In the preproteomic age, one of the best approaches for deciphering the physiological role of an unknown gene product was to examine the phenotype of mutant strains and take educated guesses for its function that would guide biochemical experimentation. Such a process could take months to years to accomplish. In the proteomic age, the best initial approach is to use a computer to compare the deduced amino acid sequence of the gene product with those of proteins of known function, and with luck, one might get to the same place in seconds to minutes! But what happens when the results of these two approaches don't appear to make any sense with each other?
This issue of PNAS presents an article by Nakahigashi et al. (1) that shows how a gene product originally identified by its mutant phenotype as an enzyme of heme biosynthesis and then by sequence similarity as a possible DNA adenine-N-6-methyltransferase actually functions as a protein glutamine methyltransferase modulating the termination activity of release factors (RFs) in ribosomal protein synthesis! It provides a nice case study for why one can't rest until the biochemistry is done.
The hemK gene of Escherichia coli originally was described in 1995 from a genetic screen designed to reveal new types of heme synthesis mutants (2). Here, one mutation in a light-resistant revertant of a light-sensitive hemH− porphyrin-deficient strain was mapped to a locus in an operon consisting of the hemA gene encoding glutamyl-tRNA reductase, catalyzing the first committed step of the heme synthesis pathway, and the apparently unrelated prfA gene encoding peptidyl RF1, with a transcriptional order of hemA-prfA-hemK (2, 3). HemK mutant cells were found to be unable to make heme from 5-aminolevulinate (2). The presence of 5-aminolevulinate dehydratase and porphobilinogen deaminase activities in the mutant, as well as the accumulation of porphyrin intermediates in the mutants, led to the suggestion that the hemK product might be a subunit of protoporphyrinogen oxidase, the enzyme catalyzing the final formation of protoporphyrin IX in the heme biosynthetic pathway (2). The authors made it clear, however, that such an assignment was tentative and that other roles were possible (2). Despite this caution, many database entries for the hemK product from E. coli and for the homologous protein in related organisms have been apparently upgraded “in silico” and are now designated as protoporphyrinogen oxidase (see below). At the time of the initial analysis, no clear homologous species were found in the databases, although the correct sequence was known only for the N-terminal 203 residues of a total of 277 residues (2).
The presence of the hemA gene in the operon with hemK might have been a red herring in determining its function.
By 1999, biochemical and genetic studies revealed that the hemK product appeared to have no protoporphyrinogen activity at all (4). So what is the protein doing? With the correct full-length amino acid sequence of HemK, and with the tremendous growth of protein sequence databases, two things were made clear. First, HemK homologs are present in a wide variety of species from Gram-negative and Gram-positive eubacteria to mice and humans. Second, the HemK sequence is in fact related to a large family of S-adenosylmethionine-dependent methyltransferases (4). Particularly intriguing was the presence in HemK of an NPPY sequence, up to then largely restricted to members of a subfamily of DNA methyltransferases that modify the 6-amino group of adenine bases, generally in bacterial modification methyltransferases (5). This result suggested that HemK may be a novel type of DNA adenine methyltransferase (5). However, considerable similarity (29% identity over 204 residues, including the NPPY motif) also was noted with an unrelated 4-amino-phenylalanine methyltransferase from a Streptomyces species involved in the biosynthesis of the antibiotic pristinamycin (6). At this point, it seemed that one needed to go back to square one to see exactly what function HemK actually plays in cell physiology. The state of confusion in the field at this point is amply reflected in the variety of designations of hemK homologs that come up in a blast search of the E. coli gene product. For species that have at least 23% identity over the entire sequence and are possible orthologs, 16 are designated protoporphyrin oxidases, seven are probable protoporphyrin oxidases, 16 are putative (or possible) protoporphyrin oxidases, four are adenine-specific methyltransferases, and four are putative adenine-specific methyltransferases. It turns out that all 47 of these designations are most likely to be incorrect!
The clue that led to the discovery of the real function of HemK came from independent work in an entirely new arena —the termination process of ribosomal protein synthesis and the structure and function of protein RFs that are required for the process. In E. coli, there are two RFs whose structures mimic tRNAs as they recognize the UGA, UAA, and UAG terminator codons in the A site of the ribosome and allow the hydrolysis of the nascent peptide chain from the last tRNA at the peptidyl transferase center (7). The most highly conserved feature of RFs from eukaryotic and prokaryotic organisms is a GGQ sequence, located near the site of the peptidyl-tRNA ester bond that is cleaved in the termination reaction (7, 8). In 2000, Dincbas-Renqvist et al. (9) did a careful chemical, chromatographic, and mass spectral analysis of RF2 from E. coli and provided evidence that the conserved glutamine residue at position 252 in this sequence is posttranslationally modified by a methylation reaction to give the N-5-methylglutamine derivative (Fig. 1; Table 1). The presence of the methyl group on the glutamine residue is correlated with a large increase in the efficiency of the termination reaction (9).
