Abstract
Wnt signaling plays a key role in cell proliferation and development. Recently, casein kinase I (CKI) and protein phosphatase 2A (PP2A) have emerged as positive and negative regulators of the Wnt pathway, respectively. However, it is not clear how these two enzymes with opposing functions regulate Wnt signaling. Here we show that both CKIδ and CKIɛ interacted directly with Dvl-1, and that CKI phosphorylated multiple components of the Wnt-regulated β-catenin degradation complex in vitro, including Dvl-1, adenomatous polyposis coli (APC), axin, and β-catenin. Comparison of peptide maps from in vivo and in vitro phosphorylated β-catenin and axin suggests that CKI phosphorylates these proteins in vivo as well. CKI abrogated β-catenin degradation in Xenopus egg extracts. Notably, CKI decreased, whereas inhibition of CKI increased, the association of PP2A with the β-catenin degradation complex in vitro. Additionally, inhibition of CKI in vivo stabilized the β-catenin degradation complex, suggesting that CKI actively destabilizes the complex in vivo. The ability of CKI to induce secondary body axes in Xenopus embryos was reduced by the B56 regulatory subunit of PP2A, and kinase-dead CKIɛ acted synergistically with B56 in inhibiting Wnt signaling. The data suggest that CKI phosphorylates and destabilizes the β-catenin degradation complex, likely through the dissociation of PP2A, providing a mechanism by which CKI stabilizes β-catenin and propagates the Wnt signal.
The Wnt signaling pathway is a critical regulator of embryonic development and cellular proliferation (1, 2). In the absence of Wnt, β-catenin is phosphorylated by glycogen synthase kinase 3β (GSK3β) in a complex that also contains adenomatous polyposis coli (APC) and axin, targeting β-catenin for ubiquitination and degradation. When Wnt binds to the frizzled receptor, disheveled (Dvl/Dsh) is hyperphosphorylated and activated. Dvl activation leads to inhibition of GSK3β, decreased phosphorylation of axin, APC, and β-catenin, and stabilization of β-catenin. β-catenin is then translocated to the nucleus where it binds to Lef/Tcf proteins and stimulates expression of Wnt-responsive genes.
Multiple proteins have recently been identified as regulators of β-catenin signaling. Protein phosphatase 2A (PP2A), a heterotrimeric serine/threonine phosphatase (3), interacts via its B56 regulatory subunit with APC, Dsh, and axin (4–7), whereas its catalytic C subunit interacts with axin (8, 9). Inhibition of PP2A by okadaic acid increases, whereas B56 expression decreases, β-catenin abundance in 293 cells. In Xenopus, B56 reduces both Xwnt-8-induced secondary axes and siamois and Xnr-3 expression. PP2A:B56 is likely to be a component of the β-catenin degradation complex because epistasis positions B56 downstream of GSK3β and axin but upstream of APC and β-catenin, and axin coimmunoprecipitates PP2A:B56, C, and structural A subunits. PP2A C and A also rescue Wnt-induced secondary axes, and PP2A activity is required for β-catenin degradation in Xenopus egg extracts (4, 7).
Recent studies have implicated casein kinase I (CKI), a serine/threonine protein kinase, as a positive regulator of β-catenin signaling. Two independent screens found that CKI induces axis duplication in Xenopus embryos (10, 11). CKI functions downstream of Dsh and upstream of GSK3β, interacts with Dsh, and coimmunoprecipitates with axin, GSK3β, and Dvl-3. CKI increases Dsh phosphorylation in Xenopus oocytes (10, 12), and direct phosphorylation of Dsh by CKI was demonstrated by mobility shift and 32P incorporation (12). CKIδ and CKIɛ are closely related, because their kinase domains are 98% identical and each contain a 53% identical C-terminal tail that inhibits CKIδ/ɛ when autophosphorylated. Sakanaka et al. (11) found that CKIɛ lacking the C-terminal tail does not induce a secondary axis in Xenopus embryos or coimmunoprecipitate with axin, suggesting that this domain is important for Wnt signaling. The role of CKIα, which is 77% identical to CKIɛ and lacks a C-terminal tail, in Wnt signaling is not clear, because one group reported that CKIα induces a secondary axis in Xenopus embryos (10, 12), whereas another found that it does not function in Wnt signaling (11). The physiological target(s) of CKI in the Wnt pathway are not yet known.
To investigate the mechanism by which CKI activates Wnt signaling, we explored the interaction of CKI with various regulators of β-catenin signaling. We find that CKIδ and CKIɛ, but not CKIα, interact directly with Dvl-1. CKI phosphorylates several components of the β-catenin degradation complex in vitro. In addition, CKI phosphorylates axin and β-catenin on major in vivo phosphorylation sites. A principal function of CKI may be to regulate the interaction of PP2A with the β-catenin degradation complex, because CKI decreases, whereas inhibition of CKI increases, PP2A bound to axin and β-catenin. Supporting the interplay between CKI and PP2A, epistasis analyses show that CKI functions upstream of B56, and B56 acts synergistically with kinase-dead CKI to rescue Xwnt-8-induced secondary axes. The data suggest that CKI is recruited to the β-catenin degradation complex, perhaps by Dvl, where it phosphorylates Dvl-1, APC, axin, and β-catenin, resulting in the dissociation of PP2A from the β-catenin degradation complex and decreased β-catenin degradation.
Materials and Methods
Materials.
