Abstract
The mammalian testis determining factor SRY and its related Sox factors are critical developmental regulators. They share significant similarity in their high mobility group (HMG) domain and display discrete patterns of tissue-specific expression. Here we show that SRY and the Sox protein SOX6 colocalize with splicing factors in the nucleus and are dynamically redistributed following the blockage of splicing in living cells. Anti-SOX6 antibodies supershift the spliceosomal complex from assembled splicing reactions and inhibit splicing in vitro of multiple pre-mRNA substrates. Most importantly, SOX6-depleted nuclear extracts have impaired splicing activity, which is efficiently restored by addition of the recombinant SOX6 HMG domain and also by recombinant SRY and the SOX9 HMG domain. These results reveal an unexpected biological function of the SRY, SOX6, and SOX9 gene products and provide a functional link to the biochemical mechanisms operating in mammalian sex determination and in other developmental processes regulated by Sox genes.
The high mobility group (HMG)-type DNA binding domain was originally described as a domain in the RNA polymerase I transcription factor hUBF homologous to two regions in the chromatin HMG protein 1 (1). Proteins containing HMG domains have been found in a large variety of species and are grouped into two families, based on evolutionary and functional conservation (reviewed in ref. 2). One family comprises the chromosomal HMG domain group of proteins, which are provided with two or more HMG domains and bind DNA with low to moderate affinity and little sequence specificity. The other family includes sequence-specific transcription factors, which are provided with only one HMG domain and bind to DNA sequences in the promoter of target genes to elicit their transcriptional activation. HMG domain factors bind DNA in the minor groove, induce a relevant DNA bending on binding, and are capable of binding to special or distorted DNA structures, such as four-way junctions, cis-platinum adducts, and base bulges (reviewed in ref. 3). Given these properties, HMG domain proteins are postulated to act as architectural elements, promoting formation of contacts between factors bound at distant sites on DNA or recruiting proteins that by themselves do not bind to DNA and facilitating the assembly of higher-order complexes involved in transcriptional control, gene recombination, and DNA repair.
Among sequence-specific HMG-domain proteins, a distinct position is occupied by the Sox (SRY box) protein family. A common feature of these proteins is to share more than 50% similarity in their HMG domains to SRY, the testis-determining factor located on human and mouse Y chromosomes. Sox proteins have been identified in a number of vertebrates and invertebrates, including Drosophila melanogaster and Caenorhabditis elegans. Outside the HMG box, Sox proteins are poorly conserved, and many are expressed with a tissue-specific pattern. These factors play a crucial role in development, as also shown by their involvement in several human congenital syndromes (reviewed in refs. 4 and 5). Here we demonstrate a function for SRY, SOX6, and SOX9 in the process of pre-mRNA splicing. These results suggest that SOX proteins operate through a common biochemical mechanism in regulating mammalian sex determination and other developmental processes.
Materials and Methods
DNA Clones.
pSG.Sox6 and pSG.SRY harbor the full-length Sox6 and SRY cDNAs, respectively, in the mammalian expression vector pSG5. Sox6 N- and C-terminal sequences encoding amino acids 1–476 and amino acids 475–827, respectively, were PCR amplified from pSG.Sox6 and cloned into pSG5. pGEX.Sox6 HMG, pGEX.SRY, and pGEX.SOX9 HMG harbor sequences encoding the Sox6 HMG box (amino acids 612–715), full-length SRY, and the SOX9 HMG box (amino acids 103–182), respectively, in the bacterial expression vector pGEX 4T-3.
Cell Culture and Transfections.
HeLa and COS-1 cells were grown in DMEM supplemented with 5% FCS and antibiotics. NT2/D1 cells were grown in DMEM supplemented with 10% FCS and antibiotics. Transient transfections were performed by the calcium phosphate method.
Antibodies.
Antisera were raised in rabbits directed against peptides located at the very C terminus (CKSDYSSENEAPEPVSAN) (αSox6C) and N terminus (CMSSKQATSPFACTADGEE) (αSox6N) of the mouse Sox6 protein and affinity-purified. These peptides are conserved in human SOX6. Initial immunolocalization experiments of SRY in NT2/D1 cells were performed by using a previously characterized rabbit antiserum (6), which was a kind gift from P. Berta (Montpellier, France). Subsequent experiments involving detection of SRY were performed by using a SRY-specific antiserum produced exactly as described (6). Anti-2,2,7-trimethylguanosine antibody was obtained from Oncogene Research (Darmstadt, Germany), anti-SC-35 and anti-Flag M2 antibody from Sigma, and anti-U1–70K from Eurodiagnostica (Arnhem, The Netherlands). Anti-U2AF (65) (clone MC3), anti Sm (clone Y12), and anti-phosphatidylinositol 4,5-bisphosphate (clone 2C11) antibodies were kind gifts of M. Carmo-Fonseca (Univ. Lisboa, Lisboa, Portugal), A. Lamond (Univ. Dundee, Dundee, Scotland), and G. Schiavo (London), respectively.
