Abstract
Nonribosomal peptide synthetases responsible for the production of macrocyclic compounds often use their C-terminal thioesterase (TE) domain for enzymatic cyclization of a linear precursor. The excised TE domain from the nonribosomal peptide synthetase responsible for the production of the cyclic decapeptide tyrocidine A, TycC TE, retains autonomous ability to catalyze head-to-tail macrocyclization of a linear peptide thioester with the native sequence of tyrocidine A and can additionally cyclize peptide analogs that incorporate limited alterations in the peptide sequence. Here we show that TycC TE can catalyze macrocyclization of peptide substrates that are dramatically different from the native tyrocidine linear precursor. Several peptide thioesters that retain a limited number of elements of the native peptide sequence are shown to be substrates for TycC TE. These peptides were designed to integrate an Arg-Gly-Asp sequence that confers potential activity in the inhibition of ligand binding by integrin receptors. Although enzymatic hydrolysis of the peptide thioester substrates is preferred over cyclization, TycC TE can be used on a preparative scale to generate both linear and cyclic peptide products for functional characterization. The products are shown to be inhibitors of ligand binding by integrin receptors, with cyclization and Nα-methylation being important contributors to the nanomolar potency of the best inhibitors of fibrinogen binding to αIIbβ3 integrin. This study provides evidence for TycC TE as a versatile macrocyclization catalyst and raises the prospect of using TE catalysis for the generation of diverse macrocyclic peptide libraries that can be probed for novel biological function.
Macrocyclic structure is a desirable feature for biologically or pharmaceutically active compounds. Macrocyclization stabilizes products from degradation and decreases the conformational flexibility of a molecule compared with its linear analog by constraining it to a biologically active conformation (1). In the biosynthesis of macrocyclic natural products, such as the macrolactam antibiotic tyrocidine, the macrolactone antifungal fengycin, and the macrolactone siderophore enterobactin by nonribosomal peptide synthetases (NRPSs), a C-terminal thioesterase (TE) domain introduces this constraint by catalyzing cyclization of a linear precursor (2, 3). Analogous cyclases in polyketide synthetase assemblies generate erythromycin and ketolide antibiotics as well as other bioactive macrolactones (4, 5).
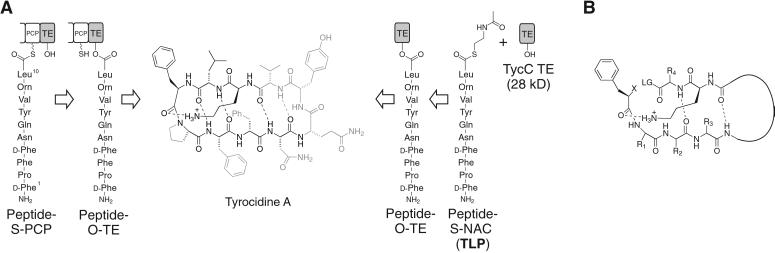
We have recently shown that several 28- to 35-kDa C-terminal TE domains excised from megadalton NRPS subunit proteins retain autonomous macrocyclization activity with peptide thioester substrates that mimic their natural protein-bound peptide phosphopantetheinyl substrates (6, 7). As a representative example, TycC TE, the C-terminal TE domain from the synthetase responsible for the biosynthesis of tyrocidine, macrocyclizes a soluble decapeptide thioester with the native peptide sequence of the antibiotic tyrocidine A (Fig. 1A). We have investigated the substrate specificity of TycC TE by site-specific alterations in the soluble substrate. Substitution of single residues of the peptide sequence by alanine, as well as targeted substitution of ester for amide bonds in the linear precursor, suggested that the TycC TE could serve as a permissive macrocyclization catalyst (Fig. 1A) (6–8). The elements of the substrate that appeared necessary for cyclization included the potential for preorganization of the substrate in an antiparallel β-sheet, as well as side-chain requirements for an N-terminal d-phenylalanine and a penultimate ornithine. The sum of our site-specific alterations have led us to propose a model for the minimal substrate requirements for cyclization and raised the question of the effect on cyclization of simultaneous alterations in potentially substitutable positions (Fig. 1B) (8).
Figure 1.
(A) The natural reaction of the C-terminal TE domain from the NRPS responsible for production of the cyclic decapeptide antibiotic tyrocidine A is shown (Left). The terminal peptidyl carrier protein (PCP) is shown with its phosphopantetheine arm represented by SH. A linear decapeptide is transferred to the active-site serine (OH) of the TE domain, followed by head-to-tail cyclization giving the product tyrocidine A. (Right) The experimental design for studying the isolated TE domain. The protein-bound substrate is replaced by a decapeptide SNAC (TLP), which is cyclized by the isolated TE domain, TycC TE. The portions of the substrate that have been shown to tolerate substitution without leading to a more than 3-fold drop in the rate of cyclization are shown in gray (6, 8). (B) The proposed model for the minimal substrate, based on examination of substrates with site-specific alterations (6–8). The curved line represents a variable length linker. R represents variable side chains, X represents NH2 or OH, and LG represents a leaving group.
