Abstract
The structure of RNA polymerase II in a complex with the inhibitor α-amanitin has been determined by x-ray crystallography. The structure of the complex indicates the likely basis of inhibition and gives unexpected insight into the transcription mechanism.
The structure of 10-subunit 0.5-MDa yeast RNA polymerase II (pol II), recently determined at 2.8 Å resolution, reveals the architecture and key functional elements of the enzyme (1). The two largest subunits, Rpb1 and Rpb2, lie at the center, on either side of a nucleic acid-binding cleft, with the many smaller subunits arrayed around the outside. Rpb1 and Rpb2 interact extensively in the region of the active site and also through a domain of Rpb1 that lies on the Rpb2 side of the cleft, connected to the body of Rpb1 by an α-helix that bridges across the cleft.
Proof that nucleic acids bind in the channel comes from the molecular replacement solution of a transcribing pol II complex at 3.3 Å resolution (2). This structure shows the template DNA unwinding some three residues before the active site, followed by nine base pairs of DNA–RNA hybrid. Adjacent regions of Rpb1 and Rpb2 form a highly complementary surface, resulting in extensive DNA–RNA hybrid–protein interaction. The “bridge” helix seems to play an important role, binding to both the second and third unpaired DNA bases and also to the coding base, paired with the first residue of the RNA. Comparison of the pol II structure in different crystal forms shows a division of the enzyme in several mobile elements that my facilitate DNA and RNA movement during transcription. Comparison of the pol II structure with that of the related bacterial RNA polymerase (3) suggests mobility of the bridge helix as well (2).
The pol II structures open the way to many lines of investigation. Structures of cocrystals of pol II with interacting molecules can be solved, the full power of site-directed mutagenesis can be brought to bear on the transcription mechanism, and so forth. Here we report the structure of a cocrystal of pol II with the most potent and specific known inhibitor of the enzyme, α-amanitin. The active principle of the “death cap” mushroom, α-amanitin blocks both transcription initiation and elongation (4–6). The structure of the cocrystal suggests that α-amanitin interferes with a protein conformational change underlying the transcription mechanism.
Materials and Methods
Crystals of yeast pol II were grown as described and were soaked in cryoprotectant solution containing 50 μg/ml α-amanitin and 1 mM MgSO4 for 1 week before freezing and x-ray data collection to 2.8 Å resolution (Table 1; ref. 7). Data collection was carried out at 100 K by using 0.5° oscillations with an Area Detector Systems Quantum 4 charge-coupled device (CCD) detector at Stanford Synchrotron Radiation Laboratory beamline 11-1. Diffraction data were processed with denzo and reduced with scalepack (8). The previous 2.8-Å pol II structure was subjected to rigid body refinement against the cocrystal data. The R-free test set from the native form 2 pol II data was used for the pol II α-amanitin refinement (1). Refinement of the cocrystal structure was preformed by using cns (9). A σA-weighted difference electron density map was consistent with the known structure of amanitin toxins (Fig. 1A). After positional and B-factor refinement of the pol II model and minor adjustments to the model, an α-amanitin model was placed. The α-amanitin model was generated from 6′-O-methyl-α-amanitin (S)-sulfoxide methanol solvate monohydrate as obtained from the Cambridge Structure Database [accession code 3384082 (10)]. To conform to the known composition and stereochemistry of α-amanitin, the 6′-O-methyl group was removed from the 6′-O-methyltryptophan residue (α-amanitin position 4) and the stereochemistry of the sulfoxide was modified to R. Topology and refinement parameter files for use in cns for the α-amanitin structure were generated by using hic-up (11). Rigid body refinement was performed on the α-amanitin alone, followed by positional and B-factor refinement of the entire pol II-α-amanitin complex and further minor adjustment of the model, giving a final free-R factor of 28% (Table 2). The refined σA-weighted 2Fobs − Fcalc map (Fig. 1B) clearly shows density for the main chain atoms. Some of the side chains, however, such as that of the 4,5-dihydroxyisoleucine residue, are only partially visible (ordered) in the map. The stereo chemistry of the 4,5-dihydroxyisoleucine γ hydroxyl is important in amanitin inhibition, suggestive of a role in hydrogen bonding (12). Poor ordering in our cocrystal indicates that at least in yeast, the proposed hydrogen bond is not formed. This may partially explain the lesser sensitivity of Saccharomyces cerevisiae to α-amanitin compared with other eukaryotes (4).
Table 1.
