Abstract
The nonconserved, hydrophilic N-terminal domain of eukaryotic DNA topoisomerase I (topo I) is dispensable for catalytic activity in vitro but essential in vivo. There are at least five putative nuclear localization signals and a nucleolin-binding signal within the first 215 residues of the topo I N-terminal domain. We have investigated physiological functions of the topo I N-terminal domain by fusing it to an enhanced green fluorescent protein (EGFP). The first 170 residues of the N-terminal domain allow efficient import of chimeric proteins into nuclei and nucleoli. The nucleolar localization of this protein does not depend on its interaction with nucleolin, whereas ongoing rDNA transcription clearly is crucial. Immunoprecipitation experiments reveal that the topo I N terminus (topoIN)-EGFP fusion protein associates with the TATA-binding protein in cells. Furthermore, DNA damage results in extensive nuclear redistribution of the topoIN-EGFP chimeric product. The redistribution is also p53-dependent and the N terminus of topo I appears to interact with p53 in vivo. These results show that the topo I localization to the nucleolus is related to the p53 and DNA damage, as well as changes in transcriptional status. Nucleolar release of topo I under conditions of cellular duress may represent an important, antecedent step in tumor cell killing by topoisomerase active agents.
Keywords: nucleolar localization‖DNA damage‖camptothecin
DNA topoisomerase I (topo I) affects DNA topology by making a single strand break in DNA, followed by one or more cycles of controlled rotation, then resealing and releasing the DNA (1). This is an enzyme that participates in a variety of DNA templating activities, such as transcription (2–5) and DNA replication (6, 7), presumably to reduce and modulate torsional stress in DNA. Topo I is also the primary target of a group of anticancer drugs related to the natural product camptothecin (CPT) (for reviews, see refs. 8 and 9). These drugs stabilize topo I-DNA covalent complexes causing single-stranded DNA breaks, which are thought to be converted into lethal double-strand DNA breaks. Moreover, topo I can indirectly influence genomic instability through illegitimate recombination (10–13). Although topo I is not necessary in yeast (14), it is required for embryonic development in Drosophila melanogaster (15) and mice (16); therefore, topo I is essential in the context of a multicellular organism. Recently, it was proposed that topoisomerase I and p53 might cooperate to eliminate damaged genomes from the whole organism (17).
Limited proteolysis and crystallographic data indicate that topo I is comprised of (i) a highly charged N-terminal domain (≈24 kDa), (ii) a conserved core domain (≈54 kDa), (iii) a linker region (≈3 kDa), and (iv) a highly conserved C-terminal domain, which contains the active site tyrosine (1, 18). In vitro studies reveal that two highly conserved globular domains (the core and the C-terminal domain) are crucial for catalytic activity, and two other regions (N terminus and linker) that are not required for catalytic activity (1, 18). Despite the fact that the nonconserved N-terminal domain is dispensable for the relaxation activity, it may still have important roles in vivo. For example, the lethality of human topo I overexpression in yeast is dependent on the N-terminal domain, which carries the nuclear localization signal sequence (NLS) (19, 20). Furthermore, studies in Drosophila also show that the hydrophilic N terminus can target topo I to transcriptionally active loci, suggesting additional roles for the domain in vivo (21, 22).
In this work, we have used enhanced green fluorescent protein (EGFP)–topo I fusion constructs to evaluate the dynamics of topo I distribution in living cells. We demonstrate that topo I fusion proteins localize to the nucleolus presumably at sites of rDNA transcription. The localization can be significantly altered by drugs that poison topo I–DNA reaction complexes or by selective inhibition of ribosomal gene transcription. The N-terminal domain of topo I is required for this response. These findings suggest a model whereby the N-terminal domain of topo I might serve as a primary targeting device for the enzyme. Moreover, localization of the fusion protein is clearly dynamic and can rapidly change in response to changes in transcription, DNA damage, and topoisomerase inhibition. Finally, we report that the p53 status of the cell influences how endogenous topo I is shuffled around following DNA damage.
Materials and Methods
Plasmid Construction.
Plasmids containing the human topo I N terminus fused with EGFP and the deletion constructs used in this study are shown in Fig. 1. The topo I N terminus cDNA (residues 1–215) fragment was synthesized with PCR. The primers used in this PCR were 5′-CCGGGAATTCATGAGTGGGGACCACCTC-3′ and 5′-CGGGATCCACTTGATGCCTTCAGGATAG-3′. After digestion with restriction enzymes EcoRI and BamHI sites, the PCR fragment was inserted into the EcoRI-BamHI sites of expression vector pEGFPN2 (CLONTECH) and sequenced to confirm the DNA junctions. The other three shorter constructs were also constructed from PCR products with appropriate oligonucleotide primers.
Figure 1.
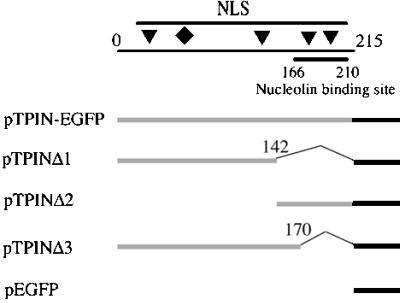
Schematic description of topoIN-EGFP constructs. The N-terminal domain of topo I and its deleted fragments were cloned into pEGFPN2. The top line shows the topo I N terminus (residues 1–215). Four putative NLSs (triangles) are located at residues 59–65 (KKHKEKE), residues 150–156 (KKIKTED), residues 174–180 (KKPKNKED), and residues 192–198 (KKKPKKE) (18). A novel NLS is marked by the diamond at residues 117–146 (DEDDAD or KDEPEDDG) (20). The nucleolin-binding region is located from residues 166 to 210 as shown. The lower collection of lines represent fusion constructs of N-terminal segments of topo I (light line) and EGFP (heavy line), and numbers correspond to amino acid residues of topo I deletions in the fusion constructs.