Recombinant Proteins.
Recombinant Sox6 HMG box, SRY, and SOX9 HMG box were produced as glutathione S-transferase (GST)-fusion proteins and purified on glutathione-Sepharose (Amersham Pharmacia), following the manufacturer's procedures.
Immunofluorescence Microscopy and Microinjection.
Immunofluorescence was carried out after fixing the cells with 4% paraformaldehyde for 15 min at room temperature and permeabilized with 0.1% Triton X-100 in PBS for 10 min at room temperature. Cells were analyzed with a Leica confocal microscope. Cytoplasmic microinjection of oligonucleotides was performed as described (7, 8) using an antisense U6 oligonucleotide and an irrelevant oligonucleotide (C2; ref. 8) as a control. Lysine-fixable Texas red- and Cascade blue-conjugated dextrans (Molecular Probes) were coinjected along with the oligonucleotides as a marker for the injected cells.
In Vitro Splicing and Spliceosome Analysis.
HeLa nuclear extracts were prepared by the Dignam method (9). For in vitro splicing, δEX14-15, derived from the chicken δ-crystallin gene (10), human β-globin (11), and D. melanogaster fushi-tarazu (12) pre-mRNAs were used. Each splicing reaction contained 60 μg of HeLa nuclear extract, 3% poly(vinyl alcohol), 1.8 mM MgCl2, 0.67 mM ATP, 27 mM creatine phosphate, 8 units of RNasin, and 2 × 104 cpm of the pre-mRNA substrate. After a 10-min preincubation at 30°C of the HeLa nuclear extract (5–7 mg/ml), the other reagents were added and incubated for a further 2 h at 30°C. After incubation, the reaction was mixed with heparin (12.5 mg/ml), freeze-thawed three times, electrophoresed on a 3.75% polyacrylamide gel for spliceosome analysis (13) or purified by proteinase K treatment and phenol/chloroform extraction, precipitated, and analyzed on a polyacrylamide/urea gel. Antibody inhibition of the in vitro splicing assay was performed by adding the αSox6C antibody, αSox6N antibody, or control rabbit IgG after the preincubation period. For supershifting of the spliceosomal complexes, antibodies were added at different times, and reactions were analyzed by electrophoresis on a native polyacrylamide gel.
Immunodepletion and Reconstitution with Recombinant Proteins.
αSox6C antibody or control IgG was added on ice to nuclear extracts containing all splicing reaction reagents except for the probe. After 30 min at 30°C, protein A-Sepharose was added for 15 min at room temperature, and the supernatant was then used for in vitro splicing. As a control for depletion, the SOX6 protein was detected by immunoprecipitation of the depleted extracts by the αSox6C antibody. Reconstitution was performed by mixing nuclear extracts and recombinant proteins on ice before preincubation.
Results
SOX6 and SRY Are Localized in Nuclear Splicing Factor Speckle Domains.