Crystallographic data
| Space group | I222 |
| Unit cell, Å | 122.5 by 222.5 by 374.2 |
| Wavelength, Å | 0.965 |
| Mosaicity, ° | 0.44 |
| Resolution, Å | 20–2.8 (2.9–2.8) |
| Completeness, % | 99.8 (99.4) |
| Redundancy | 3.9 (2.9) |
| Unique reflections | 124,441 (12,292) |
| Rsym, % | 6.7 (21.6) |
Values in parentheses correspond to the highest-resolution shell. Rsym = Σi,h|I(i,h) − 〈I(h)〉|/Σi,h|I(i,h)|, where 〈I(h)〉 is the mean of the I observations of reflection h. Rsym was calculated with anomalous pairs merged; no sigma cut-off was applied.
Figure 1.
Stereo image of final α-amanitin structure. (A) σA-weighted Fobs − Fcalc electron density at 2.8 Å resolution (red) contoured at 3 sigma calculated from the initial pol II placement before α-amanitin was included in the model. The final α-amanitin structure is shown (ball and stick model). (B) σA-weighted 2Fobs − Fcalc electron density at 2.8 Å resolution (blue) contoured at 1.2 sigma, superimposed on the final α-amanitin structure (ball and stick model). Only the electron density around α-amanitin is shown. This figure was generated by using BOBSCRIPT and RASTER3D (21–23).
Table 2.
Refinement statistics
| Nonhydrogen atoms | 27,906 |
| Protein residues | 3,490 |
| Water molecules | 69 |
| Anisotropic scaling (B11, B22, B33) | −6.3, −6.9, 13.1 |
| rms deviation bonds | 0.0083 |
| rms deviation angles | 1.4 |
| Reflection test set | 3,757 (3.0%) |
| Rcryst/Rfree | 22.9/28.0 |
| Average B factor overall | 57 |
| Average B factor pol | 57 |
| Average B factor amanitin | 78 |
| Average B factor water | 35 |
Rcryst/free = ∑h∥Fobs(h)| − |Fcalc(h)∥/∑h|Fobs(h)|. Rcryst and Rfree were calculated from the working and test reflection sets, respectively.
Results and Discussion
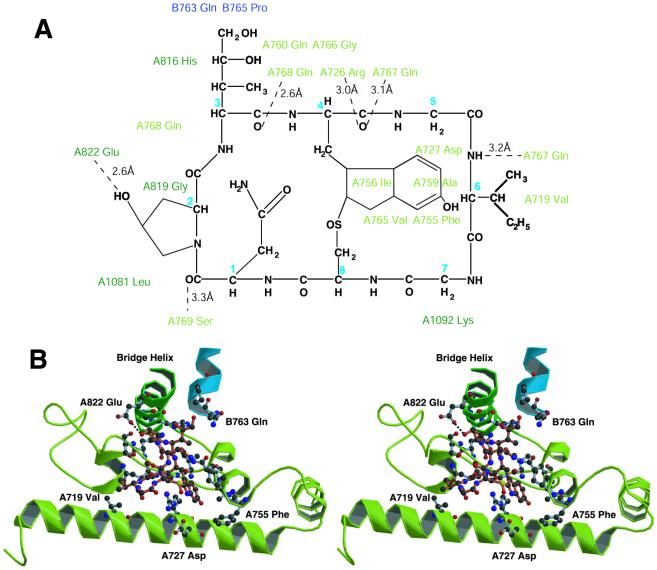
The α-amanitin binding site is beneath a “bridge helix” extending across the cleft between the two largest pol II subunits, Rpb1 and Rpb2, in a “funnel”-shaped cavity in the pol II structure (Fig. 2 A and B). Most pol II mutations affecting α-amanitin inhibition map to this site (Table 3), showing that it is functionally relevant and not an artifact of crystallization (13–15). Pol II residues interacting with α-amanitin are located almost entirely in the bridge helix (in the previously defined “cleft” region of Rpb1) and in an adjacent part of Rpb1 on the Rpb2-side of the cleft [in the previously defined funnel region of Rpb1 (Fig. 3 A and B; Table 3)]. There is a strong hydrogen bond between hydroxyproline 2 of α-amanitin and bridge helix residue Glu-A822. There is an indirect interaction involving the backbone carbonyl group of 4,5-dihydroxyisoleucine 3 of α-amanitin, hydrogen-bonded to residue Gln-A768, which is, in turn, hydrogen-bonded to bridge helix residue His-A816. Finally, there are several hydrogen bonds between α-amanitin and the region of Rpb1 adjacent to the bridge helix. Binding of α-amanitin therefore buttresses the bridge helix, constraining its position with respect to the Rpb2-side of the cleft.