Reagents.
The topo I antibody is a human antibody against topo I isolated from serum of scleroderma patients and was donated by TopoGEN (Columbus, OH). The GFP antibody was obtained from CLONTECH. CPT was also from TopoGEN. 5,6-Dichloro-1-β-d-ribofuranosylbenzimidazole (DRB), actinomycin D, and α-amanitin were purchased from Sigma.
Cell Culture.
The MCF-7 and SK-BR-3 cell lines in this study were derived from human mammary adenocarcinomas. Both cell lines are cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS (CellGro, Herndon, VA). The p53 heterozygous (MP3ab) and homozygous null mutant (MP3a) mouse cell lines were gifts from Gustavo Leone (Ohio State Univ.). Mouse cell lines were cultured in DMEM supplemented with 15% FBS.
Transfection.
Freshly plated cells (50 to 80% confluence) were transfected with 2 μg of DNAs per 35-mm dish, using SuperFect transfection reagent (Qiagen, Valencia, CA), according to the manufacturer's instructions.
Fluorescence Microscopy.
Cells were cultured to 50–80% confluence on glass cover slips. Before examination, cover slips were placed directly onto a glass slide with a drop of 50% glycerol in PBS (9.1 mM Na2HPO4/1.7 mM NaH2PO4,/50 mM NaCl, pH 7.4). Fluorescent images were captured using a Nikon Eclipse E800 microscope attached to a MicroMax camera (Princeton Instruments, Trenton, NJ).
Nuclear Protein Preparation.
To prepare samples for Western blot, cells were washed twice with cold PBS and resuspended in 1 ml of buffer A (100 mM NaCl/50 mM KCl/20 mM Tris⋅HCl, pH 7.5/0.1 mM EDTA/0.1 mM PMSF/10% glycerol/0.2% Nonidet P-40/0.1% Triton X-100). Following incubation on ice (10 min), nuclei were centrifuged (2,000 × g, 10 min) and lysed in 100 μl of 1× electrophoresis sample buffer and boiled for 2–3 min.
Immunoprecipitation.
Cells were washed twice with PBS, resuspended in 3 ml of ice cold RIPA (1× PBS/1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS/10 μg/ml PMSF), disrupted by repeated aspiration through a 21-gauge needle, and centrifuged (10,000 × g, 10 min, 4°C). The supernatant was precleared by adding 1.0 μg of rabbit IgG, together with 20 μl of protein A-agarose at 4°C for 30 min. After centrifugation, supernatants (cell lysates) were incubated with 10 μg of agarose-conjugated TATA box-binding protein (TBP) antibody (Santa Cruz Biotechnology) at 4°C overnight with mixing. Immunoprecipitates were collected by centrifugation (2,500 × g, 5 min, 4°C) and washed with 1.0 ml PBS four times. After the final wash, pellets were resuspended in 40 μl of electrophoresis sample buffer, boiled for 2–3 min, and analyzed by SDS/PAGE and Western blotting.
Western Blotting Analysis.
The nuclear protein or immunoprecipitated samples were separated by SDS/10% PAGE, followed by electroblot transfer to nitrocellulose. Topo I-specific antibodies (Scleroderma antibody from TopoGEN, www.topogen.com) were used to detect topo I with the BM Chemiluminescence Western Blotting Kit (Mouse/Rabbit) (Boehringer Mannheim). To detect the expression of fusion proteins, immunoblotting was carried out using anti-GFP antibody (CLONTECH). Goat anti-mouse antibody conjugated with horseradish peroxidase was used to illuminate primary immune complexes. Signals were visualized with the ECL Western Blot Detection System (Amersham Pharmacia).
Results
Nuclear and Nucleolar Localization of topo I N terminus (topoIN)-EGFP Fusion Proteins.
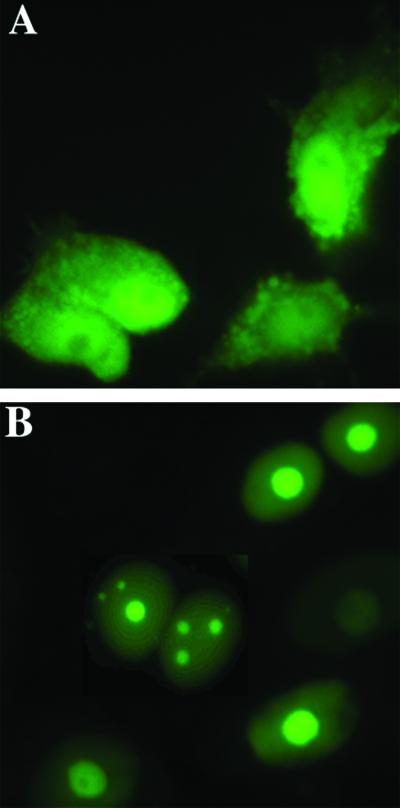
To examine the physiological and biochemical functions of the nonconserved N-terminal domain of endogenous topo I, we evaluated the nuclear distribution of topoIN-EGFP fusion proteins in living cells. A specific, intense green fluorescence signal was clearly restricted to the nuclei of cells transfected with topoIN-EGFP (Fig. 2B). In contrast, EGFP (Fig. 2A, vector control) was distributed more or less evenly throughout the whole cell. In addition, about 90% of transfected cells displayed nucleolar localization (Fig. 2B), which is consistent with the results of topo I localization (2, 23, 24). These results are also consistent with the recent report from Mo et al. (20, 25) that the N-terminal 215 residues contain signals mediating the efficient import of topo I into nuclei and nucleoli.
