Abstract
The primary target of SgrAI restriction endonuclease is a multiple sequence of the form 5′-CPu↓CCGGPyG. Previous work had indicated that SgrAI must bind two recognition sites simultaneously for catalysis [Bilcock, D. T., Daniels, L. E., Bath, A. J. & Halford, S. E. (1999) J. Biol. Chem. 274, 36379–36386]. In the present study, SgrAI is shown to cleave not only its canonical sequences, but also the sequences 5′-CPuCCGGPy(A,T,C) and 5′-CPuCCGGGG, both referred to as secondary sequences. On plasmid pSK7, SgrAI cleaves secondary sites 26-fold slower than the canonical site. However, the same plasmid, but without the canonical site, is cleaved 200-fold slower. We show that DNA termini generated by cleaving the canonical site for SgrAI assist in the cleavage of secondary sites. The SgrAI-termini in cis with respect to secondary site are markedly preferred over those in trans. The SgrAI-termini provided in a form of oligonucleotide duplex are also shown to stimulate canonical site cleavage. At a 40-fold molar excess of the SgrAI-termini over substrate, the SgrAI specificity is shown to improve by two orders of magnitude, because of concurrent 10-fold increase in the cleavage of canonical site and 50-fold decrease in the cleavage of secondary sites. The unconventional reaction pathway by which SgrAI utilizes the self-generated DNA termini to cleave its DNA targets has not been observed hitherto among type II restriction endonucleases. Based on our work and previous reports, a pathway of DNA binding and cleavage by the SgrAI restriction endonuclease is proposed.
Recent studies on type II restriction endonucleases revealed that there is a greater degree of diversity within this class of endonucleases than first considered (1–3). For example, the multimodular endonuclease FokI binds specific DNA as a monomer (4), however two FokI–DNA complexes must interact with each other to cleave DNA (5). A homodimer of either EcoRII (6) or NaeI (7) also binds two recognition sites, but catalysis occurs only at one of the two sites. In their case, one site acts as an allosteric activator for cleavage at the second site (3). Recently, a similar reaction pathway has been shown to be characteristic for Sau3AI, except that it is a monomeric protein that dimerizes in the presence of DNA (8). SfiI (9, 10), NgoMIV (11), and Cfr10I (12) are tetramers of identical subunits that need to bind to two recognition sites for catalysis, but in contrast to EcoRII or NaeI, they cleave four phosphodiester bonds in a concerted fashion (10–12). SgrAI, described here, is yet another example of the diverse mechanisms exhibited by restriction endonucleases.
SgrAI recognizes the partially degenerate sequence 5′-CPu↓CCGGPyG (13) and cleaves two recognition sites concertedly (14). It has been reported that SgrAI exhibits relaxation of sequence specificity (15), also known as “star” activity (2). Under certain reaction conditions, many restriction endonucleases cleave DNA at sites that differ from the canonical site by one base pair (2). The star activity of EcoRV and EcoRI restriction endonucleases had been investigated thoroughly (16–22). The efficiency of cleavage at different star sites varies depending on the nature of the noncanonical base and its location within the sequence (16–18). Usually, extreme reaction conditions have to be used to stimulate cleavage at star sites. For EcoRV endonuclease, the discrimination factor derived from comparing the ratio of activity at its canonical site (GATATC) over that at the best star site, GTTATC, had a value of 3 × 105 under optimal reaction conditions (22). However, in the presence of 10% dimethyl sulfoxide, the discrimination factor changes to 103 (21). In the presence of Mn2+, the discrimination factor for EcoRV changes to 6-fold when assayed on plasmid DNA (22) or to 60-fold when assayed on oligonucleotide substrates (20).
This report describes the analysis of the substrate specificity of SgrAI restriction endonuclease. The results demonstrate that under standard reaction conditions SgrAI cleaves to completion the noncanonical sequences, 5′-CPuCCGGPy(A,T,C) and 5′-CPuCCGGGG, both referred to here as “secondary” sequences/sites. DNA termini generated by cleaving the canonical site for SgrAI assist in the cleavage of secondary sites. SgrAI-specific termini provided in trans on oligonucleotide duplex also increase canonical site cleavage rate. Activation of SgrAI by the self-generated DNA termini demonstrates yet another facet in the activities of site-specific restriction endonucleases.
Materials and Methods
Enzymes and Substrates.
Enzymes and DNA substrates were from New England Biolabs. SgrAI restriction endonuclease was purified to ≈95% homogeneity (tested by SDS/PAGE) either from the Streptomyces griseus cells (13) or from recombinant Escherichia coli that carries the cloned sgrAIR gene (New England Biolabs). One unit of SgrAI activity is defined as the amount of enzyme required for complete digestion of 1 μg of λ DNA in a total reaction volume of 50 μl in 1 h at 37°C by using NEBuffer4 (20 mM Tris-acetate buffer, pH 7.9/10 mM magnesium acetate/50 mM potassium acetate/1 mM DTT/100 μg/ml BSA). The specific activity of SgrAI is 150,000 units/mg protein. All concentrations of SgrAI given here refer to the dimeric form of protein with two subunits of Mr 37,930.
Construction of Plasmid Substrates.
