Abstract
The mitochondrion of Trypanosoma brucei lacks tRNA genes. Its translation system therefore depends on the import of cytosolic, nucleus-encoded tRNAs. Thus, most trypanosomal tRNAs function in both the cytosol and the mitochondrion, and all are of the eukaryotic type. This is also the case for the elongator tRNAMet, whereas the only other trypanosomal tRNAMet, the eukaryotic initiator, is found exclusively in the cytosol. Unlike their cytosolic counterparts, organellar initiator tRNAsMet carry a formylated methionine. This raises the question of how initiation of translation works in trypanosomal mitochondria, where only elongator tRNAMet is found. Using in organello charging and formylation assays, we show that unexpectedly a fraction of elongator tRNAMet becomes formylated after import into mitochondria. Furthermore, in vitro experiments with mitochondrial extracts demonstrate that only the trypanosomal elongator and not the initiator tRNAMet is recognized by the formylation activity. Finally, RNA interference assays identify the gene encoding the trypanosomal formylase activity. Whereas the predicted protein is homologous to prokaryotic and mitochondrial methionyl-tRNAMet formyltransferases, it has about twice the mass of any of these proteins.
Keywords: mitochondrial translation‖initiator tRNAMet‖formylation‖mitochondrial tRNA import‖methionyl-tRNAMet formyltransferase
Initiation of protein synthesis occurs universally with the amino acid methionine in eukaryotes and Archea or its derivative formyl-methionine in Bacteria and organelles. All organisms have two classes of tRNAsMet, an initiator tRNAMet-i, which is used for initiation of protein synthesis, and an elongator tRNAMet-e, which functions in the insertion of methionine into internal peptidic linkages (1, 2). Whereas elongator tRNAsMet-e of all organisms look similar, there are two distinct groups of initiator tRNAsMet-i. Bacterial and organellar initiator tRNAsMet-i are only functional when carrying a formylated methionine. Addition of the formyl group to the methionine of the charged initiator tRNAMet-i is catalyzed by methionyl-tRNAMet formyltransferase (MTF), which is found in Bacteria and organelles but not in Archea or the eukaryotic cytosol. It recognizes specific features of the tRNA such as the CA mismatch at the top of the acceptor stem (position 1–72), which is conserved in bacterial initiator tRNAsMet-i. Initiator tRNAsMet-i of the eukaryotic cytosol and Archea, on the other hand, carry an A1·T72 base pair, which is not found in any other tRNA. Mitochondrial tRNAsMet are less clearly defined. Often, only a single type of tRNAMet is found, which is assumed to be used as an initiator in the formylated state and as an elongator when carrying an underivatized methionine (3). If only a single tRNAMet is found in mitochondria, it most closely resembles an initiator type; however, frequently an A1·T72 base pair replaces the typical C1·A72 mismatch. Thus, many mitochondrial tRNAsMet superficially resemble eukaryotic initiator tRNAsMet-i.
Whereas in many eukaryotes all of the tRNAs necessary for organellar translation are encoded on the mitochondrial genome, there is a large variety of organisms where a variable number of apparently essential mitochondrial tRNA genes are missing. It has been shown that the lack of these genes is generally compensated by import of the corresponding cytosolic tRNAs. In all cases known, imported tRNAs originate from nuclear genes encoding cytosolic, eukaryotic-type tRNAs involved in cytosolic translation (4). An extreme situation is found in the trypanosomatids, which have lost their entire set of mitochondrial tRNA genes and therefore must import all tRNAs necessary for organellar protein synthesis (5, 6). In these organisms, the bacterial-type translation system of mitochondria is faced with the problem that it has to function with imported, eukaryotic-type tRNAs only. This raises the question of how mitochondrial translation initiation, known to require a formylated bacterial-type initiator tRNAMet-i, can function with imported eukaryotic-type tRNAsMet. In this study, we have used the trypanosomatid Trypanosoma brucei to investigate this problem.
Materials and Methods
Cells.
Procyclic T. brucei, stock 427, was grown at 27°C in SDM-79 medium supplemented with 5% FBS. Cells were harvested at late-log phase corresponding to 2.5 to 4.5 × 107 cells/ml.
Isolation of Mitochondria.
Mitochondria used for in organello aminoacylation and formylation assays were isolated under isotonic conditions as described (7). The final fraction of mitochondria was resuspended at high protein concentration (30–50 mg/ml) in SoTE (0.6 M sorbitol/20 mM Tris⋅HCl, pH 7.5/2 mM EDTA) containing 20 mg/ml fatty acid-free BSA and frozen in aliquots. Mitochondria purified with this method retain an intact outer membrane and are respiration competent (8).
In Organello Aminoacylation and Formylation Assays.