Figure 2.
Nuclear and Nucleolar localization of the topoIN-EGFP fusion proteins. MCF-7 cells were transfected with pTPIN-EGFP (B) or control vector pEGFPN2 (A). Living cells were imaged 24 h posttransfection as described in Materials and Methods. The percentage of cells displaying nucleolar fluorescence was determined to be 90% (146 cells of a total of 161 displayed nucleolar localization when transfected with pTP1N-EGFP; B). Nucleolar fluorescence was not observed in controls (A).
The Nucleolin Binding Signal Is Not Necessary for Nucleolar Localization.
It has been reported that topo I can bind nucleolin (21), suggesting a possible nucleolar localization mechanism. To determine whether interaction between topo I and nucleolin is necessary and/or sufficient for localization, the N-terminal domain was dissected using various constructs fused to EGFP. Western blotting data confirmed that fusion protein molecular weights were consistent with predicted values (data not shown).
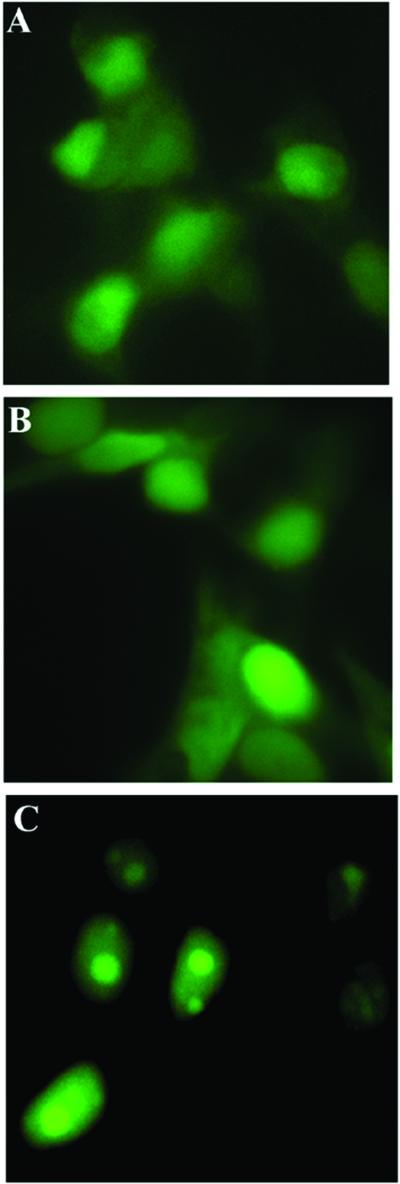
The initial two constructs involved topo I N-terminal domain fragments cut in half. The amino terminus pTPINΔ1 (residues 1–142) has three putative NLS and the carboxyl terminus pTPINΔ2 (residues 142–215) has two NLSs along with the nucleolin-binding region (residues 166–210; see also Fig. 1). Roughly 20% of cells transfected with either of these two constructs displayed nuclear localization and neither displayed nucleolar localization (Fig. 3 A and B). The results show that the nucleolin-binding region alone is not sufficient for the nucleolar localization.
Figure 3.
Subcellular localization of the shorter topoIN-EGFP fusion products. MCF-7 cells were transfected with pTPINΔ1 (residues 1–142) (A), pTPINΔ2 (residues 143–215) (B), and pTPINΔ3 (residues 1–170) (C), and living cells were imaged 24 h posttransfection as described in Materials and Methods.
To further characterize the nucleolin-binding region, we constructed a fusion protein topoINΔ3 (residues 1–170), which has a novel nuclear localization signal in human topo I (20), but lacks residues the nucleolin binding region (21). Surprisingly, 90% of cells expressing this fusion product displayed nucleolar green fluorescence (Fig. 3C). Although the fluorescent intensity in this experiment was somewhat less than that seen with the full-length N-terminal domain construct, the data nonetheless demonstrate that the nucleolin binding region is not absolutely necessary for the nucleolar localization of the bulk of topo I.
Ongoing rDNA Transcription Is Crucial for Nucleolar Localization.
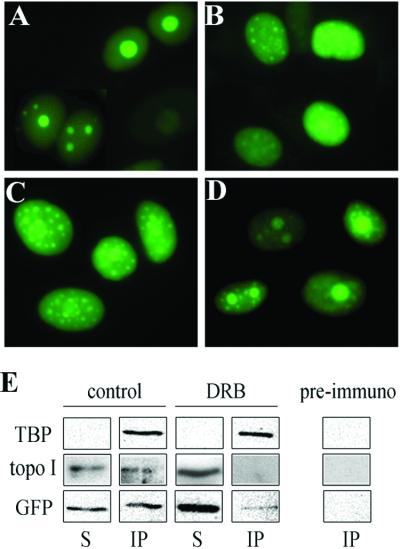
Topo I has been reported to have a predominantly nucleolar distribution and to be involved in rRNA synthesis (2, 23, 24). Furthermore, it is associated with TBP in the general transcription complex (3). To determine whether ongoing transcription is important for the nucleolar localization of topoIN-EGFP proteins, we examined whether RNA synthesis inhibitors might induce its redistribution. DRB inhibits RNA transcription driven by all three RNA polymerases (26), whereas actinomycin D specifically inhibits RNA polymerase I at low concentrations (27, 28), and α-amanitin inhibits RNA polymerase II and III specifically (28, 29). MCF-7/topoIN-EGFP cells were treated with the drug DRB at concentrations that inhibit total uridine incorporation into RNA by 50% (26). After treatment for 2 h with 30 μM DRB, the topoIN-EGFP green fluorescence signal appeared to be dispersed evenly throughout the nuclei and was clearly more punctiform as a pattern of small spheres (Fig. 4B), relative to the DMSO control cells (Fig. 4A). Inhibition of transcription by DRB did not destroy or morphologically alter the nucleoli, at least as seen in phase microscopy (data not shown) and in agreement with other reports (26).