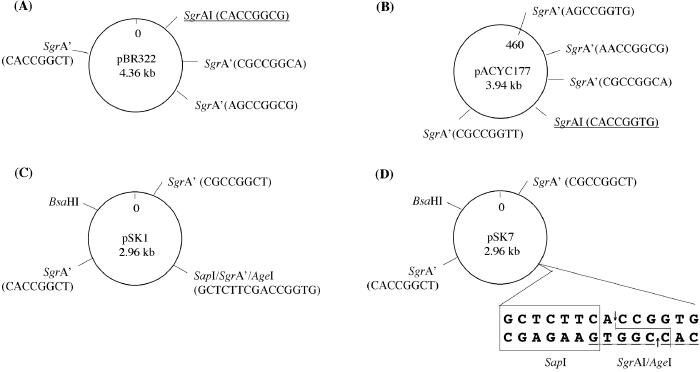
Four pACYC177 derivatives, each containing one variant of the sequence 5′-CPuCCGGPuG at position 460 nt, were constructed as follows. An 860-bp BsaI-AatII fragment of pBR322 was amplified in four PCR reactions. In each PCR, the BsaI-site primer had a pBR322 sequence AGCCGGTG replaced by a CGCCGGGG, CACCGGGG, CGCCGGAG, or CACCGGAG. The AatII and BsaI cleaved PCR products were ligated into the AatII and BsaI sites of pACYC177. The resulting constructs were designated as p177/GG (site CGCCGGGG), p177/AG (site CACCGGGG), p177/GA (site CGCCGGAG), and p177/AA (site CACCGGAG). The plasmids pSK1 and pSK7 are derivatives of pBluescript SK(−) (Stratagene). To construct them, either the secondary site GACCGGTG (pSK1) or the canonical site CACCGGTG (pSK7) was introduced at position 1037 nt of pBluescript SK(−) downstream the SapI site by the overlapping PCR technique (23).
Restriction maps of plasmid substrates used in this study are shown in Fig. 1.
Figure 1.
Schematic restriction maps of plasmids used in this study. SgrAI canonical sites are underlined. Secondary sites are marked as SgrA′. The sequences at the respective sites are shown in parentheses. pACYC177 in B was modified at position 460 as described in Materials and Methods. In D, SapI recognition sequence is boxed and its cleavage site is shown by arrows; SgrAI/AgeI cleavage site is shown as a solid line.
Secondary-Site Cleavage Assay.
The linear forms of plasmid pSK7 were prepared by digestion with the appropriate restriction endonuclease followed by phenol-chloroform extraction and alcohol precipitation. Secondary-site cleavage reactions were carried out at 37°C in either 50 μl or 100 μl of NEBuffer4 containing 1–2 μg of DNA and 5–50 units of SgrAI. The incubation time varied and is indicated elsewhere. The reactions were quenched by adding 0.3 volume of “stop” buffer (60 mM EDTA, pH 8.0/50% glycerol/0.2% SDS/0.02% bromophenol blue) and the products were analyzed by electrophoresis in either 0.7% or 1% agarose gels. The ethidium bromide-stained gels were analyzed as described (5).
Results
Analysis of Substrate Specificity of SgrAI Restriction Endonuclease.
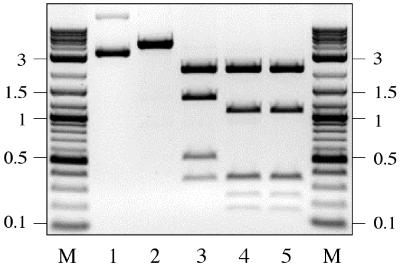
pBR322 DNA carries a single SgrAI canonical site (Fig. 1A). As expected, after 1-h incubation with 5 units of SgrAI, pBR322 was converted into the linear form (Fig. 2, lane 2). However, after 20-h incubation with 50 units of SgrAI, pBR322 was cleaved at three additional sites (Fig. 2, lane 3). Identical results were obtained using SgrAI purified from the S. griseus strain as well as from a recombinant E. coli source. The SgrAI digestion of pBR322 DNA that was precleaved with BsrFI (Pu↓CCGGPy) did not alter the BsrFI cleavage pattern, indicating that the additional SgrAI cleavage sites carry the sequence PuCCGGPy (Fig. 2, lanes 4 and 5). The fragment sizes obtained experimentally with SgrAI and with SgrAI+BsrFI were compared with the computer-derived mapping data for the sequence PuCCGGPy. The mapping data indicated that the sequences CGCCGGCA, CGCCGGCT, and CACCGGCT were cleaved by SgrAI. The first seven bases of all three sequences are equivalent to the SgrAI canonical site, but the last base is either adenine or thymine in place of the canonical guanine. In the view of these results, the sequence CPuCCGGPyN was further tested as an additional SgrAI target. This sequence represents 15 unique sequences including 3 SgrAI canonical sites (Table 1). Of these, only four sites are present on plasmid pBR322 (Fig. 2). To test whether SgrAI was cleaving the other CPuCCGGPyN sequences, three commonly used DNA substrates, plasmid pACYC184, bacteriophage lambda (λ), and adenovirus 2 (Ad2), were digested with SgrAI as described for pBR322 (data not shown). The obtained cleavage patterns were compared with the computer-derived mapping data for the sequence CPuCCGGPyN. Seven cleavage sites produced by SgrAI on pACYC184, 17 sites on Ad2 DNA, and 18 sites on λ DNA were mapped and determined to be consistent with the sequence CPuCCGGPyN (Table 1). The mapped sites included 14 unique CPuCCGGPyN sequences out of 15 (Table 1). The last unique site, CGCCGGCC, which was not present on the above tested DNA substrates, was determined to be cleaved by SgrAI by using a derivative of pBR322 that contained two copies of this sequence within the 3.0-kb Pseudomonas alcaligenes genomic DNA insert (R. Vaisvila, New England Biolabs).