Anti-β-catenin and anti-GSK3β antibodies were from Transduction Laboratories (Lexington, KY); anti-Flag M2 and anti-Flag M2-agarose were from Sigma; and anti-HA mAb F-7 was from Santa Cruz Biotechnology. Anti-PP2A C and A antibodies have been described (13). Anti-CKIα antibodies were made to an N-terminal peptide. Anti-CKIδ/ɛ antibodies recognize their shared N terminus (14). CKI proteins were expressed in Escherichia coli and purified as described (14). CKI-7 was from Seikagaku Kogyo (Tokyo) and IC261 was a gift from G. Keesler, Aventis Pharmaceuticals, Bridgewater, NJ.
Yeast Two-Hybrid Assay.
CKI genes were cloned into pBTM116-URA3 to express LexA fusion proteins. Dvl-1, axin, β-catenin, β-TrCP, APCRI (amino acids 1–1121), APC25 (amino acids 1014–2032), GSK3β, and B56α were cloned into pGAD-GH (15) to express GAL4 transcriptional activation domain (GAL4AD) fusion proteins. LexA plasmids were transformed into Saccharomyces cerevisiae strain DY151 (MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3) and GAL4AD plasmids into D5735 (MATa LYS2:lexA:HIS3 ura3:ADE2:lexA:lacZ). The mating assay was performed as described (16, 17). Diploids were grown on SD-Ura−Leu−His− plates with 1 or 3 mM 3-aminotriazole (3-AT).
In Vitro Transcription/Translation and Coimmunoprecipitation.
Templates were amplified from pGAD-GH by using the following primers: 5′-GGATCAATTAACCCTCACTAAAGGGAACGCCGCCA CCATGGATGATGTATATAACTATCTATTCGATG-3′ and 5′-GAGCGCGCGTAATACGACTCAC TATAGGGCGAAAT-3′. Proteins were synthesized separately by using the TNT System (Promega; ref. 18) and then mixed and incubated at room temperature for 1.5 h in the presence of anti-HA-mAb agarose. Immunoprecipitates were washed four times with IP buffer (50 mM Tris⋅HCl, pH 7.5/137 mM NaCl/10% glycerol/0.1% Nonidet P-40). Bound proteins were eluted with SDS-loading buffer, separated by SDS/PAGE, and dried and analyzed using a Molecular Dynamics PhosphorImager.
32P-Labeling and Phosphopeptide Mapping.
Cell culture, transfection, and lysate preparation was performed as described (4, 19), except in the case of β-catenin, where cells were incubated with 40 μM lactacystin for 3 h before harvest. Immunoprecipitations were performed with 300 μg of lysate protein. Immunoprecipitates were washed three times with IP buffer, once with KR buffer (50 mM Tris⋅HCl, pH 7.5/10% glycerol/10 mM MgCl2) and the volume adjusted to 30 μl with KR buffer. The kinase reaction was initiated by addition of 100 nM CKIδΔ319 and 10 μM [γ-32P]ATP (5 μCi/nmol; 1 Ci = 37 GBq) and incubated for 20 min at 30°C with gentle shaking. The reaction was terminated by addition of SDS-loading buffer and heating at 100°C for 3 min. The samples were analyzed by SDS/PAGE and immunoblotted on poly(vinylidene difluoride) (PVDF) membranes. The membranes were then air dried, and 32P-incorporation was determined by PhosphorImager analysis. Tryptic phosphopeptide maps were prepared as described (20, 21). For in vivo 32P labeling, cells were labeled with 2 mCi [32P]orthophosphate in 1 ml of phosphate- and serum-free DMEM for 3 h. Wnt3a-conditioned medium (0.35 ml) was added for the last 15 min of labeling. Immunoprecipitates were analyzed as described above.
Stability of Axin and β-Catenin Complexes.
For in vitro assays, 450 μg of lysate protein from adenovirus-transformed human embryonic kidney (293) cells expressing Myc:axin or Myc:β-catenin were incubated with 30 μl of 50% anti-Myc-Sepharose in IP buffer supplemented with 10 mM MgCl2, 170 μM ATP, and Complete protease inhibitors (Boehringer Mannheim). Either 480 nM His6-CKIɛK38R (K38R) and 250 μM CKI-7 (22), or 180 nM CKIɛ were added to the incubations. Immunoprecipitations were carried out as described. Samples were analyzed by SDS/PAGE and immunoblotting. The membranes were stripped and reprobed with the indicated antibodies. For in vivo assays, 293 cells expressing Myc:β-catenin were incubated in media containing 40 μM IC261, a membrane-permeable CKIδ/ɛ-specific inhibitor (23), or DMSO, for 1 h before lysis. Cells were homogenized in IP buffer supplemented with 40 μM IC261 or DMSO. Immunoprecipitations and immunoblotting were performed as described above.
RNA Microinjections.