Figure 4.
Ongoing rDNA transcription and topoIN-EGFP nucleolar localization. MCF-7 cells transfected with topoIN-EGFP were grown at 37°C under the following conditions: (A) Control (DMSO) for 5 h; (B) 30 μM DRB for 2 h; (C) 0.04 μg/ml actinomycin D for 3 h; (D) 300 μg/ml α-amanitin for 5 h. Live cells were imaged exactly as described in Materials and Methods. Results are representative of three separate experiments. (E) Analysis of TBP, topo I, and topoIN-EGFP polypeptides in anti-TBP immunoprecipitates from MCF-7/topoIN-EGFP extracts. Following immunoprecipitation, Western blots were analyzed of the supernatants (S) or precipitates (IP), using the three probes shown on the left of the figure.
Because human rRNA genes are transcribed by RNA polymerase I, actinomycin D was used to block rRNA transcription in the MCF-7/topoIN-EGFP cells (27, 28). Fig. 4C shows that actinomycin D treatment gives an identical result obtained with DRB treatment. To determine whether the nucleolar delocalization was specific for RNA polymerase I, we also tested α-amanitin to inhibit RNA polymerase II and III specifically (28, 29). Although α-amanitin treatment altered the distribution and localization of topo I (Fig. 4D), the effects were clearly less dramatic compared with actinomycin D (compare Fig. 4 C and D) and α-amanitin did not result in the loss of nucleolar topo I. Because topo I has also been implicated in transcription by RNA polymerase II, some influence on topo I distribution by α-amanitin was expected. These data suggest that the nucleolar localization of topoIN-EGFP is selectively disrupted by inhibiting RNA polymerase I transcription.
Because the nucleolar topoIN-EGFP fusion proteins delocalize after inhibition of rDNA transcription, we used an immunoprecipitation assay to examine whether the N-terminal fusion protein is a part of a macromolecular transcriptional complex. We first immunoprecipitated the TBP from MCF-7/topoIN-EGFP extracts. As expected, a portion of endogenous topo I protein (40–50%, three experiments) was coimmunoprecipitated with anti-TBP antibody (Fig. 4E). This result is consistent with the FAR-Western experiments reported by Merino et al. (3). Similar amounts of TopoIN-EGFP were coimmunoprecipitated with anti-TBP antibody (Western probed with anti-GFP IgG), suggesting that the N terminus of topo I was interacting with the transcriptional complex in vivo. This result suggests that the EGFP-topo I fusion protein is behaving similarly to the intact topo I protein, at least with regard to interaction with TBP. As a control, EGFP without topo I sequence was not coimmunoprecipitated with anti-TBP antibody by using MCF-7/EGFP cell extracts (data not shown). Moreover, when immunoprecipitations were carried in the MCF-7/topoIN-EGFP extracts from cells treated with 30 μM DRB for 2 h, we found only trace amounts (typically less than 3–8%, three different experiments) of topoIN-EGFP protein bound to transcriptional complex. No endogenous topo I signal was detected in these same immunoprecipitates (Fig. 4E). In summary, the immunoprecipitation experiments indicate that the RNA synthesis inhibitors can abrogate the physical interaction between topo I and the transcriptional complex (TBP), suggesting that ongoing transcription is required for topo I nucleolar localization.
Nucleolar Redistribution and p53.
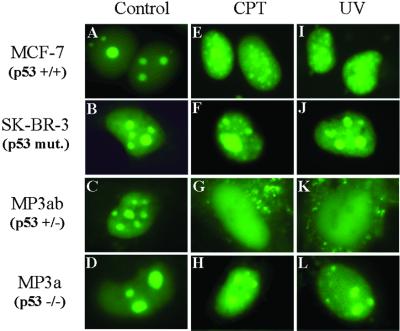
Treating cells with either topo I poisons or UV irradiation causes a redistribution of topo I whereby the enzyme is demobilized away from nucleolar centers (30, 31). Because the recruitment of topo I onto the genome after UV treatment is p53-dependent (17), p53 may also play a role in the redistribution phenomenon with topo I. We explored this possibility by testing the influence of CPT or UV on the subnuclear localization of the fusion proteins in p53 mutant or wild-type cell lines (Fig. 5). In p53 wild-type MCF-7 cells, following a 30-min treatment of CPT (10 μM) or 4 h after a UV pulse (20 J/m2), topoIN-EGFP nucleolar egress was detected as a loss of nucleolar signal and a clear increase in nucleoplasmic fluorescence in 90% of transfected cells (compare Fig. 5 A with E and I). In contrast, with the mutant p53 cell line (SK-BR-3), the majority (>90%) of the cells looked the same before and after drug or UV exposure. These cells do not appear to release topoIN-EGFP from their nucleoli after CPT or UV treatment (representative cells shown in before treatment in Fig. 5B, and after in Fig. 5 F and J). Note that there were no detectable changes in topoIN-EGFP polypeptide levels following CPT or UV treatment (data not shown), which is consistent with our prior studies on intact topo I (17). We repeated these experiments by using isogenic mouse cell lines from animals that were heterozygous for p53 (one allele deleted, but still wild type for p53) compared with homozygous null mutants. In the mouse fibroblasts used in these experiments, the topoIN-EGFP was concentrated in the nucleolar regions (as with human cells); however, nucleoplasmic-associated fluorescence is somewhat higher (Fig. 5C) in control mouse fibroblasts compared with human cells (Fig. 5A). The difference between man and rodent represents a species-specific effect probably related to the nonconserved nature of the N-terminal region of topo I, as noted above. A single wild-type p53 allele resulted in a clear and unambiguous release of topo IN-EGFP from the nucleolus following CPT or UV treatment in the majority or cells examined (Fig. 5 G and K, respectively). The effect was particularly dramatic in the p53 wild-type mouse lines where we detected large amounts of topo I in the cytoplasm (Figs. 5 G and K). As with the SK-BR-3 mutant line, significant amounts of nucleolar fluorescence remained after CPT/UV in the mouse p53 knockout cells, although some leakage of the topoIN-EGFP into the nucleoplasm was evident (compare Fig. 5 D with H and L). However, we did not detect any release of topo I into the cytoplasm in the p53 null line relative to the MP3ab (p53+/−) cells (compare Fig. 5 H and L with G and K). These results suggest that functional, wild-type p53 is required to effect the complete and full release of topo I from the nucleolus following DNA damage.
