5.48, df = 11, p < .001, DM25 vs. DM250, ts = 14.08, df = 11, p < .001). However, all populations were more fit than the ancestor in both DM2.5 and DM250; that is, adaptation to the DM25 environment brought about correlated improvements in fitness, rather than tradeoffs, at other glucose concentrations (Figure 2). In addition, populations were uniformly more fit in the DM2.5 environment than in the DM250 environment (paired t-test, ts = 6.37, df = 11, p < .001).
Table 2.
Nested ANOVA for relative fitness obtained for the 12 evolved populations in three different glucose concentrations
| Source | df | MS | F | P |
| Concentration | 2 | 1.187 | 35.12 | < .0001 |
| Population | 11 | .0678 | 1.967 | .098 |
| Clone (Population) | 23 | .015 | 1.042 | .439 |
| Concentration * Population | 22 | .0339 | 2.345 | .007 |
| Concentration * | 46 | .0144 | 1.684 | .015 |
| Clone (Population) | ||||
| Error | 105 | .00858 |
Table 3.
Nested ANOVA for relative fitness obtained for the 12 evolved populations in the experimental environment plus bile salts
| Source | df | MS | F | P |
| Population | 11 | .0716 | 2.118 | .062 |
| Clone (Population) | 23 | .0338 | 1.817 | .054 |
| Error | 35 | .0186 |
Figure 2.
Mean fitness (± SE) relative to the ancestor of evolved populations when measured in the experimental environment at varying concentrations of glucose. Each point is the mean of 3 random clones, each replicated twice. Open squares: 2.5 μg/ml glucose; closed circles: 25 μg/ml glucose; closed triangles: 250 μg/ml glucose.
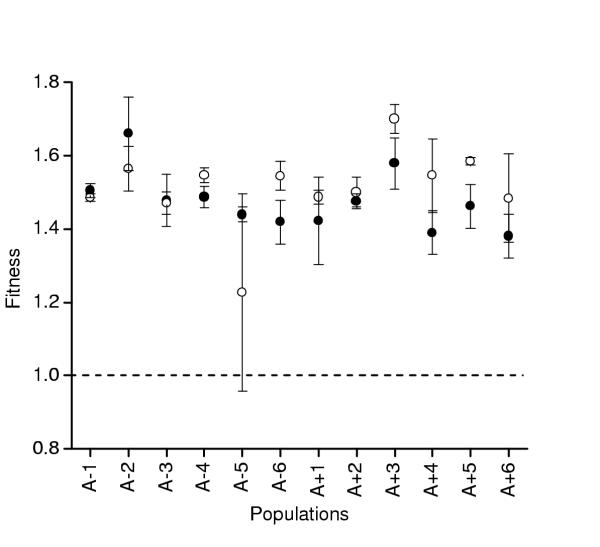
In the second experiment, I evaluated the effect of adding bile salts to the experimental medium (Figure 3). The evolved populations performed nearly equivalently with or without bile salts, relative to the ancestor (1-tailed paired ts = 1.10, df = 11, p = .138). A nested ANOVA on the fitness data in DM25 + bile salts did reveal marginally significant variation among and within populations (Table 3). A single clone from population A-5 caused nearly all of this variation within populations; this same clone was also deficient on numerous carbon sources on Biolog plates. When I omitted this clone from the nested ANOVA, there was more significant variation among populations for fitness in bile salts (F11,22 = 2.615, p = .026) but not among clones within populations (F22,34 = .718, p = .790)
Figure 3.
Mean fitness (± SE) relative to the ancestor of evolved populations when measured in the experimental environment with (closed circles) and without the addition of 1.5 g/L bile salts (open circles). Each point is the mean of 3 random clones, each replicated twice.