We now address the ability of TycC TE to catalyze the cyclization of peptide thioesters with different therapeutic potential, switching from antibiotic endpoints to peptide inhibitors of integrin receptors. The molecules studied, as they differ dramatically from the natural tyrocidine substrate, allow for examination of the effects of aggregate changes in the substrate on TycC TE catalysis of cyclization.
Integrins are α/β heterodimeric receptors that mediate numerous cell–cell and cell–matrix interactions (9, 10). Many natural ligands for integrins contain an Arg-Gly-Asp (RGD) sequence that is believed to be important for recognition by the integrins, including fibrinogen (ligand for αIIbβ3 and αvβ3), vitronectin (ligand for αvβ3), and fibronectin (ligand for α5β1) (11). Peptides containing the RGD motif are able to inhibit receptor-ligand binding and therefore are potentially useful therapeutic agents. With the introduction of a conformational constraint, peptides cyclized by means of a disulfide linkage have been shown to have a higher affinity and increased specificity when compared with their linear counterparts (12, 13). Specificity also is linked to the structural presentation of the RGD motif, as presentation on a β type II′ turn gives αIIbβ3 specificity, whereas presentation on a β type I turn gives specificity for the integrin αvβ3 (14, 15).
In this work, we explore the ability of TycC TE to catalyze cyclization of rationally designed peptide thioesters and evaluate the integrin inhibition activity of the macrolactam enzymatic products. The utility of the excised TE domain for the synthesis of macrocyclic molecules with novel bioactivity is revealed by our characterization of the cyclization of these integrin binding peptides.
Materials and Methods
Substrate Synthesis.
Peptide N-acetylcysteamine thioester (SNAC) substrates were synthesized by solid-phase peptide synthesis followed by solution-phase thioester formation. Peptide synthesis was carried out on a Perseptive Biosystems 9050 synthesizer (0.3 mmol scale) by using 2-chlorotrityl resin derivatized with Leu (Novabiochem). 9-Fluorenylmethoxycarbonyl (Fmoc)-Nα-Me-Arg(Mtr) was purchased from Bachem. All other amino acids and coupling reagents were obtained from Novabiochem. Couplings were carried out with Fmoc-protected amino acids, except for the N-terminal d-Phe, which was Boc-protected. Side-chain protecting groups were: Boc for Orn and Trp, Mtr for Nα-Me-Arg, Pbf for Arg, and t-Bu for Asp. For all couplings not involving Nα-Me-Arg, 30-min single coupling using diisopropylcarbodiimide (DIPCDI)/1-hydroxybenzotriazole (HOBt) chemistry was conducted. Coupling of Fmoc-Nα-Me-Arg(Mtr) to the growing peptide was carried out with DIPCDI/HOBt for 4 h. Coupling of the amino acid subsequent to Nα-Me-Arg(Mtr) was carried out by double coupling (3 h each) with DIPCDI and 1-hydroxy-7-azabenzotriazole (HOAt). Cleavage of the protected peptide from resin, solution-phase thioester formation, deprotection, and purification were conducted as described (7), with the exception of increasing to 24 h the length of deprotection of the peptide thioesters containing Mtr-protected Nα-Me-Arg. The purity and identity of peptide-SNACs were confirmed by analytical HPLC and matrix-assisted laser desorption ionization–time of flight mass spectroscopy (MALDI-TOF MS) (Table 1).
Table 1.
Characterization of substrates and reaction products by MALDI-TOF MS
| Compound | Species | Mass, calculated | Mass, observed |
|---|---|---|---|
| Substrates | |||
| IT1-SNAC | [M+H]+ | 1,221.6 | 1,221.5 |
| IT2-SNAC | [M+H]+ | 1,207.6 | 1,207.2 |
| IT3-SNAC | [M+H]+ | 836.5 | 836.6 |
| Products | |||
| IT1-COOH | [M+H]+ | 1,120.6 | 1,120.5 |
| cyclo-IT1 | [M+H]+ | 1,102.6 | 1,102.6 |
| IT2-COOH | [M+H]+ | 1,106.6 | 1,106.5 |
| cyclo-IT2 | [M+H]+ | 1,088.6 | 1,088.5 |
| IT3-COOH | [M+H]+ | 735.5 | 735.5 |
| cyclo-IT3 | [M+H]+ | 717.5 | 717.5 |
Kinetic Assays of Peptide Thioester Substrates.