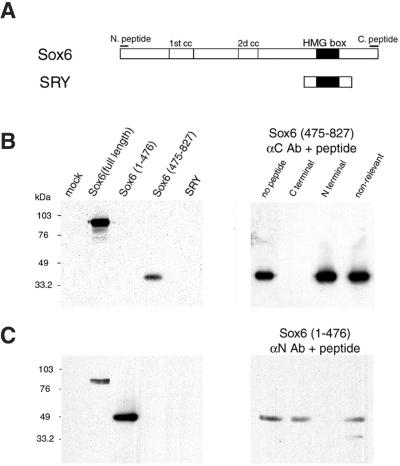
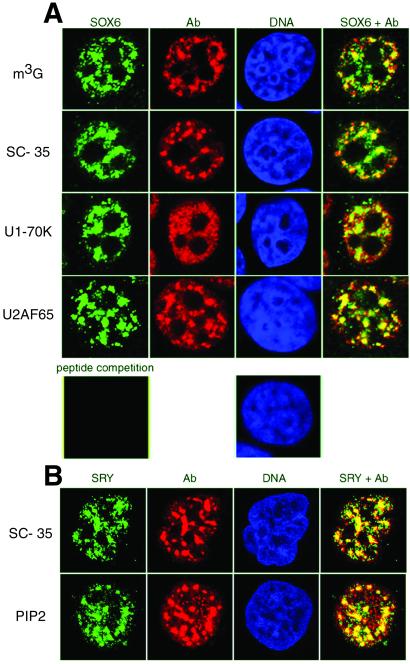
Although several studies have revealed the expression pattern of SRY and Sox genes in embryonal and adult tissues, little is known about the subcellular distribution of the proteins encoded by these genes. We have raised two antibodies against distinct peptides located at the very N and C termini of the Sox6 (14) protein. These peptide sequences are conserved in both mouse Sox6 and human SOX6 (15), and are uniquely present in Sox6 among all members of the Sox subfamily of HMG domain factors. The two antibodies are highly specific and recognize either the N- or the C-terminal portion of Sox6, respectively, whereas both readily recognize the full-length protein (Fig. 1 B and C). We used our Sox6-specific antibodies, and have been able to detect widespread expression of the protein in various adult mouse tissues (data not shown). In HeLa cells, independently from the fixation method used (either paraformaldehyde, Fig. 2A, or methanol, data not shown), SOX6 is concentrated in a few speckles in the nucleus and is also diffusely distributed in the nucleoplasm. This pattern of staining was consistently found in various cell lines and in mouse embryonal tissues (data not shown) and is reminiscent of the characteristic localization of components of the pre-mRNA splicing machinery (16, 17). Indeed, double staining immunofluorescence reveals that SOX6 colocalizes with splicing factors and U small nuclear RNAs in most of the nuclear speckle domains (18) and in the connecting regions between speckles (Fig. 2A). The specificity of SOX6 staining was confirmed by peptide competition (Fig. 2A). As shown in Fig. 2B, and as also reported previously (6), SRY is concentrated in nuclear speckles in NT2/D1 human embryonic carcinoma cells. The distribution of these nuclear speckles is analogous to the one of the SC-35 splicing factor and of the phospholipid phosphatidylinositol 4,5-bisphosphate (PIP2), known to be localized in splicing factor speckle domains in the nucleus (19, 20).
Figure 1.
Generation of anti-SOX6 specific antibodies. (A) Diagram showing the structure of the Sox6 and SRY proteins. The HMG domain is indicated with a black box and coiled-coil domains in Sox6, with a white box. The N- and C-terminal epitopes recognized by anti-Sox6 antibodies are indicated with bars. (B Left) COS-1 cells were mock-transfected or transfected with expression vectors encoding full-length Sox6, Sox6-(1–476), Sox6-(475–827), and SRY. Western blotting using the anti-Sox6 C-terminal antibody was performed on whole cell extracts. (Right) C-terminal antibody binding is blocked by preincubation with a 500-fold molar excess of the specific peptide used for immunization, but not by the same amount of the N-terminal Sox6 peptide or by an unrelated peptide. (C) As in B, but the anti-Sox6 N-terminal antibody was used for Western blotting.
Figure 2.
SOX6 and SRY colocalize with splicing factors in nuclear speckle domains. (A) HeLa cells were stained for SOX6 (green), U small nuclear RNAs 2,2,7-trimethylguanosine cap (m3G), SC-35, U1–70K, U2AF65 (all in red), and DNA (blue), and were analyzed by confocal microscopy. Overlay is shown on Right. Preincubation of the anti-Sox6 antibody with the specific peptide eliminates staining (Bottom Left). (B) NT2/D1 cells were stained for SRY (green), SC-35 and phosphatidylinositol 4,5-bisphosphate (PIP2) (both in red), and DNA (blue). Overlay is shown on Right. Cells were analyzed by confocal microscopy.
Redistribution of SRY and SOX6 with Splicing Factors.
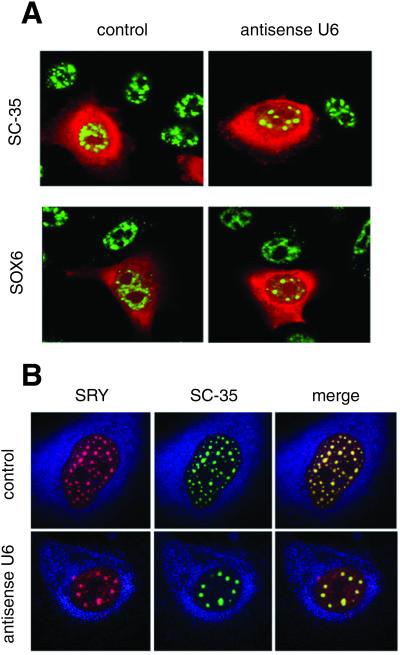
Blockage of splicing in living cells by antisense U6 small nuclear RNA oligonucleotides is known to produce the accumulation of the SC-35 splicing factor into enlarged speckles (7, 8). In our experiments, this redistribution of SC-35 is clearly evident (Fig. 3A). Importantly, antisense U6 injection into HeLa cells caused also SOX6 to redistribute into larger and rounder nuclear speckles (Fig. 3A). Similarly, antisense U6 injection into NT2/D1 cells also triggered the redistribution of SRY and SC-35 into enlarged speckles (Fig. 3B). In both cases, injection of control oligonucleotides (8) had no effect (Fig. 3). Collectively these results demonstrate that SOX6 and SRY are functionally colocalized with splicing factors in the nucleus and thereby suggest that these proteins may be directly involved in the biochemical process of splicing.