All assays were done in 20 mM Tris⋅HCl, pH 7.2/15 mM KH2PO4/0.6 M sorbitol/20 mM MgSO4/5 mg/ml fatty acid-free BSA. Aminoacylation reactions were performed in 50 μl containing 750 μg of isotonically purified mitochondria and 20–40 μCi (1 Ci = 37 GBq) of [35S]methionine/[35S]cysteine mixture (>1,000 Ci/mmol) (from Hartmann Analytic or from Amersham Pharmacia). The labeled [35S]cysteine (ca. 25% of total) was competed out by the addition of 0.5 mM unlabeled cysteine. Intramitochondrial ATP production was induced by the addition 5 mM α-ketoglutarate/5 mM glycerol 3-phosphate/2 mM ADP (8). In organello formylation reactions were done as described for the aminoacylation assays, except that where applicable (see Fig. 3A) 5 μCi of [14C]formate (55 mCi/mmol) (Hartmann Analytic) was used and that the [35S]methionine/[35S]cysteine mixture was replaced by 1 mM unlabeled methionine. The reactions were incubated for 15 min at 30°C, and the mitochondria were reisolated by centrifugation (8,000 × g, 5 min at 4°C). Finally, RNAs were isolated from the mitochondrial pellets by using the acid guanidinium method (10), and 35S- or 14C-labeled tRNAsMet were analyzed on 8 M urea/10% polyacrylamide gels and visualized by fluorography. It was important to prevent deacylation of the labeled tRNAsMet during electrophoresis by either running the gel under acidic condition (11) or by subjecting the sample to a deamination procedure (5) before electrophoresis. Alternatively to the gel electrophoresis, aminoacylation and formylation of tRNAsMet were also analyzed by the TLC-based procedure described below.
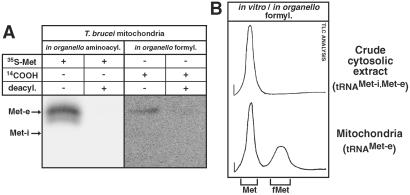
Figure 3.
Formylation activity in mitochondria of T. brucei. (A) In organello aminoacylation assays (Left) and formylation assays (Right) are shown. Isolated mitochondria were incubated with [35S]methionine (35S-Met) or [14C]-formate (14COOH) and subjected to deacylation (deacyl.) as indicated. The deamidated samples were separated on a high resolution 8 M urea/10% polyacrylamide gel at pH 8.0 and visualized by fluorography. The double band (Left) is an artifact of the deamination procedure (5). The positions of tRNAMet-e and tRNAMet-i as determined by Northern analyses of total RNA are indicated. (B) Tracings of the [35S]methionine signals from TLC analyses of formylation reactions (see text for details). (Upper) In vitro formylation assay performed with crude cytosolic extract and [35S]methionine. (Lower) 35S signals obtained from an in organello formylation assay (Mitochondria). The tRNAsMet known to be present in the two fractions are indicated in parentheses. The positions of unlabeled methionine (Met) and formylmethionine (fMet) separated under the same conditions in a parallel TLC lane are indicated. The origin is marked by the short vertical line on the left.
In Vitro Formylation Assays.
Preparation of RNA-free cytosolic and mitochondrial matrix fraction was done as described (12). Substrate tRNAs were prepared as follows. Total or mitochondrial RNA was isolated according to the acid guanidinium method (10) and separated on a preparative 8 M urea/10% polyacrylamide gel run at pH 8.0. The region of the gel containing the tRNAs was visualized by ethidium bromide staining and cut out. After an overnight elution, the tRNAs were ethanol-precipitated and quantified by A260 measurements.
In vitro charging and formylation assays were performed in 50 μl with 30–60 μg of RNA-free cytosolic or mitochondrial matrix fraction in 1× acylation buffer (50 mM Tris⋅HCl, pH 7.5/25 mM KCl/8 mM MgCl2) containing 2 mM ATP/0.1 mM 10-formyl-tetrahydrofolate (THF)/1 mM cysteine/10 mM 2-mercaptoethanol/0.3% (wt/vol) CHAPS and the same amount of [35S]methionine that was used for the in organello assays. The following amounts of substrate tRNAs were used: 7 μg for isolated cytosolic tRNA, 3 μg for isolated mitochondrial tRNAs, and 1.5 μg each of gel-eluted cytosolic fraction containing either tRNAMet-e or tRNAMet-i only (see Fig. 4). After incubation for 15 min at 30°C, the tRNAs were reisolated by using the acid guanidinium procedure and analyzed as described for the in organello assays.
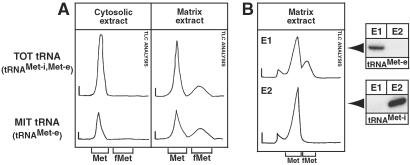
Figure 4.
Mitochondrial formylation activity selectively recognizes elongator tRNAMet-e. Tracings of the 35S signals from TLC analyses of in vitro formylation reactions using RNA-free cytosolic and RNA-free mitochondrial matrix extract are shown. (A) Gel-eluted total tRNAs (TOT tRNA, Upper) and gel-eluted mitochondrial tRNAs (MIT tRNA, Lower) were used as substrates in the experiments. (B) Gel-purified tRNAMet-e (E1) or tRNAMet-i (E2) originating from total cellular RNA were tested. The low intensity signal migrating slower than methionine was specific for the isotope mixture from Hartmann Analytic and was not found if the Amersham Pharmacia product was used. It coincides with the position of a cysteine marker. The positions of unlabeled methionine (Met) and formyl-methionine (fMet) are indicated as in Fig. 3. The purity of the eluted tRNA fractions, regarding the initiator and elongator tRNAsMet, was tested by the Northern blots shown (Right), which were probed with the same oligonucleotides that were used in Fig. 1.