Figure 5.
Effect of camptothecin and UV irradiation on the subnuclear localization of the topoIN-EGFP fusion products. MCF-7 cells (p53 wild type), SK-BR-3 (p53 mutant) cells, and transgenic mouse cell lines MP3ab (p53+/−), MP3a (p53−/−) were transfected with pTPIN-EGFP (topo IN-EGFP) as described in Materials and Methods. At 24 h posttransfection, cells were treated with CPT (10 μM, 30 min) or UV (20 J/m2 followed by a 4-h growth/recovery period). Live cells were then imaged directly as described in Materials and Methods: (A–D) no treatment controls; (E–H) cells treated with CPT. (I–L) cells treated with UV. In the MCF-7 cells, 161 of 184 total displayed redistribution of topoIN-EGFP following CPT treatment compared with 12 of 247 total in SK-BR-3 cells. Similarly, 250 MCF-7 cells of 266 total displayed nucleolar delocalization compared with only 19 of 131 SK-BR-3 cells (combined results with both CPT and UV experiments). Consistent results were obtained with the wild-type and mutant mouse cell lines (see text).
Physical Association of the topo1 N-Terminal Domain and p53.
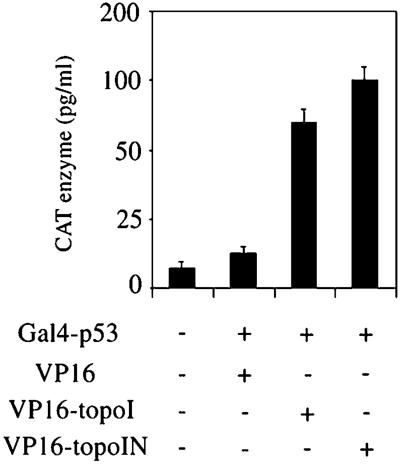
Both in vitro and in vivo association assays revealed that topo I and p53 form a molecular complex (17, 32–34). The experiments in the current work further demonstrate that redistribution of topoIN-EGFP after CPT and UV treatment is influenced by p53. We next investigated whether topo I N-terminal domain itself can bind directly to p53 in vivo. The VP16 activation domain (from HSV-1) was fused to either the topo I N-terminal domain or whole-length topo I cDNA and cloned into a mammalian expression plasmid (Fig. 6A). The interaction of these two fusion proteins with p53 was assayed by cotransfecting cells with a plasmid containing p53 sequences fused to the DNA binding domain of the yeast activator protein GAL4. The activity of the fusion protein was then measured using a chloramphenicol acetyltransferase (CAT) reporter controlled by a promoter containing five GAL4 DNA-binding sites.
Figure 6.
Interaction of topo I and topo I N-terminal domain with p53 in vivo. The three activator derivatives of VP16 were: VP16 alone, VP16 activation domain fused to intact topo I, or fused to the topo I N-terminal domain (residues 1–170). The GAL4 construct was the p53 gene fused to the GAL4 DNA-binding domain. The reporter used in the transfections was a GAL4 fused 5′ of E1bCAT. The reading frames of all fusions were confirmed by DNA sequencing. MCF-7 cells were transfected with 1 μg E1bCAT reporter, 5 μg of the GAL4 construct, and 5 μg of the different activator derivatives. Extracts were prepared at 48 h posttransfection. CAT protein was directly measured using the CAT ELISA Kit (Boehringer Mannheim) and corrected for internal variability by using β-galactosidase. Results are representative of four separate experiments.
These expression constructs were cotransfected into MCF-7 cells. As expected, the p53 construct had no stimulatory effect on the activity of the VP16-activating domain alone (Fig. 6B, lane 2); however, cotransfection of VP16-topo I and GAL4-p53 results in a 5-fold increase in reporter gene expression (Fig. 6B, lane 3). This result is consistent with the immunoprecipitation data we have published (see ref. 15, Figure 7C), demonstrating the in vivo interaction between topo I and p53 (17, 32–34). Similar results were observed when VP16-topoIN was cotransfected with GAL4-p53 (Fig. 6B, lane 4). These data suggest that topo I N terminus alone is sufficient to interact with p53 in vivo.