RNA was synthesized using mMessage mMachine (Ambion) and purified using RNeasy (Qiagen, Chatsworth, CA). Xenopus embryos were microinjected at the four-cell stage near the equatorial midline in one ventral cell, and scored after 3 days at room temperature. The following amounts of RNA were microinjected: CKIδ, 2.5–6.5 ng; CKIɛ, 1.75–4.75 ng; CKIɛK38A (K38A), 1.13–2 ng; B56α, 200–250 pg; Xwnt-8, 15 pg. The phenotypes of the embryos were categorized by degree of secondary axis formation: complete, each head possesses a complete complement of eyes and cement glands; incomplete, one head possesses an incomplete complement of cement gland and eyes; or vestigial, the secondary axis lacks a cement gland and eyes. The B56/CKI data sets were analyzed using a stratified Wilcoxon rank-sum test with exact inference (STATXACT software, Cytel, San Diego). This is a nonparametric test for difference in the median of two ordered categorical responses. The B56/K38A data set was analyzed using a continuation ratio model that was fitted (using SAS software, SAS Institute, Cary, NC) with separate covariates for the applied treatments and an interaction term. Stratification by the day of experiment was used to adjust for day-to-day variability of embryos and two-sided P values are reported.
Results
Direct Interaction of CKI with Wnt Signaling Components.
To address the mechanism by which CKI enhances β-catenin stability, we first investigated its interactions with other components of the Wnt pathway. Directed two-hybrid assays were performed using CKIɛ and various Wnt pathway candidates. CKIɛ interacted with Dvl-1, but no interactions were detected with axin, β-catenin, β-TrCP, APCRI, APC25, B56α, or GSK3β (data not shown).
The characteristics required for the CKI/Dvl-1 interaction were determined using two-hybrid assays (Table 1). Kinase-dead CKIɛ (K38R) interacted with Dvl-1 about as efficiently as wild type. Dvl-1 interacted with CKIɛ lacking the C-terminal tail (K38RΔ319), although with decreased strength. CKIδ interacted almost as strongly with Dvl-1 as did CKIɛ, although CKIα did not (Table 1). Therefore, there is a kinase activity-independent interaction between CKIδ/ɛ and Dvl-1 that is enhanced by the C-terminal tail.
Table 1.
Specificity of the CKI/DvI-1 interaction
| 3-AT | CKIɛ | K38R | K38RΔ319 | CKIδ | CKIα |
|---|---|---|---|---|---|
| 1 mM | ++++ | +++ | ++ | ++ | − |
| 3 mM | +++ | ++ | + | + | − |
Yeast containing the indicated LexA:CKI constructs were mated to yeast containing a DvI-1/GAL4AD fusion construct. The ability of the resulting diploids to grow on synthetic media lacking histidine and supplemented with 1 or 3 mM 3-AT was assessed. Yeast colonies were scored − to ++++, indicating the level of growth after 3 days.
Coimmunoprecipitation of CKI and Dvl-1.
To confirm the two-hybrid results, specific components of the Wnt signaling pathway were expressed in reticulocyte lysates and then mixed with His6-CKIɛ. His6-CKIɛ strongly coimmunoprecipitated Dvl-1 and weakly coimmunoprecipitated axin; β-catenin, β-TrCP, APCRI, APC25, B56α, and GSK3β were not coimmunoprecipitated (data not shown). To determine whether CKI kinase activity was required for the interaction, CKIɛ, K38R, Dvl-1, and axin were expressed separately in reticulocyte lysates and then mixed. CKIɛ coimmunoprecipitated Dvl-1, and there was a low but reproducible immunoprecipitation of axin. Consistent with the results of the two-hybrid assay, coimmunoprecipitation was equally effective with K38R (Fig. 1A).
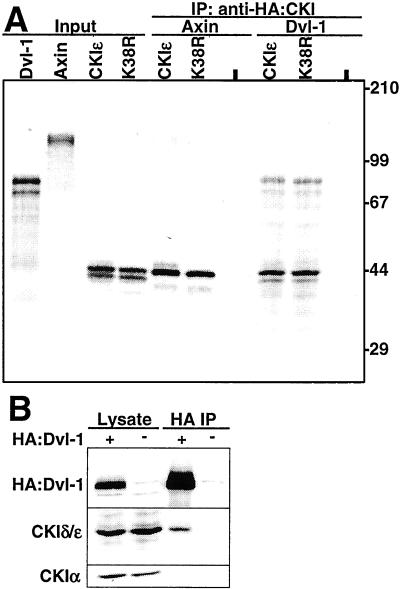
Figure 1.
CKIδ/ɛ interacts with Dvl-1. (A) CKI coimmunoprecipitates Dvl-1. In vitro translated proteins were loading directly (Input), or incubated either alone (-), or with CKIɛ or K38R. The samples were immunoprecipitated with anti-HA and coimmunoprecipitating proteins were detected by SDS/PAGE and PhosphorImager analysis. (B) Dvl-1 coimmunoprecipitates CKIδ/ɛ, but not CKIα. HA: Dvl-1 was immunoprecipitated from 293 cells with anti-HA and the lysate and pellet were probed with anti-HA, anti-CKIδ/ɛ, and anti-CKIα antibodies. The experiments were repeated twice with similar results.
To confirm these results in vivo, HA:Dvl-1 was expressed in 293 cells, immunoprecipitated, and the immunoprecipitate was analyzed for the presence of coimmunoprecipitating proteins. As Fig. 1B shows, HA:Dvl-1 coimmunoprecipitated endogenous CKIδ/ɛ, whereas neither CKIα, nor other components of the Wnt pathway, including PP2A A and C, GSK3β, and β-catenin (data not shown), were found in the immunoprecipitate. Therefore, CKIδ/ɛ interacts with Dvl-1 both in vitro and in vivo.
CKIδ/ɛ Phosphorylates Dvl-1, APC, Axin, and β-Catenin in Vitro.