To distinguish methionylated from formylmethionylated tRNAsMet, the following procedure was developed. Isolated 35S-labeled tRNAs from an in organello or an in vitro assay were deacylated in 15 μl of 1 M Tris⋅HCl, pH 9.2, for 1–2 h at 30°C. In a next step, the tRNAs were precipitated by the addition of 2.5 vol of cold ethanol, and the sample was pelleted at 16,000 × g for 20 min at 4°C. The supernatant containing the released [35S]methionine or formyl[35S]methionine was dried down and resuspended in 15 μl of water. Finally, 2 μl of each reaction was spotted on a cellulose TLC plate, which was developed in a 13:3:1 mixture of diethylether, acetic acid, and water. Unlabeled methionine and formylmethionine (2 μl each) dissolved in 1 M Tris⋅HCl, pH 9.2, were used as markers and visualized by UV. The fractionated labeled amino acids were detected by using a gas flow Geiger Müller counter (TLC-linear analyzer, Berthold, Nashua, NH) and displayed on the monitor of a multichannel analyzer (system BS 27/N, Berthold). The data shown in Figs. 3, 4, and 6 are the tracings reproduced from the pictures on the monitor. Exposure times varied from 30 min to 4 h. For all experiments, 10% of the reaction before and after the deacylation step was analyzed on an acidic 8 M urea/10% polyacrylamide gel. This served to confirm the identity of the labeled tRNAsMet and the completeness of the deacylation as well as the integrity of the tRNA.
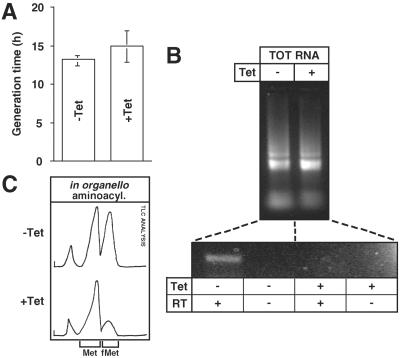
Figure 6.
Depletion of trypanosomal MTF by tetracycline-inducible RNA interference. (A) Mean values of the generation times for cells grown in the absence (−Tet) and the presence (+Tet) of tetracycline over an observed growth period of 14 days. (B Upper) Ethidium bromide-stained agarose gel of 3 μg each of total RNA (TOT RNA) isolated from uninduced (−) and induced (+) cells. (Lower) RT-PCR analyses of RNA from both cell lines using primers specific for the MTF mRNA (see Materials and Methods). Samples without RT serve as controls to show that MTF mRNA was specifically amplified. (C) TLC analyses of in organello formylation reactions by using digitonin-purified mitochondria from cells grown in the absence (ratio of the two peaks, 50:50) and the presence of tetracycline (ratio of the two peaks 78:22). For explanation of the additional slow migrating peak, see Fig. 4.
3′-End Splint Labeling.
Quantitative 3′-end modification of distinct RNA species in a complex mixture of RNAs can be achieved with specific oligonucleotides and Sequenase 2.0 (United States Biochemical) by using the 3′-end splint-labeling protocol as originally described by Hausner et al. (13). In this method, the oligonucleotide hybridizes to the 3′ end of the RNA, where it leaves a 5′ overhang that allows the enzyme to add the corresponding complementary dNTP to the 3′ end of the selected RNA. Quantitative 3′-end modification of the elongator tRNAMet-e by dCTP addition was done by using the oligonucleotide (GTGGTGCGATCGGTGAGGCTCGAACTCA) and 100 μM dCTP as described (14). The control sample was treated identically except that an oligonucleotide known to modify the tRNALys was used (14). After 3′-end modification, both tRNA fractions were subjected to in vitro aminoacylations as described above.
RNA Interference Assay.
A fragment corresponding to the 5′ part of trypanosomal MTF (nucleotide 16–509 from the ATG) was amplified by using the 5′ primer (CCGCTCGAGGTGTGGACATAGTG) and the 3′ primer (GGCAAGCTTTTCGCCATAGACGTCG) having flanking XhoI and HindIII sites, respectively. The resulting 493-bp fragment was cloned into the corresponding restriction sites of the pZJM vector (15). The linearized vector was then transfected into the procyclic T. brucei strain 29-13, which expresses T7 RNA polymerase and the tetracycline repressor (16). Selection and induction with tetracycline were done as described (15). To estimate the MTF–mRNA quantities in cells grown in the presence and absence of tetracycline, total RNA was isolated from an equal number of uninduced and induced cells and used for a reverse transcriptase (RT)–PCR analysis by using avian myeloblastosis virus reverse transcriptase (Promega) and the same set of primers as above. Previous experiments have demonstrated that this method yields semiquantitative results if analyzed in the linear range of the amplification. Finally, in organello formylation activity was compared in both cell lines. Washed cells (108 cells each) of cultures grown in the presence and the absence of tetracycline were resuspended in 0.5 ml of SoTE; 0.5 ml of SoTE containing 0.025% (wt/vol) of digitonin was added, and the sample was mixed by pipetting and incubated on ice for 5 min. After centrifugation (8,000 × g for 5 min at 4°C), the supernatant was discarded, and the resulting mitochondrial pellet was resuspended in 250 μl of the buffer used for the in organello formylation assays. Control experiments have shown that the mitochondrial fraction obtained by digitonin treatment is capable of respiratory substrate-driven ATP production. The labeling reaction and the analysis of the labeled products was done as described for the in organello assays.