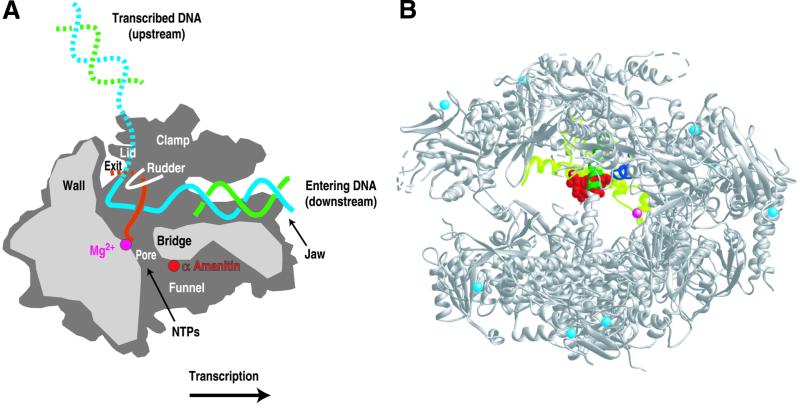
Figure 2.
Location of α-amanitin bound to pol II. (A) Cutaway view of a pol II-transcribing complex showing the location of α-amanitin binding (red dot) in relation to the nucleic acids and functional elements of the enzyme. Adapted from ref. 24. (B) Ribbons representation of the pol II structure (top view in refs. 1 and 7). Eight zinc atoms are shown in light blue, the active site magnesium is magenta, the region of Rpb1 around α-amanitin is light green (funnel) and dark green (bridge helix), the region of Rpb2 near α-amanitin is dark blue, and α-amanitin is red. This figure was prepared by using RIBBONS (25).
Table 3.
Hydrogen bonds, buried surface area, and known amanitin mutants
| Residue in yeast | Δ surface area, Å2 | H-bond | Residue in human | Mutations |
|---|---|---|---|---|
| Val-A719 | −32 | Asn-A742 | ||
| Leu-A722 | 0 | Leu-A745 | Mouse L745F (13) | |
| Asn-A723 | −22 | Asn-A746 | ||
| Arg-A726 | −63 | NH1 to AMA pos. 4 O 3.0 Å | Arg-A749 | Mouse R749P (14) Drosophila melanogaster R741H(15) |
| Asp-A727 | −7 | Asp-A750 | ||
| Phe-A755 | −8 | Lys-A778 | ||
| Ile-A756 | −48 | Ile-A779 | Mouse I779F (14) | |
| Ala-A759 | −7 | Ser-A782 | ||
| Gln-A760 | −33 | Gln-A783 | ||
| Cys-A764 | 0 | Val-A787 | Caenorhabditis elegans C777Y(15) | |
| Val-A765 | −2 | Val-A788 | ||
| Gly-A766 | −1 | Gly-A789 | ||
| Gln-A767 | −34 | N to AMA pos. 4 O 3.1 Å | Gln-A790 | |
| O to AMA pos. 5 N 3.2 Å | ||||
| Gln-A768 | −16 | OE1 to AMA pos. 3 O 2.6 Å | Gln-A791 | |
| Ser-A769 | −37 | N to AMA pos. 2 O 3.3 Å | Asn-A792 | Mouse N792D (14) |
| Gly-A772 | −24 | Gly-A795 | C. elegans G785E (15) | |
| Lys-A773 | −4 | Lys-A796 | ||
| Arg-A774 | −2 | Arg-A797 | ||
| Tyr-A804 | −2 | Tyr-A827 | ||
| His-A816 | −13 | His-A839 | ||
| Gly-A819 | −19 | Gly-A842 | ||
| Gly-A820 | −8 | Gly-A843 | ||
| Glu-A822 | −15 | OE2 to AMA pos. 2 OD2 2.6 Å | Glu-A845 | |
| Gly-A823 | −13 | Gly-A846 | ||
| Asp-A826 | −2 | Asp-A849 | ||
| Thr-A1080 | −1 | Thr-A1103 | ||
| Leu-A1081 | −63 | Leu-A1104 | ||
| Lys-A1092 | −37 | Lys-A1115 | ||
| Lys-A1093 | −1 | Asn-A1116 | ||
| Gln-B763 | −16 | Gln-B718 | ||
| Pro-B765 | −11 | Pro-B720 | ||
| Total | −541 |
Δ surface area (Å2) is the change in solvent-exposed surface as calculated with program areaimol, using a standard probe radius of 1.4 Å. Potential hydrogen bonds with a donor-acceptor distance below 3.3 Å were included. Residues that are different between yeast and human are in bold. Mutations are changes in Rpb1 in eukaryotes that are known to affect α-amanitin inhibition. α-Amanitin also seems to make a contact with part of the disordered loop between A1081 and A1092. Unfortunately, only density for ∼1 amino acid appears, preventing placement of this loop or even reliable determination of which amino acid in the disordered loop is responsible for this interaction.