Figure 3.
Block of splicing in living cells redistributes SOX6 and SRY into enlarged nuclear speckles. (A) HeLa cells were injected with an antisense U6 or a control oligonucleotide. SC-35 (Left) and SOX6 (Right; both in green) were revealed by immunofluorescence. Injected cells are labeled in red by coinjected Texas red-dextran. (B) NT2/D1 cells injected with an antisense U6 (Lower) or a control oligonucleotide (Upper). SRY (Left; in red) and SC-35 (Center; in green) were revealed by immunofluorescence. Overlay is shown on the right. The cytoplasm is labeled in blue by coinjected Cascade blue-dextran.
Anti-SOX6 Antibodies Inhibit Splicing of Multiple Substrates in Vitro.
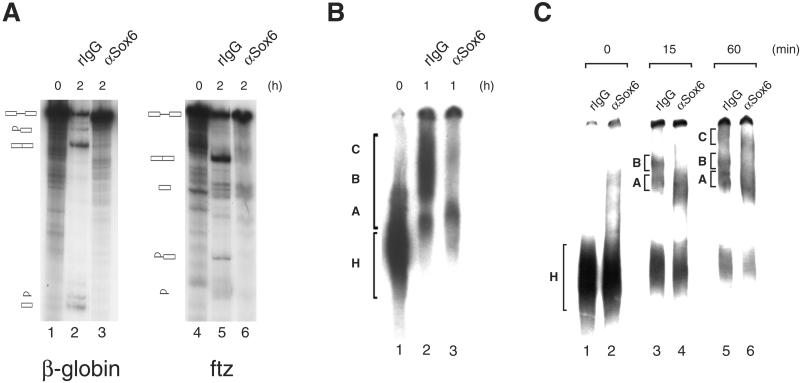
We used antibody inhibition as a first approach to investigate the involvement of SOX6 in the pre-mRNA splicing process by using HeLa nuclear extracts. Anti-SOX6 antibodies are able to inhibit both steps of splicing of β-globin and fushi-tarazu (Fig. 4A) pre-mRNAs, whereas no effect was produced by nonimmune rabbit IgG. Furthermore, formation of the active spliceosomal complexes B and C when a chicken δ-crystallin substrate was used was also inhibited by anti-SOX6 antibodies (Fig. 4B). In addition, when spliceosomal complexes preassembled on a β-globin pre-mRNA substrate were analyzed by native gel electrophoresis, a specific and drastic reduction in the amount of the active B and C complexes after the addition of anti-Sox6 antibodies was observed (Fig. 4C). In conclusion, because SOX6 targeting affects the splicing of multiple pre-mRNA substrates, this molecule appears to have the features of a general splicing factor.
Figure 4.
Effect of anti-SOX6 antibodies on in vitro splicing. (A) Splicing of β-globin (lane 1) and fushi-tarazu (lane 4) pre-mRNAs is inhibited after 2 h of incubation by 25 μg of αSox6N antibody (lanes 3 and 6, respectively), whereas the same concentration of rabbit IgG (lanes 2 and 5, respectively) has no effect. Products and intermediates were analyzed on a 8% polyacrylamide/8 M urea gel. (B) Spliceosome formation on a δ-crystallin pre-mRNA was analyzed on a 3.75% polyacrylamide native gel at time 0 (lane 1) and 1 h after the start of the splicing reaction in the presence of 25 μg of rabbit IgG (lane 2) or αSox6C antibody (lane 3). The position of the inactive H complex and of the active A, B, and C complexes is indicated. (C) Spliceosomal complexes preassembled on a β-globin pre-mRNA substrate were incubated with nonimmune rabbit IgG (lanes 1, 3, and 5) or anti-Sox6 C-terminal antibody (lanes 2, 4, and 6) and analyzed by native gel electrophoresis. The position of the inactive H complex and active A, B, and C complexes is indicated at various times of incubation.