Discussion
The N-terminal region of topo I from amino acids 1–215 is highly charged, largely disordered, and hypersensitive to protease digestion (18). Although this domain is dispensable for catalytic activity of the enzyme in vitro (19), our data as well as other key papers suggest a number of critical intracellular functions. Studies in Drosophila show that the N terminus targets to transcriptionally active loci and is important in responding to cellular processes during the reprogramming of gene expression that attends development (21, 22). In yeast, overexpression of human topo I is lethal but also depends on the N-terminal domain (19). The primary function of the N-terminal region of topo I is probably nuclear/nucleolar targeting. Moreover, because the 166–210 amino acid region of human topo I can form a complex with nucleolin, a possible mechanism for nucleolar localization of topo I had been proposed (21).
Green fluorescent protein (GFP) is a powerful genetic reporter and localization system. Because the detection of intracellular GFP requires only irradiation by near UV (without fixation, addition of substrates/cofactors), it provides an excellent means for monitoring protein localization in living cells as well as the dynamics in response to different cellular processes or drug treatment (35, 36). Using this system to track the localization of topoIN-EGFP fusion, we show that the N-terminal 215 residues are able to direct the fusion protein into the nucleus and nucleolus. As noted above, the interaction between topo I and nucleolin suggests that this association may be the mechanism by which topo I enters the nucleus and concentrates in the nucleoli. The EGFP fusion approach allows us test this hypothesis, and using fusion constructs to dissect of topo I N terminus, we show that the 166–210 nucleolin binding region of topo I is neither sufficient nor necessary for the nuclear and nucleolar localization of topo I.
The nucleolus is the apparatus for ribosomal RNA synthesis and transcription proceeds at a high rate in this region. Prior reports indicated that topo I is enriched in the nucleolus and catalytically active on ribosomal DNA, suggesting that topo I may relieve positive and negative supercoiling that attends high transcriptional rates associated with rDNA templating (2, 23, 24). Nucleolar localization data on topoIN-EGFP before and after treatment with transcriptional inhibitors suggest that retention of topo I within the nucleolar milieu depends on ongoing rRNA synthesis. DRB inhibits RNA transcription driven by all three RNA polymerases (26), whereas actinomycin D specifically inhibits RNA polymerase I at low concentrations (27, 28). Note also that these lower concentrations of actinomycin D greatly reduce topo I/DNA cleavage complexes (37, 38), arguing against the idea that the actinomycin D result simply mimics the CPT response. Consistently, both DRB and actinomycin D treatments result in a rapid redistribution of chimeric proteins; however, selective inhibition of RNA polymerases II and III gave a much less dramatic redistribution of topo I with little nucleolar loss. A possible mechanism for the recruitment of topo I to the transcriptional complex is that topo I might recognize specific DNA structures; for example, positive or negative supercoiling associated with active chromatin (39). However, based on our results, this explanation alone seems unlikely, because the topoIN-EGFP construct lacks the conserved core domains that mediate the preferential binding of topo I to supercoiled DNA (39).
Immunoprecipitation experiments reveal that the topo I N-terminal domain interacts with the TATA-binding protein (which among other things is involved in rDNA gene transcription driven by RNA polymerase I). Moreover, RNA synthesis inhibitors can abrogate the physical interaction between topo I and TBP. Thus, nucleolar localization could be driven partially or wholly by protein–protein interactions between topo I and the transcriptional machinery. The hydrophilic N-terminal region may well provide the surface for the interactions. Indeed, topo I has been reported to be a transcription factor that represses basal transcription, and DNA relaxation activity of the enzyme was dispensable for this transcriptional repression (3). These results suggest topo I might have other functions during transcription besides its relaxation activity.
It has been previously shown that treatment of cells with a topo I poison (camptothecin), or UV irradiation causes redistribution of nucleolar topo I into nucleoplasm (30, 31); however, little is known about this mechanism. By using topoIN-EGFP imaging, we show that this redistribution is p53-dependent. It has been suggested that the redistribution of topo I in response to CPT treatment is associated with action of drug, which stabilizes topo I on genomic DNA elsewhere other than nucleoli (30); however, this seems unlikely because CPT treatment also leads to topoIN-EGFP nucleolar delocalization and none of the EGFP constructs in this study possess catalytic activity. Instead, it appears that the altered topo I patterns observed in p53 wild-type cells reflects a true alteration in topo I localization that accompanies the cellular response to general DNA damage (note that stabilization of topo I onto genomic DNA by CPT will generate single-stranded DNA breaks or genomic DNA damage). Several p53-dependent biological processes would be activated in p53 wild-type cells: (i) enhanced transcriptional activity (e.g., of DNA damage response genes other than rDNA), (ii) DNA repair, and/or (iii) initiation of apoptosis. One can envision that involvement of topo I in any/all of these processes could drive the redistribution of topo I. An essentially identical pattern of redistribution of topoIN-EGFP after UV damage further rules out a purely drug-like mechanism of nucleolar delocalization. Thus, the topoIN-EGFP constructs should provide useful tools to investigate possible mechanisms of cellular response to topo I drug treatment, providing new insights into mechanisms of cytotoxicity with these drugs. For example, following the kinetics of topoIN-EGFP redistribution after drug treatment might have utility in distinguishing between patients with drug-sensitive and drug-resistant tumors when applied to clinical samples.