Miscellaneous.
Northern blots were done as described (17). The indicated oligonucleotides directed against the T-stem loop region were used for the specific detection of tRNAMet-i (GTTGGTTTCGATCCAACG) and tRNAMet-e (GTGAGGCTCGAACTCACG).
Results
tRNAsMet in T. brucei.
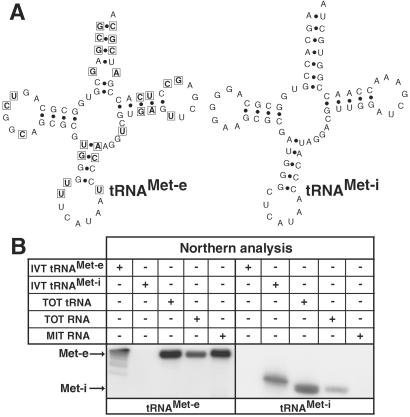
Analysis of the nearly complete genomic sequence of T. brucei (kindly provided by The Institute for Genomic Research and the Sanger Centre) has identified genes for two distinct but homologous tRNAsMet whose predicted secondary structures are shown in Fig. 1A. One resembles a standard elongator tRNAMet-e; the other one has an A1·U72 base pair and three consecutive G·C base pairs at the end of the anticodon stem and therefore corresponds to the cytosolic initiator tRNAMet-i (1). Northern analysis by using oligonucleotides specifically hybridizing to either of the two tRNAsMet is depicted in Fig. 1B. The left panel shows that a band is detected in total cellular RNA, which in size corresponds closely to the in vitro transcript of the elongator tRNAMet-e. The band was detected in both RNA isolated from whole cells as well as from the mitochondrial fraction, indicating that the tRNA is in part imported into mitochondria. The right panel shows the analogous experiment for the initiator tRNAMet-i, which is found in the cytosol only. In this case, the in vitro transcript migrates a bit slower than the molecules detected in isolated RNA fractions, which is due to the fact that it is one nucleotide longer than the expected mature tRNA.
Figure 1.
Elongator and initiator tRNAsMet in T. brucei. (A) Predicted secondary structures of the homologous elongator and cytosolic initiator tRNAsMet. Nucleotides that are different between the two molecules are boxed in the elongator tRNAMet. (B) Duplicate Northern blots were analyzed with oligonucleotide probes specific for elongator tRNAMet-e (Left) and initiator tRNAMet-i (Right). The following RNA fractions were tested: IVT tRNAMet-e/Met-i (5 ng each), in vitro transcripts of the corresponding tRNAsMet; TOT RNA, RNA isolated from total cell (3 μg each); TOT tRNA, gel-eluted tRNAs from a total RNA fraction (2.1 μg each); and MIT RNA, RNA isolated from mitochondria (3 μg each).
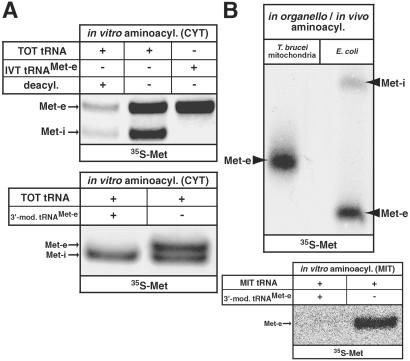
Are there other as yet undetected tRNAsMet in T. brucei? This is clearly possible, as in a recent genome-wide analysis it was concluded that only 80% of all trypanosomal tRNA genes have been identified (R. Pach and A.S., unpublished results). To detect all existing tRNAsMet, we performed aminoacylation assays by using RNA-free cytosol as source of enzyme and total cellular tRNAs as substrate. Fig. 2A (Upper) shows that two charged tRNAMet species are present in the cell. The slower migrating one corresponds in size to the charged in vitro transcript of the elongator tRNAMet-e, whereas the other one comigrates with the corresponding signal for initiator tRNAMet-i (data not shown). Separate experiments confirmed that the signals of both charged tRNA species coincide with the signals obtained on Northern blots (data not shown). To exclude that a second, as yet unidentified tRNAMet, which comigrates with the known elongator tRNAMet, is contributing to the observed aminoacylation signal, further charging assays were performed. In these assays, we used a total tRNA fraction in which the 3′-end of the elongator tRNAMet-e was quantitatively modified by 3′-end splint labeling (13, 14, 18) as a substrate (see Material and Methods). Fig. 2A (Lower) shows that this treatment completely abolishes aminoacylation of the elongator tRNAMet-e, whereas charging of the homologous initiator tRNAMet-i is not affected. These results therefore strongly suggest that [35S]methionine-labeled band corresponding to the elongator tRNAMet-e is caused by this tRNA only. Should another as yet unknown tRNAMet contribute to the observed signal, it would not only need to comigrate with the elongator tRNAMet-e (even on high resolution gels) but also to share the same 3′ sequence. Thus, the only two bands detected by aminoacylation (Fig. 2A) can be assigned to the two known trypanosomal tRNAsMet, indicating that no other tRNAsMet exist in T. brucei.