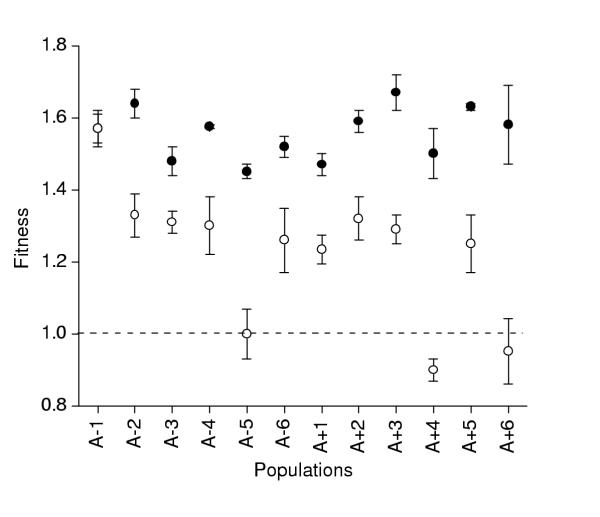
In the final experiment, I altered the competitive environment in a more pronounced manner by using a much more complex medium, LB, instead of minimal salts supplemented with glucose. I anticipated the greatest difference in fitness between this environment and the control. Indeed, mean fitness in LB was significantly lower than that in the control environment (paired t-test, ts = 12.278, df = 11, p < .001). However, it was surprising that nine of 12 populations had nevertheless increased in fitness in LB relative to the ancestor, and that the fitness of one population (A-1) was statistically indistinguishable when grown in LB or DM25 (Figure 4). On the other hand, three populations were statistically equivalent to the ancestor in LB fitness (i.e. the 95% confidence intervals of these means (not shown) did not exclude the ancestral fitness, which equals 1.0). In addition, a nested ANOVA using fitness in LB revealed highly significant variation among populations but not within populations (Table 4).
Figure 4.
Mean fitness (± SE) relative to the ancestor of evolved populations when measured in the minimal experimental environment (DM25, closed circles) and a novel, complex environment (LB, open circles). Each point is the mean of 3 random clones, each replicated twice.
Table 4.
Nested ANOVA for relative fitness obtained for the 12 evolved populations in the foreign environment, LB
| Source | df | MS | F | P |
| Population | 11 | .196 | 13.072 | <.001 |
| Clone (Pop) | 23 | .015 | .896 | .693 |
| Error | 35 | .168 |
Most populations become sensitive to novobiocin
Assays that examined the ability of each population to grow on agar plates containing novobiocin demonstrated that most populations became sensitive to this antibiotic (not shown). Nine of 12 populations were completely sensitive to the antibiotic; no colonies were observed for any of the three clones tested per population. Only one of the 12 populations (Ara+3) retained a level of resistance comparable to that of the ancestor, and two other populations (Ara-3 and Ara+5) were intermediately sensitive.
Discussion
Spiegelman and collaborators [34,35] asked bacteriophage Qβ only to "go forth and replicate" in the medium containing Qβ replicase, and numerous functions were lost. We have issued a similar injunction to these E. coli populations, except in a glucose and minimal salts medium. Following this precedent, these long-term evolving E. coli populations should have become streamlined glucose scavengers and evolved a substantially reduced niche. This was not the case, at least not nearly to the extent observed in Qβ. Only one absolute catabolic loss (D-ribose) [36] was observed in all twelve lineages, and one additional function (resistance to novobiocin) was lost in most populations. These populations performed no worse on average than the ancestor in most functions assayed, and the significant losses that were found tended to be small in absolute magnitude (not shown, see also [27]).
Nonetheless, the fundamental niche approximated by Biolog plates declined by an average of 25%, and no systematic gains in function occurred that might have compensated for other losses of function. In these populations, specialization did not involve numerous wholesale losses of function, but rather reduced performance on many substrates not used during the evolution experiment. The degree of decay varied both among and within populations for growth on different substrates (Table 1), and a substantial amount of functional decay occurred in two of the 36 clones sampled. Thus, the fundamental catabolic niche of these populations became narrower while adapting to this simple laboratory environment.
However, I arrived at a strikingly different conclusion when I measured the relative fitness, which is analogous to the realized niche, of the evolved populations in four different foreign environments. In these assays, performance was quantified by directly competing the evolved genotypes with the ancestors, and in nearly every case the evolved genotype was the superior competitor. These correlated improvements in fitness in novel environments suggest that adaptation to the laboratory environment was nonspecific and multifaceted, and seem to downplay the significance of the compromised fundamental niche.