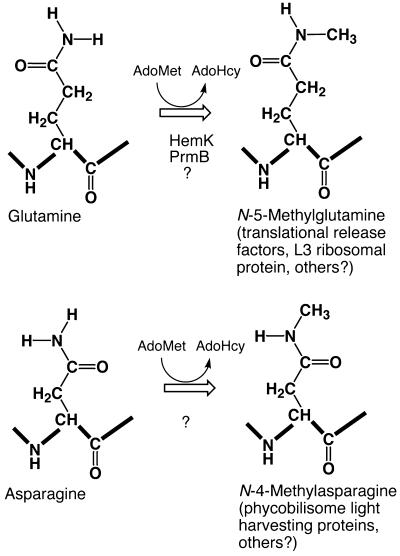
Figure 1.
Methyltransferase reactions that result in the formation of N-methylated amides in protein-bound glutamine and asparagine residues. The N-5 and N-4 designations refer to the number of the carboxyl carbon atom that the methyl amine is attached to; heavy lines show the polypeptide backbone. AdoMet, S-adenosyl-l-methionine; AdoHcy, S-adenosyl-l-homocysteine. HemK catalyzes the methylation of RFs; PrmB catalyzes the methylation of the L3 ribosomal protein. See text for references.
Table 1.
Known sites of glutamine/asparagine side-chain methylation in proteins (see text for references)
| Protein | Source | Sequence (modified site with *) | Methyltransferase gene |
|---|---|---|---|
| RF1 | E. coli | *SSGAGGQHVNTTD 235 | hemK |
| RF2 | E. coli | *TSGAGGQHVNRTE 252 | hemK |
| Allophycocyanin, beta chain | Anabaena variabilis | *ITRPGGNMYTTRR 72 | ? |
| Ribosomal protein L3 | E. coli | *GSIGQNQTPGKVF 150 | prmB (yfcB?) |
With these results, Nakahigashi et al. (1) then realized that the presence of the hemA gene in the operon with hemK might have been a red herring in determining its function and they turned their attention to the presence of what was originally thought to be the unrelated prfA gene that encodes RF1 in the hemA-prfA-hemK operon. RF1 is a close homolog of RF2 and the sequence around the conserved glutamine residue is almost identical in both proteins (Table 1). With this connection to translational RFs, the knowledge of the methylation modification of these factors, and sequence similarities of HemK with methyltransferases, they were able to put all of the pieces together. In their article, they clearly show that mutants in hemK lack the modification of the glutamine residue in both RF1 and RF2 and that the HemK protein is sufficient to catalyze the methylation reaction in vitro (1). So, how can one now rationalize the original heme-defective phenotype seen? From microarray analysis, Nakahigashi et al. (1) point out that the lack of HemK methylation has global effects on the expression of genes involved in aerobic and anaerobic metabolism that may explain the light sensitivity of the mutant, but it is still unclear exactly what caused the defects in heme metabolism observed, including the apparent lack of heme synthesis and the accumulation of intermediates in its biosynthetic pathway (2).
Does the methylation of the side chain of glutamine residues occur in proteins other than the translational RFs? In fact, the presence of N-5-methylglutamine derivatives in proteins was first reported by Lhoest and Colson in 1977 (10). Here, it was found that the E. coli ribosomal protein L3 contains a modification that gives methylamine upon acid hydrolysis and a product comigrating with free N-5-methylglutamine after extensive proteolysis (10). The site of methylation was found to be glutamine-152 of the E. coli large ribosomal subunit protein L3 (11) (Table 1). The gene encoding the methyltransferase was designated prmB and mapped to a position between aroC at 52.67 min and purF at 52.29 min (12). Mutants in the prmB gene displayed a cold-sensitive phenotype and accumulated unstable and abnormal ribosomal particles (11). From these results, it is clear that the HemK methyltransferase (whose gene maps at 27.7 min) is a distinct enzyme from the PrmB methyltransferase. Additionally, there appears to be no sequence similarity in the methyl-accepting site of L3 and the RFs (Table 1). Nevertheless, a HemK homolog, designated YfcB, is present at 52.70 min on the E. coli chromosome very close to the reported position of the prmB gene. The YfcB protein, labeled a hypothetical adenine-specific methylase in GenBank, is 32% identical to E. coli HemK over 194 residues and would appear to be an excellent candidate for the PrmB L3 ribosomal protein glutamine methyltransferase (Table 1). It will now be of considerable interest to ask whether additional proteins containing methylated glutamine residues are present in cells.