Figure 2.
Aminoacylation assays by using [35S]methionine. (A Upper) In vitro aminoacylation (aminoacyl.) of gel-eluted total tRNA (TOT tRNA) and in vitro transcribed elongator tRNAMet (IVT tRNAMet-e) with [35S]methionine (35S-Met) by using RNA-free cytosolic extract (CYT). Incubation at pH 9.2 leads to deacylation (deacyl.) of the charged tRNAsMet. The positions of the corresponding signals obtained on duplicate Northern blots are indicated by arrows. (Lower) Aminoacylation assays using cytosolic extract and total tRNA in which the 3′-end of the elongator tRNAMet-e was modified by splint labeling (3′-mod., left lane) or mock-treated (right lane). All samples were subjected to a deamination reaction before they were separated on an 8 M urea/10% polyacrylamide gel at pH 8.0. (B Upper) In organello and in vivo aminoacylation assay of isolated T. brucei mitochondria or E. coli cells, respectively, using [35S]methionine. Samples were analyzed on a high resolution 8 M urea/10% polyacrylamide gel run at pH 5.0 (11) and visualized by fluorography. The positions of mitochondrial elongator tRNAMet-e as obtained on a duplicate Northern blot and of eubacterial initiator and elongator tRNAsMet are indicated. (Lower) Aminoacylation assays using matrix extract (MIT) and mitochondrial tRNA (MIT tRNA) in which the 3′-end of the elongator tRNAMet-e was modified by splint labeling (3′-mod., left lane) or mock-treated (right lane). All samples were subjected to a deamination reaction before they were separated on an 8 M urea/10% polyacrylamide gel at pH 8.0.
To look at the situation in mitochondria, we performed in organello aminoacylation experiments that are expected to label all possible mitochondrial tRNAsMet. Isolated mitochondria were incubated with respiratory substrates, ADP and [35S]methionine. The ATP needed by the mitochondrial methionyl–tRNA synthetase is provided by respiratory substrate-driven oxidative phosphorylation (8). Fig. 2B shows that only a single band comigrating with in vitro transcribed elongator tRNAMet-e is detected. On the right lane, an in vivo charging experiment with Escherichia coli is shown to demonstrate the resolving power of the gel system, which readily separates bacterial initiator and elongator tRNAsMet that are identical in size. As with total tRNA, modification of the mitochondrial elongator tRNAMet-e by 3′-end splint labeling results in a complete loss of the observed aminoacylation signal (Fig. 2B Lower). Thus, the imported elongator tRNAMet-e seems to be the only tRNAMet that is present in mitochondria.
Formylation Activity in T. brucei Mitochondria.
No initiator tRNAMet exists in T. brucei mitochondria, which raises the question of which tRNA is used for translation initiation and whether it becomes formylated. To detect formylated trypanosomal methionyl-tRNAMet, we used two approaches. First, analogous to the aminoacylation assay described in the previous paragraph, we performed in organello formylation assays. The formyl group in formylated initiator tRNAMet-i is derived from 10-formyl-THF. The compound is synthesized by formyl-THF synthase, which can use formate as a formyl donor. Isolated mitochondria were therefore incubated with respiratory substrates, ADP and [14C]formate, and processed as described in Materials and Methods. Fig. 3A (Right) shows a [14C]formate-labeled band that, in size, corresponds to the imported elongator tRNAMet-e visualized (Left) by in organello aminoacylation with [35S]methionine. In both experiments, the radioactive label is sensitive to deacylation at pH 9.2, indicating that the label is on the amino acid. Thus, it is concluded that formylation of a tRNA species that, in size, corresponds to the elongator tRNAMet-e occurs in T. brucei mitochondria. In the second approach, we established a method that allows the direct detection of formyl-methionine. Aminoacylation assays by using [35S]methionine were performed either in energized mitochondria or in vitro in the presence of ATP and the formyl donor 10-formyl-THF. After incubation, [35S]methionine-labeled RNA was isolated and incubated at pH 9.2. This condition results in the release of the charged amino acid, which is then analyzed by TLC optimized to separate methionine from its formylated derivative. Using this protocol, it was shown that from crude cytosolic extract containing RNA and proteins, the only 35S-labeled molecule released during the deacylation reaction was methionine (Fig. 3B Upper). In an in organello formylation assay, however, not only [35S]methionine but also formyl-[35S]methionine is detected (Fig. 3B Lower). The obtained results are in full agreement with the formate-labeling experiment discussed before (Fig. 3A) and show that a significant fraction (40–50%, depending on the experiment) of the elongator methionyl-tRNAMet-e is formylated in mitochondria of T. brucei.