Figure 3.
Interaction of α-amanitin with pol II. (A) The chemical structure of α-amanitin, with residues of pol II that lie within 4 Å [determined by using CONTACT (26)] placed near the closest contact. The Cαs of α-amanitin are labeled with blue numbers. Hydrogen bonds are shown as dashed lines with the distances indicated. (B) Stereoview of the α-amanitin binding pocket. Ball and stick models of α-amanitin (red bonds) and of pol II residues within 4 Å (gray bonds) are shown. Rpb1 from A700 to A809 (funnel region) is light green. Rpb1 from A810 to A825 (bridge helix) is dark green. Rpb2 from B760 to B769 is blue. This figure was generated by using BOBSCRIPT and RASTER3D (21–23).
This mode of α-amanitin interaction can account for the biochemistry of inhibition. There is little if any influence of α-amanitin binding on the affinity of pol II for nucleoside triphosphates (5, 16). Moreover, after the addition of α-amanitin to a transcribing pol II complex, a phosphodiester bond can still be formed (17, 18). The rate of translocation of pol II on DNA is, however, reduced from several thousand to only a few nucleotides per minute (5, 6). These findings are consistent with binding of α-amanitin too far from the active site to interfere with nucleoside triphosphate entry or RNA synthesis (or its reversal) (Fig. 2A; ref. 5). They may be explained by a constraint on bridge helix movement. It was previously suggested that such movement is coupled to DNA translocation. The suggestion was based on two observations. First, in the structure of a pol II-transcribing complex, bridge helix residues directly contact the DNA base paired with the first base in the RNA strand. Second, although the sequence of the bridge helix is well conserved, the conformation is different in a bacterial RNA polymerase structure, with bridge helix residues in position to contact the second base in the DNA strand (1, 2, 19). Movement of bridge helix residue Glu-A822 by as little as 1 Å would extend the length of the donor-acceptor pair for the hydrogen bond to hydroxyproline 2 of α-amanitin beyond 3.3 Å, effectively breaking the bond.
Structural derivatives of α-amanitin show the importance of bridge helix interaction for inhibitory activity. The derivative proamanullin, which lacks the hydroxyl group of hydroxyproline 2, involved in hydrogen bonding to bridge helix residue Glu-A822, and which also lacks both hydroxyl groups of 4,5-dihroxyisoleucine 3, is about 20,000-fold less inhibitory than α-amanitin. This effect is caused almost entirely by the alteration of hydroxyproline 2, because alteration of 4,5-dihydroxyisoleucine 3 alone, in the derivative amanullin, reduces inhibition only about 4-fold (4, 20). Other changes in α-amanitin structure may affect inhibition indirectly, by diminishing the overall affinity for pol II. For example, shortening the side chain of isoleucine-6 of α-amanitin reduces inhibition by about 1,000-fold. This side chain inserts in a hydrophobic pocket of pol II in the cocrystal structure.
Thus three lines of evidence on α-amanitin inhibition, coming from biochemical studies of transcription, from structure-activity relationships, and from cocrystal structure determination, converge on a simple picture. Binding of α-amanitin to pol II permits nucleotide entry to the active site and RNA synthesis but prevents the translocation of DNA and RNA needed to empty the site for the next round of synthesis. The inhibition of translocation is caused by interaction of α-amanitin with the pol II bridge helix, whose movement is required for translocation.