Figure 2.
Mapping of the SgrAI secondary sites on pBR322. Digestions were carried out in 40 μl of NEBuffer4 (buffer composition is listed in Materials and Methods) containing 2 μg of pBR322 DNA. Lanes: 1, uncut DNA; 2, DNA digested with 5 units of SgrAI for 1 h; 3, DNA digested with 50 units of SgrAI for 20 h; 4, BsrFI-cleaved DNA sequentially digested with 50 units of SgrAI for 20 h; 5, DNA digested with BsrFI; M, 0.1–10-kb DNA ladder.
Table 1.
Frequency of CPuCCGGPyN sites on different DNAs
| No. | Sequence* | pBR322† | pACYC184 | Ad2 | Lambda |
|---|---|---|---|---|---|
| 1 | CGCCGGCG | 0 | 0 | 2 (2) | 0 |
| 2 | CGCCGGCA | 1 (1) | 1 (1) | 3 (2) | 0 |
| 3 | CGCCGGCT | 1 (1) | 1 (1) | 2 (2) | 1 (0) |
| 4 | CGCCGGCC | 0 | 0 | 0 | 0 |
| 5 | CGCCGGTG | 1 (1) | 1 (1) | 3 (3) | 5 (5) |
| 6 | CGCCGGTA | 0 | 0 | 0 | 4 (2) |
| 7 | CGCCGGTT | 0 | 1 (1) | 1 (1) | 3 (1) |
| 8 | CGCCGGTC | 0 | 0 | 0 | 2 (2) |
| 9 | CACCGGTG | 0 | 1 (1) | 1 (1) | 1 (1) |
| 10 | CACCGGTT | 0 | 1 (1) | 3 (2) | 1 (0) |
| 11 | CACCGGTA | 0 | 1 (1) | 0 | 1 (1) |
| 12 | CACCGGTC | 0 | 0 | 0 | 2 (1) |
| 13 | CACCGGCT | 1 (1) | 0 | 3 (2) | 1 (0) |
| 14 | CACCGGCC | 0 | 0 | 3 (2) | 3 (2) |
| 15 | CACCGGCA | 0 | 0 | 0 | 10 (3) |
| CPuCCGGPyN | 4 (4) | 7 (7) | 21 (17) | 34 (18) |
Canonical SgrAI sequences are underlined.
Number of experimentally mapped sites is given in parentheses.
Cleavage of either Ad2 or λ DNA also yielded a few fragments that were inconsistent with the computer-derived mapping data for the sequence CPuCCGGPyN, however. Therefore, either Ad2 or λ DNA was double-digested with BsrFI and SgrAI and the obtained cleavage pattern was compared with the BsrFI cleavage pattern (data not shown). The comparison revealed that SgrAI cleaved at sites that were not a subset of the sequence PuCCGGPy. The mapping data predicted that the sequence CPuCCGGPuG is the most likely candidate. Wild-type pACYC177 (Fig. 1B) and four derivatives of pACYC177, each carrying one of the four CPuCCGGPuG sequences, were digested with SgrAI for 20 h (data not shown). The sites CGCCGGGG (p177/GG) and CACCGGGG (p177/AG) were cleaved with a rates that were similar to the rate of cleavage at secondary site AGCCGGTG on pACYC177. However, only a few percent of p177/GA was cleaved at the site CGCCGGAG, indicating that it was cleaved ≈50-fold slower than the sequence AGCCGGTG. No cleavage at the site CACCGGAG on p177/AA was detected, indicating that this site was cleaved less than 100-fold of the rate of AGCCGGTG. Taken together, the data indicate that SgrAI cleaves sequences with guanine, but not adenine, at the seventh position with an efficiency that is equivalent to the sites CPuCCGGPy(A,T,C).
Both sets of noncanonical sequences, CPuCCGGPy(A,T,C) and CPuCCGGGG, are distinguishable from star sites by the rates at which they are cleaved and are referred to here as “secondary” sequences/sites.
Secondary-Site Cleavage Is Efficient in the Presence of Canonical Site.
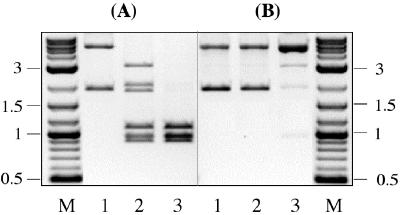
Plasmid pSK7 carries a single canonical site and two secondary sites (Fig. 1D). On plasmid pSK1 the canonical site is replaced by the third secondary site (Fig. 1C). Each plasmid was digested with 50 units of SgrAI for 1 or 20 h. After 1-h incubation more than 50% of pSK7 was cleaved at both secondary sites, and 100% of the sites were cleaved after 20 h (Fig. 3A). In contrast, no DNA cleavage was observed on pSK1 after 1-h incubation (Fig. 3B, lane 2). After 20 h, pSK1 DNA was fully converted to the relaxed form, indicating that plasmid was cut at one strand of the duplex. But less than 5% of the relaxed form of pSK7 was cleaved at both strands, indicating that double-strand cleavage at secondary sites was markedly reduced (Fig. 3B, lane 3). The nucleotide sequence of pSK1 is identical to the sequence of pSK7, except for the position 1037 nt where pSK1 carries a secondary site GACCGGTG, whereas pSK7 carries a canonical site CACCGGTG. The observed differences in cleavage of two plasmids thus indicate that SgrAI activity on the secondary sites depends on the presence of a canonical site.