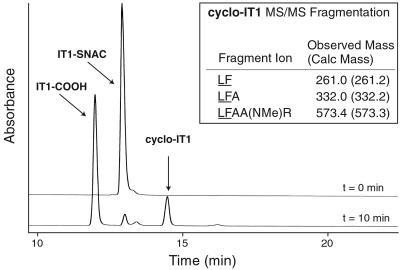
TycC TE was expressed with a C-terminal hexahistidine tag and purified as described (6). Reactions were carried out in total volume of 200 μl with 25 mM Mops, pH 7.0 at 24°C. Reactions were initiated with addition of TycC TE and quenched by addition of 12.5 μl of 1.7% trifluoroacetic acid (TFA)/water followed by flash-freezing in liquid N2. Product analysis was carried out by HPLC (Beckman System Gold) with reverse-phase C18 column (Vydac) using a gradient from 20% to 65% of 0.1% TFA/acetonitrile into 0.1% TFA/water over 35 min. Product formation with peptide thioester substrates was shown to be linear to 2 min (data not shown). The identities of linear acid and cyclic peptide products were confirmed by MALDI-TOF MS (Table 1). The regiochemistry of cyclic peptide products was further confirmed by HPLC MS-MS analysis of the purified product with a Hewlett–Packard 1100 HPLC coupled to a Finnigan-MAT LCQ ion trap mass spectrometer at the Biopolymers Facility of the Howard Hughes Medical Institute/Harvard Medical School. For each cyclic product, multiple fragment ions were found that contained the d-Phe-1 to C-terminal Leu linkage expected from head-to-tail cyclization. For cyclo-IT1, see observed peptides in Fig. 3. For cyclo-IT2, detected peptide fragments included the sequence LF observed 261.0 (calculated 261.2), LFA 332.1 (332.2), and LFAAR 559.3 (559.3). For cyclo-IT3, detected fragments included the a-ion of LF 233.1 (233.2) and the b-ion of LF(NMe)RGD 603.4 (603.3). For kinetic analysis of substrates, peptide thioester concentration was varied, and product formation at 1 min was quantified and fitted to the Michaelis–Menten kinetic model.
Figure 3.
The reaction of TycC TE with IT1-SNAC followed by HPLC. Reactions initially contained 15 μM IT1-SNAC, 200 nM TycC TE, 25 mM Mops, pH 7.0 at 24°C and were quenched at time 0 min or 10 min. The labeled hydrolyzed and cyclic products, IT1-COOH and cyclo-IT1, respectively, were identified by MALDI-TOF MS. (Inset) Shown are peptide fragment ions generated by MS-MS that confirm the presence of the underlined d-Phe-1 to Leu-10 linkage formed by cyclization.
Preparative Scale Generation of Linear Peptide Acid and Cyclic Peptide Products.
The reactions studied kinetically were carried out on the preparative scale to generate products for bioactivity studies. Reactions were conducted with 200 nM TycC TE, 150 μM peptide-SNAC in a total volume of 30–90 ml, with 25 mM Mops, pH 7.0. After 30-min incubation with gentle agitation at 24°C, the substrates were >95% converted to products (data not shown). A second aliquot of enzyme was added to bring TycC TE to 400 nM final concentration. After an additional 30-min incubation, trifluoroacetic acid was added to a final concentration of 0.1%, and the reactions were frozen in liquid N2. Lyophilization of the reaction afforded a white solid that was redissolved in 5 ml water. Preparative HPLC was carried out as described for purification of the peptide-SNAC substrates, and the products corresponding to the linear peptide acid and cyclic peptide were purified. The purity of the products was more than 99% by analytical HPLC, and the product identity was confirmed by MALDI-TOF MS (Table 1).
Preparation of Recombinant Soluble Integrins and Their Ligands.
Soluble recombinant α5β1, αVβ3, and αIIbβ3 integrins were purified from the culture supernatants from CHO-lec 3.2.8.1 cells stably tranfected with corresponding subunits (J.T., unpublished work). The design for all constructs was essentially the same as for the soluble α5β1 reported previously (16), in that all subunits are truncated before the transmembrane domain and fused to a BASE-p1 (for β) and ACID-p1 (for α) peptide (17) with one residue mutated to Cys. To facilitate purification, a hexahistidine tag was attached to the C terminus of the β subunit. The recombinant integrins were purified by Ni-NTA agarose and gel filtration, and the purities were confirmed to be >95% by SDS/PAGE. Human fibrinogen (Enzyme Research Laboratories, South Bend, IN) was labeled with Sulfo-NHS-LC-biotin (Pierce) according to the manufacturer's recommendations. Recombinant fibronectin fragment (Fn9–10) was prepared and biotinylated as described (16).
Ligand Binding Assay.
Linear and cyclic peptide products were assayed for inhibition of fibrinogen binding to αVβ3 and αIIbβ3 or fibronectin fragment binding to α5β1. Purified integrins (250 ng) were coated onto microtiter wells followed by blocking of nonspecific binding sites with 1% BSA. Biotinylated ligands (9 nM for fibrinogen, 50 nM for Fn9–10) were added to wells together with varying concentrations of peptides in 50 mM Tris (pH 7.5), 150 mM NaCl, 1 mM MnCl2, 0.1% BSA and incubated for 1 h at room temperature. After washing three times with buffer, the bound ligands were quantitated by peroxidase-conjugated streptavidine. Assays were performed in duplicate and repeated at least three times. Binding values were fitted to a sigmoidal curve used to derive IC50 values.