Restoration of Splicing by SRY and by the SOX6 and SOX9 HMG Domains.
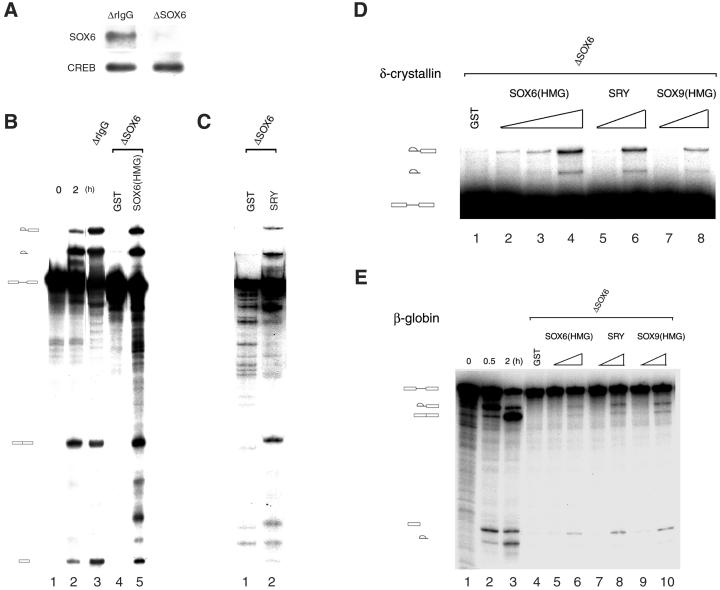
We used our specific anti-SOX6 antibodies to fully deplete HeLa cell extracts of SOX6 (Fig. 5A). A mock-depleted HeLa nuclear extract was able to process a δ-crystallin pre-mRNA substrate (Fig. 5B, lane 3), whereas splicing activity of a SOX6-depleted nuclear extract was impaired at the level of the first step of the splicing reaction (Fig. 5B, lane 4). Consistent with this result, a SOX6-depleted nuclear extract was unable to form active spliceosomal complexes when analyzed on native gels (data not shown). Addition of a recombinant GST-SOX6 HMG domain fusion protein to a nuclear extract that was previously depleted of SOX6 can reconstitute its in vitro splicing activity, whereas addition of GST had no effect (Fig. 5B, lanes 4 and 5). This result with reconstituted extracts shows that SOX6 is the only factor depleted by the anti-SOX6 antibody that is essential for splicing activity in vitro. HeLa cells did not express endogenous SRY, and nuclear extracts from the SRY-expressing cell line NT2/D1 were not functional in in vitro splicing assays (data not shown). We then took advantage of the impaired splicing activity of SOX6-depleted HeLa nuclear extracts to test the capacity of other members of the SOX family to restore splicing in vitro. GST-SRY (Fig. 5 C, lane 2, and D, lanes 5 and 6) and GST-SOX9 HMG (Fig. 5D, lanes 7 and 8) were also able to restore splicing activity of SOX6-depleted nuclear extracts. All these recombinant proteins showed a dose-dependent activity in splicing restoration already at nanomolar concentrations, although their GST fusion partner is inactive, even at higher doses (Fig. 5D). Remarkably, splicing of a β-globin substrate by HeLa nuclear extract was also impaired by SOX6 depletion (Fig. 5E). Also in this case, addition of nanomolar amounts of GST-SOX6 HMG, GST-SRY, and GST-SOX9 HMG was sufficient to restore splicing activity of the SOX6-depleted extract.
Figure 5.