Figure 3.
Efficiency of secondary-site cleavage on (A) pSK7 and (B) pSK1 plasmid DNA. The locations and sequences of the SgrAI canonical and secondary sites are shown in Fig. 1 C and D. One microgram of DNA (lane 1) was digested with 50 units of SgrAI either for 1 h (lane 2) or 20 h (lane 3). Lane M, 0.1–10-kb DNA ladder.
DNA Termini Generated by Cleaving the Canonical Site Assist in the Cleavage of Secondary Sites.
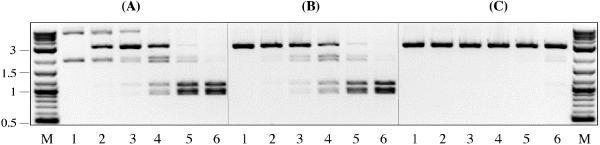
The progress of SgrAI cleavage at the canonical and secondary sites with time was monitored on the supercoiled form of plasmid pSK7 (Fig. 4A). The reaction was carried out in 100 μl of NEBuffer4 containing 5 nM of DNA and 88 nM of SgrAI. The samples were withdrawn from the reaction after 1, 5, 25, 125, and 1,200 min and immediately mixed with the “Stop” buffer. The reaction products were separated by electrophoresis and quantified using NIH IMAGE V. 1.61 program. The canonical and secondary-site cleavage rates were determined from the increase in the amounts of the corresponding DNA products with time. The rate at which SgrAI cleaved the canonical site (ν = 2.9 nM⋅min−1) was 26-fold faster than the rate of cleavage at the secondary sites (ν = 0.11 nM⋅min−1). After 5 min from the start of reaction, ≈90% of DNA was cleaved at the canonical site, but only 5% of DNA was cleaved at the secondary sites (Fig. 4A, lane 3). Because SgrAI continued to cleave the remaining secondary sites to completion (Fig. 4A, lanes 4–6), it is not likely that the uncut canonical sites assist in the cleavage of secondary sites.
Figure 4.
SgrAI cleavage of pSK7 DNA: (A) supercoiled form of DNA, (B) AgeI-linearized form of DNA, (C) SapI-linearized form of DNA (see Fig. 1D for restriction map). The SgrAI reaction was carried out at 37°C in 100 μl of NEBuffer4 containing 5 nM of DNA and 88 nM of SgrAI. Lane 1, DNA before adding SgrAI to the reaction. Lanes 2–6: 15-μl samples were withdrawn after 1, 5, 25, 125, and 1,200 min from the start of reaction and immediately quenched by adding 7.5 μl of “stop” buffer. Lane M, 0.1–10-kb DNA ladder.
The secondary site cleavage was monitored on pSK7 DNA that was linearized at the SgrAI canonical site by digestion with AgeI (Fig. 4B). AgeI cleaves the sequence A ↓ CCGGT, which on pSK7 overlaps with the SgrAI canonical site, CA ↓ CCGGTG. Therefore, DNA termini yielded by AgeI are equivalent to the SgrAI-generated termini (Fig. 1D). The rate at which SgrAI cleaved secondary sites on the AgeI-linearized DNA (ν = 0.13 nM⋅min−1) was very similar to that observed on the supercoiled form of pSK7 (ν = 0.11 nM⋅min−1). To further investigate the role of the SgrAI-specific DNA termini, the secondary-site cleavage was tested on the SapI-linearized form of pSK7 (Fig. 4C). SapI cleaves DNA outside its recognition sequence: GCTCTTC(N)1/4. On plasmid pSK7, the SgrAI canonical site was engineered into the SapI cleavage site (Fig. 1D). After SapI digestion, one DNA strand of pSK7 is cleaved at the identical to SgrAI phosphodiester bond (5′-CA ↓ CCGGTG). However, the other DNA strand is cleaved one nucleotide closer: 3′-GTGGC ↓ CAC instead of a 3′-GTGGCC ↓ AC that represents a canonical SgrAI cleavage. The reactions profile on SapI-linearized pSK7 DNA (Fig. 4C) is clearly distinct from that observed in Fig. 4 A and B, because only 5% of DNA was cleaved by SgrAI after 20 h. Therefore, one misplaced phosphodiester bond break within the canonical recognition sequence has an inhibitory effect on the SgrAI secondary-site activity similar to that observed on pSK1 without the canonical site.
Taken together, the data presented in Fig. 4 indicate that the SgrAI-canonical termini are essential for the SgrAI cleavage at secondary sites.
Cis-Oriented Termini with Respect to Secondary Site Are Favored.
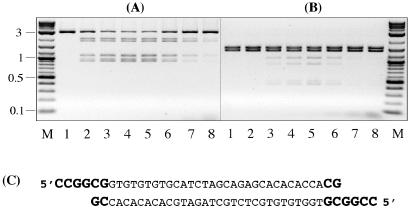
To test whether the secondary-site cleavage can be enhanced by addition of the SgrAI-canonical termini in trans, the SgrAI reaction was carried out in the presence of short DNA duplex that was flanked by the SgrAI-canonical termini (Fig. 5C). AgeI-linearized pSK7 DNA (5 nM) was cleaved with SgrAI for 30 min in the presence of various concentrations of oligonucleotide duplex (Fig. 5A). With increasing concentrations of the oligo, the secondary-site cleavage slightly increased from the control level without oligo (lane 2) to a maximum registered at 10 nM oligo (lane 4) and then decreased to below control level at higher concentrations of oligo (lanes 7 and 8). At the optimal concentration (10 nM), the rate of enhancement was 2-fold. No effect on secondary-site cleavage was observed in control experiments with a fully double-stranded DNA duplex that was prepared by filling-in the 5′ protruding single-stranded termini with DNA Polymerase I Klenow Fragment (data not shown). These results indicate that SgrAI-specific termini provided by the oligo can increase secondary-site cleavage efficiency even though DNA itself is flanked by the canonical termini.