To look at the substrate specificity of the activity in some more detail, we performed further types of in vitro formylation assays. As a source of enzyme we used RNA-free cytosolic and mitochondrial matrix fractions (see Materials and Methods). These fractions were incubated with ATP, [35S]methionine, and 10-formyl-THF by using both crude total tRNA or isolated mitochondrial tRNAs as substrates. Fig. 4A Left shows that cytosolic extract is able to charge both the cytosolic as well as the mitochondrial tRNAsMet but is devoid of formylation activity. The mitochondrial matrix, on the other hand, shows both methionyl-tRNA synthetase as well as formylation activity, as evidenced by the two peaks that are detected by TLC (Fig. 4A Right). Interestingly, the mitochondrial activity not only formylates isolated mitochondrial tRNAs but, to a lesser extent, also cytosolic tRNAs. The cytosolic tRNA fraction contains both the cytosol-specific initiator tRNAMet-i as well as the nonimported fraction (ca. 94% of total) of elongator tRNAMet-e (R. Pach and A.S., unpublished results). To see which of the two molecules in the cytosolic fraction is formylated in the in vitro reaction, the two tRNAsMet were separated from each other and eluted from a denaturing polyacrylamide gel. Fig. 4B Left shows that elongator tRNAMet-e from the cytosolic fraction is formylated by the matrix-localized activity with similar efficiency to the elongator tRNAMet-e of mitochondrial origin. The formylation activity therefore does not discriminate between cytosolic and mitochondrial isotype of elongator tRNAMet-e. Indeed, extensive RT-PCR analysis suggests that elongator tRNAsMet-e isolated from mitochondria and from the cytosol have identical nucleotide sequences (not shown). The cytosol-specific initiator tRNAMet-i, however, cannot be formylated. This is surprising, as the eukaryotic-type initiator tRNAMet-i more closely resembles the mitochondrial initiator tRNAMet-i of other organisms than does the trypanosomal elongator tRNAMet-e. In vitro formylation assays were also performed by using in vitro transcribed elongator tRNAMet-e as substrates. Whereas the synthetic tRNAMet-e could be charged efficiently (Fig. 2A, right lane), the in vitro transcript could not be formylated, indicating an essential role for a nucleotide or base modification for the recognition by the trypanosomal MTF (data not shown).
The Gene for Mitochondrial MTF of T. brucei.
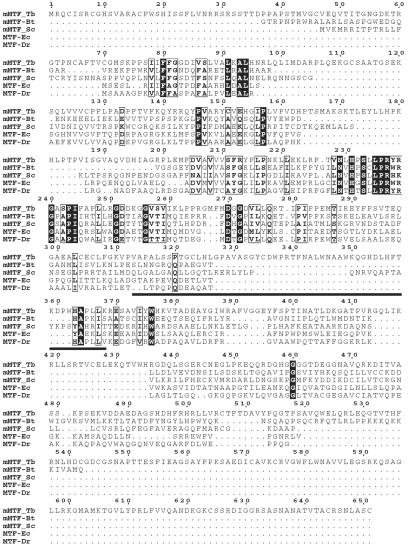
To identify the gene encoding the formylation activity, we searched the T. brucei genomic databases (kindly provided by The Institute of Genomic Research and the Sanger Centre) with the default BLAST algorithm by using bacterial and mitochondrial MTF amino acid sequences as templates. The obtained genomic fragments allowed us to assemble a contig corresponding to a complete ORF. Expression of this ORF was confirmed by amplification of the corresponding cDNA by using an oligo(dT) primer and a gene-specific primer hybridizing to the 5′-untranslated region of the mRNA. Furthermore, analysis of the cDNA showed that its sequence is identical to the genomic DNA. An alignment of the predicted protein sequence of this ORF with two mitochondrial (cattle, yeast) and two bacterial MTFs (Deinococcus radiodurans, E. coli) is shown in Fig. 5. If the ORF of the trypanosomal gene is compared with the complete nonredundant database, the only homologous proteins detected are bacterial and mitochondrial MTFs. The amino terminus of the putative trypanosomal MTF is predicted to have a mitochondrial targeting sequence when analyzed with the iPSORT or MITOPROT programs. Interestingly, however, the predicted trypanosomal protein is approximately twice the size of all other MTFs.
Figure 5.
Multiple sequence alignment of mitochondrial and bacterial MTF polypeptides. The sequences of three mitochondrial MTFs (mMTF) from T. brucei (Tb), Bos taurus (Bt), and Saccharomyces cerevisiae (Sc) and two bacterial MTFs from E. coli (Ec) and D. radiodurans (Dr) were aligned by using the CLUSTAL W program with default parameters. Strictly conserved residues are shown in black boxes. Conservative replacements are boxed. The regions corresponding to the linker region (position 313–368) as defined in E. coli are underlined.