Results
Design of Peptide Thioester Substrates.
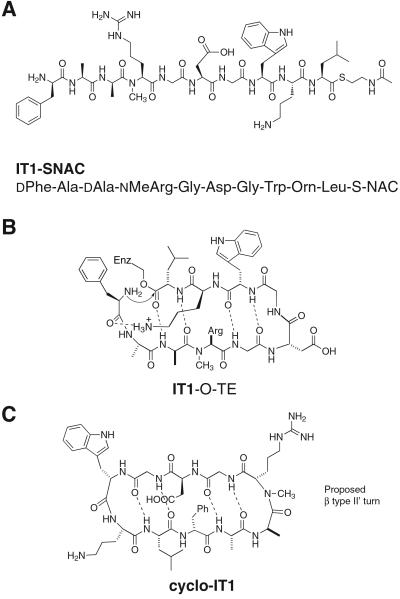
The peptide thioester substrate, IT1-SNAC, was designed for this study to satisfy two sets of requirements (Fig. 2A). To test the effect of a large number of changes in the peptide thioester substrate on enzymatic cyclization, we wanted to retain only the elements present in the minimal peptide model (Fig. 1B). To this end, when compared with the linear peptide sequence of the natural substrate tyrocidine, IT1-SNAC had an alteration in the side chains of seven of the 10 residues. Further, it incorporates two changes in stereochemistry at positions 3 and 4 and the introduction of an Nα-methyl residue in the backbone, making it a stringent test of the minimal peptide model (Fig. 2B).
Figure 2.
(A) Structure of peptide substrate IT1-SNAC. (B) The intermediate in the reaction of IT1-SNAC with the active-site serine of TycC TE modeled in the proposed cyclizing conformation of the natural substrate tyrocidine A. The substrate meets the criteria of the minimal substrate model and introduces numerous amino acid side-chain and stereochemical alterations relative to the natural substrate. (C) The proposed structure in solution of cyclo-IT1, the product of enzymatic cyclization of IT1-SNAC. The β type II′ turn-inducing d-Ala-Nα-Me-Arg sequence could position Arg at the i+2 position of the turn.
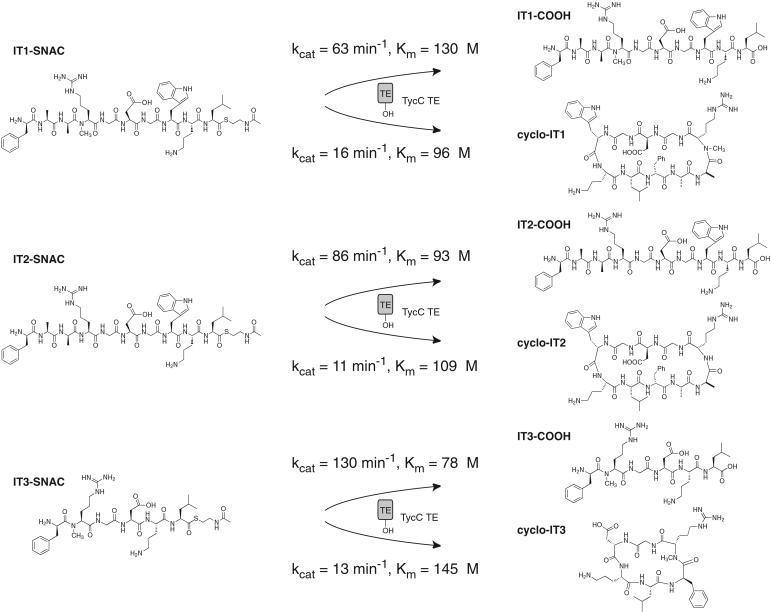
The sequence of the variant positions was set to create a potential cyclic integrin inhibitor. It is known that an RGD sequence situated with the Arg at the i+2 position of a β type II′ turn can give specificity for the αIIbβ3 integrin (14). As d-amino acids followed by Nα-methyl amino acids are known to be strong β type II′ turn-inducing elements (14, 18), we incorporated d-Ala-3-Nα-Me-Arg-4 into the linear sequence of IT1-SNAC (Fig. 2C). Further, it is known that the residues C terminal to the RGD pharmacophore can affect specificity. For the integrin α5β1, although little is known about the preferred spatial presentation of the RGD sequence, a Gly-Trp or Gly-Phe sequence appended to the C terminus of RGD has been shown to increase potency of inhibition (13, 19). To introduce potential potency for α5β1 inhibition, we introduced Gly-Trp after RGD in IT1-SNAC. The des-Nα-Me substrate IT2-SNAC and hexapeptide IT3-SNAC allow for examination of the effect of two features of the designed molecule, Nα-methylation and the length of the peptide, respectively, on both peptide cyclization and bioactivity (see Fig. 4).