Figure 5.
Secondary-site cleavage on (A) AgeI-linearized and (B) SapI+BsaHI-cleaved pSK7 DNA in the presence of oligonucleotide duplex. Lane M, 0.1–10-kb DNA ladder. Lane 1, DNA control (no SgrAI added). Lanes 2–8: 0, 5, 10, 20, 50, 100, and 200 nM of oligo was added to the 50-μl reactions that contained 5 nM of DNA and 88 nM of SgrAI and incubated either (A) 30 min or (B) 4 h at 37°C. (C) Nucleotide sequence of the oligonucleotide duplex. SgrAI canonical termini are shown in bold.
Next, the effect of oligo was studied using the SapI+BsaHI-cleaved pSK7 DNA that does not carry canonical termini in cis with respect to secondary sites. After 4-h incubation, DNA cleavage in control sample (without oligo) was still undetectable (Fig. 5B, lane 2), but 15% of DNA was cleaved in the presence of 20 nM of oligo (Fig. 5B, lane 5). Therefore, SgrAI is capable of mediating trans interactions between canonical DNA termini provided by oligo and secondary sites provided by SapI-BsaHI fragments, although with a 37-fold reduced efficiency compared with that observed on substrate with canonical termini in cis (Fig. 5A, lane 4).
Specificity of SgrAI Is Increased at 200 nM Oligo Concentration.
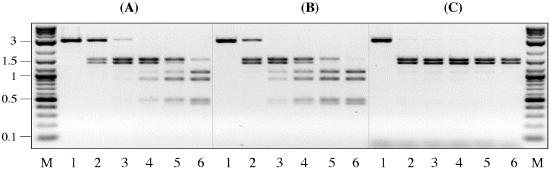
SgrAI cleavage at the canonical and secondary sites was monitored with time in the presence of 0, 20, or 200 nM oligo. The reactions were carried out with the BsaHI-linearized form of pSK7 (Fig. 1D). Compared with the control without oligo (Fig. 6A), SgrAI cleaved canonical site 2-fold faster in the presence of 20 nM of oligo (Fig. 6B), and 10-fold faster in the presence of 200 nM of oligo (Fig. 6C). Therefore, SgrAI-specific termini provided in a form of oligonucleotide duplex stimulate canonical site cleavage even though the oligo itself can be considered a product of the SgrAI-catalyzed reaction.
Figure 6.
SgrAI cleavage of the BsaHI-linearized pSK7 DNA in the absence (A) or in the presence of (B) 20 nM or (C) 200 nM of oligonucleotide duplex. The oligonucleotide sequence is shown in Fig. 5C. SgrAI reactions were carried out as described in Fig. 4. Lane 1, DNA before adding SgrAI to the reaction. Lanes 2–6: 15-μl samples were withdrawn after 1, 5, 25, 125, and 1,200 min from the start of reaction and immediately quenched by adding 7.5 μl of “stop” buffer. Lane M, 0.1–10-kb DNA ladder.
The profile of secondary-site cleavage on the BsaHI-linearized form of pSK7 is distinct from that observed in Fig. 4 A and B. The rates at which two secondary sites were cleaved on the BsaHI-linearized form of pSK7 were no longer identical to each other: one secondary site was cleaved 3.5-fold faster than the other site (Fig. 6A, lanes 4 and 5). The reason why two sites are cleaved with different rates, even though the reaction involves exactly the same sites that were cleaved at very similar rates on either the supercoiled or AgeI-linearized form of pSK7 (Fig. 4 A and B), is not yet completely investigated. One explanation is that after the BsaHI-linearized DNA is cleaved at the canonical site the secondary sites are left on separate DNA fragments (Fig. 1D). Because each fragment is flanked by one cis canonical terminus, the intermolecular interactions between the canonical termini and secondary site must be different for two fragments. It was expected then that in the presence of oligo the differences between cleavage rates of two sites would diminish, because the oligo termini would compensate for the loss of one cis-oriented canonical terminus. However, this expectation was only partly fulfilled. In the presence of 20 nM oligo, both secondary sites were cleaved faster than without oligo, but the difference between the cleavage rates of two sites was only reduced from 3.5-fold without oligo to 2-fold with a 20 nM oligo (Fig. 6B). These results indicate that the other factors besides DNA termini, perhaps DNA fragment conformation itself, may influence the rate of secondary-site cleavage.
Strikingly, in the presence of 200 nM oligo, DNA cleavage at secondary sites was no longer detected even after 20 h (Fig. 6C). The effect is even more striking if a 10-fold activation of canonical site cleavage is taken into account. At a 40-fold molar excess of the SgrAI-canonical termini over substrate DNA, due to the concurrent 10-fold increase in a rate of canonical site cleavage and at least 50-fold decrease in a rate of secondary-site cleavage, the SgrAI specificity was improved by more than two orders of magnitude.