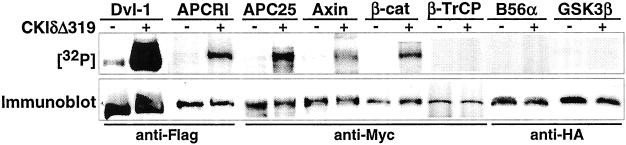
To directly determine which components of the Wnt pathway are in vitro substrates of CKI, various epitope-tagged proteins were expressed in 293 cells, immunoprecipitated, and then incubated with [γ-32P]ATP with or without added CKI. For these experiments, CKIδ lacking the C-terminal tail (CKIδΔ319) was used, because the C-terminal tail of CKI reduces its activity in in vitro assay systems (14). As shown in Fig. 2, Dvl-1, APCRI, APC25, axin, and β-catenin were all phosphorylated by CKIδΔ319. CKI did not phosphorylate GSK3β, B56α, or β-TrCP. Phosphorylation was not due to coprecipitating kinases, because there was minimal phosphorylation of the immunoprecipitated proteins when [γ-32P]ATP was added without CKI. In the case of Dvl-1, however, there is a somewhat higher level of background phosphorylation, presumably due to coimmunoprecipitation of endogenous CKI (Fig. 1B). These results suggest CKI is capable of directly phosphorylating multiple components of the Wnt pathway in vitro.
Figure 2.
CKI phosphorylates multiple components of the Wnt pathway. The indicated epitope-tagged Wnt pathway components were expressed in 293 cells and then immunoprecipitated and incubated with [γ-32P]ATP with or without CKIδΔ319. Phosphorylation was assessed by SDS/PAGE and PhosphorImager analysis, and tagged proteins in the pellet were assessed by immunoblotting. The experiments were repeated twice with similar results.
CKI Phosphorylates β-Catenin and Axin on Major in Vivo Sites.
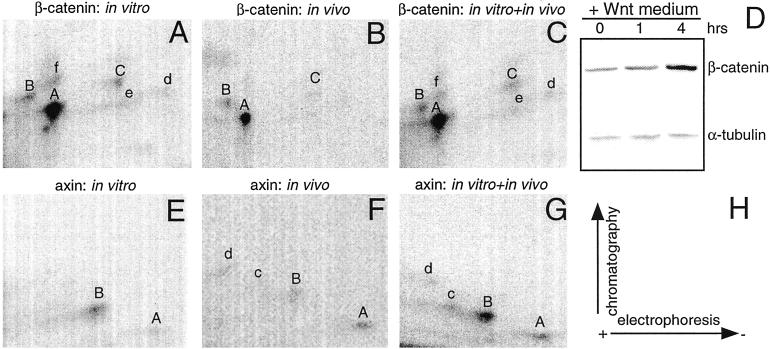
To further explore the finding that multiple Wnt pathway proteins may be physiological substrates of CKI, we compared the in vivo and in vitro CKI-phosphorylated tryptic phosphopeptide maps of β-catenin and axin. 32P-labeled cells were stimulated with Wnt3a conditioned medium before lysis. Consistent with previous reports, the Wnt3a-conditioned medium increased β-catenin abundance (Fig. 3D). In vitro, CKI phosphorylated multiple β-catenin peptides, whereas in vivo, β-catenin accumulated phosphate on only three peptides (labeled A, B, and C in Fig. 3 A–C). Mixing equal amounts of radioactive peptides from in vivo and in vitro phosphorylated β-catenin demonstrated that CKI phosphorylates β-catenin in vitro on sites that are phosphorylated in vivo, namely peptides A, B, and C. In the case of axin, the in vivo and in vitro labeled protein shared phosphopeptides A and B. These results are consistent with the hypothesis that CKI phosphorylates β-catenin and axin in vivo.
Figure 3.
CKI phosphorylates β-catenin and axin on major in vivo sites. Tryptic phosphopeptide maps of β-catenin (A–C) and axin (E–G). The major phosphopeptides are indicated with letters; uppercase letters denote shared peptides, whereas lowercase letters denote unique peptides. (D) The immunoblot confirms the effects of Wnt3a on endogenous β-catenin accumulation in 293 cells. α-Tubulin serves as a loading control. (H) The diagram represents the directions of electrophoresis and chromatography. The experiments were repeated twice with similar results.
CKI Abrogates β-Catenin Degradation in Xenopus Egg Extracts.
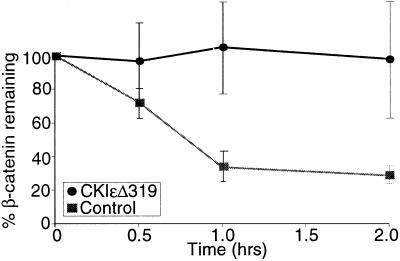
CKI activates Wnt signaling, phosphorylates multiple components of the β-catenin degradation complex, and increases β-catenin abundance in in vivo systems (10, 11). To test whether CKI can directly increase β-catenin abundance, we used an in vitro assay of β-catenin degradation in Xenopus egg extracts (7, 24). β-catenin was degraded in egg extracts with a half-life of less than 1 h (Fig. 4). Addition of CKIɛΔ319 abrogated β-catenin degradation. CKIɛΔ319 was not a general inhibitor of proteasome-mediated degradation, because it did not block the degradation of 125I-ubiquitinated lysozyme (data not shown). K38RΔ319 had no effect on β-catenin degradation (data not shown). These results suggest that CKI can directly increase β-catenin abundance, that the kinase activity of CKI is required for this effect, and that CKI is present in limiting quantities in Xenopus egg extracts.