Figure 4.
Kinetics of hydrolysis and cyclization of peptide thioester substrates by TycC TE.
Test of the Minimal Substrate Model for TycC TE.
The peptide SNAC substrates were synthesized and tested as substrates for enzymatic transformation by TycC TE. An initial test of IT1-SNAC revealed the formation of two products that were identified by MALDI-TOF MS as the linear free decapeptide acid IT1-COOH and the head-to-tail cyclized decapeptide cyclo-IT1 (Fig. 3). The regiochemistry of the cyclic product was confirmed by MS-MS fragmentation as peptide fragment ions containing the predicted dipeptide linkage from d-Phe-1 to Leu-10 can be identified (Fig. 3). The kinetics of hydrolysis and cyclization were studied and revealed that hydrolysis is preferred over cyclization in a ratio of ≈4:1, as compared with a ratio of 1:6 for the native tyrocidine sequence (Fig. 4) (6). The absolute reduction in kcat of cyclization is only 4-fold from the kcat = 60 min−1 for the native tyrocidine decapeptide thioester (6), which is remarkable given the large number of alterations incorporated in the substrate. The flux to cyclization was sufficient to allow the reaction to be carried out on the preparative scale, generating milligram quantities of the linear and cyclic products for characterization of integrin binding activity.
In the proposed cyclizing conformation of IT1-SNAC, the Nα-methyl Arg is situated with the methyl group directed away from the core of the β-sheet (Fig. 2B). The effect on cyclization of deletion of the Nα-methyl group from IT1-SNAC was addressed by kinetic characterization of IT2-SNAC (Fig. 4). The rate for cyclization is moderately decreased and that for hydrolysis slightly increased, giving a ratio of hydrolysis to cyclization of ≈8:1. The results suggest that TycC TE is largely insensitive to the presence or absence of the Nα-methyl residue.
We have previously shown that TycC TE can be used to generate cyclic peptides from 6–14 residues in length (7). The substrate IT3-SNAC was designed with the Nα-methyl RGD sequence inserted after d-Phe-1 and upstream of Orn-5–Leu-6 as a hexapeptide substrate that met the requirements of the minimal substrate model and could potentially position the RGD sequence on a β type II′ turn. The hexapeptide substrate was cyclized at a rate comparable to the decapeptide, although the hexapeptide thioester was a better substrate for hydrolysis (Fig. 4).
Assay of Enzymatic Products as Integrin Inhibitors.
The purified linear and cyclic enzymatic reaction products were tested for their ability to inhibit integrin-ligand binding. Soluble recombinant integrins αIIbβ3, αVβ3, and α5β1 were purified and used with biotinylated fibrinogen, for αVβ3 and αIIbβ3 integrins, or a biotinylated fibronectin fragment, for α5β1, in a competition binding experiment. All linear and cyclic enzymatic peptide products studied showed measurable inhibition activity against all integrin receptor subtypes. The linear and cyclic peptides were tested in comparison to a standard integrin inhibitor, a linear GRGDSP peptide derived from the native fibronectin sequence (20). The control peptide has a 1.7 μM IC50 in the binding displacement assay against αIIbβ3, 6.7 nM against the αvβ3, and 30 μM against the α5β1 for a 10,000-fold range in potency against these three integrin receptor subtypes. Given interassay variability, using normalized IC50 values (Q = IC50[peptide]/IC50[GRGDSP]) allows for comparison between integrin subtypes as well as intra-assay results (Table 2).
Table 2.
Integrin inhibition activity of RGD-containing peptides
| IC50, nM (Q = ratio versus control peptide)
|
|||
|---|---|---|---|
| αIIbβ3 | αvβ3 | α5β1 | |
| IT1-COOH | 350 (0.21) | 380 (57) | 30,000 (1.0) |
| cyclo-IT1 | 210 (0.12) | 120 (18) | 400,000 (13) |
| IT2-COOH | 26,000 (15) | 230 (34) | 25,000 (0.83) |
| cyclo-IT2 | 2,300 (1.4) | 90 (24) | 600,000 (20) |
| IT3-COOH | 3,700 (2.2) | 40 (6.0) | 300,000 (10) |
| cyclo-IT3 | 460 (0.27) | 6.5 (0.97) | 300,000 (10) |
| GRGDSP (control) | 1,700 (1.0) | 6.7 (1.0) | 30,000 (1.0) |
With the αIIbβ3 integrin, three of the peptides were more active than the control peptide in inhibition of fibrinogen binding. In agreement with the predictions incorporated in our peptide design, the cyclization and Nα-methylation constraints contributed to the potency of the best inhibitors. The most active inhibitor, cyclo-IT1, was both cyclic and contained Nα-methylated Arg. Cyclization also leads to an 8-fold increase in the potency of the hexapeptide, cyclo-IT3, relative to IT3-COOH. The des-Nα-methyl peptides cyclo-IT2 and IT2-COOH both were at least 10-fold less potent than their Nα-methylated analogs.