Different Sox factors may complement SOX6 function in splicing. (A) HeLa nuclear extracts were immunodepleted by using either nonimmune rabbit IgG or anti-Sox6 antibodies. SOX6 in the depleted extract was detected by Western blot after immunoprecipitation (Upper). The levels of another nuclear protein (CREB) are not altered by the immunodepletion procedure (Lower). (B) A δ-crystallin pre-mRNA substrate (lane 1) is efficiently spliced by a HeLa nuclear extract (lane 2). Nonimmune IgG-depleted nuclear extract (lane 3) retains full splicing activity. SOX6-depleted nuclear extract is functional only when supplemented with GST-SOX6 HMG domain (8 μM; lane 5), but not with GST (11.5 μM; lane 4). Time of incubation (hours) is indicated. Reactions were analyzed on a 6% polyacrylamide/8 M urea gel. (C) Splicing of a δ-crystallin pre-mRNA substrate by a SOX6-depleted nuclear extract supplemented with either GST (11.5 μM; lane 1) or GST-SRY (6 μM; lane 2) was analyzed at 2 h after the start of the splicing reaction. (D) Titration of the restoration of the splicing activity of a SOX6-depleted nuclear extract by GST-SOX6 HMG domain (lanes 2–4: 8, 80 and 800 nM), GST-SRY (lanes 5 and 6: 60 and 600 nM), and GST-SOX9 HMG domain (lanes 7 and 8, 65 and 650 nM) by using a δ-crystallin pre-mRNA substrate. GST (1.2 μM) was added to the reaction in lane 1. (E) Splicing of a β-globin pre-mRNA substrate by HeLa nuclear extract (lanes 1–3). A SOX6-depleted nuclear extract is also not functional in splicing of this substrate (lane 4). Splicing activity can be restored by addition of increasing amounts of GST-SOX6 HMG domain (lanes 5 and 6, 1.5 and 15 nM), GST-SRY (lanes 7 and 8, 2.3 and 23 nM), and GST-SOX9 HMG domain (lanes 9 and 10, 1.7 and 17 nM). GST (40 nM) was added to the reaction in lane 4.
Discussion
More than 20 different genes encoding members of the Sox family of HMG domain factors have been identified up to date after the cloning of the founding member of the family, the mammalian testis-determining gene SRY (21). Mutations causing a number of human diseases characterized by the perturbation of critical developmental processes have been identified in specific SOX genes (reviewed in refs. 4 and 5). Based on the structural similarity of the Sox HMG domain to the DNA-binding domain of a family of DNA-binding transcriptional activators, a number of studies have described their transcriptional properties. However, Sox proteins are atypical transcription factors under several points of view. They have a fairly high affinity of binding to nonspecific DNA, and mutations in their binding site are often well tolerated (22, 23). In addition, like other HMG box domain factors, they bind and bend linear DNA by partial intercalation in the minor groove, and can also bind to four-way junctions (23, 24). Paradoxically, human SRY lacks a transcriptional activation domain, which is present in mouse Sry and is necessary to produce male sex determination in transgenic mice (25). An attractive model to explain how Sox proteins control gene expression is that they function as architectural factors bound to DNA, influencing local chromatin structure through their capacity to bend DNA and to assemble multiprotein transcriptional complexes (26).
Here we have shown a role for SRY, SOX6, and SOX9 in the process of pre-mRNA splicing. Both the first and the second step in the splicing reaction are impaired in a SOX6-depleted nuclear extract, but both can be restored by addition of recombinant SRY, SOX6, and SOX9 HMG domains. We are aware of the possibility that other splicing factors associated with SOX6 could be codepleted during the immunodepletion. However, considering the mild conditions used, the fact that the addition of SOX and SRY recombinant proteins suffices to restore splicing activity shows that these factors are the only limiting components in the nuclear extract removed during the immunodepletion procedure. Considering the unusual DNA-binding properties of Sox proteins, it is interesting to speculate that these factors may also bind to RNA and thereby induce RNA bending, thus affecting the structure of pre-mRNAs during splicing and favor protein–RNA and RNA–RNA interactions. This alteration could facilitate the rearrangements that take place in the spliceosome during the splicing process (reviewed in refs. 27 and 28).
The widespread expression of Sox6 (not shown) and the effect of antibodies targeting SOX6 on the splicing of different pre-mRNA substrates suggest that it may function as a general splicing factor. This suggestion can explain the postnatal lethal phenotype of homozygote mutant mice where a chromosomal inversion affects the Sox6 gene (29) and of mice with a targeted mutation of both Sox6 alleles (30). The striking finding that SRY can complement splicing activity of a nuclear extract that has been depleted of SOX6 indicates a role in splicing also for the testis-determining factor. This role is confirmed by the colocalization of SRY with markers of the subnuclear domain occupied by splicing factors in the Sertoli-like NT2/D1 cell line and by its redistribution after inhibition of splicing in living cells. SRY is also localized in nuclear speckles in cells of the developing male gonad (6). SOX9 is another important regulator of the male sex-determination pathway (reviewed in ref. 31), and its HMG domain is able to restore splicing activity of SOX6-depleted nuclear extracts in our assay. Because SOX9 is also distributed in nuclear speckles in NT2/D1 cells (32), it will be interesting to assess whether these are colocalized with splicing factor speckle domains.