Figure 4.
CKI abrogates β-catenin degradation in Xenopus egg extracts. Reticulocyte lysate-expressed [35S]β-catenin and luciferase were incubated in Xenopus egg extracts with or without 1.15 μM CKIɛΔ319. Protein degradation was assessed by SDS/PAGE and autoradiography. Data from three separate experiments are plotted as the ratio of β-catenin to luciferase ± SEM.
CKIɛ Destabilizes the β-Catenin Degradation Complex.
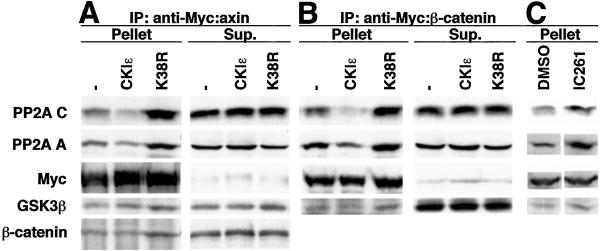
To determine whether the ability of CKI to phosphorylate multiple components of the β-catenin degradation complex leads to a longer β-catenin half-life through destabilization of the complex, we assessed the effect of CKI on the integrity of the β-catenin degradation complex. Initial assays using transient transfection of wild-type and kinase-dead CKI were complicated by nonspecific effects on expression of cotransfected plasmids, a result also seen by others (P. Polakis, personal communication); therefore, a biochemical approach was taken. Lysates from 293 cells expressing Myc:axin or Myc:β-catenin were incubated with anti-Myc-Sepharose either alone or with 180 nM CKIɛ. Myc:axin and Myc:β-catenin coimmunoprecipitated endogenous PP2A C, PP2A A, GSK3β, and β-catenin (Fig. 5 A and B). After incubation of the lysates with CKIɛ, however, there was a decrease in PP2A C and A subunits coimmunoprecipitating with both axin and β-catenin (Fig. 5 A and B). The quantity of coprecipitating GSK3β and β-catenin did not vary substantially. The decrease in PP2A in the immunoprecipitate was not due to variation in the efficiency of the immunoprecipitation, because the immunoprecipitation of axin and β-catenin was comparable in the presence or absence of CKI. In addition, it was not due to a global decrease in PP2A abundance, because CKIɛ caused no discernible change in the amount of PP2A present in the lysate or supernatant.
Figure 5.
CKIɛ destabilizes the β-catenin degradation complex in vitro and in vivo. Lysates from 293 cells expressing (A) Myc:axin or (B) Myc:β-catenin were incubated alone (-), with 180 nM CKIɛ (CKIɛ), or with 480 nM K38R and 250 μM CKI inhibitor CKI-7 (K38R) in the presence of anti-Myc-Sepharose. The pellet and 10% of the supernatant were analyzed by SDS/PAGE and immunoblotting. (C) 293 cells expressing Myc:β-catenin were incubated with DMSO or 40 μM IC261 before harvest and anti-Myc-Sepharose immunoprecipitation. The immunopellets were analyzed by SDS/PAGE and immunoblotting. Each experiment was repeated twice with similar results.
Because CKI decreased the association of PP2A C and A subunits with axin and β-catenin, we determined whether inhibiting CKI enhances PP2A binding. Lysates from 293 cells expressing Myc:axin or Myc:β-catenin were incubated with both 480 nM K38R and 250 μM CKI inhibitor CKI-7 to completely block endogenous CKI activity present in the extract. Inhibition of CKI increased the amount of endogenous PP2A C and A subunits coimmunoprecipitating with axin and β-catenin (Fig. 5 A and B). Additionally, there was a reproducible increase in endogenous GSK3β and β-catenin coprecipitating with Myc:axin, and in endogenous GSK3β coprecipitating with Myc:β-catenin.
To examine this phenomenon under more physiological conditions, the consequence of CKI inhibition on the stability of the β-catenin degradation complex was investigated in vivo. 293 cells expressing Myc:β-catenin were incubated with 40 μM IC261, a membrane-permeable CKIδ/ɛ-specific inhibitor, and the stability of the β-catenin degradation complex was determined by coimmunoprecipitation. Inhibition of CKI increased the amount of endogenous PP2A C, PP2A A, and GSK3β coimmunoprecipitating with Myc:β-catenin (Fig. 5C), providing an in vivo confirmation of this phenomenon. In summary, the decreased association of PP2A C and PP2A A with the β-catenin degradation complex observed in in vitro assays in the presence of elevated CKI activity, and the increased association of PP2A C, PP2A A, GSK3β, and β-catenin observed in both in vitro and in vivo assays with CKI inhibition, suggest that CKI destabilizes the β-catenin degradation complex.
Epistasis Places CKI Upstream of the B56 Regulatory Subunit of PP2A.