Because p53 can directly bind to topo I (17, 32, 33), the p53-dependent subcellular redistribution of topo I might be a direct result of interaction between these two proteins. The mammalian two-hybrid experiment (Fig. 6) also demonstrated that topo I N-terminal domain is sufficient to interact with p53 in vivo. Both topo I drug treatment and UV irradiation can relocalize and stabilize wild-type p53 in the nuclei (40–42). The physical association between p53 and topoIN-EGFP proteins could simply disrupt the dynamic equilibrium between nucleolar and nonnucleolar populations of topoIN-EGFP proteins, and the shift in equilibrium may be reflected by the redistribution of the proteins from the nucleoli. Functional p53 is required for activation of a G1 checkpoint and the resulting growth arrest is thought to allow cells time to repair DNA before replication (43–45) or in some cells eradicate DNA damage laden cells that may be precancerous (46). The dynamic localization of topoIN-EGFP in response to transcription inhibitors, camptothecin, and UV irradiation may reflect the cellular functions of topo I involving transcription, DNA repair, and apoptosis. Previously we proposed two models to explain how endogenous topo I might be recruited onto the genome in a p53- and cell cycle-dependent manner following DNA damage (17). First, topo I may simply be an active participant in excision repair and is recruited directly into repair sites. A second model is that topo I contributes to the general demise of the cell by introducing genomic damage and subsequent p53-dependent elimination through apoptosis. These two models are not mutually exclusive and it is possible that DNA damaged cells exist in a balance between repair (or resurrection) and apoptosis. Double staining cells with specific antibodies against other proteins, for example, Gadd45 and proliferating cell nuclear antigen (PCNA), which are involved in these processes, may help us better understand topo I functions in response to DNA damage. Gadd45, a p53-responsive factor, might drive local chromatin modifications to facilitate topo I accessibility after DNA damage (47), and PCNA plays an essential role in both transcription and nucleotide excision repair (48, 49).
In summary, we have generated several constructs of human topo I N-terminal domain sequences linked to EGFP and have used them to demonstrate that the topo I fusion protein can be localized to nucleoli. Moreover, localization of topoIN-EGFP is clearly dynamic and can rapidly change in response to transcription, DNA repair, topoisomerase inhibition, and p53 status. Therefore, the ability of topo I to respond to cellular processes may reside primarily in the N-terminal domain, which could be important in cell killing by topo active agents. The dynamic shuttling of topo I in and around the genome may be relevant to tumor-cell killing by camptothecins in several ways. For example, if the majority of cellular topo I is located in the nucleolus and active on ribosomal DNA (2, 23, 24), in p53 mutant cells, little of this sequestered topo I would be released following CPT treatment (Fig. 5). Consequently, the DNA damage would be localized to repeat gene sequences in the rRNA cistrons that might be readily repaired by recombination pathways. In a p53 wild-type cells, nucleolar topo I efflux might pepper the genome with widespread damage leading to lesions in many expressed genes. Downstream consequences of global topo I/DNA lesions might be more dramatic leading to greater cell killing.
Acknowledgments
We thank Dr. Gustavo Leone for helpful discussions and providing mouse cell lines. This study was supported by National Institutes of Health Grant RO1-AG16692.
Abbreviations
- TBP
TATA box-binding protein
- EGFP
enhanced green fluorescent protein
- NLS
nuclear localization signal sequence
- CPT
camptothecin
- topo
topoisomerase
- topoIN
topo I N terminus
- DRB
5,6-dichloro-1-β-d-ribofuranosylbenzimidazole
References
- 1.Stewart L, Redinbo M R, Qiu X, Hol W G J, Champoux J J. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 2.Muller M T, Pfund W P, Mehta V B, Trask D K. EMBO J. 1985;4:1237–1243. doi: 10.1002/j.1460-2075.1985.tb03766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Merino A, Madden K R, Lane W S, Champoux J J, Reinberg D. Nature (London) 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- 4.Wu H-Y, Liu L F. J Mol Biol. 1991;219:615–622. doi: 10.1016/0022-2836(91)90658-s. [DOI] [PubMed] [Google Scholar]
- 5.Kertzschmarr M, Meisterernst M, Roeder R G. Proc Natl Acad Sci USA. 1993;90:11508–11512. doi: 10.1073/pnas.90.24.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holm C, Covey J M, Kerrigan D, Pommier Y. Cancer Res. 1989;49:6365–6368. [PubMed] [Google Scholar]
- 7.D'Arpa P, Beardmore C, Liu L F. Cancer Res. 1990;50:6919–6924. [PubMed] [Google Scholar]
- 8.Liu L F. Annu Rev Biochem. 1989;58:351–375. doi: 10.1146/annurev.bi.58.070189.002031. [DOI] [PubMed] [Google Scholar]
- 9.Pommier Y, Pourquier P, Fan Y, Strumberg D. Biochim Biophys Acta. 1998;1400:83–106. doi: 10.1016/s0167-4781(98)00129-8. [DOI] [PubMed] [Google Scholar]
- 10.Wang J C, Caron P R, Kim R A. Cell. 1990;62:403–406. doi: 10.1016/0092-8674(90)90002-v. [DOI] [PubMed] [Google Scholar]