Recently, it was shown that the phenomenon of RNA interference does occur in T. brucei and can be used to inhibit expression of specific genes (15, 19, 20). We therefore used this technique to test whether the identified ORF indeed encodes the trypanosomal MTF. To do so, we established a stable cell line expressing sense and antisense strands of the 5′-segment of the putative MTF gene under the control of two opposing tetracycline-inducible T7 RNA polymerase promoters (16). The RT-PCR experiments in Fig. 6B show the mRNA level of the trypanosomal MTF homologue drops below detectable levels after addition of the antibiotic. Equal numbers of cells grown in the presence or the absence of tetracycline were subjected to digitonin treatment to obtain a crude mitochondrial fraction (see Materials and Methods). Both organellar fractions were then used for in organello formylation assays identical to the ones described before (Fig. 3B), and the formylation state of the imported elongator tRNAMet-e was analyzed by TLC (Fig. 6C). The experiment showed that methionyl-tRNA synthetase activity was comparable in both samples, but the formylation activity was greatly reduced in mitochondria from the cells grown in the presence of tetracycline. The formylated fraction of the elongator tRNAMet-e (ca. 50%) in the cells grown in the absence of tetracycline was larger than in the experiments shown in Fig. 3B (ca. 40%), which could be due to the fact that different mitochondrial isolation procedures and cell lines were used in the two experiments. The cells with reduced formylation activity did not show a growth phenotype (Fig. 6A), which might be because there is still enough of the enzyme left to support normal growth. Thus, we conclude that the characterized ORF indeed encodes the trypanosomal MTF and that it is this enzyme that is responsible for the formylation of the imported elongator methionyl-tRNAMet-e in T. brucei.
Discussion
As expected for a eukaryote, two distinct tRNAsMet are found in the cytosol of T. brucei. One shows all of the properties of a eukaryotic initiator tRNAMet, whereas the other one is an elongator tRNAMet. However, unlike in other eukaryotes, there are no mitochondrial tRNA genes in T. brucei. The two nucleus-encoded tRNAsMet therefore have to be sufficient for two translation systems, one in the cytosol and one in mitochondria. Moreover, only the elongator tRNAMet-e is imported into mitochondria, indicating that initiation and elongation of organellar translation must use a single tRNAMet. This is not unusual, as many mitochondria have only a single tRNAMet, which, however, shows at least some structural features of typical initiator tRNAsMet (3). In T. brucei, the situation is very different because the imported tRNAMet is apparently identical to the one that is used in cytosolic elongation. A hallmark of bacterial-type translation is that it uses formylated tRNAMet-i. Interestingly, formylation of tRNAMet still occurs in T. brucei mitochondria, even though an initiator-type tRNAMet is absent. T. brucei therefore has a MTF that naturally recognizes elongator tRNAMet-e. Thus, the substrate specificity of the trypanosomal MTF is diametrically opposed to bacterial or mitochondrial MTFs, which in vivo only formylate initiator and not elongator tRNAsMet. The bovine mitochondrial MTF, which is the only mitochondrial MTF characterized so far (21), has a relaxed substrate specificity when compared with the bacterial enzyme, especially concerning the acceptor stem region of the substrate tRNAMet (3). The trypanosomal elongator tRNAMet-e, however, is devoid of all of the known structural features that are recognized by the mitochondrial MTF in the bovine system. Neither the first three acceptor stem base pairs of the bovine substrate nor the purine-11·pyrimidine-24 base pairs conserved in most initiator tRNAsMet are found in trypanosomal tRNAMet-e. In agreement with this, it was shown that the bovine MTF has only residual activity (0.2% when compared with the natural substrate) when tested with elongator tRNAMet of E. coli (3).
The features of the trypanosomal elongator tRNAMet-e that are recognized by the mitochondrial MTF are also present in the cytosolic fraction of the elongator tRNAMet-e. A mitochondrial tRNA-editing event as prerequisite for formylation can therefore be excluded. Furthermore, in vitro experiments have shown that the enzyme can discriminate between the elongator tRNAMet-e and the homologous cytosol-specific initiator tRNAMet-i. In vitro transcribed elongator tRNAMet-e, on the other hand, can be aminoacylated but not formylated when treated with mitochondrial extract. This suggests that a nucleotide or base modification is required as a recognition element for the trypanosomal MTF.
Mining the genome database and RNA interference assay allowed us to identify the gene responsible for the mitochondrial MTF activity in T. brucei. An alignment of three mitochondrial and two bacterial MTFs is shown in Fig. 5. The general structure of MTFs consisting of two domains that are separated by a linker region seems to be retained in trypanosomes (22, 23). Interestingly, the linker region (at position 313–368) is longer than in any of the other enzymes. Most strikingly, however, is the fact that the protein is approximately twice the size of any other MTFs. The larger size of the trypanosomal protein is mainly because of unique amino-terminal (ca. 40 aa) and carboxyl-terminal (ca. 110 aa) extensions as well as to four internal insertions (13–22 aa). As for the bovine mitochondrial MTF (3), no obvious sequence element corresponding to the previously described 16-aa insertion sequence in the amino-terminal domain of the E. coli enzyme (22) is found in the trypanosomal enzyme. This element is responsible for interaction with the acceptor stem of the tRNAMet-i (24); its absence might therefore by explained by the fact that the trypanosomal enzyme must recognize the acceptor stem of an elongator-type tRNAMet-e.
In summary, we have shown that in T. brucei, a single gene product, the nucleus-encoded elongator tRNAMet-e, has three distinct functions in two cellular compartments. It is required for translation elongation in the cytosol and for both the initiation and elongation step in mitochondrial translation. A fraction of the imported elongator methionyl-tRNAMet-e becomes formylated in mitochondria. This formylation is carried out by an unusual MTF, which compared with the corresponding enzymes in other cells, has an inversed specificity concerning the tRNA substrate and selectively recognizes elongator tRNAMet-e.