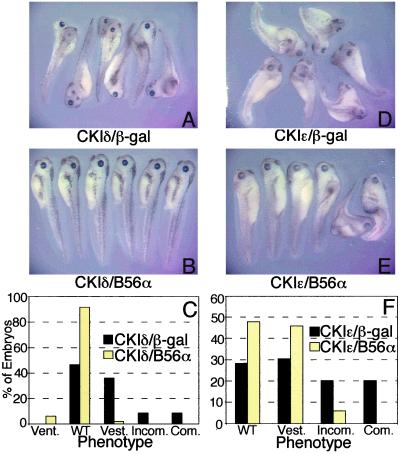
CKI and PP2A functions appear to be linked through their association with the β-catenin degradation complex (7). The ability of CKI to decrease, and the inhibition of CKI to increase, PP2A association with the β-catenin degradation complex suggests that these proteins may be proximate to one another in the Wnt pathway. To further dissect their points of action, we positioned their activities in the Wnt pathway by using molecular epistasis analyses. Xenopus embryos were ventrally microinjected with CKI and either B56α or β-galactosidase (β-gal) RNA. The microinjection of CKIδ resulted in complete, incomplete, and vestigial secondary axes at a frequency of 9%, 9%, and 34%, respectively. B56α blocked CKIδ-induced secondary axes, because 92% of the embryos were now wild type and only 2% had secondary axes, all of which were vestigial (Fig. 6 A–C). The microinjection of CKIɛ resulted in complete, incomplete, and vestigial secondary axes at a frequency of 20%, 20%, and 31%, respectively. B56α reduced the extent of CKIɛ-induced secondary axes; none had a complete axis, 6% had an incomplete axis, and 46% had vestigial axes, whereas 48% were now wild type (Fig. 6 D–F). Therefore, B56 rescues secondary axes induced by CKI, placing B56 downstream of CKI.
Figure 6.
B56 acts downstream of CKI. Xenopus embryos were injected with CKIδ and (A) β-gal or (B) B56α; or CKIɛ and (D) β-gal or (E) B56α RNA. (C and F) Diagrams depicting the degree of axis formation in CKIδ and CKIɛ injected embryos, respectively. Vent., ventralized; WT, wild type; Vest., vestigial secondary axis or slightly dorsalized; Incom., incomplete secondary axis; Com., complete secondary axis. The data shown are from a representative experiment which was performed five times for an n of 236 (CKIδ/β-gal) or 231 (CKIδ/B56α), or six times for an n of 290 (CKIɛ/β-gal) or 288 (CKIɛ/B56α). The statistical analysis was carried out on the cumulative experiments, and the difference between the phenotypes resulting from β-gal and B56α RNA injections in both the CKIδ and CKIɛ experiments is highly significant (P value < 10−4).
Kinase-Dead CKIɛ and B56 Synergistically Inhibit Wnt Signaling.
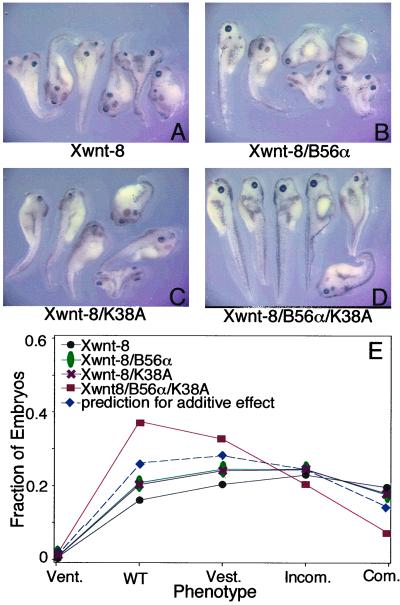
Kinase-dead forms of CKI inhibit Wnt signaling (10, 11), presumably by competing with wild-type CKI for binding to the β-catenin degradation complex. Because we found that inhibition of CKI increases the association of PP2A with the β-catenin degradation complex, the presence of both ectopic kinase-dead CKI and B56 may cooperatively enhance PP2A bound to the complex. Therefore, we tested the ability of B56 and K38A to cooperatively reduce secondary axes induced by Xwnt-8 in Xenopus embryos (25). B56 and K38A each reduced secondary axes when coinjected singly with Xwnt-8 (Fig. 7 A–C). Moreover, Xwnt-8/B56/K38A-injected embryos displayed a further reduction in secondary axes (Fig. 7D). As shown in Fig. 7E, the Xwnt-8/B56/K38A-injected embryos showed a greater than additive shift toward a wild-type phenotype, revealing that B56 and K38A act synergistically in reducing Xwnt-8 signaling.
Figure 7.
B56 and kinase-dead CKI synergistically inhibit Wnt signaling. Xenopus embryos were injected with Xwnt-8 and (A) β-gal, (B) B56α, (C) K38A, or (D) B56α/K38A RNA. Equal amounts of each RNA were injected per embryo, except for β-gal, which was used to normalize total RNA injected per embryo. (E) The graph is a plot of the fitted distribution of responses across the phenotypes from a representative experiment. The injections were performed eight times for an n of 365 (Xwnt-8), 365 (Xwnt-8/B56α), 369 (Xwnt-8/K38A), or 368 (Xwnt-8/K38A/B56α). The statistical analysis shows that there is a synergistic interaction between K38A and B56α, because the phenotypic shift is greater than that predicted for an additive interaction (P value = 0.0066).
Discussion
CKI and PP2A are positive and negative regulators of Wnt signaling, respectively. CKI increases β-catenin abundance and induces secondary axes in Xenopus embryos through its activation of Wnt signaling (10, 11). PP2A promotes β-catenin degradation and blocks Wnt signaling (4, 7). We have found that the roles of CKI and PP2A in Wnt signaling may be intertwined. CKI may activate Wnt signaling by phosphorylating key members of the β-catenin degradation complex and dissociating PP2A from the complex.