Each of these novel environments was not entirely foreign to the evolved populations, however. Though glucose concentration was both raised and lowered tenfold, conditions remained otherwise identical to those of the evolution experiment. These populations were clearly superior at the lower concentration, which, in hindsight, likely results from their familiarity with this glucose level. Because glucose concentration in the experimental medium drops below 25 μg/ml as the population grows, competitive ability at lower concentrations was likely favored, even if only briefly [21]. Yet the mechanism behind the greater fitness gains in 2.5 μg/ml than in 250 μg/ml glucose need not be the glucose concentration itself. Vasi et al. [21] argued that selection in these lines for improved resource affinity at low concentrations was weak, which perhaps implicates some other factor in the medium (oxygen concentration, different concentrations of metabolic by-products, toxicity of increased nutrients, etc) as the cause of the lesser fitness gains in 250 μg/ml glucose.
Bile salts are an important component of the natural environment of E. coli, so much so that resistance to these compounds is often used to define enteric bacteria in microbiological assays. Bile salts are like novobiocin in that both are antibiotics that interact with the outer membrane, but their mechanisms of interaction and their effects on these populations are rather different. Most populations completely lost the ability to grow in the presence of novobiocin, which suggests an increase in membrane permeability [33]. However, no significant differences in fitness were found when bile salts were added to the selective medium, indicating that the mechanisms of increased outer membrane permeability were not detrimental to bile resistance. This may reflect an ability of E. coli to actively export bile salts that was unaffected by this experiment [31]. Thus, if changes in membrane permeability (perhaps through modified expression of genes like cls[33]) were responsible for the loss of novobiocin resistance, they apparently had specific effects that did not compromise other membrane traits like tolerance to bile salts. Experiments are underway to establish the mechanism that links adaptation, increased membrane permeability, and increased novobiocin sensitivity.
I found no systematic cost of adaptation to the simple laboratory environment when I modified the glucose concentration or added bile salts, but rather found correlated improvements in fitness that were sometimes nearly equivalent to the direct adaptation. The simple explanation is that each of these environments differed by only one factor from the minimal medium in which the populations have evolved. However, the complex LB environment is certainly different enough from minimal glucose medium that any improvements in fitness in the LB environment are surprising. One interpretation of all of the correlated adaptations in novel environments is that natural selection favored beneficial mutations that are unaffected by the nutrient medium itself. Whereas I altered the carbon source and media composition, I did not change other conditions like the speed of shaking (aeration), temperature, or the batch culture protocol. Perhaps more significantly, the populations remained sheltered from the potential onslaught of competing bacteria and predatory viruses, which could conceivably drastically alter their performance. It is likely that the 12 populations have adapted in part to several of components of the feast-and-famine, minimal, laboratory environment, and changing only one of them may not affect the superiority of other genetic adaptations. Thus, detecting "adaptive decay" may require more dramatic modifications to the environment.
Evidently, these evolving populations, by acquiring mutations that were beneficial under several environmental conditions, expanded the niche they could potentially realize. More specifically, mutations that improved fitness in 25 μg/ml glucose may have also improved fitness in 250 μg/ml glucose (because of the common resource) as well as in dilute LB (because of the equivalent cell density allowed by the medium). The same genotypes examined here were also tested in undiluted LB, which is a medium that is roughly 100-fold more concentrated. More losses of fitness were found, which suggests some specialization based on resource concentration, but replicates of the same genotype were too inconsistent to be considered reliable (V.S. Cooper, unpublished data). In general, these competitions in novel environments suggest that mutations that were beneficial in the selective environment may sometimes have positive pleiotropic effects.
These contrasting conclusions about how the niche of these bacterial populations has evolved may actually be reconciled if we consider the potential mechanisms of adaptation – in fact, they might inform our hunt for them as well. The contrary findings of novobiocin sensitivity and bile salt resistance have already been presented, but these results, along with other catabolic phenotypes, potentially limit the gene candidates upon which selection may have acted to improve life in a simple glucose-limited environment. The fact that evolved clones appear to respire several resources poorly, yet are at least as fit as the ancestor in the diverse LB environment, suggest that enhancements in the general growth potential of these populations may compensate for these deficiencies. This argues indirectly that the mutations responsible for reduced fundamental diet breadth are either completely silent in environments similar to the evolution model, or are in fact beneficial [36].