With the αvβ3 integrin, relative to the control peptide GRGDSP, all linear and cyclic peptides tested were less active with the exception of equipotent cyclo-IT3. In contrast to the αIIbβ3 integrin, Nα-methylation slightly decreases the potency of inhibition of cyclo-IT1 and IT1-COOH relative to their des-Nα-methyl analogs. The finding that the cyclic hexapeptide is a reasonably potent inhibitor at αvβ3 was surprising given the β type II′ turn-stabilizing d-Phe-Nα-Me-Arg sequence, which has been seen to confer αIIbβ3 specificity (14). However, based on Q values, cyclo-IT3 shows a 3.5-fold specificity preference for the αIIbβ3 integrin.
The study of inhibition of the fibronectin fragment binding to the α5β1 integrin by the enzymatically synthesized linear and cyclic peptide products reveals that cyclization need not increase the potency of an inhibitor. The linear decapeptide acids IT1-COOH and IT2-COOH are more than 10-fold more potent than their cyclized analogs and equipotent to the control peptide. Notably, these inhibitors also contain the Gly-Trp sequence C terminal to RGD. Given the recent finding of a synergistic binding site in α5β1 that recognizes the Gly-Trp sequence (19), it is feasible that cyclization constrains the peptide such that an active conformation allowing for interaction at this synergistic site with the linear peptide is no longer accessible with the cyclic peptide. As cyclo-IT1 and cyclo-IT3 both are proposed to present the RGD sequence in the context of a β type II′ turn, the lack of inhibitory potency of these molecules suggests that the conformational preference favored by αIIbβ3 is not shared by α5β1.
Discussion
In the biosynthesis of most macrocyclic compounds by NRPSs and polyketide synthetases, chain termination and macrocyclization is catalyzed by a C-terminal TE domain. The TE domains are α,β hydrolase family members that use an active-site serine as part of a catalytic triad (21). A linear substrate activated as a thioester is attacked by the active-site serine forming an acyl-O-TE intermediate (Fig. 1A) (22). Deacylation occurs by attack of the acyl-O-TE intermediate by an intramolecular nucleophile, with minor flux to hydrolyzed, linear product by attack of water. When excised from their native context, at the terminus of large multidomain synthetase proteins, the isolated TE domains retain their ability to act as enzymatic macrocyclization catalysts when probed with peptide SNACs (6, 7). Our initial biochemical studies of the TE domain from the synthetase responsible for the production of the antibiotic tyrocidine established that TycC TE could cyclize substrates with site-specific alterations in the native peptide sequence (8). Studies with substrates containing single side chains substituted by alanine, alteration in the length of the peptide substrate, and substitution of specific amide linkages for ester bonds led us to propose a minimal set of requirements for a cyclization substrate for TycC TE (Fig. 1B).
TycC TE Is a Permissive Cyclization Catalyst.
The versatility of TycC TE as a peptide thioester cyclase has been addressed in this study by the synthesis and characterization of substrates dramatically different from the native tyrocidine sequence. Seven side chains that had previously been shown to be tolerant to individual substitution by alanine were simultaneously altered in the designed substrate IT1-SNAC. The side-chain alterations in some positions resulted in radically different substituents, such as l-Arg for d-Phe-4 and l-Trp for l-Val-8. Additionally, the substrate contained two alterations in stereochemistry and an Nα-methylated residue.
When IT1-SNAC was assayed with TycC TE, the enzymatic macrocyclization producing the head-to-tail cyclized decapeptide cyclo-IT1, occurred with a kcat only 4-fold reduced from that of the wild-type tyrocidine sequence (kcat = 60 min−1) (6). As a test of the minimal substrate model, IT1-SNAC lends support to the notion that only a small number of elements of the substrate are necessary for cyclization to be a productive outcome. The importance of these minimal substrate elements is revealing with regard to the mechanism of action of TycC TE. After formation of the acyl-O-TE intermediate, the enzyme could recognize and position the important side chains to aid in bringing the reactive termini together. The minimal requirements could also reflect the ability of the tethered peptide intermediate to take on a product-like conformation by the formation of a stable antiparallel β-sheet. The cyclization of the substrates IT2-SNAC and IT3-SNAC by TycC TE are further demonstrations of the permissive cyclization ability of the TE domain. The TE domain is largely insensitive to the presence of an Nα-methylated amino acid in the substrate and can catalyze cyclization of the shortened, hexapeptide thioester, satisfying the requirements of the minimal substrate model. Further, the cyclization reaction was regiospecific, forming exclusively monomeric head-to-tail cyclized molecules as evidenced by MS-MS fragmentation of the purified cyclic products.