Our studies show that CKIδ and CKIɛ, but not CKIα, interact with Dvl-1 both by two-hybrid analyses and by coimmunoprecipitation. We were unable to demonstrate a direct interaction between CKI and axin, but did see a low but reproducible coimmunoprecipitation of axin with CKI. Because CKI phosphorylates axin in vitro on major in vivo sites, CKI and axin may interact transiently in a manner that is difficult to detect by two-hybrid analysis or coimmunoprecipitation. Two-hybrid data suggest that the CKIδ/ɛ C-terminal tail may stabilize the Dvl-1 interaction. However, the C-terminal tail was not essential for CKI to phosphorylate Wnt pathway substrates in vitro, or to inhibit β-catenin degradation in Xenopus egg extracts. Interestingly, CKIδ did not interact with Dvl-1 as strongly as CKIɛ, suggesting that the differential interaction of CKIδ and CKIɛ with Dvl-1 may be due to their divergent C termini.
CKI phosphorylates Dvl-1, APC, axin, and β-catenin in vitro. Because CKI binds most strongly to Dvl, the binding of CKI to Dvl-1 may recruit it to phosphorylate these proteins, each of which are members of the β-catenin degradation complex. Dvl is a likely in vivo target of CKI, given that it is hyperphosphorylated in response to Wnt signaling. Another kinase has recently been identified that phosphorylates Dvl, PAR-1 (26). PAR-1 is a Wnt-stimulated kinase that is required for Wnt signal transduction, and is therefore also a likely regulator of Dvl. However, it has been shown that the activation of Dvl through hyperphosphorylation may not be required for Wnt signal transduction (27). The phosphorylation of APC, axin, and β-catenin by GSK3β inhibits Wnt signaling. Because CKI activates Wnt signaling, if APC, axin, and/or β-catenin are in vivo targets of CKI, the sites phosphorylated by CKI are likely to be distinct from those phosphorylated by GSK3β. Preliminary data suggest that CKIɛ phosphorylation of β-catenin increases its affinity for Lef-1 (Z.-H.G. and D.M.V., unpublished results), suggesting that CKI may also propagate the Wnt signal by promoting an interaction between β-catenin and Lef-1.
APC and axin anchor β-catenin and GSK3β in the β-catenin degradation complex. This complex also contains CKI and PP2A. In vitro or in vivo inhibition of CKI increases the association of endogenous PP2A, GSK3β, and β-catenin with the complex, likely increasing GSK3β-dependent β-catenin phosphorylation and reducing Wnt signaling. In addition, CKI decreases the association of PP2A C and A with the β-catenin degradation complex. Therefore, complexes that contain both CKI and PP2A are likely to be transient. These results suggest that in the absence of CKI activity, the β-catenin degradation complex contains PP2A, resulting in low β-catenin levels, whereas in the presence of CKI activity, the complex is destabilized and the Wnt signal is transduced. Moreover, the finding that in vivo inhibition of CKI stabilizes the β-catenin degradation complex suggests that CKI actively destabilizes the β-catenin degradation complex and propagates the Wnt signal in 293 cells.
The relationship between CKI and PP2A in Wnt signaling was further examined by characterizing their interplay in early Xenopus development. Epistasis analysis shows that B56 acts downstream of CKI. The ability of K38A to act synergistically with B56 in reducing Xwnt-8-induced secondary axes suggests that there is an intimate mechanistic interaction between CKI and PP2A in Wnt signaling. Thus, both biochemical and phenotypic observations suggest that these two enzymes with opposing functions are intimate antagonists in the regulation of Wnt signaling.
How does CKI propagate the Wnt signal? CKI may mediate the phosphorylation of specific β-catenin degradation complex components (e.g., APC, axin, and/or β-catenin) reducing their affinity for other members of the complex (e.g., PP2A, GSK3β, and β-catenin), and releasing them from the complex. The loss of PP2A, a counterregulatory phosphatase, may further enhance CKI-mediated phosphorylation of complex components, amplifying CKI's signaling activity. Therefore, the CKI-induced dissociation of the β-catenin degradation complex would reduce GSK3β-mediated phosphorylation and degradation of β-catenin, and propage of the Wnt signal.
Acknowledgments
We thank K. Thomas and A. Branscomb for critically reading the manuscript, B. Smiley for RNA preparation, M. Schlesinger and E. Vielhaber for protein purification, X. Li for assistance in preparation of egg extracts, A. Szabo and the Biostatistics Shared Resource for statistical analyses, and G. Keesler, H. Yuan, P. Polakis, F. Costantini, R. Moon, A. Kikuchi, J. Woodgett, D. Stillman, and R. Takada for reagents. This work was supported by National Institutes of Health Grants R01CA80809 and R01CA71074, and the Huntsman Cancer Foundation. Statistical analyses, oligonucleotide synthesis, and DNA sequencing were supported by Cancer Center Support Grant 2P30CA42014.