Discussion
In this study we show that SgrAI endonuclease under standard reaction conditions cleaves 14 noncanonical sequences, referred to as secondary sequences. The secondary sequences are of two distinct types. The sequence CPuCCGGPy(C,A,T), where the canonical guanine at the eighth position is substituted by any other base, represents 12 unique sequences (Table 1). Two secondary sites are represented by the sequence CPuCCGGGG, where the canonical pyrimidine at the seventh position is substituted with guanine, but other bases are strictly canonical. The sequence CGCCGGAG was also cleaved, but at a 50-fold slower rate than CPuCCGGGG, so that complete digestion was not achieved. Under reaction conditions used in this study no DNA cleavage at any other noncanonical sequence has been observed in a total 96,984 base pairs of sequence space on five DNA substrates. Although unlikely, there may exist other SgrAI target sequences that are not present on DNA substrates used here.
The rates at which SgrAI cleaves secondary sequences on pSK7 DNA are 26-fold slower than the rate of cleavage of the canonical sequence. However, on DNA that lacks a canonical site, secondary-site cleavage is reduced by two orders of magnitude. In this study, the canonical site cleavage products are shown to be an essential factor in the secondary-site cleavage. Canonical termini in cis configuration are 37-fold more efficient in stimulating secondary-site cleavage over those in trans configuration. Moreover, the SgrAI-specific termini at a 40-fold molar excess over the canonical site increase the velocity of canonical site cleavage by 10-fold. The peak velocity occurs at termini concentration, where the secondary-site cleavage is virtually inhibited. Therefore, the specificity of SgrAI may be increased by two orders of magnitude by simply manipulating the ratio of the canonical termini concentration relative to the substrate concentration. The phenomenon of SgrAI activation by the self-generated reaction products has not been observed previously among the type II restriction endonucleases, including those that exhibit a star activity.
In vivo, the host DNA is protected from the endogenous restriction endonuclease cleavage by site-specific methylation. However, when a restriction endonuclease exhibits star activity in vitro, a question that always arises is how the bacterial cell protects its DNA from the cleavage at star sites. It has been proposed that either the cognate methyltransferase can methylate star sites in vivo (24), or the cell has an efficient repair system that can seal the strand breaks produced at star sites (25, 26). For SgrAI, canonical site methylation should abolish the production of the SgrAI-termini required for secondary-site cleavage, so that secondary sites, even when unmethylated, would be much more resistant to SgrAI. In contrast, if the SgrAI restriction-modification system acts as a defense mechanism, the cleavage of invading DNA at canonical sites will trigger secondary-site cleavage, so that the efficiency of foreign DNA degradation might be effectively increased.
Based on kinetic studies, it has been proposed that SgrAI cleaves two copies of its canonical sequence in a concerted manner (14). Such behavior of SgrAI resembles the mode of action of tetrameric endonuclease SfiI that binds two copies of its recognition sequence before cleaving concertedly at four phosphodiester bonds (9, 10). Other examples of restriction endonucleases that cleave two recognition sites in a concerted fashion are Cfr10I and NgoMIV (11, 12). The SgrAI recognition sequence (CPu↓CCGGPyG) is a subset of the sequences cleaved by either Cfr10I (Pu↓CCGGPy) or NgoMIV (G↓CCGGC). The NgoMIV three-dimensional structure consists of a protein tetramer and two DNA molecules cleaved at their recognition sites (11). Cfr10I was also shown to exist as a homotetramer (12). The structural comparison of NgoMIV and Cfr10I suggests that both enzymes probably use similar residues to contact the central CCGG part of their recognition sequence and share similar structural mechanism for catalysis (11, 27). The amino acid residues that have been shown to be functionally and structurally conserved in NgoMIV and Cfr10I are also highly conserved in SgrAI. Thus, SgrAI may share with NgoMIV and Cfr10I the catalytic mechanism and the mechanism by which it interacts with the conserved part of the recognition sequence. However, although the subunit composition of the SgrAI protein has yet to be determined, the preliminary data suggest that in solution SgrAI may exist in a different form than the catalytically active tetrameric form (S. Halford, personal communication).