Acknowledgments
We thank N. Thompson and R. Burgess (McArdle Laboratory for Cancer Research, Univ. of Wisconsin) for generously providing antibody for protein purification. We thank B. Freedman (North American Mycological Association) for providing samples of Amanita phalloides. We thank COMPAQ (Houston) for providing a Unix workstation. This research is based in part on work done at SSRL, which is funded by the U.S. Department of Energy Office of Basic Energy Sciences. The structural biology program is supported by the National Institutes of Health National Center for Research Resources Biomedical Technology Program and the Department of Energy Office of Biological and Environmental Research. P.C. was supported by a postdoctoral fellowship of the Deutsche Forschungsgemeinschaft. D.A.B. was supported by postdoctoral fellowship no. PF-00-014-01-GMC from the American Cancer Society. This research was supported by National Institutes of Health Grant GM49985 (to R.D.K.).
Abbreviation
- pol II
polymerase II
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code 1K83).
References
- 1.Cramer P, Bushnell D A, Kornberg R D. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 2.Gnatt A L, Cramer P, Fu J, Bushnell D A, Kornberg R D. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 3.Zhang G, Campbell E A, Minakhin L, Richter C, Severinov K, Darst S A. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 4.Wieland T, Faulstich H. Experientia. 1991;47:1186–1193. doi: 10.1007/BF01918382. [DOI] [PubMed] [Google Scholar]
- 5.Chafin D R, Guo H, Price D H. J Biol Chem. 1995;270:19114–19119. doi: 10.1074/jbc.270.32.19114. [DOI] [PubMed] [Google Scholar]
- 6.Rudd M D, Luse D S. J Biol Chem. 1996;271:21549–21558. doi: 10.1074/jbc.271.35.21549. [DOI] [PubMed] [Google Scholar]
- 7.Cramer P, Bushnell D A, Fu J, Gnatt A L, Maier-Davis B, Thompson N E, Burgess R R, Edwards A M, David P R, Kornberg R D. Science. 2000;288:640–649. doi: 10.1126/science.288.5466.640. [DOI] [PubMed] [Google Scholar]
- 8.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 9.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 10.Wieland T, Gotzendorfer C, Dabrowski J, Lipscomb W N, Shoham G. Biochemistry. 1983;22:1264–1271. doi: 10.1021/bi00274a043. [DOI] [PubMed] [Google Scholar]
- 11.Kleywegt G J, Jones T A. Acta Crystallogr D. 1998;54:1119–1131. doi: 10.1107/s0907444998007100. [DOI] [PubMed] [Google Scholar]
- 12.Zanotti G, Wieland T, Benedetti E, Di Blasio B, Pavone V, Pedone C. Int J Pept Protein Res. 1989;34:222–228. doi: 10.1111/j.1399-3011.1989.tb00234.x. [DOI] [PubMed] [Google Scholar]
- 13.Bartolomei M S, Corden J L. Mol Cell Biol. 1987;7:586–594. doi: 10.1128/mcb.7.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartolomei M S, Corden J L. Mol Gen Genet. 1995;246:778–782. doi: 10.1007/BF00290727. [DOI] [PubMed] [Google Scholar]
- 15.Chen Y, Weeks J, Mortin M A, Greenleaf A L. Mol Cell Biol. 1993;13:4214–4222. doi: 10.1128/mcb.13.7.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cochet-Meilhac M, Chambon P. Biochim Biophys Acta. 1974;353:160–184. doi: 10.1016/0005-2787(74)90182-8. [DOI] [PubMed] [Google Scholar]
- 17.Vaisius A C, Wieland T. Biochemistry. 1982;21:3097–3101. doi: 10.1021/bi00256a010. [DOI] [PubMed] [Google Scholar]
- 18.Gu W, Powell W, Mote J, Jr, Reines D. J Biol Chem. 1993;268:25604–25616. [PMC free article] [PubMed] [Google Scholar]
- 19.Darst S A. Curr Opin Struct Biol. 2001;11:155–162. doi: 10.1016/s0959-440x(00)00185-8. [DOI] [PubMed] [Google Scholar]
- 20.Zanotti G, Mohringer C, Wieland T. Int J Pept Protein Res. 1987;30:450–459. doi: 10.1111/j.1399-3011.1987.tb03353.x. [DOI] [PubMed] [Google Scholar]
- 21.Esnouf R M. J Mol Graph Model. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 22.Esnouf R M. J Mol Graph Model. 1997;15:112–113. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 23.Merrit E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 24.Klug A. Science. 2001;292:1844–1846. doi: 10.1126/science.1062384. [DOI] [PubMed] [Google Scholar]
- 25.Carson M. In: Methods in Enzymology. Sweet R M, Carter C W, editors. Vol. 277. New York: Academic; 1997. pp. 493–505. [PubMed] [Google Scholar]
- 26.Collaborative Computational Project, No. 4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]