Abbreviations
- CKI
casein kinase I
- PP2A
protein phosphatase 2A
- GSK3β
glycogen synthase kinase 3β
- APC
adenomatous polyposis coli
- Dvl/Dsh
disheveled
- K38A
CKIɛK38A
- K38R
CKIɛK38R
- β-gal
β-galactosidase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bienz M, Clevers H. Cell. 2000;103:311–320. doi: 10.1016/s0092-8674(00)00122-7. [DOI] [PubMed] [Google Scholar]
- 2.Polakis P. Genes Dev. 2000;14:1837–1851. [PubMed] [Google Scholar]
- 3.Virshup D M. Curr Opin Cell Biol. 2000;12:180–185. doi: 10.1016/s0955-0674(99)00074-5. [DOI] [PubMed] [Google Scholar]
- 4.Seeling J M, Miller J R, Gil R, Moon R T, White R, Virshup D M. Science. 1999;283:2089–2091. doi: 10.1126/science.283.5410.2089. [DOI] [PubMed] [Google Scholar]
- 5.Ratcliffe M J, Itoh K, Sokol S Y. J Biol Chem. 2000;275:35680–35683. doi: 10.1074/jbc.C000639200. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto H, Hinoi T, Michiue T, Fukui A, Usui H, Janssens V, Van Hoof C, Goris J, Asashima M, Kikuchi A. J Biol Chem. 2001;276:26875–26882. doi: 10.1074/jbc.M100443200. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Yost H J, Virshup D M, Seeling J M. EMBO J. 2001;20:4122–4131. doi: 10.1093/emboj/20.15.4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu W, Zeng L, Costantini F. J Biol Chem. 1999;274:3439–3445. doi: 10.1074/jbc.274.6.3439. [DOI] [PubMed] [Google Scholar]
- 9.Ikeda S, Kishida M, Matsuura Y, Usui H, Kikuchi A. Oncogene. 2000;19:537–545. doi: 10.1038/sj.onc.1203359. [DOI] [PubMed] [Google Scholar]
- 10.Peters J M, McKay R M, McKay J P, Graff J M. Nature (London) 1999;401:345–350. doi: 10.1038/43830. [DOI] [PubMed] [Google Scholar]
- 11.Sakanaka C, Leong P, Xu L, Harrison S D, Williams L T. Proc Natl Acad Sci USA. 1999;96:12548–12552. doi: 10.1073/pnas.96.22.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McKay R M, Peters J M, Graff J M. Dev Biol. 2001;235:388–396. doi: 10.1006/dbio.2001.0308. [DOI] [PubMed] [Google Scholar]
- 13.McCright B, Brothman A R, Virshup D M. Genomics. 1996;36:168–170. doi: 10.1006/geno.1996.0438. [DOI] [PubMed] [Google Scholar]
- 14.Cegielska A, Gietzen K F, Rivers A, Virshup D M. J Biol Chem. 1998;273:1357–1364. doi: 10.1074/jbc.273.3.1357. [DOI] [PubMed] [Google Scholar]
- 15.Hannon G J, Demetrick D, Beach D. Genes Dev. 1993;7:2378–2391. doi: 10.1101/gad.7.12a.2378. [DOI] [PubMed] [Google Scholar]
- 16.Brent R, Finley R L., Jr Annu Rev Genet. 1997;31:663–704. doi: 10.1146/annurev.genet.31.1.663. [DOI] [PubMed] [Google Scholar]
- 17.McCright B, Virshup D M. In: Methods In Molecular Biology. Ludlow J, editor. Totowa, NJ: Humana Press; 1997. pp. 263–277. [Google Scholar]
- 18.Gao Z H, Metherall J, Virshup D M. Biochem Biophys Res Commun. 2000;268:562–566. doi: 10.1006/bbrc.2000.2168. [DOI] [PubMed] [Google Scholar]
- 19.Vielhaber E, Eide E, Rivers A, Gao Z-H, Virshup D M. Mol Cell Biol. 2000;20:4888–4899. doi: 10.1128/mcb.20.13.4888-4899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gietzen K F, Virshup D M. J Biol Chem. 1999;274:32063–32070. doi: 10.1074/jbc.274.45.32063. [DOI] [PubMed] [Google Scholar]
- 21.Hardie D G. In: Practical Approach Series 211. Hardie D G, editor. Oxford: Oxford Univ. Press; 1999. pp. 97–126. [Google Scholar]
- 22.Chijiwa T, Hagiwara M, Hidaka H. J Biol Chem. 1989;264:4924–4927. [PubMed] [Google Scholar]
- 23.Mashhoon N, DeMaggio A J, Tereshko V, Bergmeier S C, Egli M, Hoekstra M F, Kuret J. J Biol Chem. 2000;275:20052–20060. doi: 10.1074/jbc.M001713200. [DOI] [PubMed] [Google Scholar]
- 24.Salic A, Lee E, Mayer L, Kirschner M W. Mol Cell. 2000;5:523–532. doi: 10.1016/s1097-2765(00)80446-3. [DOI] [PubMed] [Google Scholar]
- 25.Christian J L, McMahon J A, McMahon A P, Moon R T. Development (Cambridge, UK) 1991;111:1045–1055. doi: 10.1242/dev.111.4.1045. [DOI] [PubMed] [Google Scholar]
- 26.Sun T-Q, Lu B, Feng J-J, Reinhard C, Jan Y N, Fantl W J, Williams L T. Nat Cell Biol. 2001;3:628–636. doi: 10.1038/35083016. [DOI] [PubMed] [Google Scholar]
- 27.Rothbacher U, Laurent M N, Deardorff M A, Klein P S, Cho K W, Fraser S E. EMBO J. 2000;19:1010–1022. doi: 10.1093/emboj/19.5.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]