Besides the unusual MTF described in this study, there are other examples for adaptations of the trypanosomatid mitochondrial translation system to eukaryotic-type tRNAs. In Leishmania mitochondria, a C to U RNA-editing event in the anticodon of the imported tRNATrp(CCA) allows the decoding of UGG and UGA codons. The unedited tRNATrp(CCA) in the cytosol, however, as required reads the standard UGG tryptophan codon only (25). Finally, whereas organelles are known to be devoid of glutaminyl-tRNA synthetases, as their tRNAs are charged in a two-step process requiring a tRNA-dependent transamidase (9), such enzymes exist in trypanosomatid mitochondria (12). Thus, exploring the limits of adaptation of a bacterial-type translation system to eukaryotic components may help to reveal fundamental requirements of translation.
Acknowledgments
We thank Drs. P. Englund and G. Cross for providing us with the pZJM vector and the T. brucei 29-13 strain, respectively. This study was supported by Swiss National Foundation Grants 31-056825.99 and 4037-55154 and by a fellowship of the Prof. Dr. Max Cloëtta Foundation.
Abbreviations
- MTF
methionyl-tRNAMet formyltransferase
- THF
tetrahydrofolate
- RT
reverse transcriptase
Footnotes
References
- 1.RajBhandary U L. J Bacteriol. 1994;176:547–552. doi: 10.1128/jb.176.3.547-552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanquet S, Mechulam Y, Schmitt E. Curr Opin Struct Biol. 2000;10:95–101. doi: 10.1016/s0959-440x(99)00055-x. [DOI] [PubMed] [Google Scholar]
- 3.Takeuchi N, Vial L, Panvert M, Schmitt E, Watanabe K, Mechulam Y, Blanquet S. J Biol Chem. 2001;276:20064–20068. doi: 10.1074/jbc.M101007200. [DOI] [PubMed] [Google Scholar]
- 4.Schneider A, Marechal-Drouard L. Trends Cell Biol. 2000;10:509–513. doi: 10.1016/s0962-8924(00)01854-7. [DOI] [PubMed] [Google Scholar]
- 5.Hancock K, Hajduk S L. J Biol Chem. 1990;265:19208–19215. [PubMed] [Google Scholar]
- 6.Simpson A M, Suyama Y, Dewes H, Campbell D A, Simpson L. Nucleic Acids Res. 1989;17:5427–5445. doi: 10.1093/nar/17.14.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hauser R, Pypaert M, Häusler T, Horn E K, Schneider A. J Cell Sci. 1996;109:517–523. doi: 10.1242/jcs.109.2.517. [DOI] [PubMed] [Google Scholar]
- 8.Allemann N, Schneider A. Mol Biochem Parasitol. 2000;111:87–94. doi: 10.1016/s0166-6851(00)00303-0. [DOI] [PubMed] [Google Scholar]
- 9.Schön A, Kannangara C G, Gough S, Söll D. Nature (London) 1988;331:187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- 10.Chomczyinski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 11.Varshney U, Lee C-P, RajBhandary U L. J Biol Chem. 1991;266:24712–24718. [PubMed] [Google Scholar]
- 12.Nabholz C E, Hauser R, Schneider A. Proc Natl Acad Sci USA. 1997;94:7903–7908. doi: 10.1073/pnas.94.15.7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hausner T P, Giglio L M, Weiner A M. Genes Dev. 1990;4:2146–2156. doi: 10.1101/gad.4.12a.2146. [DOI] [PubMed] [Google Scholar]
- 14.Schneider A, McNally K P, Agabian N. Nucleic Acids Res. 1994;22:3699–3705. doi: 10.1093/nar/22.18.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, Morris J C, Drew M E, Englund P T. J Biol Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- 16.Wirtz E, Leal S, Ochatt C, Cross G A. Mol Biochem Parasitol. 1999;99:89–101. doi: 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 17.Hauser R, Schneider A. EMBO J. 1995;14:4212–4220. doi: 10.1002/j.1460-2075.1995.tb00095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang Z, Szostak J W. Nucleic Acids Res. 1996;24:4360–4361. doi: 10.1093/nar/24.21.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ngo I, Tschudi C, Gull K, Ullu E. Proc Natl Acad Sci USA. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shi H, Djikeng A, Mark T, Wirtz E, Tschudi C, Ullu E. RNA. 2000;6:1069–1076. doi: 10.1017/s1355838200000297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takeuchi N, Kawakami M, Omori A, Ueda T, Spremulli L L, Watanabe K. J Biol Chem. 1998;273:15085–15090. doi: 10.1074/jbc.273.24.15085. [DOI] [PubMed] [Google Scholar]
- 22.Schmitt E, Blanquet S, Mechulam Y. EMBO J. 1996;15:4749–4758. [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitt E, Panvert M, Blanquet S. EMBO J. 1998;17:6819–6826. doi: 10.1093/emboj/17.23.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramesh V, Gite S, RajBhandary U L. Biochemistry. 1998;37:15925–15932. doi: 10.1021/bi981873x. [DOI] [PubMed] [Google Scholar]
- 25.Alfonzo J D, Blanc V, Estevez A M, Rubio M A T, Simpson L. EMBO J. 1999;18:7056–7062. doi: 10.1093/emboj/18.24.7056. [DOI] [PMC free article] [PubMed] [Google Scholar]