Conclusions
The expansion of the potential niche of these populations into different media formulations is indeed remarkable, but should not be construed as an evolutionary trend towards generalism. Travisano and Lenski [22,23] demonstrated that 2,000 generations of evolution was sufficient for these populations to lose fitness when grown on alternative sugars, and the reductions in maximum growth rate at extreme temperatures suggest a trend towards specialization in thermal niche [28]. Together, these investigations of the correlated responses to adaptation to a specific environment demonstrate the mismatch between the arbitrary scales that we use to measure performance and real biological alternatives. Genetic variation need not project phenotypes along a Celsius scale or among an assortment of carbon sources. As a result, generalizations about the nature of the evolution of specialization, and the clarification of the "folk wisdom," may elude us until the mechanisms and tendencies of evolutionary change are better revealed. Thus, despite the inclination to add complexity to these microbial environments so as to better simulate "the real world," it remains evident that we as evolutionary geneticists still have our hands full.
Methods
Measurement of diet breadth
Following closely the protocol described in Cooper and Lenski [27], I used Biolog® ES (designed for E. coli and Salmonella spp.) microtiter plates to obtain estimates of catabolic diet breadth. These plates have 95 different carbon sources and a tetrazolium indicator dye whose intensity is proportional to the amount of respiration on that substrate. I measured optical density immediately after inoculation and then after 24 hours of incubation; the difference in these figures, less the reading from the blank well, estimated functionality on each resource. Diet breadth is expressed as the average performance, relative to the ancestor, on all foreign resources. Mathematical transformations required to generate this average are described in detail in Cooper and Lenski [27]. Experiments were conducted in two complete blocks of 36 evolved 10,000-generation clones (12 populations × 3 clones) and 6 ancestral genotypes (2 ancestors × 3 replicates).
Competition in foreign environments
All experiments were conducted in 50 ml Erlenmeyer flasks in a 37°C shaking incubator, conditions identical to the long-term evolutionary environment (see [19] for specific culture conditions). Cultures were founded from -80° freezer isolates and grown in Davis minimal (DM) medium supplemented with 1000 μg/ml of glucose. Cultures were then acclimated to the test environment via a 1/4,000 dilution into fresh medium and incubated for 24 h. Each evolved genotype and the ancestor of the opposite Ara marker were each diluted 1/200 into a common flask containing 9.9 ml of the test media, and then incubated 1 day. (Certain clones generated unusually low yields in the LB environment, so competitions were founded with a 2:1 starting ratio of evolved to ancestral genotypes, rather than the typical 1:1. All other details of competitions involving these clones were unchanged.) Initial and final densities of each competition culture were determined by plating aliquots onto TA indicator plates, which allow Ara- and Ara+ competitors to be distinguished by their colony color. Relative fitness was quantified by calculating the ratio of the number of doublings for the derived and ancestral competitors (see [19] for details). Competition experiments versus the ancestor were conducted in two complete blocks of 36 evolved 10,000-generation clones (12 populations × 3 clones) in each foreign environment and the paired control DM25 environment. The concentration for the bile salt treatment was selected to mimic the concentration of bile salts in MacConkey Agar (1.5 g/L of Sigma Bile Salts, consisting of 50% sodium cholate, 50% sodium deoxycholate), which is typically used to identify enteric bacteria.
For each competition experiment I conducted one or more of the following statistical tests using SPSS version 8.0 (SPSS, Inc., Chicago, IL): 1) a nested ANOVA on the fitness data in the novel environment designed to identify significant population- and clone-level variance (population and clone are random factors); 2) a mixed-model nested ANOVA designed to identify the effect of environment (fixed factor) on population and clone-level variance (both random factors); and 3) a paired t-test to differentiate fitness in each experimental environment relative to control. In addition, I inspected the data for potential outlier clones within populations that caused significant effects at the level of clone. I identified one clone from one population (A + 6, clone A) that was an outlier in nearly all environments (novel and control). Given that it was also inferior in the control environment, this clone was omitted and analyses 1–3 were recalculated.
Assay for novobiocin sensitivity
Novobiocin resistance is an ancestral character that is sometimes dependent upon mutations in the cell envelope, including the structural component lipopolysaccarides as well as outer membrane porins [32]. Assays were conducted by distributing approximately 250 cells on LB plates containing 400 μg/ml novobiocin. Replicates of the ancestors produced an average of 40 colonies after 48 hours. If 2 colonies emerged after 72 hours, the evolved clone was deemed to be sensitive. Three clones for each of the twelve populations were assayed in this manner and replicated twice each.