What about the possibilities for methylating the side chain of asparagine residues, having one less methylene group in the side chain than glutamine residues? Reactions resulting from the modification of asparagine residues leading to N-4-methylasparagine (or γ-N-methylasparagine) are well known in the light-gathering phycobilisome proteins of the photosynthetic apparatus of cyanobacteria and red algae (13, 14) (Fig. 1). Asparagine residues at position 72 in the β-chain of many C- and R-phycocyanins, allophycocyanin, and B-, C-, and R-phycoerythrins are methylated on the side-chain amide nitrogen (13, 14). This modification appears to fine-tune the interaction of the protein with the chromatophore to minimize energy losses as these proteins gather light for the photosynthetic apparatus of these cells. Mutant cells containing unmethylated phycobilisomes demonstrate lower efficiencies of energy transfer than their methylated counterparts (15, 16). Interestingly, when the asparagine residue was replaced with a glutamine residue in these proteins, significant amounts of methylation were still detected, suggesting the methyltransferase might recognize either an asparagine or glutamine after the two glycine residues or that a second enzyme activity is present (17). Of considerable interest is the fact that the asparagine residue methylated in phycobiliproteins is located in a similar sequence context to that of the glutamine residues in the RFs methylated by the HemK methyltransferase, where the amide residue in both cases is preceded by a pair of glycine residues (Table 1).
What new chemistry might be affected by the methylation of amides of asparagine and glutamine residues? The methylation reaction certainly adds bulk to the side-chain amide group and also removes the possible participation of one of the two amide hydrogen atoms in hydrogen bonding schemes (Fig. 1). Studies on synthetic peptides have suggested that methylation can slow the spontaneous deamidation of the side chain of asparagine residues by 45-fold (18).
This work now extends our knowledge of the extensive posttranslational modification of proteins. Many of these modifications represent permanent changes that serve to expand the chemical diversity of amino acid side chains beyond that offered by the 20 amino acids used in ribosomal synthesis. In addition to the modifications described here, methylation reactions are known to create new amino acids by modifying the α-amino group at the N terminus, as well as the side-chain residues of histidine, lysine, and arginine residues (19). Some modifications are reversible and represent possible modes of regulation. For methylation reactions, these include the formation of hydrolysable methyl esters on the side-chain carboxyl groups of glutamate residues as well as the C-terminal carboxyl groups of isoprenylcysteine residues, leucine residues, and lysine residues (20). It is unclear whether the formation of the methylated amides in glutamine and asparagine residues is reversible in the cell; it would be interesting to ask whether enzymatic activities exist that can catalyze the regeneration of the original residues or the formation of the corresponding acid.
Finally, this article raises issues of just how well protein functions can be assigned by sequence comparison. S-adenosylmethionine (AdoMet)-dependent methyltransferases would appear to be an ideal place to use sequence comparisons. These enzymes have the very simple function of transferring a methyl group from the sulfonium atom of AdoMet to a variety of nucleophiles, including oxygen, sulfur, nitrogen, and carbon atoms on proteins, nucleic acids, carbohydrates, lipids, and small molecules in all organisms. Evolution has been kind to the workers in this field because most of these enzymes have retained four “signature motifs,” short stretches of sequence that have been apparently conserved over billions of years of evolution and that can allow the identification of candidate methyltransferases with some success (21, 22). It was originally hoped that examination of sequence similarity outside of the signature motifs would allow one to start characterizing families of methyltransferases specific for N-DNA, C-DNA, protein carboxyl, protein amino, protein guanidino, etc., methyl-accepting substrates. For methyltransferase families that have been highly conserved, this approach does appear to work. For instance, it has been possible to predict new members of the protein methyltransferase family that catalyze the formation of symmetric and asymmetric dimethylarginine derivatives (23). But for many methyltransferases (including the HemK methyltransferase seen here), it doesn't seem to be so simple. For example, sequence analysis of the 13 best methyltransferase candidates in yeast led to probably incorrect identifications of 12 of these gene products and only one useful “hit,” where a novel protein arginine methyltransferase was matched with a guanidinoacetate methyltransferase of higher cells (22).
It is clear that much caution is needed in interpreting the results of sequence searches from databases that are contaminated with entries where putative or possible functions somehow become functions. It is also clear that a mechanism needs to be put in place to correct annotations of the databases once the biochemistry is experimentally established. Lastly, what should be done when gene designations such as hemK are themselves misleading—should history prevail (as in dnaK) or should an effort be made to provide a name more directly associated with the function?