For all substrates examined, whereas cyclization occurs at rates only 4- to 6-fold reduced from the natural peptide sequence, hydrolysis predominates over cyclization. The increased flux of hydrolysis to cyclization, in the face of only moderately decreased rates of cyclization, suggests that the acyl-O-TE intermediate is more accessible to water with the peptides examined in this study. To build on this finding that cyclization is a productive outcome with substrates that retain the minimal substrate elements requires addressing the problem of hydrolysis of peptide thioester substrates to make NRPS TE domains more effective synthetic tools to make diverse macrocyclic structures. Potential solutions to this problem may come from an exploration of TE catalysis in nonaqueous solvents, structure-driven mutagenesis to suppress acyl-O-TE hydrolysis, or enzymatic production of an activated peptide substrate from the peptide acid (23, 24).
TycC TE Catalysis Can Generate Integrin-Inhibiting Peptides.
Certain classes of integrins recognize RGD motifs in their protein ligands, and RGD-containing peptides can inhibit ligand binding either directly or indirectly. These peptides are drug candidates against αIIbβ3 in thrombosis (25) and against αvβ3 in tumor angiogenesis (26) and osteoporosis (27). The peptide thioester substrates described in this study contain the RGD sequence in various contexts. TE domain enzymatic conversion generated the linear peptide acid and cyclic peptide products that all showed varying degrees of measurable integrin inhibition activity against the integrins αIIbβ3, αVβ3, and α5β1.
The introduction of constraints can affect the potency and specificity of a bioactive peptide. Cyclization of a peptide, for example, limits a molecule to a subset of the conformational space available to its linear analog. If the bioactive conformational space is accessible to the cyclic molecule, it has an entropic advantage for binding. With the αIIbβ3 integrin, cyclization led to better inhibition of fibrinogen binding as evidenced by cyclo-IT1 and cyclo-IT3. Alternately, cyclization can preclude a molecule from taking on a bioactive conformation, as seen with loss of potency against the α5β1 integrin for cyclo-IT1 and cyclo-IT2 compared with their linear analogs IT1-COOH and IT2-COOH. A cyclization constraint can further be useful in the design of bioactive molecules as it stabilizes the molecule from degradation. The second constraint explored in this study was Nα-methylation of a residue in the peptide. The most active inhibitor of αIIbβ3, cyclo-IT1, was designed with the strong β type II′ turn-inducing d-Ala-Nα-Me-Arg sequence. The Nα-methylation constraint by itself makes IT1-COOH and cyclo-IT1 more than 40-fold and 10-fold more potent against αIIbβ3 than their respective des-Nα-Me versions IT2-COOH and cyclo-IT2.
It is notable that TycC TE, excised from a synthetase involved in the biosynthesis of a cyclic peptide antibiotic, is a useful tool for the generation of cyclic molecules with bioactivity against an unrelated therapeutic target, the integrin receptors. The TE domain is able to catalyze cyclization of dramatically different substrates that retain a small number of important structural elements. The potential diversity of macrocyclic compounds attainable by TE catalysis is further amplified by the fact that a myriad of macrocyclization reactions are performed by TE domains from different NRPS and polyketide synthetase assembly lines (2). The utility of such terminal TE domains can be exploited in efforts to engineer synthetases to generate novel compounds (28–31). Additionally, it is feasible that these small (25–35 kDa) enzymatic domains in isolation could be used in the generation of diverse libraries of macrocyclic compounds with possible biological activity in areas distinct from the natural synthetase product.
Acknowledgments
We thank Dr. Timothy Springer for advice and assistance. This work was supported by National Institutes of Health Grants GM-20011 (to C.T.W.) and HL-48675 (to J.T.).