- 11.Wang J C. J Biol Chem. 1991;266:6659–6662. [PubMed] [Google Scholar]
- 12.Zhu J, Schiestl R H. Mol Cell Biol. 1996;16:1805–1812. doi: 10.1128/mcb.16.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J C. Annu Rev Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 14.Yanagida M, Sternglanz R. In: DNA Topology and Its Biological Effects. Cozzarelli N R, Wang J C, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. pp. 299–320. [Google Scholar]
- 15.Lee M P, Brown S D, Chen A, Hsieh T-S. Proc Natl Acad Sci USA. 1993;90:6656–6660. doi: 10.1073/pnas.90.14.6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morham S G, Kluckman K D, Voulomanos N, Smithies O. Mol Cell Biol. 1996;16:6804–6809. doi: 10.1128/mcb.16.12.6804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao Y, Okada S, Chang L-S, Muller M T. Cancer Res. 2000;60:4538–4543. [PubMed] [Google Scholar]
- 18.Stewart L, Ireton G C, Champux J J. J Biol Chem. 1996;271:7602–7608. doi: 10.1074/jbc.271.13.7602. [DOI] [PubMed] [Google Scholar]
- 19.Alsner J, Svejstrup J Q, Kjeldsen E, Sorensen B S, Westergaard O. J Biol Chem. 1992;267:12408–12411. [PubMed] [Google Scholar]
- 20.Mo Y, Wang C, Beck W T. J Biol Chem. 2000;275:41107–41113. doi: 10.1074/jbc.M003135200. [DOI] [PubMed] [Google Scholar]
- 21.Bharti A K, Olson M O J, Kufe D M, Rubin E H. J Biol Chem. 1996;271:1993–1997. doi: 10.1074/jbc.271.4.1993. [DOI] [PubMed] [Google Scholar]
- 22.Shaiu W-L, Hsieh T-S. Mol Cell Biol. 1998;18:4358–4367. doi: 10.1128/mcb.18.7.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fleischmann G, Pflugfelder G, Steiner E K, Javaherian K, Howard G C, Wang J C, Elgin S C. Proc Natl Acad Sci USA. 1984;81:6958–6962. doi: 10.1073/pnas.81.22.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Wang J, Liu L. Proc Natl Acad Sci USA. 1988;85:1060–1064. doi: 10.1073/pnas.85.4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mo Y-Y, Wang P, Beck W T. Exp Cell Res. 2000;256:480–490. doi: 10.1006/excr.2000.4864. [DOI] [PubMed] [Google Scholar]
- 26.Buckwalter C A, Lin A H, Tanizawa A, Pommier Y G, Cheng Y-C, Kaufmann S H. Cancer Res. 1996;56:1674–1681. [PubMed] [Google Scholar]
- 27.Perry R P. Exp Cell Res. 1963;29:400–406. [Google Scholar]
- 28.Huang S, Deerinck T J, Ellisman M H, Spector D L. J Cell Biol. 1998;143:35–47. doi: 10.1083/jcb.143.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weinmann R, Raskas H J, Roeder R G. Cold Spring Harbor Symp Quant Biol. 1975;34:495–500. doi: 10.1101/sqb.1974.039.01.061. [DOI] [PubMed] [Google Scholar]
- 30.Danks M K, Garrett K E, Marion R C, Whipple D O. Cancer Res. 1996;56:1664–1673. [PubMed] [Google Scholar]
- 31.Thielmann H W, Popanda O, Staab H J. J Cancer Res Clin Oncol. 1999;125:193–208. doi: 10.1007/s004320050263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gobert C, Bracco L, Rossi F, Olivier M, Tazi J, Lavelle F, Larsen A K, Riou J F. Biochemistry. 1996;35:5778–5786. doi: 10.1021/bi952327w. [DOI] [PubMed] [Google Scholar]
- 33.Albor A, Kaku S, Kulesz-Martin M. Cancer Res. 1998;58:2091–2094. [PubMed] [Google Scholar]
- 34.Gobert C, Skladanowski A, Larsen A K. Proc Natl Acad Sci USA. 1999;96:10355–10369. doi: 10.1073/pnas.96.18.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chalfie M, Tu Y, Euskirchen G, Ward W, Prasher D. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 36.Chalfie M. Photochem Photobiol. 1995;62:651–656. doi: 10.1111/j.1751-1097.1995.tb08712.x. [DOI] [PubMed] [Google Scholar]
- 37.Trask D K, Muller M T. Proc Natl Acad Sci USA. 1988;85:1417–1421. doi: 10.1073/pnas.85.5.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wassermann K, Markovits J, Jaxel C, Capranico G, Kohn K W, Pommier Y. Mol Pharmacol. 1990;38:38–45. [PubMed] [Google Scholar]
- 39.Liu L F, Wang J C. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maltzman W, Czyzyk L. Mol Cell Biol. 1984;4:1689–1694. doi: 10.1128/mcb.4.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kastan M B, Okywere O, Sidransky D, Vogelstein B, Craig R W. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- 42.Lu X, Lane D P. Cell. 1993;75:765–778. doi: 10.1016/0092-8674(93)90496-d. [DOI] [PubMed] [Google Scholar]
- 43.Lane D. Int J Cancer. 1994;57:623–627. doi: 10.1002/ijc.2910570502. [DOI] [PubMed] [Google Scholar]
- 44.Levine A J, Perry M E, Chang A, Silver A, Dittmer D, Wu M, Welsh D. Br J Cancer. 1994;69:409–416. doi: 10.1038/bjc.1994.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linke S P, Klarkin K C, Wahl G M. Cancer Res. 1997;57:1171–1179. [PubMed] [Google Scholar]
- 46.Hill L L, Ouhtit A, Loughlin S M, Kripke M L, Ananthaswamy H N, Owen-Schaub L B. Science. 1999;285:898–900. doi: 10.1126/science.285.5429.898. [DOI] [PubMed] [Google Scholar]
- 47.Carrier F, Georgel P T, Pourquier P, Blake M, Kontny H U, Antinore M J, Gariboldi M, Myers T G, Weinstein J N, Pommier Y, Fornace A J., Jr Mol Cell Biol. 1999;19:1673–1685. doi: 10.1128/mcb.19.3.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aboussekhra A, Wood R D. Exp Cell Res. 1995;221:326–332. doi: 10.1006/excr.1995.1382. [DOI] [PubMed] [Google Scholar]
- 49.Smith M, Ford J M, Hollander M C, Bortnick R A, Amundson S A, Seo Y R, Deng C-X, Hanawalt P C, Fornace A J J. Mol Cell Biol. 2000;20:3705–3714. doi: 10.1128/mcb.20.10.3705-3714.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]