It is tempting to speculate that during embryonal life, SRY controls the splicing of pre-mRNA substrates whose products act in the gonadal ridge of XY embryos to trigger the male differentiation program. The function of SRY in splicing identifies an unexpected link with the sex-lethal and transformer genes, whose products play key roles in Drosophila sex determination through their role as splicing regulators (reviewed in ref. 33). Furthermore, SRY may continue to work as a splicing factor in Sertoli and germ cells in the adult testis (34). A role in regulated splicing for SRY is not incompatible with its activity as a constitutive splicing factor in our experiments, as shown by the example of the SR proteins. These factors play essential roles in constitutive splicing, but are also important for 5′ and 3′ splice site selection through sequence-specific binding to splicing enhancers (35). Recent studies show that functional complementation among the members of the Sox family also occurs in vivo. In fact, the HMG domains of Sox3 and Sox9 can substitute for the Sry HMG domain in producing male sex determination in transgenic XX mice (36), and the Sox9 gene can induce testis development in XX transgenic mice (37), similarly to what was shown earlier with Sry (38). These results provide compelling evidence that Sox factors may be functionally redundant in producing specific phenotypic effects.
Acknowledgments
We thank M. Ohno for his very helpful suggestions; B. Bardoni and J. Stevenin for discussions; A. Dierich for help with microinjection; E. Heitz, M. Rastegar, J.-L. Weickert, and the Institut de Génétique et de Biologie Moléculaire et Cellulaire cell culture, oligonucleotide-peptide synthesis and sequencing facilities for technical assistance; G. Duval, K. Tamai, and V. Caussanel for antibody production; J.-L. Vonesch, D. Hentsch, and M. Boeglin for help with confocal microscopy; and P. Berta, M. Carmo-Fonseca, A. Lamond, A. Krainer, I. Mattaj, M. Ohno, G. Scherer, and G. Schiavo for gifts of reagents. This work was supported by Centre National de la Recherche Scientifique, Institut National de la Santé et de la Recherche Médicale, Centre Hospitalier Universitaire Régional, Fondation pour la Recherche Médicale, Association pour la Recherche sur le Cancer, Human Frontiers Science Program, and Organon (Akzo/Nobel).
Abbreviations
- HMG
high mobility group
- GST
glutathione S-transferase
References
- 1.Jantzen H-M, Admon A, Bell P B, Tjian R. Nature (London) 1990;344:830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- 2.Laudet V, Stehelin D, Clevers H. Nucleic Acids Res. 1993;21:2493–2501. doi: 10.1093/nar/21.10.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Travers A. Curr Opin Struct Biol. 2000;10:102–109. doi: 10.1016/s0959-440x(99)00056-1. [DOI] [PubMed] [Google Scholar]
- 4.Pevny L H, Lovell-Badge R. Curr Opin Genet Dev. 1997;7:338–344. doi: 10.1016/s0959-437x(97)80147-5. [DOI] [PubMed] [Google Scholar]
- 5.Wegner M. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poulat F, Girard F, Chevron M-P, Gozé C, Rebillard X, Calas B, Lamb N, Berta P. J Cell Biol. 1995;128:737–748. doi: 10.1083/jcb.128.5.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Keefe R T, Mayeda A, Sadowski C L, Krainer A R, Spector D L. J Cell Biol. 1994;124:249–260. doi: 10.1083/jcb.124.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larsson S H, Charlieu J-P, Miyagawa K, Engelkamp D, Rassoulzadegan M, Ross A, Cuzin F, van Heynegen V, Hastie N D. Cell. 1995;81:391–401. doi: 10.1016/0092-8674(95)90392-5. [DOI] [PubMed] [Google Scholar]
- 9.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohno M, Shimura Y. Genes Dev. 1996;10:997–1007. doi: 10.1101/gad.10.8.997. [DOI] [PubMed] [Google Scholar]
- 11.Krainer A R, Maniatis T, Ruskin B, Green M R. Cell. 1984;36:993–1005. doi: 10.1016/0092-8674(84)90049-7. [DOI] [PubMed] [Google Scholar]