Abbreviations
- NRPS
nonribosomal peptide synthetase
- TE
thioesterase
- RGD
Arg-Gly-Asp
- SNAC
N-acetylcysteamine thioester
- MALDI-TOF MS
matrix-assisted laser desorption ionization–time of flight MS
References
- 1.Rizo J, Gierasch L M. Annu Rev Biochem. 1992;61:387–418. doi: 10.1146/annurev.bi.61.070192.002131. [DOI] [PubMed] [Google Scholar]
- 2.Keating T A, Ehmann D E, Kohli R M, Marshall C G, Trauger J W, Walsh C T. ChemBioChem. 2001;2:99–107. doi: 10.1002/1439-7633(20010202)2:2<99::AID-CBIC99>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 3.Marahiel M A, Stachelhaus T, Mootz H D. Chem Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 4.Cane D E, Walsh C T, Khosla C. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 5.Cane D E, Walsh C T. Chem Biol. 1999;6:R319–R325. doi: 10.1016/s1074-5521(00)80001-0. [DOI] [PubMed] [Google Scholar]
- 6.Trauger J W, Kohli R M, Mootz H D, Marahiel M A, Walsh C T. Nature (London) 2000;407:215–218. doi: 10.1038/35025116. [DOI] [PubMed] [Google Scholar]
- 7.Kohli R M, Trauger J W, Schwarzer D, Marahiel M A, Walsh C T. Biochemistry. 2001;40:7099–7108. doi: 10.1021/bi010036j. [DOI] [PubMed] [Google Scholar]
- 8.Trauger J W, Kohli R M, Walsh C T. Biochemistry. 2001;40:7092–7098. doi: 10.1021/bi010035r. [DOI] [PubMed] [Google Scholar]
- 9.Hynes R O. Cell. 1992;69:11–25. doi: 10.1016/0092-8674(92)90115-s. [DOI] [PubMed] [Google Scholar]
- 10.Springer T A. Cell. 1994;76:301–314. doi: 10.1016/0092-8674(94)90337-9. [DOI] [PubMed] [Google Scholar]
- 11.Ruoslahti E. Annu Rev Cell Dev Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 12.O'Neil K T, Hoess R H, Jackson S A, Ramachandran N S, Mousa S A, DeGrado W F. Proteins. 1992;14:509–515. doi: 10.1002/prot.340140411. [DOI] [PubMed] [Google Scholar]
- 13.Koivunen E, Gay D A, Ruoslahti E. J Biol Chem. 1993;268:20205–20210. [PubMed] [Google Scholar]
- 14.Bach A C I, Espina J R, Jackson S A, Stouten P F W, Duke J L, Mousa S A, DeGrado W F. J Am Chem Soc. 1996;118:293–294. [Google Scholar]
- 15.Dechantsreiter M A, Planker E, Matha B, Lohof E, Holzemann G, Jonczyk A, Goodman S L, Kessler H. J Med Chem. 1999;42:3033–3040. doi: 10.1021/jm970832g. [DOI] [PubMed] [Google Scholar]
- 16.Takagi J, Erickson H P, Springer T A. Nat Struct Biol. 2001;8:412–416. doi: 10.1038/87569. [DOI] [PubMed] [Google Scholar]
- 17.O'Shea E K, Lumb K J, Kim P S. Curr Biol. 1993;3:658–667. doi: 10.1016/0960-9822(93)90063-t. [DOI] [PubMed] [Google Scholar]
- 18.Chalmers D K, Marshall G R. J Am Chem Soc. 1995;117:5927–5937. [Google Scholar]
- 19.Humphries J D, Askari J A, Zhang X P, Takada Y, Humphries M J, Mould A P. J Biol Chem. 2000;275:20337–20345. doi: 10.1074/jbc.M000568200. [DOI] [PubMed] [Google Scholar]
- 20.Hautanen A, Gailit J, Mann D M, Ruoslahti E. J Biol Chem. 1989;264:1437–1442. [PubMed] [Google Scholar]
- 21.Lawson D M, Derewenda U, Serre L, Ferri S, Szittner R, Wei Y, Meighen E A, Derewenda Z S. Biochemistry. 1994;33:9382–9388. doi: 10.1021/bi00198a003. [DOI] [PubMed] [Google Scholar]
- 22.Li J, Szittner R, Derewenda Z S, Meighen E A. Biochemistry. 1996;35:9967–9973. doi: 10.1021/bi9605292. [DOI] [PubMed] [Google Scholar]
- 23.Klibanov A M. Nature (London) 2001;409:241–246. doi: 10.1038/35051719. [DOI] [PubMed] [Google Scholar]
- 24.Carrea G, Riva S. Angew Chem Int Ed. 2000;39:2226–2254. [PubMed] [Google Scholar]
- 25.Topol E J, Byzova T V, Plow E F. Lancet. 1999;143:227–231. doi: 10.1016/S0140-6736(98)11086-3. [DOI] [PubMed] [Google Scholar]
- 26.Brooks P C, Montgomery A M, Rosenfeld M, Reisfeld R A, Hu T, Klier G, Cheresh D A. Cell. 1994;79:1157–1164. doi: 10.1016/0092-8674(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 27.Engleman V W, Nickols G A, Ross F P, Horton M A, Griggs D W, Settle S L, Ruminshi P G, Teitelbauin S L. J Clin Invest. 1997;99:2284–2292. doi: 10.1172/JCI119404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stachelhaus T, Schneider A, Marahiel M A. Science. 1995;269:69–72. doi: 10.1126/science.7604280. [DOI] [PubMed] [Google Scholar]
- 29.Stachelhaus T, Schneider A, Marahiel M A. Biochem Pharmacol. 1996;52:177–186. doi: 10.1016/0006-2952(96)00111-6. [DOI] [PubMed] [Google Scholar]
- 30.McDaniel R, Thamchaipenet A, Gustafsson C, Fu H, Betlach M, Ashley G. Proc Natl Acad Sci USA. 1999;96:1846–1851. doi: 10.1073/pnas.96.5.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cortes J, Wiesmann K E, Roberts G A, Brown M J, Staunton J, Leadlay P F. Science. 1995;268:1487–1489. doi: 10.1126/science.7770773. [DOI] [PubMed] [Google Scholar]