The results presented in this paper indicate that SgrAI cleaves secondary sites by a mechanism that involves long-range interactions between the sequence-specific DNA termini, representing cleaved canonical site, and the secondary site. Taking into account the structure of NgoMIV (11) and the kinetic and biochemical studies performed on SgrAI (14), the following SgrAI reaction pathway is proposed. SgrAI binds its canonical sequence as a dimer. This primary complex is not active. However, binding to recognition site triggers dimer–dimer association that yields a functionally active tetrameric complex. After a tetrameric SgrAI–DNA complex is formed, the DNA is cleaved simultaneously at all four phosphodiester bonds. After cleavage, the SgrAI tetramer dissociates into two primary dimers, which may remain bound to the cleaved DNA termini. At this point, the SgrAI dimer either dissociates from cleaved DNA or is capable of re-associating with another SgrAI dimer bound to the uncut target, yielding an intermediate SgrAI complex composed of one cleaved and one intact DNA molecule. The complex is catalytically active to produce a double-stranded break within the uncut site. A tetrameric SgrAI complex may form, composed of one SgrAI dimer bound to the cleaved canonical site and a second SgrAI dimer bound to cis-oriented secondary site. In this complex, the secondary site is cleaved at a 26-fold slower rate. A tetrameric complex bound to two secondary sites is at least two orders of magnitude less efficient in cleaving secondary sites. Most likely, it dissociates before DNA is cleaved at both strands. However, if the initial double-stranded cut is produced at a few secondary sites, SgrAI dimer may form a synapse between two canonical termini generated at two different cuts. This complex then might be efficiently used to cleave the cis-oriented secondary sites. During the reaction, as more canonical termini are generated, secondary site cleavage becomes more efficient. The SgrAI-generated termini in cis with respect to the secondary site are 37-fold more efficient over those in trans. Thus, self-activation is most effective on DNA carrying multiple secondary sites. When cleaving at the canonical sites, SgrAI also prefers the sites in cis, but operates on sites in trans at a 5-fold reduced rate (14). It has been shown that SfiI operates on sites either in cis or in trans, but cleaves sites in cis more efficiently simply because the sites in cis are closer to each other in the three-dimensional space (10). To cleave secondary sites in trans to canonical termini, SgrAI has to form a synapse of three elements—i.e., two termini and a secondary site—across three-dimensional space. When one terminus is in cis to secondary site, the task is simpler, because SgrAI has to bring one terminus across three-dimensional space. The other factors, such as sequence-specificity of secondary site itself, the length of spacer sequence between canonical termini and secondary site, number of cis-oriented secondary sites on the same DNA molecule, and/or their orientation to each other, may also influence the efficiency of secondary-site cleavage. However, this has yet to be demonstrated experimentally.
SgrAI differs from the prototypical type II restriction endonucleases by binding and cleaving two target sequences in a concerted manner (14). Here it is shown that SgrAI differs from the SfiI-like endonucleases by employing self-generated DNA termini to assist the cleavage at both canonical and secondary sites. Intriguingly, these properties resemble the initial steps of a wide range of genetic processes that are aimed to perform programmed DNA rearrangements (28–31). During this process the distant target sites are brought together by synapsis, then cleaved at both strands, exchanged and eventually resealed as new DNA junctions. There is growing evidence of an evolutionary link between the restriction endonucleases that require two target sites for catalysis and some of the proteins that are involved in programmed DNA rearrangements. The restriction endonuclease NaeI, for example, has been transformed to a DNA topoisomerase by a single amino acid substitution (32, 33). NaeI displays a modular structure consisting of the N-terminal endonuclease domain and a C-terminal domain that possesses a catabolite activator protein (CAP) motif present in many type IA and type II DNA topoisomerases (34). EcoRII reveals sequence homology with the highly conserved region of the integrase-family recombinases and also requires two sites for catalysis (35). Based on comparative sequence analysis, the Holliday junction resolvase Hjc was classified as a distantly related member of the type II restriction endonuclease family (36). Tn7 transposase consists of two proteins: TnsA is responsible for cleavage at the 5′ ends, whereas TnsB is responsible for the cleavage and joining at the 3′ ends of the transposon (37). Recently the crystal structure of TnsA has been solved displaying a restriction endonuclease-like fold of TnsA (38). Moreover, the spatial locations of the active site residues of TnsA are very close to those of Cfr10I and NgoMIV (11, 27). Most likely, SgrAI is another member that shares similar catalytic site organization; however, its structure remains to be determined. The continued combined approaches from genetic, biochemical, and structural analyses hopefully will provide more detailed knowledge about the mode of action of SgrAI endonuclease and reveal whether it is evolutionarily linked to the proteins that perform programmed DNA rearrangements.
Acknowledgments