- 12.Rio D C. Proc Natl Acad Sci USA. 1988;85:2904–2908. doi: 10.1073/pnas.85.9.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konarska M M, Sharp P A. Cell. 1986;46:845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- 14.Connor F, Wright E, Denny P, Koopman P, Ashworth A. Nucleic Acids Res. 1995;23:3365–3372. doi: 10.1093/nar/23.17.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen-Barak O, Hagiwara N, Arlt M F, Horton J P, Brilliant M H. Gene. 2001;265:157–164. doi: 10.1016/s0378-1119(01)00346-8. [DOI] [PubMed] [Google Scholar]
- 16.Carmo-Fonseca M, Tollervey D, Pepperkok R, Barabino S M, Merdes A, Brunner C, Zamore P D, Green M R, Hurt E, Lamond A I. EMBO J. 1991;10:195–206. doi: 10.1002/j.1460-2075.1991.tb07936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spector D L, Fu X-D, Maniatis T. EMBO J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spector D L. Exp Cell Res. 1996;229:189–197. doi: 10.1006/excr.1996.0358. [DOI] [PubMed] [Google Scholar]
- 19.Boronenkov I V, Loijens J C, Umeda M, Anderson R A. Mol Biol Cell. 1998;9:3547–3560. doi: 10.1091/mbc.9.12.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Osborne S L, Thomas C L, Gschmeissner S, Schiavo G. J Cell Sci. 2001;114:2501–2511. doi: 10.1242/jcs.114.13.2501. [DOI] [PubMed] [Google Scholar]
- 21.Sinclair A H, Berta P, Palmer M S, Hawkins J R, Griffiths B L, Smith M J, Foster J W, Frischauf A M, Lovell-Badge R, Goodfellow P N. Nature (London) 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 22.Harley V R, Jackson D I, Hextall P J, Hawkins J R, Berkovitz G D, Sockanathan S, Lovell-Badge R, Goodfellow P N. Science. 1992;255:453–456. doi: 10.1126/science.1734522. [DOI] [PubMed] [Google Scholar]
- 23.Ferrari S, Harley V R, Pontiggia A, Goodfellow P N, Lovell-Badge R, Bianchi M E. EMBO J. 1992;11:4497–4506. doi: 10.1002/j.1460-2075.1992.tb05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pontiggia A, Rimini R, Harley V R, Goodfellow P N, Lovell-Badge R, Bianchi M E. EMBO J. 1994;13:6115–6127. doi: 10.1002/j.1460-2075.1994.tb06958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bowles J, Cooper L, Berkman J, Koopman P. Nat Genet. 1999;22:405–408. doi: 10.1038/11981. [DOI] [PubMed] [Google Scholar]
- 26.Kamachi Y, Uchikawa M, Kondoh H. Trends Genet. 2000;16:182–187. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- 27.Green M. Annu Rev Cell Biol. 1991;7:559–599. doi: 10.1146/annurev.cb.07.110191.003015. [DOI] [PubMed] [Google Scholar]
- 28.Staley J P, Guthrie C. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 29.Hagiwara N, Klewer S E, Samson R A, Erickson D T, Lyon M F, Brilliant M H. Proc Natl Acad Sci USA. 2000;97:4180–4185. doi: 10.1073/pnas.97.8.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smits P, Li P, Mandel J, Zhang Z, Deng J M, Behringer R R, de Crombrugghe B, Lefebvre V. Dev Cell. 2001;1:277–290. doi: 10.1016/s1534-5807(01)00003-x. [DOI] [PubMed] [Google Scholar]
- 31.Swain A, Lovell-Badge R. Genes Dev. 1999;13:755–767. doi: 10.1101/gad.13.7.755. [DOI] [PubMed] [Google Scholar]
- 32.de Santa Barbara P, Bonneaud N, Boizet B, Desclozeaux M, Moniot B, Südbeck P, Scherer G, Poulat F, Berta P. Mol Cell Biol. 1998;18:6653–6665. doi: 10.1128/mcb.18.11.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schütt C, Nöthiger R. Development (Cambridge, UK) 2000;127:667–677. doi: 10.1242/dev.127.4.667. [DOI] [PubMed] [Google Scholar]
- 34.Salas-Cortés L, Jaubert F, Barbaux S, Nessmann C, Bono M R, Fellous M, McElreavey K, Rosemblatt M. Int J Dev Biol. 1999;43:135–140. [PubMed] [Google Scholar]
- 35.Tacke R, Manley J L. Curr Opin Cell Biol. 1999;11:358–362. doi: 10.1016/S0955-0674(99)80050-7. [DOI] [PubMed] [Google Scholar]
- 36.Bergstrom D E, Young M, Albrecht K H, Eicher E M. Genesis. 2000;28:111–124. [PMC free article] [PubMed] [Google Scholar]
- 37.Vidal V P I, Chaboissier M-C, de Rooij D G, Schedl A. Nat Genet. 2001;28:216–217. doi: 10.1038/90046. [DOI] [PubMed] [Google Scholar]
- 38.Koopman P, Gubbay J, Vivian N, Goodfellow P N, Lovell-Badge R. Nature (London) 1991;351:117–121. doi: 10.1038/351117a0. [DOI] [PubMed] [Google Scholar]