Footnotes
See companion article on page 1473.
References
- 1.Nakahigashi K, Kubo N, Narita S-I, Shimaoka T, Goto S, Oshima T, Mori H, Maeda M, Wada C, Inokuchi H. Proc Natl Acad Sci USA. 2002;99:1473–1478. doi: 10.1073/pnas.032488499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakayashiki T, Nishimura K, Inokuchi H. Gene. 1995;153:67–70. doi: 10.1016/0378-1119(94)00805-3. [DOI] [PubMed] [Google Scholar]
- 3.Elliott T. J Bacteriol. 1989;171:3948–3960. doi: 10.1128/jb.171.7.3948-3960.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Guen L, Santos R, Camadro J M. FEMS Microbiol Lett. 1999;173:175–182. doi: 10.1111/j.1574-6968.1999.tb13499.x. [DOI] [PubMed] [Google Scholar]
- 5.Bujnicki J M, Radlinska M. IUBMB Life. 1999;48:247–249. doi: 10.1080/713803519. [DOI] [PubMed] [Google Scholar]
- 6.Blanc V, Gil P, Bamas-Jacques N, Lorenzon S, Zagorec M, Schleuniger J, Bisch D, Blanche F, Debussche L, Crouzet J, Thibaut D. Mol Microbiol. 1997;23:191–202. doi: 10.1046/j.1365-2958.1997.2031574.x. [DOI] [PubMed] [Google Scholar]
- 7.Song H, Mugnier P, Das A K, Webb H M, Evans D R, Tuite M F, Hemmings B A, Barford D. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 8.Frolova L Y, Tsivkovskii R Y, Sivolobova G F, Oparina N Y, Serpinsky O I, Blinov V M, Tatkov S I, Kisselev L L. RNA. 1999;5:1014–1020. doi: 10.1017/s135583829999043x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dincbas-Renqvist V, Engstrom A, Mora L, Heurgue-Harnard V, Buckingham R, Ehrenberg M. EMBO J. 2000;19:6900–6907. doi: 10.1093/emboj/19.24.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lhoest J, Colson C. Mol Gen Genet. 1977;154:175–180. doi: 10.1007/BF00330833. [DOI] [PubMed] [Google Scholar]
- 11.Lhoest J, Colson C. Eur J Biochem. 1981;121:33–37. doi: 10.1111/j.1432-1033.1981.tb06425.x. [DOI] [PubMed] [Google Scholar]
- 12.Colson C, Lhoest J, Urlings C. Mol Gen Genet. 1979;169:245–250. doi: 10.1007/BF00382270. [DOI] [PubMed] [Google Scholar]
- 13.Klotz A V, Leary J A, Glazer A N. J Biol Chem. 1986;261:15891–15894. [PubMed] [Google Scholar]
- 14.Klotz A V, Glazer A N. J Biol Chem. 1987;262:17350–17355. [PubMed] [Google Scholar]
- 15.Swanson R V, Glazer A N. J Mol Biol. 1990;214:787–796. doi: 10.1016/0022-2836(90)90293-u. [DOI] [PubMed] [Google Scholar]
- 16.Thomas B A, Bricker T M, Klotz A V. Biochim Biophys Acta. 1993;1143:104–108. [Google Scholar]
- 17.Thomas B A, McMahon L P, Klotz A V. Biochemistry. 1995;34:3758–3770. doi: 10.1021/bi00011a034. [DOI] [PubMed] [Google Scholar]
- 18.Klotz A V, Thomas B A. J Org Chem. 1993;58:6985–6989. [Google Scholar]
- 19.Clarke S. Curr Opin Cell Biol. 1993;5:977–983. doi: 10.1016/0955-0674(93)90080-a. [DOI] [PubMed] [Google Scholar]
- 20.Zobel-Thropp P, Yang M C, Machado L, Clarke S. J Biol Chem. 2000;275:37150–37158. doi: 10.1074/jbc.M001005200. [DOI] [PubMed] [Google Scholar]
- 21.Kagan R M, Clarke S. Arch Biochem Biophys. 1994;310:417–427. doi: 10.1006/abbi.1994.1187. [DOI] [PubMed] [Google Scholar]
- 22.Niewmierzycka A, Clarke S. J Biol Chem. 1999;274:814–824. doi: 10.1074/jbc.274.2.814. [DOI] [PubMed] [Google Scholar]
- 23.Frankel, A., Yadav, N., Lee, J., Branscombe, T. L., Clarke, S. & Bedford, M. (2002) J. Biol. Chem.277, in press. [DOI] [PubMed]