We thank Dr. H. Kong and L. Higgins for providing the nucleotide and amino acid sequence of SgrAI as well as SgrAI protein purified from the native strain. We thank Dr. R. Roberts for critical reading of the manuscript and Dr. S. Halford for constructive discussions and sharing unpublished results.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Halford S E, Bilcock D T, Stanford N P, Williams S A, Milsom S E, Gormley N A, Watson M A, Bath A J, Embleton M L, Gowers D M, et al. Biochem Soc Trans. 1999;27:696–699. doi: 10.1042/bst0270696. [DOI] [PubMed] [Google Scholar]
- 2.Roberts R J, Halford S E. In: Nucleases. Linn S M, Lloyd R S, Roberts R J, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 35–88. [Google Scholar]
- 3.Bickle T A, Kruger D H. Microbiol Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wah D A, Hirsch J A, Dorner L F, Schildkraut I, Aggarwal A K. Nature (London) 1997;388:97–100. doi: 10.1038/40446. [DOI] [PubMed] [Google Scholar]
- 5.Bitinaite J, Wah D A, Aggarwal A K, Schildkraut I. Proc Natl Acad Sci USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reuter M, Kupper D, Meisel A, Schroeder C, Kruger D H. J Biol Chem. 1998;273:8294–8300. doi: 10.1074/jbc.273.14.8294. [DOI] [PubMed] [Google Scholar]
- 7.Baxter B K, Topal M D. Biochemistry. 1993;32:8291–8298. doi: 10.1021/bi00083a033. [DOI] [PubMed] [Google Scholar]
- 8.Friedhoff P, Lurz R, Luder G, Pingoud A. J Biol Chem. 2001;276:23581–23588. doi: 10.1074/jbc.M101694200. [DOI] [PubMed] [Google Scholar]
- 9.Wentzell L M, Nobbs T J, Halford S E. J Mol Biol. 1995;248:581–595. doi: 10.1006/jmbi.1995.0244. [DOI] [PubMed] [Google Scholar]
- 10.Nobbs T J, Szczelkun M D, Wentzell L M, Halford S E. J Mol Biol. 1998;281:419–432. doi: 10.1006/jmbi.1998.1966. [DOI] [PubMed] [Google Scholar]
- 11.Deibert M, Grazulis S, Sasnauskas G, Siksnys V, Huber R. Nat Struct Biol. 2000;7:792–799. doi: 10.1038/79032. [DOI] [PubMed] [Google Scholar]
- 12.Siksnys V, Skirgaila R, Sasnauskas G, Urbanke C, Cherny D, Grazulis S, Huber R. J Mol Biol. 1999;291:1105–1118. doi: 10.1006/jmbi.1999.2977. [DOI] [PubMed] [Google Scholar]
- 13.Tautz N, Kaluza K, Frey B, Jarsch M, Schmitz G G, Kessler C. Nucleic Acids Res. 1990;18:3087. doi: 10.1093/nar/18.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bilcock D T, Daniels L E, Bath A J, Halford S E. J Biol Chem. 1999;274:36379–36386. doi: 10.1074/jbc.274.51.36379. [DOI] [PubMed] [Google Scholar]
- 15.Laue F, Ankenbauer W, Schmitz G G, Kessler C. Nucleic Acids Res. 1990;18:3421. doi: 10.1093/nar/18.11.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thielking V, Alves J, Fliess A, Maass G, Pingoud A. Biochemistry. 1990;29:4682–4691. doi: 10.1021/bi00471a024. [DOI] [PubMed] [Google Scholar]
- 17.Alves J, Selent U, Wolfes H. Biochemistry. 1995;34:11191–11197. doi: 10.1021/bi00035a026. [DOI] [PubMed] [Google Scholar]
- 18.Lesser D R, Kurpiewski M R, Jen-Jacobson L. Science. 1990;250:776–786. doi: 10.1126/science.2237428. [DOI] [PubMed] [Google Scholar]
- 19.Engler L E, Welch K K, Jen-Jacobson L. J Mol Biol. 1997;269:82–101. doi: 10.1006/jmbi.1997.1027. [DOI] [PubMed] [Google Scholar]
- 20.Sam M D, Horton N C, Nissan T A, Perona J J. J Mol Biol. 2001;306:851–861. doi: 10.1006/jmbi.2000.4434. [DOI] [PubMed] [Google Scholar]
- 21.Taylor J D, Halford S E. Biochemistry. 1989;28:6198–6207. doi: 10.1021/bi00441a011. [DOI] [PubMed] [Google Scholar]
- 22.Vermote C L, Halford S E. Biochemistry. 1992;31:6082–6089. doi: 10.1021/bi00141a018. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Russell D W. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 24.Woodbury C P, Jr, Downey R L, von Hippel P H. J Biol Chem. 1980;255:11526–11533. [PubMed] [Google Scholar]
- 25.Heitman J, Zinder N D, Model P. Proc Natl Acad Sci USA. 1989;86:2281–2285. doi: 10.1073/pnas.86.7.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor J D, Goodall A J, Vermote C L, Halford S E. Biochemistry. 1990;29:10727–10733. doi: 10.1021/bi00500a003. [DOI] [PubMed] [Google Scholar]
- 27.Bozic D, Grazulis S, Siksnys V, Huber R. J Mol Biol. 1996;255:176–186. doi: 10.1006/jmbi.1996.0015. [DOI] [PubMed] [Google Scholar]
- 28.Polard P, Chandler M. Mol Microbiol. 1995;15:13–23. doi: 10.1111/j.1365-2958.1995.tb02217.x. [DOI] [PubMed] [Google Scholar]
- 29.Landy A. Proc Natl Acad Sci USA. 1999;96:7122–7124. doi: 10.1073/pnas.96.13.7122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gopaul D N, Van Duyne G D. Curr Opin Struct Biol. 1999;9:14–20. doi: 10.1016/s0959-440x(99)80003-7. [DOI] [PubMed] [Google Scholar]
- 31.Redinbo M R, Champoux J J, Hol W G. Curr Opin Struct Biol. 1999;9:29–36. doi: 10.1016/s0959-440x(99)80005-0. [DOI] [PubMed] [Google Scholar]
- 32.Jo K, Topal M D. Science. 1995;267:1817–1820. doi: 10.1126/science.7892605. [DOI] [PubMed] [Google Scholar]
- 33.Jo K, Topal M D. Nucleic Acids Res. 1998;26:2380–2384. doi: 10.1093/nar/26.10.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huai Q, Colandene J D, Chen Y, Luo F, Zhao Y, Topal M D, Ke H. EMBO J. 2000;19:3110–3118. doi: 10.1093/emboj/19.12.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Topal M D, Conrad M. Nucleic Acids Res. 1993;21:2599–2603. doi: 10.1093/nar/21.11.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daiyasu H, Komori K, Sakae S, Ishino Y, Toh H. Nucleic Acids Res. 2000;28:4540–4543. doi: 10.1093/nar/28.22.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Craig N L. Curr Top Microbiol Immunol. 1996;204:27–48. doi: 10.1007/978-3-642-79795-8_2. [DOI] [PubMed] [Google Scholar]
- 38.Hickman A B, Li Y, Mathew S V, May E W, Craig N L, Dyda F. Mol Cell. 2000;5:1025–1034. doi: 10.1016/s1097-2765(00)80267-1. [DOI] [PubMed] [Google Scholar]