Acknowledgments
Acknowledgements
I thank Richard Lenski for his tireless experimental and editorial advice. I thank J. Conner, A. de Visser, A. Jarosz, T. Marsh, P. Moore, S. Remold, and D. Rozen for many helpful discussions, K. Ritahlati for Biolog instruction, and L. Ekunwe for technical assistance. This research was supported by grants from the NSF to the author (DEB-9801538) and to Richard Lenski (DEB-9981397), and by the Michigan Society of Fellows.
References
- Holt RD. Demographic constraints in evolution: towards unifying the evolutionary theories of senescence and niche conservatism. Evolutionary Ecology. 1996;10:1–11. [Google Scholar]
- MacArthur RH, Levins R. Competition habitat selection, and character displacement in a patchy environment. PNAS. 1964;51:1207–10. doi: 10.1073/pnas.51.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levins R. In: Evolution in Changing Environments. Princeton NJ, editor. Princeton University Press; 1968. [Google Scholar]
- Charlesworth B. Evolution in age-structured populations. Cambridge: Cambridge University Press;; 1980. [Google Scholar]
- Lynch M, Gabriel W. Environmental Tolerance. American Naturalist. 1987;129:283–303. doi: 10.1086/284635. [DOI] [Google Scholar]
- Futuyma DJ, Moreno G. The evolution of ecological specialization. Annual Review of Ecology and Systematics. 1988;19:207–233. doi: 10.1146/annurev.es.19.110188.001231. [DOI] [Google Scholar]
- Hutchinson GE. Concluding remarks. Cold Spring Harbor Symp Quant Biol. 1958;22:415–427. [Google Scholar]
- Fong DW, Kane TC, Culver DC. Vestigialization and loss of nonfunctional characters. Annual Review of Ecology and Systematics. 1995;26:249–268. doi: 10.1146/annurev.es.26.110195.001341. [DOI] [Google Scholar]
- Gould F. Rapid host range evolution in a population of the phytophagous mite Tetranychus urticae Koch. Evolution. 1979;33:791–802. doi: 10.1111/j.1558-5646.1979.tb04735.x. [DOI] [PubMed] [Google Scholar]
- Futuyma DJ, Wasserman SS. Food plant specialization and feeding efficiency in the tent caterpillars Malacosoma disstria and Malacosoma americanum. Entomologia Experimentalis Et Applicata. 1981;30:106–110. [Google Scholar]
- Futuyma DJ, Cort RP, Vannoordwijk I. Adaptation to host plants in the fall cankerworm (Alsophila pometaria) and its bearing on the evolution of host affiliation in phytophagous insects. American Naturalist. 1984;123:287–296. doi: 10.1086/284204. [DOI] [Google Scholar]
- Bennett AF, Lenski RE. Evolutionary adaptation to temperature. II. Thermal niches of experimental lines of Escherichia coli. Evolution. 1993;47:1–12. doi: 10.1111/j.1558-5646.1993.tb01194.x. [DOI] [PubMed] [Google Scholar]
- Lenski RE, Bennett AF. Evolutionary response of Escherichia coli to thermal stress. The American Naturalist. 1993;142:S47–S64. doi: 10.1086/285522. [DOI] [PubMed] [Google Scholar]
- Mongold JA, Bennett AF, Lenski RE. Evolutionary adaptation to temperature. IV. Adaptation of Escherichia coli at a niche boundary. Evolution. 1996;50:35–43. doi: 10.1111/j.1558-5646.1996.tb04470.x. [DOI] [PubMed] [Google Scholar]
- Filchak KE, Feder JL, Roethele JB, Stolz U. A field test for host-plant dependent selection on larvae of the apple maggot fly, Rhagoletis pomonella. Evolution. 1999;53:187–200. doi: 10.1111/j.1558-5646.1999.tb05344.x. [DOI] [PubMed] [Google Scholar]
- Fry JD. Trade-offs in fitness on different hosts: Evidence from a selection experiment with a phytophagous mite. American Naturalist. 1990;136:569–580. doi: 10.1086/285116. [DOI] [Google Scholar]
- Jaenike J. Host specialization in phytophagous insects. Annual Review of Ecology and Systematics. 1990;21:243–273. doi: 10.1146/annurev.es.21.110190.001331. [DOI] [Google Scholar]
- Rausher MD. Tradeoffs in performance on different hosts: Evidence from within-site and between-site variation in the beetle Deloyala guttata. Evolution. 1984;38:582–595. doi: 10.1111/j.1558-5646.1984.tb00324.x. [DOI] [PubMed] [Google Scholar]
- Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. The American Naturalist. 1991;138:1315–1341. doi: 10.1086/285289. [DOI] [Google Scholar]
- Lenski RE, Travisano M. Dynamics of adaptation and diversification: A 10,000-generation experiment with bacterial populations. PNAS. 1994;91:6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasi F, Travisano M, Lenski RE. Long-term experimental evolution in Escherichia coli. II. Changes in life-history traits during adaptation to a seasonal environment. The American Naturalist. 1994;144:432–456. doi: 10.1086/285685. [DOI] [Google Scholar]
- Travisano M, Vasi F, Lenski RE. Long-term experimental evolution in Escherichia coli. III. Variation among replicate populations in correlated responses to novel environments. Evolution. 1995;49:189–200. doi: 10.1111/j.1558-5646.1995.tb05970.x. [DOI] [PubMed] [Google Scholar]
- Travisano M, Lenski RE. Long-term experimental evolution in Escherichia coli. IV. Targets of selection and the specificity of adaptation. Genetics. 1996;143:15–26. doi: 10.1093/genetics/143.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elena SF, Cooper VS, Lenski RE. Punctuated evolution caused by selection of rare beneficial mutations. Science. 1996;272:1802–1804. doi: 10.1126/science.272.5269.1802. [DOI] [PubMed] [Google Scholar]
- Elena SF, Lenski RE. Long-term experimental evolution in Escherichia coli. VII. Mechanisms maintaining genetic variability within populations. Evolution. 1997;51:1058–1067. doi: 10.1111/j.1558-5646.1997.tb03953.x. [DOI] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- Cooper VS, Lenski RE. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature. 2000;407:736–739. doi: 10.1038/35037572. [DOI] [PubMed] [Google Scholar]
- Cooper VS, Bennett AF, Lenski RE. Evolution of thermal dependence of growth rate of Escherichia coli populations during 20,000 generations in a constant environment. Evolution. 2001;55:889–896. doi: 10.1554/0014-3820(2001)055[0889:eotdog]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Papadopoulos D, Schneider D, Meier-Eiss J, Arber W, Lenski RE, Blot M. Genomic evolution during a 10,000-generation experiment with bacteria. PNAS. 1999;96:3807–3812. doi: 10.1073/pnas.96.7.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen DE, Lenski RE. Long-term experimental evolution in Escherichia coli. VIII. Dynamics of a balanced polymorphism. American Naturalist. 2000;155:24–35. doi: 10.1086/303299. [DOI] [PubMed] [Google Scholar]
- Thanassi DG, Cheng LW, Nikaido H. Active efflux of bile salts by Escherichia coli. J Bacteriol. 1997;179:2512–2518. doi: 10.1128/jb.179.8.2512-2518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido H. Outer membrane. In: Neidhardt FC et al, editor. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 2. Washington, D.C.: ASM Press;; 1996. [Google Scholar]
- Tropp BE, Ragolia L, Xia WM, Dowhan W, Milkman R, Rudd KE, Ivanisevic R, Savic DJ. Identity of the Escherichia coli cls and nov genes. Journal of Bacteriology. 1995;177:5155–5157. doi: 10.1128/jb.177.17.5155-5157.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills DR, Peterson RL, Spiegelman S. An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule. PNAS. 1967;58:217. doi: 10.1073/pnas.58.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffhill R, Scheiner-Bernloehr H, Orgel LE, Spiegelman S. In vitro selection of bacteriophage Qμ ribonucleic acid variants resistanat to ethidium bromide. Journal of Molecular Biology. 1970;51:531–539. doi: 10.1016/0022-2836(70)90006-9. [DOI] [PubMed] [Google Scholar]
- Cooper VS, Schneider D, Blot M, Lenski RE. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of Escherichia coli B. Journal of Bacteriology. 2001;183:2834–2841. doi: 10.1128/JB.183.9.2834-2841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]